Abstract
The Saccharomyces cerevisiae APN1 gene that participates in base excision repair has been localized both in the nucleus and the mitochondria. APN1 deficient cells (apn1Δ) show increased mutation frequencies in mitochondrial DNA (mtDNA) suggesting that APN1 is also important for mtDNA stability. To understand APN1-dependent mtDNA repair processes we studied the formation and repair of mtDNA lesions in cells exposed to methyl methanesulfonate (MMS). We show that MMS induces mtDNA damage in a dose-dependent fashion and that deletion of the APN1 gene enhances the susceptibility of mtDNA to MMS. Repair kinetic experiments demonstrate that in wild-type cells (WT) it takes 4 hr to repair the damage induced by 0.1% MMS, whereas in the apn1Δ strain there is a lag in mtDNA repair that results in significant differences in the repair capacity between the two yeast strains. Analysis of lesions in nuclear DNA (nDNA) after treatment with 0.1% MMS shows a significant difference in the amount of nDNA lesions between WT and apn1Δ cells. Interestingly, comparisons between nDNA and mtDNA damage show that nDNA is more sensitive to the effects of MMS treatment. However, both strains are able to repair the nDNA lesions, contrary to mtDNA repair, which is compromised in the apn1Δ mutant strain. Therefore, although nDNA is more sensitive than mtDNA to the effects of MMS, deletion of APN1 has a stronger phenotype in mtDNA repair than in nDNA. These results highlight the prominent role of APN1 in the repair of environmentally induced mtDNA damage.
Keywords: base excision repair, mitochondrial DNA, alkylating agent
INTRODUCTION
Mitochondrial dysfunction due to oxidative damage has been associated with certain neurodegenerative disorders, cancer, and aging [Esposito et al., 1999]. A major target of oxidative damage within mitochondria is mitochondrial DNA (mtDNA). Therefore, elucidating how cells maintain the stability of the mtDNA is of great relevance to understanding the etiology of such diseases. DNA lesions can arise in several ways, most importantly by methylation, oxidation, and deamination of normal bases yielding a variety of lesions [Friedberg et al., 2006]. These lesions can occur by spontaneous processes within the cells or may be induced by environmental agents. If left unrepaired, oxidative and alkylation DNA damage may constitute potent replication blocks, causing cytoxicity and enhanced mutagenesis [Prakash et al., 2005; Johnson et al., 2007]. Oxidative and alkylating DNA damage is mainly repaired by base excision repair (BER) [Sung and Demple, 2006].
There is evidence that mammalian mitochondria can remove alkylating and oxidative lesions [Bohr, 2002]. Some of these lesions include O6-alkylguanines, 7,8-dihydro-8-oxodG, formamidopyrimidine, and abasic sites [Pinz and Bogenhagen, 1998; de Souza-Pinto et al., 2001]. These observations lead to the proposal that BER is active in mitochondria. This proposal is supported by biochemical experiments, which indicate that mitochondrial BER occurs via a short-patch mechanism [Stierum et al., 1999]. Recent evidence supports the notion that long-patch BER can also occur in mammalian mitochondria [Szczesny et al., 2008]. The following components of the BER machinery have been identified in mammalian mitochondria: the glycosylases Ogg1, Ung, Ntg1; the AP endonuclease Ape1; and DNA ligase III [Mandavilli et al., 2002]. The replicative mtDNA polymerase Pol γ has also been shown to participate in BER [Longley et al., 1998].
The components of the BER machinery have also been identified in the yeast Saccharomyces cerevisiae. These include the genes for the DNA glycosylases NTG1 [You et al., 1999] and OGG1 [Singh et al., 2001], the AP endonuclease APN1 [Vongsamphanh et al., 2001], and CDC9 which encodes a DNA ligase [Donahue et al., 2001]. In yeast, the MIP1 gene encodes the replicative mtDNA Pol γ and it is highly likely that this gene product also participates in BER [Foury, 1989; Stuart et al., 2006]. Genes that participate in other DNA repair pathways have also been identified in the maintenance of mitochondrial genomic stability. For example, MHR1, which is involved in recombination, has been shown to be required for the maintenance of mtDNA stability [Ling et al., 1995, 2000]. Another gene product required for the repair of oxidative damaged mtDNA is Mgm101p, a mitochondrial nucleoid protein whose role is not understood but has been proposed to be involved in recombination-mediated mtDNA repair [Meeusen et al., 1999]. DIN7, a homolog of the endonuclease RAD2, localizes in mitochondria and is also required for mtDNA stability [Fikus et al., 2000]. Translesion synthesis components such as Rev1, Rev3, and Rev7 have also been identified in yeast mitochondria [Zhang et al., 2006]. Deletion of most of these genes results in increased mtDNA lesions and mtDNA mutations, suggesting that mtDNA repair is important for the maintenance of mtDNA stability.
Notwithstanding the relatively large group of genes associated with mtDNA stability, there is little evidence on how cells respond to environmentally induced mtDNA damage. In this study, we focus our attention on the APN1 gene because, within BER, the actions of Apn1p represent a central step as this protein acts upon substrates generated by either the action of DNA glycosylases (AP sites) or on substrates (3′-α,β-unsaturated aldehydes) generated by the actions of glycosylases containing AP lyase activity. Because of the central role of Apn1p in BER, we hypothesize that deletion of the APN1 gene will strongly affect the sensitivity and repair kinetics of mtDNA damage induced by DNA damaging agents, parameters that are unknown in the yeast S. cerevisiae. To test this hypothesis we applied a quantitative polymerase chain reaction (QPCR) assay, which can detect a variety of DNA lesions such as AP sites, strand breaks, and base modifications, to measure mtDNA damage induced by the simple alkylating agent methylmethanesulfonate (MMS). By using this approach we were able to determine dose–response curves and repair kinetics of mtDNA and nuclear DNA (nDNA) damage induced by MMS. Our results show that yeast strains harboring a null mutation in the APN1 gene (apn1Δ) are more susceptible to mtDNA damage and have slower repair kinetics than wild-type (WT) cells. In addition, we report that although nDNA is more sensitive than mtDNA to the effects of MMS treatment in yeast cells, mtDNA repair has a stronger requirement for Apn1p than nDNA repair. These results indicate that BER is an important mechanism for the maintenance of mtDNA stability in response to an environmentally induced insult.
MATERIALS AND METHODS
Yeast Strains, Media, and MMS Treatment
The S. cerevisae strains EMY74.7 (MATa his3-Δ1 leu2-3,−112 trp1Δ ura3-52) and its isogenic apn1Δ derivative YRP191 were used in this study. Cells were grown in YPD medium (1% yeast extract, 2% peptone, and 2% dextrose) (Sigma Chemical Co. (St. Louis, Missouri) overnight. Cells (1 × 109 cells per MMS dose) were washed with water and resuspended in 0.05 M KPO4 buffer (pH 7.0). Cells were subsequently treated with different concentrations MMS concentrations: 0.025% (2.95 mM), 0.050% (5.9 mM), 0.075% (8.85 mM), 0.10% (11.8 mM), and 0.150% (17.7 mM) for 20 min. The reaction was terminated by the addition of 10 ml of 10% sodium thiosulfate. Cells were collected by centrifugation, washed with 5% sodium thiosulfate and then with water. Cells were resuspended in 1 ml of water and, when needed, appropriate dilutions of cells were plated on YPD medium for viability determinations. Spheroplasts were prepared as follows: cells (1 × 109 cells/ml) were resuspended in 1 ml H2O followed by the addition of 0.1 ml of 0.5 M EDTA (Sigma Chemical Co. (St. Louis, Missouri) (pH 9.0) and 25 μl β-mercaptoethanol (Sigma Chemical Co. (St. Louis, Missouri). After a 10-min incubation at room temperature, cells were centrifuged for 1 min in a microcentrifuge and then resuspended in 1 ml of 1.1 M Sorbitol −0.1 M EDTA (pH 7.5). Zymolase 100T (ICN Biomedicals, Costa Mesa, CA) (0.1 ml) at 10 mg/ml was added to the cells followed by a 10-min incubation at 37°C. For repair kinetics experiments, cells were treated with 0.1% MMS for 20 min as described earlier. After the treatment period, cells were resuspended in YPD media and grown at 30°C for up to 4 hr. Aliquots (1 × 109 cells) were removed at different time points and spheroplasts were prepared as described earlier.
