Summary
Menin is a tumor suppressor that is mutated in patients with multiple endocrine neoplasia type I (MEN1), an inherited tumor-prone syndrome. Since there is no obvious conserved structural domain in menin that suggests a function, little is known as to how menin suppresses tumorigenisis. Although menin interacts with a variety of nuclear proteins including transcription factors, it is unknown whether menin itself can directly bind DNA. Here we show that menin directly binds to double stranded DNA in a sequence-independent manner. It also binds a variety of DNA structures, including Y-structures, branched structures, and 4-way junction structures. The C-terminus of menin mediates binding to DNA, but MEN1 disease-derived mutations in the C-terminus abolish the ability of menin to bind DNA. Importantly, these MEN1 disease-related menin mutants also fail to repress cell proliferation as well as cell cycle progression at G2/M phase. Furthermore, detailed mutagenesis studies indicate that positively charged residues in two nuclear localization signals mediate direct DNA binding as well as repression of cell proliferation. Collectively, these results demonstrate, for the first time, a novel biochemical activity of menin, binding to DNA, and link its DNA binding to the regulation of cell proliferation.
Keywords: menin, MEN1, DNA binding, cell proliferation, inhibition
INTRODUCTION
Menin, a tumor suppressor consisting of 610 amino acid residues, is primarily a nuclear protein (1). Men1, the gene encoding menin, is frequently mutated in patients with multiple endocrine neoplasia type I (MEN1), an inherited tumor syndrome characterized by multiple endocrine tumors (2,3). There are no obvious motifs in menin that suggest a tumor-suppressing function. Consequently, how menin suppresses tumorigenesis is poorly understood (2,3). Identification of menin-interacting proteins has provided new insights into the function of menin (4). For example, menin interacts with a diverse of group of transcription factors and co-regulators, including JunD, NF-κB, Pem, Smad3 and mSin3A-histone deacetylase (5–9). These reports suggest a role for menin in the regulation of gene transcription.
Menin has also been shown to participate in the regulation of cell proliferation and genome stability. For instance, ectopic expression of menin in Ras-transformed NIH3T3 cells represses cell proliferation as well as tumorigenesis (10). In addition, overexpression of menin inhibits the proliferation of a human insulinoma cell line (11). Numerous reports also indicate a potential role for menin in the regulation of DNA repair and genome stability (12–15). Consistent with these reports, menin has recently been shown to interact with FancD2, a protein that participates in DNA repair (16,17), as well as with RPA2, a critical component of the single stranded DNA binding protein RPA (18,19). Many tumor suppressors are involved in the regulation of multiple cellular processes. For example, Brca1, a tumor suppressor mutated in patients with inherited breast cancer, is also involved in the regulation of gene transcription, cell proliferation, and genome stability (20,21). Recently, Brca1 has been shown to directly bind DNA in a sequence-independent manner (22).
Given the fact that menin associates with various DNA binding proteins as well as chromatin (4,18), we sought to test whether menin directly binds DNA and, if it does, whether binding to DNA is crucial for menin’s function in regulating cell proliferation. In the current studies, we demonstrate that menin directly binds DNA. Menin also binds a variety of other DNA structures and the C-terminus of menin is responsible for direct binding to DNA. Importantly, MEN1-patient-derived mutations in the C-terminus abolish the ability of menin to bind DNA and also fail to repress cell proliferation. These results uncover a mechanistic link between DNA binding by menin and its role in repressing cell proliferation.
EXPERIMENTAL PROCEDURES
Plasmid construction
Oligonucleotides were synthesized by Integrated DNA Technologies, Inc. To generate GST-F1, GST-F2, and GST-F3, which encode amino acid residues 1–218, 219–395, and 396–610, respectively, these menin fragments were amplified by PCR and cloned into the BamHI/Not I site of pGEX-6P (Amersham-Pharmacia Biotech). To generate mutations in the carboxyl terminus of menin, GST-F3 was mutated by site-directed mutagenesis to generate GST-F3-527X, GST-F3-558X, GST-F3-NLS1pm, GST-F3-NLS2pm, and GST-F3-NLS1+2pm, using a Quick Exchange site-directed mutagenesis kit (Stratagene). To express the full-length menin, the PCR amplified cDNA was cloned into pET21b (Novagen), and the protein was expressed in in E. coli strain BL21 (DE3) as a C-terminal His6-tagged proteins. The retroviral expression construct for either menin or Flag epitope-tagged menin was generated as previously described (18), and its various NLS mutants were also generated by site-directed mutagenesis.
Gelshift assays
The DNA probes for gelshift assays were radio-labeled in the presence of γ-32P-ATP and T4 polynucleotide kinase (23). The four different structures of oligonucleotide DNA were as follows: linearized double strand (ds) DNA (5’-CTGCAGGGTTTTTGTTCCAGTCTGTAGCACTGTGTAAGACAGGCCA-3’ and its antisense sequence), Y-structure (5’-GGCTTAGACACTGTGCACAGTGCTACAGACTGGAACAAAAACCCTGCAG-3’ and 5’CTGCAGGGTTTTTGTTCCAGTCTGTAGCACTGTGGAAGACAGGCCAGATC-3’), branched structure (the Y-structure annealed with 5’-CACAGTGTCTAAGCC-3’), and 4-way junction structure (5’TGCATGCTGAGACTTCTCATTACACAGTGCTACAGACTGGAACAAAAACCCTGCA G-3’, GACCTGGCACGTAGGACAGCATGGGATCTGGCCTGTCTTACAGTACAATGCATTGTA CATGAACGTAGCATC-3, 5-GATGCTACGTTCATGTACAATGCATTGTACTGTAATGAGAAGTCTCAGCATGCA-3, and 5’-CTGCAGGGTTTTTGTTCCAGTCTGTAGCACTGTGTAAGACAGGCCAGATCCCATGCT GTCCTACGTGCCAGGTC-3), as previously described (22). The sequence of the DNA for generating probes with random sequences was 5’-CACTCGAGGGATCCGAATTC-N(25)-TCTAGAAAGCTTGTCGACGC-3’, as previously described (24). The plasmid DNA, puc19 (Invitrogen), was linearized with BamH1, dephosphorylated with calf intestine phosphatase, and end-labeled with γ-32P-ATP in the presence of T4 polynucleotide kinase. Gelshift assays were carried out in 1 × gelshift buffer containing the following components, as previously described (22): 20 mM Tris-Hcl (pH 7.4), 60 mM NaCl, 4 mM DTT, 0.1 mg/ml BSA, 0.1% Triton X-100, and radio-labeled DNA probe (105 dpm) in a volume of 10–14 µl. Reactions were incubated at room temperature for 10 min before separation on 0.7% agarose gels in 0.5×TBE buffer at 5 V/cm for 2–4 hrs. The gels were dried onto 3 MM Whatman paper and exposed to films and Phosphor Imager for quantitation.
Cell lines and tissue culture
Human embryonic kidney (HEK) 293 cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% (v/v) fetal calf serum, penicillin (100 units/ml), and streptomycin (100 µg/ml) as previously described (25). Menin-null and menin-complemented mouse embryonic fibroblasts (MEFs) were generated and maintained as previously described (18).
Protein expression and purification
Exponentially growing DH5α E.coli cells harboring various GST-menin constructs were induced with 150 µM IPTG at room temperature. The bacteria were collected and lysed at 4°C in 1×PBS buffer with protease inhibitors and Triton-X-100 (1%), followed by sonication. The supernatant from the lysates was incubated with Glutathione Sepharose™ 4B beads (Amersham Parmacia Biotech) and the beads were washed extensively with 1×PBS buffer with protease inhibitors prior to elution with the elution buffer containing 10 mM glutathione. To express the full-length menin, with or without small deletions, the corresponding constructs were transformed in E. coli strain BL21 (DE3) and the proteins were expressed as C-terminal His6-tagged proteins. Proteins were purified to homogeneity over a Ni-NTA agarose resin (Qiagen), followed by anion exchange chromatography (Source-15Q) and gel filtration (Superdex-200, Amersham-Biotech).
