Abstract
Factors that drive prostate cancer progression remain poorly defined, thus hindering the development of new therapeutic strategies. Disseminated tumors are treated through regimens that ablate androgen signaling, as prostate cancer cells require androgen for growth and survival. However, recurrent, incurable tumors that have bypassed the androgen requirement ultimately arise. This study reveals that the Brm ATPase, a component of selected SWI/SNF complexes, has significant antiproliferative functions in the prostate that protect against these transitions. First, we show that targeted ablation of Brm is causative for the development of prostatic hyperplasia in mice. Second, in vivo challenge revealed that Brm−/− epithelia acquire the capacity for lobe-specific, castration-resistant cellular proliferation. Third, investigation of human specimens revealed that Brm mRNA and protein levels are attenuated in prostate cancer. Fourth, Brm down-regulation was associated with an increased proliferative index, consistent with the mouse model. Lastly, gene expression profiling showed that Brm loss alters factors upstream of E2F1; this was confirmed in murine models, wherein Brm loss induced E2F1 deregulation in a tissue-specific manner. Combined, these data identify Brm as a major effector of serum androgen–induced proliferation in the prostate that is disrupted in human disease, and indicate that loss of Brm confers a proliferative advantage in prostate cancer.
Introduction
SWI/SNF chromatin remodeling complexes are critical mediators of transcriptional control, as manifest through their ability to mobilize nucleosomes (1, 2). SWI/SNF complexes are recruited to chromatin by sequence-specific transcription factors, and therein, regulate gene expression by either promoting nucleosome dispersion or condensation (3, 4). To control this process, it is increasingly apparent that the specificity of SWI/SNF function is dictated by biochemical diversity in subunit composition, and that imbalances in subunit expression can induce significant, often tissue-specific biological consequence (3, 5–8).
SWI/SNF complexes consist of a central ATPase (Brg1 or Brm), the function of which is required for nucleosome repositioning, a cohort of ‘‘core’’ subunits that are required to reconstitute SWI/SNF function in vitro (BAF47/INI1, BAF155, and BAF170), and combinatorial assembly of six to eight additional BAF (Brg1-associated factor) subunits that confer specificity of function to individual SWI/SNF complexes (3–5). Despite the general requirement of the central ATPase for SWI/SNF function, genetic analyses revealed remarkable functional heterogeneity between Brm and Brg1. For example, loss of Brg1 is an embryonic lethal event (9), whereas Brm−/− mice are viable and develop normally (10). Subsequent studies using conditional deletion of Brg1 revealed divergent roles of this ATPase for skin development, limb morphogenesis, erythropoiesis, vasculature, gliogenesis, neural stem/progenitor cells, and T-cell development (9, 11–15). Although few phenotypes have been detected in Brm−/− mice, these animals are up to 15% heavier than their wild-type littermates, and show the enlargement of selected organs (e.g., liver, kidney, heart) but reductions in others (spleen, testes; ref. 16). Thus, despite the shared capacity of Brg1 and Brm to confer SWI/SNF activity, the ATPases seem to serve distinct biological functions.
In cells of prostatic origin, SWI/SNF proved capable of regulating the androgen receptor (AR), a ligand-dependent transcription factor that is a major effector of prostate cancer growth and progression. AR is the first-line therapeutic target for disseminated prostate cancer, as prostate cancer cells are exquisitely dependent on AR signaling (17, 18). Ligand (androgen) activation of AR induces rapid dimerization and nuclear translocation of the protein, which binds to DNA at androgen-responsive elements and induces a program of gene transcription in prostate cancer cells that results in cell survival and proliferation (19). In addition, AR directs the expression of secretory products that are used clinically to monitor prostate cancer growth and progression (i.e., prostate-specific antigen; refs. 20, 21). Using in vitro models, it was previously shown that AR requires SWI/SNF to induce gene expression on prostate-specific target genes, and that the receptor shows a preference in transient assays for Brm-induced SWI/SNF action (22, 23). Similarly, it was shown in cell-based assays that Brm is the predominant ATPase recruited to sites of AR action after androgen stimulation (22, 23). Through these actions, it was hypothesized that Brm-mediated AR signaling supports androgen-dependent transcriptional activation in the context of the prostate. In prostate cancer therapy, engagement of AR signaling is targeted through the use of regimens that either deprive the receptor of ligand (androgen ablation) or through the use of direct AR antagonists (17, 19, 24). Although these strategies result in tumor remission, recurrent tumors arise within 2 to 3 years in which AR action is restored despite the absence of the ligand (25). Transition to this therapy-resistant phase of tumor progression represents the incurable stage of disease; thus, it is imperative to discern the signaling pathways that influence AR signaling in the prostate. Given the importance of SWI/SNF in controlling AR action in vitro, the present study interrogated the consequence of the Brm ATPase in the prostate using genetically defined in vivo models and analyses of human prostate cancer tumors.
Materials and Methods
Mice, xenografts, and human specimens
Targeted deletion of Brm was previously described (16). Animals were bred and maintained at the Laboratory Animal Medical Services of University of Cincinnati, and tracked by the Institutional Animal Care and Use Committee. Mice were obtained from (Brm+/− × Brm+/−) offspring. Genotypes were determined as previously described (16). Prostates were resected after sacrifice using standard methods, and immediately dissected into the anterior, ventral, and dorsolateral lobes. Dissected tissues were snap-frozen (for protein or RNA collection) or were suspended in 10% neutral buffered formalin and paraffin embedded. For acquisition of human tissue, unidentified specimens from radical prostatectomy were obtained from the University of Cincinnati, Department of Pathology tissue bank, in accordance with Institutional Review Board approval. Tissue microarrays containing benign and malignant tissue cores were obtained from U.S. Biomax (PR801).
Castration/re-supplementation studies
Castration/re-supplementation studies were performed as indicated using standard procedures. Briefly, 12-week-old male mice were castrated under anesthesia, recovered for 14 days, then randomized into cohorts that received a daily subcutaneous injection of testosterone propionate (3 mg/mL, 100 µL) dissolved in sesame oil or vehicle control. On the third day, mice received two i.p. injections of 5-bromo-2-deoxyuridine [150 mg/kg bromodeoxyuridine (BrdUrd); Sigma]. Mice were sacrificed after 7 days of injection and prostates immediately resected.
Histology and immunohistochemistry
H&E staining was performed using standard methodologies. For Brm immunohistochemistry, tissue samples were deparaffinized, antigens retrieved using antigen unmasking solution (Vector Laboratories), incubated in peroxidase blocking reagent (DakoCytomation) and stained with antisera for Brm (AbCam). Visualization was performed using the Vectastain Elite ABC system and 3,3′-diaminobenzidine (Vector Laboratories) and hematoxylin. Specimens were evaluated by a clinical pathologist according to established guidelines, and the immunoreactivity of Brm was scored according to intensity [0 (none), + (low), ++ (moderate), +++ (high)] and extent of tumor staining [0 (none), 1 (<25%), 2 (>25%, <50%), 3 (>50%)]. The final Brm score is displayed as a composite (intensity + extent). For proliferative indices, antisera against Ki67 (DakoCytomation) or BrdUrd (Accurate Chemical) was used according to the guidelines of the manufacturer. The DeadEnd Colorimeteric TUNEL System (Promega) was employed to detect apoptotic cells. For each variable, indices were quantified by counting the total positive and negative nuclei in the epithelial layer. A minimum of 1,000 cells were scored per mouse for the anterior and ventral prostate lobes, whereas at least 500 cells per mouse (due to small size) were tallied for the dorsolateral prostate lobes. Data are presented as the mean ± SD. Statistical analyses were performed using ANOVA. Sample size per condition and P values are as indicated.
