Abstract
We discuss the heterogeneity of autosomal dominant cone and cone-rod dystrophies (adCD, and adCORD, respectively). As one of the best characterized adCD genes, we focus on the GUCA1A gene encoding guanylate cyclase activating protein 1 (GCAP1), a protein carrying three high affinity Ca2+ binding motifs (EF hands). GCAP1 senses changes in cytoplasmic free [Ca2+] and communicates these changes to GC1, by either inhibiting it (at high free [Ca2+]), or stimulating it (at low free [Ca2+]). A number of missense mutations altering the structure and Ca2+ affinity of EF hands have been discovered. These mutations are associated with a gain of function, producing dominant cone and cone rod dystrophy phenotypes. In this article we review these mutations and describe the consequences of specific mutations on GCAP1 structure and GC stimulation.
31.1. Heterogeneity of autosomal dominant cone and cone-rod dystrophies
Cone and cone-rod dystrophies (CD and CRD, respectively) are rare diseases and highly heterogeneous. Major hallmarks are photophobia, reduced central visual acuity, achromatopsia, but preserved peripheral vision mediated by rod photoreceptors at early stages (Hamel, 2007). CD and CRD are diagnosed mainly on the basis of changes in the photopic and scotopic electroretinogram, but also by fundoscopy and optical coherence tomography (Wissinger et al., 2008; Wolfing et al., 2006). CD and CRD are most often caused by mutations in genes expressed in photoreceptors, such as CRX, RIM1, PITPNM3, UNC119, GUCY2D and GUCA1A associated with multiple functions, including gene regulation, regulation of cGMP synthesis, and regulation of Ca2+ entry at the photoreceptor synapse (Table 21.1).
Table 31.1.
Genetic loci associated with dominant cone dystrophies. Column 1, chromosomal localization. Column 2, disease nomenclature according to RetNet. Column 3, Online Mendelian Inheritance in Man (OMIM) nomenclature. Column 4, gene symbol. Column 5, function of the gene product. Column 6, references.
| gene | Chromosome | OMIM | Function | Defect | References |
|---|---|---|---|---|---|
| CRX (CORD3) |
19q13.3 | 120970 | Transcription factor | multiple missense mutations |
(Swain et al., 1997; Freund et al., 1997) |
| GUCA1A (CORD3) |
6p21.1 | 602093 | Guanylate cyclase activator |
Y99C;N104K; I143NT; |
Review: (Baehr & Palczewski, 2007) |
| GUCY2D (CORD6) |
17p13.1 | 601777 | Photoreceptor guanylate cyclase |
E837D, R838A, R838H, R838C, T839M |
(Perrault et al., 1996); review: (Baehr & Palczewski, 2007) |
| PITPNM3 (CORD5) |
17p13.2 | 600977 | involved in photoreceptor membrane renewal; Drosophila homolog is retinal degeneration B (rdgB) |
Q626H | (Kohn et al., 2007) |
| QRX (CORD11) |
18q21.1-q21.3 | 600624 | Transcription factor | R87Q R137G |
(Wang et al., 2004) |
| RIMS1 (CORD7) |
6q13 | 603649 | Function unknown; ribbon synapse- associated |
R844H | (Johnson et al., 2003) |
| UNC119 | 17q11.2 | 604011 | Function unknown; localizes to rod and cone cytoplasm and ribbon synapses |
K57ter | (Kobayashi et al., 2000) |
The transcription factor CRX, a factor essential for the maintenance of mammalian photoreceptors, regulates the expression of several outer segment proteins such as visual pigments and arrestin (Furukawa et al., 1999). A number of missense mutations and truncations, presumably null alleles, have been discovered in families with CORD3, but is unclear whether the phenotype is caused by a dominant negative effect of a truncated protein or by haploinsufficiency (Freund et al., 1997; Swain et al., 1997). Human RIM1, a putative effector protein for the small GTPase rab3 involved in synaptic exocytosis was shown to be associated with CORD7 (Johnson et al., 2003; Wang et al., 1997). RIM1 is a large multidomain protein localizing to the synaptic ribbon and possibly involved in regulation of glutamate release (Schoch et al., 2002). The CORD7 missense mutation was identified in a region interacting with synaptic proteins like the a1D subunit of L-type Ca2+ channels (Johnson et al., 2003). UNC119 encodes a protein termed UNC119/RG4, related to PrBP/δ (PDE6d), a prenyl binding protein (Zhang et al., 2007). The mutation, UNC119/RG4(K57ter), was incorporated into a transgene producing a slowly progressing rod/cone dystrophy (Kobayashi et al., 2000). It was recently discovered that UNC119/RG4 interacts with CaBP4, a synaptic Ca2+-binding protein in photoreceptors (Haeseleer, 2008) and with ribeye (Alpadi et al., 2008), suggesting a possible function in synaptic transmission (Kobayashi et al., 2000).The membrane-associated phosphatidylinositol transfer protein (PITPNM3), a human homolog of the D. melanogaster rdgB gene, is expressed ubiquitously, and also in retina, particularly the OPL and Müller cells (Tian & Lev, 2002). The CORD5 mutation is located in the C-terminal region of PITPNM3, but the functional consequences of the mutation are unclear (Kohn et al., 2007).
