Abstract
The recognition of carbon sources and the regulatory adjustments to recognized changes are of particular importance for bacterial survival in fluctuating environments. Despite a thorough knowledge base of Escherichia coli's central metabolism and its regulation, fundamental aspects of the employed sensing and regulatory adjustment mechanisms remain unclear. In this paper, using a differential equation model that couples enzymatic and transcriptional regulation of E. coli's central metabolism, we show that the interplay of known interactions explains in molecular-level detail the system-wide adjustments of metabolic operation between glycolytic and gluconeogenic carbon sources. We show that these adaptations are enabled by an indirect recognition of carbon sources through a mechanism we termed distributed sensing of intracellular metabolic fluxes. This mechanism uses two general motifs to establish flux-signaling metabolites, whose bindings to transcription factors form flux sensors. As these sensors are embedded in global feedback loop architectures, closed-loop self-regulation can emerge within metabolism itself and therefore, metabolic operation may adapt itself autonomously (not requiring upstream sensing and signaling) to fluctuating carbon sources.
Keywords: computational model, metabolism, regulation, sensing, systems biology
Introduction
Adaptations to fluctuating carbon source availability are of particular importance for bacteria. These adaptations are realized by systems of molecular regulations, which (1) recognize carbon sources and (2) adjust metabolic operation to the recognized changes. To understand system behavior, molecular knowledge alone is often not sufficient (Kitano, 2002a). Instead, it needs to be understood how a system's behavior emerges from the interactions between the characterized molecules (Kitano, 2002b). To attain such a system understanding of bacterial metabolic adaptations to carbon source availability, the coupling between the recognition and adjustment aspects and between the enzymatic and genetic regulatory layers must be understood.
Recent studies in Escherichia coli have focused on these couplings to improve our understanding of such adaptations in terms of general, topological motifs (Balázsi et al, 2005; Martinez-Antonio et al, 2006; Reece et al, 2006; Seshasayee et al, 2006; Janga et al, 2007; Krishna et al, 2007; Werner et al, 2009). However, these studies do not link the topological motifs to the molecular-level details of specific adaptations; therefore, the molecular-level interplay of enzymatic and genetic regulation is to date only understood for comparatively simple carbon source adaptations, such as the adaptation to lactose (Cohen and Monod, 1957; Ozbudak et al, 2004). For more complex adaptations involving many operons, such as the adaptations between the glycolytic substrate glucose and the gluconeogenic substrate acetate that require an extensive remodeling of central metabolism, it remains unclear how recognition and regulatory adjustment function in molecular-level detail, and how these processes are coupled to a coordinated adaptation.
Bacteria typically use two major means to recognize carbon sources. Some sugars (e.g. fructose, galactitol, mannitol, mannose, sorbitol) are recognized through the phosphotransferase sugar uptake system (PTS) (Janausch et al, 2002; Plumbridge, 2002; Schlegel et al, 2002; Bijlsma and Groisman, 2003; Bettenbrock et al, 2007) other sugars (e.g. arabinose, glycerol, galactose, lactose, maltose, melibiose, fucose) are recognized intracellularly through regulatory proteins (transcription factors, in short TFs) (Cohen and Monod, 1957; Ozbudak et al, 2004; Martinez-Antonio et al, 2006; Seshasayee et al, 2006). However, for organic acids such as acetate, succinate, or malate as well as for many other carbon sources, neither transmembrane sensors nor regulatory proteins with sensing function have been identified. It thus remains unclear how these carbon sources are recognized.
Concerning the adjustment of metabolic operation between growth on glycolytic and gluconeogenic carbon sources, only local aspects are understood in molecular-level detail. Examples of such local aspects are the branch point effect at the diversion of carbon flux through the glyoxylate (GLX) shunt (LaPorte et al, 1984), the PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution (Sauer and Eikmanns, 2004), or the regulation of cAMP levels by the PTS (Bettenbrock et al, 2007). What remains unclear is how these local regulations work together to accomplish a coordinated adaptation on the systems level.
In this paper, we show that (1) the interplay of the known interactions in E. coli's central metabolism is capable of recognizing carbon sources indirectly through a mechanism we termed distributed sensing of intracellular metabolic fluxes, and that (2) these molecular-level interactions can adjust E. coli's metabolic operation between growth on glycolytic and gluconeogenic carbon sources, and that (3) this adaptation is governed by general principles. We derived these results with a simulation-based approach that rests on a differential equation model of E. coli's central metabolism including its enzymatic, transcriptional, and posttranslational regulation.
Results
Model
When we screened the available molecular knowledge of central metabolism to understand E. coli's adaptations from growth on glucose to acetate and vice versa, we noted that:
Four TFs have been identified that regulate the expression of central metabolic enzymes and whose activities are modulated by binding of central metabolites: • Cra-fructose-1,6-bisphosphate (Cra-FBP) (Ramseier et al, 1993) • Crp-cyclic AMP (Crp-cAMP) (Botsford and Harman, 1992) • IclR-glyoxylate (IclR-GLX) and IclR-pyruvate (IclR-PYR) (Lorca et al, 2007) • PdhR-pyruvate (PdhR-PYR) (Quail and Guest, 1995).
