Abstract
Despite progress with triplex-forming oligonucleotides or helix-invading peptide nucleic acids (PNAs), there remains a need for probes facilitating sequence-unrestricted targeting of double stranded DNA (dsDNA) at physiologically relevant conditions. Invader LNA probes, i.e., DNA duplexes with “+1 interstrand zipper arrangements” of intercalator-functionalized 2’-amino-α-L-LNA monomers, are demonstrated herein to recognize short mixed sequence dsDNA targets. This approach, like pseudo-complementary PNA (pcPNA), relies on relative differences in stability between probe duplexes and the corresponding probe:target duplexes for generation of a favourable thermodynamic gradient. Unlike pcPNA, Invader LNA probes take advantage of the “nearest neighbour exclusion principle”, i.e., intercalating units of Invader LNA monomers are poorly accommodated in probe duplexes but extraordinarily well tolerated in probe-target duplexes (ΔTm/modification up to +11.5 °C). Recognition of isosequential dsDNA-targets occurs: a) at experimental temperatures much lower than the thermal denaturation temperatures (Tm’s) of Invader LNAs or dsDNA-targets, b) at a wide range of ionic strengths, and c) with good mismatch discrimination. dsDNA recognition is monitored in real-time using inherent pyrene-pyrene excimer signals of Invader LNA probes, which provides insights into reaction kinetics and enables rational design of probes. These properties render Invader LNAs as promising probes for biomedical applications entailing sequence-unrestricted recognition of dsDNA.
INTRODUCTION
Extensive efforts have been made in the past two decades to develop agents for site-specific targeting of double stranded DNA (dsDNA). These efforts have been stimulated by the prospect of developing enabling tools for functional genomics, and discovering novel classes of therapeutic and diagnostic agents1. dsDNA-targeting agents have been used in a variety of applications including site-specific modulation of gene expression2,3, induction of site-specific mutagenesis, recombination or repair of genomic DNA2,3, and direct detection of dsDNA in living cells4. Triplex forming oligonucleotides (TFOs)1–3 and peptide nucleic acids (PNAs)5,6 are the most widely used dsDNA-targeting probes despite:
the necessity for long homopurine tracts (>15 nt) in dsDNA targets to ensure sufficient binding affinity/specificity of TFOs/PNAs1,
weak pH-dependent Hoogsteen base pairs between TC-motif TFOs and dsDNA, although this has been addressed by modification of TFOs with affinity-enhancing building blocks such as 2’-deoxy-5-methylcytidine, C5-functionalized pyrimidines7–9, Locked Nucleic Acid (LNA)10–12, α-L-LNA13, Bridged Nucleic Acids (BNAs)14,15, C5-alkynyl functionalized LNA16 and non-nucleosidic monomers17,
formation of secondary structures by G-rich TFOs in the presence of physiological concentrations of potassium ions, which affects dsDNA target binding18,19, and
the need for non-physiological ionic strengths to facilitate strand invasion by PNA5,6.
Accordingly, there has been an intense search for alternative probe technologies that facilitate recognition of mixed sequence dsDNA targets20. Examples include minor groove binding polyamides21, TFOs capable of recognizing all four Watson-Crick base pairs via the major groove22,23, optimized PNAs24,25, antigene (ag) PNA26,27, agLNA28,29, “Zorro” LNA30, pseudo-complementary (pc) DNA31,32, pcPNA33–38, and other approaches39–41. While these emerging probe strategies are promising, there remains a need for a probe technology that allows fast, efficient and specific dsDNA targeting at physiologically relevant ionic strengths with minimal sequence restrictions and uncomplicated “DNA-like” handling (adequate aqueous solubility, compatibility with delivery agents).
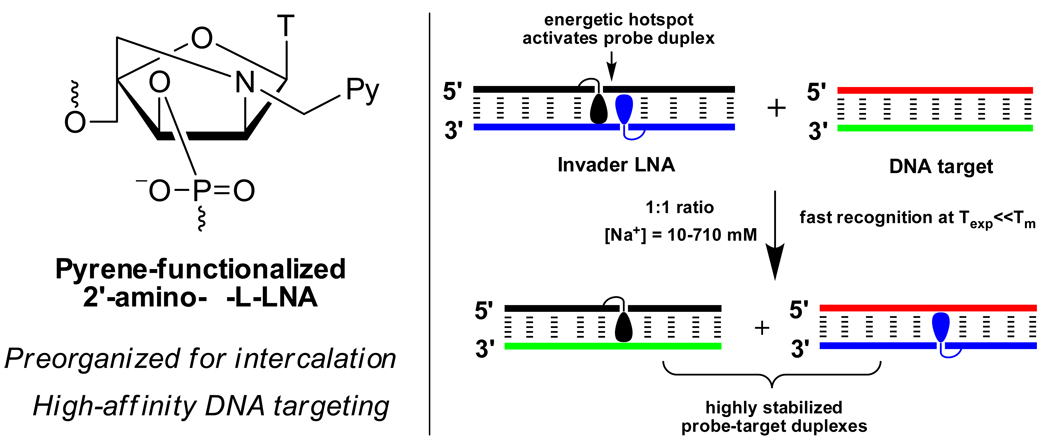
We have recently introduced N2’-intercalator-functionalized 2’-amino-α-L-LNA and have demonstrated their ability to precisely direct the appended intercalators into the core of DNA duplexes42–47. As a result, extraordinary increases in thermal affinity toward DNA complements of up to +19.5 °C per modification are observed relative to unmodified reference strands. This has facilitated the development of probes for detection of single nucleotide polymorphisms45 and stabilization of abasic sites46.
Herein, we demonstrate recognition of isosequential dsDNA targets using double stranded 2’-N-(pyren-1-yl)methyl-2’-amino-α-L-LNA probes dubbed Invader LNA€,42 in buffers of varying ionic strength (Fig. 1). Important insights into the thermodynamics and kinetics of the recognition events are gained from thermal denaturation, steady-state fluorescence and fluorescence decay experiments, allowing the identification of key design principles.
Figure 1.
The Invader LNA concept.
RESULTS AND DISCUSSION
Thermal denaturation properties of 2’-N-(pyren-1-yl)methyl-2’-amino-α-L-LNA
Singly modified 2’-N-(pyren-1-yl)methyl-2’-amino-α-L-LNA form extraordinarily stable duplexes with DNA complements compared to the corresponding unmodified reference DNA duplex ON1:ON2 (ΔTm = +9.5 to +11.5 °C and ΔΔG293 = −9 to −14 kJ/mol, entries 3–9, Table 1). Slightly less stabilized duplexes are observed when monomer X is incorporated close to the 5’-terminus (entry 2, Table 1). The corresponding duplexes with RNA targets are invariably less stable, but still significantly stabilized (ΔTm/mod-values between +0.5 and +6.0 °C, Table S7†).
Table 1.