31.2. Guanylate cyclase 1 (GC1) and GCAP1
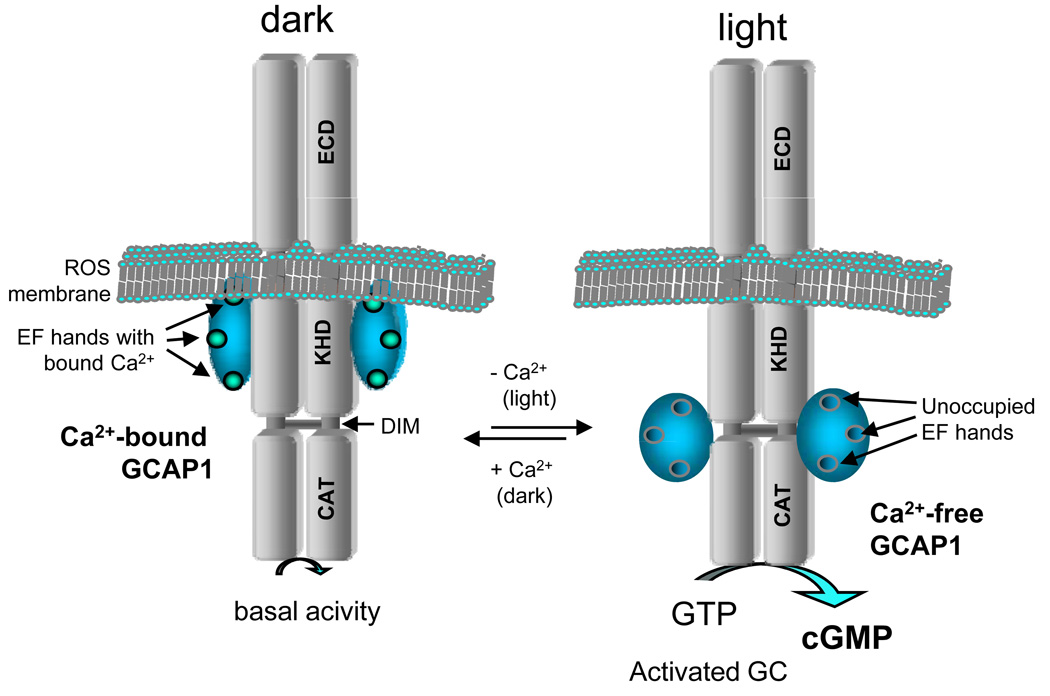
By far the best characterized genes associated with dominant CD/CRD are GUCY2D (CORD6), encoding photoreceptor guanylate cyclase 1 (retGC-1 or GC1), and GUCA1A, encoding the Ca2+-binding protein GCAP1 (CORD3). Both of these genes are expressed at high levels in photoreceptors and locate to the outer segments where phototransduction takes place. GC1 is a transmembrane protein with a large amino-terminal ECD of unknown function, a kinase-like homology domain possibly involved in autophosphorylation, a dimerization domain, and a catalytic domain. GC1 is Ca2+-insensitive in its purified form. Its Ca2+ sensitivity on the outer segment disk membrane is mediated by guanylate cyclase-activating proteins (GCAPs) (Figure 31.1). GCAPs are Ca2+-binding proteins belonging to the calmodulin superfamily equipped with four EF hand motifs. GC1 and GCAP1 interact intracellularly since GCAP1 is present in the cytoplasm (deletion of the extracellular domain does not affect GC stimulation by GCAP1). The side of contact at GC1 involves the kinase-like domain because its deletion diminished the stimulation by GCAP1. GCAP1 in turn interacts with GC1 through the N-terminal region around the EF1 motif. A number of mutations causing dominant cone-rod dystrophy in GUCY2D are restricted to the dimerization domain. Some of the important missense mutations of the dimerization domain are E837D, R838A, R838H, R838C, T839M (Payne et al., 2001; Downes et al., 2001; Wilkie et al., 2000). Interestingly, the three disease mutations at residue 838 are non-equivalent. They exhibit GC activity equal or superior to WT GC at low free [Ca]free in the order R838C< R838H< R838A and showed a higher affinity for GCAP1 than WT GC (Wilkie et al., 2000).