Analysis of apoptosis and cell cycle using flourescence activating cell scanning (FACS)
For PI/BrDU staining, on day 0, cells were seeded at a density of 2 × 105 cells/100 mm dish. On day 2, cells were pulsed for 45 minutes with 10 µM 5’-Bromo-2’-Deoxyuridine-5’-Triphosphate (BrDU, Sigma). Following harvesting, 10 6 cells per sample were fixed in 70% ethanol while vortexing. Cell pellets were washed in washing buffer (Phosphate Buffered saline plus 0.5% Bovine Serum Albumin), pelleted, and denatured in denaturing solution (3N HCl and 0 .5% Tween-20) for 10 minutes. After pelleting, cells were then incubated with anti-BrDU antibody (Pharmingen) for 20 minutes, followed by washing and pelleting. Next, FITC-conjugated goat anti-mouse IgG (Pharmingen) antibody was added for 15 minutes. After another washing and pelleting, cells were resuspended in 0.5 mL propidium iodide (Sigma, 10 µg/mL in PBS) and incubated for 30 minutes. Samples were analyzed on a FACSCalibur (Becton-Dickinson).
For Annexin V staining, cells were seeded at a density of 2 × 105 cells/100 mm dish on day 0. On day 2, cells and supernatant were collected and stained with Annexin-V-fluorescein isothiocyanate (FITC), using the Annexin V-FITC Apoptosis Detection Kit (MBL, Inc., Woburn, MA). Duplicate samples were analyzed on a FACSCalibur (Becton-Dickinson).
Western blotting and immunofluorescent staining
Proteins from cell lysates or various cellular fractions were separated on SDS-PAGE gels, transferred to Hybond C+ membranes (Amersham-Pharmacia Biotech), and blotted with various primary antibodies, followed by incubation with appropriate horseradish peroxidase-conjugated secondary antibodies. The blotted proteins were visualized using the enhanced chemiluminescence detection system (Amersham-Pharmacia Biotech). For immunofluorescent staining, cells were set up on coverslips and processed for incubation with the primary antibody (M2) and the FITC-conjugated secondary antibody (Jackson ImmunoResearch Laboratories, Inc.) as previously described (18) and the stained cells were photographed under a fluorescence microscope (Nikon).
3H-Thymidine incorporation assay
On day 0, cells in 2ml of normal medium were seeded in each well of a 6-well plate. On day 4, 3H-thymidine (ICN Biomedicals Inc) was added to the medium. 3H-thymidine was first diluted in Opti-MEM (Gibco-BRL) and then was added to each well. Triplicate samples were incubated at 37 °C, 5% CO2 for 4 hours before being trypsinized and harvested. Incorporation of 3H-thymidine into cells was determined using a Model 1450 Microbeta Trillux Scintillation Counter (Wallac), as previously described (26).
RESULTS
Menin binds double stranded DNA
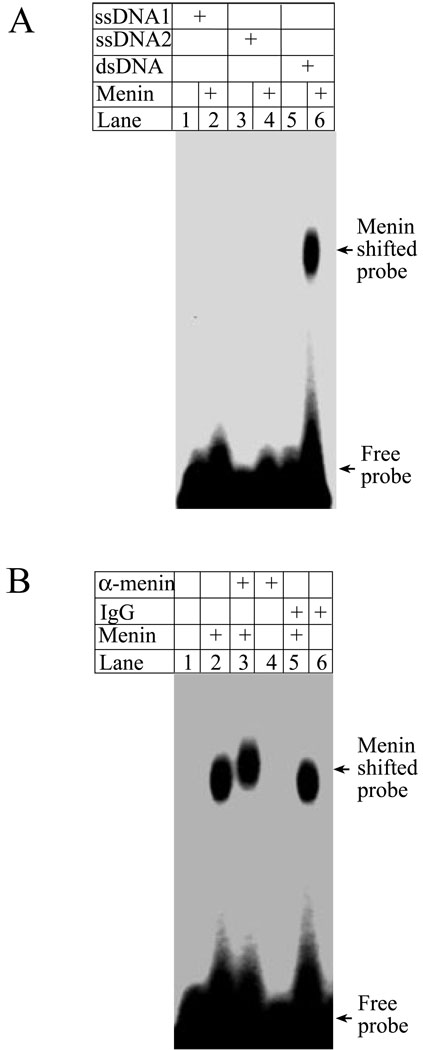
Menin is primarily a nuclear protein that interacts with a variety of other DNA binding proteins as well as chromatin (1,18). The tumor suppressor gene Brca1, which also represses cell proliferation (27), binds DNA in a sequence-independent manner (22). However, it is unclear whether menin itself binds DNA, either in a sequence-specific or non-specific manner. To examine whether menin binds DNA directly, we incubated full-length menin with either a radio-labeled single stranded DNA (ssDNA) probe or a double stranded DNA (dsDNA) probe, each of which is 65 base pairs (bp) in length. These probes were used to characterize the DNA binding by tumor suppressor Brca1 (22). To detect the potential menin-DNA interaction, the reaction was performed in the absence of non-specific competitive DNA, and separated in agarose gels. Figure 1A shows that neither of the single stranded DNA probes binds to menin (lanes 2 and 4); however, the dsDNA probe is shifted by menin (lane 6). These results indicate that menin binds dsDNA. Consistent with this, an affinity-purified anti-menin antibody supershifts the menin-bound DNA probe (Fig. 1B, lane 3), although the control IgG fails to supershift menin-bound DNA (lane 6). Collectively, these experiments demonstrate that menin binds dsDNA. As a note, menin-DNA complexes were unable to migrate out from the wells of polyacrylamide gels (data not shown, A. Silva), which have small pore sizes, whereas they were able to do so in agarose gels, which have larger pores. This is similar to what was described for the DNA binding by another tumor suppressor gene, Brca1 (22). Collectively, these results suggest that multiple molecules of menin may bind to a DNA probe.
Figure 1. Menin binds to double stranded DNA.
(A) Full-length menin protein (3 µg) was incubated with 32P-labeled single stranded DNA or double stranded DNA, as described in EXPERIMENTAL PROCEDURES, and subjected to gelshift assays. (B) Anti-menin antibody specifically supershifts menin that binds double stranded DNA. Anti-menin antibody (1 µg) or control IgG (1 µg) was used in the reactions for gelshift assays.
Menin binds DNA in a sequence-independent manner
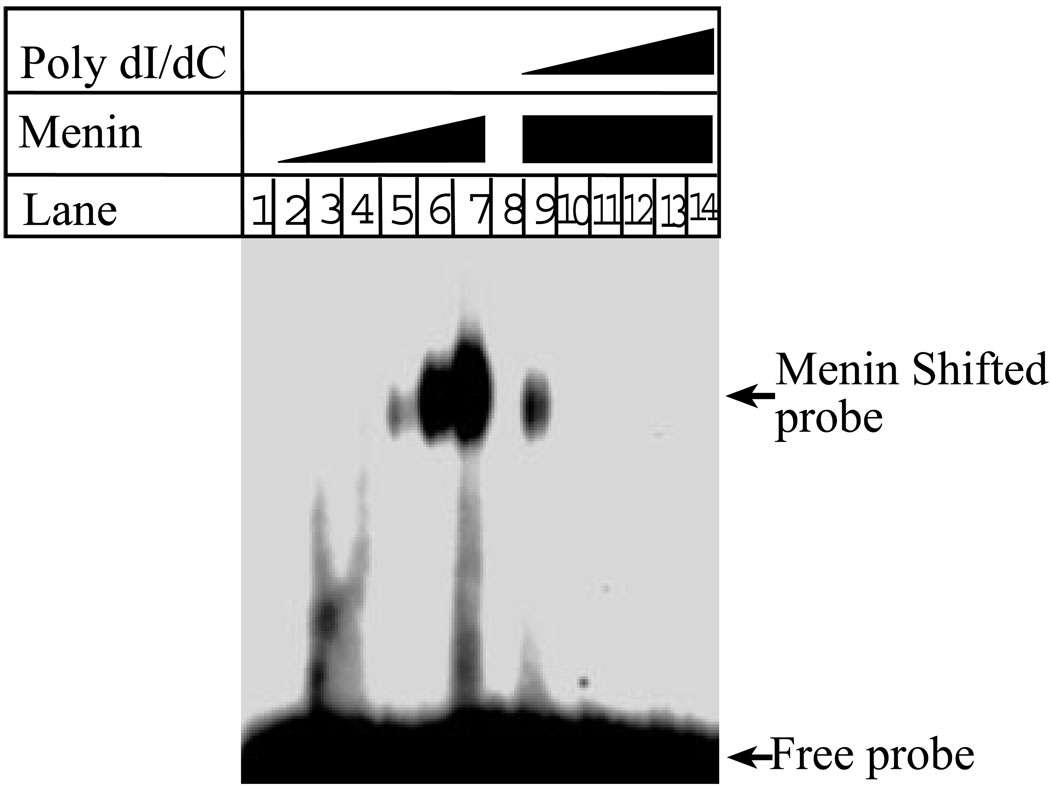
There are various DNA binding proteins with functions ranging from the regulation of gene transcription to the regulation of DNA replication. For example, transcription factors usually bind to sequence-specific DNA (24). However, proteins involved in DNA replication or repair, including human replication origin binding proteins, Brca1 and Rad50, bind DNA in a sequence-non-specific manner (22,28,29). To determine whether menin binds DNA in a DNA sequence-specific or non-specific fashion, we synthesized a pool of 65-mer oligonucleotides that harbor random nucleotide sequence in the middle region. The oligonucleotides were converted to dsDNA, and then used as a DNA probe for gelshift assays. Figure 2 shows that menin binds to the DNA with random sequences (lanes5–7). Notably, the ability of menin to bind to the random DNA is competed by poly dI/dC, a homopolymer of dsDNA (lanes 9–14). These results indicate that menin not only binds to DNA in a non-sequence specific fashion, but that the homopolymer of DNA competes with this non-sequence specific binding, further suggesting that the DNA binding by menin is not sequence-specific in this condition. To further demonstrate that menin is indeed able to bind DNA in sequence-independent manner, we incubated full-length menin with linearized and radio-labeled Puc19, a plasmid of 2.6 kb (kilobases), and examined whether menin is also able to bind this long stretch of DNA. The results show that menin indeed shifts the plasmid DNA probe (data not shown, A. Silva).