Each of the four involved metabolites assumes distinct levels during glycolytic and gluconeogenic growth across available published experimental data sets (Lowry et al, 1971; Botsford and Harman, 1992; Bettenbrock et al, 2007; Bennett et al, 2009).
The expression levels of central metabolic enzymes regulated by these TFs are markedly distinct for growth on glycolytic and gluconeogenic carbon sources (Oh et al, 2002).
The levels of these enzymes correlate with the activities of the regulating TFs due to distinct levels of their activating/inhibiting effectors (during glycolytic and gluconeogenic growth). For instance, on glucose, the PYR dehydrogenase (Pdh) level is high (compared with acetate), which is consistent with that enzyme's transcriptional repressor PdhR being inhibited by a comparatively high PYR concentration.
The fact that this set of differentially expressed pathways covers central metabolism (excluding the pentose phosphate pathway), and the promising results of earlier work on the role of TF–metabolite interactions in cellular recognition and adjustments (Martinez-Antonio et al, 2006; Reece et al, 2006; Seshasayee et al, 2006) let us to hypothesize that the current knowledge of E. coli's metabolism can already explain the molecular adaptations between glycolytic and gluconeogenic carbon sources. Specifically, we hypothesized that these adaptations are accomplished by a system-wide regulation architecture that emerges when the known enzymatic and transcriptional regulations become coupled through the five listed TF–metabolite interactions. To (1) assess whether such coupled molecular interactions can indeed work together to adapt metabolic operation, and if yes, (2) to understand this system-level adaptation in molecular-level detail, we constructed a comprehensive differential equation model that is centered on the coupling of enzymatic and transcriptional regulation, which is accomplished by the five above listed TF–metabolite interactions.
Model topology
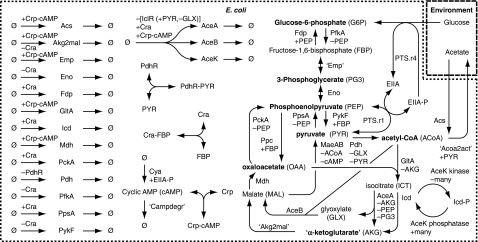
The model topology, shown in Figure 1, comprises the Embden–Meyerhoff pathway (EMP) (also known as the glycolysis or gluconeogenesis pathway, depending on flux direction), the tricarboxylic acid (TCA) cycle, the GLX shunt, the anaplerotic reactions, the diversion of carbon flux to the GLX shunt through phosphorylation of isocitrate dehydrogenase, the uptake of glucose through phosphorylation of PTS proteins, the uptake and excretion of acetate, the allosteric regulation of enzymes in the mentioned pathways and their transcriptional regulation by the above listed TFs, and the regulation of these TFs' activities through the above listed TF–metabolite interactions. Overall, the model comprises two compartments, the cell and its environment. The cellular compartment contains 12 metabolites, 22 enzymes and 2 PTS proteins, 4 TFs, 17 transcriptional regulations, 28 enzymatic regulations, 26 metabolic reactions, 2 kinase and 2 phosphatase reactions, 5 TF–metabolite interactions, the expression of 16 genes, and the degradation of the produced proteins as well as their dilution due to cell growth. The environmental compartment contains two carbon sources. The cell and the environment are coupled through glucose uptake as well as acetate uptake and excretion reactions.
Figure 1.
Topology of the model. The model comprises two compartments, the cell and its environment. Bold metabolite names are biomass precursors and as such substrates for first-order reactions to void. Reactions and metabolites in quotation marks represent lumped reactions and metabolites, respectively. Regulation of enzyme activity through small molecule effectors is indicated below the enzyme name, transcriptional regulation above the protein-producing reaction.
This model topology is centered on the above listed TF–metabolite interactions, as illustrated in Figure 2, and includes the known molecular interactions in E. coli's central metabolism, retrieved from the EcoCyc database (Keseler et al, 2009). The followed strategy for the systematic assembly of these interactions (see Supplementary information) (1) ensures the inclusion of all known interactions between modeled compounds, (2) minimizes the number of omitted interactions across the system boundary, which occur, for instance, when a modeled reaction is regulated by a metabolite outside the system boundary, and (3) omits energy, cofactor, oxygen, and proton balances. These measures establish a system boundary that cuts out the studied adaptation mechanism from the rest of the cell. This approach rests on the modularity of cellular regulation (Wolf and Arkin, 2003), which increases the autonomy of cellular subsystems. Because of this modularity, it is very likely that the in silico dynamics of the modeled regulatory subsystem, which adapts metabolism between glycolytic and gluconeogenic growth, are in vivo only modulated but not fundamentally changed by the activity of omitted regulations across the system boundary, which, for instance, balance the energy, cofactor, oxygen, and proton requirements.
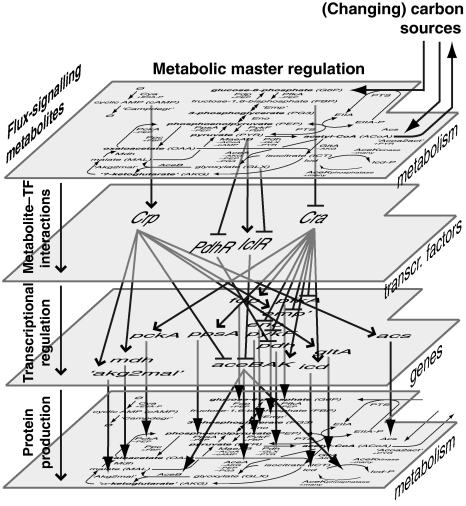
Figure 2.