Figure 31.1.
Cartoon of the activation of GC1 by GCAP1. At high free Ca2+ (dark), GCAP and GC form a complex, but enzymatic activity is very low (basal activity is needed to maintain micromolar cGMP in the cytoplasm). At low free Ca2+ (light), GCAP1 converts into an activator of GC and GC activity accelerates.
31.3. The EF hand motifs of GCAP1
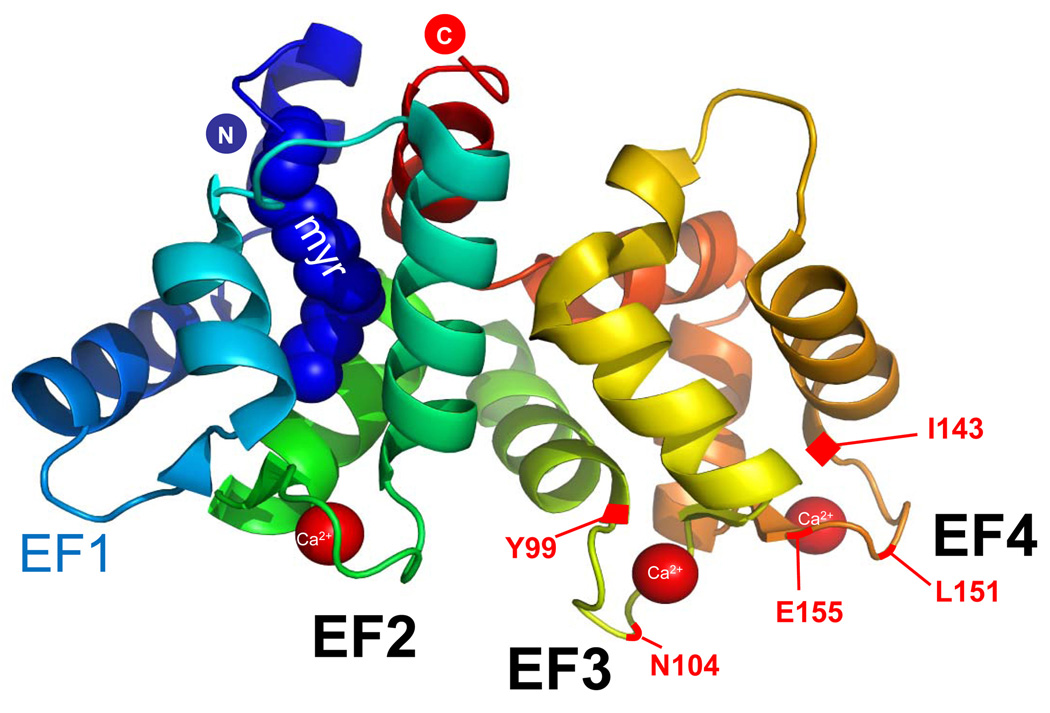
GCAPs are N-myristoylated neuronal Ca2+ sensors with three functional high affinity Ca2+-binding sites termed EF hands. The structure of GCAP1 in its Ca2+-bound form has been determined recently in high resolution (Stephen et al., 2007). The structure shows that in the Ca2+-bound state, the N-terminal acyl side chain is buried deeply between an N-terminal and a C-terminal helix, in contrast to the closely related recoverin, where the myristoyl group is exposed to the solvent (Figure 31.2). The EF hands consist of a helix-loop-helix secondary structure that chelates Ca2+ ions. EF hands also have affinity for Mg2+ ions but the interaction is several orders of magnitude weaker (Gifford et al., 2007). The loop consists of 12 amino acids rich in acidic residues providing oxygen ligands for Ca2+ coordination. The N-terminal region contains a EF hand motif (EF1) in which Ca2+ coordination is prevented by lack of acidic side chains (Palczewski et al., 2004). EF hands 2–4 are fully functional, canonical EF-hand Ca2+-binding sites. Their individual roles have been explored mostly by site-directed mutagenesis, and recording of conformational changes in the absence and presence of Ca2+ and/or Mg2+ (Rudnicka-Nawrot et al., 1998; Otto-Bruc et al., 1997; Peshenko & Dizhoor, 2007; Sokal et al., 1999).
Figure 31.2.
Structure of GCAP1 (adapted from (Baehr & Palczewski, 2009). N-terminal (blue) and C-terminal (red) helices bury the myristoyl group attached to Gly-2. The EF hands are solvent exposed and shown with bound Ca2+. The EF1 motif is incompetent for Ca2+ binding. Approximate locations of residues associated with cone dystrophy are depicted in red.
