Summary
The Hippo signaling pathway regulates organ size and tissue homeostasis from Drosophila to mammals. Central to this pathway is a kinase cascade wherein Hippo (Hpo), in complex with Salvador (Sav), phosphorylates and activates Warts (Wts), which in turn phosphorylates and inactivates the Yorkie (Yki) oncoprotein, known as the YAP coactivator in mammalian cells. The FERM domain proteins Merlin (Mer) and Expanded (Ex) are upstream components that regulate Hpo activity through unknown mechanisms. Here we identify Kibra (Kbr) as another upstream component of the Hippo signaling pathway. We show that Kbr functions together with Mer and Ex in a protein complex localized to the apical domain of epithelial cells, and that this protein complex regulates the Hippo kinase cascade via direct binding to Hpo and Sav. These results shed light on the mechanism of Ex and Mer function, and implicate Kbr as a potential tumor suppressor with relevance to neurofibromatosis.
Introduction
In multicellular organisms, cell growth, proliferation and death must be coordinated in order to attain proper organ size during development and to maintain tissue homeostasis in adult life. Recent studies in Drosophila have led to the discovery of the Hippo signaling pathway as a key mechanism that controls organ size by impinging on cell growth, proliferation and apoptosis. Central to the Hippo pathway is a kinase cascade comprised of four tumor suppressors, including the Ste20-like kinase Hippo (Hpo) (Wu et al., 2003; Udan et al., 2003; Harvey et al., 2003; Pantalacci et al., 2003; Jia et al., 2003) and its regulatory protein Salvador (Sav) (Tapon et al., 2002; Kango-Singh et al., 2002), the NDR family kinase Warts (Wts) (Justice et al., 1995; Xu et al., 1995) and its regulatory protein Mats (Lai et al., 2005). The Hpo-Sav complex phosphorylates and activates the Wts-Mats complex (Wu et al., 2003; Wei et al., 2007), which in turn, phosphorylates and inactivates the oncoprotein Yki (Huang et al., 2005), which normally functions as a coactivator for the TEAD/TEF family transcription factor Scalloped (Sd) (Wu et al., 2008; Zhang et al., 2008; Goulev et al., 2008). Recent studies have also implicated the atypical cadherin Fat (Ft) (Silva et al., 2006; Willecke et al., 2006; Bennett and Harvey, 2006; Cho et al., 2006), as well as the membrane-associated FERM-domain proteins Expanded (Ex) and Merlin (Mer) (Maitra et al., 2006; Hamaratoglu et al., 2006; Pellock et al., 2006; Tyler and Baker, 2007), as upstream components of the Hippo pathway. How these proteins are biochemically linked to the Hippo kinase cascade remains largely unknown, although Ex can at least partially regulate the Hippo pathway by directly binding and sequestering Yki in the cytoplasm (Badouel et al., 2009). Ft differs from Ex, Mer and core components of the Hippo kinase cascade in that besides tissue growth, Ft also regulates planar cell polarity (PCP), for which it interacts with another cadherin Dachsous (Ds) (Yang et al., 2002; Matakatsu and Blair, 2006; Casal et al., 2006). Most recently, it was shown that a gradient of Ds activity in imaginal discs can modulate Hippo-mediated growth regulation (Rogulja et al., 2008; Willecke et al., 2008), thus potentially linking PCP to the Hippo kinase cascade, although the biochemical mechanism of this linkage remains to be determined.
The physiological function of the Hippo pathway is best understood in Drosophila imaginal discs, where inactivation of the Hippo pathway tumor suppressors, or overexpression of the Yki oncoprotein, results in tissue overgrowth characterized by excessive cell proliferation, diminished apoptosis, and increased transcription of Hippo pathway target genes such as the cell death inhibitor diap1 (Huang et al., 2005) and the microRNA bantam (Thompson and Cohen, 2006; Nolo et al., 2006), as well as ex and mer as part of a negative feedback regulatory loop (Hamaratoglu et al., 2006). Recent studies further implicated the Hippo pathway as a conserved mechanism of organ size control and tissue homeostasis in mammals. Thus, the mammalian homologues of Hpo (Mst1/2), Sav (WW45), Wts (Lats1/2) and Yki (YAP) constitute an analogous kinase cascade (Dong et al., 2007; Zhao et al., 2007), and transgenic overexpression of YAP or inactivation of Mst1/2 led to massive organomegaly and rapid progression to tumorigenesis (Dong et al., 2007; Camargo et al., 2007; Zhou et al., 2009). Furthermore, NF2, the mammalian homologue of mer, is a well-established tumor suppressor gene whose mutations lead to neurofibromatosis (McClatchey and Giovannini, 2005; Okada et al., 2007).
Besides its prominent role in controlling imaginal disc growth, the Hippo pathway is required during Drosophila oogenesis for the proper maturation of posterior follicle cells (PFCs) (Polesello and Tapon, 2007; Meignin et al., 2007; Yu et al., 2008). In the absence of Hippo signaling, the PFCs fail to undergo a Notch-mediated mitotic cycle-endocycle switch and accumulate in extra layers of follicular epithelium. The PFC maturation defects, in turn, lead to a disruption of the anterior-posterior (AP) polarity of the underlying oocyte, which manifests itself as mislocalization of the oocyte nucleus and AP axis determinants such as the RNA-binding protein Staufen (Stau) (Polesello and Tapon, 2007; Meignin et al., 2007; Yu et al., 2008). Interestingly, the oocyte polarity defect is observed in mutants for components of the Hippo kinase cascade as well as ex and mer, but not ft (Polesello and Tapon, 2007; Meignin et al., 2007; Yu et al., 2008), suggesting that the canonical Hippo pathway may integrate different signals in different developmental contexts.
In this study, we identify Kibra (Kbr) as an upstream component of the Hippo pathway. Loss of kbr leads to oogenesis defects, imaginal disc overgrowth and aberrant gene expression characteristic of defective Hippo signaling. We show that Kbr functions together with Mer and Ex in an apical protein complex, which through direct binding to the Hpo-Sav complex, regulates the Hippo kinase cascade and thus Yki phosphorylation. These findings uncover an important missing link in the Hippo signaling pathway and shed light on the molecular mechanism of the Ex and Mer tumor suppressor proteins.
Results
Identification of kbr as a gene required for oocyte polarity
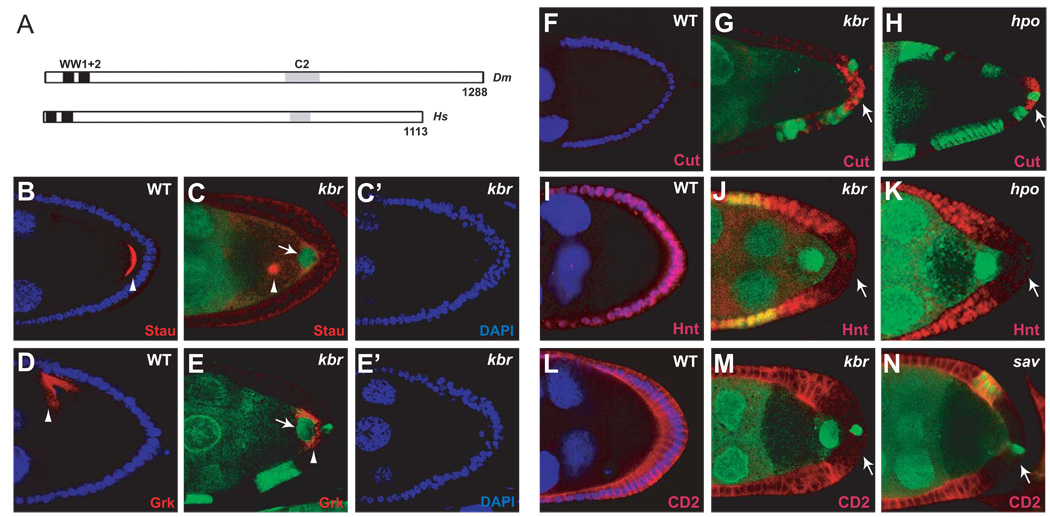
In a genetic screen for oocyte polarity mutants based on FRT/FLP-induced mitotic clones in follicle cells, we identified four lethal P-element insertion lines on chromosome 3R that caused mislocalization of Stau-GFP and Stau to the center of the oocyte when the PFCs were made homozygous mutant for the P-element insertions (Figure S1B–E). This polarity defect was observed with variable penetrance depending on the specific P-element line analyzed, likely due to their hypomorphic nature. These lethal lines (264/09, 1156/7, f06952 and EP3494) fail to complement each other and all carry a P-element insertion near the 5’ UTR or within the first intron of CG33967 (Figure S1A). CG33967 encodes a 1288 amino acid protein that shares 39% identity with KIBRA, a cytoplasmic protein named after its predominant expression in kidney and brain in humans (Kremerskothen et al., 2003). Both CG33967 and KIBRA contain two N-terminal WW domains and one C-terminal C2 domain (Figure 1A). For simplicity, we shall refer to CG33967 as kibra (kbr) to distinguish it from its human orthologue KIBRA.
Figure 1. Loss of kbr results in PFC defects similar to those caused by loss of canonical Hippo pathway genes.
(A) Schematic diagram of the Drosophila Kbr protein (top; Dm) and its human orthologue KIBRA (bottom; Hs). The conserved WW domains and C2 domains are indicated.
(B-E’) kbr is required for oocyte polarity. Stage-9 wildtype (B and D) and mosaic egg chambers containing large kbrdel PFC clones (GFP-negative, C-C’ and E-E’) were stained for Stau (red) or Grk (red), and counterstained with DAPI (blue). Note the mislocalization of Stau from the posterior pole to the center of the oocyte (arrowheads in B and C), the mislocalization of Grk from the dorsal anterior corner to the posterior pole of the oocyte (arrowheads in D and E), and the multiple cell layers associated with kbr PFC clones (C’ and E’). Also note the mislocalization of the oocyte nucleus to the posterior pole (arrows in C and E).