The model topology is centered around the known TF–metabolite interactions and establishes a feedback loop from the metabolic layer through the transcriptional regulatory layer and the gene expression layer back to the metabolic layer. The metabolic layer is directly upstream of the transcriptional regulatory layer, enabling it to perform the coordinating function of metabolic master regulation.
Kinetic equations
The focus of this study is to investigate whether the coupling of enzymatic and transcriptional regulation can accomplish dynamic adaptations between different steady-state growth conditions. Because of the importance of metabolite levels for controlling transcriptional regulation (see above), we needed to follow a modeling approach that accounts for metabolite dynamics shaped by enzyme kinetics and enzymatic regulation. These dynamics can only be adequately captured by a kinetic model, as, for example, existing constraint-based models with Boolean regulatory architecture such as developed and used by Covert et al (2004) and Shlomi et al (2007) ignore enzymatic regulation. Specifically, these models assume a rigid, positive correlation between metabolic flux and metabolite levels (neglecting the fact that this correlation can also be negative, see Kremling et al (2008), and nuanced, see below). The problem of many uncertain parameter values of kinetic models is mitigated by the fact that the structure of mechanistic systems biology models already tightly constrains the possible system behaviors, leaving most parameter values unimportant (Brown et al, 2004; Gutenkunst et al, 2007).
Following a kinetic modeling approach, we translated the topology into differential equations by assigning the most appropriate rate law to each interaction (see Supplementary Tables S1 and S2). To formulate the rate equation for the biomass-generating reaction, we back-calculated the requirements for metabolites outside the system boundary to their respective precursors inside the system boundary. We included the growth-rate dependency of the gene expression rates due to growth rate-dependent levels of DNA polymerases and ribosomes (Bremer and Dennis, 2008).
The resulting 47 ordinary differential equations contain 193 parameters and are of the form
with x the vector of dynamic state variables (the concentrations of intracellular compounds and extracellular carbon sources), f the vector of kinetic rate equations, p the vector of parameters, and S the stoichiometric matrix. Refer to the Supplementary information for the full-model equations as well as for a MATLAB and SBML version of the model.
Parameter values for the rate equations were estimated through application of the ‘divide-and-conquer approach’ (Kotte and Heinemann, 2009) on published experimental steady state-omics data sets for balanced growth on either glucose or acetate (see Supplementary information).
Supplementary Table S5 lists the values of a single parameter vector that excellently reproduces the steady state data (see Supplementary Tables S3 and S4) for balanced growth on either glucose or acetate with the coefficient of determination R2≈0.999, and with the sensitivities shown in Supplementary Figure S2. These parameter values are used for all simulations presented in this paper.
Reproduction of known physiological behavior
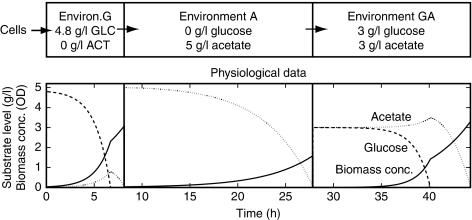
To test whether the model is indeed capable of adapting to changing carbon sources, we subjected it to a sequence of three consecutive environments. The first environment contains glucose as the sole carbon source and is ‘inoculated’ with glucose-adapted in silico cells, meaning that the initial conditions of the in silico cells were set to the steady state values of glucose growth. Figure 3 shows that the in silico cells grow on glucose, produce acetate from glucose, and after glucose depletion commence their adaptation to acetate, which they re-consume until it is depleted. The in silico cells are then ‘transferred’ to a second environment that contains acetate as the sole carbon source, which the cells consume to successfully complete their adaptation to the acetate-adapted steady state. Finally, the in silico cells are ‘transferred’ to a third environment that contains both glucose and acetate as carbon sources. Although at this point the cells are adapted to the present acetate, they quickly adapt to glucose instead, produce acetate from glucose, and then re-adapt to acetate following glucose depletion.
Figure 3.
The model reproduces E. coli's known physiological behaviors of preferential glucose uptake and of acetate production from glucose.
These simulations show that the model reproduces E. coli's known physiological behavior of glucose repression, which is the preferential uptake of glucose over acetate, and of an overflow metabolism, which is the production of acetate from glucose. Further, Supplementary Figures S4–S8 show that (1) throughout the transitions, all intracellular metabolite and enzyme levels remain within physiologically reasonable bounds and (2) all compound levels and reaction rates approach their measured steady state values for balanced growth. Whereas the measured steady states and the physiological behavior are correctly reproduced, the simulated trajectories of the intracellular compound levels during the transitions from one of the two steady states to the other (see Supplementary information) are uncertain. The uncertainty of the simulated trajectories arises from (1) uncertainty in the parameter values, (2) uncertainty in the model structure due to ambiguity in the selection of rate laws, and (3) possible effects of not modeled cellular regulations, that is, those ensuring the omitted energy, cofactor etc. balances, onto the simulated trajectories.