Immunoblots and mRNA analyses
Immunoblots were performed using standard methods that have been previously described (26). Briefly, equal protein was separated by SDS-PAGE, transferred to Immobilon PVDF, and protein detected using antisera directed against β-actin (sc-1616), probasin (sc-17126), E2F1 (sc-193), and E2F4 (sc-1082). Protein was visualized using the Odyssey IR system (LiCoR). For mRNA analyses, total RNA was isolated from mouse tissues and fibroblast cells using TRIZOL reagent (Invitrogen) and reverse transcribed into cDNA. Amplifications were performed with primers directed against E2F1 (sense, 5′-GACCCTGCAGAACAGATGG-3′; antisense, 5′-GATGTCTCCTGGCATGAGG-3′) and 18SRNA.
Bioinformatics
Analyses of Brm (SMARCA2) expression were performed with gene expression array data from a cohort of nonneoplastic and malignant prostates and the HG-U133A Affymetrix platform, as previously described (27). Samples were normalized using the Robust Multichip analysis algorithm as implemented in Bioconductor/R (28). Normalized gene expression values for each transcript in each sample were set to its ratio relative to the expression of that transcript’s measurement in nonneoplastic tissue. Genes that correlated (Pearson coefficients, 0.6–1.0) or anticorrelated (Pearson coefficients, −1.0 to −0.6) with the average expression of four probesets that map to SMARCA2 in benign tissue, primary prostate cancer, and metastatic deposits were determined using GeneSpring GX v7.3.1 (Agilent Technologies). Statistically overrepresented (P < 0.05) gene ontologies in both KEGG and BioCarta pathways were identified using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) 2007 (29).
Results
Brm loss causes lobe-specific prostatic hyperplasia
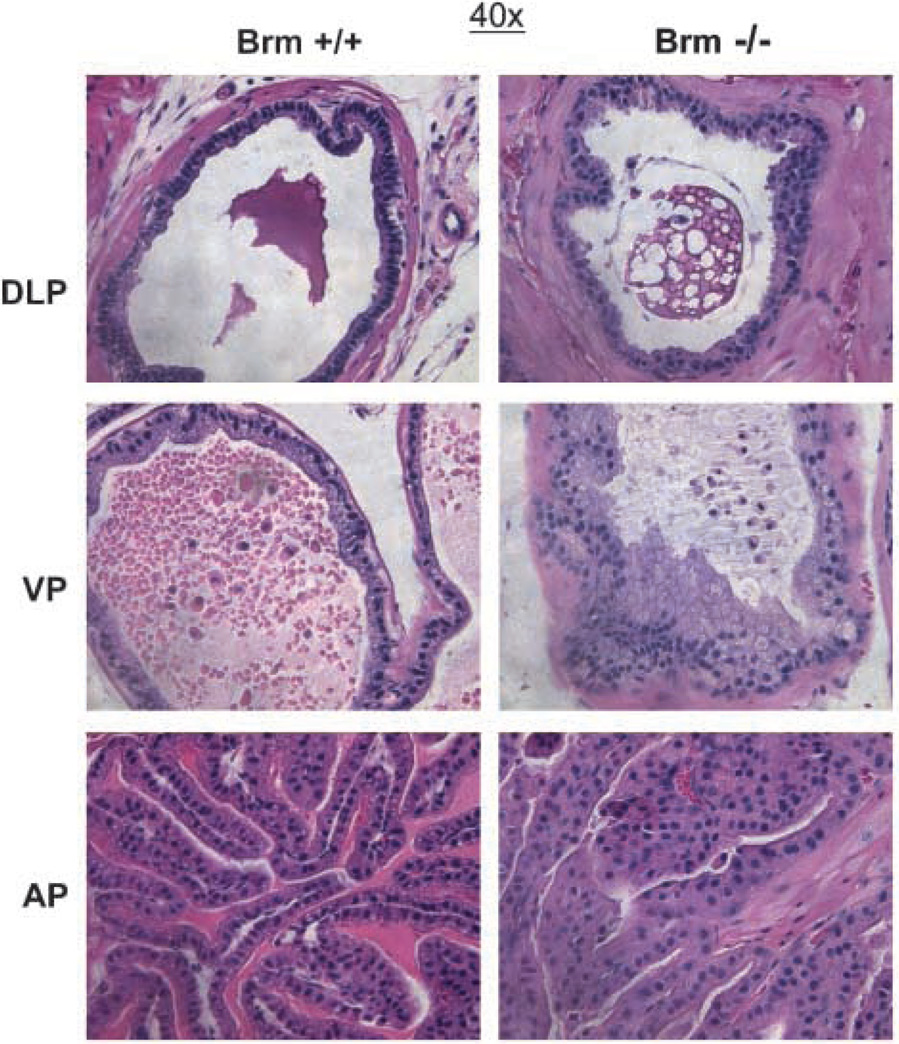
As SWI/SNF-dependent Brm function has been shown in cultured prostate cancer cells to potentially modulate AR activity (22, 23), the importance of Brm in the murine prostate was examined. Immunohistochemistry analyses showed that Brm is expressed in all three prostate lobes (dorsolateral, DLP; ventral, VP, and anterior, AP), with the highest levels of expression observed in the luminal epithelia (Supplementary Fig. S1). To determine the effect of Brm status on the prostatic epithelia, mice harboring targeted deletion of the Brm (SMARCA2) locus were used. Consistent with previous reports, Brm−/− mice generated offspring with approximate Mendelian frequency (16), and the colony was maintained through breeding of heterozygous animals. First, 6-month-old littermates were examined for histoarchitecture of the prostate after resection of discrete lobes and H&E staining (Fig. 1). As shown, no visible distinctions were noted in the DLP, irrespective of genotype (top). Minimal alteration was noted in the VP, although Brm−/− lobes showed sporadic regions of epithelial cell tufting (middle). By contrast, marked hypercellularity and loss of organized glandular structure with cribriform appearance was observed in the AP (bottom). These morphologic changes were compatible with hyperplasia, but no nuclear atypia was observed (Supplementary Fig. S2). Thus, Brm loss is sufficient to induce lobe-specific expansion of the luminal epithelia.
Figure 1.
Brm loss causes lobe-specific disruption of glandular structures in the murine prostate. H&E sections of the DLP, VP, and AP from 6-month-old littermates.