31.4. GUCA1A mutations associated with adCD and adCRD
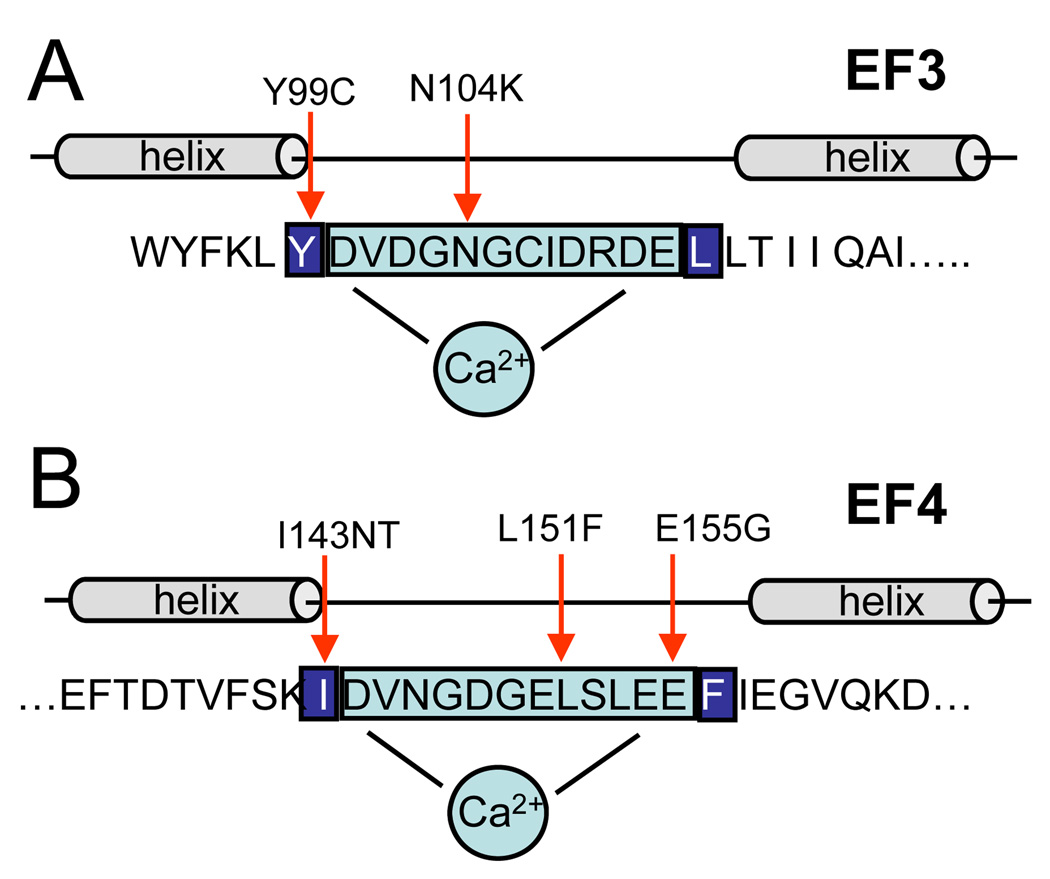
Pathogenic mutations of residues flanking EF3 and EF4, as well as within the EF3 and EF4 loop of GCAP1 are associated with autosomal dominant cone or cone-rod dystrophy (Baehr & Palczewski, 2007). These mutations are Y99, N104, I143, L151, E155 (Sokal et al., 1998; Dizhoor et al., 1998; Nishiguchi et al., 2004; Wilkie et al., 2001; Jiang et al., 2005; Sokal et al., 2005). The residue Y99 is located adjacent to the EF3 hand and I143 adjacent to the EF4 hand. N104 is located in EF3, and E155G, L151F are located in EF4 (Fig. 31.2, Fig 31.3). When replaced by amino acids with different chemical properties, the mutant residues can disrupt coordination of Ca2+ to the mutant loop and change the Ca2+ sensitivity of GCAP1. As a result, mutant GCAPs are not fully inactivated at dark Ca2+ levels, leading to the persistent stimulation of GC1 in the dark, elevated cGMP and Ca2+ levels, and cell death.
Figure 31.3.
Cartoon of the EF3 and EF4 hand motifs in GCAP1. The 12 amino acids comprising the Ca2+-binding loop are boxed and shaded blue, flanking hydrophobic amino acids are highlighted on dark blue background. Mutations linked to adCD are identified by red arrows.