The simulated trajectories of the intracellular compounds is thus one among many possible responses that reproduce the data. However, the possible behaviors of systems biology models are already tightly constrained by the model structure and generally rest on few crucial parameters (Brown et al, 2004; Gutenkunst et al, 2007)—alterations of most rate laws and variations of parameters only modulate but do not fundamentally change the overall system behavior. Exploiting this property, we investigate one possible response that has the remarkable property of adapting central metabolism to changing carbon sources. We aim at (1) understanding in detail how the interplay of molecular interactions accomplishes the adaptation, to (2) identify the general conceptual functions realized by these molecular interactions.
Recognition of extracellular carbon sources
As the in silico cells successfully adapt to fluctuating levels of glucose and acetate, they must have a mechanism to recognize these carbon sources. But how does an in silico cell recognize acetate without a transmembrane sensor for extracellular acetate or a TF binding to intracellular acetate? Similarly, it is unclear whether the glucose sensing function of the PTS is the exclusive mechanism to recognize glucose, or whether this sensing function is integrated into a larger sensing architecture.
As a sensing mechanism translates environmental information into TF activity, either through phosphorylation or effector binding, the sensing mechanism of the in silico cell is identified once it is understood how an extracellular carbon source affects intracellular TF activities. Whereas phosphorylation of the TFs Cra, Crp, IclR and PdhR has as yet not been reported, these TFs' activities are known to respond to concentration changes of their effectors (Botsford and Harman, 1992; Ramseier et al, 1993; Quail and Guest, 1995; Lorca et al, 2007). Therefore, we here exclude the possibility of phosphorylation; the only remaining possibility to modulate these TFs' activities are concentration changes of their effectors. Our model simulations revealed that the TFs' effectors indeed respond to changes in the availability of extracellular carbon sources with a concentration change that modulates the TFs' activities (see Supplementary Figure S8). But what links the levels of the intracellular effectors to the presence of extracellular carbon sources? The only entity capable of providing this missing link is metabolic flux, which must therefore form an integral part of the sensing mechanism. With this constraint, we deduced the following tripartite sensing mechanism.
First, the cell ensures a basal expression of the relevant carbon source transporter and uptake pathway. Hence, when that carbon source enters the cell's environment, it is taken up at least at a basal rate. In the in silico cell, such basal uptake is realized by a constitutive expression of the glucose-transporting PTS, and by a basal expression of the acetate-transporting ‘super-enzyme’ Acs, which in the model lumps the acetate transport reaction and the subsequent conversion to acetyl-CoA (note that on glucose, acetate is both produced through the ‘Acoa2act’ reaction and re-consumed through the Acs reaction at a lower rate). As the uptake of a carbon source propagates as intracellular flux through downstream metabolic pathways, the ensured uptake of a carbon source whenever present causes intracellular fluxes to respond to the presence of this carbon source.
Second, the enzyme kinetics and enzymatic regulation in pathways affected by the uptake flux is such that a certain metabolic intermediate in these pathways responds to the uptake flux in a characteristic way. The function of the enzyme kinetics and enzyme regulations is to establish a measurable signal for metabolic flux, which, being a rate, is an entity that the cell cannot measure directly. The in silico cell uses two distinct motifs to establish such flux-signaling metabolites.
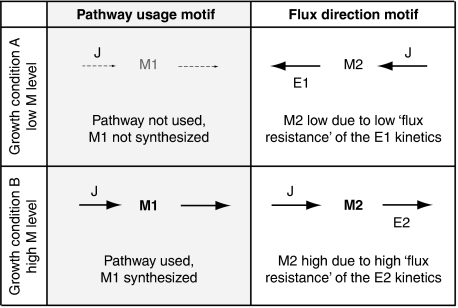
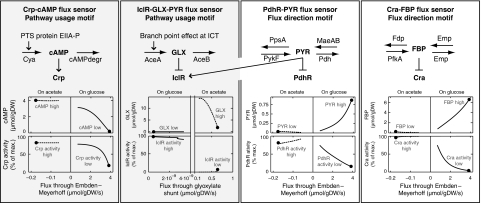
The motif pathway usage, illustrated in the left panel of Figure 4, places the flux-signaling metabolite in a pathway that is only used in one growth condition but not in the other; hence, synthesis of the metabolite, that is a high metabolite concentration, signals pathway usage. The in silico cell uses this motif to establish distinct levels of cAMP and GLX (see Figure 5). In both cases, the differential pathway induction is realized through fast changes in protein phosphorylation states. The phosphorylation of the PTS protein EIIA, which induces the formation of cAMP, is directly coupled to glucose uptake; the coupling of acetate uptake to flux through the GLX shunt is achieved by the regulation of the phosphorylation of isocitrate dehydrogenase, which diverts flux from the TCA cycle to the GLX shunt.
The motif flux direction, illustrated in the right panel of Figure 4, places the flux-signaling metabolite in a reversible pathway that is operated in different directions depending on carbon source availability. As different flux directions mean that different enzymes ‘consume’ the metabolite, these enzymes' kinetics can have different ‘flux resistances’ such that the metabolite assumes high levels for one flux direction and low levels for the other; hence, the metabolite level signals flux direction. The in silico cell uses this motif to create distinct levels of the flux-signaling metabolites FBP and PYR (see Figure 5).