Figure 2. Menin binds DNA in a sequence-independent manner.
Increasing concentrations of full-length menin (0, 0.063, 0.125, 0.25, 0.5, 1.0, 2.0 µg per 10 µl) were incubated with the short dsDNA probe, and then subjected to gelshift assays (lanes 1–7). The binding to DNA by menin was competed with increasing concentrations of sequence-non-specific poly dI/dC (lanes 9–14) in the gelshift assay.
The C-terminus of menin mediates binding to DNA
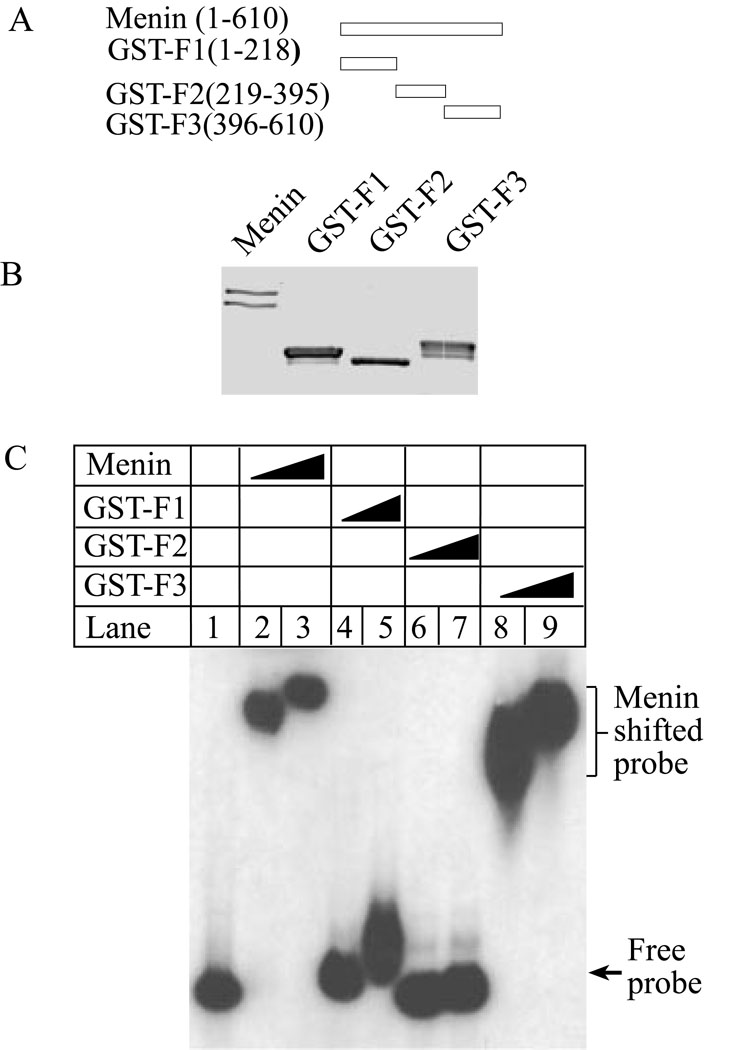
Since there are no motifs or domains in menin that suggest any obvious DNA binding activity, we sought to determine whether such a domain exists in menin. To this end, we generated constructs to express various GST-menin fusion proteins that span three various regions-the N-terminus, the middle region, and the carboxyl terminus of menin (Fig. 3A). The full-length menin and its various fragments were expressed and purified from E. coli, and their sizes and purity were determined on SDS-PAGE by Comassie blue staining (Fig. 3B). These various menin fusion proteins were incubated with a plasmid DNA probe for gelshift assays (Fig. 3C). As expected, full-length menin binds the probe DNA (Fig. 3C, lanes 2–3). The N-terminus does not appear to dramatically shift the DNA probe (lanes 4–5). The middle region of menin, spanning from amino acid residues 219–395, does not shift the DNA probe (Fig. 3C, lanes 6–7). Notably, even at the lowest concentration of the protein used the carboxyl terminus of menin shiftes the DNA probe completely, (Fig. 3C, lanes 8–9). These results demonstrate that the carboxyl terminus is the major domain in menin that mediates binding to DNA in a sequence-independent fashion.
Figure 3. The carboxyl-terminus of menin is responsible for binding to DNA.
(A) A diagram of menin and its fragments used in the experiment. (B) A Coomassie Blue staining of menin and various GST-menin fragment fusion proteins (5 µg). The low band in lane 1 represents the form partially proteolyzed during purification. (C) The carboxyl terminus of menin binds to DNA. Full-length menin, GST-F1, GST-F2, and GST-F3 (0.5 and 1.0 µg for each) were incubated with puc19 plasmid DNA probe for gelshift assays.
Menin binds to various DNA structures
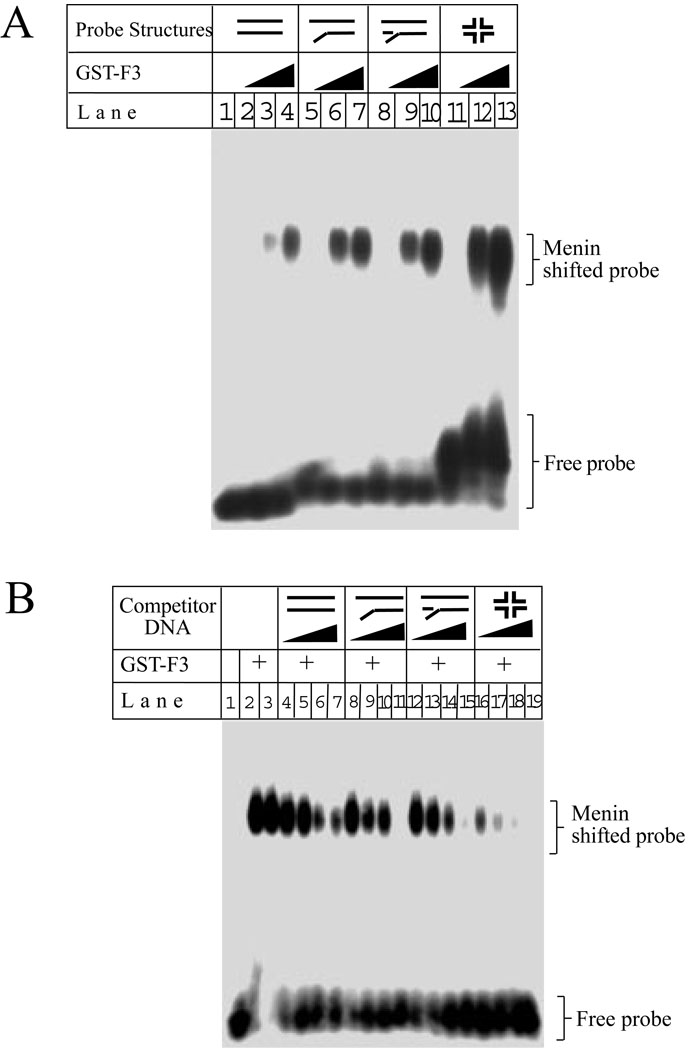
DNA structures encountered in DNA metabolism include regular dsDNA, Y-structures, branched structures, and 4-way junction structures, a 4-strand cross structure analogous to Holiday structure (30,31). BRAF35, a protein associating with the tumor suppressor Brca2, preferentially binds to 4-way structures (32) and plays a critical role in regulating cell proliferation (32). Thus, we examined whether menin binds to these various DNA structures. Figure 4A shows that the carboxyl terminus of menin containing amino acid residues 396–610, binds linearized dsDNA (lanes 3–4), Y-structures (lanes 6–7), branched structures (lanes 9–10), and 4-way structures (lanes 12–13). Similary, full-length menin also binds these four DNA structures (data not shown). To further support the binding of menin to these various structures, a GST- menin (F3) fusion protein was incubated with the Y structure DNA probe and its binding to the probe was competed by each of the following 4 various DNA structures: linearized, Y-shaped, branched, and 4-way junction structures. Figure 4B shows that each of the four structures competed with the DNA probe for binding to menin. However, at the lowest concentration of the competitor DNA used (36 nM), the 4-way structure DNA competed away approximately 60% of the bound probe DNA, while the other three structures only competed away 20–25% of the bound DNA probe (Fig. 4B, lanes 4, 8, 12, and 16). Similar results were obtained when the full-length menin was used in the gelshift assays with various competitive DNA structures (data not shown). The stronger competition for binding to the probe by the 4-way structure may reflect the higher mass of the 4 way structure at an equal molar concentration, since the molecular weight for the 4 way structure is more than twice that of the Y structure probe (78839 vs 30540). Together, these results indicate that menin can bind to various DNA structures.