Hybridization data for duplexes between 2’-N-(pyren-1-yl)methyl-2’-amino-α-L-LNA ON3–ON16 and complementary DNA.[a]
| Entry | ON | Duplex |
Tm [ΔTm] (°C) |
ΔH [ΔΔH] (kJ/mol) |
(T293ΔS) [Δ(T293ΔS)] (kJ/mol) |
ΔG293 [ΔΔG293] (kJ/mol) |
|---|---|---|---|---|---|---|
| 1 | ON1 | 5’-d(GGT ATA TAT AGG C) | 37.5 | −382 | −325 | −57 |
| ON2 | 3’-d(CCA TAT ATA TCC G) | - | - | - | - | |
| 2 | ON3 | 5’-d(GGX ATA TAT AGG C) | 44.5 | −316 | −256 | −60 |
| ON2 | 3’-d(CCA TAT ATA TCC G) | [+7.0] | [+66] | [+69] | [−3] | |
| 3 | ON4 | 5’-d(GGT AXA TAT AGG C) | 47.5 | −356 | −290 | −66 |
| ON2 | 3’-d(CCA TAT ATA TCC G) | [+10.0] | [+26] | [+35] | [−9] | |
| 4 | ON5 | 5’-d(GGT ATA XAT AGG C) | 49.0 | −380 | −311 | −69 |
| ON2 | 3’-d(CCA TAT ATA TCC G) | [+11.5] | [+2] | [+14] | [−12] | |
| 5 | ON6 | 5’-d(GGT ATA TAX AGG C) | 47.0 | −358 | −293 | −66 |
| ON2 | 3’-d(CCA TAT ATA TCC G) | [+9.5] | [+24] | [+32] | [−9] | |
| 6 | ON1 | 5’-d(GGT ATA TAT AGG C) | 48.5 | −409 | −338 | −71 |
| ON7 | 3’-d(CCA XAT ATA TCC G) | [+11.0] | [−27] | [−13] | [−14] | |
| 7 | ON1 | 5’-d(GGT ATA TAT AGG C) | 48.5 | −383 | −314 | −70 |
| ON8 | 3’-d(CCA TAX ATA TCC G) | [+11.0] | [−1] | [+11] | [−13] | |
| 8 | ON1 | 5’-d(GGT ATA TAT AGG C) | 48.0 | −390 | −320 | −70 |
| ON9 | 3’-d(CCA TAT AXA TCC G) | [+10.5] | [−8] | [+5] | [−13] | |
| 9 | ON1 | 5’-d(GGT ATA TAT AGG C) | 47.0 | −392 | −325 | −67 |
| ON10 | 3’-d(CCA TAT ATA XCC G) | [+9.5] | [−10] | [0] | [−10] | |
| 10 | ON11 | 5’-d(GGX AXA TAT AGG C) | 53.5 | −300 | −233 | −66 |
| ON2 | 3’-d(CCA TAT ATA TCC G) | [+16.0] | [+82] | [+92] | [−9] | |
| 11 | ON12 | 5’-d(GGX ATA XAT AGG C) | 57.5 | −340 | −266 | −74 |
| ON2 | 3’-d(CCA TAT ATA TCC G) | [+20.0] | [+42] | [+59] | [−17] | |
| 12 | ON13 | 5’-d(GGX ATA TAX AGG C) | 56.5 | −399 | −319 | −80 |
| ON2 | 3’-d(CCA TAT ATA TCC G) | [+19.0] | [−17] | [+6] | [−23] | |
| 13 | ON1 | 5’-d(GGT ATA TAT AGG C) | 57.5 | −385 | −306 | −79 |
| ON14 | 3’-d(CCA XAX ATA TCC G) | [+20.0] | [−3] | [+19] | [−22] | |
| 14 | ON1 | 5’-d(GGT ATA TAT AGG C) | 61.0 | −414 | −328 | −86 |
| ON15 | 3’-d(CCA XAT AXA TCC G) | [+23.5] | [−32] | [−3] | [−29] | |
| 15 | ON1 | 5’-d(GGT ATA TAT AGG C) | 59.0 | −405 | −322 | −83 |
| ON16 | 3’-d(CCA XAT ATA XCC G) | [+21.5] | [−23] | [+3] | [−26] |
Tm-values (ΔTm = change in Tm-value relative to ON1:ON2) and Gibbs free energy of duplex formation at 293 K, ΔG293, determined by baseline fitting of optical melting curve (A260 vs T) recorded in medium salt buffer ([Na+] = 110 mM, [Cl-] = 100 mM, pH 7.0 (adjusted with NaH2PO4/Na2HPO4)), using 1.0 µM of each strand. Values are averages of at least two measurements. ΔΔG293 = difference in Gibbs free energy of duplex formation relative to reference duplex ON1:ON2. For structure of monomer X, see Fig. 1.
ONs modified with two 2’-N-(pyren-1-yl)methyl-2’-amino-α-L-LNA X monomers positioned as next-nearest neighbours exhibit slightly less-than-additive increases in thermal affinity (compare e.g. entries 2, 3 and 10, Table 1). Conversely, when two X monomers in an ON are separated by greater distances, more-than-additive increases in thermal affinity are observed (compare e.g. entries 2, 5 and 12, Table 1), resulting in the formation of highly stabilized duplexes with ΔTm-values up to +23.5 °C.
The increased thermal affinity of 2’-N-(pyren-1-yl)methyl-2’-amino-α-L-LNA ON3–ON16 toward complementary DNA primarily arises from less unfavourable entropic contributions that either are not fully counterbalanced by less favourable enthalpic contributions (entries 2–5 and 10–11, Table 1) or are augmented by marginally more favourable enthalpic contributions (entries 7–8, 12–13 and 15, Table 1). This most likely reflects the conformational preorganization of the bicyclic 2’-amino-α-L-LNA skeleton, which directs the appended pyrene moiety to the duplex core for favourable intercalation (Fig. 1) 42–47.
Thermal denaturation properties of Invader LNA probes
Double stranded 2’-N-(pyren-1-yl)methyl-2’-amino-α-L-LNA probes with a single “+1 interstrand zipper arrangement” of X monomers are relatively unstable (ΔTm = −1.5 to +3.5 °C and ΔΔG293 = −3.0 to +9.0 kJ/mol, entries 1–4, Table 2). For the sake of simplicity, probes with +1 zipper arrangements of X monomers are henceforth dubbed “Invader LNA”, and these particular zipper arrangements are dubbed “energetic hotspots”42,€. The low stability of Invader LNA probes most likely reflects a violation of the ‘nearest neighbour exclusion principle’, which states that intercalators at most bind to every second base pair of a DNA duplex due to constraints in the local expandability of duplexes49. Invader LNA probes feature one intercalator per base pair (Fig. 1). Two additional observations support this hypothesis. First, the less favourable enthalpy of Invader LNA probes indicates intercalator-mediated perturbation of local duplex geometry and base pairing (see ΔΔH for entries 1–4, Table 2). Secondly, DNA duplexes with other interstrand zipper arrangements of X monomers and therefore lower local densities of intercalating units, are extraordinarily stable (ΔTm = +16.5 °C to +22.0 °C for −3, −1, +3 and +5 zippers, Table S8†).
Table 2.
Hybridization data for Invader LNA probes and their free energy, , for recognition of dsDNA target ON1:ON2 at 293 K.a
| Entry | ON | Duplex |
Tm [ΔTm] (°C) |
ΔH[ΔΔH] (kJ/mol) |
(T293ΔS)[Δ(T293ΔS)] (kJ/mol) |
ΔG293[ΔΔG293] (kJ/mol) |
(kJ/mol) |
|---|---|---|---|---|---|---|---|
| 1 | ON3 | 5’-d(GGX ATA TAT AGG C) | 36.5 | −262 | −214 | −48 | −26 |
| ON7 | 3’-d(CCA XAT ATA TCC G) | [−1.0] | [+120] | [+111] | [+9] | ||
| 2 | ON4 | 5’-d(GGT AXA TAT AGG C) | 41.0 | −360 | −301 | −60 | −19 |
| ON8 | 3’-d(CCA TAX ATA TCC G) | [+3.5] | [+22] | [+24] | [−3] | ||
| 3 | ON5 | 5’-d(GGT ATA XAT AGG C) | 40.5 | −375 | −315 | −60 | −22 |
| ON9 | 3’-d(CCA TAT AXA TCC G) | [+3.0] | [+7] | [+10] | [−3] | ||
| 4 | ON6 | 5’-d(GGT ATA TAX AGG C) | 36.0 | −311 | −260 | −51 | −24 |
| ON10 | 3’-d(CCA TAT ATA XCC G) | [−1.5] | [+71] | [+65] | [+6] | ||
| 5 | ON11 | 5’-d(GGX AXA TAT AGG C) | 48.0 | −219 | −165 | −55 | −34 |
| ON14 | 3’-d(CCA XAX ATA TCC G) | [+10.5] | [+163] | [+160] | [+2] | ||
| 6 | ON12 | 5’-d(GGX ATA XAT AGG C) | 53.0 | −209 | −152 | −56 | −47 |
| ON15 | 3’-d(CCA XAT AXA TCC G) | [+15.5] | [+173] | [+173] | [+1] | ||
| 7 | ON13 | 5’-d(GGX ATA TAX AGG C) | 48.0 | −230 | −175 | −55 | −50 |
| ON16 | 3’-d(CCA XAT ATA XCC G) | [+10.5] | [+152] | [+150] | [+2] |
Conditions as specified in Table 1.
