Abstract
Although most metazoan mitochondrial genomes are highly streamlined and encode little noncoding DNA outside of the “AT” region, the accumulation of mitochondrial pseudogenes and other types of noncoding DNA has been observed in a growing number of animal groups. The nematode species Caenorhabditis briggsae harbors two mitochondrial DNA (mtDNA) pseudogenes, named Ψnad5-1 and Ψnad5-2, presumably derived from the nad5 protein-coding gene. Here, we provide an in-depth analysis of mtDNA pseudogene evolution in C. briggsae natural isolates and related Caenorhabditis species. Mapping the observed presence and absence of the pseudogenes onto phylogenies suggests that Ψnad5-1 originated in the ancestor to C. briggsae and its recently discovered outcrossing relative species Caenorhabditis sp. 5 and Caenorhabditis sp. 9. However, Ψnad5-1 was not detected in Caenorhabditis sp. 9 natural isolates, suggesting a lineage-specific loss of this pseudogene in this species. Our results corroborated the previous finding that Ψnad5-2 originated within C. briggsae. The observed pattern of mitochondrial pseudogene gain and loss in Caenorhabditis was inconsistent with predictions of the tandem duplication–random loss model of mitochondrial genome evolution and suggests that intralineage recombination–like mechanisms might play a major role in Caenorhabditis mtDNA evolution. Natural variation was analyzed at the pseudogenes and flanking mtDNA sequences in 141 geographically diverse C. briggsae natural isolates. Although phylogenetic analysis placed the majority of isolates into the three previously established major intraspecific clades of C. briggsae, two new and unexpected haplotypes fell outside of these conventional groupings. Ψnad5-2 copy number variation was observed among C. briggsae isolates collected from the same geographic site. Patterns of nucleotide diversity were analyzed in Ψnad5-1 and Ψnad5-2, and confidence intervals were found to overlap values from synonymous sites in protein-coding genes, consistent with neutral expectations. Our findings provide new insights into the mode and tempo of mitochondrial genome and pseudogene evolution both within and between Caenorhabditis nematode species.
Keywords: Caenorhabditis, mitochondrial genome, nucleotide diversity, phylogenetics, pseudogene
Introduction
The mitochondrial genomes of animals are generally minimized in size, usually encoding the same set of 13 protein-coding and 24 structural RNA genes, with little duplicated or noncoding DNA outside of the control (a.k.a. “AT”) region (Lynch et al. 2006; Gissi et al. 2008). Accumulation of noncoding DNA is thought to have a negative impact, increasing the susceptibility of these genomes to certain mutation types, such as repeat-associated deletions (Townsend and Rand 2004; Howe and Denver 2008). The high mutation rates of animal mitochondrial DNA (mtDNA) are thought to provide an evolutionary environment prohibitive to the accumulation of noncoding DNA in animal mitochondrial genomes (Lynch et al. 2006). Some incidences of mtDNA noncoding elements preserved across long evolutionary timescales have been attributed to selection for their continued maintenance for hypothesized functional reasons (Kumazawa et al. 1996; Bakke et al. 1999) or secondary structure features that prevent their loss (McKnight and Shaffer 1997). Despite their overall rarity, pseudogenes have been described in a number of animal mitochondrial genomes, including certain species of amphibians (Mueller and Boore 2005; San Mauro et al. 2006; Kurabayashi et al. 2008), reptiles (Macey et al. 2004; Fujita et al. 2007), fish (Mabuchi et al. 2004), and nematodes (Tang and Hyman 2007; Howe and Denver 2008). Although in most of these cases the pseudogenes are short sequences derived from transfer RNAs (tRNAs), nematode species such as Caenorhabditis briggsae have been found to contain relatively large (>100 bp) pseudogenes derived from protein-coding genes (Howe and Denver 2008). Most previous studies involving mtDNA pseudogenes have been centered on understanding their association with mitochondrial genome rearrangement events. Much remains to be understood about the evolution of animal mtDNA pseudogenes themselves, particularly within species.
Caenorhabditis briggsae is a hermaphroditic species in the Elegans group of Caenorhabditis nematodes (Kiontke and Fitch 2005) and has been developed as a comparative development, genomics, and evolution model for C. elegans (Stein et al. 2003; Cutter et al. 2006; Zhao et al. 2008). Caenorhabditis briggsae natural isolates have a worldwide distribution characterized by distinct latitudinal phylogeographic population structure with isolates falling into three major clades (Cutter et al. 2006; Dolgin et al. 2008). Although the deeper relationships of major C. briggsae intraspecific clades are well understood, knowledge on more local patterns of genetic diversity and phylogeography in this species remains enigmatic. Caenorhabditis sp. 5, a gonochoristic species found only in southeast Asia thus far, is one of many Caenorhabditis nematode species discovered over the last few years as a consequence of a renewed push to identify more species in the genus and a closely related sister species to the famous model C. elegans. Caenorhabditis sp. 9 is another recently discovered gonochoristic species that is a close relative to C. briggsae.