Figure 4. Menin binds to dsDNA, Y-structures, branched structures, and 4-way junction structures.
(A) GST-menin (GST-F3, 0.0, 0.5, and 1.0 µg as indicated) binds to the four distinct DNA structures in gelshift assay. (B) GST-menin (GST-F3, 0.5 µg) was incubated with a Y-structure probe, in the presence of increasing concentrations (0, 0.036, 0.071, 0.143, and 0.286 µM) of each of the following DNA structures: dsDNA, Y-structures, branched structures, and 4-way structure for gelshift assays.
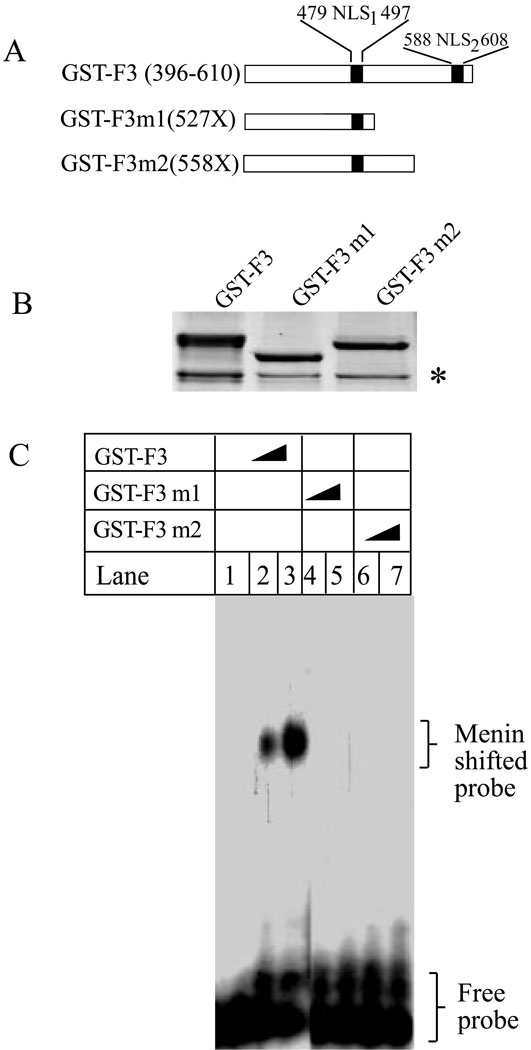
MEN1-related mutations in the C-terminus compromise menin’s ability to bind DNA as well as to inhibit cell proliferation
Over 63% of mutations in the Men1 gene eventually lead to loss of the carboxyl terminus (33). Thus, we sought to determine whether a common disease-related mutation at the carboxyl terminus, R527X, and a second mutation, M558X, interferes with the ability of menin to bind to DNA. Both mutants retain the first nuclear localization signal (NLS1, 479–497), but lose NLS2 (588–608). The wild type C-terminus of menin (GST-F3) and its two truncation mutants, 527X and 558X, were expressed and purified from E. Coli (Fig. 5B). The purified menin and its mutants were incubated with a short dsDNA probe. Figure 5C shows that, as expected, the wild type C-terminus binds the probe (lanes 2–3). However, the two mutants fail to bind to the DNA (lanes 4–7). These results indicate that two common mutations from MEN1 patients lose the ability to bind DNA, suggesting that disruption of menin-DNA binding may at least in part related to the development of MEN1.
Figure 5. MEN1-related menin mutations, menin 527X and 558X, fail to bind DNA.
(A) A diagram of the two MEN1-related mutations. (B) Coomassie Blue staining of GST-F3 , GST-F3-527X, and GST-F3-558X (3.8µg). The asterisk (*) denotes a non-specific band . (C) GST-F3 (0, 0.5, and 1.0 µg), GST-F3-527X, and GST-F3-558X (0, 0.5 and 1.0 µg) were incubated with a Y-structure probe for gelshift assays.
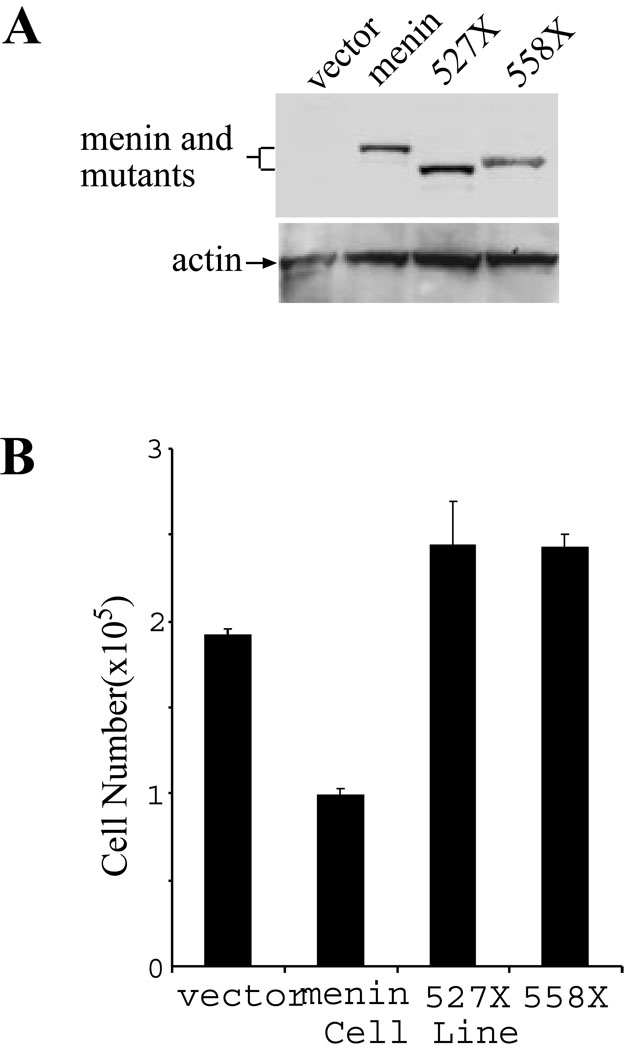
To further examine the effects of these MEN1-related menin mutants on the regulation of cell proliferation, we introduced wild type menin or the two mutants into a menin-null cell line (18) via retroviral infection. Expression of the wild type and mutant menin was confirmed by Western blotting analysis (Fig. 6A, top panel). Equal loading of the cell lysates is confirmed by detection of the levels of actin by Western blot analysis (Fig. 6A, bottom panel). These various cell lines were seeded for culture and the number of cells for each of the cell lines was counted on day 4 of culture, to determine the rate of cell proliferation (Fig. 6B). The results indicate that complementation of the menin-null cells with wild type menin represses cell proliferation. However, the complementation of the menin-null cells with the MEN1-related menin mutants, 527X and 558X, fails to repress cell proliferation (Fig. 6B). These results suggest that that DNA binding by menin may play a crucial role in repressing cell proliferation.
Figure 6. MEN1-related menin mutations abrogate the ability of menin to repress cell proliferation.