DNA Isolation and Quantification
DNA was isolated from spheroplasts using a Qiagen (Valencia, CA) DNA extraction kit following the manufacturer’s instructions with minor modifications such as an extended incubation (overnight) during the DNA precipitation step and a longer centrifugation (45 min). DNA was quantified using the PicoGreen dsDNA Quantitation Kit (Invitrogen, CA) (Molecular Probes), which employs a fluorescent dye that interacts specifically with dsDNA. PicoGreen fluorescence was measured using a microplate reader (Wallac 1420 VICTOR F) with a 485-nm excitation filter and a 535-nm emission filter. Lambda DNA was used to construct a standard curve to determine the concentration of the unknown samples.
Integrity of the isolated DNA was confirmed by 0.8% agarose gel electrophoresis, staining with ethidium bromide, and visualizing under UV-light using a gel documentation system (UVP). Alkaline agarose gels were prepared according to Sambrook et al. [1989] and Sutherland et al. [2006]. Briefly, agarose gels were prepared using 50 mM NaCl, 0.5 mM EDTA (pH 8.0) and equilibrated in electrophoresis buffer (30 mM NaOH, 1 mM EDTA) for 30 min before samples were loaded. Before electrophoresis, samples were precipitated with ethanol and dissolved in 20 μL of alkaline loading buffer containing 50 mM NaOH, 1 mM EDTA, 2.5% Ficoll (Type 400; Fisher Scientific, Fairlawn, NJ), and 0.025% bromocresol purple. Gels were resolved using voltages up to 7.5 V/cm until adequate migration was accomplished. Gels were then neutralized using two 15-min washes with 0.1 M Tris HCl (pH 7.5). DNA on the gels was stained with ethidium bromide for 30 min, followed by destaining with two 15-min washes with water. DNA was visualized in a gel documentation system and the intensities of the high-molecular-weight DNA bands and the low-molecular-weight DNA smears on the gel were quantified using the Quantity One software package (Bio-Rad Laboratories, Hercules, CA). The band intensities were compared with the intensity of the control (no MMS) lane, which was arbitrarily assigned a value of 1 and expressed as the ratio of the intensities of the high-molecular-weight DNA band to the low-molecular-weight DNA smear.
Quantitative Polymerase Chain Reaction
The QPCR assay was performed as previously described [Ayala-Torres et al., 2000] with the following modifications: the quantification of PCR products was performed using Picogreen and the PCR amplification was done using the Master Amp XL Polymerase (Epicentre, Madison, WI) with the appropriate premixes. All the PCR reactions were done using 7.5 ng of starting material. The large PCR products (see below) were resolved on 1% agarose gels, while the small PCR products were resolved on 15% polyacrylamide gels. Ethidium bromide-stained gels were visualized under UV-light. Experiments that included template concentration, salt concentration, and PCR cycle tests were performed to ensure that the PCR conditions were optimal for the exponential amplification of both the large and small mtDNA fragments and the large nDNA fragment (data not shown).
Large mtDNA Fragment
The PCR amplification profile for a 6.9-kb yeast mitochondrial fragment was as follows: an initial denaturation for 45 sec at 94°C, followed by 16 cycles of denaturation for 15 sec, and annealing/extension at 56°C for 12 min. A final extension at 72°C was performed for 10 min at the completion of the profile. The primer nucleotide sequences used for the amplification of the yeast mitochondrial fragment were the following: 5′-GTG CGT ATA TTT CGT TGA TGC GT-3′ (sense) and 5′-GTC ACC ACC TCC TGC TAC TTC AA-3′ (antisense). These primers amplify a 6.9-kb fragment encompassing the mitochondrial gene COX1. The PCR reaction was carried out using Premix 5 (Epicentre, Madison, WI). PCR products were quantified using the PicoGreen dsDNA Quantitation Kit.
Small mtDNA Fragment
The PCR amplification profile for a 112-bp yeast mitochondrial fragment was as follows: An initial denaturation for 45 sec at 94°C, followed by 22 cycles of denaturation for 15 sec, and annealing/extension at 64°C for 45 sec. A final extension at 72°C was performed for 10 min at the completion of the profile. The primer nucleotide sequences used for the amplification of the small mtDNA fragment were the following: 5′-CCA CTT GGG GAT TGT GAT TC-3′ (sense) and 5′-AAC TCG TAC AGC CCT CCA AA-3′ (antisense). These primers amplify a 112-bp fragment within the mitochondrial gene COX1. The PCR reaction was carried out using Premix 2 (Epicentre, Madison, WI). PCR products were quantified using the PicoGreen dsDNA Quantitation Kit.
Large nDNA Fragment
The PCR amplification profile for a 6.9-kb yeast nDNA fragment was as follows: An initial denaturation for 45 sec at 94°C, followed by 20 cycles of denaturation for 15 sec, and annealing/extension at 56°C for 12 min. A final extension at 72°C was performed for 10 min at the completion of the profile. The primer nucleotide sequences used for the amplification of the yeast nuclear fragment were the following: 5′-ATG CGG GAC TGG CTG CTG AAA CG-3′ (sense) and 5′-TTC AAG CCC TCC ACT GGG AAA-3′ (antisense). These primers amplify a 6.9-kb fragment encompassing the PFK2 gene. The PCR reaction was carried out using Premix 4 (Epicentre). PCR products were quantified using the PicoGreen dsDNA Quantitation Kit.
Lesion Frequency Estimation
DNA lesion frequencies were calculated using the Poisson equation as previously described [Yakes and Van Houten, 1997; Ayala-Torres et al., 2000]. Briefly, assuming a random distribution of lesions, and using the Poisson equation which is defined as f(x) = e−λ λx/x! for the zero class molecules (x = 0) (molecules exhibiting no damage), amplification is directly proportional to the fraction of undamaged DNA templates. Therefore, the average lesion frequency per strand can be calculated as λ = −ln AD/AU, where AD represents the amount of amplification of the damaged template and AU is the amount of amplification product from undamaged DNA. The results were expressed as a relative amplification ratio (AD/AU) and lesion frequency per 10 kb per strand. To estimate lesions per mitochondrial genome, the lesion number per 10 kb per strand was extrapolated using the size of the S. cerevisiae mitochondrial genome (85.5 kb).