(F–H) Stage 9 wildtype (F) and mosaic egg chambers containing kbr (G) or hpo (H) PFC clones (GFP-negative) were stained for Cut (red). Cut expression was undetectable in stage 9 wildtype egg chambers (F), but persisted in kbr or hpo PFC clones (arrows).
(I–K) Similar to (F–H) except that stage 8 egg chambers were stained for Hnt (red). While Hnt expression was detected in all columnar follicle cells in wildtype egg chambers (I), it was abolished in kbr (J) or hpo (K) PFC clones (arrows).
(L–N) Stage 8 wildtype (L) and mosaic egg chambers containing kbr (M) or sav (N) PFC clones (GFP-negative) were stained for E(spl):CD2 (red). Note the reduction of E(spl):CD2 signal in kbr (M) or sav (N) PFC clones (arrows) compared to the wildtype (L).
See Figure S1 for supplemental data to Figure 1.
Using the FRT/FLP-mediated genomic deletion strategy (Parks et al., 2004), we generated a null allele of kbr that deletes its entire coding sequence (Figure S1A). kbrdel caused a similar Stau mislocalization phenotype as the P-element insertion lines described above, but with 100% penetrance (n= 72) (Figure 1B–C’). Therefore, all subsequent studies have been conducted with the kbrdel allele. Besides Stau mislocalization, PFC clones of kbrdel caused mislocalization of oocyte nucleus and Gurken (Grk) protein in the oocyte (94%, n= 68) (Figure 1D–E’), as well as the formation of multilayered follicular epithelium (72%, n=55) (Figure 1B–E’). Both the oocyte polarity and the multilayered PFC phenotypes were rescued by expression of a wildtype Kbr or Kbr:GFP transgene in the kbrdel mutant PFC cells (Figure S1F–I’), further confirming that kbr is responsible for the observed oogenesis defects.
Loss of kbr produces Notch signaling defects in PFCs similar to those of canonical Hippo pathway mutants
The oocyte polarity defects and the multilayered follicular epithelium observed in kbr mosaic egg chambers grossly resemble those caused by loss of Hippo signaling in PFCs (Polesello and Tapon, 2007; Meignin et al., 2007; Yu et al., 2008). To corroborate a similarity between kbr mutants and loss of Hippo signaling at a molecular level, we compared the expression of several Notch target genes in mosaic egg chambers bearing PFC clones of kbr or hpo. The expression of Cut, a marker of mitotic follicle cells that is normally inhibited by Notch signaling at the mitotic-endocycle switch at stage 7 of oogenesis (Sun and Deng, 2005), was maintained in both kbr and hpo mutant PFCs up to stage 10 (Figure 1F–H). Conversely, the expression of Hindsight (Hnt), which is normally induced by Notch signaling at stage 7 (Sun and Deng, 2007), was compromised in both kbr and hpo mutant PFCs (Figure 1I–K). Consistent with a defect in mitotic-endocycle transition, kbr mutant PFCs had smaller nuclei than wildtype PFCs, a phenotype that resembles that of canonical Hippo pathway mutants (Polesello and Tapon, 2007; Meignin et al., 2007; Yu et al., 2008). Of note, the persistent expression of Cut in kbr PFCs can be rescued by a wildtype kbr transgene (Figure S1H–I’).
To distinguish whether loss of kbr has a direct impact on Notch signaling or influences the mitotic-endocycle switch in a more indirect manner, we monitored the expression of E(spl)-CD2, a direct Notch signaling reporter (de Celis et al., 1998). We found that E(spl)-CD2 expression was compromised in kbr mutant PFC clones during stages 7–10, as in sav mutant PFC clones (Figure 1L–N). We conclude that kbr, like known Hippo pathway components, is required for proper transcriptional response of the PFCs to Notch signaling.
Loss of kbr results in tissue overgrowth characteristic of Hippo signaling defects
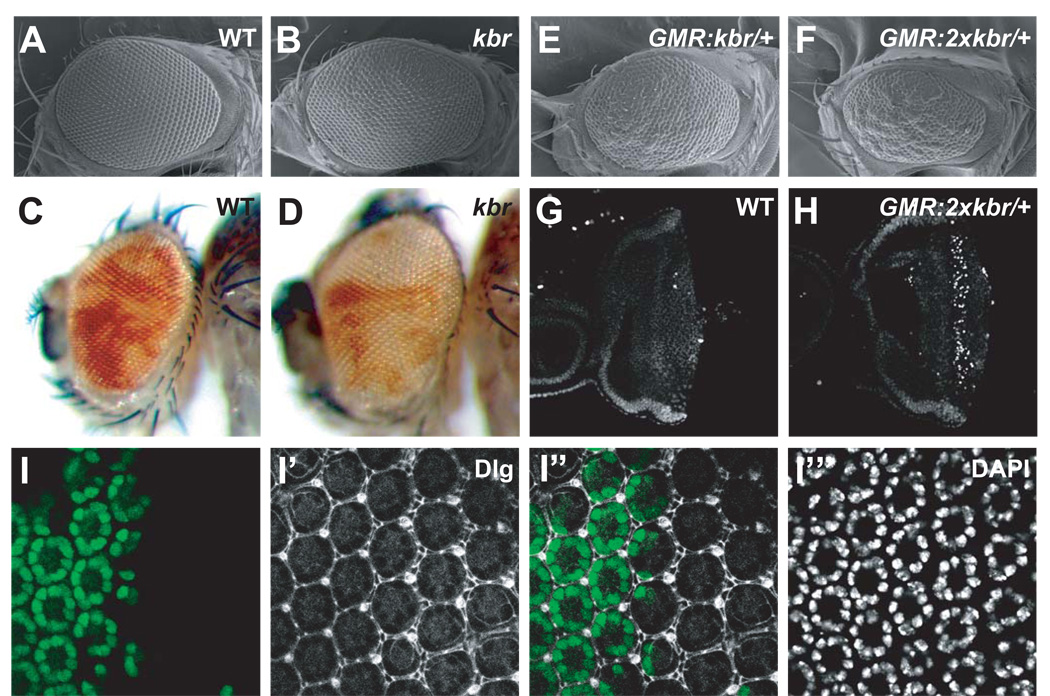
The multitude of similarities between kbr and canonical Hippo pathway mutants during Drosophila oogenesis implicate Kbr as a component of the Hippo signaling pathway. We further corroborated this hypothesis in Drosophila imaginal discs, where the canonical Hippo pathway plays a prominent role in controlling tissue size through coordinated regulation of cell proliferation and cell death. Induction of kbr mutant clones using an eyeless-FLP (ey-flp) source resulted in adult eyes that were larger than normal (Figure 2A–D). Notably, the kbr mosaic eyes showed an over-representation of mutant over wild type tissues (Figure 2C–D), and kbr mutant clones contained an average of 5.8 extra interommatidial cells per ommatidial cluster (Figure 2I–I”‘), suggesting that loss of kbr, like the canonical Hippo pathway components, results in increased cell proliferation and/or survival. Conversely, overexpression of kbr with the GMR-Gal4 driver led to ectopic apoptosis and a decrease in eye size (Figure 2E–H), which was largely suppressed by co-expression of the baculovirus cell death inhibitor P35 (Hay et al., 1994) (Figure S2) and which resembled those caused by hyper-activation of the Hippo pathway.
Figure 2. kbr is a negative regulator of imaginal disc growth.
(A–B) Scanning electron micrographs (SEM) of a wildtype eye (A) and an eye composed predominantly of kbr mutant cells (B). Note the increased eye size in (B). Genotypes: (A) y w ey-flp; FRT82B/FRT82B Ubi-GFP, (B) y w ey-flp; FRT82B kbrdel/FRT82B Ubi-GFP.
(C–D) Same as in (A–B) except that stereomicroscopic images are shown. Note that adult eyes mosaic for kbr (D) contained predominantly mutant tissues (white), whereas eyes mosaic for a control chromosome contained far less white tissues (C).
(E–F) SEM images of fly eyes in which kbr was overexpressed by the GMR-Gal4 driver. Genotypes: (E) GMR-Gal4/UAS-kbr, (F) GMR-Gal4 UAS-kbr/UAS-kbr.
(G–H) TUNEL staining of wildtype (G) and GMR-Gal4 UAS-kbr/UAS-kbr (H) eye imaginal discs. Note the ectopic cell death in kbr-overexpressing eye disc (H).
(I–I’’’) A mid-pupal retina containing kbr mutant clones, marked by the lack of GFP (I), and stained for Discs-Large (Dlg, I’) and DAPI (I’’’). Superimposed GFP and Dlg are shown in (I’’). Note the increased number of interommatidial cells in kbr clones.
See Figure S2 for supplemental data to Figure 2.