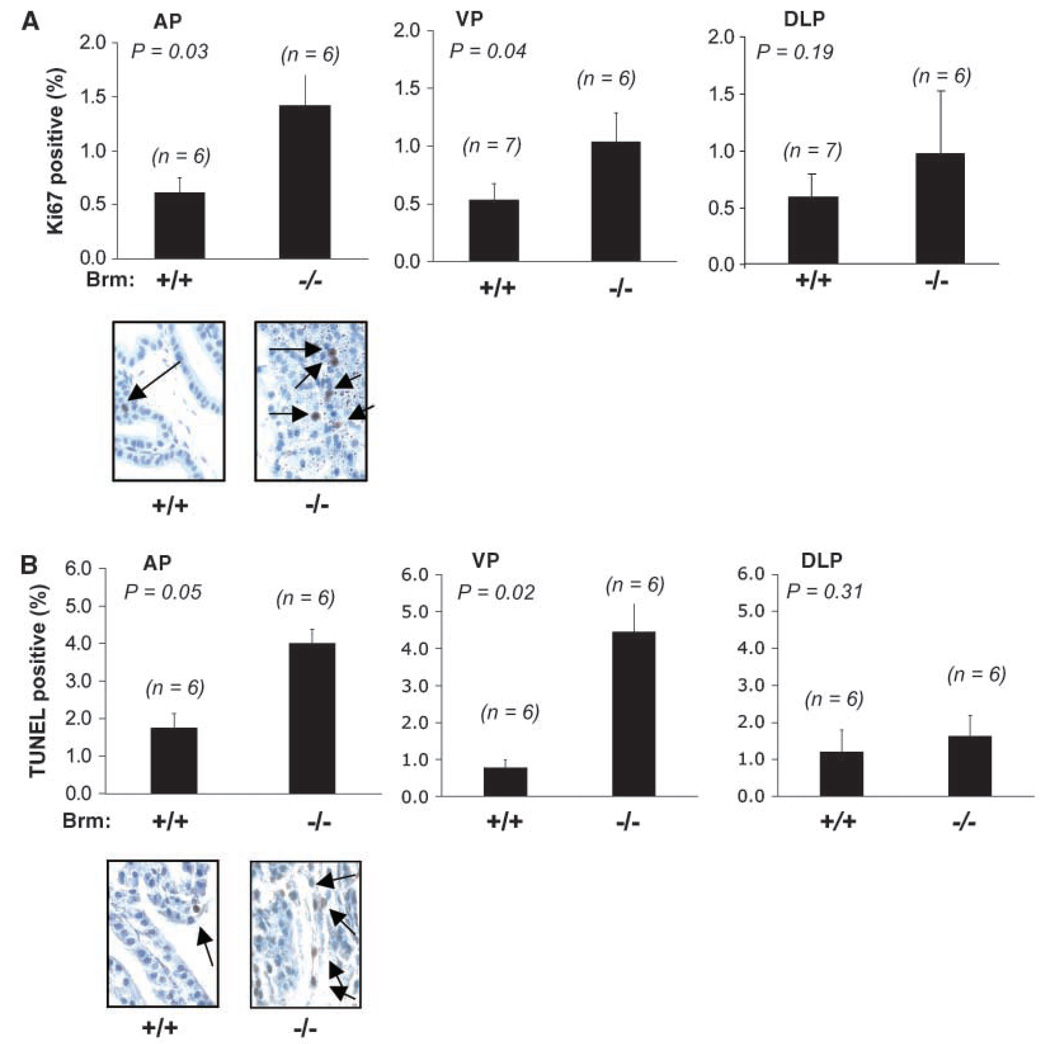
Given these observations, the underpinning cause of luminal cell alterations was investigated by assessing proliferative and apoptotic indices (Fig. 2). As shown in A, the proliferative (Ki67) indices of the epithelial compartment within the AP and VP were significantly enhanced in glands obtained from Brm−/− mice as compared with Brm+/+ littermates (inductions of 2.4-fold and 2.0-fold, respectively). A trend of enhanced proliferation was observed in the DLP, but these data did not reach statistical significance, consistent with the absence of a consistent pathologic phenotype in this lobe. Together, these data suggest that the observed hypercellularity in the AP and modest alterations in VP epithelial cell compartments are likely the result of a hyperplastic phenotype. This phenotype did not consistently progress to neoplasia over the time period of monitoring (up to 12 months of age), as only 1 of more than 20 mice analyzed developed a neoplastic lesion (data not shown). Because genetic alterations which induce epithelial cell hyperplasia in the murine prostate are often accompanied by an induction in the apoptotic index (30, 31), terminal nucleotidyl transferase–mediated nick end labeling (TUNEL) positivity was also quantified (Fig. 2B). In these studies, the apoptotic index was elevated in both the AP (2.3-fold) and VP (5.7-fold) lobes, whereas no alterations were observed in the DLP. Together, these data indicate that Brm loss induces lobe-specific hyperplasia that does not consistently progress to neoplasia, likely as a result of a concomitant induction in the apoptotic rate.
Figure 2.
Brm loss induces hyperplasia of the prostatic epithelia. A, the proliferative indices of Brm+/+ and Brm−/− littermates were determined in unchallenged 6-month-old animals by immunostaining for Ki67. At least 1,000 cells were tallied per animal by a blinded observer for each AP and VP. In the DLP, at least 500 cells were scored per animal, due to the small size of this lobe. Averages, SD, and n numbers are as indicated. A representative example of the AP is provided (bottom). B, lobes in A were processed to detect apoptotic indices by TUNEL staining.
Brm−/− prostatic epithelia remain androgen-responsive but acquire androgen independence
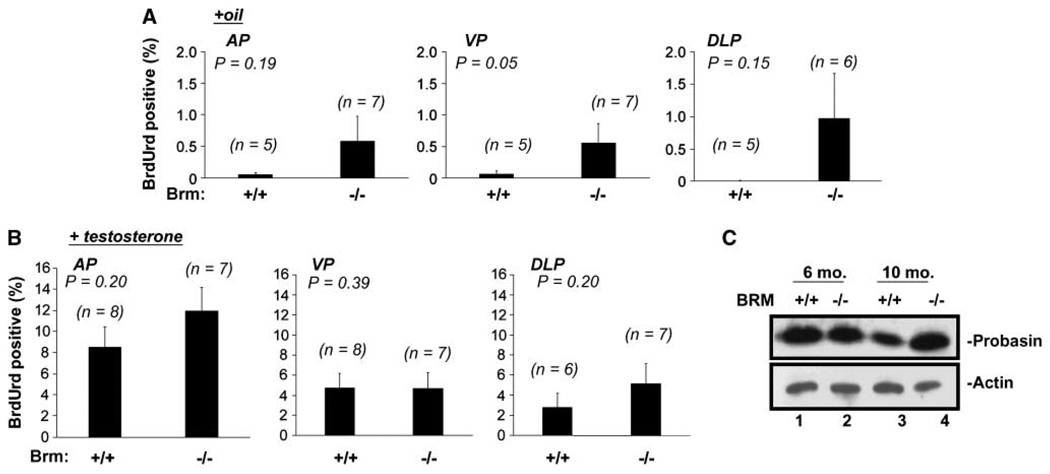
It has been previously shown that androgen ablation causes cell death and/or quiescence of the murine prostatic luminal epithelia (32). Cell death induced by androgen ablation is complete within 14 days, but repopulation of the luminal epithelia can be induced by re-supplementation with testosterone (33). Therefore, the effect of Brm disruption on serum androgen dependence and androgen responsiveness was examined using the schema outlined in Supplementary Fig. S3. Briefly, 3-month-old littermates of each genotype were castrated (to induce androgen ablation), and after 14 days, mice were randomized into two different cohorts. The first cohort received daily injections of vehicle (to monitor serum androgen dependence) for 7 days preceding sacrifice, whereas the second cohort received testosterone injections (to monitor androgen responsiveness). In both, BrdUrd was administered on the 3rd day of injection, so as to capture the proliferative index of cells after castration or re-supplementation. As expected, little BrdUrd incorporation was observed in the epithelia of Brm+/+ animals treated with vehicle alone (Fig. 3A, left bars for each lobe), consistent with the serum androgen dependence of these cells. By contrast, there was a trend of enhanced BrdUrd incorporation in the Brm−/− epithelia of the AP and DLP, which reached statistical significance in the VP (Fig. 3A, right bars), wherein Brm−/− epithelia incurred a 9.1-fold induction of BrdUrd incorporation over Brm+/+ controls. These data indicate that Brm loss can unexpectedly induce castration-resistant proliferation in a subset of epithelia.
