Abstract
Interleukin-8 (IL-8 or CXCL8) plays a critical role in orchestrating the immune response by binding and activating the receptor CXCR1 that belongs to the GPCR class. IL-8 exists as both monomers and dimers, and both bind CXCR1 but with differential affinities. It is well established that the monomer is the high-affinity ligand and that the interactions between the ligand N-loop and receptor N-domain play a critical role in determining binding affinity. In order to characterize the structural basis of differential binding of the IL-8 monomer and dimer to the CXCR1 N-domain, we analyzed binding-induced NMR chemical shift and peak intensity changes and show that they are exquisitely sensitive and can provide detailed insights into the binding process. We used three IL-8 variants, a designed monomer, a trapped disulfide-linked dimer, and WT at dimeric concentrations. NMR data for the monomer show that nonsequential residues that span the entire N-loop are involved in the binding process and that the binding is mediated by a network of extensive direct and indirect coupled interactions. Interestingly, in the case of WT, binding induces dissociation of the dimer–receptor complex to the monomer-receptor complex, and in the case of the trapped dimer, binding results in increased global conformational flexibility. Increased dynamics is evidence of unfavorable interactions, indicating that binding of the WT dimer triggers conformational changes that disrupt dimer–interface interactions, resulting in its dissociation. These results together provide evidence that binding is not a localized event but results in extensive coupled interactions within the monomer and across the dimer interface and that these interactions play a fundamental role in determining binding affinity.
Chemokines constitute the largest subfamily of cytokines and mediate various biological processes from organogenesis and homeostasis to recruitment and activation of leukocytes during host defense against infection (1, 2). Chemokines exert their function by binding and activating 7-transmembrane G protein coupled receptors (GPCR)1 on the cell surface. An imbalance in chemokine function can be detrimental and has been attributed to the pathophysiology of several inflammatory and autoimmune diseases (3–5). All chemokines share the fundamental property of reversibly existing as monomer and dimer, and therefore knowledge of the structural and molecular basis of receptor binding of both monomeric and dimeric species is essential to understand how chemokines mediate function.
Interleukin-8 (IL-8, also known as CXCL8), one of the best studied chemokines, plays a critical role in host immune response by binding and activating the CXCR1 receptor on neutrophils. Structures of a trapped IL-8 monomer and WT dimer are known (6, 7). The structure of the monomer is similar to that of the monomeric subunit in the dimer, except the C-terminal helical residues that are structured in the dimer are unstructured in the monomer. Using trapped monomeric and trapped dimeric IL-8 variants, it has been shown that both forms can bind the CXCR1 receptor, but the monomer is the high-affinity ligand (8). Site-specific mutagenesis and generation of chimeric chemokines by swapping similar domains between IL-8 and other chemokines have shown that IL-8 N-terminal and N-loop residues play a critical role in mediating affinity, activity, and specificity (9–14). On the basis of these data and similar studies with other chemokines, it has been proposed that IL-8 receptor binding involves two interactions: between the IL-8 N-loop and the receptor N-domain residues (site I) and between the IL-8 N-terminal and the receptor extracellular loop residues (site II) (12–20).
In this study, we have characterized the structural basis of CXCR1 N-domain binding of three different IL-8 variants, a designed monomer, a trapped dimer, and WT dimer, using NMR spectroscopy. We used an IL-8(1–66) deletion mutant which is a monomer at millimolar concentrations and an IL-8(1–72) R26C mutant which is a trapped dimer that contains a disulfide across the dimer interface at the 2-fold symmetry. Binding-induced NMR chemical shift perturbation and peak intensity changes in the HSQC spectra of the monomer indicate global structural changes and suggest that direct and indirect interactions mediate the binding process. In particular, these data show that nonsequential residues dispersed through the entire span of the N-loop are involved in docking to the receptor N-domain. Most interestingly, NMR data of the WT dimer show binding-induced dissociation of the dimer–receptor N-domain complex. NMR data including 15N-relaxation measurements for the trapped dimer show evidence of increased conformational flexibility in the bound form providing a structural rationale for the binding-induced dissociation of the WT dimer. These results together indicate that binding is not a localized event but a global event and is mediated by extensive coupled interactions within the monomer and across the dimer interface and that these interactions play a fundamental role in determining binding affinity.