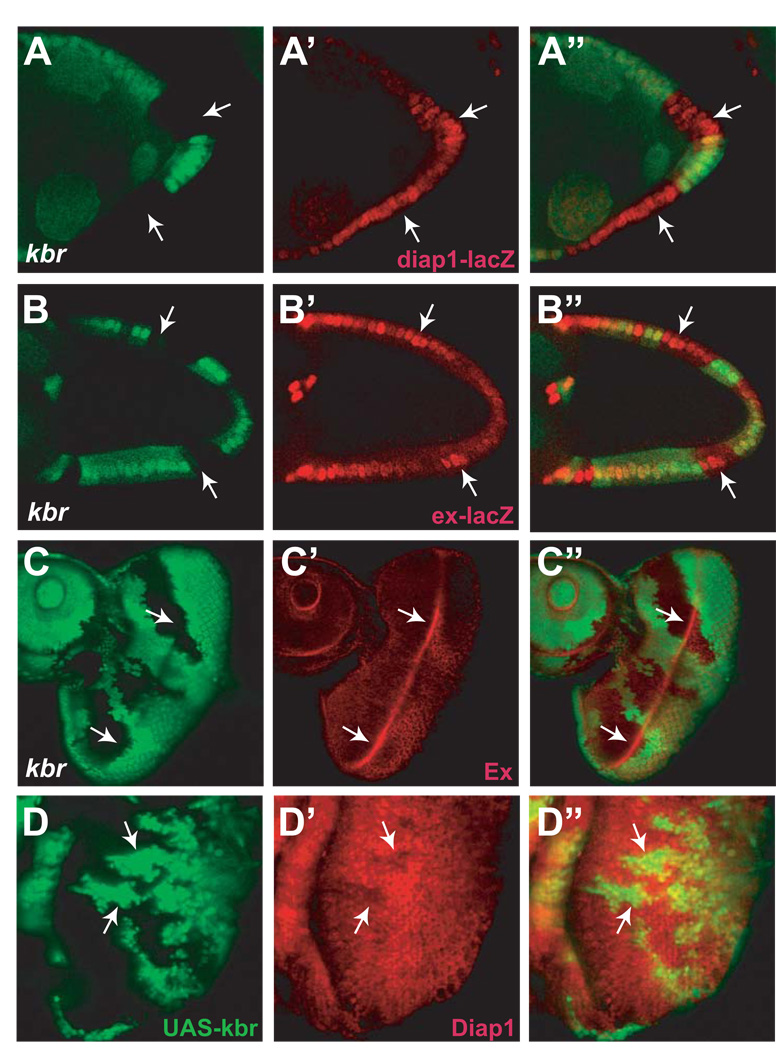
To link Kbr to the Hippo signaling pathway further, we examined the expression of diap1 and ex, two of the most commonly used Hippo pathway target genes. Transcription of both diap1 and ex, monitored with the thj5c8 and the exe1 enhancer trap lines respectively, was up-regulated in kbr mutant PFC clones (Figure 3A–B”). In the eye imaginal disc, however, loss of kbr has a much milder effect – kbr mutant clones showed a modest increase in Ex protein levels along the morphogenetic furrow (Figure 3C–C”), and no visible up-regulation of diap1-lacZ expression (Figure 6M–M”). Interestingly, kbr-overexpressing clones in the eye imaginal disc did show a cell-autonomous decrease in Diap1 levels, especially along the morphogenetic furrow (Figure 3D–D”). Overall, the modest effect of kbr on Ex and Diap1 levels in the eye resembles that of mer (Pellock et al., 2006), but is weaker than canonical Hippo pathway mutants such as hpo, sav or wts. These molecular defects correlate with their relative severity of overgrowth phenotypes, with the canonical Hippo pathway mutants showing multiply folded eye surface and >40 extra interommatidial cells per cluster (Hamaratoglu et al., 2006), whereas the kbr and mer mutants showing smooth eye surface and much fewer interommatidial cells (Figure 2 and Figure 6).
Figure 3. kbr regulates Hippo pathway target genes.
(A-B”) Egg chambers containing kbr mutant clones (GFP-negative) and stained for diap1-lacZ (A-A”) or ex-lacZ (B-B”) reporter expression (red). Note the elevated levels of diap1-lacZ and ex-lacZ in kbr PFC clones (arrows).
(C-C’’) An eye disc containing kbr mutant clones (GFP-negative) and stained with α-Ex antibody (red). Note the upregulation of Ex levels in kbr mutant cells along the morphogenetic furrow (arrows).
(D-D’’) An eye disc containing kbr-overexpressing clones (GFP-positive) and stained with α-Diap1 antibody (red). Note the decreased levels of Diap1 in kbr-overexpressing clones (arrows) close to the morphogenetic furrow.
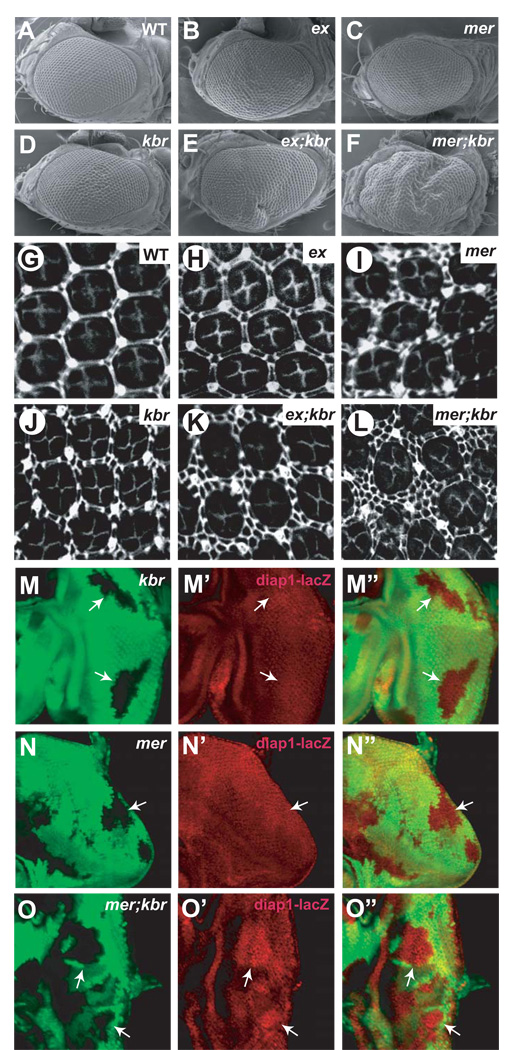
Figure 6. Kbr functions together with Mer and Ex to regulate tissue growth and Hippo signaling in vivo.
(A–F) SEM images of compound eyes from the following genotype: (A) wildtype, (B) y w ey-flp; FRT40A exel/FRT40A Ubi-GFP, (C) y w ey-flp, Ubi-GFP FRT19A/mer4 FRT19A, (D) y w ey-flp; FRT82B kbrdel/FRT82B Ubi-GFP, (E) y w ey-flp; FRT40A exel/FRT40A Ubi-GFP; FRT82B kbrdel/FRT82B Ubi-GFP, (F) y w ey-flp, Ubi-GFP FRT19A/mer4 FRT19A; FRT82B kbrdel/FRT82B Ubi-GFP. Note the smooth eye surface in (B–D) and the deformed eye surface with folded eye tissues in (E–F).
(G–L) Mid-pupal retina of the indicated genotype and stained for Dlg. Twenty ommatidial clusters of each genotype were used for counting interommatidial cells.
(M-O’) Third instar eye discs containing kbr (M-M”), mer (N-N”) or mer; kbr (O-O”) clones and diap1-lacZ reporter. Note the elevated levels of diap1-lacZ (red) in mer; kbr, but not kbr or mer clones (arrows).
Kbr regulates Yki and Hpo phosphorylation
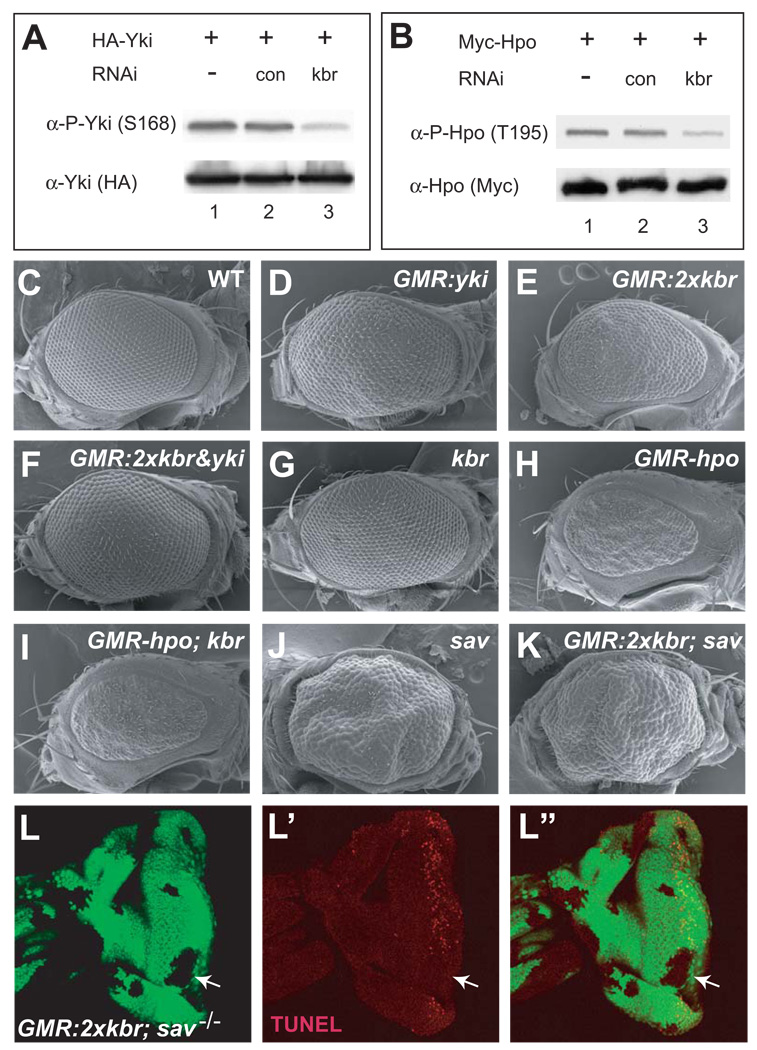
Independent of the genetic screen for oocyte polarity mutants, we identified kbr as a putative Hippo pathway component in a genome-wide RNAi screen in Drosophila S2 cells using the Hippo-responsive Yki S168 phosphorylation (Dong et al., 2007) as pathway readout (F. Yin and D. Pan, unpublished results). RNAi knockdown of kbr not only resulted in a decrease of Yki S168 phosphorylation (Figure 4A), but also a decrease of Hpo kinase activity (Figure 4B), as measured by a phospho-specific antibody against Hpo’s autophosphorylation site (Colombani et al., 2006). These molecular data directly implicate kbr in the Hippo pathway, and further place Kbr upstream of Hpo activation.
Figure 4. Kbr regulates Yki and Hpo phosphorylation and functions upstream of Hpo-Sav.
(A–B) S2 cells were transfected with HA-Yki (A) or Myc-Hpo (B) along with no dsRNA (lane 1), control dsRNA (lane 2), or kbr dsRNA (lane 3), and probed with α-P-Yki(S168) (A) or and α-P-Hpo(T195) (B). Note the suppression of Yki and Hpo phosphorylation by kbr RNAi.