Invader LNA with two energetic hotspots form surprisingly stable duplexes (ΔTm = +10.5 to +15.5 °C, entries 5–7, Table 2). Interestingly, this trend is not paralleled in the change in free energy upon hybridization at 293K (ΔΔG293 ~ 1–2 kJ/mol relative to unmodified ON1:ON2, entries 5–7, Table 2). This is a result of extraordinarily favourable entropic contributions rendering ΔG highly temperature dependent (see Δ(ΔT293S) for entries 5–7, respectively, Table 2).
Thermodynamic dsDNA-targeting potential of Invader LNA probes
The abovementioned thermal denaturation studies suggest that it is energetically favourable for Invader LNA probes to dissociate and bind to complementary DNA targets (Fig. 1). To quantitatively assess the thermodynamic dsDNA-targeting potential of different Invader LNA probe designs, the available free energy for room temperature recognition of isosequential DNA was estimated as ΔG293rec = ΔG293 (probe strand 1 : target strand 1) + ΔG293 (probe strand 2 : target strand 2) - ΔG293 (dsDNA target) - ΔG293 (Invader LNA probe) (Table 2)50. Probes with a single energetic hotspot provide ΔG293rec = −19.0 to −26.0 kJ/mol (entries 1–4, Table 2), while probes with two energetic hotspots exhibit considerably more inherent free energy for dsDNA-recognition (ΔG293rec = −34.0 kJ/mol to −50.0 kJ/mol, entries 5–7, Table 2). Invader LNA probes with two hotspots separated by two or four base pairs exhibit additive increases in ΔG293rec-values relative to the corresponding single hotspot Invader LNA probes (e.g., compare ΔG293rec-values for entries 1, 3 and 6, Table 2). This contrasts Invader LNA probes with sequentially incorporated hotspots. This suggests that Invader LNA probes should feature two separated energetic hotspots for maximized thermodynamic dsDNA-targeting potential.
Fluorescence properties of Invader LNA probes
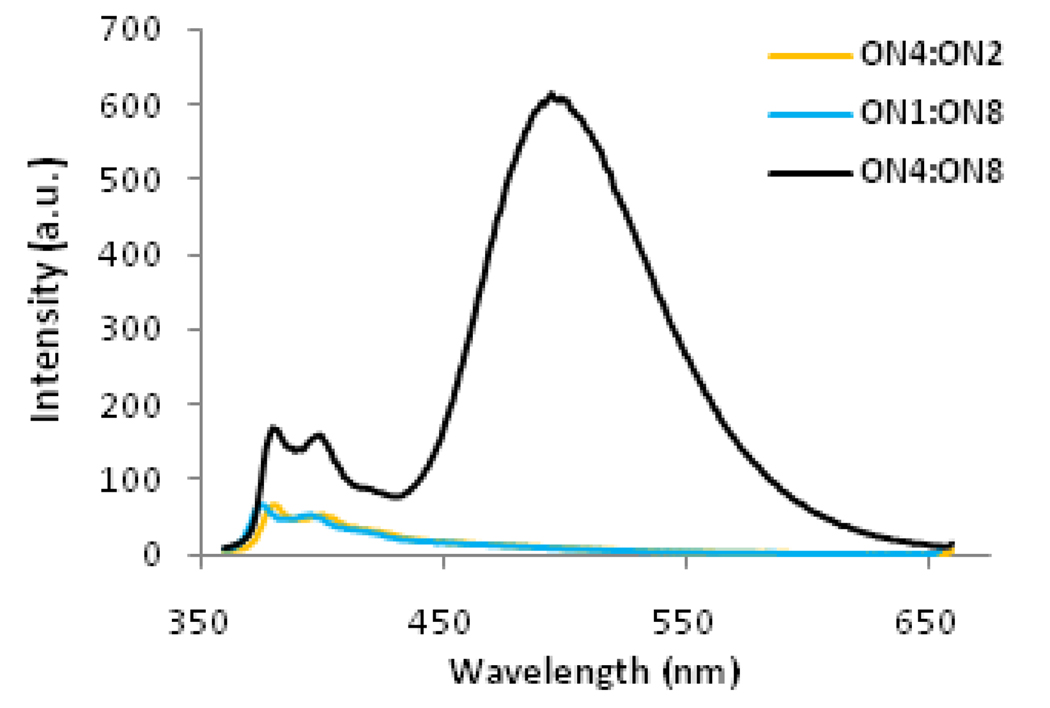
Steady-state fluorescence emission spectra of all Invader LNA probes obtained using an excitation wavelength λex = 335 nm, exhibit structured pyrene monomer peaks at λmax = 380 nm and 400 nm along with an intense unstructured peak centered at λmax = 495 nm (Fig. 2 and Fig S2†). Duplexes with other interstrand zipper arrangements of X monomers only exhibit marginal signal intensity at 495 nm (results not shown). Pyrene emission signals at 495 nm are routinely attributed to pyrene-pyrene excimers41,42,45,51–55, suggesting that the pyrene moieties of the two X monomers comprising the energetic hotspots in Invader LNA probes are separated by ~3.4 Å56 (Fig. 1). In contrast, emission spectra of duplexes between 2’-N-(pyren-1-yl)methyl-2’-amino-α-L-LNA and complementary DNA exhibit pyrene monomer fluorescence and only negligible excimer signals (Fig. 2 for ON4:ON2 and ON1:ON8; see also Fig. S2†). Given these experimental observations, the signal intensity at λem = 495 nm can accordingly be used as an inherent optical readout to monitor dsDNA recognition by Invader LNA probes in real-time.
Figure 2.
Steady-state fluorescence emission spectra (λex = 335 nm, 20 °C) of Invader LNA probe ON4:ON8 and probe:target duplexes ON4:ON2 and ON1:ON8 recorded in thermal denaturation buffer (all ONs at 1.0 µM concentration). Corresponding spectra for other Invader LNA probes are shown in Fig. S2†.