Mitochondrial Mass Estimation
10-N-Nonyl acridine orange (NAO; Invitrogen, CA) was used to estimate mitochondrial mass [Petit et al., 1992; Metivier et al., 1998]. Cells were treated with MMS as described earlier and resuspended in 1 ml of water. A 100 μl aliquot of cells was added to 885 μl of 0.02 M HEPES (pH 7.4) and 0.05 M glucose, followed by loading with 15 μl of a 0.21 mM stock solution of NAO. The optimal NAO concentration used was determined by performing a titration curve (data not shown). Samples were incubated for 30 min at room temperature in the dark. After incubation with NAO, cells were centrifuged, washed, and resuspended in 100 μL PBS. The fluorescence of NAO was detected by using a multilabel counter Victor2 Wallac 1420 with excitation at 485 nm and emission at 535 nm.
Statistical Analysis
Data obtained by the QPCR and the mitochondrial mass estimation assays were analyzed using one way analysis of variance (ANOVA). Differences were considered significant if P was < 0.05.
RESULTS
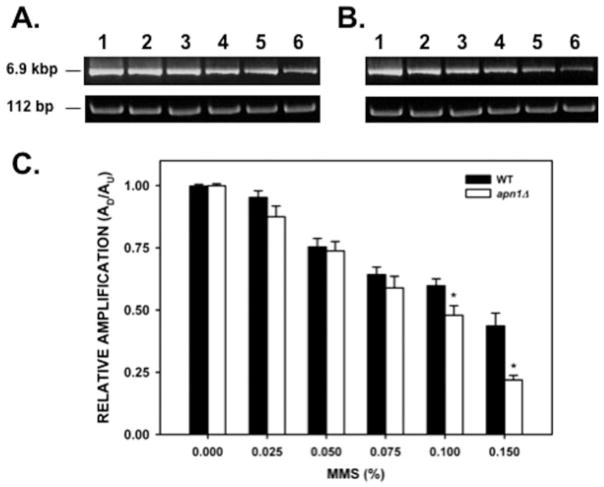
Genetic evidence suggests a role for the APN1 gene in the repair of mtDNA damage [Vongsamphanh et al., 2001]. We defined the role of APN1 in the repair of environmentally induced mtDNA damage in WT and apn1Δ yeast strains by studying the formation and repair of mtDNA lesions induced by the simple alkylation agent MMS. MtDNA lesions were detected by a gene-specific PCR-based assay (QPCR) that has been used extensively to detect a variety of DNA lesions [Yakes and Van Houten, 1997; Ayala-Torres et al., 2000; Karthikeyan et al., 2003; Santos et al., 2003, 2006; Trushina et al., 2004]. The rationale of the QPCR assay for quantitation of DNA damage is based on the ability of certain DNA lesions such as base modifications, AP sites, and strand breaks to prevent the movement of the PCR polymerase, resulting in decreased amplification of the template. By using this approach we determined the optimal conditions for the amplification of 6.9-kb and 112-bp fragments from the yeast mtDNA genome (data not shown). The larger (6.9 kb) fragment was used to detect the formation and repair of DNA lesions, whereas the smaller (112 bp) fragment was used to correct for variations in the steady-state levels of mtDNA. We performed dose–response experiments by treating WT and apn1Δ cells with increasing MMS concentrations. Experiments using MMS doses ranging from 0.2% up to 0.6% could not be analyzed, since the frequency of mtDNA lesions was so high that it precluded the PCR amplification of the mtDNA target sequence (data not shown). Therefore, we reduced the range of treatment doses to a low dose of 0.025% up to 0.15% MMS. The cell survival after MMS treatment is summarized on Table I. The survival data show that there were no differences in viability between the WT and apn1Δ strains even at the highest MMS dose used (0.150% MMS, 17.7 mM; 67.3% and 62.4% survival for the WT and apn1Δ strains, respectively). Differences in viability between both strains were seen with doses of 0.2% MMS or higher [Johnson et al., 1998]. QPCR analysis of DNA isolated from these cells indicated that treatment of the WT strain with increasing concentrations of MMS (0.025, 0.050, 0.075, 0.100, and 0.150%) caused a dose-dependent decrease (4.5, 24.4, 35.5, 40.0, and 56.2%, respectively) in the amplification of the large mtDNA amplicon (Fig. 1). Analysis of the apn1Δ yeast strain indicated that the reduction in the amplification of the mtDNA fragment was not significantly different than the WT strain when low doses (0.025, 0.050, and 0.075%) of MMS were used (Fig. 1C). However, we detected a statistically significant decrease in the amplification of the larger mtDNA amplicon when the apn1Δ cells were treated with 0.100% and 0.150% MMS (P = 0.028 and P < 0.001, respectively). We estimated the number of lesions per 10 kb per strand (Table II) and found that at the highest MMS dose used (0.150%), the WT strain harbored 1.32 lesions per 10 kb per strand, whereas the apn1Δ harbored 2.25 lesions per 10 kb per strand. These lesion frequencies translate into 22.6 lesions per mitochondrial genome in the WT strain and 38.4 lesions per mitochondrial genome in the apn1Δ strain. The higher number of lesions present in the apn1Δ strain represents the combined effect of N-alkylpurine lesions (primarily, 3-methylguanine) and inefficient repair of AP sites produced by removal of 3-methylguanine generated by the MMS treatment.
TABLE I.
Cell Survival After MMS Treatmenta
| Yeast strain | Cell survival |
||||
|---|---|---|---|---|---|
| 0.025b | 0.050b | 0.075b | 0.100b | 0.150b | |
| WT | 86.3 | 85.7 | 83.8 | 79.5 | 67.3 |
| apn1Δ | 88.8 | 87.2 | 81.1 | 73.9 | 62.4 |
Values represent the average of three independent experiments done in duplicate. Standard deviation was less than 20% for all the values.
MMS dose (%).
Fig. 1.
Mitochondrial DNA damage induced by MMS. Yeast cells (A, wild type; B, apn1Δ mutant) were treated with increasing concentrations of MMS for 20 min, followed by DNA isolation. QPCR analysis was performed as described in the “Materials and Methods” section. (A, B) Representative gel electrophoresis indicating the expected sizes of the large (6.9 kb) and small (112 bp) mitochondrial DNA fragments. Lanes 1–6: PCR products from yeast cells treated with 0, 0.025, 0.05, 0.075, 0.1, 0.15% MMS, respectively. (C) Relative levels of amplification of the 6.9-kb mitochondrial fragment after normalization with the small mtDNA fragment (n = 3 independent experiments performed in triplicate). Asterisks (*) denote statistical significance (P < 0.05). Error bars represent SEM.
TABLE II.
Mitochondrial DNA Lesion Number After MMS Treatmenta
| Yeast strain | Mitochondrial DNA lesion number |
|||||
|---|---|---|---|---|---|---|
| 0.0b | 0.025b | 0.050b | 0.075b | 0.100b | 0.150b | |
| WT | 0.004 | 0.08 | 0.43 | 0.67 | 0.77 | 1.32 |
| apn1Δ | 0.01 | 0.21 | 0.46 | 0.86 | 1.12 | 2.25 |
Lesion numbers were calculated per 10.0 kb per strand using the Poisson equation as described in the “Materials and Methods” section.
MMS dose (%).