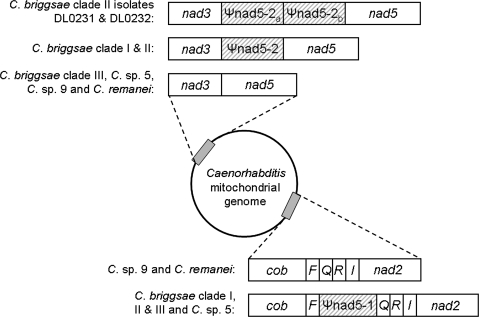
Large noncoding elements have been described in the mitochondrial genomes of C. briggsae and Caenorhabditis sp. 5 (Howe and Denver 2008), providing an excellent model system to examine animal mtDNA pseudogene evolution. The two pseudogenes present in the mitochondrial genome of C. briggsae are both derived from the nad5 protein-coding gene (88–89% identity between each pseudogene and nad5) and were named Ψnad5-1 and Ψnad5-2 (Howe and Denver 2008). Note that we are here switching to the more commonly used, standardized mtDNA gene naming scheme to facilitate easier comparisons to other mtDNA studies; Ψnad5-1 and Ψnad5-2 are the same elements that Howe and Denver (2008) referred to as ΨND5-1 and ΨND5-2, respectively. These mtDNA pseudogene elements are homologous to a central region of the nad5 gene. Ψnad5-1 (214–223 bp) is present in all three intraspecific clades of C. briggsae as well as Caenorhabditis sp. 5, whereas Ψnad5-2 (325–344 bp) is only present in two C. briggsae intraspecific clades. Ψnad5-1 is located between trnQ and trnF, whereas Ψnad5-2 is located directly upstream of nad5, on the opposite side of the mitochondrial genome (fig. 1). Direct repeats in Ψnad5-2 and nad5 are associated with large, function-disrupting, and deleterious nad5 gene deletions in the mtDNA of C. briggsae natural isolates (Howe and Denver 2008). The nad5 deletions are heteroplasmic, and isolate-specific levels vary from 0% to ∼50% of the total mtDNA pool within a nematode. Howe and Denver (2008) also described the presence of putative compensatory alleles in the Ψnad5-2 direct repeat of some isolates that renders the Ψnad5-2 repeat an imperfect match to the downstream repeat in nad5—isolates with these alleles showed significantly lower nad5 deletion levels than those without the compensatory mutations. Although the putative compensatory alleles were initially identified exclusively in one C. briggsae intraspecific subclade (the “temperate” clade), the Howe and Denver (2008) study considered only 24 C. briggsae natural isolates. There are currently >100 geographically diverse C. briggsae natural isolates available for study—the patterns of mtDNA pseudogene conservation and distribution patterns of the putative Ψnad5-2 compensatory alleles in the larger set of C. briggsae isolates, and Caenorhabditis sp. 9, remain unknown. Furthermore, the patterns of intraspecific molecular genetic diversity of mtDNA pseudogenes have not been well studied in C. briggsae or any other animal species.
FIG. 1.
Positions of pseudogenes in Caenorhabditis mitochondrial genomes. Genes are indicated by white rectangles (single-letter abbreviation used for tRNA genes), and dashed boxes show pseudogenes. The Caenorhabditis species and/or intraspecific groups in which each arrangement is observed is indicated on the left of each displayed mtDNA region.
In the present study, we analyze the evolution of mtDNA pseudogenes and flanking sequence in 141 global natural isolates of C. briggsae, as well as related Caenorhabditis species. We report on the gain, conservation, and loss of pseudogenes in C. briggsae and its close relatives Caenorhabditis sp. 5 and Caenorhabditis sp. 9. The pseudogene and flanking coding sequences are also used to reconstruct phylogenetic relationships among the nematodes analyzed. mtDNA pseudogene nucleotide diversity (π) is analyzed among different C. briggsae clades and compared with diversity patterns in mtDNA protein-coding sequences. Our analysis reveals mtDNA pseudogene gain and loss events in Caenorhabditis, high levels of molecular genetic diversity in C. briggsae mtDNA pseudogenes, new insights into global and local patterns of C. briggsae genetic diversity, and divergent C. briggsae mtDNA haplotypes that suggest substantial genetic diversity remains to be discovered in this species.
Materials and Methods
Nematode Isolate Culturing and Species Diagnosis
Natural geographic isolates of 141 C. briggsae, 5 Caenorhabditis sp. 5, and 2 Caenorhabditis sp. 9 were collected and examined (supplementary table 1, Supplementary Material online). Isolates were obtained from our own collection efforts and as gifts from helpful colleagues (see Acknowledgments). All isolates were expanded in culture, prepared for cryogenic storage, and collected for DNA extraction using standard techniques (Wood 1988; Denver et al. 2003). Species diagnoses in Caenorhabditis are generally performed by testcrossing unknown hermaphrodites or females with males of a known species (Barriere and Felix 2006); although certain crosses of Caenorhabditis sp. 9 and C. briggsae isolates yield viable progeny (Félix M-A, unpublished data), they are considered different species due to strong hybrid sterility and their different reproductive modes.
Polymerase Chain Reaction and DNA Sequencing
Polymerase chain reaction (PCR), product purification, and direct DNA sequencing were performed as previously described (Denver et al. 2003; Howe and Denver 2008). Supplementary table 2 (Supplementary Material online) provides all PCR and sequencing primers used for this study. For all isolates analyzed here, two mtDNA regions were amplified: the first was an ∼810-bp amplicon containing trnQ, Ψnad5-1, trnF, and the 5′ end of cob; the second was an ∼1,150-bp amplicon containing the 3′ end of nad3, Ψnad5-2, and the 5′ end of nad5. A single nuclear locus, primarily composed of the third intron of the Cbr-polh-1 gene (DNA polymerase eta homolog) and ∼850–1,000 bp in length (depending on isolate), was also amplified for comparative purposes with the mtDNA data. For Caenorhabditis sp. 5 and Caenorhabditis sp. 9, one additional region was analyzed for phylogenetic analysis: an ∼400-bp segment composed of a small subunit ribosomal RNA gene segment, trnS(ucn), trnN, and trnY. Sequence data were submitted to GenBank under accession numbers (GU451901-GU452323).
Phylogenetic and Nucleotide Diversity Analyses
Multiple alignment and phylogenetic analyses were performed using the MEGA4 software package (Tamura et al. 2007). DNA sequence multiple alignments were performed using the ClustalW function of MEGA4; the IUB DNA weight matrix was used and the gap opening and extension penalties were set to 15 and 6.66, respectively (default settings). Reliabilities of resultant multiple alignments were evaluated by visual inspection after running ClustalW. For a first phylogenetic analysis involving multiple nematode species, amplified regions from Caenorhabditis sp. 5 and Caenorhabditis sp. 9 were aligned with the homologous sequences from 1 C. remanei (strain EM464; complete mtDNA sequence provided as a gift from W. K. Thomas) and 24 C. briggsae sequenced mitochondrial genomes (Howe and Denver 2008); 1,995 bp of aligned DNA sequence was used. For the first phylogenetic analysis, concatenated gene sequences were used and pseudogene sequences were excluded. The mtDNA sequence from C. remanei strain EM464 was used as the outgroup based on results from a previous phylogenetic analysis of Caenorhabditis using nuclear genes (Kiontke et al. 2007). For a second phylogenetic analysis involving only C. briggsae isolates, alignments were performed for the two amplified mtDNA regions that included pseudogene sequences—the two regions were concatenated for subsequent analysis (1,694 bp in the alignment). Redundant haplotypes were removed prior to phylogenetic analysis. In MEGA4, neighbor joining (NJ) and maximum parsimony methods were employed for phylogenetic analyses. For NJ analyses, the maximum composite likelihood model of sequence evolution was implemented. For maximum parsimony analyses, the close-neighbor interchange method was used for tree searching. All substitution types were included in all phylogenetic analyses. The reliabilities of tree topologies resulting from phylogenetic analyses were evaluated using bootstrap testing (1,000 replicates performed for each analysis).