Targeting of dsDNA by Invader LNA probes
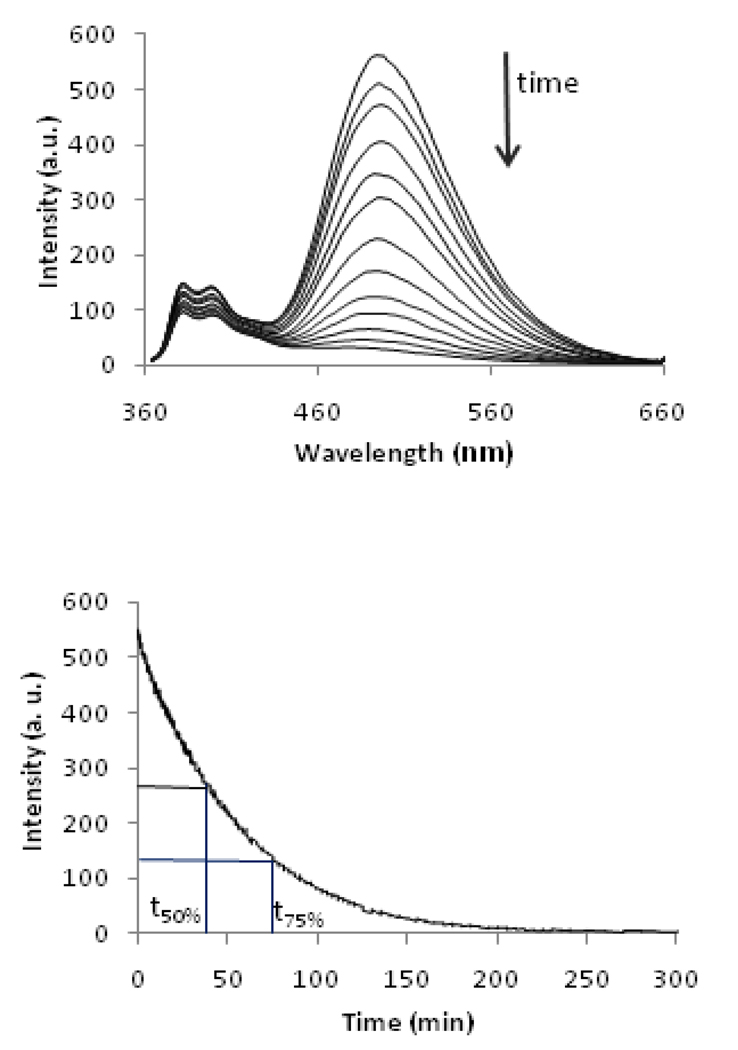
Pre-annealed Invader LNA probes were added to an equimolar quantity of their pre-annealed isosequential 13-mer dsDNA target ON1:ON2 (all strands at 1.0 µM). The recognition experiments were performed in thermal denaturation buffer ([Na+] = 110 mM, pH 7) at an experimental temperature (Texp = 20 °C) markedly below the Tm-values of dsDNA target ON1:ON2 (37.5 °C, Table 1), Invader LNA probes (36–53 °C, Table 2) and the corresponding probe-target duplexes (44.5–61.0 °C, Table 2). Addition of Invader LNA probes results in a rapid decrease in excimer intensity (λem = 495 nm) as the two stacking pyrene moieties of the probes are forced apart (Fig. 1) during recognition of the dsDNA target (Fig. 3 and Fig S3†). The excimer signals decay according to a first order rate equation when equimolar quantities between Invader LNA and dsDNA targets are used, but become more complex when non-equimolar quantities of probes and targets are used (results not shown).
Figure 3.
Upper panel: time course of steady-state fluorescence spectra upon addition of pre-annealed Invader LNA ON4:ON8 to pre-annealed isosequential dsDNA target ON1:ON2 (λex = 335 nm, 20 °C, all strands 1.0 µM). Lower panel: time course of fluorescence signal decay at λem = 495 nm. Corresponding spectra for other Invader LNA are shown in Fig. S3†.
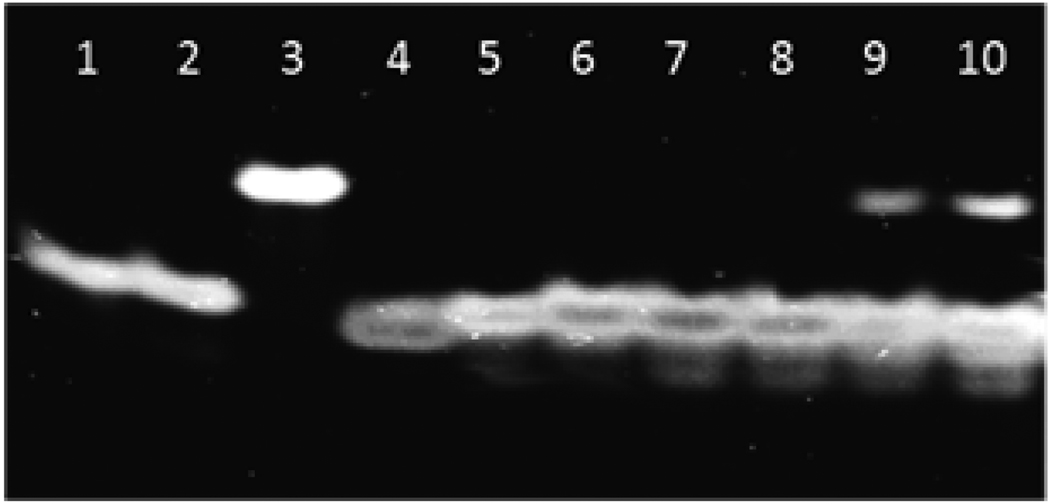
Electrophoretic mobility shift assays (EMSAs, 16 % non-denaturing PAGE) were performed to validate the fluorescence-based assay as a convenient method to monitor dsDNA-recognition by Invader LNA probes. The representative single hotspot Invader LNA ON4:ON8 exhibits significantly lower electrophoretic mobility than isosequential dsDNA target ON1:ON2 and probe-target duplexes ON1:ON8 and ON4:ON2 (Fig. 4). The difference in electrophoretic mobility between the dsDNA target and probe-target duplexes is much smaller although discernible (compare lanes 1, 2, and 4, Fig. 4). We attribute the observed differences to a) an extensively distorted duplex geometry of Invader LNA probe ON4:ON8 as suggested by thermodynamic data (Table 2) and preliminary results from molecular modelling (results not shown), and b) slightly perturbed probe-target duplex geometry (elongation) upon intercalation of a single pyrene moiety, as previously reported47.
Figure 4.
Electrophoretic mobility shift assay, 16% nondenaturing PAGE run at 4 °C, demonstrating dsDNA-recognition by Invader LNA probes. Probe-target duplex ON1:ON8 (lane 1); probe-target duplex ON4:ON2 (lane 2); Invader LNA probe ON4:ON8 (lane 3); dsDNA target ON1:ON2 (lane 4); equimolar mixture of Invader LNA probe ON4:ON8 + dsDNA target ON1:ON2 which were annealed (lane 5), or incubated at: 37 °C for 4h (lane 6), rt for 4h (lane 7), rt for 8 h (lane 8), 4 °C for 8h (lane 9) or 4 °C for 12h (lane 10), respectively.
Incubation of equimolar quantities of Invader LNA ON4:ON8 and dsDNA target ON1:ON2 at room temperature or 37 °C results in disappearance of the bands corresponding to the Invader LNA probe and dsDNA target. Instead, a single band with identical electrophoretic mobility as probe-target duplexes ON1:ON8 and ON4:ON2 emerges (compare lanes 6–8 with lanes 1, 2 and 5). Incubation of Invader LNA and dsDNA at 4 °C results in incomplete recognition as indicated by the presence of weak bands corresponding to ON4:ON8 and ON1:ON2. These observations are in line with the results from the fluorescence assay (Fig. 3) and demonstrate that the decrease in excimer signal intensity upon addition of Invader LNA probes to isosequential dsDNA targets is attributable to dsDNA-recognition.
The time period resulting in a 50% or 75% decrease of the original excimer signal (t50% or t75%) was determined for four representative Invader LNA to assess recognition kinetics. The selected Invader LNA probes include two single hotspot Invader LNAs (ON4:ON8 and ON6:ON10), and two double hotspot Invader LNAs where the hotspots are separated by zero or four base pairs (ON11:ON14 and ON13:ON16, respectively). Recognition of ON1:ON2 occurred quickly at 20 °C with all studied Invader LNA probes (t50% = 29–48 min, entry 1, Table 3), but was particularly fast with ON11:ON14 having two sequentially positioned hotspots. Interestingly, Invader LNA probe ON13:ON16 exhibits the most favourable ΔG293rec-value for recognition of dsDNA ON1:ON2 (Table 2) and yet displays the slowest recognition kinetics (Table 3). Increases in experimental temperatures result in faster dsDNA-recognition (t50% = 37.5/29.0, 14.2/11.1, 4.8/4.3, 1.5/1.6 and 0.5/0.6 min at Texp = 20, 22.5, 25, 27.5 and 30 °C for ON4:ON8/ON11:ON14, respectively).
Table 3.