Third, the flux-signaling metabolites bind to target TFs and modulate their activities. These TF–metabolite interactions can be interpreted as a measurement of metabolite levels by TFs. The joint systems of TF–metabolite interactions and either of the above-described motifs, which ensure that the flux-signaling metabolite levels carry information about metabolic fluxes, conceptually form (indirect) sensors for metabolic fluxes. A molecular sensor for intracellular metabolic flux is thus defined as a system of regulations and enzyme kinetics, comprising (1) either of the two motifs pathway usage or flux direction and (2) the binding of the thus established flux-signaling metabolites to TF(s). Note that this definition makes a clear distinction between the direct physical measurement, that is of the flux-signaling metabolite levels by TFs, and the flux sensor's conceptually measured entity, that is the metabolic fluxes, which are not directly measurable and hence translated into a measurable signal. Note that metabolic flux sensors are conceptually analogous to man-made sensors—for instance, the not directly measurable velocity of an aircraft (a rate, like metabolic flux) is commonly inferred from physical measurements of the (velocity signaling) air pressure in so-called pitot tubes, which are constructed such that the air pressure inside the tubes carries velocity information.
The in silico cell is equipped with four flux sensors. Three of these sensors ultimately measure at different positions of the flux direction (and possibly its magnitude) through the EMP. Cra-FBP measures the flux through the upper EMP where FBP is located. On glucose, PdhR-PYR measures the lower glycolytic flux that is fed into the TCA cycle through PYR, whereas on acetate, PdhR-PYR measures the flux through the malic enzymes that is fed into the lower EMP. When glucose is taken up, the PTS-coupled Crp-cAMP sensor reports the glucose uptake and thus the glycolytic flux into the upper EMP; on acetate, the PTS-coupled cAMP level equilibrates with the PEP/PYR ratio that results from gluconeogenic flux into the lower EMP. The fourth of these flux sensors, IclR-GLX, senses the flux through the GLX shunt and is further modulated by the PYR signal (through IclR-PYR).
Figure 5 shows that the levels of the in silico cell's four flux-signaling metabolites as well as the activities of the four target TFs are distinct not only for balanced growth on either glucose or acetate, but also for lower uptake rates of these substrates. Thus, the property of distinct TF activities is robust to changes in the substrate uptake rates, which may arise, for example, from fluctuating concentrations of the carbon sources. Because of this robustness, the TFs reliably sense the presence of extracellular glucose and acetate through binding ‘endogeneous’ (Martinez-Antonio et al, 2006; Seshasayee et al, 2006) metabolite signals. Note that in some cases (e.g. Cra-FBP on glucose, see the fourth column in Figure 4), the gradual differences in TF activity resulting from variations in substrate uptake rates vary sensitively and monotonously with the magnitude of the sensed flux, which conceptually enables the sensing of intracellular flux magnitudes.
Figure 4.
Two general motifs to establish flux-signaling metabolites. The first motif, pathway usage, places the flux-signaling metabolite in a pathway that is used in one growth condition but not in the other. The second motif, flux direction, places the flux-signaling metabolite in a reversible pathway that is used in different directions depending on the growth conditions.
Figure 5.
The four flux sensors of the in silico cell. The four flux-signaling metabolites are established through enzyme kinetics and regulations that realize the motifs ‘pathway usage’ and ‘flux direction.’ The simulated dependencies of the flux-signaling metabolites on the fluxes they measure show that these metabolites exhibit markedly distinct levels not only for balanced growth (filled dots), but also for up to 80% lower glucose (regular lines) or acetate (dashed lines) uptake rates. The levels of these metabolites thus report extracellular glucose and acetate. These distinct metabolite levels propagate into distinct activities of their target TFs, which adjusts the transcriptional regulation exerted by these TFs to the present carbon sources.
As the in silico cell establishes and uses sensors for several intracellular metabolic fluxes, the overall sensing architecture infers the present carbon sources from a pattern of metabolic fluxes and is as such of a distributed nature. We therefore termed this architecture distributed sensing of intracellular metabolic fluxes. The core of this sensing architecture is formed not by transmembrane sensors but by the four flux sensors shown in Figure 4, which establish flux-signaling metabolites according to the two presented general motifs. These flux sensors use intracellular metabolic flux as a means to correlate the presence of extracellular carbon sources with the levels of intracellular metabolites. The recognition of glucose through the PTS transmembrane complex is embedded as one flux sensor in this distributed sensing architecture; the other three flux sensors function without the help of transmembrane complexes.
Coupling of recognition and adjustment
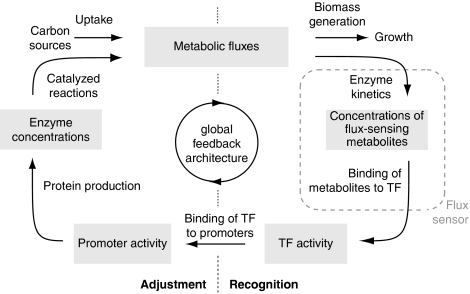
Having identified the sensing mechanism, it still needs to be understood how the recognition of carbon sources is followed by an adjustment of metabolic operation to the recognized changes.