π Values were calculated for mitochondrial protein-coding sequences and both pseudogenes using DnaSP v4.0 (Rozas et al. 2003). Protein-coding sequence flanking Ψnad5-1 and Ψnad5-2 (portions of cob, nad3, and nad5) was concatenated in-frame—this data set was used to calculate nonsynonymous- and synonymous-site nucleotide diversity (πN and πS, respectively); 327 codons were analyzed. Populations were defined according to clades observed from the phylogenetic trees. Approximate 95% confidence intervals (CIs) for π estimates were determined in DnaSP v4.0 using coalescent simulations, following the methods reported in a previous analysis of mtDNA and nucleotide diversity in Caenorhabditis nematode species (Graustein et al. 2002). The simulations computed π value estimates and CIs given input theta estimates (values empirically calculated from corresponding data sets in DnaSP) and the assumption of no recombination. Ten thousand replicates were performed for each simulation.
Results and Discussion
Gain, Conservation, and Loss of Caenorhabditis mtDNA Pseudogenes
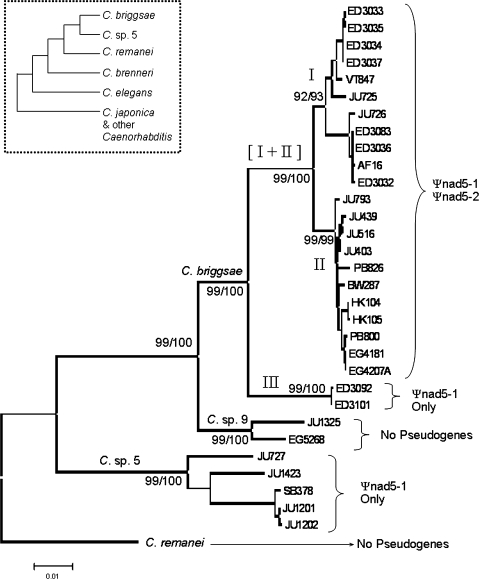
To examine the evolutionary origins and fates of pseudogene sequences in the mitochondrial genomes of Caenorhabditis nematodes, we analyzed two mtDNA regions: one containing Ψnad5-1 (in C. briggsae and Caenorhabditis sp. 5) and a second containing Ψnad5-2 (in C. briggsae)—see figure 1. mtDNA analysis of crosses between divergent natural strains of C. elegans showed that mtDNA is inherited exclusively through the hermaphrodite oocyte lineage in this species (Morris K, Thomas WK, personal communication). Thus, we applied a phylogenetic approach to investigate the evolution of Caenorhabditis mtDNA pseudogenes under the assumptions that mtDNA was inherited through the hermaphrodite lineages and that there is no heterologous mtDNA recombination. To construct a reference phylogeny for mapping pseudogene gain and loss events in Caenorhabditis (fig. 2), 1,995 bp of aligned mtDNA protein-coding, tRNA, and ribosomal RNA gene sequence was analyzed from 24 isolates of C. briggsae, 5 isolates of Caenorhabditis sp. 5, and 2 isolates of Caenorhabditis sp. 9. Ψnad5-1 and Ψnad5-2 sequences were excluded from these analyses. We also included homologous sequence from C. remanei strain EM464 mtDNA as an outgroup sequence based on a previous phylogenetic analysis of Caenorhabditis nuclear DNA (Kiontke et al. 2007). Our phylogeny was consistent with the previous nuclear analysis (Kiontke et al. 2007), and Caenorhabditis sp. 9 was placed as a closely related sister taxon to C. briggsae (fig. 2). Natural populations of C. briggsae have been subdivided into three major clades in previous analyses of both nuclear DNA and mtDNA (Dolgin et al. 2008; Howe and Denver 2008). Here, we propose and apply a new, simplified, and generic clade naming scheme for C. briggsae: Clade I (previously “tropical” clade, contains AF16 reference nuclear genome strain), Clade II (previously temperate clade), and Clade III (previously “equatorial” and “Kenya” clade). We propose this new scheme because the present study reveals a greater degree of latitudinal overlap between C. briggsae clades than was observed in previous smaller scale studies (discussed further below). Caenorhabditis sp. 5 is a recently discovered, unnamed, outcrossing species found only in southeast Asia thus far (see supplementary table 1, Supplementary Material online) that is more closely related to C. briggsae than C. remanei and C. elegans (Kiontke et al. 2007; Cutter 2008). Caenorhabditis sp. 9 is an even more recently discovered, unnamed, outcrossing species that is capable of crossing with C. briggsae to make fertile offspring. Two isolates of this species are currently available: one from India and the other from the Democratic Republic of the Congo (supplementary table 1, Supplementary Material online).
FIG. 2.
Presence and absence of mtDNA pseudogenes in Caenorhabditis. Main display is an NJ phylogram for Caenorhabditis briggsae, Caenorhabditis sp. 9, Caenorhabditis sp. 5, and C. remanei using 1,995 bp of mtDNA sequence. All gene sequences amplified for this study were used though pseudogene sequences were excluded so that their presence/absence could be independently mapped onto the phylogeny. The presence/absence of Ψnad5-1 and Ψnad5-2 in different species and intraspecific clades of C. briggsae is indicated on the right. Bootstrap values for maximum parsimony (left) and NJ (right) methods (1,000 replicates performed for each) are indicated to the left of the corresponding node. Scale bar shows 0.01 substitutions per site. The cladogram in the dashed box on the top left shows evolutionary relationships in the Elegans clade of Caenorhabditis nematodes based on nuclear DNA data (Kiontke et al. 2007).