Figure 3.
AR signaling is retained in Brm−/− epithelia, and is accompanied by castration-resistant proliferation. A, androgen-independent proliferative indices were determined by quantifying BrdUrd incorporation rates after castration (oil cohort). At least 500 epithelial cells per AP and VP or 150 per DLP were scored per animal. One DLP was excluded from analyses due to its inability to obtain sufficient cell numbers for counting. Columns, mean; bars, SE. B, androgen dependent (AR dependent) proliferation indices were determined in the testosterone cohort, as described in A. C, probasin (AR target gene) expression was monitored by immunoblot from the prostates of 6-month-old or 10-month-old intact animals, as indicated. Actin was included as a loading control.
Because the transition to castration resistance is frequently associated with a heightened sensitivity to androgen, the effect of Brm loss on androgen responsiveness was challenged through re-supplementation studies (Fig. 3B). After testosterone re-supplementation, BrdUrd indices in each lobe of the Brm−/− mouse were indistinguishable from that observed in Brm+/+ littermates. These data indicate that androgen signaling remains intact in the absence of Brm, and that the proliferative response observed in unchallenged animals is not a result of heightened androgen sensitivity. The concept that Brm disruption is sufficient to support evidence of castration resistance in vivo was surprising, as studies in cultured cells have suggested that AR activity primarily uses Brm-dependent SWI/SNF chromatin remodeling activity to support its transactivation function (22, 23), and cells of prostatic origin require AR activity. Therefore, to further test this conclusion, AR activity was monitored in Brm−/− prostates by assessing levels of a well-defined, prostate-specific AR target gene in the mouse, probasin. As shown, no reduction in probasin expression was observed by immunoblot (Fig. 3C) or immunohistochemistry (data not shown) in Brm−/− prostates from unchallenged mice. Together, these data indicate that Brm loss does not compromise the AR response to testosterone, but can induce castration-resistant proliferation in the murine prostate.
Brm expression is attenuated in prostate cancer
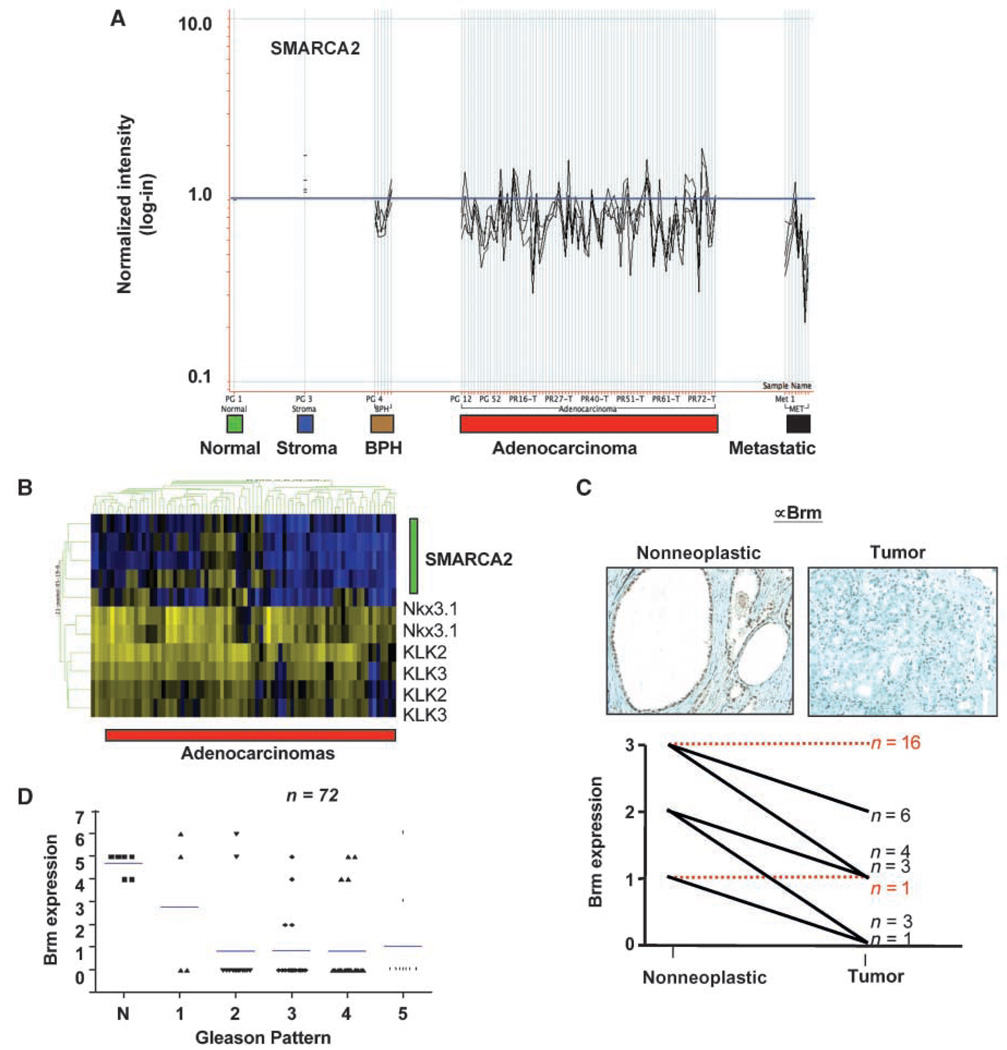
Because targeted disruption of Brm resulted in hyperplasia and castration-resistance in a subset of prostatic epithelia, the status of Brm expression in human prostate cancer was initially determined by gene expression profiling of tumor samples that have been previously characterized (27). Using four probe sets that map to the 3′-end of the SMARCA2 transcript, Brm expression was evaluated relative to nonneoplastic tissue (Fig. 4A). As shown, a significant decrease in Brm levels was observed in tumor specimens, with the lowest expression observed in advanced (metastatic) disease. These findings were validated in a second available data set (34), wherein Brm expression was significantly reduced in tumor versus nonneoplastic tissue (Supplementary Fig. S4). Analyses revealed that the levels of AR target gene expression (KLK3, KLK2, and Nkx3.1) remained unchanged, irrespective of Brm levels (Fig. 4B), consistent with observations in the prostates of Brm−/− mice. Together, these data indicate that reduced Brm expression is not sufficient to alter AR activity in vivo, but is an unexpectedly frequent event in prostate cancer.
Figure 4.