Kinetic parameters (t50%/t75% [min]) for recognition of isosequential dsDNA-target ON1:ON2 by selected Invader LNA.a
| Buffer | ON4:ON8 | ON6:ON10 | ON11:ON14 | ON13:ON16 | ||||
|---|---|---|---|---|---|---|---|---|
| t50% | t75% | t50% | t75% | t50% | t75% | t50% | t75% | |
| MS | 38 | 74 | 34 | 67 | 29 | 60 | 48 | 103 |
| HS | 126 | 257 | - | - | 31 | 68 | - | - |
| KMST | 158 | 296 | - | - | 19 | 42 | - | - |
[probe] = [target] = 1.0 µM; t50% and t75% = time in min required to reduce fluorescence intensity at λem = 495 nm by 50% and 75%, respectively. MS = medium salt, HS = high salt, KMST-buffer specified in main text
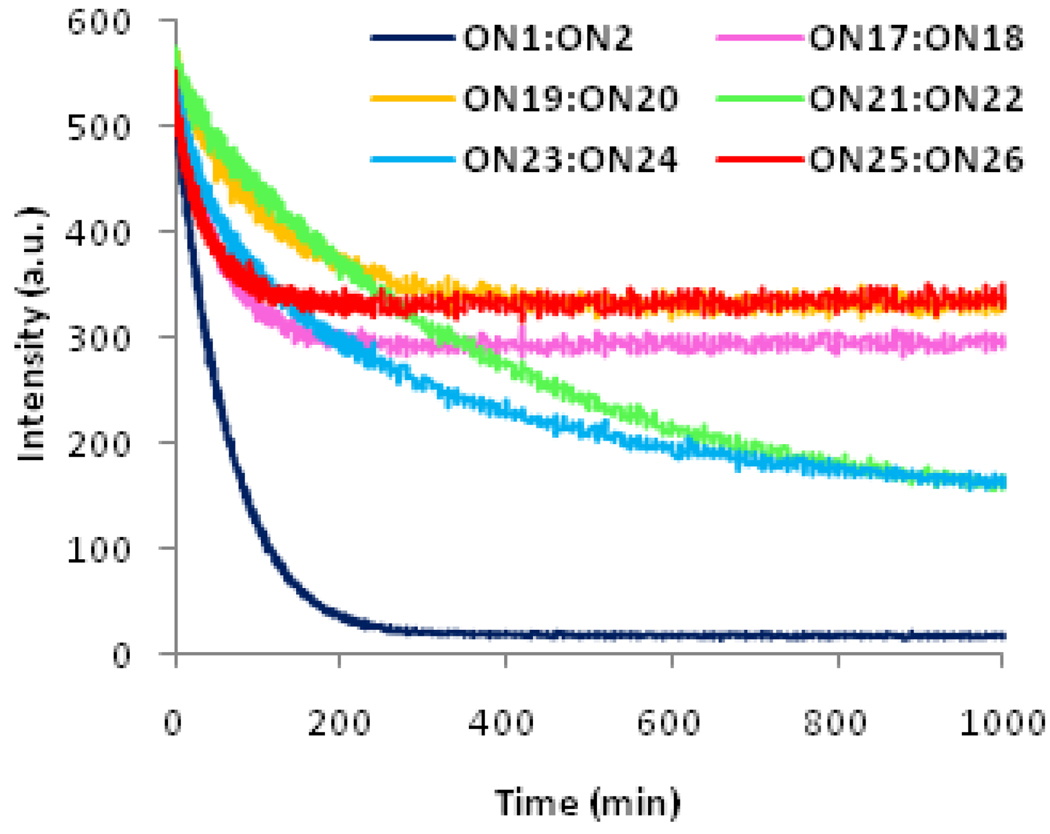
Next, the ability of Invader LNA probes to discriminate between correct and incorrect dsDNA targets was evaluated. Addition of representative Invader LNA probe ON4:ON8 to fully base paired but non-isosequential dsDNA targets generally resulted in a slower and less pronounced decay of the excimer signal (Fig. 5). More specifically, the signal intensity at 495 nm decreased by 35–45% to reach a plateau ~200 min after addition of ON4:ON8 to 13-mer dsDNA targets containing a single sequence disparity at positions outside the hotspot region (ON17:ON18 or ON25:ON26, Fig. 5; sequences shown in Table 4). Slower but more pronounced decreases in excimer signal (~70% decrease after >16 h) were observed upon addition of Invader LNA probes ON4:ON8 to 13-mer dsDNA targets ON21:ON22 or ON23:ON24 with single sequence disparities within the hotspot regions. Thus, the data suggest that Invader LNA probes discriminate mismatched dsDNA targets well$.
Figure 5.
Time course of fluorescence signal decay at λem = 495 nm upon addition of pre-annealed Invader LNA ON4:ON8 to pre-annealed isosequential dsDNA ON1:ON2 or fully base paired non-isosequential dsDNA targets. Sequences of targets ON17–ON26 are shown in Table 4. Conditions are as described in Figure 2.
Table 4.
Hybridization data for non-isosequential dsDNA targets and the available free energy, ΔG293rec for their recognition by Invader LNA probe ON4:ON8 at 293 K.a
| ON | Duplex |
Tm (°C) |
ΔG293 (kJ/mol) |
(kJ/mol) |
|---|---|---|---|---|
| ON1 | 5’-d(GGT ATA TAT AGG C) | 37.5 | −57 | −26 |
| ON2 | 3’-d(CCA TAT ATA TCC G) | |||
| ON17 | 5’-d(GGA ATA TAT AGG C) | 38.5 | −59 | +8 |
| ON18 | 3’-d(CCT TAT ATA TCC G) | |||
| ON19 | 5’-d(GGT ATC TAT AGG C) | 41.0 | −60 | +6 |
| ON20 | 3’-d(CCA TAG ATA TCC G) | |||
| ON21 | 5’-d(GGT ATG TAT AGG C) | 42.0 | −60 | −1 |
| ON22 | 3’-d(CCA TAC ATA TCC G) | |||
| ON23 | 5’-d(GGT ATT TAT AGG C) | 38.5 | −57 | 0 |
| ON24 | 3’-d(CCA TAA ATA TCC G) | |||
| ON25 | 5’-d(GGT ATA TAA AGG C) | 38.0 | −58 | +7 |
| ON26 | 3’-d(CCA TAT ATT TCC G) |
Conditions as specified in Table 1. Underlined positions denote sites of sequence disparity relative to Invader LNA ON4:ON8.
In agreement with these observations, unfavourable or marginally favourable ΔG293rec-values are observed for recognition of non-isosequential dsDNA targets by Invader LNA probe ON4:ON8 (Table 4). This reflects the energetic penalty upon hybridization of 2’-N-(pyren-1-yl)methyl-2’-amino-α-L-LNA ON4 or ON8 to mismatched DNA strands (see Δ(ΔG293)-values, Table 5). ON4 and ON8 discriminate DNA strands with mismatches opposite of deoxyribonucleotides well (compare ΔTm-values of entries 1 and 5–10 in Table 5 and Table S9†). In contrast, mismatched DNA targets with thymine or guanine moieties opposite of thymine monomer X are less efficiently discriminated (see entries 3–4, Table 5). This translates in marginally favourable ΔG293rec-values for recognition of ON21:ON22 and ON23:ON24 by Invader LNA probe ON4:ON8 (entries 4–5, Table 4) and the distinct signal decay profile during dsDNA recognition (Fig. 5).
Table 5.