(C–K) SEM images of eyes from the following genotypes: (C) wildtype, (D) GMR-Gal4; UAS-yki, (E) GMR-Gal4 UAS-kbr/UAS-kbr, (F) GMR-Gal4 UAS-kbr/UAS-kbr; UAS-yki, (G) y w ey-flp; FRT82B kbrdel/FRT82B Ubi-GFP, (H) GMR-hpo, (I) y w ey-flp; GMR-hpo; FRT82B kbrdel/FRT82B Ubi-GFP, (J) y w ey-flp; FRT82B savshrp1/FRT82B Ubi-GFP, (K) y w ey-flp; GMR-Gal4 UAS-kbr/UAS-kbr; FRT82B savshrp1/FRT82B Ubi-GFP.
(L–L”) TUNEL staining of an eye disc containing sav mutant clones (GFP-negative) and simultaneously overexpressing kbr posterior to the morphogenetic furrow. Note the diminished TUNEL staining in sav mutant clones (arrow). Genotype: y w hs-flp; GMR-Gal4 UAS-kbr/UAS-kbr; FRT82B savshrp1/FRT82B Ubi-GFP.
To substantiate this molecular model, we investigated the genetic relationship between kbr and core components of the Hippo kinase cascade. Overexpression of yki by the GMR-Gal4 driver results in increased eye size, whereas overexpression of kbr results in an opposite phenotype. In agreement with our molecular data placing kbr upstream of yki, co-overexpression of kbr and yki resulted in eye size similar to that caused by yki overexpression alone (Figure 4C–F). Next, we examined genetic epitasis between kbr and hpo, taking advantage of the opposite eye size phenotypes caused by loss of kbr and hpo overexpression. Notably, kbr mutant eyes that overexpressed hpo showed a small eye phenotype similar to that caused by hpo overexpression (Figure 4G–I), consistent with our molecular data placing kbr upstream of hpo. We further investigated the genetic epistasis between kbr and sav, a component of the Hpo-Sav kinase complex, by combining kbr overexpression with loss of sav. As shown above, overexpression of kbr during eye development results in ectopic cell death and severely reduced eye size. Significantly, both defects were reversed by loss of sav – sav mutant eyes that overexpressed kbr showed overgrowth similar to that of sav mutant alone (Figure 4J–K), and ectopic cell death induced by kbr overexpression was suppressed in sav mutant clones, but not in the neighbouring sav+ cells (Figure 4L–L”). Taken together, these results place Kbr upstream of the Hpo-Sav kinase complex in the Hippo signalling pathway.
Kbr functions together with Mer and Ex in a protein complex localized to apical membrane domain of epithelial cells
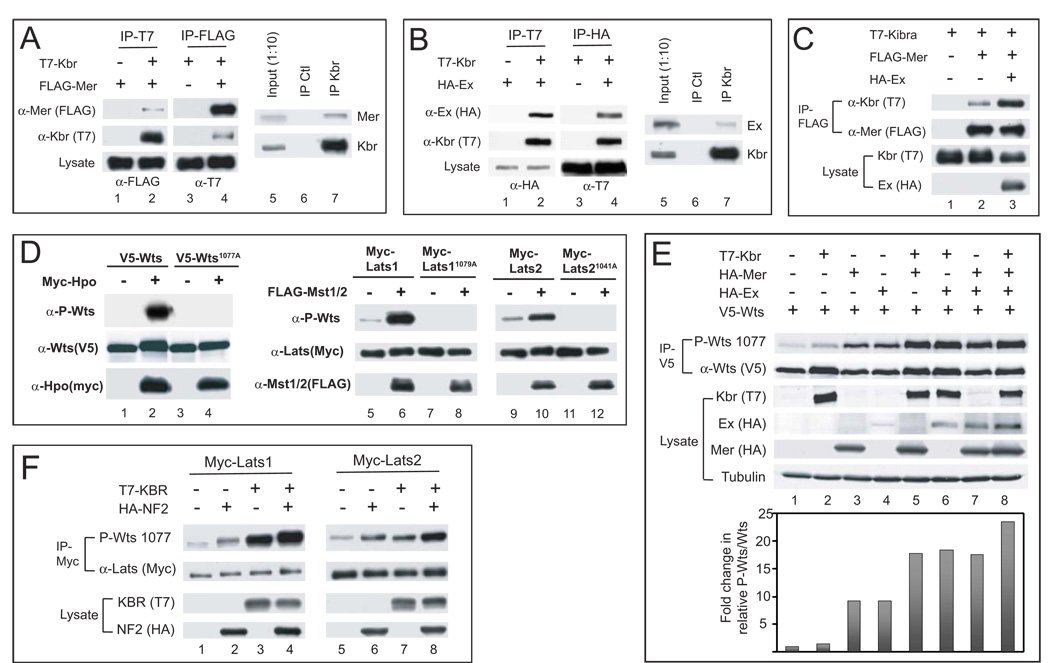
To understand the molecular mechanism by which Kbr regulates Hippo signaling, we searched the available protein interaction map (PIM) database, and found that Kbr was isolated as a Mer-binding protein in a yeast two-hybrid (Y2H)-based global PIM analysis conducted by Hybrigenics (Formstecher et al., 2005). Notably, multiple overlapping Kbr clones were isolated using two independent Mer baits, demonstrating that the interaction is of high-confidence (Figure S3A). In agreement with the Y2H result, epitope-tagged Kbr and Mer, as well as endogenous Kbr and Mer, coimmunoprecipitated with each other in Drosophila S2 cells (Figure 5A). We also tested interactions between Kbr and Ex, a related FERM domain protein known to associate with and act synergistically with Mer (McCartney et al., 2000; Hamaratoglu et al., 2006), and found that Kbr and Ex coimmunoprecipitated with each other, at both overexpressed and endogenous levels (Figure 5B). Interestingly, Ex potentiated Kbr-Mer interaction – in the presence of transfected Ex, significantly more Kbr was coimmunoprecipitated by Mer (Figure 5C).
Figure 5. Kbr forms a protein complex and acts synergistically with Mer and Ex to promote Wts phosphorylation.
(A) Physical association between Kbr and Mer. Lanes 1–4: S2 cell lysates expressing the indicated combination of T7-Kbr and FLAG-Mer constructs were immunoprecipitated (IP) and probed with the indicated antibodies. FLAG-Mer was detected in T7-IP in the presence (lane 2), but not the absence (lane 1), of T7-Kbr. Conversely, T7-Kbr was detected in FLAG-IP in the presence (lane 4), but not the absence (lane 3), of FLAG-Mer. Lanes 5–7: α-Kbr was used to IP endogenous Kbr from untransfected S2 cells and probed with α-Mer or α-Kbr antibody. Mer was detected in Kbr-IP, not in IP with control IgG.
(B) Physical association between Kbr and Ex. Lanes 1–4: similar to (A) except that T7-Kbr and HA-Ex were tested for co-IP. HA-Ex was detected in T7-IP products in the presence (lane 2), but not the absence (lane 1), of T7-Kbr. Conversely, T7-Kbr was detected in HA-IP products in the presence (lane 4), but not the absence (lane 3), of HA-Ex. Lanes 5–7: α-Kbr was used to IP endogenous Kbr from untransfected S2 cells. Ex was detected in Kbr-IP, but not in IP with control IgG.
(C) Ex potentiates Kbr-Mer interaction. S2 cells expressing the indicated constructs were analyzed by co-IP. Note that in the presence of HA-Ex, significantly more T7-Kbr was detected in FLAG-Mer IP (compare lanes 3 and 2).
(D) Phospho-specific antibody against the hydrophobic motif of Drosophila Wts and human Lats1/2. The indicated V5-Wts or Myc-Lats1/2 constructs were expressed in S2 cells or HEK293 cells respectively, immunoprecipitated and probed with antibodies against P-Wts (top gels) and the respective epitopes (middle gels). A fraction of cell lysate was probed with the indicated antibodies to evaluate expression levels of Myc-Hpo or FLAG-Mst1/2 (bottom gels). Hpo induced Wts T1077 phosphorylation, which was abolished by a T1077A mutation. Mst1/2 induced Lats1 T1079 or Lats2 T1041 phosphorylation, which was abolished by a Lats1 T1079A mutation or a Lats2 T1041A mutation.
(E) Kbr promotes Wts phosphorylation in conjunction with Mer and Ex. V5-IP from S2 cells expressing the indicated constructs were probed with α-P-Wts(T1077) and α-V5 using the Odyssey Infrared Imaging System. A fraction of the cell lysate was probed with indicated antibodies to evaluate protein expression levels. The P-Wts signal relative to the V5-Wts signal, expressed in arbitrary units, is plotted in the graph.
(F) The human KBR protein promotes Lats1/Lats2 phosphorylation in conjunction with human NF2. Myc-IP from HEK293 cells expressing the indicated constructs probed with α-P-Wts and α-Myc. A fraction of the cell lysate was probed with the indicated antibodies to evaluate protein expression levels. Note that accompanying the induced hydrophobic motif phosphorylation, KBR and NF2 also caused retarded mobility of Lats proteins, which is more obvious for Lats1 than Lats2.
See Figure S3 for supplemental data to Figure 5.
The protein-protein interactions among Kbr, Mer and Ex suggest that these proteins function together in a protein complex to regulate the Hippo pathway. For simplicity, we shall refer to this protein complex as KEM (Kbr, Ex and Mer) complex hereafter. To corroborate this model further, we examined the subcellular localization of Kbr. Unfortunately, while our home-made α-Kbr antibody was of sufficient quality for immunoprecipitation and western blotting (Figures 5A–B and S3L), it did not work on immunostaining. As an alternative, we examined the subcellular localization of a Kbr:GFP fusion protein. We found that Kbr:GFP was localized to the cytoplasm and enriched in the apical membrane domain of ovarian follicle cells (Figure S3B), reminiscent of that of Mer and Ex. Interestingly, the apical membrane localization of Kbr:GFP was not noticeably affected by loss of ft, ex or mer (Figure S3B–E), and conversely, loss of kbr did not affect the apical membrane localization of Ex and Mer (Figure S3F–K’). Combined with the fact that Mer and Ex are localized to apical membrane domain independent of each other (McCartney et al., 2000), these results suggest that components of the KEM complex are targeted to the apical membrane domain independent of each other.