Tumor-specific loss of Brm mRNA and protein is observed in human prostate cancers. A, Brm (SMARCA2) expression was analyzed in benign prostatic hyperplasia (n = 6), primary prostatic adenocarcinomas (n = 79), and metastatic samples of prostate cancer (n = 8; bottom). The relative expression levels as compared with nonneoplastic controls are shown, as determined using all SMARCA2 probe sets that map to the 3′-end of the gene. B, using gene expression analyses for tumors in A, the relative association between SMARCA2 status (using four probe sets, green bar) and AR target gene expression (Nkx3.1, KLK2, and KLK3) was assessed. C, analyses of Brm protein levels in matched nonneoplastic and primary prostate cancer specimens. Top, representative immunostaining. Bottom, quantification of expression from 34 matched samples using a 0 to 3 scale based on the intensity of immunohistochemistry. Relative expression and n for each category. Red dotted lines, the cohorts in which Brm expression was unchanged relative to nonneoplastic; black lines, tumor specimens for which Brm levels were reduced. D, relative Brm expression as a function of Gleason grade, as obtained by tissue microarray. Bars, mean.
To validate these findings, immunohistochemical analyses were first performed on radical prostatectomy specimens, wherein matched tumor and nonneoplastic tissues were available (n = 34). Although Brm expression levels were variable in nonneoplastic tissue, the protein was readily detected in all specimens (Fig. 4C). In matched tumor tissue, attenuation of Brm expression was observed in 47% (16 of 34) of the specimens, wherein Brm levels were reduced as compared with matched tissue. Representative immunostaining is provided (top), and quantification revealed that tumor-specific reductions in Brm expression occurred irrespective of the relative Brm levels in paired nonneoplastic tissue. Finally, Brm levels were examined in a larger cohort of tumor specimens using a tissue microarray (n = 72). These analyses confirmed that Brm is significantly reduced in prostate cancer as compared with nonneoplastic tissues, and that this event can occur even in low-grade disease (Fig. 4D). Collectively, these data are consistent with observations from gene expression analyses, and strongly suggest that Brm down-regulation is a frequent event in prostate cancer.
Low Brm expression induces E2F1 deregulation and confers a proliferative advantage
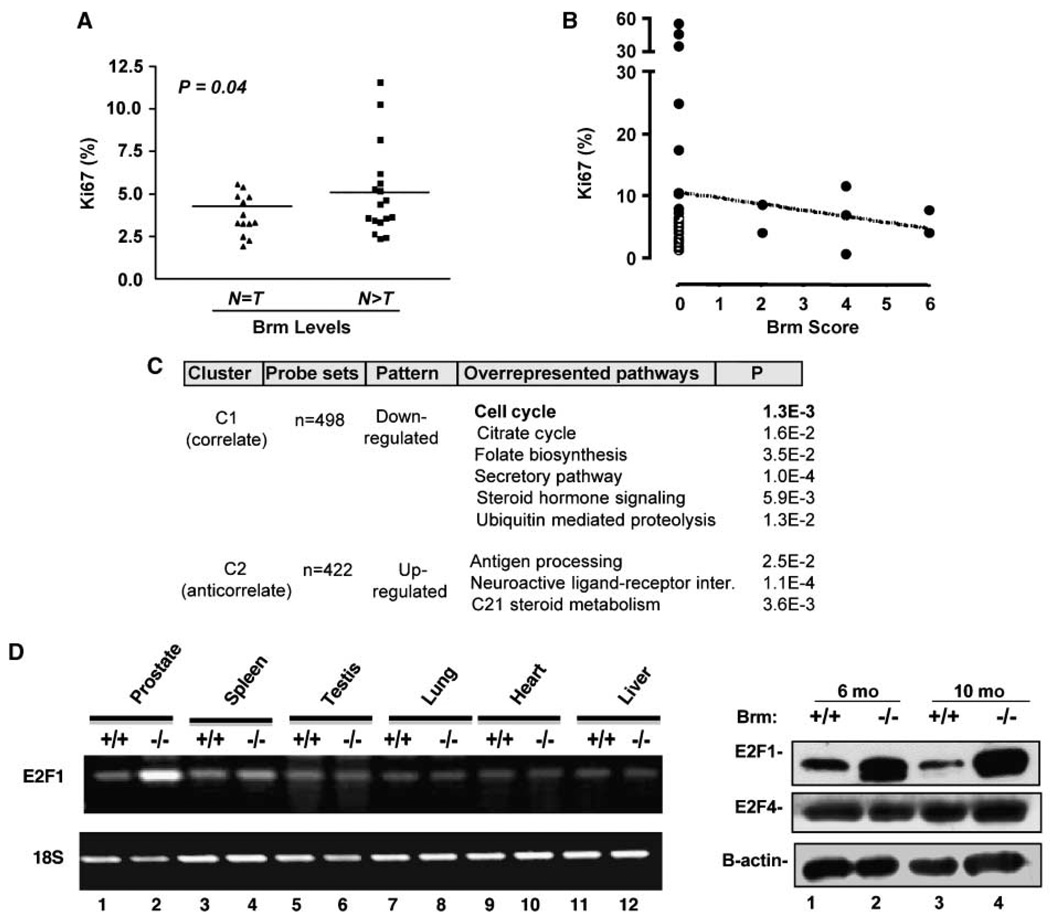
Because Brm expression was lost or reduced with substantive frequency in prostate cancer, the consequence of this event was determined in human disease. Initially, tumors quantified in Fig. 4C for relative Brm levels were assessed for proliferative indices. As shown in Fig. 5A, tumors demonstrating reduced Brm expression showed significant induction of the Ki67 index, as compared with those tumors for which Brm levels were indistinguishable from matched nonneoplastic tissue. To validate these results, the effect of Brm status on the proliferative index in prostate cancer specimens was determined using a tissue microarray, wherein Brm status was inversely proportional to the Ki67 index (Fig. 5B). Together, these data are consistent with observations in the Brm−/− mice, and show that Brm attenuation is associated with a higher proliferative index in prostate cancer.
Figure 5.
Brm loss induces E2F1 deregulation and confers a proliferative advantage in the prostate. A, specimens determined in Fig. 4C as having a tumor-specific loss of Brm expression (N > T) or no change in Brm expression relative to matched nonneoplastic samples (N = T) were analyzed in serial sections for Ki67 indices. B, serial sections from tissue microarray(n = 36) were analyzed to quantify relative Brm and Ki67 expression. C, representation of statistically overrepresented gene pathways that are altered as a function of Brm status (determined using the samples described in Fig. 4A and identified as per Supplementary Figs . S5 and S6 using DAVID). D, relative mRNA expression of E2F1 in tissues from Brm+/+ and Brm−/− littermates (left); protein expression of E2F1, E2F4, and β-actin (loading control) was monitored in prostates from 6-month-old or 10-month-old Brm+/+ and Brm−/− littermates by immunoblot (right).