(A). Menin-null cells (#21) were infected with control retroviruses or retroviruses expressing each of the following forms of menin: menin (wild type), 527X, and 558X. The expression of the wild type and mutant menin, as indicated, was detected by Western blot analysis using an anti-menin antibody (#77). As a control, the amount of actin in each lysates was detected using an anti-actin antibody (bottom panel). (B). The MEFs expressing the wild type or mutant menin were seeded at a density of 2 × 105 cells per 100 mm dish in duplicate on day 0, and counted on day 4.
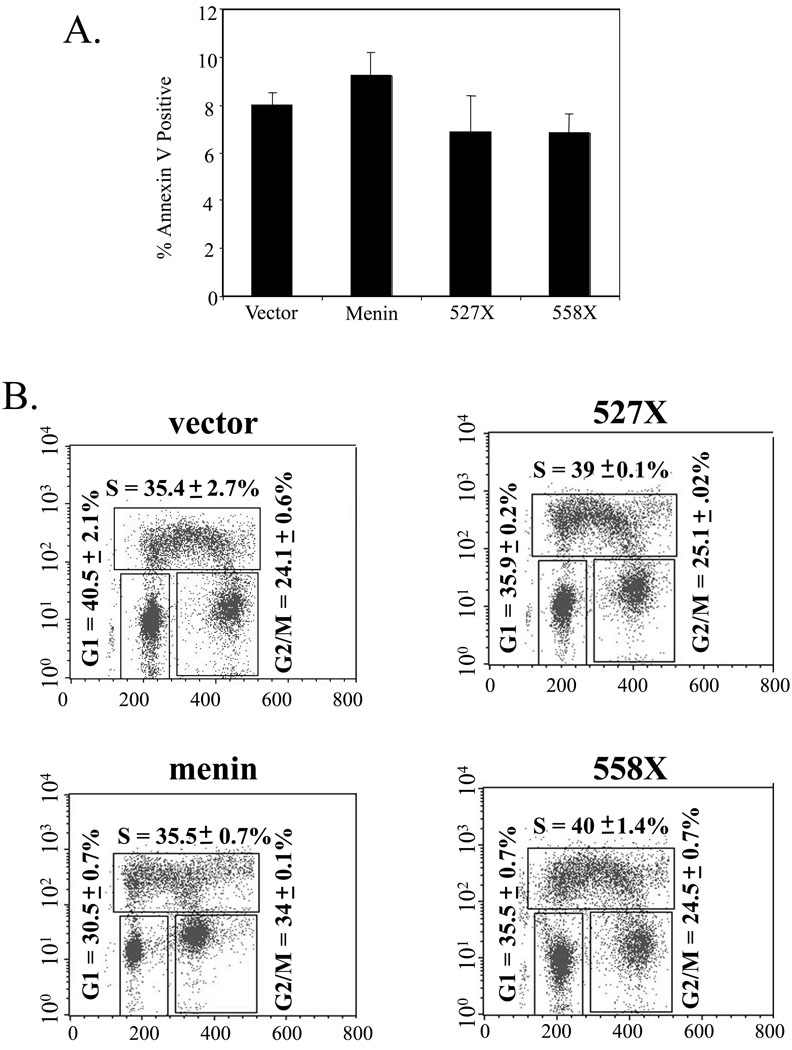
We previously observed that menin potentiates TNF-α-induced apoptosis (34). Thus we sought to determine whether menin and its MEN1-related mutants alter the basal level of apoptosis and steady state of cell cycle distribution in our culture conditions. To this end, control cells or cells expressing wild type menin or mutants were examined for apoptotic cells, using Annexin V staining followed flow cytometry analysis. Figure 7A shows that control cells (vector) contain 8% apoptotic cells, menin-expressing cells contain 9% apoptotic cells, while both mutant 527X and 558X-expressing cells contain 7% apoptotic cells. These results suggest that menin does not obviously upregulate the basal level of apoptosis. However, we cannot completely rule out the possibility that a slight increase of apoptosis in menin-expressing cells could in part contribute to the menin-mediated inhibition of accumulation of cell numbers, because a low level of rapidly degrading apoptotic cells may escape detection.
Figure 7. Analysis of apoptosis and cell cycle profiles in cells expressing wild type or mutant menin.
(A) Annexin V staining of menin-null MEFs expressing vector, menin, 527X, or 557X. Cells were seeded at a density of 2 × 105 cells/100 mm dish and allowed to grow for 48 hours. Cells and supernatant were then harvested and stained with Annexin V as described in Materials and Methods. Data are presented as an average of two independent experiments. (B) Cell cycle profiles of menin-null MEFs expressing vector, menin, 527X, or 557X. Cells were seeded at a density of 2 × 105 cells/100 mm dish and allowed to grow for 48 hours. Cells were then processed and stained with PI and anti-BrDU antibody as described in Materials and Methods. This is representative of two independent experiments.
We further determined the steady-state distribution of various cell cycle phases in the above cell lines. Control cells or cells expressing wild type or mutant menin were pulsed with BrdU and processed for analysis with flow cytometry. Figure 7B shows that control cells in G1, S, and G2/M phases account for 40.5%, 35.4%, and 24.1%, respectively. Distribution of menin-expressing cells is 30.5%, 35.5%, and 34%, respectively. These results show that menin decreases the number of cells in G1 but increases the number of cells in G2/M, suggesting a potential block in G2/M phase. Consistent with this, both menin 527X and 558X mutants have a cell cycle distribution similar to that in menin-null cells-36%, 39–40%, and 25% for G1, S, and G2/M, respectively (Fig. 7B). We also sought to determine the cell cycle progression in these cell lines by synchronizing the cells using serum starvation or various cell cycle blocking drugs. However, serum starvation failed to synchronize the cells and various drugs including mimosine and hydroxyurea were highly toxic to the cells. Collectively, these results suggest that menin affects cell proliferation by affecting cell cycle progression, and its ability to bind DNA is crucial for menin's role in suppressing cell proliferation and MEN1.
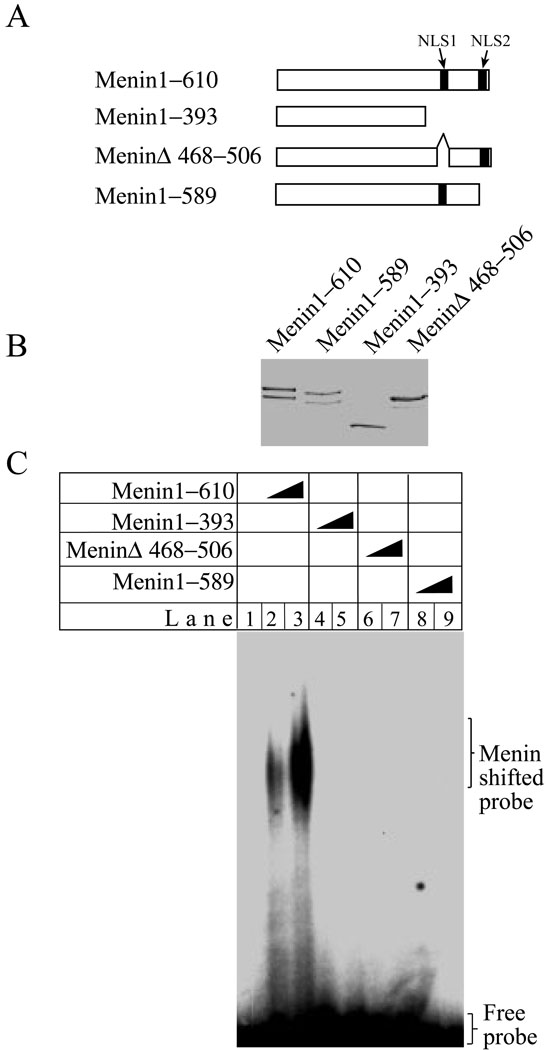
Positively charged residues in the two NLSs in the C-terminus of menin are essential for direct DNA binding as well as repression of cell proliferation
Since the two nuclear localization signals (NLS) are located in the C-terminus (1), which binds DNA, we examined whether the two NLSs play a role in binding to DNA. Thus, menin proteins containing either a truncation of the C-terminus, or internal deletions of small regions of the NLSs were expressed and purified from E. Coli, and quantity and purity of the proteins were verified by Coomassie Blue staining (Fig. 8A and B). These various menin proteins were incubated with a DNA probe for gelshift assays. As expected, the full-length menin binds to the DNA probe (Fig. 8C, lanes 2–3). In contrast, truncation of the C-terminus (lanes 4–5), deletion of NLS1 (lanes 6–7) and deletion of NLS2 (8–9), abolish the ability of menin to bind DNA.