The decrease in PCR amplification of the large mtDNA fragment after MMS treatment could be due to MMS treatment causing changes in the steady-state levels of mtDNA molecules. This could occur via selective replication of undamaged mtDNA molecules or, alternatively, by degradation of damaged mtDNA molecules. The amplification of the small mtDNA fragment provides for convenient normalization between possible changes in the steady-state levels of mtDNA molecules and therefore is informative in terms of differentiating between DNA repairs when compared with selective replication of undamaged mitochondrial genomes. If the latter were occurring, it would have been reflected as an increase in the amplification of the small mtDNA fragment. We observed that the amplification of the small 112-bp fragment was unaffected after MMS treatment (Figs. 1A and 1B, lower panels). These results suggest that the decrease in amplification of the 6.9-kb fragment in both strains was not due to changes in the steady-state level of mtDNA molecules.
As additional controls, we analyzed the quality of the DNA obtained after MMS treatment by means of native and alkaline agarose gel electrophoresis. The results of the native agarose gel electrophoresis showed that all the DNA samples obtained were in high-molecular-weight form (Fig. 2A). When these samples were analyzed by alkaline agarose gel electrophoresis, a dose-dependent decrease in the high-molecular-weight DNA form and a concomitant increase of low-molecular-weight DNA smear were observed, indicating the presence of alkali-labile sites induced by the MMS treatment (Fig. 2B). The ratio in the intensities of the high-molecular-weight DNA band to the low-molecular-weight DNA smear was higher in the WT strain than in the apn1Δ strain, indicating that the repair-deficient strain harbored more alkali-labile sites. These results indicate that the MMS treatment effectively introduced lesions in the DNA and that the DNA extraction protocol yields DNA of high quality.
Fig. 2.
Genomic DNA integrity obtained from cells treated with MMS. DNA (0.75 μg) isolated from WT and apn1Δ cells after MMS treatment was analyzed by native and alkaline agarose gel electrophoresis. (A) Native agarose gel electrophoresis. Samples were loaded according to increasing MMS dosage. (B) Alkaline agarose gel electrophoresis. Samples were loaded as in A. The ratio of the intensities of the high-molecular-weight DNA band to the low-molecular-weight DNA smear was determined as described in the “Materials and Methods” section. MW: molecular weight marker containing λ DNA digested with Hind III. DNA from the untreated WT or apn1Δ strains was digested with Bgl I/Hind III and included as low-molecular-weight DNA controls.
Another alternative explanation for the decrease in PCR amplification after MMS treatment could be that damaged mitochondria were selectively degraded by mitophagy [Kim et al., 2007]. To determine whether the steady-state levels of mitochondrial mass changed after MMS treatment we performed experiments by staining mitochondria with NAO. This dye specifically interacts with mitochondrial cardiolipin so that changes in the mitochondrial mass of a cell can be monitored by fluorescence [Petit et al., 1992; Metivier et al., 1998]. Therefore, if damaged mitochondria were being selectively eliminated immediately after MMS treatment, then this should be reflected by a reduction in the NAO fluorescence. The results of these experiments show that MMS treatment did not cause a decrease in the mitochondrial mass in either the WT strain or the apn1Δ strain (Fig. 3). If anything, MMS treatment causes a small increase in the mitochondrial mass within the apn1Δ mutant strain. We therefore concluded that the loss in PCR amplification was due to increased lesion number in the mtDNA but not due to a loss of damaged mitochondria.
Fig. 3.
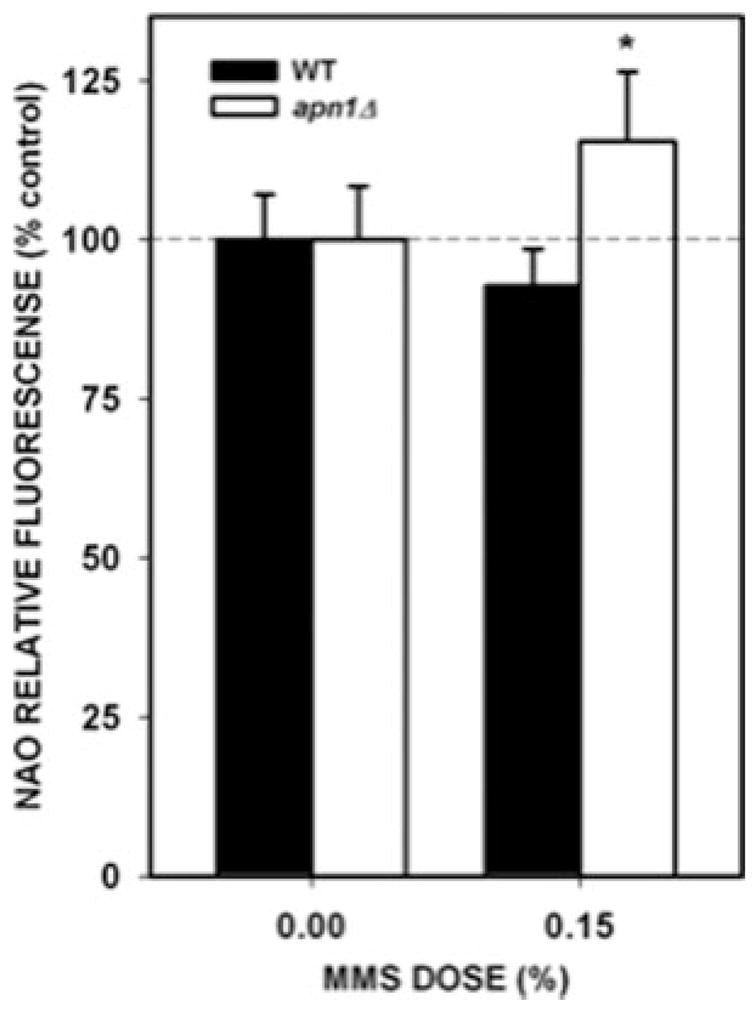
Estimation of mitochondrial mass after MMS treatment. Yeast cells were treated with 0.15% MMS for 20 min. After MMS treatment, cells were loaded with NAO and the fluorescence intensity was determined as described in the “Materials and Methods” section. The values represent the result of three independent experiments performed in triplicate. Asterisks (*) denote statistical significance (P < 0.05). Error bars represent SEM.