We found that Ψnad5-1 is present in all surveyed C. briggsae and Caenorhabditis sp. 5 isolates but was not found in either Caenorhabditis sp. 9 isolate. This pseudogene is also not present in C. remanei or C. elegans mitochondrial genomes (Denver et al. 2003; Howe and Denver 2008). By contrast, Ψnad5-2 is present only in Clade I and Clade II of C. briggsae and absent in all other surveyed Caenorhabditis species, suggesting a single origin event within C. briggsae. Given our results, we infer that an absence of Ψnad5-1 and Ψnad5-2 was the ancestral mitochondrial genome architecture state in Caenorhabditis. We mapped the Ψnad5-1 presence/absence data onto our phylogeny (fig. 2), which revealed two possible paths of Ψnad5-1 evolution. First, it is possible that there were two independent origins of Ψnad5-1 on the phylogeny: one gain in C. briggsae and a separate gain in Caenorhabditis sp. 5. A second possibility is that there was a single origin of Ψnad5-1 in the ancestor to (C. briggsae + Caenorhabditis sp. 9 + Caenorhabditis sp. 5) and a loss in Caenorhabditis sp. 9. We observed that Ψnad5-1 occupies identical junctions between tRNAQ and tRNAF in both C. briggsae and Caenorhabditis sp. 5. Although independent duplications of the same sequence are not unheard of in the mitochondrial genome (Fujita et al. 2007), it is unlikely that such events would share the same junctions. We thus conclude that the most likely explanation for the observed pattern of Ψnad5-1 presence/absence is a single gain in the ancestor to (C. briggsae + Caenorhabditis sp. 9 + Caenorhabditis sp. 5), with a subsequent loss in the Caenorhabditis sp. 9 branch. Caenorhabditis sp. 9 was found to possess 6 bp of unassignable intergenic sequence between tRNAQ and tRNAF, where Ψnad5-1 resides in C. briggsae and Caenorhabditis sp. 5. Although we cannot determine its origin with certainty, it is possible that this small intergenic spacer is all that remains of Ψnad5-1 in Caenorhabditis sp. 9.
The probable loss of Ψnad5-1 from Caenorhabditis sp. 9, but not other lineages, raises questions regarding the evolutionary forces acting on these pseudogenes. Differences in effective population size may provide an apt explanation because this factor determines the relative strengths of selection versus drift (Charlesworth 2009). However, there are currently too few isolates of Caenorhabditis sp. 5 and Caenorhabditis sp. 9 to estimate effective population size for these species. Differences in mating system (C. briggsae is hermaphroditic, whereas the two unnamed species are gonochoristic) might also be hypothesized to have an effect, though Ψnad5-1 is present in both C. briggsae and Caenorhabditis sp. 5. Lower mtDNA mutation rates in species with mtDNA pseudogenes relative to those without the elements provide another possible contributing factor. Although the mtDNA mutation process in C. elegans has been well characterized (Denver et al. 2000), little is known about mtDNA mutation in other nematode species. The evolutionary reasons why Ψnad5-1 was eliminated entirely from one species and preserved in two others, one hermaphroditic and one gonochoristic, remain unknown.
The tandem duplication–random loss (TDRL) model of mitochondrial genome evolution (Boore and Brown 1998) posits that tandem duplication of mtDNA stretches followed by random loss (via pseudogenization then degeneration) of redundant genes is responsible for between-lineage differences in mtDNA gene order. The TDRL model has gained wide empirical support, primarily in deuterostomes, with the observation of requisite “transition state” duplicated and pseudogenized sequences in a number of test cases (Paabo et al. 1991; Arndt and Smith 1998; Yu et al. 2008). For the Caenorhabditis mtDNA pseudogenes (homologous to a central region of nad5), however, no instances of longer “precursor” pseudogene regions were detected for either Ψnad5-1 or Ψnad5-2 in any of the Caenorhabditis species and strains analyzed here. Ψnad5-1 occurs in a tRNA cluster on the opposite side of the mitochondrial genome relative to its presumed coding gene ancestor (nad5); a very large mtDNA duplication event involving 19 genes would be required to explain the presence of Ψnad5-1 under the TDRL model. Furthermore, mtDNA gene order is identical in ancestral species lacking Ψnad5-1 (e.g., C. remanei, C. elegans) and those harboring the pseudogene (C. briggsae, Caenorhabditis sp. 5). Thus, the large duplication event required to invoke TDRL would also have to be followed by a duplicate gene pseudogenization/loss process that resulted in an mtDNA gene order identical to the ancestral order—highly unlikely considering the involvement of 19 duplicated genes. Although Ψnad5-2 is directly upstream of nad5 which on the surface might seem to make the TDRL model more applicable to this element as compared with Ψnad5-1, Ψnad5-2 originated within C. briggsae (fig. 2) and no natural isolates of this species show any evidence of a longer, more intact pseudogene copy predicted by the TDRL model. More specifically, if Ψnad5-2 originated as a consequence of a tandem duplication, sequences homologous to the 5′ region of nad5 would be a required evolutionary intermediate that we would expect to observe in some C. briggsae strain.
We hypothesize that the sudden and punctuated appearance and disappearance of these Caenorhabditis mtDNA pseudogene elements are driven by intra- and/or intergenomic mtDNA recombination-like processes that occur within hermaphrodite oocyte lineages—molecular exchange between nonallelic direct repeat sequence tracts from different molecules in the population of mitochondrial genomes within an organelle might lead to the gain or loss of sequences in a given mtDNA molecule. mtDNA recombination of this type has been documented in other nematodes species (Lunt and Hyman 1997) but not in Caenorhabditis. Further studies targeting predicted recombination intermediates (e.g., mtDNA subgenomic circles) will be required to evaluate the potential role of recombination-like mechanisms in shaping mtDNA evolution in Caenorhabditis and other animal taxa.