Kbr, Ex and Mer act synergistically to regulate the Hippo kinase cascade
Since Mer and Ex can synergistically activate Wts phosphorylation in Drosophila S2 cells (Hamaratoglu et al., 2006), we examined the effect of Kbr, Mer and Ex on Wts phosphorylation. To facilitate this analysis, we generated a phospho-specific antibody against T1077 of the Wts hydrophobic motif, based on previous reports that Lats1/2 are phosphorylated by Mst1/2 at a conserved site (Chan et al., 2005; Praskova et al., 2008). Indeed, the phospho-Wts antibody detected a specific signal that was induced by Hpo and abolished by a T1077A mutation (Figure 5D). This antibody also showed cross-reactivity with the highly conserved hydrophobic motif in mammalian Lats1/2 (Figures 5D and S3M). When transfected individually into S2 cells, Kbr, Mer and Ex could each stimulate Wts T1077 phosphorylation, although the effect of Kbr was more modest than that of Mer or Ex (Figure 5E). Interestingly, despite the relatively modest effect of Kbr on Wts T1079 phosphorylation, co-expression of Kbr with Mer or Ex synergistically activated Wts phosphorylation to a similar extent as the Mer-Ex combination (Figure 5E). Furthermore, the Kbr-Mer-Ex combination led to greater Wts phosphorylation than the Mer-Ex combination did (Figure 5E). Thus, Kbr, Mer and Ex can act synergistically to induce Wts phosphorylation in cultured cells.
If Kbr, Mer and Ex act synergistically to regulate the Hippo pathway, we should expect mer; kbr or ex; kbr double mutants to show a more severe phenotype than any of the single mutants, in much the same way that mer; ex double mutants cause a stronger phenotype than either single mutant (Hamaratoglu et al., 2006). Indeed, both ex; kbr and mer; kbr eyes displayed a stronger overgrowth phenotype and a greater number of interommatidial cells than did the respective single mutants (Figure 6A–L). For example, although ex, mer and kbr mutant adult eyes all had smooth exterior surface, ex; kbr and mer; kbr adult eyes were deformed with extra folds of tissues (Figure 6A–F). Likewise, although ex, mer and kbr mutant eyes had an average of 1.4, 8.4 and 5.8 extra interommatidial cells, respectively, ex; kbr and mer; kbr eyes had 23 and 51 extra interommatidial cells per cluster, respectively (Figure 6G–L). The synergy between kbr and mer could be further seen at the level of Hippo pathway target gene expression – although neither mer nor kbr mutant showed visible upregulation of diap1 transcription, mer; kbr double mutant clones showed significantly elevated levels of diap1 expression (Figure 6M–O”). These genetic interactions suggest that Kbr functions together with Ex and Mer in at least a partially redundant manner to regulate the Hippo pathway.
The KEM complex regulates the Hippo kinase cascade via direct binding to the Hpo-Sav complex
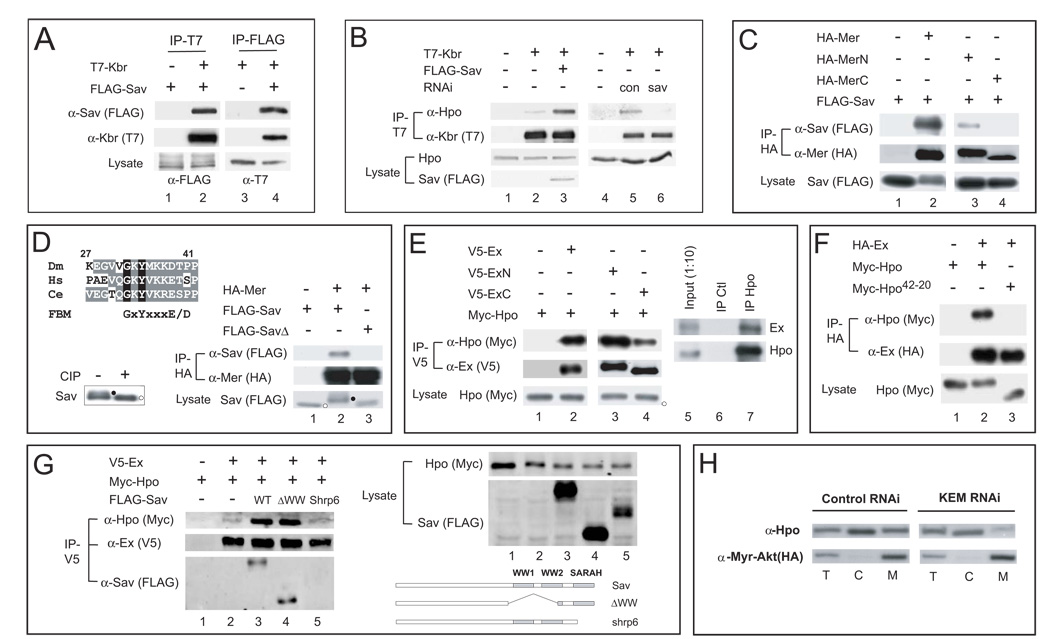
Having established that Kbr functions upstream of Hpo together with Mer and Ex, we investigated how Kbr is linked to the canonical Hippo kinase cascade. We were particularly intrigued by the presence of two WW domains within Kbr, given that it is the third Hippo pathway component containing two WW domains. Interestingly, while WW1 and WW2 of Yki are canonical WW domains containing two signature tryptophan residues, the WW2 domain of Kbr and Sav has a single tryptophan. Recent structural studies showed that the atypical WW2 domain of the mouse Sav homologue can form a homodimer (Ohnishi et al., 2007). The presence of the atypical WW2 domain in Kbr prompted us to investigate whether Kbr could hetero-dimerize with Sav. Indeed, we found that Kbr and Sav bind to each other in an Y2H assay (Figure S4B). Consistent with our Y2H result, epitope-tagged Kbr and Sav (Figure 7A), as well as epitope-tagged Sav and endogenous Kbr (Figure S4C), co-immunoprecipitated with each other in S2 cells, whereas epitope-tagged Kbr and Yki proteins did not (data not shown). Interestingly, we found that epitope-tagged Kbr immunoprecipitated endogenous Hpo protein in S2 cells in a Sav-dependent manner – the amount of endogenous Hpo immunoprecipitated by Kbr was significantly increased by co-transfection of Sav and diminished by RNAi knockdown of endogenous Sav (Figure 7B). These observations are consistent with our genetic epistasis analysis placing Kbr upstream of Sav (Figure 4), and further suggest that Sav may link Kbr to the Hpo kinase complex.
Figure 7. Multiple protein-protein interactions link the KEM complex to the Hpo-Sav complex.
(A) Physical association between Kbr and Sav. S2 cell lysates expressing the indicated constructs were immunoprecipitated (IP) and probed with the indicated antibodies. FLAG-Sav was detected in T7-Kbr IP. Conversely, T7-Kbr was detected in FLAG-Sav IP.
(B) Sav potentiates association between Kbr and Hpo. T7-IP from S2 cells transfected with the indicated constructs or dsRNA was probed with antibody against endogenous Hpo. Note that the amount of endogenous Hpo detected in T7-Kbr IP was increased by co-expression of FLAG-Sav (compare lanes 3 and 2), and diminished by sav RNAi (compare lanes 6 and 5).
(C) Physical association between Mer and Sav. FLAG-Sav was detected in HA-IP from S2 cells co-expressing HA-Mer, HA-MerN (N-terminal half of Mer), but not HA-MerC (C-terminal half of Mer).
(D) Sav contains a FERM binding motif (FBM) that is required for binding to Mer. Top: alignment of FBM sequence from Sav orthologues in Drosophila (Dm), human (Hs) and worm (Ce). The consensus FBM is also shown. Lower right: HA-IP from S2 cells expressing HA-Mer with FLAG-Sav or FLAG-SavΔGKY (SavΔ) was probed with the indicated antibodies. FLAG-Sav, but not FLAG-SavΔGKY, was detected in HA-IP. Also note the mobility shift of Sav, but not SavΔGKY, induced by Mer co-expression. Lower left: phosphatase (CIP) treatment of FLAG-Sav IP from cells expressing HA-Mer and FLAG-Sav. Hyper- and hypo-phosphorylated Sav is indicated by black and white circles next to the protein bands, respectively.
(E) Physical association between Ex and Hpo. Lanes 1–4: Myc-Hpo was detected in V5-IP from S2 cell co-expressing V5-Ex, V5-ExN (N-terminal half of Ex), or V5-ExC (C-terminal half of Ex). Lanes 5–7: α-Hpo was used to IP endogenous Hpo from untransfected S2 cells. Endogenous Ex was detected in Hpo-IP, but not in IP with control IgG.
(F) The SARAH domain of Hpo is required for binding to Ex. HA-IP from S2 cells expressing HA-Ex with Myc-Hpo or Myc-Hpo42-20 was analyzed. Hpo42-20 mimics a hypomorphic hpo allele that deletes just the SARAH domain (Wu et al., 2003). Myc-Hpo (lane 2), but not Myc-Hpo42-20 (lane 3), was detected in HA-Ex IP.
(G) Ex, Sav and Hpo can co-exist in the same complex. V5-IP from S2 cells expressing the indicated constructs was analyzed. Note that in the presence of FLAG-Sav or FLAG-SavΔWW (deleting the WW domains), but not FLAG-Savshrp6 (mimicking a sav allele that deletes just the SARAH domain (Kango-Singh et al., 2002)), significantly more Myc-Hpo was detected in V5-Ex IP (compare lanes 2–5). A small fraction of the cell lysate was probed to evaluate protein expression levels (top right). The schematic structure of Sav mutants used in the experiment is also shown (lower right).