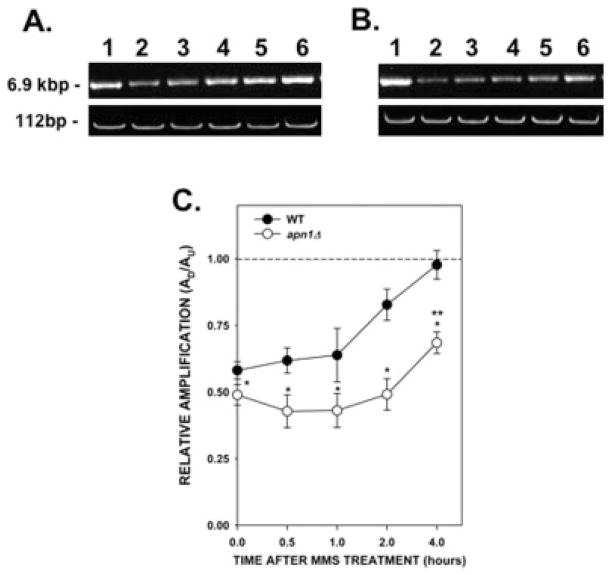
To determine the repair kinetics of mtDNA damage, WT and apn1Δ cells were treated with 0.100% MMS and, after MMS inactivation, cells were resuspended in YPD media and allowed to grow for several hours. Aliquots were removed at various times after treatment and DNA from these cells was isolated and analyzed using QPCR. Analysis of the large mtDNA fragment immediately after MMS treatment (0 time) showed a decrease in the PCR product when compared with that of the untreated cells (Figs. 4A and 4B, upper panels). This decrease in amplification was similar to that observed in the dose–response experiments (Fig. 1). We observed a gradual increase in the amplification of the 6.9-kb PCR fragment that reached the level of untreated cells 4 hr after treatment. This increase in amplification suggests that lesions present in the mtDNA from WT cells were being repaired. We observed no changes in the small mtDNA fragment indicating that there were no changes in the steady-state levels of mtDNA molecules (Figs. 4A and 4B, lower panels). Analysis of the apn1Δ yeast strain showed no significant increase in the repair of mtDNA during the first 2 hr after treatment. However, we observed a small, but statistically significant, increase (P = 0.007) in the PCR fragment 4 hr after treatment, suggesting that the apn1Δ yeast strain was not totally defective in mtDNA repair. Nevertheless, this repair activity did not match the activity found in the WT strain, which is significantly different at all the time points tested. The lesion frequencies in the WT and apn1Δ strains 4 hr after MMS treatment were 0.04 and 0.56 lesions per 10 kb per strand, respectively (Table III). These lesion frequencies correspond to 0.68 lesions per mitochondrial genome in the WT strain and 9.58 lesions per mitochondrial genome in the apn1Δ strain. The results of these experiments indicate that deletion of the APN1 gene results in cells that are compromised in the repair of mtDNA damage induced by MMS.
Fig. 4.
Kinetics of repair of mtDNA damage induced by MMS. Yeast cells were treated with 0.1% MMS for 20 min. After MMS inactivation, cells were washed with water, resuspended in fresh YPD media, and incubated for up to 4 hr at 30°C. Aliquots of cells were removed at the indicated times. DNA isolation and QPCR were performed as described. (A) Wild type; (B) apn1Δ mutant. (A, B) Representative gel electrophoresis indicating the expected sizes of the mtDNA fragments. Lane 1: PCR products from untreated yeast cells; lanes 2–6: PCR products from yeast cells after 0, 0.5, 1, 2, and 4 hr after MMS, respectively. (C) Relative levels of amplification of the 6.9-kb mitochondrial fragment after normalization with the small mtDNA fragment (n = 2 independent experiments performed in triplicate). Asterisks (*) denote statistical significance (P < 0.05) between WT (closed circles) and apn1Δ (open circles) cells. Double asterisk (**) denotes statistical significance in the apn1Δ cells between the 0 and 4 hr after treatment time points. Error bars represent SEM.
TABLE III.
Mitochondrial DNA Lesion Number During Repair Kinetics Experimentsa
| Yeast strain | Mitochondrial DNA lesion number |
||||
|---|---|---|---|---|---|
| 0b | 0.5b | 1.0b | 2.0b | 4.0b | |
| WT | 0.81 | 0.71 | 0.68 | 0.29 | 0.04 |
| apn1Δ | 1.11 | 1.28 | 1.27 | 1.06 | 0.56 |
Lesion numbers were calculated per 10.0 kb per strand using the Poisson equation as described in the “Materials and Methods” section.
Time after MMS treatment (hr).
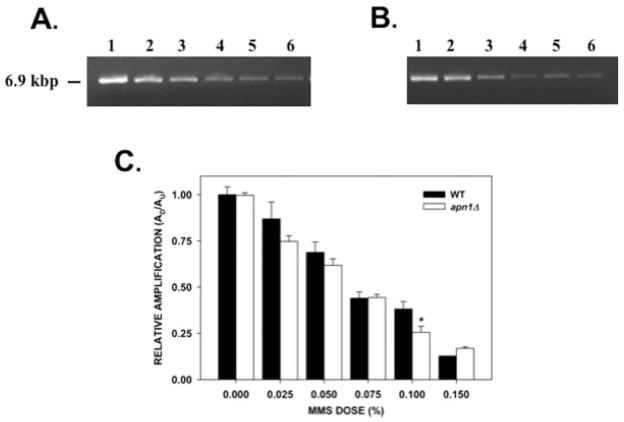
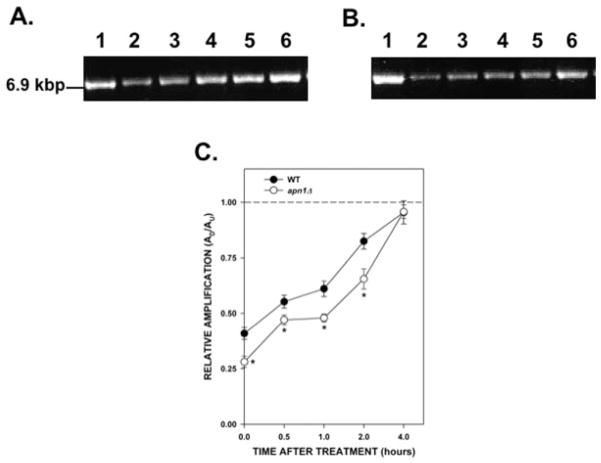
Experiments using human fibroblasts have shown that mtDNA is more susceptible than nDNA to the effects of hydrogen peroxide (H2O2) treatment [Yakes and Van Houten, 1997; Santos et al., 2003]. We decided to test if the same was true in yeast cells but with respect to alkylation damage. Amplification of a fragment encompassing the PFK2 gene showed that MMS treatment induced DNA lesions in a dose-dependent fashion in both the WT and the apn1Δ strain (Fig. 5). The relative amplification of the nDNA fragment is not statistically different between the WT and the apn1Δ strains except in cells treated with 0.100% MMS. At this dose, there was a statistically significant difference (P = 0.003) in the relative amplification of the nDNA fragment, with DNA from the apn1Δ strain showing less amplification. This is consistent with a previous report showing that apn1Δ cells have reduced capacity in the repair of AP sites induced by MMS treatment [Johnson et al., 1998]. When we compared the lesion frequencies between the mtDNA and nDNA (Tables II and IV, respectively) we observed that nDNA harbored more DNA lesions than mtDNA after MMS treatment. Therefore, these results suggest that nDNA is more susceptible to alkylation damage than mtDNA. However, in contrast to mtDNA repair, both strains were able to repair the majority of the nDNA lesions induced by MMS in 4 hr (Fig. 6). The lesion frequencies 4 hr after treatment were 0.09 lesions per 10 kb per strand for the WT strain and 0.07 lesions per 10 kb per strand for the apn1Δ strain (Table V). These experiments suggest that deletion of the APN1 gene is more detrimental to mtDNA repair than to nDNA repair.
Fig. 5.
nDNA damage induced by MMS. Relative levels of amplification of a 6.9-kb nDNA fragment (from the PFK2 gene) in yeast cells treated with MMS. Yeast cells (A, wild type; B, apn1Δ mutant) were treated with increasing concentrations of MMS for 20 min, followed by DNA isolation. QPCR analysis was performed as described in the “Materials and Methods” section. (A, B) Representative gel electrophoresis indicating the expected sizes of the 6.9-kb nDNA fragment. Lanes 1–6: PCR products from yeast cells treated with 0, 0.025, 0.05, 0.075, 0.1, 0.15% MMS, respectively; (C) Relative levels of amplification of the 6.9-kb nDNA fragment (n = 3 independent experiments performed in triplicate). Asterisks (*) denote statistical significance (P < 0.05). Error bars represent SEM.