Natural mtDNA Variation in C. briggsae
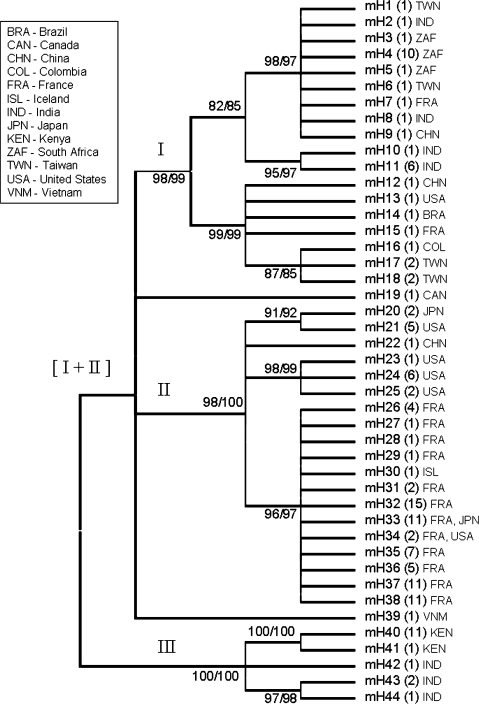
In addition to analyzing pseudogene gain and loss events in the genus Caenorhabditis, we used the pseudogene and flanking sequences to investigate within-species mtDNA variation in 141 geographically diverse C. briggsae isolates (supplementary table 1, Supplementary Material online). Phylogenetic relationships of C. briggsae hermaphrodite lineages were analyzed using a combined sequence set of 1,694 bp containing Ψnad5-1; Ψnad5-2; and sequences from nad3, nad5, trnQ, trnF, and cob. These regions were amplified and sequenced for 117 isolates and combined with homologous regions from published mitochondrial genome sequences for 24 additional C. briggsae isolates (Howe and Denver 2008). Among the 141 isolate-specific sequences analyzed, 44 unique haplotypes were observed and used in subsequent phylogenetic analyses. The three major intraspecific clades of C. briggsae identified in previous studies (Dolgin et al. 2008; Howe and Denver 2008) were again identified here with strong bootstrap support (fig. 3). Furthermore, Ψnad5-1 was observed in all C. briggsae natural isolates, whereas Ψnad5-2 was present only in isolates falling into the (I + II) superclade, also consistent with our previous analysis of 24 isolates (Howe and Denver 2008). The worldwide geographic distribution patterns of the growing collection of C. briggsae natural isolates are available on an online GoogleMap resource we have made with help from our colleague A. D. Cutter (supplementary note 1, Supplementary Material online).
FIG. 3.
Phylogenetic analysis of Caenorhabditis briggsae mitochondrial haplotypes. Unrooted 80% bootstrap consensus cladogram was constructed using 1,694 bp of mtDNA, including both Ψnad5-1 and Ψnad5-2, analyzed in all 141 isolates. The 44 unique mtDNA haplotypes were used for phylogenetic analysis. Ψnad5-2 is absent in Clade III isolates—the gapped region in Clade III isolate sequences was treated as a single event in phylogenetic analyses (complete deletion parameters in MEGA4). Bootstrap values for maximum parsimony (left) and NJ (right) methods (1,000 replicates performed for each) are indicated to the left of the corresponding node. Numbers in parentheses next to haplotype designators show the numbers of isolates with that haplotype. Three-letter country code designators (United Nations designators) are also provided for general geographic information—see supplementary table 1 (Supplementary Material online) for more detailed information on haplotypes and geographic locations, especially for isolates collected from islands. Complete phylograms from NJ and maximum parsimony analyses are shown in supplementary figs. 1 and 2 (Supplementary Material online), respectively.
Our analysis of mtDNA pseudogene regions in 141 C. briggsae natural isolates enabled the study of intraspecific patterns of genetic diversity in this species at both global and more local scales and revealed new and unexpected C. briggsae haplotypes and isolate groupings. One unexpected finding was that a new set of isolates from southern India were placed in Clade III, whereas in previous studies, only equatorial isolates from Kenya composed this group (Dolgin et al. 2008; Howe and Denver 2008). This observation suggests that Clade III isolates have a wider geographic range than previous smaller scale studies suggested. Furthermore, both Clade III and Clade I isolates were identified at the same sites in Ponmudi, Kerala, India (supplementary table 1, Supplementary Material online), whereas in previous studies, only isolates from a single major intraspecific clade were observed at any given location.
Although most haplotypes were placed within the three previously established clades of C. briggsae, two haplotypes, each unique to one isolate, were found to fall outside of these groupings. Our bootstrap consensus phylogenies placed isolate QR24 (Quebec, Canada) inside the (I + II) superclade but not within either Clade I or Clade II (fig. 3). NJ and maximum parsimony analyses yielded conflicting results in terms of the placement of QR24, though both arrangements were poorly supported by bootstrap analysis (supplementary figs. 1 and 2, Supplementary Material online). Similarly, isolate JU1424 (northeast Vietnam) was placed inside the (I + II) superclade but not within either Clade I or Clade II; NJ and maximum parsimony again yielded conflicting and poorly supported results. We also analyzed a nuclear region (primarily composed of the Cbr-polh-1 third intron) in the 141 C. briggsae isolates. Whereas this nuclear region yielded similar results to mtDNA for QR24, JU1424 was placed within Clade I with strong bootstrap support in both NJ and maximum parsimony analyses (supplementary fig. 3, Supplementary Material online). The incongruous results for JU1424 suggest that this strain’s nuclear genome has derived, at least in part, from a typical Clade I genotype. Both QR24 and JU1424 mitochondrial genomes contained Ψnad5-1 and Ψnad5-2. These findings suggest that substantial genetic diversity in C. briggsae might still remain undiscovered, especially in light of the still patchy sampling from parts of the world other than North America and Europe. The uncertain phylogenetic placement of QR24 and JU1424 in either Clade I or Clade II along with the increased extent of geographic overlap of isolates placed in different major intraspecific clades motivated the new generic C. briggsae clade naming scheme implemented here.
In addition to yielding new insights into the global distribution patterns of C. briggsae natural isolates, our study also provided the opportunity to study more localized patterns of genetic diversity in this species. France has been very well sampled (69/141 isolates analyzed here are from this country), and a previous analysis of six nuclear loci in the C. briggsae natural isolates (Cutter et al. 2006) showed that all isolates from France except one (JU516) shared identical alleles across all six loci, indicating that the France isolates are closely related to one another. However, knowledge on patterns of genetic variation within and among geographic sites in France has remained enigmatic. The more rapidly evolving mtDNA sequences examined here revealed 12 distinct haplotypes, all very similar to one another (Clade II), among the 69 France isolates analyzed. Ten different geographic locations in France were surveyed (supplementary table 1, Supplementary Material online) and either one or two haplotypes were found at each of these locations (supplementary fig. 4, Supplementary Material online). Haplotypes mH32 and mH37 were each observed at three different locations; all other France haplotypes were specific to a single location in this country. However, some France haplotypes were also observed in other parts of the world: mH33 was observed in France and Japan; mH34 was observed in France and the United States. This pattern of identical mtDNA haplotypes appearing in isolates from disparate geographic locations—also observed in C. elegans (Denver et al. 2003)—suggests that C. briggsae nematodes are capable of migrating large distances, perhaps facilitated by associations with humans or other animals (Kiontke and Sudhaus 2006).