As Brm regulates a number of sequence-specific transcription factors, underlying causes of the observed hyperplastic phenotypes are likely to be complex. Therefore, gene expression analyses were performed using data from the human prostate cancers assessed for Brm status in Fig. 4A, in which samples that exhibited either high or low Brm expression were used to identify genes that were coordinately regulated. In these analyses, genes were identified that correlated (cluster C2, 498 features) or anticorrelated (cluster C1, 422 features) with Brm mRNA levels (Supplementary Fig. S5A). Functional enrichment analyses of each group were performed, and a summary of overrepresented biological pathway alterations and gene lists are provided in Supplementary Fig. S5B. Gene lists are provided in Supplementary Fig. S6. Given the alterations of proliferative rate observed in Brm−/− mice, it was noteworthy that alterations in cell cycle control were observed in tumors with low Brm−/− expression. Analyses of corresponding alterations revealed a significant pattern of gene regulation that would be predicted to confer a proliferative advantage (summarized in Supplementary Fig. S5C) associated with E2F1 deregulation. For example, activation of transforming growth factor-β suppresses cell cycle control, mediated in part through the ability of Smad2/3 and Smad4 transcriptional complexes to induce the p15ink4b and p21cip1 CDK inhibitors (35, 36). Notably, reductions in Smad2 and Smad4 expression were associated with Brm loss; conversely, induction of tumor necrosis factor-α (which can suppress Smad signaling) also correlated with low Brm expression. These collective data indicate that loss of Brm likely relieves the suppression of the cell cycle by facilitating G1 CDK activity, and it would be predicted that this event would induce excessive RB phosphorylation and promote G1 progression. More proximal to this pathway, it was also observed that Brm down-regulation significantly correlated with reduced expression of RB itself, and down-regulation of the p27kip1 tumor suppressor. Despite these observations, cross-comparison with published data sets of RB target genes revealed that only a subset of RB targets were deregulated upon Brm loss (data not shown). Paramount among these was E2F1, a critical mediator of both cell cycle progression and cell death, which was induced in tumors with low Brm mRNA levels. Indeed, E2F1 anticorrelated with Brm expression (two independent probe sets with Pearson correlations of −0.628 and −0.625); in the second data set (Supplementary Fig. S4), Pearson coefficients were −0.931 and −0.844. As these data potentially implicated Brm as an effector of E2F regulation, E2F1 levels were assessed in the murine prostate.
Consistent with expression analyses in human tumors, E2F1 was enhanced in the prostates of Brm−/− animals at both the mRNA (Fig. 5D, left) and protein levels (right). Alteration in E2F1 activity was specific in this tissue, as no alterations were observed with E2F4. Notably, no E2F1 deregulation was observed in tissues reported to be enlarged in Brm−/− mice (liver and heart). In addition, no change was observed in tissues not reported to be hyperplastic (lung), in tissues that are slightly reduced in size upon Brm loss (testes), or in derived murine embryonic fibroblasts (Fig. 5D; Supplementary Fig. S7). Unexpectedly, a modest induction of E2F1 was observed in the spleen, which is slightly reduced in size in Brm−/− animals. However, the most striking alteration observed was increased E2F1 expression in the prostate. Thus, E2F1 deregulation in response to Brm loss seems to be highly context-specific, and is not required in other tissue types for a hyperproliferative phenotype. Together, these data indicate that loss of Brm expression is sufficient to induce E2F1 deregulation and hyperproliferation in cells of prostatic origin. In summary, the studies herein implicate Brm as a novel suppressor of androgen-dependent proliferation in the prostate, and show that Brm loss is sufficient to induce a tissue-specific and E2F1-associated proliferative advantage in this tissue type.
Discussion
The present study shows that loss of the Brm ATPase is sufficient to induce a hyperplastic phenotype in murine models of the prostate and that Brm down-regulation is observed with high frequency in prostate cancer. Although, Brm function has been proposed to influence AR function in vitro, functional studies in murine models show that AR signaling is retained in Brm−/− animals. By contrast, and consistent with a role in prostate cancer progression, Brm loss is associated with the transition to castration resistance. These outcomes are underpinned by observations in both the Brm−/− prostates and analyses of gene expression profiles in human prostate cancer, wherein attenuation of Brm was associated with a high proliferative index and E2F1 deregulation. Combined, these data identify Brm as a critical modulator of E2F1 expression and growth control in this tissue, and suggest that loss of Brm in prostate cancer confers a proliferative advantage.
The demonstration that Brm down-regulation occurs with high frequency in prostate cancer was not expected, as Brm−/− mice have not been reported to develop tumors in tissues examined previously (16). However, challenge of Brm−/− mice with lung carcinogens was reported to increase adenoma formation (37), and loss of Brm expression has been decisively demonstrated in human lung cancer (38). This latter event was shown to occur through epigenetic regulation and silencing of Brm mRNA expression. By contrast, recently reported Brm loss in gastric cancers occurs as a result of posttranscriptional Brm mRNA regulation (39). Although the mechanism by which Brm is down-regulated in human prostate cancer remains unexplored, gene expression arrays are suggestive of mRNA regulation. While this study was in preparation, a report emerged which suggested that Brg1 mRNA levels are elevated in prostate cancer, especially under conditions wherein Brm expression levels were low (40). Ectopic expression of Brg1 (but not Brm) in cultured cell models increased the invasive capacity of AR-negative PC3 cells, therefore further suggesting that Brg1 and Brm serve differential functions in cells of prostatic origin. In the present study, no significant alteration in Brg1 mRNA was observed in Brm-deficient prostate tissues, either in the murine epithelia or in human prostate cancer (data not shown). Thus, it is not anticipated that the proliferative phenotypes observed result from Brg1 deregulation.
The result of Brm down-regulation in both human disease and targeted deletion in murine prostates seems to be the induction of a hyperplastic phenotype. Although observed only in the VP and AP, lobe-specific hyperplasia is a frequently observed event after oncogenic insult in the murine prostate (30, 31). Although the process that contributes to lobe-specific outcomes are of interest, our data showing that Brm expression is inversely correlated to the proliferative index in prostate cancer and the murine prostate provide compelling evidence that Brm loss confers a growth advantage to this tissue. This outcome seems to be tissue-specific, as previous analyses of Brm−/− animals revealed both organ-specific increases and decreases in weight (16). More detailed analyses of proliferative defects in Brm−/− have been previously investigated in MEFs, which retain serum dependence, show little change in cell cycle kinetics after serum stimulation, and do not form multilayers, but are partially compromised in the cellular response to contact inhibition or UV-induced DNA damage (16). Whether these phenotypes contribute to the proliferative advantage observed in the prostate, and the cause of observed tissue-specific effects in Brm−/− animals, will be the focus of future studies.
Particular to prostate cancer, it was surprising to observe that AR signaling was refractory to Brm loss, both in the murine prostates and as correlated with Brm down-regulation in human disease. Based on studies performed in vitro (22, 23), it was expected that AR activity might be compromised upon perturbation of this SWI/SNF subunit. Whether retained AR signaling in the absence of Brm is attributed to developmental plasticity remains an unexplored but clinically relevant question because AR target gene expression was unaltered in tumors with reduced Brm expression. Although Brg1 expression was not enhanced in Brm−/− epithelia, it is possible that Brg1-containing complexes are sufficient to sustain AR activity, and/or that the chromatin remodeling needs of AR are satisfied by other chromatin remodeling complexes. Interestingly, down-regulation of SMARCA3 (HLTF) and SMARCA5 (ISWI/SNF2H) was significantly observed in conjunction with Brm down-regulation in cancer specimens (Supplementary Fig. S6), thus indicating that loss of Brm likely induces the perturbation of other chromatin remodeling pathways that may facilitate AR signaling.