Hybridization data for duplexes involving ON4 or ON8 and mismatched DNA.a
| Entry | ON | Sequence |
Tm [ΔTm] (°C) |
ΔG293 [Δ(ΔG293)] (kJ/mol) |
|---|---|---|---|---|
| 1 | ON17 | 5’-d(GGA ATA TAT AGG C) | 39.5 | −57 |
| ON8 | 3’-d(CCA TAX ATA TCC G) | [−9.0] | [+13] | |
| 2 | ON19 | 5’-d(GGT ATC TAT AGG C) | 39.0 | −58 |
| ON8 | 3’-d(CCA TAX ATA TCC G) | [−9.5] | [+12] | |
| 3 | ON21 | 5’-d(GGT ATG TAT AGG C) | 44.5 | −63 |
| ON8 | 3’-d(CCA TAX ATA TCC G) | [−4.0] | [+7] | |
| 4 | ON23 | 5’-d(GGT ATT TAT AGG C) | 46.0 | −64 |
| ON8 | 3’-d(CCA TAX ATA TCC G) | [−2.5] | [+6] | |
| 5 | ON25 | 5’-d(GGT ATA TAA AGG C) | 37.0 | −55 |
| ON8 | 3’-d(CCA TAX ATA TCC G) | [−11.5] | [+15] | |
| 6 | ON4 | 5’-d(GGT AXA TAT AGG C) | 37.5 | −54 |
| ON18 | 3’-d(CCT TAT ATA TCC G) | [−10.0] | [+12] | |
| 7 | ON4 | 5’-d(GGT AXA TAT AGG C) | 38.0 | −56 |
| ON20 | 3’-d(CCA TAG ATA TCC G) | [−9.5] | [+10] | |
| 8 | ON4 | 5’-d(GGT AX A TAT AGG C) | 39.5 | −58 |
| ON22 | 3’-d(CCA TAC ATA TCC G) | [−8.0] | [+8] | |
| 9 | ON4 | 5’-d(GGT AX A TAT AGG C) | 35.0 | −53 |
| ON24 | 3’-d(CCA TAA ATA TCC G) | [−12.5] | [+13] | |
| 10 | ON4 | 5’-d(GGT AXA TAT AGG C) | 38.0 | −56 |
| ON26 | 3’-d(CCA TAT ATT TCC G) | [−9.5] | [+10] |
Conditions as specified in Table 1. Underlined positions denote position of mismatched base pairs. ΔTm = change in Tm value and Δ(ΔG293) = difference in ΔG293 value, relative to fully matched ON1:ON8 (entries 1–5) or ON4:ON2 (entries 6–10).
Discrimination of non-isosequential dsDNA targets can likely be improved in next-generation Invader LNA probes using recently reported N2’-intercalator-functionalized 2’-amino-α-L-LNA building blocks, which exhibit significantly improved thermal discrimination of mismatched DNA44,47.
Finally, the influence of buffer composition on dsDNA-recognition by Invader LNA probes was studied. Representative Invader LNA probes ON4:ON8 or ON11:ON14 were added to isosequential dsDNA target ON1:ON2 as described above except for the use of buffers that either represent physiological conditions more accurately (KMST-buffer: pH 7.2, 140 mM KCl, 10 mM MgCl2, 1 mM spermine and 40 mM Tris-Cl) or are of very high ionic strength ([Na+] = 710 mM). Recognition of dsDNA targets was observed in both buffers at 20 °C, and particularly rapid with Invader LNA probe ON11:ON14 featuring two sequentially incorporated energetic hotspots (Table 3). These observations suggest that Invader LNA probes may be applicable over a significantly wider range of conditions than PNA-based dsDNA-targeting approaches 5,6,33–38.
Interestingly, Invader LNA probe ON11:ON14 has less free energy available for targeting of dsDNA at high salt conditions than ON4:ON8 (see ΔG293rec-values, Table S6†). Thus, a favourable energetic gradient (ΔG293rec ≪ 0 kJ/mol) is a prerequisite for successful dsDNA-targeting by Invader LNA probes, but reaction kinetics must be taken into consideration for practical applications. Both aspects strongly depend on by the number and position of energetic hotspots within the probes. Invader LNA probes with two sequentially incorporated energetic hotspots are thermodynamically as well as kinetically activated toward recognition of dsDNA.
The successful targeting of dsDNA target ON1:ON2 with Invade r LNA ON11:ON14 at high salt conditions is remarkable considering that the experimental temperature (Texp = 20 °C) is markedly lower than the thermal denaturation temperatures of the dsDNA target, Invader LNA probe, and probe:target duplexes ON1:ON14 and ON11:ON2 (Tm = 45.5 °C, 59.0 °C, 59.0 °C and 63.5 °C, respectively, Tables S2 and S4†, respectively).
While additional studies will be needed to elucidate the recognition mechanism, the data presented herein seem inconsistent with a dissociative pathway57. This pathway would involve dissociation of Invader LNA probes and dsDNA targets into four single strands (which is not likely to occur at Texp ≪ Tm), followed by energetically favourable self-assembly of probe-target duplexes. We speculate that a sequential displacement pathway57 may be involved where partial melting of dsDNA targets and Invader LNA probes reveals nucleation sites serving as initiation sites for formation of probe-target duplexes. Within this hypothesis, kinetic activation of Invader LNA probes with two sequential hotspots could be related to more efficient formation of nucleation sites.
CONCLUSION AND OUTLOOK
Important features and design principles of Invader LNA probes have been identified during the course of this study:
The Invader LNA approach is general. Every investigated Invader LNA probe displays fast recognition of isosequential dsDNA targets in a variety of buffers, including buffers mimicking physiological conditions.
Recognition of mixed sequence dsDNA targets is conveniently followed in real-time by decreases in the pyrenepyrene excimer signal inherently exhibited by Invader LNA probes.
Invader LNA probes with two sequentially incorporated hotspots exhibit more pronounced thermodynamic gradients and are kinetically more activated toward dsDNA-recognition than single hotspot probes.
Invader LNA probes discriminate non-isosequential dsDNA targets.
The ability of Invader LNA probes to thermodynamically and kinetically discriminate mismatched dsDNA-targets can be envisioned to result in the development of sensors for detection of single nucleotide polymorphism58,59. Moreover, recent reports on the use of Zorro LNA30 and agLNA28,29 for recognition of accessible target regions within plasmids or chromosomal DNA, warrant evaluation of optimized Invader LNA probes as site-specific modulators of gene expression60. Studies along these lines are currently underway in our laboratory.
Experimental
MATERIALS
Synthesis of 2’-N-(pyren-1-yl)methyl-2’-amino-α-L-LNA
The corresponding phosphoramidite of monomer X was obtained and incorporated into 13-mer AT-rich oligodeoxyribonucleotides (ONs) using established protocols42,47. Briefly described, automated synthesis of ONs was performed in 0.2 µmol scale applying standard procedures except for extended coupling times (30 min, using 1H-tetrazole as catalyst), which resulted in stepwise coupling yields of ~99% for monomers X (Fig. 1). Following standard workup and purification, the composition and purity (>80%) of modified ONs was verified by MALDI-MS (Table S1†) and ion-exchange HPLC, respectively.
Thermal denaturation studies
Concentrations of all ONs were estimated using the following extinction coefficients (L×mmol−1×cm−1) at 260 nm: dA (15.20), dC (7.05), dG (12.01), T (8.40); rA (15.40), rC (9.00), rG (13.70), rU (10.00); pyrene (22.40). ONs (1.0 µM each strand) were thoroughly mixed in Tm-buffer (see below), denatured by heating and subsequently cooled to the starting temperature of the experiment. Thermal denaturation curves (A260 vs. T) were recorded using a Cary 100 UV/VIS spectrometer equipped with a Peltier temperature programmer. The temperature was varied from at least 15 °C below to 15 °C above the thermal denaturation temperature (Tm) using a ramp of 0.5 °C/min. Quartz optical cells with a path length of 10 mm were used. In general, a medium salt Tm-buffer was used (100 mM NaCl, 0.1 mM EDTA, pH 7.0 adjusted with 10 mM NaH2PO4/5 mM Na2HPO4). Other utilized buffers include low and high salt buffers (composition as for medium salt buffer except that 0 mM and 700 mM NaCl were used, respectively) and a KMST-buffer (140 mM KCl, 10 mM MgCl2, 1 mM spermine and 40 mM Tris-Cl, pH 7.2). Thermal denaturation temperatures and thermodynamic parameters for duplex formation were determined by baseline fitting of melting curves using software provided with the UV/VIS spectrometer. Bimolecular reactions, two-state melting behaviour and a heat capacity change ΔCp = 0 upon hybridization were assumed48. The validity of these assumptions for N2’-functionalized 2’-amino-α-L-LNA have been previously discussed47. Reported thermal denaturation temperatures were determined as an average from two separate experiments within ±1.0 °C. For determination of thermodynamic parameters, a minimum of two experimental denaturation curves were each analyzed at least three times to minimize errors arising from baseline choice, and average values are listed. Changes in Gibbs free energy and entropy upon duplex formation were determined at the temperatures of the dsDNA-targeting experiments (Texp= 293.15 K).