We also discovered that two isolates from St Joseph, MO (DL0231, DL0232—Clade II) produced much larger PCR amplicons at the Ψnad5-2 locus as compared with other Clade I and Clade II C. briggsae isolates. These two isolates were collected along with six other C. briggsae isolates from the same rotting apples sampling site—the latter six produce smaller amplicons similar to those observed in other isolates. In DL0231 and DL0232, however, DNA sequencing analysis revealed that these two isolates each contained identical tandem duplicate copies of Ψnad5-2 (referred to as Ψnad5-2a and Ψnad5-2b—see fig. 1). The DNA sequence was identical in Ψnad5-2a and Ψnad5-2b, also the same as the sequence observed in the single Ψnad5-2 copies analyzed in the other six St Joseph isolates. PCR analysis (supplementary fig. 5, Supplementary Material online) of the Ψnad5-2 region in DL0231 and DL0232 suggests that the duplication-bearing genome is not fixed but rather coexists in a heteroplasmic state with genomes containing single copies of Ψnad5-2 and nad5 deletion-bearing molecules. This finding shows that the Ψnad5-2 region is subject not only to DNA deletion dynamics (Howe and Denver 2008) but also to tandem duplication events.
Nucleotide Diversity of C. briggsae mtDNA Pseudogenes
Pseudogene sequences are often assumed to be nonfunctional and expected to evolve in a neutral fashion (Li et al. 1981). Although we have no a posteriori reasons to expect that Ψnad5-1 has evolved in a nonneutral fashion, for Ψnad5-2, there are biological reasons why we might expect selection to have affected its evolution. Direct repeats present in Ψnad5-2 and downstream in nad5 are associated with large heteroplasmic mtDNA deletions that eliminate the 5′ end of nad5; isolate-specific deletion levels were shown to be negatively correlated with nematode fitness (Howe and Denver 2008). Furthermore, two putative compensatory alleles (named DRSeq2 and DRSeq3) have been characterized in C. briggsae natural isolates; the alleles occur in the Ψnad5-2 direct repeat copy and render the Ψnad5-2 repeat an imperfect match to the downstream nad5 repeat; isolates with these putative compensatory alleles have significantly lower deletion levels than those where the Ψnad5-2 and nad5 direct repeats are perfect matches. The compensatory alleles were found exclusively in Clade II isolates in our previous small-scale study (Howe and Denver 2008); this pattern was also observed in the current analysis of 141 C. briggsae isolates. The frequency of compensatory allele DRSeq2 in the Clade II isolates analyzed was 0.06 (encoded by a few isolates from North America), and the frequency of allele DRSeq3 was 0.82 (present in all the numerous isolates from France). These allele frequencies should be interpreted with caution, however, given the highly biased worldwide sampling of C. briggsae isolates. It is unknown why the putative compensatory alleles evolved in Clade II but not in Clade I. Although our lab-based fitness assays suggested that the deletions are deleterious, it is possible that in nature the deletions are somehow beneficial to the Clade I nematodes.
To evaluate whether the C. briggsae mtDNA pseudogenes are evolving in a fashion consistent with neutral expectations, we analyzed π for Ψnad5-1 and Ψnad5-2 and compared the diversity estimates with nonsynonymous- and synonymous-site values (πN and πS, respectively) from concatenated protein-coding sequences (nad3, nad5, and cob partial sequences). Synonymous sites in protein-coding genes are frequently used to approximate patterns of neutral molecular evolution, whereas nonsynonymous sites in mtDNA are generally under strong stabilizing selection associated with protein-coding function. We calculated π values for the four sequence categories (Ψnad5-1, Ψnad5-2, nonsynonymous sites, and synonymous sites) in each of C. briggsae Clade I and Clade II using DnaSP v4.0 (Rozas et al. 2003). For Clade III, Ψnad5-2 is absent and therefore not included in π calculations. Approximate 95% CIs for each π calculation were estimated using coalescent simulations in DnaSP following the approach used in Graustein et al. (2002).
Consistent with results from previous studies (Cutter et al. 2006; Howe and Denver 2008), we observed greater levels of genetic diversity in Clade I versus Clade II isolates regardless of the sequence category analyzed (table 1), though πS values were more similar between the clades as compared with the results of Howe and Denver (2008) that analyzed fewer isolates but nearly complete mitochondrial genome sequences. Clade II nematodes have been isolated throughout the northern temperate latitudes of North America, Europe, and Asia yet display very little genetic diversity—this observation led Cutter et al. (2006) to hypothesize that C. briggsae might have started colonizing temperate latitudes in just the last few thousand years. Our study also provided the opportunity to compare patterns of genetic diversity in Clade III with that observed in the other two major intraspecific clades. π Estimates were similar between Clade III and Clade I isolates at synonymous sites and Ψnad5-1 (Ψnad5-2 is not present in Clade III nematodes). When considered in light of the position of Clade III in the C. briggsae intraspecific phylogeny (fig. 3), this result suggests that Clade III is a deeply ancestral and diverse group. We also calculated nucleotide diversity specific to the Ψnad5-1 element in the five Caenorhabditis sp. 5 isolates and found the π estimate to be similar to those observed in C. briggsae Clade I and Clade III.
Table 1.
π Estimates for mtDNA Pseudogenes and Protein-Coding Sequences.
| Ψnad5-1 | Ψnad5-2 | PC Syn. Sites | PC Nonsyn. Sites | |
| Cb Clade I | 0.0422 (0.0106–0.0781)a | 0.0218 (0.0051–0.0411) | 0.0151 (0.0027–0.0326) | 0.0031 (0.0007–0.0089) |
| Cb Clade II | 0.0035 (0.0004–0.0149) | 0.0079 (0.0018–0.0202) | 0.0115 (0.0027–0.0306) | 0.0012 (0.0001–0.0043) |
| Cb Clade III | 0.0399 (0.0120–0.0967) | NAb | 0.0268 (0.0085–0.0728) | 0.0003 (0–0.0026) |
| C. sp. 5 | 0.0299 (0.0057–0.0951) | NA | ND | ND |
PC syn. sites, synonymous sites in protein-coding genes; PC nonsyn. sites, nonsynonymous sites in protein-coding genes; Cb, Caenorhabditis briggsae; C. sp. 5, Caenorhabditis sp. 5; ND, not determined.