31.5. EF3: The GCAP1(Y99C) and GCAP1(N104K) mutations
Y99 is a hydrophobic amino acid that does not distort the helix N-terminal to EF3. However, replacement of Y99 by a Cys residue (Y99C), the first mutation in GCAP1 linked to dominant cone dystrophy, had adverse effects on the structure of EF3 and Ca2+-binding (Sokal et al., 1998; Dizhoor et al., 1998). When Y99 was replaced by Trp, a hydrophobic residue, biological activity of mutant GCAP1 was unchanged (Sokal et al., 1999). Analysis of the EF3-hand motif Ca2+ binding kinetics with the Y99W mutant (W3Cys−), exploiting the intrinsic Trp fluorescence of Trp99, showed a significant increase in the Trp fluorescence intensity of W3-GCAP1(w−) in the presence of high Ca2+, reflecting a conformational change (Sokal et al., 1999). Thus, the EF3-hand motif is a key region for conversion of GCAP1 from activator to inhibitor, consistent with mutations in this region being causative of cone dystrophy.
N104 occupies a position in the EF3 loop that is critical for Ca2+ coordination by providing oxygen of the amide side chain for coordination. N104K likely weakens Ca2+ coordination at EF3 under physiological conditions preventing formation of the Ca2+-bound structure that is essential for inhibiting GC catalytic activity. The GCAP1 crystal structure implies that mutations in EF3 may distort contacts of the kinked C-terminal helix with the N-terminal helix of GCAP1. In contrast to wild-type GCAP1, GCAP1(N104K) is more susceptible to proteolysis at 1 mM Ca2+. The reason for this distinction is the inability of the mutant GCAP1 to assume the tight Ca2+-bound form of GCAP1 which is less accessible to trypsin. We conclude that the N104K mutation introduces a structural change that is irreversible even at 1 mM Ca2+.
31.6. EF4: The GCAP1(I143NT), GCAP1(L151F) and GCAP1(E155G) mutations
Another pathogenic mutation of a flanking hydrophobic residue (I143NT) (Nishiguchi et al., 2004) was observed in EF4, emphasizing the importance of an intact N-terminal helix for Ca2+ binding. Substitution of Ile143, positioned at the N-terminal end of EF4, by two polar residues changes the orientation of the N-terminal α-helix, distorting the loop conformation that is essential for Ca2+ binding, and decreasing the affinity for Ca2+. Changes in these positions among other Ca2+-binding proteins have also been shown to impair Ca2+ coordination (Falke et al., 1994). Biochemical analysis showed that the GCAP1(I143NT) mutant adopted a conformation susceptible to proteolysis, and its properties suggest that it is incompletely inactivated by high Ca2+ concentrations as should occur with dark adaptation.
An A464G transition in the GUCA1A gene (Wilkie et al., 2001) changed amino acid Glu155 in the EF4-hand motif to Gly. Ca2+ binding at the EF4-hand motif does not affect structural changes of GCAP1 to the same extent as at the EF3-hand motif, as shown measuring intrinsic fluorescence as a function of Ca2+ using a Trp at position 142 (Sokal et al., 1999). However, it exerts similar dominant effects on GC1 stimulation as does GCAP1(Y99C). The residue Glu155 of GCAP1 is invariant in all GCAPs (Palczewski et al., 2004); an invariant Glu at position 12 of the EF-hand loop, contributing both of its side-chain oxygen atoms to the metal-ion coordination, has been shown to be essential for Ca2+ coordination (Nakayama et al., 1992; Falke et al., 1994). In the L151F mutation, one hydrophobic residue (L) replaces another (F) which is not much bulkier than L. The resulting phenotype of adCD is therefore surprising. However, the pathogenic properties of the GCAP1(L151F) mutations described in this article are supported by several independent observations. First, the mutation decreases the Ca2+ sensitivity of GC stimulation, an effect also seen in other EF hand mutations. Second, the recombinant GCAP1-L151F is susceptible to proteolysis. Third, molecular dynamics of WT GCAP1 and GCAP1(L151F) confirmed that a significant change in the structure of mutant GCAP1 influences the binding of Ca2+ in EF4 and EF2. Fourth, the L151F mutations have been independently identified in a large Utah pedigree with dominant cone dystrophy. The reason for the discrepancy in phenotype (adCD versus adCORD) is unclear, but anomalies in the rod response may be slow in developing and may depend on the genetic background.