TABLE IV.
Nuclear DNA Lesion Number After MMS Treatmenta
| Yeast strain | Nuclear DNA lesion number |
|||||
|---|---|---|---|---|---|---|
| 0.0b | 0.025b | 0.050b | 0.075b | 0.100b | 0.150b | |
| WT | 0.03 | 0.06 | 0.56 | 1.20 | 1.48 | 2.98 |
| apn1Δ | 0.01 | 0.43 | 0.70 | 1.18 | 2.09 | 2.58 |
Lesion numbers were calculated per 10.0 kb per strand using the Poisson equation as described in the “Materials and Methods” section.
MMS dose (%).
Fig. 6.
Kinetics of repair of nDNA damage induced by MMS. Yeast cells were treated with 0.1% MMS for 20 min. Cells were washed with water, resuspended in fresh YPD media, and incubated for up to 4 hr at 30°C. Aliquots of cells were removed at the indicated times. DNA isolation and QPCR were performed as described. (A) Wild type; (B) apn1Δ mutant. (A, B) Representative gel electrophoresis indicating the expected sizes of the nDNA fragments. Lane 1: PCR products from untreated yeast cells; lanes 2–6: PCR products from yeast cells after 0, 0.5, 1, 2, and 4 hr after MMS, respectively. (C) Relative levels of amplification of the 6.9-kb nDNA fragment. The dashed line represents the amplification of untreated cells, which was set to a value of 1.0. Results are from two independent experiments with each PCR performed in triplicate. Asterisks (*) denote statistical significance (P < 0.05) between WT (closed circles) and apn1Δ cells (open circles). Error bars represent SEM.
TABLE V.
Nuclear DNA Lesion Number During Repair Kinetics Experimentsa
| Yeast strain | Nuclear DNA lesion number |
||||
|---|---|---|---|---|---|
| 0b | 0.5b | 1.0b | 2.0b | 4.0b | |
| WT | 1.36 | 0.88 | 0.74 | 0.29 | 0.09 |
| apn1Δ | 1.94 | 1.11 | 1.07 | 0.65 | 0.07 |
Lesion numbers were calculated per 10.0 kb per strand using the Poisson equation as described in the “Materials and Methods” section.
Time after MMS treatment (hr).
DISCUSSION
The results presented in this work establish dose–response as well as repair kinetics of mtDNA and nDNA damage induced by an alkylating agent (MMS). Our results indicate that MMS induces mtDNA lesions in a dose-dependent fashion and that WT cells are capable of repairing those mtDNA lesions (Figs. 1 and 4). We were able to detect significant differences in the MMS sensitivity and mtDNA repair capacity between a WT strain and a strain carrying a null mutation in the BER gene APN1. Our studies also show that although nDNA is more susceptible to MMS-induced lesions than mtDNA in yeast cells, repair of mtDNA has a stronger requirement for the presence of APN1 than nDNA.
The majority of MMS-induced lesions are N-methylpurine adducts with only a very small proportion being O6-methylguanine (0.3%) [Pegg, 1984]. Since 3-methylade-nine blocks the progression of polymerases, the QPCR assay is probably detecting these adducts directly, and the apparent increase in lesions in the apn1Δ strain when compared with the WT strain shown in Figure 1 is likely due to repair during the 20-min incubation period in the WT strain. We cannot totally exclude direct reversal of lesions via O6-methylguanine methyltransferase, which is encoded in S. cereviase by the MGT1 gene [Xiao et al., 1991; Xiao and Samson, 1992]. However, since these lesions constitute a small subset of the lesions induced by MMS, we believe that this is an unlikely event. Our results are in agreement with studies performed in rats treated with a different alkylating agent, N-ethyl-N-nitrosourea (ENU); as is the case with MMS, ENU generates a small proportion of lesions recognized by MGMT, the mammalian homolog of yeast MGT1 [Satoh et al., 1988].
BER-dependent repair of alkylation damage in yeast cells is initiated by the action of the glycosylase encoded by the MAG1 gene [Chen et al., 1989; Xiao et al., 2001]. However, the product of this gene shows no mitochondrial localization signal and experiments using GFP-tagged Mag1 reveal a primarily nuclear and cytoplasmic localization (yeastgfp.ucsf.edu) [Huh et al., 2003]. Thus, we are uncertain of the nature of the enzymatic activity that generates the AP sites recognized by Apn1p. One possibility is the glycosylase/lyase enzyme Ntg1p, which has been proposed to act as a backup in the repair of alkylation damage [Hanna et al., 2004]. Another alternative could be an unidentified N-alkylpurine glycosylase in mitochondria. Finally, since alkylated purines are chemically unstable (which could lead to increased spontaneous generation of AP sites), we cannot rule out nonenzymatic mechanisms for the generation of Apn1p substrates after MMS treatment in mitochondria [O’Brien and Ellenberger, 2004]. Further experiments to determine the mtDNA lesion frequency after MMS treatment in ntg1Δ cells will help understand the role of specific glycosylases in the repair of mtDNA.
An important observation from our work is the fact that, even though the apn1Δ strain shows a compromised mtDNA repair capacity, this strain is not totally deficient in the repair of mtDNA lesions induced by MMS (Fig. 4). One possible explanation could be the action of APN1-independent repair processes. For example, the AP lyase activity of Ntg1 [You et al., 1999; Alseth et al., 2004; Hanna et al., 2004] or another unidentified mitochondrial AP endonuclease could incise at the AP sites allowing BER to proceed and finish the repair process. In fact, it has been proposed that Ntg1 and Apn1 constitute alternate mechanisms for the generation of mutagenic intermediates in the mtDNA [Phadnis et al., 2006]. An alternate explanation could be that some mitochondria containing damaged mtDNA are eliminated by mitophagy [Kim et al., 2007]. This would lead to a smaller population of damaged mtDNA copies and better amplification of the mtDNA fragment. Experiments using a Pol γ mutant (mip1ts) have shown that the half-life of mitochondria is ~4 hr [Zhang et al., 2007]. Therefore, the time scale of our repair kinetics experiments would allow enough time for mitophagy to occur. Note that this situation is different to our dose–response experiments (Fig. 1) in which the time scale was only 20 min. Further experiments are necessary to elucidate the role of mitophagy in eliminating mitochondria containing damaged mtDNA.
Yakes and Van Houten [1997] showed that mtDNA is more susceptible to oxidative damage than nDNA. With respect to alkylation damage in yeast cells, nDNA seems to be more sensitive to the effects of MMS treatment (Fig. 5). However, our results are in agreement with the proposal that mtDNA repair capacity is limited, since under the same conditions, apn1Δ cells were able to repair the nDNA lesions induced by MMS treatment, whereas the repair of mtDNA lesions was severely delayed (although not totally abolished) (Fig. 6).
Both forms of human AP endonucleases (APE1/APEX1 and APE2) have been linked to mitochondria suggesting a role for these enzymes in mitochondrial BER in humans [Tsuchimoto et al., 2001; Chattopadhyay et al., 2006]. In the particular case of APE1, several single nucleotide polymorphisms have been reported to cause a decrease in enzymatic reactivity toward AP-site containing oligonucleotides [Hadi et al., 2000]. It may be possible that individuals carrying these alleles may be more susceptible to mtDNA damage. This is an important point to consider not only in the etiology of certain types of cancer but in the chemotherapeutic outcomes of patients, since novel forms of treatment may be directed toward mitochondria [Fishel et al., 2003; Cai et al., 2005]. Studies directed to understand the relationship between APE1 and mtDNA repair may shed light on the relationship between the repair of environmentally induced DNA damage and pathological conditions such as cancer and neurodegeneration.