Calculated π values are indicated on top with approximate 95% CIs shown below their respective values in parentheses.
NA indicates not applicable (Clade III and C. sp. 5 lack Ψnad5-2).
If the C. briggsae pseudogenes have evolved in neutral or nearly neutral fashions, then genetic diversity at these loci would be expected to be similar to that observed at other sites presumed to be evolving neutrally, such as synonymous sites in protein-coding genes. The π estimates specific to mtDNA pseudogenes were similar to those observed in corresponding clade-specific πS estimates (95% CIs overlap in all cases—see table 1). As expected, the πN estimates were all much lower than their corresponding clade-specific πS estimates, and the pseudogene-specific π estimates, presumably due to the effects of purifying selection associated with protein-coding function. In addition to the values reported in table 1, we also calculated π values for all 141 C. briggsae natural isolates in Ψnad5-1 (Ψnad5-2 is absent from Clade III isolates) and observed its value (π = 0.0570; 95% CI = 0.0224–0.1423) to be similar to the πS value calculated from all isolates (πS = 0.0721; 95% CI = 0.0224–0.1501). Given these results, we cannot rule out the hypothesis that the C. briggsae mtDNA pseudogenes have evolved in a mostly neutral fashion similar to synonymous sites in mtDNA protein-coding genes. However, the small sizes of Ψnad5-1 and Ψnad5-2 posed inherent limitations on the power of our analyses, and some large magnitudinal differences in the π estimates were observed that are worthy of discussion. For example, π values were greater for the pseudogenes than synonymous sites in Clade I, whereas the opposite was true for Clade II. Assuming synonymous sites are neutral, these patterns might reflect diversifying selection acting on the pseudogenes in Clade I and purifying selection acting on the pseudogenes in Clade II. For Ψnad5-2, the compensatory alleles DRSeq2 and DRSeq3 are observed only in Clade II; thus, purifying selection to maintain the compensatory alleles might be acting in Clade II. Although we were unable to rule out neutral evolution of the pseudogenes, our analyses characterized evolutionary processes across entire aligned pseudogene regions where the potential selective effects on a few nucleotide sites (e.g., the DRSeq2 and DRSeq3 putative compensatory alleles) might have been overwhelmed by neutral evolution across the majority of sites.
Conclusions
This study provides important insights into both the between- and the within-species evolution of Caenorhabditis mtDNA pseudogenes and shows that these elements harbor high levels of nucleotide diversity in C. briggsae natural populations. The punctuated appearance and disappearance of nad5-derived pseudogenes on the Caenorhabditis phylogeny are inconsistent with the widely accepted TDRL model for animal mitochondrial genome evolution and suggest that recombination-like processes might play a major role in Caenorhabditis mitochondrial genome evolution. The discovery of the two unusual new C. briggsae mtDNA haplotypes (JU1424 and QR24) suggests that there is still much to learn about the levels and sources of genetic diversity in this nematode species. We are optimistic that the ongoing discovery of new C. briggsae natural isolates and Caenorhabditis species, coupled with the application of new DNA sequencing technologies to nematode mitochondrial genomics, will provide further insights into the evolution of Caenorhabditis mtDNA pseudogenes in the future.
Supplementary Material
Supplementary note 1, tables 1 and 2, and figures 1–5 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We are grateful to M. Ailion, F. Back, D. Baïlle, A. Barrière, A. Choe, E. Dolgin, E. Hoyos, M. Leroy, V. Robert, and the Caenorhabditis Genetics Center for providing nematode strains and to Mark Dasenko at the Oregon State University Center for Genome Research and Biocomputing for DNA sequencing support. Thanks to W. K. Thomas for providing the C. remanei mtDNA sequence and to A. D. Cutter for insightful comments on the manuscript and help with the C. briggsae GoogleMap resource. D.R.D. was supported by National Institutes of Health Grant GM087678 and M.-A.F. was supported by the Centre National de la Recherche Scientifique.
References
- Arndt A, Smith MJ. Mitochondrial gene rearrangement in the sea cucumber genus Cucumaria. Mol Biol Evol. 1998;15:1009–1016. doi: 10.1093/oxfordjournals.molbev.a025999. [DOI] [PubMed] [Google Scholar]
- Bakke II, Shields GF, Johansen S. Sequence characterization of a unique intergenic spacer in gadiformes mitochondrial DNA. Mar Biotechnol (NY). 1999;1:411–415. doi: 10.1007/pl00011797. [DOI] [PubMed] [Google Scholar]
- Barriere A, Felix MA. Isolation of C. elegans and related nematodes. WormBook. 2006;17:1–9. doi: 10.1895/wormbook.1.115.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore JL, Brown WM. Big trees from little genomes: mitochondrial gene order as a phylogenetic tool. Curr Opin Genet Dev. 1998;8:668–674. doi: 10.1016/s0959-437x(98)80035-x. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. Fundamental concepts in genetics: effective population size and patterns of molecular evolution and variation. Nat Rev Genet. 2009;10:195–205. doi: 10.1038/nrg2526. [DOI] [PubMed] [Google Scholar]
- Cutter AD. Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Mol Biol Evol. 2008;25:778–786. doi: 10.1093/molbev/msn024. [DOI] [PubMed] [Google Scholar]
- Cutter AD, Felix MA, Barriere A, Charlesworth D. Patterns of nucleotide polymorphism distinguish temperate and tropical wild isolates of Caenorhabditis briggsae. Genetics. 2006;173:2021–2031. doi: 10.1534/genetics.106.058651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Morris K, Lynch M, Vassilieva LL, Thomas WK. High direct estimate of the mutation rate in the mitochondrial genome of Caenorhabditis elegans. Science. 2000;289:2342–2344. doi: 10.1126/science.289.5488.2342. [DOI] [PubMed] [Google Scholar]