31.7. Conclusion
All EF hand mutations alter the Ca2+ sensitivity of GCAP1, leading to the constitutive stimulation of GC1 at high [Ca2+] limiting its ability to fully inactivate GC1 under physiological dark conditions. Persistent stimulation of GC by the mutant proteins is predicted to lead to elevated levels of cGMP in the dark-adapted retina, which in turn causes a higher percentage of cGMP-gated channels in the plasma membrane to be opened. The altered physiological cGMP levels may be subtle and thus cause relatively slow retinal degeneration. The reason for the mostly cone-specific degeneration in response to this physiological defect is not understood. GCAP1 may be more active in cones than rods, or, alternatively, it may reflect other differences in cGMP metabolism between rods and cones. Animal models with homologous dominant GCAP1 mutations will be helpful to address this uncertainty. A mouse line expressing GCAP1(Y99C) was generated and shown to shift the Ca2+ sensitivity of GCs in photoreceptors, keeping it partially active at 250 nM free Ca2+, the normal resting Ca2+ concentration in darkness (Olshevskaya et al., 2004).
Reference List
- Alpadi K, Magupalli VG, Kappel S, Koblitz L, Schwarz K, Seigel GM, Sung CH, Schmitz F. RIBEYE recruits Munc119, a mammalian ortholog of the Caenorhabditis elegans protein unc119, to synaptic ribbons of photoreceptor synapses. J Biol.Chem. 2008;283:26461–26467. doi: 10.1074/jbc.M801625200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baehr W, Palczewski K. Guanylate cyclase-activating proteins and retina disease. Subcell.Biochem. 2007;45:71–91. doi: 10.1007/978-1-4020-6191-2_4. [DOI] [PubMed] [Google Scholar]
- Baehr W, Palczewski K. Focus on Molecules: Guanylate cyclase-activating proteins (GCAPs) Exp Eye Res. 2009 doi: 10.1016/j.exer.2008.12.016. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dizhoor AM, Boikov SG, Olshevskaya E. Constitutive activation of photoreceptor guanylate cyclase by Y99C mutant of GCAP-1. J.Biol.Chem. 1998;273:17311–17314. doi: 10.1074/jbc.273.28.17311. [DOI] [PubMed] [Google Scholar]
- Downes SM, Payne AM, Kelsell RE, Fitzke FW, Holder GE, Hunt DM, Moore AT, Bird AC. Autosomal dominant cone-rod dystrophy with mutations in the guanylate cyclase 2D gene encoding retinal guanylate cyclase-1. Arch.Ophthalmol. 2001;119:1667–1673. doi: 10.1001/archopht.119.11.1667. [DOI] [PubMed] [Google Scholar]
- Falke JJ, Drake SK, Hazard AL, Peerson OB. Molecular tuning of ion binding to calcium signaling proteins. Quantative Review Biophysics. 1994;27:219–290. doi: 10.1017/s0033583500003012. [DOI] [PubMed] [Google Scholar]
- Freund CL, Gregory EC, Furukawa T, Papaioannou M, Looser J, Ploder L, Bellingham J, Ng D, Herbrick JA, Duncan A, Scherer SW, Tsui LC, Loutradis AA, Jacobson SG, Cepko CL, Bhattacharya SS, McInnes RR. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell. 1997;91:543–553. doi: 10.1016/s0092-8674(00)80440-7. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Li T, Davis FC, Cepko CL. Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat.Genet. 1999;23:466–470. doi: 10.1038/70591. [DOI] [PubMed] [Google Scholar]
- Gifford JL, Walsh MP, Vogel HJ. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem.J. 2007;405:199–221. doi: 10.1042/BJ20070255. [DOI] [PubMed] [Google Scholar]
- Haeseleer F. Interaction and Colocalization of CaBP4 and Unc119 (MRG4) in Photoreceptors. Invest Ophthalmol.Vis.Sci. 2008;49:2366–2375. doi: 10.1167/iovs.07-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel CP. Cone rod dystrophies. Orphanet.J Rare.Dis. 2007;2:7. doi: 10.1186/1750-1172-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Katz BJ, Yang Z, Zhao Y, Faulkner N, Hu J, Baird J, Baehr W, Creel DJ, Zhang K. Autosomal dominant cone dystrophy caused by a novel mutation in the GCAP1 gene (GUCA1A) Mol.Vis. 2005;11:143–151. [PubMed] [Google Scholar]