Although AR activity was retained in Brm−/− epithelia (evidenced by sustained probasin expression in unchallenged animals and uncompromised response to androgen re-supplementation), a major implication of the present study is that Brm loss was sufficient to induce castration-resistant proliferation in a subset of prostatic epithelia. This may be of clinical importance, as the transition to serum androgen independence typically represents the development of incurable disease. Based on gene expression profiling of Brm-deficient tumors, at least one underlying mechanism is hypothesized to occur through the regulation of the retinoblastoma tumor suppressor (RB)/E2F1 axis. Prior studies showed that RB directly requires SWI/SNF function to exert negative control over E2F target genes that are critical for Sphase progression (41–43); indeed, E2F1 itself is a known E2F target gene. Moreover, pathway analyses from human specimens revealed that several upstream alterations that would result in RB inactivation (down-regulation of Smad2/3, Smad4, p27kip1, and RB itself) likely contribute to the observed induction of E2F1. In the prostate, loss of RB function has been shown to convey resistance to androgen ablation and AR antagonist therapies in multiple in vitro systems (44), and targeted deletion of RB in prostatic epithelia provides a proliferative advantage in vivo (45). Thus, Brm down-regulation may represent a mechanism to partially suppress RB function in this tissue. Because elevated E2F1 is associated with both the induction of cellular proliferation and apoptosis (46), these data likely explain the observed outcomes in Brm−/− prostatic epithelia. Based on these findings, it is hypothesized that these downstream effects of E2F1 deregulation may actually hinder the progression of the hyperplastic phenotype, and the relevance of this supposition for human disease is being explored.
One additional mechanism that could contribute to the castration-resistant phenotype is alterations in local hormone synthesis. Although there are multiple mechanisms that contribute to resurgent AR activity in therapy-resistant tumors (25), it is now apparent that a substantive percentage of castration-resistant prostate cancers may arise from the activation of intracrine de novo androgen synthesis within the tumor microenvironment (47–49). Indeed, analysis of patients with recurrent tumors after hormone therapy supports this contention, and the recent observation that an irreversible Cyp17 inhibitor, abiraterone, can reduce prostate-specific antigen in patients that failed hormone therapy highlights the clinical relevance of this supposition (50). It is of interest that four effectors of C21 steroid hormone metabolism (Cyp11A1, which generates pregnenolone from cholesterol; Cyp11B2, which converts corticosterone to aldosterone; Cyp17A1, which assists pregnenolone and DHEA production, but has also been recently implicated in a backdoor pathway for steroid production; and Cyp21A7, which hydroxylates progesterone) were anticorrelated with Brm levels in gene expression arrays from human tumors. Thus, alterations in Brm may affect steroid hormone synthesis. However, serum testosterone levels were indistinguishable from Brm-positive animals in castration/re-supplementation studies (Supplementary Fig. S3B). As such, the current data do not support the contention that Brm loss increases serum testosterone levels; however, the local effect within the prostate cannot be excluded and is the focus of ongoing study.
In summary, the data presented herein show that Brm loss results in prostate-specific hyperplasia, the transition to androgen independence, and deregulated E2F1 expression after targeted ablation in murine tissues. These events hold consequence in human disease, wherein down-regulation of Brm is associated with a high proliferative index. Together, these data reveal a putative tumor suppressor function for Brm in the prostate, and suggest that loss of this protein is sufficient to confer a proliferative advantage in this tissue.
Supplementary Material
Acknowledgments
Grant support: R01 CA116777 and R21 CA121402 (K.E. Knudsen), CA91048 and CA102848 (B. Weissman), and Canceropole Ile-de-France (C. Muchardt).
We thank Drs. R. Matusik and S. Hayward for instruction on prostate resection and pathology. We also recognize the contributions of M. Schiewer and Dr. E. Knudsen for critical commentary and ongoing discussions.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Muchardt C, Yaniv M. ATP-dependent chromatin remodelling: SWI/SNF and Co. are on the job. J Mol Biol. 1999;293:187–198. doi: 10.1006/jmbi.1999.2999. [DOI] [PubMed] [Google Scholar]
- 2.Vignali M, Hassan AH, Neely KE, Workman JL. ATP-dependent chromatin-remodeling complexes. Mol Cell Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang W. The SWI/SNF family of ATP-dependent chromatin remodelers: similar mechanisms for diverse functions. Curr Top Microbiol Immunol. 2003;274:143–169. doi: 10.1007/978-3-642-55747-7_6. [DOI] [PubMed] [Google Scholar]
- 4.Trotter KW, Archer TK. Nuclear receptors and chromatin remodeling machinery. Mol Cell Endocrinol. 2007;265–266:162–167. doi: 10.1016/j.mce.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trotter KW, Archer TK. The BRG1 transcriptional coregulator. Nucl Recept Signal. 2008;6:e004. doi: 10.1621/nrs.06004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kadam S, Emerson BM. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol Cell. 2003;11:377–389. doi: 10.1016/s1097-2765(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 7.Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 8.Moshkin YM, Mohrmann L, van Ijcken WF, Verrijzer CP. Functional differentiation of SWI/SNF remodelers in transcription and cell cycle control. Mol Cell Biol. 2007;27:651–661. doi: 10.1128/MCB.01257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bultman SJ, Gebuhr TC, Magnuson T. A Brg1 mutation that uncouples ATPase activity from chromatin remodeling reveals an essential role for SWI/SNF-related complexes in β-globin expression and erythroid development. Genes Dev. 2005;19:2849–2861. doi: 10.1101/gad.1364105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sumi-Ichinose C, Ichinose H, Metzger D, Chambon P. SNF2β-BRG1 is essential for the viability of F9 murine embryonal carcinoma cells. Mol Cell Biol. 1997;17:5976–5986. doi: 10.1128/mcb.17.10.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lessard J, Wu JI, Ranish JA, et al. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffin CT, Brennan J, Magnuson T. The chromatin-remodeling enzyme BRG1 plays an essential role in primitive erythropoiesis and vascular development. Development. 2008;135:493–500. doi: 10.1242/dev.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gebuhr TC, Kovalev GI, Bultman S, Godfrey V, Su L, Magnuson T. The role of Brg1, a catalytic subunit of mammalian chromatin-remodeling complexes, in T cell development. J Exp Med. 2003;198:1937–1949. doi: 10.1084/jem.20030714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Indra AK, Dupe V, Bornert JM, et al. Temporally controlled targeted somatic mutagenesis in embryonic surface ectoderm and fetal epidermal keratinocytes unveils two distinct developmental functions of BRG1 in limb morphogenesis and skin barrier formation. Development. 2005;132:4533–4544. doi: 10.1242/dev.02019. [DOI] [PubMed] [Google Scholar]