Steady-state fluorescence emission experiments
Fluorescence measurements were performed on a Cary Eclipse fluorimeter equipped with a Peltier temperature controller using quartz optical cells with a path length of 10 mm. A concentration of 1.0 µM of each strand in medium salt Tm-buffer was used. No corrections for the minimal solvent background or attempts to eliminate dissolved oxygen from the buffer solution were made. Mixtures were kept at 20 °C (±0.2 °C) at all times. Steady-state fluorescence emission spectra (360–600 nm) were obtained using an excitation wavelength of 335 nm, excitation and emission slits of 5.0 nm and photomultiplier voltage set at 800V. The setup for dsDNA-targeting experiments is detailed in the Results and Discussion section.
Electrophoretic mobility shift assays
Stock solutions (4 µM) of dsDNA targets, Invader LNA probes and probe-target duplexes were prepared by annealing appropriate complementary single strands in a pH 7.2 incubation buffer (70 mM HEPES, 15 mM MgCl2, 15% sucrose, 0.15% spermine, 300 mM NaCl) and storing the solutions at 4°C. Control duplexes (4 µL) or reaction mixtures (4 µL of each duplex) were incubated as described in Figure 5, mixed with 6× DNA loading dye solution (2 µL), loaded onto a 16% native polyacrylamide gel in 1× TBM buffer (89 mM Tris, 89 mM boric acid, 10 mM MgCl2 and run for 24 h at 10 V/cm and 4 °C. Gels were stained with SYBR Gold and documented on a Flour S multiimager.
Supplementary Material
ACKNOWLEDGEMENT
The input of Prof. Jesper Wengel (Nucleic Acid Center, Univ. Southern Denmark) during initial stages of this project is highly appreciated. We thank Dr. Rod Hill and Dr. Madhusudhan Papasani (AVS, Univ. Idaho) for access to and assistance with the Flour S multiimager.
FUNDING
This study was supported by Award Number R01 GM088697 from the National Institute Of General Medical Sciences, National Institutes of Health [grant number P20 RR016448 from the COBRE Program of the National Center for Research Resources]; Idaho NSF EPSCoR; the BANTech Center at Univ. Idaho; a Univ. Idaho Research Office and Research Council Seed Grant; The Danish National Research Foundation; and the Nucleic Acid Based Drug Design Ph.D. school (NAC DRUG; supported by the Danish Agency for Science Technology and Innovation).
Footnotes
Invader LNA” are defined as double stranded DNA probes with one or more +1 interstrand zipper arrangements of intercalator-modified 2’-amino-α-L-LNA monomers. This particular zipper arrangement is also termed energetic hotspot, for simplicity. For a formal definition of the ‘zipper’ nomenclature see Table S4†.
For steady-state fluorescence emission spectra of duplexes between ON4 or ON8 and mismatched DNA targets and additional discussion, see Fig. S4.
Electronic Supplementary Information (ESI) available: MALDI-MS of 2’-N-(pyren-1-yl)methyl-2’-amino-α-L-LNA ON3–ON16 (Table S1), representative thermal denaturation profiles (Fig. S1), hybridization data for ON3–ON16 vs complementary DNA and for Invader LNA probes at different ionic strengths (Tables S2–S5), ΔG293rec for recognition of ON1:ON2 by Invader LNA probes at different ionic strengths (Table S6), additional discussion of thermodynamic parameters, hybridization data for ON3–ON16 vs RNA complements (Table S7), hybridization data for duplexes with various interstrand zipper arrangements of X monomers (Table S8), steady-state fluorescence emission spectra of representative Invader LNA probes and probe-target duplexes (Fig. S2), signal decay profile during recognition of dsDNA by representative Invader LNA probes (Fig. S3), steady-state fluorescence emission spectra of duplexes between ON4 or ON8 and mismatched DNA targets (Fig. S4), hybridization data for duplexes between ON1 or ON2 and mismatched DNA targets (Table S9). See DOI: 10.1039/b000000x/
References
- 1.Duca M, Vekhoff P, Oussedik K, Halby L, Arimondo PB. Nucleic Acids Res. 2008;36:5123–5138. doi: 10.1093/nar/gkn493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rogers FA, Lloyd JA, Glazer PM. Curr. Med. Chem. Anti-Cancer Agents. 2005;5:319–326. doi: 10.2174/1568011054222300. [DOI] [PubMed] [Google Scholar]
- 3.Besch R, Giovannangeli C, Degitz K. Curr. Drug Targets. 2004;5:691–703. doi: 10.2174/1389450043345100. [DOI] [PubMed] [Google Scholar]
- 4.Ghosh I, Stains CI, Ooi AT, Segal DJ. Mol. BioSyst. 2006;2:551–560. doi: 10.1039/b611169f. [DOI] [PubMed] [Google Scholar]
- 5.Kaihatsu K, Janowski BA, Corey DR. Chem. Biol. 2004;11:749–758. doi: 10.1016/j.chembiol.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 6.Nielsen PE. In: Sequence-Specific DNA Binding Agents. Michael J, editor. Cambridge, UK: Warring, Royal Society of Chemistry; 2006. pp. 96–108. [Google Scholar]
- 7.Bijapur J, Keppler MD, Bergqvist S, Brown T, Fox KR. Nucleic Acids Res. 1999;27:1802–1809. doi: 10.1093/nar/27.8.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brazier JA, Shibata T, Townsley J, Taylor BF, Frary E, Williams NH, Williams DM. Nucleic Acids Res. 2005;33:1362–1371. doi: 10.1093/nar/gki254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Miller PS, Seidman MM. Org. Biomol. Chem. 2008;6:4212–4217. doi: 10.1039/b810709b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torigoe H, Hari Y, Sekiguchi M, Obika S, Imanishi T. J. Biol. Chem. 2001;276:2354–2360. doi: 10.1074/jbc.M007783200. [DOI] [PubMed] [Google Scholar]
- 11.Sun B-W, Babu BR, Sørensen MD, Zakrzewska K, Wengel J, Sun J-S. Biochemistry. 2004;43:4160–4169. doi: 10.1021/bi036064e. [DOI] [PubMed] [Google Scholar]
- 12.Brunet E, Alberti P, Perrouault L, Babu BR, Wengel J, Giovannangeli C. J. Biol. Chem. 2005;280:20076–20085. doi: 10.1074/jbc.M500021200. [DOI] [PubMed] [Google Scholar]
- 13.Kumar N, Nielsen KE, Maiti S, Petersen M. J. Am. Chem. Soc. 2006;128:14–15. doi: 10.1021/ja055483r. [DOI] [PubMed] [Google Scholar]
- 14.Koizumi M, Morita K, Daigo M, Tsutsumi S, Abe K, Obika S, Imanishi T. Nucleic Acids Res. 2003;31:3267–3273. doi: 10.1093/nar/gkg416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rahman SMA, Seki S, Obika S, Yoshikawa H, Miyashita K, Imanishi T. J. Am. Chem. Soc. 2008;130:4886–4896. doi: 10.1021/ja710342q. [DOI] [PubMed] [Google Scholar]
- 16.Sau SP, Kumar P, Anderson BA, østergaard ME, Deobald L, Paszczynski A, Sharma PK, Hrdlicka PJ. Chem. Commun. 2009:6756–6758. doi: 10.1039/b917312a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filichev VV, Pedersen EB. J. Am. Chem. Soc. 2005;127:14849–14858. doi: 10.1021/ja053645d. [DOI] [PubMed] [Google Scholar]