- Johnson S, Halford S, Morris AG, Patel RJ, Wilkie SE, Hardcastle AJ, Moore AT, Zhang K, Hunt DM. Genomic organisation and alternative splicing of human RIM1, a gene implicated in autosomal dominant cone-rod dystrophy (CORD7) Genomics. 2003;81:304–314. doi: 10.1016/s0888-7543(03)00010-7. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Higashide T, Hamasaki D, Kubota S, Sakuma H, An W, Fujimaki T, McLaren MJ, Weleber RG, Inana G. HRG4 (UNC119) mutation found in cone-rod dystrophy causes retinal degeneration in a transgenic model. Invest Ophthalmol.Vis.Sci. 2000;41:3268–3277. [PubMed] [Google Scholar]
- Kohn L, Kadzhaev K, Burstedt MS, Haraldsson S, Hallberg B, Sandgren O, Golovleva I. Mutation in the PYK2-binding domain of PITPNM3 causes autosomal dominant cone dystrophy (CORD5) in two Swedish families. Eur.J Hum.Genet. 2007;15:664–671. doi: 10.1038/sj.ejhg.5201817. [DOI] [PubMed] [Google Scholar]
- Nakayama S, Moncrief ND, Kretsinger RH. Evolution of EF-hand calcium-modulated proteins II. Domains of several subfamilies have diverse evolutionary histories. J.Mol.Evol. 1992;34:416–448. doi: 10.1007/BF00162998. [DOI] [PubMed] [Google Scholar]
- Nishiguchi KM, Sokal I, Yang L, Roychowdhury N, Palczewski K, Berson EL, Dryja TP, Baehr W. A Novel Mutation (I143NT) in Guanylate Cyclase-Activating Protein 1 (GCAP1) Associated with Autosomal Dominant Cone Degeneration. Invest Ophthalmol.Vis.Sci. 2004;45:3863–3870. doi: 10.1167/iovs.04-0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshevskaya EV, Calvert PD, Woodruff ML, Peshenko IV, Savchenko AB, Makino CL, Ho YS, Fain GL, Dizhoor AM. The Y99C mutation in guanylyl cyclase-activating protein 1 increases intracellular Ca2+ and causes photoreceptor degeneration in transgenic mice. Journal of Neuroscience. 2004;24:6078–6085. doi: 10.1523/JNEUROSCI.0963-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto-Bruc A, Buczylko J, Surgucheva I, Subbaraya I, Rudnicka-Nawrot M, Crabb J, Arendt A, HArgrave PA, Baehr W, Palczewski K. Functional reconstitution of photoreceptor guanylate cyclase with native and mutant forms of guanylate cyclase activating protein 1. Biochemistry. 1997;36:4295–4302. doi: 10.1021/bi963000d. [DOI] [PubMed] [Google Scholar]
- Palczewski K, Sokal I, Baehr W. Guanylate cyclase-activating proteins: structure, function, and diversity. Biochem.Biophys.Res.Commun. 2004;322:1123–1130. doi: 10.1016/j.bbrc.2004.07.122. [DOI] [PubMed] [Google Scholar]
- Payne AM, Morris AG, Downes SM, Johnson S, Bird AC, Moore AT, Bhattacharya SS, Hunt DM. Clustering and frequency of mutations in the retinal guanylate cyclase (GUCY2D) gene in patients with dominant cone-rod dystrophies. J.Med.Genet. 2001;38:611–614. doi: 10.1136/jmg.38.9.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrault I, Rozet J-M, Calvas P, Gerber S, Camuzat A, Dollfus H, Chatelin S, Souied E, Ghazi I, Leowski C, Bonnermaison M, Le Paslier D, Frezal J, Dufier J-L, Pittler SJ, Munnich A, Kaplan J. Retinal-specific guanylate cyclase gene mutations in Leber's congenital amaurosis. Nature Genet. 1996;14:461–464. doi: 10.1038/ng1296-461. [DOI] [PubMed] [Google Scholar]
- Peshenko IV, Dizhoor AM. Activation and inhibition of photoreceptor guanylyl cyclase by guanylyl cyclase activating protein 1 (GCAP-1): the functional role of Mg2+/Ca2+ exchange in EF-hand domains. J Biol.Chem. 2007;282:21645–21652. doi: 10.1074/jbc.M702368200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicka-Nawrot M, Surgucheva I, Hulmes JD, Haeseleer F, Sokal I, Crabb JW, Baehr W, Palczewski K. Changes in biological activity and folding of guanylate cyclase-activating protein 1 as a function of calcium. Biochemistry. 1998;37:248–257. doi: 10.1021/bi972306x. [DOI] [PubMed] [Google Scholar]
- Schoch S, Castillo PE, Jo T, Mukherjee K, Geppert M, Wang Y, Schmitz F, Malenka RC, Sudhof TC. RIM1alpha forms a protein scaffold for regulating neurotransmitter release at the active zone. Nature. 2002;415:321–326. doi: 10.1038/415321a. [DOI] [PubMed] [Google Scholar]