Figure 8. Mutations in each of two nuclear localization signals (NLS) of the C-terminus of menin abolish binding of menin to DNA.
(A) A diagram of the mutations in the two NLSs of menin. (B) Full-length menin and various mutants (5 µg each) as shown in (A) were stained with Coomassie Blue. (C) Menin (0, 0.5, and 1.0 µg) and its various mutants (0.5, and 1.0 µg) were incubated with a Y-structure probe for gelshift assays.
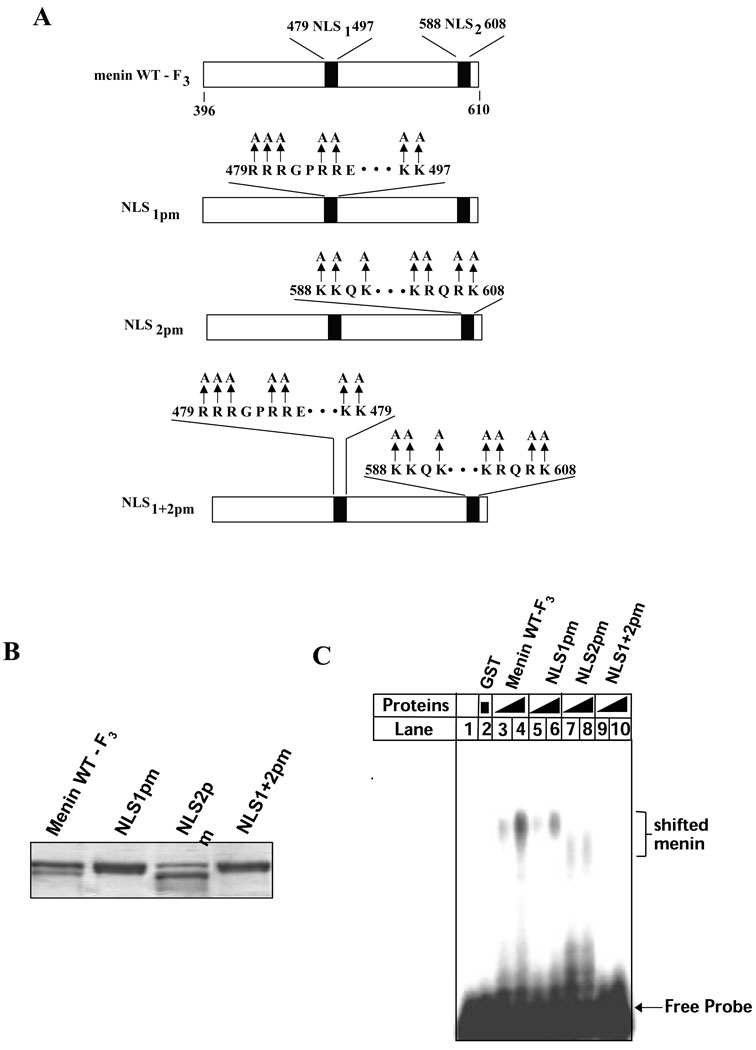
Since the deletion of several amino acid residues in the NLS sequences could affect the conformation of the C-terminus that alters DNA binding, we sought to introduce point mutations, replacing the positively charged residues arginines or lysines with alanines, into the NLSs as shown in Figure 9A. The point mutations should generate more subtle changes to the protein. The resulting GST-fusion proteins with various point mutations were expressed and purified for gelshift assays (Fig. 9B). Figure 9C shows that the wild type C-terminus of menin binds the DNA probe, as expected (lanes3–4), but introduction of point mutations into either NLS1 (479–497) (lanes 5-) or NLS2 (588–608) (lanes7–8) dramatically decreases its ability to bind DNA. Combined point mutations in both NLS1 and NLS2 abolish the ability of menin to bind DNA. These results demonstrate that the NLS1 and NLS2 are essential for the role of menin in direct DNA binding, and these short sequences may directly contact DNA structure.
Figure 9. Positively charged resides in NLS1 and NLS2 are essential for direct DNA binding.
(A) A diagram for wild type or mutant GST-menin (F3, C-terminus) fusion proteins. (B) Wild type or mutant GST-menin fusion proteins (3 ug) were separated on SDS-PAGE and stained with Commassie Blue. (C) Various GST-menin fusion proteins (0.5 or 1.0 µg per lane), as indicated, were used for gelshift assays.
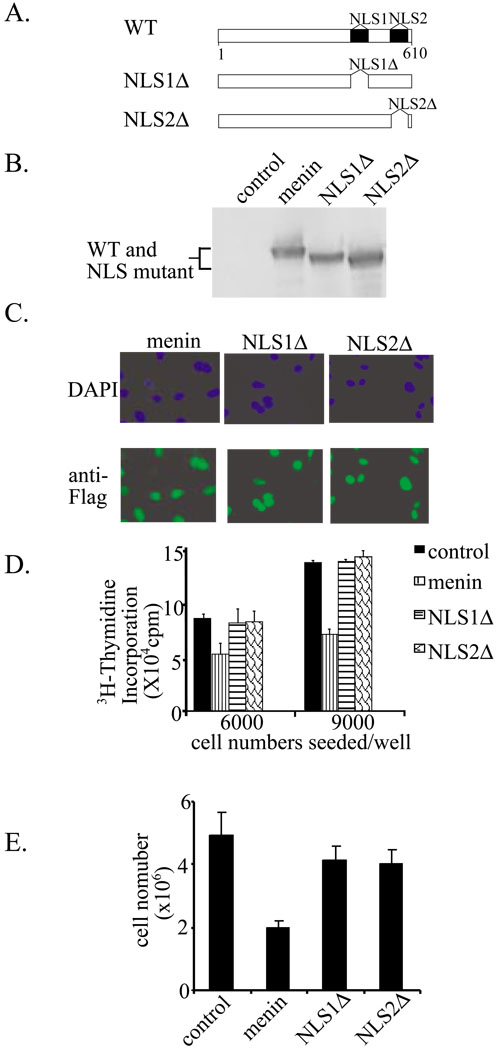
We further determined the effect of the NLS alterations on menin’s nuclear localization and its role in regulation of cell proliferation. Menin-null cells were infected with retroviruses expressing wild type menin or its two mutants (Fig. 10A). The expression of the various proteins was confirmed by Western blotting analysis (Fig. 10B). Additionally, nuclear localization of the wild type and menin mutants was examined by immunofluoresence staining (Fig. 10C). Mutations in either NLS1 or NLS2 do not affect the nuclear localization of menin (Fig. 10C). We examined whether mutation of these NLSs, which compromise DNA binding by menin, affects menin’s role in repression of cell proliferation. To this end, 3H-thymidine incorporation was used to examine the rate of cell proliferation in menin-null MEFs complemented with either wild type menin or the various mutants described above. Figure 10D indicates that complementation of the menin-null cells with wild type menin suppresses cell proliferation. Conversely, mutation in either NLS1 or NLS2, although still localizing to the nucleus (Fig. 10C), fails to suppress cell proliferation. To further strengthen this conclusion, the same numbers of cells for each of these cells were seeded, and the number of the cells from each of the 4 cell lines was counted on day 4 of culture (Fig. 10E). These results indicate that loss of either the NLS1 or NLS2 sequence results in failure of menin to repress cell proliferation. Therefore, although mutation in one of the two positively charged NLSs does not disrupt targeting to the nucleus, it does deprive menin's ability of binding DNA as well as suppressing cell proliferation. Collectively, these results indicate that association of menin with DNA, at least in part, plays a crucial role in suppressing cell proliferation.
Figure 10. Mutations of two nuclear localization signals (NLS) in the C-terminus of menin abolish menin-induced inhibition of cell proliferation.
(A) A diagram of menin and its various NLS mutants. NLS1Δ, deletion of amino acids 479–497. NSLΔ, deletion of amino acids 588–608. (B) Western blot analysis of wild type and various menin mutants from the menin-null MEFs that were infected with retroviruses expressing these proteins. Anti-Flag M2 antibody was used. (C) Nuclear localization of menin and its NLS mutants. Menin-null cells complemented with menin or its various NLS mutants were seeded and processed for immunofluorescence staining with an anti-Flag antibody. (D) The NLS mutants fail to repress cell proliferation. The menin-null MEFs complemented with wild type menin or its various NLS mutants. These cells were seeded in 6-well plates and cell proliferation was assayed by 3H-thymidine incorporation at day 4 of culture. (E) Menin-null cells infected were with various retroviruses as indicated, seeded on day 0, (2×105), and counted on day 4.