EXPERIMENTAL PROCEDURES
Design, Expression, and Purification of IL-8 Variants
WT IL-8 dimerizes at micromolar concentrations (21–23) and therefore is a dimer at millimolar concentrations used in NMR studies. We observe from NMR structural and relaxation studies that the IL-8(1–66) deletion mutant is a monomer at millimolar concentrations (P. R. B. Joseph and K. Rajarathnam, unpublished results). The trapped R26C dimer was designed by substituting a disulfide across the dimer interface at the site of 2-fold symmetry. This mutation does not perturb the dimer interface, and the structure of this trapped dimer is similar to that of the WT dimer (8).
The recombinant IL-8(1–66) monomer, trapped R26C dimer, and the WT were generated as described previously (24). All IL-8 variants were expressed in Escherichia coli strain BL21(DE3), and 15N-labeled proteins were produced by growing cells in minimal medium containing 15NH4Cl as the nitrogen source. Transformed E. coli BL21(DE3) cells were grown in minimal medium containing ampicillin to an A600 of 0.8 and induced with 1 mM isopropyl β-D-thiogalactopyranoside (IPTG) for 8 h at 37 °C. Purity and molecular weight of the proteins were confirmed using analytical high-pressure liquid chromatography (HPLC) and matrix-assisted laser desorption/ionization mass spectrometry (MALDI MS), respectively. The CXCR1 N-domain 24-mer (LWTWFEDEFANATGMPPVEKDYSP) is the same as that used in our previous studies and was synthesized using solid-phase peptide synthesis (24).
NMR Spectroscopy
For titration experiments, 15N-labeled WT, trapped R26C dimer, and IL-8(1–66) monomer were prepared in 50 mM sodium acetate buffer, pH 6.0, containing 1 mM 2,2-dimethyl-2-silapentanesulfonic acid (DSS), 1 mM sodium azide, and 10% 2H2O (v/v). Chemical shifts were referenced to DSS (25). All H-15NHSQC spectra were acquired at 30 °C using Varian Unity Plus 600 or INOVA 800 MHz spectrometers equipped with field gradient accessories. A stock solution of the CXCR1 N-domain peptide prepared in the same buffer was used for titration experiments, and NMR spectra of all IL-8 variants were collected until essentially no change in chemical shifts was observed. The final molar ratios for monomer, trapped R26C dimer, and WT dimer are 3.4,17.8, and 11.2, respectively. Two-dimensional 1H–15N HSQC spectra were acquired with 2048 complex points for direct 1H and 128 complex points for indirect 15N dimensions. The spectra were processed using NMRPipe (26) and analyzed using NMRView (27). The chemical shifts of the WT dimer and the trapped R26C dimer were assigned using previously available assignments (8,18), and the chemical shifts of the IL-8(1–66) monomer were assigned using 15N-edited NOESY and TOCSY, CBCANH, and CBCA-(CO)NH experiments (P. R. B. Joseph and K. Rajarathnam, unpublished results).
The NMR 15N-T2 relaxation experiments were carried out using the gradient 2D 1H–15N correlation pulse sequences (28) on both the free and the receptor-peptide-bound trapped R26C dimer at 30 °C on the Varian Unity Plus 600 spectrometer. The data were acquired at delays of τ = 10, 30, 50, 70, 90, 110, 130, 150, and 170 ms, and the delays were scrambled to avoid any systematic errors. We used 16 transients per increment for the free form and 128 transients per increment for the bound form due to the relatively weak and broadened signals. The T2 values were calculated by weighted nonlinear least-squares fits of the peak heights in the 2D spectra to a two-parameter exponential decay using the Mathematica suite of programs (29). The uncertainties in T2 values were taken to be the standard error of the fitted parameters.