The in silico cell achieves the coupling between recognition and adjustment through its TFs, whose activities respond to the available carbon sources and at the same time regulate the expression of target genes. This combined recognition and adjustment, centered on the four TFs, closes four global feedback loops that overarch the metabolic and genetic layers as illustrated in Figure 6. The first half of these four loops forms the recognition function and is established by the flux-signaling metabolites binding to their target TFs, creating flux sensors (the four columns in Figure 5, and the upper of the three layers of arrows in Figure 2). The second half of these four loops forms the adjustment function and is established by (1) the transcriptional regulation of the four flux-sensing TFs (the middle layer of arrows in Figure 2), which causes the regulated enzymes to approach their measured steady state values, and by (2) the impact of this transcriptional regulation on metabolic operation (the lower layer of arrows in Figure 2), which, together with allosteric enzyme regulation, adjusts the metabolic fluxes.
Figure 6.
Global feedback loop architecture. This architecture, which overarches the metabolic and genetic layers, ties the recognition of carbon sources and the adjustment of metabolic operation to the recognized changes together. The dashed box indicates those regulations that form conceptual flux sensors.
To sum up, the adaptation of the in silico cell arises from the global feedback loop-embedded, flux sensor-adjusted transcriptional regulation of the four TFs, with each TF performing one part of the overall adaptation. This adaptation incorporates both the influence of the metabolic on the genetic layer, achieved through TF–metabolite interactions, and of the genetic on the metabolic layer, achieved through the impact of adjusted enzyme levels on metabolic fluxes. Remarkably, the thus formed global feedback loops follow the general logic of the recently proposed consumer motif, which has been suggested to be ideal for the regulation of carbon source uptake (Krishna et al, 2007).
Coherence through network topology
We have shown that the transcriptional response of the in silico cell to carbon source fluctuations is realized by four global feedback loops. As the four involved TFs are not regulated by a common transcriptional master regulator, it remains unclear how the apparently independent adjustments carried out by these TFs produce a coherent response of the overall system.
As the in silico cell can modulate its four TF activities only through the levels of the TFs' effectors, coherent TF activities can only result from coherent levels of the flux-signaling effectors. Such coherence arises on the metabolic layer, which is one step upstream of the TF regulatory layer (see Figure 2). We identified two means through which network topology encourages such coherence.
First, the responses of flux-signaling metabolites are coupled to each other because the metabolic fluxes to which they respond are connected to each other through the flux balances at network nodes. Therefore, when the local flux signaled by one of the flux-signaling metabolites changes, then the local fluxes signaled by the other metabolites are also likely to change.
Second, the responses of flux-signaling metabolites are coupled to each other because they mutually regulate each other's adjacent enzymes. Therefore, a change in one flux-signaling metabolite level propagates into changes of the other flux-signaling metabolite levels. Specifically,
The levels of cAMP, PYR, PEP and the phosphorylation state of the EIIA protein are coupled through the phosphorylations of the PTS transporter (Bettenbrock et al, 2007). The levels of FBP and PEP are coupled through a dual-time switch, a motif that has been shown to be rapidly inducible yet robust to noise (Brandman et al, 2005). This switch ensures a high level of FBP and a low level of the downstream PEP on glucose, and the reverse behavior on acetate. The fast switch is formed by PEP and FBP mutually activating each other's consuming enzymes (Fdp, PykF, Ppc) through feed-forward loops, a motif that enables a high level of the upstream metabolite to lower the level of the downstream metabolite (Kremling et al, 2008). The slow switch is established through the transcriptional regulation of FBP-inhibited Cra on the expression of these metabolites' producing and consuming enzymes Fdp, PfkA, PckA, PykF, and PpsA and amplifies the coupling of FBP and PEP.
The levels of PYR and GLX are coupled through GLX acting as inhibitor of the PYR-consuming Pdh reaction, and PYR acting as corepressor of the GLX shunt operon aceBAK and as effector regulating the phoshorylation of isocitrate dehydrogenase (which diverts flux from the TCA through the GLX shunt).
Through these two means, the levels of the flux-signaling metabolites become coupled, enabling a robust, coherent response of the four TFs. The coherent behavior of the overall system is therefore not established by a common transcriptional master regulator upstream of that system, but arises from the molecular interactions within the system itself.
Discussion
In this paper, we presented a differential equation model of E. coli's central metabolism. This model includes transcriptional, posttranslational, and enzymatic regulation and is centered on the coupling of the genetic and metabolic layers, which is accomplished by TF–metabolite interactions. The model offers a consistent explanation of how a multitude of known molecular interactions fit into a coherent systems picture; the interactions work together like gear wheels that mesh with one another to adapt central metabolism between growth on the glycolytic substrate glucose and the gluconeogenic substrate acetate. A detailed discussion in the Supplementary information elucidates the specific contributions of individual interactions to the overall response.
The here proposed distributed sensing mechanism emerges from the system dynamics and could not have been deduced from a recently proposed constraint-based model integrating metabolic and transcriptional regulation (Covert et al, 2004; Shlomi et al, 2007). This model replaces the unknown sensing mechanism through Boolean rules of the form ‘If carbon source X is present, TF Y is active, otherwise not,’ and is as such not suited to understand the detailed molecular processes leading to the activation of TF Y. The underlying view of the extended constraint-based model is that top-level regulatory proteins recognize environmental conditions and adjust downstream metabolic operation. This view is challenged by the global feedback architectures proposed in our study. When kinetics are considered, the capability for closed-loop self-regulation can emerge within metabolism itself and therefore, metabolic operation may adapt itself autonomously (not requiring upstream sensing and signaling) to changing carbon sources.