- 15.Matsumoto S, Banine F, Struve J, et al. Brg1 is required for murine neural stem cell maintenance and gliogenesis. Dev Biol. 2006;289:372–383. doi: 10.1016/j.ydbio.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 16.Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2α) EMBO J. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sowery RD, So AI, Gleave ME. Therapeutic options in advanced prostate cancer: present and future. Curr Urol Rep. 2007;8:53–59. doi: 10.1007/s11934-007-0021-9. [DOI] [PubMed] [Google Scholar]
- 18.Klotz L. Hormone therapy for patients with prostate carcinoma. Cancer. 2000;88:3009–3014. doi: 10.1002/1097-0142(20000615)88:12+<3009::aid-cncr17>3.3.co;2-5. [DOI] [PubMed] [Google Scholar]
- 19.Balk SP. Androgen receptor as a target in androgen-independent prostate cancer. Urology. 2002;60:132–138. doi: 10.1016/s0090-4295(02)01593-5. [discussion 8–9]. [DOI] [PubMed] [Google Scholar]
- 20.Kim J, Coetzee GA. Prostate specific antigen gene regulation by androgen receptor. J Cell Biochem. 2004;93:233–241. doi: 10.1002/jcb.20228. [DOI] [PubMed] [Google Scholar]
- 21.Hernandez J, Thompson IM. Prostate-specific antigen: a review of the validation of the most commonly used cancer biomarker. Cancer. 2004;101:894–904. doi: 10.1002/cncr.20480. [DOI] [PubMed] [Google Scholar]
- 22.Marshall TW, Link KA, Petre-Draviam CE, Knudsen KE. Differential requirement of SWI/SNF for androgen receptor activity. J Biol Chem. 2003;278:30605–30613. doi: 10.1074/jbc.M304582200. [DOI] [PubMed] [Google Scholar]
- 23.Klokk TI, Kurys P, Elbi C, et al. Ligand-specific dynamics of the androgen receptor at its response element in living cells. Mol Cell Biol. 2007;27:1823–1843. doi: 10.1128/MCB.01297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Culig Z, Klocker H, Bartsch G, Hobisch A. Androgen receptors in prostate cancer. Endocr Relat Cancer. 2002;9:155–170. doi: 10.1677/erc.0.0090155. [DOI] [PubMed] [Google Scholar]
- 25.Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 26.Knudsen KE, Arden KC, Cavenee WK. Multiple G1 regulatory elements control the androgen-dependent proliferation of prostatic carcinoma cells. J Biol Chem. 1998;273:20213–20222. doi: 10.1074/jbc.273.32.20213. [DOI] [PubMed] [Google Scholar]
- 27.Glinsky GV, Glinskii AB, Stephenson AJ, Hoffman RM, Gerald WL. Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Invest. 2004;113:913–923. doi: 10.1172/JCI20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 29.Huang da W, Sherman BT, Tan Q, et al. The DAVID gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stanbrough M, Leav I, Kwan PW, Bubley GJ, Balk SP. Prostatic intraepithelial neoplasia in mice expressing an androgen receptor transgene in prostate epithelium. Proc Natl Acad Sci U S A. 2001;98:10823–10828. doi: 10.1073/pnas.191235898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ellwood-Yen K, Graeber TG, Wongvipat J, et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4:223–238. doi: 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 32.Denmeade SR, Lin XS, Isaacs JT. Role of programmed (apoptotic) cell death during the progression and therapy for prostate cancer. Prostate. 1996;28:251–265. doi: 10.1002/(SICI)1097-0045(199604)28:4<251::AID-PROS6>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 33.Mirosevich J, Bentel JM, Zeps N, Redmond SL, D’Antuono MF, Dawkins HJ. Androgen receptor expression of proliferating basal and luminal cells in adult murine ventral prostate. J Endocrinol. 1999;162:341–350. doi: 10.1677/joe.0.1620341. [DOI] [PubMed] [Google Scholar]
- 34.Varambally S, Yu J, Laxman B, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 35.Seoane J, Le HV, Shen L, Anderson SA, Massague J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211–223. doi: 10.1016/s0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- 36.Seoane J, Pouponnot C, Staller P, Schader M, Eilers M, Massague J. TGFβ influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat Cell Biol. 2001;3:400–408. doi: 10.1038/35070086. [DOI] [PubMed] [Google Scholar]
- 37.Glaros S, Cirrincione GM, Muchardt C, Kleer CG, Michael CW, Reisman D. The reversible epigenetic silencing of BRM: implications for clinical targeted therapy. Oncogene. 2007;26:7058–7066. doi: 10.1038/sj.onc.1210514. [DOI] [PubMed] [Google Scholar]
- 38.Reisman DN, Sciarrotta J, Wang W, Funkhouser WK, Weissman BE. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: correlation with poor prognosis. Cancer Res. 2003;63:560–566. [PubMed] [Google Scholar]
- 39.Yamamichi N, Inada K, Ichinose M, et al. Frequent loss of Brm expression in gastric cancer correlates with histologic features and differentiation state. Cancer Res. 2007;67:10727–10735. doi: 10.1158/0008-5472.CAN-07-2601. [DOI] [PubMed] [Google Scholar]
- 40.Sun A, Tawfik O, Gayed B, et al. Aberrant expression of SWI/SNF catalytic subunits BRG1/BRM is associated with tumor development and increased invasiveness in prostate cancers. Prostate. 2007;67:203–213. doi: 10.1002/pros.20521. [DOI] [PubMed] [Google Scholar]
- 41.Strobeck MW, Knudsen KE, Fribourg AF, et al. BRG-1 is required for RB-mediated cell cycle arrest. Proc Natl Acad Sci U S A. 2000;97:7748–7753. doi: 10.1073/pnas.97.14.7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strobeck MW, Reisman DN, Gunawardena RW, et al. Compensation of BRG-1 function by Brm: insight into the role of the core SWI-SNF subunits in retinoblastoma tumor suppressor signaling. J Biol Chem. 2002;277:4782–4789. doi: 10.1074/jbc.M109532200. [DOI] [PubMed] [Google Scholar]
- 43.Zhang HS, Gavin M, Dahiya A, et al. Exit from G1 and Sphase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and RbhSWI/SNF. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]
- 44.Sharma A, Comstock CE, Knudsen ES, et al. Retinoblastoma tumor suppressor status is a critical determinant of therapeutic response in prostate cancer cells. Cancer Res. 2007;67:6192–6203. doi: 10.1158/0008-5472.CAN-06-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maddison LA, Sutherland BW, Barrios RJ, Greenberg NM. Conditional deletion of Rb causes early stage prostate cancer. Cancer Res. 2004;64:6018–6025. doi: 10.1158/0008-5472.CAN-03-2509. [DOI] [PubMed] [Google Scholar]
- 46.DeGregori J, Johnson DG. Distinct and overlapping roles for E2F family members in transcription, proliferation and apoptosis. Curr Mol Med. 2006;6:739–748. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- 47.Singh P, Coe J, Hong W. A role for retinoblastoma protein in potentiating transcriptional activation by the glucocorticoid receptor. Nature. 1995;374:562–565. doi: 10.1038/374562a0. [DOI] [PubMed] [Google Scholar]
- 48.Locke JA, Guns ES, Lubik AA, et al. Androgen levels increase by intratumoral de novo steroidogenesis during progression of castration-resistant prostate cancer. Cancer Res. 2008;68:6407–6415. doi: 10.1158/0008-5472.CAN-07-5997. [DOI] [PubMed] [Google Scholar]
- 49.Montgomery RB, Mostaghel EA, Vessella R, et al. Maintenance of intratumoral androgens in metastatic prostate cancer: a mechanism for castration-resistant tumor growth. Cancer Res. 2008;68:4447–4454. doi: 10.1158/0008-5472.CAN-08-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Attard G, Reid AH, Yap TA, et al. Phase I clinical trial of a selective inhibitor of CYP17, abiraterone acetate, confirms that castration-resistant prostate cancer commonly remains hormone driven. J Clin Oncol. 2008;26:4563–4571. doi: 10.1200/JCO.2007.15.9749. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.