Acknowledgments
Grant sponsor: NIGMS; Grant numbers: S06 GM008224, 5R25 GM061838-07; Grant sponsor: NINDS; Grant number: U54 NS039408-06; Grant sponsor: NCRR; Grant number: G12RR03051.
The authors are grateful to Satya Prakash and Louise Prakash for providing the yeast strains. They also thank Christi Walter and Bennett Van Houten for helpful comments on this manuscript.
References
- Alseth I, Korvald H, Osman F, Seeberg E, Bjoras M. A general role of the DNA glycosylase Nth1 in the abasic sites cleavage step of base excision repair in Schizosaccharomyces pombe. Nucleic Acids Res. 2004;32:5119–5125. doi: 10.1093/nar/gkh851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala-Torres S, Chen Y, Svoboda T, Rosenblatt J, Van Houten B. Analysis of gene-specific DNA damage and repair using quantitative polymerase chain reaction. Methods. 2000;22:135–147. doi: 10.1006/meth.2000.1054. [DOI] [PubMed] [Google Scholar]
- Bohr VA. Repair of oxidative DNA damage in nuclear and mitochondrial DNA, some changes with aging in mammalian cells. Free Radic Biol Med. 2002;32:804–812. doi: 10.1016/s0891-5849(02)00787-6. [DOI] [PubMed] [Google Scholar]
- Cai S, Xu Y, Cooper RJ, Ferkowicz MJ, Hartwell JR, Pollok KE, Kelley MR. Mitochondrial targeting of human O6-methylguanine DNA methyltransferase protects against cell killing by chemotherapeutic alkylating agents. Cancer Res. 2005;65:3319–3327. doi: 10.1158/0008-5472.CAN-04-3335. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay R, Wiederhold L, Szczesny B, Boldogh I, Hazra TK, Izumi T, Mitra S. Identification and characterization of mitochondrial abasic (AP)-endonuclease in mammalian cells. Nucleic Acids Res. 2006;34:2067–2076. doi: 10.1093/nar/gkl177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Derfler B, Maskati A, Samson L. Cloning a eukaryotic DNA glycosylase repair gene by the suppression of a DNA repair defect in Escherichia coli. Proc Natl Acad Sci USA. 1989;86:7961–7965. doi: 10.1073/pnas.86.20.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza-Pinto NC, Eide L, Hogue BA, Thybo T, Stevnsner T, Seeberg E, Klungland A, Bohr VA. Repair of 8-oxodeoxyguanosine lesions in mitochondrial DNA depends on the oxoguanine DNA glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial DNA of OGG1-defective mice. Cancer Res. 2001;61:5378–5381. [PubMed] [Google Scholar]
- Donahue SL, Corner BE, Bordone L, Campbell C. Mitochondrial DNA ligase function in Saccharomyces cerevisiae. Nucleic Acids Res. 2001;29:1582–1589. doi: 10.1093/nar/29.7.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito L, Melov S, Panov A, Cottrell B, Wallace D. Mitochondrial disease in mouse results in increased oxidative stress. Proc Natl Acad Sci USA. 1999;96:4820–4825. doi: 10.1073/pnas.96.9.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fikus MU, Mieczkowski PA, Koprowski P, Rytka J, Sledziewska-Gojska E, Ciesla Z. The product of the DNA damage-inducible gene of Saccharomyces cerevisiae, DIN7, specifically functions in mitochondria. Genetics. 2000;154:73–81. doi: 10.1093/genetics/154.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel ML, Seo YR, Smith ML, Kelley MR. Imbalancing the DNA base excision repair pathway in the mitochondria; targeting and overexpressing N-methylpurine DNA glycosylase in mitochondria leads to enhanced cell killing. Cancer Res. 2003;63:608–615. [PubMed] [Google Scholar]
- Foury F. Cloning and sequencing of the nuclear gene MIP1 encoding the catalytic subunit of the yeast mitochondrial DNA polymerase. J Biol Chem. 1989;264:20552–20560. [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. Washington, DC: ASM Press; 2006. [Google Scholar]
- Hadi MZ, Coleman MA, Fidelis K, Mohrenweiser HW, Wilson DM., III Functional characterization of Ape1 variants identified in the human population. Nucleic Acids Res. 2000;28:3871–3879. doi: 10.1093/nar/28.20.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna M, Chow BL, Morey NJ, Jinks-Robertson S, Doetsch PW, Xiao W. Involvement of two endonuclease III homologs in the base excision repair pathway for the processing of DNA alkylation damage in Saccharomyces cerevisiae. DNA Repair (Amst) 2004;3:51–59. doi: 10.1016/j.dnarep.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Torres-Ramos CA, Izumi T, Mitra S, Prakash S, Prakash L. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev. 1998;12:3137–3143. doi: 10.1101/gad.12.19.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Yu S-L, Prakash S, Prakash L. A role for yeast and human translesion synthesis DNA polymerases in promoting replication through 3-methyl adenine. Mol Cell Biol. 2007;27:7198–7205. doi: 10.1128/MCB.01079-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan G, Santos JH, Graziewicz MA, Copeland WC, Isaya G, Van Houten B, Resnick MA. Reduction in frataxin causes progressive accumulation of mitochondrial damage. Hum Mol Genet. 2003;12:3331–3342. doi: 10.1093/hmg/ddg349. [DOI] [PubMed] [Google Scholar]
- Kim I, Rodriguez-Enrique S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling F, Makishima F, Morishima N, Shibata T. A nuclear mutation defective in mitochondrial recombination in yeast. EMBO J. 1995;14:4090–4101. doi: 10.1002/j.1460-2075.1995.tb00081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling F, Morioka H, Ohtsuka E, Shibata T. A role for MHR1, a gene required for mitochondrial genetic recombination, in the repair of damage spontenously introduced in yeast mtDNA. Nucleic Acids Res. 2000;28:4956–4963. doi: 10.1093/nar/28.24.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley MJ, Prasad R, Srivastava DK, Wilson SH, Copeland WC. Identification of 5′-deoxyribose phosphate lyase activity in human DNA polymerase γ and its role in mitochondrial base excision repair in vitro. Proc Natl Acad Sci USA. 1998;95:12244–12248. doi: 10.1073/pnas.95.21.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandavilli BS, Santos JH, Van Houten B. Mitochondrial DNA repair and aging. Mutat Res. 2002;509:127–151. doi: 10.1016/s0027-5107(02)00220-8. [DOI] [PubMed] [Google Scholar]
- Meeusen S, Tieu Q, Wong E, Weiss E, Schieltz D, Yates JR, Nunnari J. Mgm101p is a novel component of the mitochondrial nucleoid that binds DNA and is required for the repair of oxidatively damaged mitochondrial DNA. J Cell Biol. 1999;145:291–304. doi: 10.1083/jcb.145.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier D, Dallaporta B, Zamzami N, Larochette N, Susin SA, Marzo I, Kroemer G. Cytofluorometric detection of mitochondrial alterations in early CD95/Fas/APO-1-triggered apoptosis of Jurkat T lymphoma cells. Comparison of seven mitochondrion-specific fluorochromes. Immunol Lett. 1998;61:157–163. doi: 10.1016/s0165-2478(98)00013-3. [DOI] [PubMed] [Google Scholar]