Although the experimental validation of the here computationally derived principles is challenging and certainly beyond the scope of this work, two facts strongly suggest that these principles do operate in vivo. First, the extent and simplicity to which the studied systems-level adaptation is explained through the known molecular interactions is stunning: E. coli's known enzymatic and transcriptional regulation, when coupled through the bindings of effectors to only four TFs, can explain the recognition of glycolytic and gluconeogenic carbon sources and the adjustments of the EMP, of the GLX shunt, of the anaplerotic reactions, and of the PYR dehydrogenase reaction between growth on glucose and acetate, incorporating the branch point effect through phosphorylation of isocitrate dehydrogenase, the uptake of glucose through PTS protein phosphorylations, the uptake and excretion of acetate, while reproducing the preferential uptake of glucose and the production of acetate from glucose.
Second, the general principles deduced from the presented model also describe the well-studied (and not modeled) adaptation of E. coli from glucose to lactose (Cohen and Monod, 1957; Ozbudak et al, 2004). In detail, the basal uptake of extracellular lactose is ensured through a basal expression of the lactose transporter LacY. Intracellular lactose is a flux-signaling metabolite for lactose uptake flux, established through the motif of pathway usage. A flux sensor is created through the binding of this flux signal to the lactose repressor protein LacI. A ‘global’ feedback loop is closed by the LacI-regulated induction of the lac operon. Hence, the here deduced distributed adaptation mechanism reduces to correctly describe the (not distributed) adaptation of one operon by one TF. As E. coli's adaptation between glucose and acetate and its adaptation to lactose can both be explained by the general principles deduced in this paper, these adaptations may differ only in complexity but not in their nature of functioning according to the here presented principles.
In this work, we deduced general functional principles that provide the missing link to understand system-level adaptations to carbon sources in molecular-level detail. The proposed principles fall under the umbrella of distributed flux sensing. The flux sensing mechanism entails the binding of TFs to flux-signaling metabolites, which are established through the motifs signaling of pathway usage and signaling of flux direction, and are embedded in global feedback loop architectures. These principles allow an autonomous adaptation of metabolic operation to growth in fluctuating environments.
Supplementary Material
Model structure, Parameter estimation, Sensitivity analysis, Simulation results, Contributions of individual interactions to the overall response
Acknowledgments
We thank ETH Zurich for financial support within the INIT SCM project; Paul F Cook for providing the binding kinetics of IclR to DNA; Henning Schmidt for help in translating the Matlab model into an SBML model; and Uwe Sauer for an excellent working environment. We also thank Uwe Sauer and Luca Gerosa for helpful comments on the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Balázsi G, Barabási AL, Oltvai ZN (2005) Topological units of environmental signal processing in the transcriptional regulatory network of Escherichia coli. Proc Natl Acad Sci USA 102: 7841–7846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD (2009) Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol 5: 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettenbrock K, Sauter T, Jahreis K, Kremling A, Lengeler JW, Gilles E-D (2007) Correlation between growth rates, EIIACrr phosphorylation, and intracellular cyclic AMP levels in Escherichia coli K-12. J Bacteriol 189: 6891–6900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlsma JJ, Groisman EA (2003) Making informed decisions: regulatory interactions between two-component systems. Trends Microbiol 11: 359–366 [DOI] [PubMed] [Google Scholar]
- Botsford JL, Harman JG (1992) Cyclic AMP in prokaryotes. Microbiol Rev 56: 100–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Ferrell JE Jr, Li R, Meyer T (2005) Interlinked fast and slow positive feedback loops drive reliable cell decisions. Science 310: 496–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer H, Dennis PP (2008) Modulation of chemical composition and other parameters of the cell at different exponential growth rates. In EcoSal—Escherichia coli and Salmonella: Cellular and Molecular Biology, Böck A et al. (eds). Module 5.2.3. Washington DC: ASM Press [DOI] [PubMed] [Google Scholar]
- Brown KS, Hill CC, Calero GA, Myers CR, Lee KH, Sethna JP, Cerione RA (2004) The statistical mechanics of complex signaling networks: nerve growth factor signaling. Phys Biol 1: 184–195 [DOI] [PubMed] [Google Scholar]
- Cohen GN, Monod J (1957) Bacterial permeases. Bacteriol Rev 21: 169–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covert MW, Knight EM, Reed JL, Herrgard MJ, Palsson BO (2004) Integrating high-throughput and computational data elucidates bacterial networks. Nature 429: 92–96 [DOI] [PubMed] [Google Scholar]