DISCUSSION
The current studies have uncovered the ability of menin to directly bind DNA, and further identified the C-terminus of menin and the positively charged residues in the NLSs as the domain responsible for direct DNA binding. Importantly, two MEN1-related mutations in this domain fail to bind DNA as well as to repress cell proliferation (Fig. 5–Fig. 6). These results link the role of menin in DNA binding to the suppression of cell proliferation. They suggest a model in which the interaction between menin and DNA allows menin to coordinate DNA metabolism and the regulation of cell proliferation.
Menin directly binds DNA
DNA binding proteins can generally be categorized as sequence-dependent or sequence-independent proteins. For example, transcription factors are usually sequence-dependent DNA binding proteins (24). On the other hand, the interaction of other DNA binding proteins with DNA is dependent on overall DNA structure, but not on the precise sequence. In other words, they bind DNA in a sequence-non-specific fashion. These include proteins that regulate DNA repair and gene transcription, such as Brca1 (20,27). Rad 50 and human DNA replication origin binding proteins, which are involved in DNA repair, also bind DNA in a manner that is independent of the precise DNA sequence (28,29). There are yet other DNA binding proteins, such as p53 which possesses domains that bind DNA in both a sequence–dependent and - independent manner (35,36). Menin has been shown to bind a variety of proteins including transcription factors, such as JunD, NF-µb, Smad3, Pem (4), as well as proteins involved in DNA repair, such as RPA and FancD2 (18,19). However, it has not previously been shown to directly bind DNA.
We show that menin binds directly to dsDNA (Fig. 1), but this binding does not favor a specific DNA sequence (Fig. 2–Fig. 3). Further experiments indicate that menin binds to a variety of DNA structures including regular dsDNA, Y-structures, branched structures, and 4-way junctions. Thus, menin may associate with various DNA structures to regulate gene transcription, cell proliferation, and DNA repair.
Although it is shown here that menin binds DNA in a sequence-independent manner, these findings do not rule out the possibility that menin also binds some DNA sequence specifically, using either the C-terminus or other regions. As described above, the tumor suppressor p53 (also a transcription factor) was originally described as a protein that bound to DNA non-specifically (35,37,38). In addition, p53 was later found to possess domains for sequence-specific and sequence-non-specific DNA binding (36). Therefore, it cannot be ruled out that under certain circumstances menin may be able to bind to specific DNA sequence to regulate gene transcription and/or cell proliferation.
MEN1-related mutations in menin abolish both the ability of menin to bind DNA and to suppress cell proliferation
The C-terminus of menin mediates binding to DNA (Fig. 3). In support of the importance of menin in DNA binding, mutations in the C-terminus of menin, R527X and M558X, abolish the ability of menin to bind to DNA (Fig. 5). These mutants, which have lost positively charged residues in the second NLS, still retain the first NLS, and thus should be able to target to the nucleus, since NLS1 alone is sufficient for targeting menin into the nucleus (Fig. 10). Given the fact that the majority of mutations in the Men1 gene from MEN1 patients leads to the loss of the carboxyl terminus, this correlation between the loss of the C-terminus and the inability for the mutants to bind DNA strengthens the importance of menin-DNA binding in menin’s function. Importantly, these mutants lose their ability to suppress cell proliferation (Fig. 6).
Menin represses cell proliferation and increases the number of G2/M cells, suggesting a potential G2/M block (Fig. 6–Fig. 7). Consistent with this, Menin does not markedly increase basal-level apoptosis (Fig. 7A) (34). Several reports show that ectopic expression of menin leads to reduced accumulation of cell numbers, but it is not clear in these experimental conditions menin affect either cell cycle progression or apoptosis, or both (7,10,11,39). A recent interesting report shows that knock-down of menin by an antisense approach in rat intestinal epithelial cell line, IEC-17, leads to increased expression of cyclin D1 and CDK4 and increased number of S-phase cells (40). This is consistent with a model that menin downregulates the CDK4/6 and pRB pathway and inhibits cell cycle transition for G1 to S phase. However, it is not clear whether ectopic expression of menin in menin-knock-down cells can truly inhibit cell cycle progression from G1 to S phase in the cells. In our MEF system, we observed that expression of menin modestly increases number of cells in G2/M, but decreases cells in G1 (Fig. 7B). This difference is likely due to the different cell lines used. However, it is noteworthy that MEN1-related menin mutants fail to increase cell numbers in G2/M (Fig. 7B). Together, these results suggest that interaction of menin with DNA is crucial for the role of menin in suppressing cell proliferation.
However, these results do not exclude the possibility that the C-terminus may also interact with other partners to suppress cell proliferation. Moreover, it is not yet clear how the menin-DNA interaction facilitates menin-mediated suppression of cell proliferation. It is possible that its proximity to cellular DNA facilitates regulation of the expression of genes that are crucial for regulation of cell proliferation. Alternatively, association of menin with chromatin DNA may enhance its role in the regulation of DNA replication.
The arginines and lysines in the two NLSs in the C-terminus of menin mediates direct DNA binding and repression of cell proliferation
To further study the ability of menin to bind DNA, we made even smaller alterations in the C-terminus sequence, namely internal deletion of NLS1 or NLS2. These minor alterations in the C-terminus indeed abolish the ability of menin to bind DNA (Fig. 8). It is possible that multiple positively charged lysine or arginine residues in the NLSs play a role in DNA binding, simply by binding the negatively charged DNA. This is consistent with the observation that the interaction between menin and DNA is not sequence-specific. Alternatively, this small deletion could also change the structure of the C-terminus, leading to loss of the DNA-binding activities. To address this concern and further pinpoint the precise amino acid residues that are responsible for direct DNA binding, point mutations were introduced to each of the NLSs (Fig. 9A and B). The resulting mutant proteins fail to optimally bind DNA (Fig. 9C). These results definitively demonstrate a crucial role for the NLSs in mediating direct binding to DNA. The positively charged residues in the NLSs may directly contact the negatively charged DNA. However, it is still unclear how menin represses cell proliferation. Perhaps by binding to the chromatin DNA (32), menin senses the status of chromatin DNA and transduces a signal to keep the cell proliferation in check. On the other hand, menin may regulate gene transcription in part by association with chromatin DNA, and some of the target genes may lead to repression of cell proliferation. No matter what the precise mechanisms are, the currently studies uncover a novel role of menin, direct binding to DNA, and link DNA binding to the regulation of cell proliferation and suppression of MEN1.
Acknowledgements
This work is in part supported by a NIH grant (R01 CA113962 to XH), a Research grant from American Cancer Society (RSG-03-055-01-LIB to XH), and a scholar award from Rita Alan Foundation (to XH).
Contributor Information
Ping La, Abramson Family Cancer Research Institute, Department of Cancer Biology and Signal Transduction Program, University of Pennsylvania, Philadelphia, PA 19104-6160, USA.
Albert C. Silva, Abramson Family Cancer Research Institute, Department of Cancer Biology and Signal Transduction Program, University of Pennsylvania, Philadelphia, PA 19104-6160, USA
Zhaoyuan Hou, Abramson Family Cancer Research Institute, Department of Cancer Biology and Signal Transduction Program, University of Pennsylvania, Philadelphia, PA 19104-6160, USA.
Haoren Wang, Abramson Family Cancer Research Institute, Department of Cancer Biology and Signal Transduction Program, University of Pennsylvania, Philadelphia, PA 19104-6160, USA.
Robert W. Schnepp, Abramson Family Cancer Research Institute, Department of Cancer Biology and Signal Transduction Program, University of Pennsylvania, Philadelphia, PA 19104-6160, USA
Nieng Yan, Department of Molecular Biology, Princeton University, Lewis Thomas Laboratory, Washington Road, Princeton, New Jersey 08544, USA.
Yigong Shi, Department of Molecular Biology, Princeton University, Lewis Thomas Laboratory, Washington Road, Princeton, New Jersey 08544, USA.
Xianxin Hua, Abramson Family Cancer Research Institute, Department of Cancer Biology and Signal Transduction Program, University of Pennsylvania, Philadelphia, PA 19104-6160, USA.