- 18.Cheng A-J, Van Dyke MW. Nucleic Acids Res. 1993;21:5630–5635. doi: 10.1093/nar/21.24.5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dagle JM, Weeks DL. Nucleic Acids Res. 1996;24:2143–2149. doi: 10.1093/nar/24.11.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simon P, Cannata F, Concordet J-P, Giovannangeli C. Biochimie. 2008;90:1109–1116. doi: 10.1016/j.biochi.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Dervan PB, Doss RM, Marques MA. Curr. Med. Chem. Anti-cancer Agents. 2005;5:373–387. doi: 10.2174/1568011054222346. [DOI] [PubMed] [Google Scholar]
- 22.Fox KR, Brown T. Quart. Rev. Biophys. 2005;38:311–320. doi: 10.1017/S0033583506004197. [DOI] [PubMed] [Google Scholar]
- 23.Rusling DA, Powers VEC, Ranasinghe RT, Wang Y, Osborne SD, Brown T, Fox KR. Nucleic Acids Res. 2005;33:3025–3032. doi: 10.1093/nar/gki625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rapireddy S, He G, Roy S, Armitage BA, Ly DH. J. Am. Chem. Soc. 2007;129:15596–15600. doi: 10.1021/ja074886j. [DOI] [PubMed] [Google Scholar]
- 25.Chenna V, Rapireddy S, Sahu B, Ausin C, Pedroso E, Ly DH. ChemBioChem. 2008;9:2388–2391. doi: 10.1002/cbic.200800441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janowski BA, Kaihatsu K, Huffman KE, Schwartz JC, Ram R, Hardy D, Mendelson CR, Corey DR. Nat. Chem. Biol. 2005;1:210–215. doi: 10.1038/nchembio724. [DOI] [PubMed] [Google Scholar]
- 27.Hu J, Corey DR. Biochemistry. 2007;46:7581–7589. doi: 10.1021/bi700230a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beane R, Ram R, Gabillet S, Arar K, Monia BP, Corey DR. Biochemistry. 2007;46:7572–7580. doi: 10.1021/bi700227g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beane R, Gabillet S, Montaillier C, Arar K, Corey DR. Biochemistry. 2008;47:13147–13149. doi: 10.1021/bi801930p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ge R, Heinonen JE, Svahn MG, Mohamed AJ, Lundin KE, Smith CIE. FASEB J. 2007;21:1902–1914. doi: 10.1096/fj.06-7225com. [DOI] [PubMed] [Google Scholar]
- 31.Kutyavin IV, Rhinehart RL, Lukhtanov EA, Gorn VV, Meyer RB, Jr, Gamper HB., Jr Biochemistry. 1996;35:11170–11176. doi: 10.1021/bi960626v. [DOI] [PubMed] [Google Scholar]
- 32.Woo J, Meyer RB, Jr, Gamper HB., Jr Nucleic Acids Res. 1996;24:2470–2475. doi: 10.1093/nar/24.13.2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lohse J, Dahl O, Nielsen PE. Proc. Natl. Acad. Sci. U.S.A. 1999:11804–11808. doi: 10.1073/pnas.96.21.11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Izvolsky KI, Demidov VV, Nielsen PE, Frank-Kamenetskii MD. Biochemistry. 2000;39:10908–10913. doi: 10.1021/bi000675e. [DOI] [PubMed] [Google Scholar]
- 35.Demidov VV, Protozanova E, Izvolsky KI, Price C, Nielsen PE, Frank-Kamenetskii MD. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5953–5958. doi: 10.1073/pnas.092127999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smolina IV, Demidov VV, Soldatenkov VA, Chasovskikh SG, Frank-Kamenetskii MD. Nucleic Acids Res. 2005;33:e146. doi: 10.1093/nar/gni151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim K-H, Nielsen PE, Glazer PM. Nucleic Acids Res. 2007;35:7604–7613. doi: 10.1093/nar/gkm666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishizuka T, Yoshida J, Yamamoto Y, Sumaoka J, Tedeschi T, Corradini R, Sforza S, Komiyama M. Nucleic Acids Res. 2008;36:1464–1471. doi: 10.1093/nar/gkm1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bryld T, Højland T, Wengel J. Chem. Commun. 2004:1064–1065. doi: 10.1039/b402414a. [DOI] [PubMed] [Google Scholar]
- 40.Filichev VV, Christensen UB, Pedersen EB, Babu BR, Wengel J. ChemBioChem. 2004;5:1673–1679. doi: 10.1002/cbic.200400222. [DOI] [PubMed] [Google Scholar]
- 41.Filichev VV, Vester B, Hansen LH, Pedersen EB. Nucleic Acids Res. 2005;33:7129–7137. doi: 10.1093/nar/gki1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hrdlicka PJ, Kumar TS, Wengel J. Chem. Commun. 2005:4279–4281. doi: 10.1039/b506986f. [DOI] [PubMed] [Google Scholar]
- 43.Kumar TS, Madsen AS, Wengel J, Hrdlicka PJ. J. Org. Chem. 2006;71:4188–4201. doi: 10.1021/jo060331f. [DOI] [PubMed] [Google Scholar]
- 44.Andersen NK, Wengel J, Hrdlicka PJ. Nucleosides Nucleotides Nucleic Acids. 2007;26:1415–1417. doi: 10.1080/15257770701539153. [DOI] [PubMed] [Google Scholar]
- 45.Kumar TS, Wengel J, Hrdlicka PJ. ChemBioChem. 2007;8:1122–1125. doi: 10.1002/cbic.200700144. [DOI] [PubMed] [Google Scholar]
- 46.Kumar TS, Madsen AS, østergaard ME, Wengel J, Hrdlicka PJ. J. Org. Chem. 2008;73:7060–7066. doi: 10.1021/jo800551j. [DOI] [PubMed] [Google Scholar]
- 47.Kumar TS, Madsen AS, østergaard ME, Sau SP, Wengel J, Hrdlicka PJ. J. Org. Chem. 2009;74:1070–1081. doi: 10.1021/jo802037v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mergny JL, Lacroix L. Oligonucleotides. 2003;13:515–537. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]
- 49.Crothers DM. Biopolymers. 1968;6:575–584. doi: 10.1002/bip.1968.360060411. [DOI] [PubMed] [Google Scholar]
- 50.For a representative calculation of ΔG293rec see Table S6†
- 51.Umemoto T, Hrdlicka PJ, Babu BR, Wengel J. ChemBioChem. 2007;8:2240–2248. doi: 10.1002/cbic.200700408. [DOI] [PubMed] [Google Scholar]
- 52.Dioubankova NN, Malakhov AD, Stetsenko DA, Gait MJ, Volynsky PE, Efremov RG, Korshun VA. ChemBioChem. 2003;4:841–847. doi: 10.1002/cbic.200300678. [DOI] [PubMed] [Google Scholar]
- 53.Hrdlicka PJ, Babu BR, Sørensen MD, Wengel J. Chem. Commun. 2004:1478–1479. doi: 10.1039/b404446k. [DOI] [PubMed] [Google Scholar]
- 54.Langenegger SM, Häner R. Chem. Commun. 2004:2792–2793. doi: 10.1039/b412831a. [DOI] [PubMed] [Google Scholar]
- 55.Nakamura M, Murakami Y, Sasa K, Hayashi H, Yamana K. J. Am. Chem. Soc. 2008;130:6904–6905. doi: 10.1021/ja801054t. [DOI] [PubMed] [Google Scholar]
- 56.Winnik FM. Chem. Rev. 1993;93:587–614. [Google Scholar]
- 57.Reynaldo LP, Vologodskii AV, Neri BP, Lyamichev VI. J. Mol. Biol. 2000;297:511–520. doi: 10.1006/jmbi.2000.3573. [DOI] [PubMed] [Google Scholar]
- 58.Kim WJ, Akaike T, Maruyama A. J. Am. Chem. Soc. 2002;124:12676–12677. doi: 10.1021/ja0272080. [DOI] [PubMed] [Google Scholar]
- 59.Yamana K, Fukunaga Y, Ohtani Y, Sato S, Nakamura M, Kim WJ, Akaike T, Maruyama A. Chem. Commun. 2005:2509–2511. doi: 10.1039/b502033f. [DOI] [PubMed] [Google Scholar]
- 60.Simonson OE, Svahn MG, Törnquist E, Lundin KE, Smith CIE. Curr. Pharmaceut. Design. 2005;11:3671–3680. doi: 10.2174/138161205774580813. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.