(H) The KEM complex is required for membrane association of Hpo. S2 transiently expressing a myristylated Akt construct (Verdu et al., 1999) were treated with control or dsRNAs against Kbr, Ex and Mer (KEM) and subjected to cell fractionation. Cytosolic (C), membrane (M), and a portion of the total lysate (T) were probed for endogenous Hpo. Note the decreased Hpo signal in membrane fraction upon KEM RNAi. Also note that myristylated-Akt was only recovered in the membrane fraction.
See Figure S4 for supplemental data to Figure 7.
If Kbr provides the only link between the KEM complex and the Hpo kinase cascade, loss of kbr should produce overgrowth phenotypes as strong as those resulting from combined loss of kbr, ex or mer. Our analysis of ex; kbr and mer; kbr double mutants clearly argued against this possibility (Figure 6), suggesting that Ex and Mer may additionally interact with the Hippo kinase cascade independent of Kbr. Consistent with the latter model, we note that the PIM analysis conducted by Hybrigenics also revealed Y2H interactions between Mer and Sav, as well as between Ex and Hpo (Figure S4A) (Formstecher et al., 2005). We confirmed these interactions in yeast (Figure S4B). In contrast, Ex and Sav do not associate with each other in yeast (Figure S4B). These interactions were also confirmed in S2 cells, as epitope-tagged Mer and Sav (Figure 7C), as well as endogenous Mer and epitope-tagged Sav (Figure S4C), coimmunoprecipitated with each other. Furthermore, epitope-tagged Ex and Hpo, as well as endogenous Ex and Hpo, coimmunoprecipitated with each other (Figure 7E).
Using co-immunoprecipitation assays, we further mapped protein domains involved in Mer-Sav and Ex-Hpo interactions. We found that the N-terminal half of Mer, which comprises mostly of the FERM domain, is required for interaction with Sav (Figure 7C). Interestingly, Sav contains a sequence near its N-terminus that matches a consensus FERM binding motif (FBM) (Figures 7D and S4A). This sequence represents the only evolutionarily conserved sequence outside Sav’s WW and SARAH domains. To test the importance of this newly recognized FBM, we mutated it by deleting a functionally critical GxY motif (Gosens et al., 2007), and found that this mutation abolished Sav-Mer association (Figure 7D). In the course of these experiments, we noted that co-expression of Mer with Sav led to phosphorylation of Sav, manifested as retarded mobility on SDS-PAGE that was reversed by phosphatase treatment (Figure 7D). Significantly, mutation of the FBM abolished not only Sav-Mer interaction, but also Mer-induced phosphorylation of Sav (Figure 7D).
In contrast to Mer-Sav interaction, both the N- (containing the FERM domain) and C-terminal half of Ex can associate with Hpo (Figure 7E). Analysis of Hpo truncation mutants revealed that the SARAH domain at its C-terminus is essential for Ex interaction (Figure 7F and data not shown). Since the SARAH domain of Hpo is known to mediate Hpo-Sav interaction (Scheel and Hofmann, 2003), we wondered whether Ex and Sav can interact with Hpo simultaneously, or in a mutually exclusive manner. Consistent with the former possibility, we found that co-expression of Sav enhanced Ex-Hpo interactions (Figure 7G). Furthermore, this enhancement requires the SARAH domain, but not the WW domains, of Sav (Figure 7G).
Given the apical membrane localization of the KEM complex and recent reports implicating mammalian MST kinases in activating NDR kinases at plasma membrane (Hergovich et al., 2005), we examined whether the KEM complex affects the membrane association of Hpo in Drosophila S2 cells. Using a subcellular fractionation protocol that separates membrane and cytosolic fractions (Hergovich et al., 2005), we found that RNAi knockdown of the KEM complex resulted in a significant decrease of endogenous Hpo protein in the membrane fraction (Figure 7H). Thus, the KEM complex regulates Hpo at least in part by bringing the latter to the membrane, where Hpo may be activated via mechanisms yet to be determined.
Conserved function of the human KIBRA protein
Although initially identified based on its predominant expression in kidney and brain (Kremerskothen et al., 2003), the KIBRA gene has garnered considerable recent interest due to the reported association between KIBRA polymorphism and individual variation in memory performance in natural human populations (Papassotiropoulos et al., 2006). At present, the biochemical function of the human KIBRA protein remains poorly understood. The human KIBRA protein has been reported to bind several proteins, including the actin cytoskeleton-associated proteins dendrin and synaptopodin, the PDZ-containing scaffolding protein PATJ, the motor protein dynein light chain 1 (DLC1), histone H3, the tyrosine kinase receptor DDR1, and PKC ζ (Duning et al., 2008; Rayala et al., 2006; Hilton et al., 2008). None of these proteins has been linked to the Hippo signaling pathway. To investigate whether Kbr may play a conserved role in regulating the Hippo pathway in mammals, we examined the effect of human KIBRA on Lats1/2 phosphorylation in HEK 293 cells. We found that, like its Drosophila counterpart, KIBRA stimulated hydrophobic motif phosphorylation of Lats1/2 (Figure 5F). Interestingly, expression of human NF2/Merlin also led to a similar increase in Lats1/2 phosphorylation and co-expression of KIBRA and NF2/Merlin synergistically induced Lats1/2 phosphorylation (Figure 5F). These results suggest that, like their Drosophila counterparts, KIBRA and NF2/Merlin may function together to regulate the Hippo pathway in mammals.
Discussion
Kbr, a new component of the emerging Hippo signaling pathway
Compared to the core kinase cascade leading from Hpo to Yki phosphorylation, signaling events upstream of Hpo are less well understood. In this study, we identify Kbr as a tumor suppressor and an essential component of the Hippo pathway. We propose a model in which Kbr functions together with Mer and Ex in an apical protein complex (the KEM complex) to transduce growth-regulatory signals to the Hpo-Sav complex, which through the canonical Hippo kinase cascade, controls Yki phosphorylation and target gene transcription. Of note, our findings do not exclude the possibility that Kbr, Ex or Mer may interact with additional Hippo pathway components besides Hpo-Sav, especially given the recent report that Ex can directly bind Yki (Badouel et al., 2009). How Kbr, Ex and Mer function together to integrate upstream signals remains to be determined. One possibility is that these proteins function redundantly in receiving signals from the same upstream regulator(s). Alternatively, each protein may be regulated by distinct upstream regulator(s).
A commonly used assay for Hippo signaling in Drosophila S2 cells involves examining mobility shifts of the Wts protein on SDS-PAGE (Wu et al., 2003). Given its large size and that not all protein phosphorylation causes discernable mobility shift on SDS-PAGE, this assay is less sensitive in detecting Wts phosphorylation than the phospho-specific antibody used in the present study. Indeed, overexpression of Ex in S2 cells has no effect on Wts mobility (Hamaratoglu et al., 2006) (our own observations), yet we demonstrate here that Ex induces robust Wts phosphorylation at its hydrophobic motif. The fact that this hydrophobic motif is a well-established direct phosphorylation site by Hpo homologues in mammalian cells (Chan et al., 2005; Praskova et al., 2008) further suggests that Ex, as well as Mer and Kbr, regulates Wts through the canonical Hippo kinase cascade. Indeed, we found that Ex-induced Wts phosphorylation is Hpo-dependent (Figure S4D). These results are not incompatible with recent report that Ex can also regulate the Hippo pathway in a kinase-independent manner (Badouel et al., 2009). Using a well-established assay for Yki transcriptional activity (Huang et al., 2005), we found that while Ex, Mer plus Kbr, or Hpo could all suppress the activity of a Yki-Gal4 fusion protein, only Ex was able to suppress the activity of a Yki-Gal4 fusion protein in which all the possible Wts-phosphorylation sites are mutated (Oh and Irvine, 2008) (Figure S4E). These observations are consistent with the view that Ex can regulate the Hippo pathway through both Wts-dependent (Hamaratoglu et al., 2006) and -independent (Badouel et al., 2009) mechanisms.
Tissue-specific differences in Hippo pathway regulation by upstream components
A comparison of the loss-of-function phenotypes of mer, ex and kbr in egg chambers and imaginal discs reveal tissue-specific differences in the relative contribution of each gene to Hippo pathway regulation. For example, loss of ex alone, but not mer or kbr, is sufficient to cause robust diap1 upregulation in imaginal discs (Pellock et al., 2006) (this study), suggesting that ex has a more essential role in diap1 transcriptional regulation. However, the converse is true in the ovary, where loss of mer or kbr results in stronger oocyte polarity and Notch signaling defects than loss of ex (Yu et al., 2008) (this study). In fact, the severity of mer or kbr mutant phenotypes in oogenesis are comparable to those of core components of the Hippo pathway such as hpo and sav (Yu et al., 2008) (this study), even though the former display much milder overgrowth than the latter in imaginal discs. Perhaps the most extreme case of tissue-specific requirement is provided by the ft tumor suppressor gene, which is required for Hippo pathway regulation in the imaginal discs (Silva et al., 2006; Willecke et al., 2006; Bennett and Harvey, 2006; Cho et al., 2006) but dispensable in developing egg chambers (Polesello and Tapon, 2007; Meignin et al., 2007; Yu et al., 2008). While the underlying molecular basis remains to be determined, such tissue-specific requirements suggest that the core Hippo kinase cascade may function as a signal integrator of multiple inputs in a dynamic and versatile manner, and that additional cell surface receptors besides Ft may signal to the Hippo pathway.
NF2, Hippo signaling and memory performance
Considerable efforts have been directed at identifying the key signaling pathways regulated by the NF2/Merlin tumor suppressor protein. These investigations have led to the identification of a number of effector mechanisms downstream of NF2/Merlin, such as growth control pathways mediated by Ras, Rac, STAT or PI3K, contact inhibition mediated by cell surface receptors or adherens junctions, and endocytosis/degradation of various membrane proteins (McClatchey and Giovannini, 2005; Okada et al., 2007). The recent identification of Mer as an upstream regulator of Hpo in Drosophila provides yet another plausible mechanism through which Mer functions as a tumor suppressor protein (Hamaratoglu et al., 2006). Our identification of Kbr, which has been independently isolated as a Mer-binding protein in an unbiased PIM study (Formstecher et al., 2005), as a novel regulator of the Hippo pathway, further strengthens the case for a functional link between NF2/Mer and the Hippo pathway. Our observation that NF2/Mer and KIBRA can synergistically stimulate Lats1/2 phosphorylation in mammalian cells (Figue 5F) not only supports an NF2/Mer-Hippo connection, but further implicates KIBRA as a potential tumor suppressor in humans with relevance to neurofibromatosis.