- Sokal I, Dupps WJ, Grassi MA, Brown J, Jr, Affatigato LM, Roychowdhury N, Yang L, Filipek S, Palczewski K, Stone EM, Baehr W. A GCAP1 missense mutation (L151F) in a large family with autosomal dominant cone-rod dystrophy (adCORD) Invest Ophthalmol.Vis.Sci. 2005;46:1124–1132. doi: 10.1167/iovs.04-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal I, Li N, Surgucheva I, Warren MJ, Payne AM, Bhattacharya SS, Baehr W, Palczewski K. GCAP1(Y99C) mutant is constitutively active in autosomal dominant cone dystrophy. Mol.Cell. 1998;2:129–133. doi: 10.1016/s1097-2765(00)80121-5. [DOI] [PubMed] [Google Scholar]
- Sokal I, Otto-Bruc AE, Surgucheva I, Verlinde CL, Wang CK, Baehr W, Palczewski K. Conformational changes in guanylyl cyclase-activating protein 1 (GCAP1) and its tryptophan mutants as a function of calcium concentration. J.Biol.Chem. 1999;274:19829–19837. doi: 10.1074/jbc.274.28.19829. [DOI] [PubMed] [Google Scholar]
- Stephen R, Bereta G, Golczak M, Palczewski K, Sousa MC. Stabilizing function for myristoyl group revealed by the crystal structure of a neuronal calcium sensor, guanylate cyclase-activating protein 1. Structure. 2007;15:1392–1402. doi: 10.1016/j.str.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain PK, Chen S, Wang QL, Affatigato LM, Coats CL, Brady KD, Fishman GA, Jacobson SG, Swaroop A, Stone E, Sieving PA, Zack DJ. Mutations in the cone-rod homeobox gene are associated with the cone-rod dystrophy photoreceptor degeneration. Neuron. 1997;19:1329–1336. doi: 10.1016/s0896-6273(00)80423-7. [DOI] [PubMed] [Google Scholar]
- Tian D, Lev S. Cellular and developmental distribution of human homologues of the Drosophilia rdgB protein in the rat retina. Invest Ophthalmol.Vis.Sci. 2002;43:1946–1953. [PubMed] [Google Scholar]
- Wang QL, Chen S, Esumi N, Swain PK, Haines HS, Peng G, Melia BM, McIntosh I, Heckenlively JR, Jacobson SG, Stone EM, Swaroop A, Zack DJ. QRX, a novel homeobox gene, modulates photoreceptor gene expression. Hum.Mol.Genet. 2004;13:1025–1040. doi: 10.1093/hmg/ddh117. [DOI] [PubMed] [Google Scholar]
- Wang Y, Okamoto M, Schmitz F, Hofmann K, Sudhof TC. Rim is a putative Rab3 effector in regulating synaptic-vesicle fusion. Nature. 1997;388:593–598. doi: 10.1038/41580. [DOI] [PubMed] [Google Scholar]
- Wilkie SE, Li Y, Deery EC, Newbold RJ, Garibaldi D, Bateman JB, Zhang H, Lin W, Zack DJ, Bhattacharya SS, Warren MJ, Hunt DM, Zhang K. Identification and functional consequences of a new mutation (E155G) in the gene for GCAP1 that causes autosomal dominant cone dystrophy. Am.J Hum.Genet. 2001;69:471–480. doi: 10.1086/323265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie SE, Newbold RJ, Deery E, Walker CE, Stinton I, Ramamurthy V, Hurley JB, Bhattacharya SS, Warren MJ, Hunt DM. Functional characterization of missense mutations at codon 838 in retinal guanylate cyclase correlates with disease severity in patients with autosomal dominant cone-rod dystrophy. Hum.Mol.Genet. 2000;9:3065–3073. doi: 10.1093/hmg/9.20.3065. [DOI] [PubMed] [Google Scholar]
- Wissinger B, Dangel S, Jagle H, Hansen L, Baumann B, Rudolph G, Wolf C, Bonin M, Koeppen K, Ladewig T, Kohl S, Zrenner E, Rosenberg T. Cone dystrophy with supernormal rod response is strictly associated with mutations in KCNV2. Invest Ophthalmol.Vis.Sci. 2008;49:751–757. doi: 10.1167/iovs.07-0471. [DOI] [PubMed] [Google Scholar]
- Wolfing JI, Chung M, Carroll J, Roorda A, Williams DR. High-resolution retinal imaging of cone-rod dystrophy. Ophthalmology. 2006;113:1019. doi: 10.1016/j.ophtha.2006.01.056. [DOI] [PubMed] [Google Scholar]
- Zhang H, Li S, Doan T, Rieke F, Detwiler PB, Frederick JM, Baehr W. Deletion of PrBP/{delta} impedes transport of GRK1 and PDE6 catalytic subunits to photoreceptor outer segments. Proc.Natl.Acad.Sci U.S.A. 2007;104:8857–8862. doi: 10.1073/pnas.0701681104. [DOI] [PMC free article] [PubMed] [Google Scholar]