- Gutenkunst RN, Waterfall JJ, Casey FP, Brown KS, Myers CR, Sethna JP (2007) Universally sloppy parameter sensitivities in systems biology models. PLoS Comput Biol 3: 1871–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janausch IG, Zientz E, Tran QH, Kršger A, Unden G (2002) C4-dicarboxylate carriers and sensors in bacteria. Biochim Biophys Acta 1553: 39–56 [DOI] [PubMed] [Google Scholar]
- Janga SC, Salgado H, Martinez-Antonio A, Collado-Vides J (2007) Coordination logic of the sensing machinery in the transcriptional regulatory network of Escherichia coli. Nucleic Acids Res 35: 6963–6972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler IM, Bonavides-Mart'nez C, Collado-Vides J, Gama-Castro S, Gunsalus RP, Johnson DA, Krummenacker M, Nolan LM, Paley S, Paulsen IT, Peralta-Gil M, Santos-Zavaleta A, Shearer AG, Karp PD (2009) EcoCyc: a comprehensive view of Escherichia coli biology. Nucleic Acids Res 37: D464–D470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H (2002a) Systems biology: a brief overview. Science 295: 1662–1664 [DOI] [PubMed] [Google Scholar]
- Kitano H (2002b) Computational systems biology. Nature 420: 206–210 [DOI] [PubMed] [Google Scholar]
- Kotte O, Heinemann M (2009) A divide-and-conquer approach to analyze underdetermined biochemical models. Bioinformatics 25: 519–525 [DOI] [PubMed] [Google Scholar]
- Kremling A, Bettenbrock K, Gilles ED (2008) A feed-forward loop guarantees robust behavior in Escherichia coli carbohydrate uptake. Bioinformatics 24: 704–710 [DOI] [PubMed] [Google Scholar]
- Krishna S, Semsey S, Sneppen K (2007) Combinatorics of feedback in cellular uptake and metabolism of small molecules. Proc Natl Acad Sci USA 104: 20815–20819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPorte DC, Walsh K, Koshland DE Jr (1984) The branch point effect. Ultrasensitivity and subsensitivity to metabolic control. J Biol Chem 259: 14068–14075 [PubMed] [Google Scholar]
- Lorca GL, Ezersky A, Lunin VV, Walker JR, Altamentova S, Evdokimova E, Vedadi M, Bochkarev A, Savchenko A (2007) Glyoxylate and pyruvate are antagonistic effectors of the Escherichia coli IclR transcriptional regulator. J Biol Chem 82: 16476–16491 [DOI] [PubMed] [Google Scholar]
- Lowry OH, Carter J, Ward JB, Glaser L (1971) The effect of carbon and nitrogen sources on the level of metabolic intermediates in Escherichia coli. J Biol Chem 246: 6511–6521 [PubMed] [Google Scholar]
- Martinez-Antonio A, Janga SC, Salgado H, Collado-Vides J (2006) Internal-sensing machinery directs the activity of the regulatory network in Escherichia coli. Trends Microbiol 14: 22–27 [DOI] [PubMed] [Google Scholar]
- Oh MK, Rohlin L, Kao KC, Liao JC (2002) Global expression profiling of acetate-grown Escherichia coli. J Biol Chem 277: 13175–13183 [DOI] [PubMed] [Google Scholar]
- Ozbudak EM, Thattai M, Lim HN, Shraiman BI, van Oudenaarden A (2004) Multistability in the lactose utilization network of Escherichia coli. Nature 427: 737–740 [DOI] [PubMed] [Google Scholar]
- Plumbridge J (2002) Regulation of gene expression in the PTS in Escherichia coli: the role and interactions of Mlc. Curr Opin Microbiol 5: 187–193 [DOI] [PubMed] [Google Scholar]
- Quail MA, Guest JR (1995) Purification, characterization and mode of action of PdhR, the transcriptional repressor of the pdhR-aceEF-lpd operon of Escherichia coli. Mol Microbiol 15: 519–529 [DOI] [PubMed] [Google Scholar]
- Ramseier TM, Negre D, Cortay JC, Scarabel M, Cozzone AJ, Saier MH Jr (1993) In vitro binding of the pleiotropic transcriptional regulatory protein, FruR, to the fru, pps, ace, pts and icd operons of Escherichia coli and Salmonella typhimurium. J Mol Biol 234: 28–44 [DOI] [PubMed] [Google Scholar]
- Reece RJ, Beynon L, Holden S, Hughes AD, Rebora K, Sellick CA (2006) Nutrient-regulated gene expression in eukaryotes. Biochem Soc Symp 73: 85–96 [DOI] [PubMed] [Google Scholar]
- Sauer U, Eikmanns BJ (2004) The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol Rev 29: 765–794 [DOI] [PubMed] [Google Scholar]
- Seshasayee AS, Bertone P, Fraser GM, Luscombe NM (2006) Transcriptional regulatory networks in bacteria: from input signals to output responses. Curr Opin Microbiol 9: 511–519 [DOI] [PubMed] [Google Scholar]
- Schlegel A, Danot O, Richet E, Ferenci T, Boos W (2002) The N terminus of the Escherichia coli transcription activator MalT is the domain of interaction with MalY. J Bacteriol 184: 3069–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomi T, Eisenberg Y, Sharan R, Ruppin E (2007) A genome-scale computational study of the interplay between transcriptional regulation and metabolism. Mol Syst Biol 3: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M, Semsey S, Sneppen K, Krishna S (2009) Dynamics of uptake and metabolism of small molecules in cellular response systems. PLoS One 4: e4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Arkin A (2003) Motifs, modules and games in bacteria. Curr Opin Microbiol 6: 125–134 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Model structure, Parameter estimation, Sensitivity analysis, Simulation results, Contributions of individual interactions to the overall response