Our identification of Kbr as an upstream regulator of the Hippo pathway has implications for understanding memory-related functions of the human KIBRA gene (Papassotiropoulos et al., 2006). Besides its well-established roles in growth control, the Hippo pathway is also required for differentiation and morphogenesis of certain postmitotic neurons in Drosophila (Mikeladze-Dvali et al., 2005; Emoto et al., 2006). We speculate that modulation of the Hippo pathway may influence the growth or differentiation of memory-related neuronal structures, a hypothesis that can be directly tested by genetic manipulation of Hippo signaling activity in animal models.
Experimental Procedures
Antibodies
Phospho-specific antibodies against Wts T1077 were produced by immunization of rabbits with the FHGFFEF(pT)FRRFFDD phosphopeptide. Antibodies reactive with the nonphosphopeptide were removed by adsorption to a nonphosphopeptide affinity column. Antibodies that flowed through this column were next passed over a column of immobilized phosphopeptide; after washing, antibodies were eluted at low pH and dialyzed. Phospho-specific antibodies against Yki S168 have been described previously (Dong et al., 2007). Antibody against Kbr was produced by immunizing rabbits with the peptides EPADAPIPVASNDAEQ. Phospho-specific antibodies against the activation loop of Mst1/Mst2/Hpo (T183 of Mst1, T180 of Mst2, T195 of Hpo) were obtained from Cell Signaling Technology. Other antibodies used include: rabbit α-Ex (Boedigheimer and Laughon, 1993) (gift of A.S. Laughon), guinea pig α-Mer (Maitra et al., 2006) (gift of R.G.Fehon), rabbit α-Hpo (Wu et al., 2003).
Yeast two-hybrid assay
Yeast two-hybrid analysis was carried out using Clontech’s Matchmaker system according to manufacture’s instructions. Ex, Mer, Kbr and Sav DNA sequences amplified by PCR were ligated into the DNA-binding domain vector pGBKT7. Hpo and Sav DNA sequences were amplified by PCR and inserted into the activation domain vector pGATT7. All the sequences were verified by DNA sequencing.
Drosophila and mammalian cell culture
Drosophila S2R+ cells were propagated in Drosophila Schneider’s Medium (GIBCO) supplemented with 10% FBS and antibiotics. For RNAi experiments, 0.5 µg of dsRNA against specific Drosophila genes or GFP (as control) was co-transfected with indicated epitope-tagged expression plasmids into S2R+ cells using Effectene reagent (Qiagen). FLAG-Mer was constructed in the pAc5.1/V5-HisB vector by adding a FLAG epitope (DYKDDDDK) to the C-terminus of Mer preceded by a triple Glycine linker. HA-Yki, Myc-Hpo, V5-Wts and FLAG-Sav have been described previously (Huang et al., 2005). HA-Ex and HA-Mer were gift from G. Halder (Hamaratoglu et al., 2006). T7-Kbr was constructed in the pAc5.1/V5-HisB vector by addition of an N-terminal T7 epitope (MASMTGGQQMG) to the full length cDNA clone RE26350. N-terminal tagged HA-MerN (aa 1–375), HA-MerC (aa 376–635), V5-Ex, V5-ExN (aa 1–709) and V5-ExC (aa 710–1427) constructs were made by PCR using pAc5.1/V5-HisB vector. Myc-Hpo42-20 was made by introducing a nonsense point mutation into Myc-Hpo removing the last 91 residues at the C terminus of Hpo (Wu et al., 2003). DNA sequence corresponding to aa 32–34 or aa 342–472 of Sav were deleted using the QuikChange site-directed mutagenesis kit (Stratagene) to make FLAG-SavΔGKY and FLAG-SavΔWW, respectively. V5-Wts1077A was also made by the same mutagenesis Kit. All constructs were verified by DNA sequencing.
Immunoprecipitation was carried out as previously described (Gao and Pan, 2001) using RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris pH8.0) or NP-40 lysis buffer (150 mM NaCl, 1% Triton X-100, 10 mM Tris pH7.4, 1 mM EGTA, 0.5% NP-40) supplemented with 1 mM PMSF and protease inhibitors (Roche). All western blotting was developed using the Amersham ECL chemilluminenscence system (GE Healthcare), except for Figure 6D, in which quantitative measurement of Wts T1077 phosphorylation was carried out using the Odyssey Infrared Imaging System (LI-COR Biosciences). Briefly, V5-Wts from transfected S2R+ cells was immunoprecipitated, separated on 8% SDS-PAGE and transferred to an Immobilon-P PVDF membrane (MILLIPORE). Blots were blocked in Odyssey blocker, then incubated with mouse anti-V5 (1:5000 dilution) and rabbit anti-phopho-Wts T1077 (1:1000 dilution). Blots were washed 3×5min in TBST buffer after primary antibody incubation then incubated with 1:10000 diluted Alexa Fluor 680 goat anti-mouse IgG (Invitrogen) and goat anti-rabbit IRDye 800 (LI-COR Biosciences). Blots were scanned with the LI-COR ODYSSEY infrared imaging system and quantified with the software provided with the system.
Fractionation of cells into cytosolic and membrane-associated proteins was carried out as described (Hergovich et al., 2005). For luciferase assay, Gal4DB-Yki (Huang et al., 2005) or Gal4DB-Yki3A (S111A, S168A, S250A) (Oh and Irvine, 2008) was transfected in triplicates in S2R+ cells with Ex, Mer and/or Kbr, and Hpo, respectively. Luciferase assay was carried out using Dual Luciferase Assay System (Promega) and a FLUOstar Lumiometer (BMG Lab Technologies).
HEK-293 cells were maintained in DMEM, 10% FBS and antibiotics (Invitrogen). HA-NF2 was constructed in the pcDNA3.1+ vector by addition of an N-terminal HA epitope (YPYDVPDYA) to the full length human NF2 cDNA clone (IMAGE 4871980, Invitrogen). For construction of T7-KBR, two overlapping EST clones, IMAGE 3570624 and 4473067 (Invitrogen), were used as PCR templates to amplify the 5’ and the remaining portion of human KIBRA cDNA, with a T7 epitope added to the N-terminus of KBR. The two PCR fragments were inserted into the pcDNA3.1+ vector in a 3-way ligation to result in T7-KBR. Myc-Lats11079A and Myc-Lats21041A were made using the QuikChange site-directed mutagenesis kit (Stratagene). All clones were verified by sequencing. All the other expression plasmids have been described previously (Dong et al., 2007).
Supplementary Material
Acknowledgments
We thank R. Fehon, G. Halder and A. Laughon for reagents, G. Barrio, C. Green, C. Ling, J. Poulton, J. Sun and F.Yin for technical assistance, and E. Chen and A.B. Thistle for critical reading of the manuscript. This work was supported by grants from the National Institutes of Health to W.M.D (GM072562) and D.J.P. (EY015708). D.J.P. is an investigator of the Howard Hughes Medical Insitute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Badouel C, Gardano L, Amin N, Garg A, Rosenfeld R, Le BT, McNeill H. The FERM-domain protein expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev. Cell. 2009;16:411–420. doi: 10.1016/j.devcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr. Biol. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- Boedigheimer M, Laughon A. Expanded: a gene involved in the control of cell proliferation in imaginal discs. Development. 1993;118:1291–1301. doi: 10.1242/dev.118.4.1291. [DOI] [PubMed] [Google Scholar]
- Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Casal J, Lawrence PA, Struhl G. Two separate molecular systems, Dachsous/Fat and Starry night/Frizzled, act independently to confer planar cell polarity. Development. 2006;133:4561–4572. doi: 10.1242/dev.02641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EH, Nousiainen M, Chalamalasetty RB, Schafer A, Nigg EA, Sillje HH. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene. 2005;24:2076–2086. doi: 10.1038/sj.onc.1208445. [DOI] [PubMed] [Google Scholar]
- Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nat.Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- Colombani J, Polesello C, Josue F, Tapon N. Dmp53 activates the Hippo pathway to promote cell death in response to DNA damage. Curr. Biol. 2006;16:1453–1458. doi: 10.1016/j.cub.2006.05.059. [DOI] [PubMed] [Google Scholar]
- de Celis JF, Tyler DM, de CJ, Bray SJ. Notch signalling mediates segmentation of the Drosophila leg. Development. 1998;125:4617–4626. doi: 10.1242/dev.125.23.4617. [DOI] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duning K, Schurek EM, Schluter M, Bayer M, Reinhardt HC, Schwab A, Schaefer L, Benzing T, Schermer B, Saleem MA, Huber TB, Bachmann S, Kremerskothen J, Weide T, Pavenstadt H. KIBRA modulates directional migration of podocytes. J. Am. Soc. Nephrol. 2008;19:1891–1903. doi: 10.1681/ASN.2007080916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto K, Parrish JZ, Jan LY, Jan YN. The tumour suppressor Hippo acts with the NDR kinases in dendritic tiling and maintenance. Nature. 2006;443:210–213. doi: 10.1038/nature05090. [DOI] [PubMed] [Google Scholar]
- Formstecher E, Aresta S, Collura V, Hamburger A, Meil A, Trehin A, Reverdy C, Betin V, Maire S, Brun C, Jacq B, Arpin M, Bellaiche Y, Bellusci S, Benaroch P, Bornens M, Chanet R, Chavrier P, Delattre O, Doye V, Fehon R, Faye G, Galli T, Girault JA, Goud B, de GJ, Johannes L, Junier MP, Mirouse V, Mukherjee A, Papadopoulo D, Perez F, Plessis A, Rosse C, Saule S, Stoppa-Lyonnet D, Vincent A, White M, Legrain P, Wojcik J, Camonis J, Daviet L. Protein interaction mapping: a Drosophila case study. Genome Res. 2005;15:376–384. doi: 10.1101/gr.2659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Pan D. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev. 2001;15:1383–1392. doi: 10.1101/gad.901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosens I, Sessa A, den Hollander AI, Letteboer SJ, Belloni V, Arends ML, Le BA, Cremers FP, Broccoli V, Roepman R. FERM protein EPB41L5 is a novel member of the mammalian CRB-MPP5 polarity complex. Exp. Cell Res. 2007;313:3959–3970. doi: 10.1016/j.yexcr.2007.08.025. [DOI] [PubMed] [Google Scholar]