- O’Brien PJ, Ellenberger T. The Escherichia coli 3-methyladenine-DNA glycosylase AlkA has a remarkably versatile active site. J Biol Chem. 2004;279:26876–26884. doi: 10.1074/jbc.M403860200. [DOI] [PubMed] [Google Scholar]
- Phadnis N, Mehta R, Meednu N, Sia EA. Ntg1p, the base excision repair protein, generates mutagenic intermediates in yeast mitochondrial DNA. DNA Repair. 2006;5:829–839. doi: 10.1016/j.dnarep.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Pegg AE. Methylation of the O6 position of guanine in DNA is the most likely initiating event in carcinogenesis by methylating agents. Cancer Invest. 1984;2:223–231. doi: 10.3109/07357908409104376. [DOI] [PubMed] [Google Scholar]
- Petit JM, Maftah A, Ratinaud MH, Julien R. 10N-nonyl acridine orange interacts with cardiolipin and allows the quantification of this phospholipid in isolated mitochondria. Eur J Biochem. 1992;209:267–273. doi: 10.1111/j.1432-1033.1992.tb17285.x. [DOI] [PubMed] [Google Scholar]
- Pinz KG, Bogenhagen DF. Efficient repair of abasic sites in DNA by mitochondrial enzymes. Mol Cell Biol. 1998;18:1257–1265. doi: 10.1128/mcb.18.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 1989. p. 6.20. [Google Scholar]
- Santos JH, Hunakova Lu, Chen Y, Bortner C, Van Houten B. Cell sorting experiments link persistent mitochondrial DNA damage with loss of mitochondrial membrane potential and apoptotic cell death. J Biol Chem. 2003;278:1728–1734. doi: 10.1074/jbc.M208752200. [DOI] [PubMed] [Google Scholar]
- Santos JH, Meyer JN, Mandavilli BS, Van Houten B. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol Biol. 2006;314:183–199. doi: 10.1385/1-59259-973-7:183. [DOI] [PubMed] [Google Scholar]
- Satoh MS, Huh N, Rajewsky MF, Kuroki T. Enzymatic removal of O6-ethylguanine from mitochondrial DNA in rat tissues exposed to N-ethyl-N-nitrosourea in vivo. J Biol Chem. 1988;263:6854–6856. [PubMed] [Google Scholar]
- Singh KK, Sigala B, Sikder HA, Schwimmer C. Inactivation of Saccharomyces cerevisiae OGG1 DNA repair gene leads to an increased frequency of mitochondrial mutants. Nucleic Acids Res. 2001;29:1381–1388. doi: 10.1093/nar/29.6.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stierum RH, Dianov GL, Bohr VA. Single-nucleotide patch base excision repair of uracil in DNA by mitochondrial protein extracts. Nucleic Acids Res. 1999;27:3712–3719. doi: 10.1093/nar/27.18.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart GR, Santos JH, Strand MK, Van Houten B, Copeland WC. Mitochondrial and nuclear DNA defects in Saccharomyces cerevisiae with mutations in DNA polymerase γ associated with progressive external ophthalmoplegia. Hum Mol Genet. 2006;15:363–374. doi: 10.1093/hmg/ddi454. [DOI] [PubMed] [Google Scholar]
- Sung J-S, Demple B. Roles of base excision repair subpathways in correcting oxidized abasic sites in DNA. FEBS J. 2006;273:1620–1629. doi: 10.1111/j.1742-4658.2006.05192.x. [DOI] [PubMed] [Google Scholar]
- Sutherland BM, Bennett PV, Sutherland SC. DNA damage quantitation by alkaline gel electrophoresis. Methods Mol Biol. 2006;314:251–273. doi: 10.1385/1-59259-973-7:251. [DOI] [PubMed] [Google Scholar]
- Szczesny B, Tann AW, Longley MJ, Copeland WC, Mitra S. Long patch base excision repair in mammalian mitochondrial genomes. J Biol Chem. 2008;283:26349–26356. doi: 10.1074/jbc.M803491200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trushina E, Dyer RB, Badger JD, II, Ure D, Eide L, Tran DD, Vrieze BT, Legendre-Guillemin V, McPherson PS, Mandavilli BS, Van Houten B, Zeitlin S, McNiven M, Aebersold R, Hayden M, Parisi JE, Seeberg E, Dragatsis I, Doyle K, Bender A, Chacko C, McMurray CT. Mutant Huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol Cell Biol. 2004;24:8195–8209. doi: 10.1128/MCB.24.18.8195-8209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimoto D, Sakai Y, Sakumi K, Nishioka K, Sasaki M, Fujiwara T, Nakabeppu Y. Human APE2 protein is mostly localized in the nuclei and to some extent in the mitochondria, while nuclear APE2 is partly associated with proliferating cell nuclear antigen. Nucleic Acids Res. 2001;29:2349–2360. doi: 10.1093/nar/29.11.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vongsamphanh R, Fortier P-K, Ramotar D. Pir1p mediates translocation of the yeast Apn1p endonuclease into the mitochondria to maintain genomic stability. Mol Cell Biol. 2001;21:1647–1655. doi: 10.1128/MCB.21.5.1647-1655.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Samson L. The Saccharomyces cerevisiae MGT1 DNA repair methyltransferease gene: Its promoter and entire coding sequence, regulation and in vivo biological functions. Nucleic Acids Res. 1992;20:3599–3606. doi: 10.1093/nar/20.14.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Derfler B, Chen J, Samson L. Primary sequence and biological functions of a Saccharomyces cerevisiae O6-methylguanine/O4-methylthymine DNA repair methyltransferase gene. EMBO J. 1991;10:2179–2186. doi: 10.1002/j.1460-2075.1991.tb07753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Chow B, Hanna M, Doetsch P. Deletion of the MAG1 DNA glycosylase gene suppresses alkylation-induced killing and mutagenesis in yeast cells lacking AP endonucleases. Mutat Res. 2001;487:137–147. doi: 10.1016/s0921-8777(01)00113-6. [DOI] [PubMed] [Google Scholar]
- Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. PNAS. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You HJ, Swanson RL, Harrington C, Corbett AH, Jinks-Robertson S, Senturker S, Wallace SS, Boiteux S, Dizdaroglu M, Doetsch PW. Saccharomyces cerevisiae Ntg1p and Ntg2p: Broad specificity N-glycosylases for the repair of oxidative DNA damage in the nucleus and mitochondria. Biochemistry. 1999;38:11298–11306. doi: 10.1021/bi991121i. [DOI] [PubMed] [Google Scholar]
- Zhang H, Chatterjee A, Singh KK. Saccharomyces cerevisiae polymerase ζ functions in mitochondria. Genetics. 2006;172:2683–2688. doi: 10.1534/genetics.105.051029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Qi H, Taylor R, Xu W, Liu LF, Jin S. The role of autophagy in mitochondria maintenance: characterization of mitochondrial functions in autophagy-deficient S. cerevisiae strains. Autophagy. 2007;3:337–346. doi: 10.4161/auto.4127. [DOI] [PubMed] [Google Scholar]