- Denver DR, Morris K, Thomas WK. Phylogenetics in Caenorhabditis elegans: an analysis of divergence and outcrossing. Mol Biol Evol. 2003;20:393–400. doi: 10.1093/molbev/msg044. [DOI] [PubMed] [Google Scholar]
- Dolgin ES, Felix MA, Cutter AD. Hakuna Nematoda: genetic and phenotypic diversity in African isolates of Caenorhabditis elegans and C. briggsae. Heredity. 2008;100:304–315. doi: 10.1038/sj.hdy.6801079. [DOI] [PubMed] [Google Scholar]
- Fujita MK, Boore JL, Moritz C. Multiple origins and rapid evolution of duplicated mitochondrial genes in parthenogenetic geckos (Heteronotia binoei; Squamata, Gekkonidae) Mol Biol Evol. 2007;24:2775–2786. doi: 10.1093/molbev/msm212. [DOI] [PubMed] [Google Scholar]
- Gissi C, Iannelli F, Pesole G. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity. 2008;101:301–320. doi: 10.1038/hdy.2008.62. [DOI] [PubMed] [Google Scholar]
- Graustein A, Gaspar JM, Walters JR, Palopoli MF. Levels of DNA polymorphism vary with mating system in the nematode genus Caenorhabditis. Genetics. 2002;161:99–107. doi: 10.1093/genetics/161.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe DK, Denver DR. Muller’s Ratchet and compensatory mutation in Caenorhabditis briggsae mitochondrial genome evolution. BMC Evol Biol. 2008;8:62. doi: 10.1186/1471-2148-8-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke K, Barriere A, Kolotuev I, Podbilewicz B, Sommer R, Fitch DH, Felix MA. Trends, stasis, and drift in the evolution of nematode vulva development. Curr Biol. 2007;17:1925–1937. doi: 10.1016/j.cub.2007.10.061. [DOI] [PubMed] [Google Scholar]
- Kiontke K, Fitch DH. The phylogenetic relationships of Caenorhabditis and other rhabditids. WormBook. 2005;11:1–11. doi: 10.1895/wormbook.1.11.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke K, Sudhaus W. Ecology of Caenorhabditis species. WormBook. 2006;9:1–14. doi: 10.1895/wormbook.1.37.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumazawa Y, Ota H, Nishida M, Ozawa T. Gene rearrangements in snake mitochondrial genomes: highly concerted evolution of control-region-like sequences duplicated and inserted into a tRNA gene cluster. Mol Biol Evol. 1996;13:1242–1254. doi: 10.1093/oxfordjournals.molbev.a025690. [DOI] [PubMed] [Google Scholar]
- Kurabayashi A, Sumida M, Yonekawa H, Glaw F, Vences M, Hasegawa M. Phylogeny, recombination, and mechanisms of stepwise mitochondrial genome reorganization in mantellid frogs from Madagascar. Mol Biol Evol. 2008;25:874–891. doi: 10.1093/molbev/msn031. [DOI] [PubMed] [Google Scholar]
- Li WH, Gojobori T, Nei M. Pseudogenes as a paradigm of neutral evolution. Nature. 1981;292:237–239. doi: 10.1038/292237a0. [DOI] [PubMed] [Google Scholar]
- Lunt DH, Hyman BC. Animal mitochondrial DNA recombination. Nature. 1997;387:247. doi: 10.1038/387247a0. [DOI] [PubMed] [Google Scholar]
- Lynch M, Koskella B, Schaack S. Mutation pressure and the evolution of organelle genomic architecture. Science. 2006;311:1727–1730. doi: 10.1126/science.1118884. [DOI] [PubMed] [Google Scholar]
- Mabuchi K, Miya M, Satoh TP, Westneat MW, Nishida M. Gene rearrangements and evolution of tRNA pseudogenes in the mitochondrial genome of the parrotfish (Teleostei: Perciformes: Scaridae) J Mol Evol. 2004;59:287–297. doi: 10.1007/s00239-004-2621-z. [DOI] [PubMed] [Google Scholar]
- Macey JR, Papenfuss TJ, Kuehl JV, Fourcade HM, Boore JL. Phylogenetic relationships among amphisbaenian reptiles based on complete mitochondrial genomic sequences. Mol Phylogenet Evol. 2004;33:22–31. doi: 10.1016/j.ympev.2004.05.003. [DOI] [PubMed] [Google Scholar]
- McKnight ML, Shaffer HB. Large, rapidly evolving intergenic spacers in the mitochondrial DNA of the salamander family Ambystomatidae (Amphibia: Caudata) Mol Biol Evol. 1997;14:1167–1176. doi: 10.1093/oxfordjournals.molbev.a025726. [DOI] [PubMed] [Google Scholar]
- Mueller RL, Boore JL. Molecular mechanisms of extensive mitochondrial gene rearrangement in plethodontid salamanders. Mol Biol Evol. 2005;22:2104–2112. doi: 10.1093/molbev/msi204. [DOI] [PubMed] [Google Scholar]
- Paabo S, Thomas WK, Whitfield KM, Kumazawa Y, Wilson AC. Rearrangements of mitochondrial transfer RNA genes in marsupials. J Mol Evol. 1991;33:426–430. doi: 10.1007/BF02103134. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- San Mauro D, Gower DJ, Zardoya R, Wilkinson M. A hotspot of gene order rearrangement by tandem duplication and random loss in the vertebrate mitochondrial genome. Mol Biol Evol. 2006;23:227–234. doi: 10.1093/molbev/msj025. [DOI] [PubMed] [Google Scholar]
- Stein LD, Bao Z, Blasiar D, et al. (36 co-authors) The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 2003;1:E45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tang S, Hyman BC. Mitochondrial genome haplotype hypervariation within the isopod parasitic nematode Thaumamermis cosgrovei. Genetics. 2007;176:1139–1150. doi: 10.1534/genetics.106.069518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend JP, Rand DM. Mitochondrial genome size variation in New World and Old World populations of Drosophila melanogaster. Heredity. 2004;93:98–103. doi: 10.1038/sj.hdy.6800484. [DOI] [PubMed] [Google Scholar]
- Wood W. The nematode Caenorhabditis elegans. 1988. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press. [Google Scholar]
- Yu Z, Wei Z, Kong X, Shi W. Complete mitochondrial DNA sequence of oyster Crassostrea hongkongensis—a case of “Tandem duplication-random loss” for genome rearrangement in Crassostrea? BMC Genomics. 2008;9:477. doi: 10.1186/1471-2164-9-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Boyle TJ, Bao Z, Murray JI, Mericle B, Waterston RH. Comparative analysis of embryonic cell lineage between Caenorhabditis briggsae and Caenorhabditis elegans. Dev Biol. 2008;314:93–99. doi: 10.1016/j.ydbio.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.