- Goulev Y, Fauny JD, Gonzalez-Marti B, Flagiello D, Silber J, Zider A. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr. Biol. 2008;18:435–441. doi: 10.1016/j.cub.2008.02.034. [DOI] [PubMed] [Google Scholar]
- Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- Hergovich A, Bichsel SJ, Hemmings BA. Human NDR kinases are rapidly activated by MOB proteins through recruitment to the plasma membrane and phosphorylation. Mol. Cell Biol. 2005;25:8259–8272. doi: 10.1128/MCB.25.18.8259-8272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton HN, Stanford PM, Harris J, Oakes SR, Kaplan W, Daly RJ, Ormandy CJ. KIBRA interacts with discoidin domain receptor 1 to modulate collagen-induced signalling. Biochim. Biophys. Acta. 2008;1783:383–393. doi: 10.1016/j.bbamcr.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Jia J, Zhang W, Wang B, Trinko R, Jiang J. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 2003;17:2514–2519. doi: 10.1101/gad.1134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- Kango-Singh M, Nolo R, Tao C, Verstreken P, Hiesinger PR, Bellen HJ, Halder G. Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development. 2002;129:5719–5730. doi: 10.1242/dev.00168. [DOI] [PubMed] [Google Scholar]
- Kremerskothen J, Plaas C, Buther K, Finger I, Veltel S, Matanis T, Liedtke T, Barnekow A. Characterization of KIBRA, a novel WW domain-containing protein. Biochem. Biophys. Res. Commun. 2003;300:862–867. doi: 10.1016/s0006-291x(02)02945-5. [DOI] [PubMed] [Google Scholar]
- Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, Ho LL, Li Y. Control of Cell Proliferation and Apoptosis by Mob as Tumor Suppressor, Mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Maitra S, Kulikauskas RM, Gavilan H, Fehon RG. The tumor suppressors Merlin and Expanded function cooperatively to modulate receptor endocytosis and signaling. Curr. Biol. 2006;16:702–709. doi: 10.1016/j.cub.2006.02.063. [DOI] [PubMed] [Google Scholar]
- Matakatsu H, Blair SS. Separating the adhesive and signaling functions of the Fat and Dachsous protocadherins. Development. 2006;133:2315–2324. doi: 10.1242/dev.02401. [DOI] [PubMed] [Google Scholar]
- McCartney BM, Kulikauskas RM, LaJeunesse DR, Fehon RG. The neurofibromatosis-2 homologue, Merlin, and the tumor suppressor expanded function together in Drosophila to regulate cell proliferation and differentiation. Development. 2000;127:1315–1324. doi: 10.1242/dev.127.6.1315. [DOI] [PubMed] [Google Scholar]
- McClatchey AI, Giovannini M. Membrane organization and tumorigenesis--the NF2 tumor suppressor, Merlin. Genes Dev. 2005;19:2265–2277. doi: 10.1101/gad.1335605. [DOI] [PubMed] [Google Scholar]
- Meignin C, varez-Garcia I, Davis I, Palacios IM. The salvador-warts-hippo pathway is required for epithelial proliferation and axis specification in Drosophila. Curr. Biol. 2007;17:1871–1878. doi: 10.1016/j.cub.2007.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikeladze-Dvali T, Wernet MF, Pistillo D, Mazzoni EO, Teleman AA, Chen YW, Cohen S, Desplan C. The growth regulators warts/lats and melted interact in a bistable loop to specify opposite fates in Drosophila R8 photoreceptors. Cell. 2005;122:775–787. doi: 10.1016/j.cell.2005.07.026. [DOI] [PubMed] [Google Scholar]
- Nolo R, Morrison CM, Tao C, Zhang X, Halder G. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr. Biol. 2006;16:1895–1904. doi: 10.1016/j.cub.2006.08.057. [DOI] [PubMed] [Google Scholar]
- Oh H, Irvine KD. In vivo regulation of Yorkie phosphorylation and localization. Development. 2008;135:1081–1088. doi: 10.1242/dev.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi S, Guntert P, Koshiba S, Tomizawa T, Akasaka R, Tochio N, Sato M, Inoue M, Harada T, Watanabe S, Tanaka A, Shirouzu M, Kigawa T, Yokoyama S. Solution structure of an atypical WW domain in a novel beta-clam-like dimeric form. FEBS Lett. 2007;581:462–468. doi: 10.1016/j.febslet.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Okada T, You L, Giancotti FG. Shedding light on Merlin's wizardry. Trends Cell Biol. 2007;17:222–229. doi: 10.1016/j.tcb.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Pantalacci S, Tapon N, Leopold P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat. Cell Biol. 2003;5:921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- Papassotiropoulos A, Stephan DA, Huentelman MJ, Hoerndli FJ, Craig DW, Pearson JV, Huynh KD, Brunner F, Corneveaux J, Osborne D, Wollmer MA, Aerni A, Coluccia D, Hanggi J, Mondadori CR, Buchmann A, Reiman EM, Caselli RJ, Henke K, de Quervain DJ. Common Kibra alleles are associated with human memory performance. Science. 2006;314:475–478. doi: 10.1126/science.1129837. [DOI] [PubMed] [Google Scholar]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, al-Herr ME, Grant D, Marcinko M, Miyazaki WY, Robertson S, Shaw KJ, Tabios M, Vysotskaia V, Zhao L, Andrade RS, Edgar KA, Howie E, Killpack K, Milash B, Norton A, Thao D, Whittaker K, Winner MA, Friedman L, Margolis J, Singer MA, Kopczynski C, Curtis D, Kaufman TC, Plowman GD, Duyk G, Francis-Lang HL. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- Pellock BJ, Buff E, White K, Hariharan IK. The Drosophila tumor suppressors Expanded and Merlin differentially regulate cell cycle exit, apoptosis, and Wingless signaling. Dev. Biol. 2006 doi: 10.1016/j.ydbio.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesello C, Tapon N. Salvador-warts-hippo signaling promotes Drosophila posterior follicle cell maturation downstream of notch. Curr. Biol. 2007;17:1864–1870. doi: 10.1016/j.cub.2007.09.049. [DOI] [PubMed] [Google Scholar]
- Praskova M, Xia F, Avruch J. MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr. Biol. 2008;18:311–321. doi: 10.1016/j.cub.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayala SK, den HP, Manavathi B, Talukder AH, Song C, Peng S, Barnekow A, Kremerskothen J, Kumar R. Essential role of KIBRA in co-activator function of dynein light chain 1 in mammalian cells. J. Biol. Chem. 2006;281:19092–19099. doi: 10.1074/jbc.M600021200. [DOI] [PubMed] [Google Scholar]
- Rogulja D, Rauskolb C, Irvine KD. Morphogen control of wing growth through the Fat signaling pathway. Dev. Cell. 2008;15:309–321. doi: 10.1016/j.devcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel H, Hofmann K. A novel interaction motif, SARAH, connects three classes of tumor suppressor. Curr. Biol. 2003;13:R899–R900. doi: 10.1016/j.cub.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr. Biol. 2006;16:2081–2089. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Sun J, Deng WM. Notch-dependent downregulation of the homeodomain gene cut is required for the mitotic cycle/endocycle switch and cell differentiation in Drosophila follicle cells. Development. 2005;132:4299–4308. doi: 10.1242/dev.02015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Deng WM. Hindsight mediates the role of notch in suppressing hedgehog signaling and cell proliferation. Dev. Cell. 2007;12:431–442. doi: 10.1016/j.devcel.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber DA, Hariharan IK. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Thompson BJ, Cohen SM. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell. 2006;126:767–774. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Tyler DM, Baker NE. Expanded and fat regulate growth and differentiation in the Drosophila eye through multiple signaling pathways. Dev. Biol. 2007;305:187–201. doi: 10.1016/j.ydbio.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- Verdu J, Buratovich MA, Wilder EL, Birnbaum MJ. Cell-autonomous regulation of cell and organ growth in Drosophila by Akt/PKB. Nature Cell Biol. 1999;1:500–506. doi: 10.1038/70293. [DOI] [PubMed] [Google Scholar]
- Wei X, Shimizu T, Lai ZC. Mob as tumor suppressor is activated by Hippo kinase for growth inhibition in Drosophila. EMBO J. 2007;26:1772–1781. doi: 10.1038/sj.emboj.7601630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, Zhang X, Halder G. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr. Biol. 2006;16:2090–2100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Willecke M, Hamaratoglu F, Sansores-Garcia L, Tao C, Halder G. Boundaries of Dachsous Cadherin activity modulate the Hippo signaling pathway to induce cell proliferation. Proc. Natl. Acad. Sci. U. S. A. 2008;105:14897–14902. doi: 10.1073/pnas.0805201105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev. Cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: The Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- Yang CH, Axelrod JD, Simon MA. Regulation of Frizzled by fat-like cadherins during planar polarity signaling in the Drosophila compound eye. Cell. 2002;108:675–688. doi: 10.1016/s0092-8674(02)00658-x. [DOI] [PubMed] [Google Scholar]
- Yu J, Poulton J, Huang YC, Deng WM. The hippo pathway promotes Notch signaling in regulation of cell differentiation, proliferation, and oocyte polarity. PLoS. ONE. 2008;3:e1761. doi: 10.1371/journal.pone.0001761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev. Cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, Zheng P, Ye K, Chinnaiyan A, Halder G, Lai ZC, Guan KL. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, Lauwers GY, Thasler W, Lee JT, Avruch J, Bardeesy N. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.