References
- 1.Guru SC, Goldsmith PK, Burns AL, Marx SJ, Spiegel AM, Collins FS, Chandrasekharappa SC. Proc Natl Acad Sci U S A. 1998;95:1630–1634. doi: 10.1073/pnas.95.4.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marx SJ, Agarwal SK, Kester MB, Heppner C, Kim YS, Skarulis MC, James LA, Goldsmith PK, Saggar SK, Park SY, Spiegel AM, Burns AL, Debelenko LV, Zhuang Z, Lubensky IA, Liotta LA, Emmert-Buck MR, Guru SC, Manickam P, Crabtree J, Erdos MR, Collins FS, Chandrasekharappa SC. Recent Prog Horm Res. 1999;54:397–438. [PubMed] [Google Scholar]
- 3.Schussheim DH, Skarulis MC, Agarwal SK, Simonds WF, Burns AL, Spiegel AM, Marx SJ. Trends Endocrinol Metab. 2001;12:173–178. doi: 10.1016/s1043-2760(00)00372-6. [DOI] [PubMed] [Google Scholar]
- 4.Poisson A, Zablewska B, Gaudray P. Cancer Letters. 2003;189:1–10. doi: 10.1016/s0304-3835(02)00509-8. [DOI] [PubMed] [Google Scholar]
- 5.Agarwal SK, Guru SC, Heppner C, Erdos MR, Collins RM, Park SY, Saggar S, Chandrasekharappa SC, Collins FS, Spiegel AM, Marx SJ, Burns AL. Cell. 1999;96:143–152. doi: 10.1016/s0092-8674(00)80967-8. [DOI] [PubMed] [Google Scholar]
- 6.Heppner C, Bilimoria KY, Agarwal SK, Kester M, Whitty LJ, Guru SC, Chandrasekharappa SC, Collins FS, Spiegel AM, Marx SJ, Burns AL. Oncogene. 2001;20:4917–4925. doi: 10.1038/sj.onc.1204529. [DOI] [PubMed] [Google Scholar]
- 7.Kaji H, Canaff L, Lebrun JJ, Goltzman D, Hendy GN. Proc Natl Acad Sci U S A. 2001;98:3837–3842. doi: 10.1073/pnas.061358098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim H, Lee JE, Cho EJ, Liu JO, Youn HD. Cancer Res. 2003;63:6135–6139. [PubMed] [Google Scholar]
- 9.Lemmens IH, Forsberg L, Pannett AA, Meyen E, Piehl F, Turner JJ, Van de Ven WJ, Thakker RV, Larsson C, Kas K. Biochem Biophys Res Commun. 2001;286:426–431. doi: 10.1006/bbrc.2001.5405. [DOI] [PubMed] [Google Scholar]
- 10.Kim YS, Burns AL, Goldsmith PK, Heppner C, Park SY, Chandrasekharappa SC, Collins FS, Spiegel AM, Marx SJ. Oncogene. 1999;18:5936–5942. doi: 10.1038/sj.onc.1203005. [DOI] [PubMed] [Google Scholar]
- 11.Sayo Y, Murao K, Imachi H, Cao WM, Sato M, Dobashi H, Wong NC, Ishida T. Endocrinology. 2002;143:2437–2440. doi: 10.1210/endo.143.6.8950. [DOI] [PubMed] [Google Scholar]
- 12.Itakura Y, Sakurai A, Katai M, Ikeo Y, Hashizume K. Biomed Pharmacother. 2000;54 Suppl 1:187s–190s. doi: 10.1016/s0753-3322(00)80041-4. [DOI] [PubMed] [Google Scholar]
- 13.Hessman O, Skogseid B, Westin G, Akerstrom G. J Clin Endocrinol Metab. 2001;86:1355–1361. doi: 10.1210/jcem.86.3.7332. [DOI] [PubMed] [Google Scholar]
- 14.Scappaticci S, Maraschio P, del Ciotto N, Fossati GS, Zonta A, Fraccaro M. Cancer Genet Cytogenet. 1991;52:85–92. doi: 10.1016/0165-4608(91)90057-2. [DOI] [PubMed] [Google Scholar]
- 15.Scappaticci S, Brandi ML, Capra E, Cortinovis M, Maraschio P, Fraccaro M. Cancer Genet Cytogenet. 1992;63:17–21. doi: 10.1016/0165-4608(92)90057-f. [DOI] [PubMed] [Google Scholar]
- 16.Timmers C, Taniguchi T, Hejna J, Reifsteck C, Lucas L, Bruun D, Thayer M, Cox B, Olson S, D'Andrea AD, Moses R, Grompe M. Mol Cell. 2001;7:241–248. doi: 10.1016/s1097-2765(01)00172-1. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 18.Jin S, Mao H, Schnepp RW, Sykes SM, Silva AC, D'Andrea AD, Hua X. Cancer Res. 2003;63:4204–4210. [PubMed] [Google Scholar]
- 19.Sukhodolets K, Hickman A, Agarwal SK, Sukhodolets M, Obungu V, Novotny E, Crabtree J, Chandrasekharappa SC, Collins FS, Spiegel AM, Burns AL, Marx SJ. Molecular and Cellular Biology. 2003;23:493–509. doi: 10.1128/MCB.23.2.493-509.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scully R, Livingston DM. Nature. 2000;408:429–432. doi: 10.1038/35044000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gadhavi PL, Greenwood MD, Strom M, King IA, Buxton RS. Biochem Biophys Res Commun. 2001;281:520–528. doi: 10.1006/bbrc.2001.4375. [DOI] [PubMed] [Google Scholar]
- 22.Paull TT, Cortez D, Bowers B, Elledge SJ, Gellert M. Proc Natl Acad Sci U S A. 2001;98:6086–6091. doi: 10.1073/pnas.111125998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Russell DW. Molecular Cloning, a laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratories Press; 2001. [Google Scholar]
- 24.Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B, Kern SE. Mol Cell. 1998;1:611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- 25.Hua X, Nohturfft A, Goldstein JL, Brown MS. Cell. 1996;87:415–426. doi: 10.1016/s0092-8674(00)81362-8. [DOI] [PubMed] [Google Scholar]
- 26.La P, Morgan T, Sykes S, Mao H, Schnepp R, Petersen C, Hua X. Oncogene. 2003;22:198–210. doi: 10.1038/sj.onc.1206100. [DOI] [PubMed] [Google Scholar]
- 27.Annab LA, Hawkins RE, Solomon G, Barrett JC, Afshari CA. Breast Cancer Res. 2000;2:139–148. doi: 10.1186/bcr45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 29.Vashee S, Cvetic C, Lu W, Simancek P, Kelly TJ, Walter JC. Genes Dev. 2003;17:1894–1908. doi: 10.1101/gad.1084203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Constantinou A, Davies AA, West SC. Cell. 2001;104:259–268. doi: 10.1016/s0092-8674(01)00210-0. [DOI] [PubMed] [Google Scholar]
- 31.Shen J, Loeb LA. Mech Ageing Dev. 2001;122:921–944. doi: 10.1016/s0047-6374(01)00248-2. [DOI] [PubMed] [Google Scholar]
- 32.Marmorstein LY, Kinev AV, Chan GK, Bochar DA, Beniya H, Epstein JA, Yen TJ, Shiekhattar R. Cell. 2001;104:247–257. doi: 10.1016/s0092-8674(01)00209-4. [DOI] [PubMed] [Google Scholar]
- 33.Pannett AA, Thakker RV. Endocr Relat Cancer. 1999;6:449–473. doi: 10.1677/erc.0.0060449. [DOI] [PubMed] [Google Scholar]
- 34.Schnepp RW, Mao H, Sykes SM, Zong WX, Silva A, La P, Hua X. J Biol Chem. 2004;279:10685–10691. doi: 10.1074/jbc.M308073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foord OS, Bhattacharya P, Reich Z, Rotter V. Nucleic Acids Res. 1991;19:5191–5198. doi: 10.1093/nar/19.19.5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolkowicz R, Rotter V. Pathol Biol (Paris) 1997;45:785–796. [PubMed] [Google Scholar]
- 37.Steinmeyer K, Deppert W. Oncogene. 1988;3:501–507. [PubMed] [Google Scholar]
- 38.Kern SE, Kinzler KW, Baker SJ, Nigro JM, Rotter V, Levine AJ, Friedman P, Prives C, Vogelstein B. Oncogene. 1991;6:131–136. [PubMed] [Google Scholar]
- 39.La P, Schnepp RW, Petersen CD, Silva AC, Hua X. Endocrinology. 2004;145:3443–3450. doi: 10.1210/en.2004-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ratineau C, Bernard C, Poncet G, Blanc M, Josso C, Fontaniere S, Calender A, Chayvialle JA, Zhang CX, Roche C. J Biol Chem. 2004 doi: 10.1074/jbc.M401835200. [DOI] [PubMed] [Google Scholar]