Calculation of Binding Constants
Apparent dissociation constants (Kd) were determined by fitting the binding-induced chemical shift changes (Δδ) to the following set of equations (30):
| (1) |
where Δδmax is the maximum chemical shift change between bound and free IL-8, CIL-8 is the total concentration of IL-8, and C is the concentration of bound IL-8.
Δδ is the observed chemical shift deviation, given by the equation:
| (2) |
where ΔδH and ΔδN are the 1H and 15N chemical shift changes (in parts per million). C is also defined by the equation:
| (3) |
where Cpep is the total concentration of the peptide.
RESULTS
Binding of the IL-8 Monomer
For all NMR experiments, unlabeled CXCR1 N-domain peptide was titrated into 15N-labeled IL-8 variants, and binding was followed by changes in 2D 15N–1H HSQC spectral characteristics. Binding-induced changes in chemical shifts are excellent probes for mapping binding surfaces involved in protein–protein interactions (31). Line broadening of resonances during titration could occur as a consequence of chemical shift differences between the free and bound forms and when the binding is in the intermediate to fast exchange regime on NMR time scale (32).
We will first describe the results of the binding of IL-8 monomer and then compare and contrast the binding of the trapped dimer and WT dimer. In the monomer, peaks corresponding to the N-loop, third β-strand, and the C-terminal helical residues showed the largest chemical shift perturbations and also substantial line broadening, indicating that these regions play a key role in the binding process (Figure 1).
Figure 1.
Binding of the IL-8 monomer to the CXCR1 N-domain. Sections of the 15N–1H HSQC spectra showing binding-induced chemical shift changes of the IL-8 monomer residues (panels A and B). Arrows indicate the direction of peak movement, and for some residues (Ile10, Tyr13, Glu48, Leu49, Arg60, Phe65) showing large chemical shift changes, the final bound peaks are labeled in red. The peaks are color coded according to peptide: protein molar ratios of 0 (black), 1.2 (green), and 3.4 (red). The peak for the bound form of residue Ile10 is not visible at the contour level shown and is marked by an asterisk. Histogram showing the binding-induced chemical shift change (Δδ) of the IL-8 monomer (panel C). The residues showing significant perturbation in the N-loop, third β-strand, and the C-terminal helix are highlighted in black. Lys15 and His18 are broadened out and are represented by an asterisk, and residues 16, 19, 32, and 53 are prolines.
Role of the N-Loop Residues
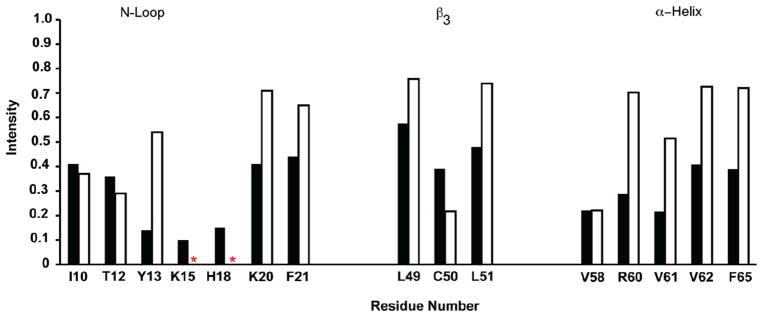
Large chemical shift perturbations were observed for N-loop residues Ile10, Thr12, and Tyr13 and to a lesser extent for residues Ser14, Lys20, and Phe21, and peaks corresponding to Lys15 and His18 completely disappear. These residues also show the largest decrease in intensity due to line broadening (Figure 2). The IL-8 structure reveals that these residues span the entire N-loop and that electrostatic and hydrophobic/packing interactions mediate binding of IL-8 to the receptor N-domain (Figure 3A).
Figure 2.
Binding-induced line broadening of the IL-8 monomer residues. A plot showing binding-induced loss of peak intensity for the N-loop, third β-strand, and the C-terminal helical residues. The unbound intensities are normalized to 1. The intensities are shown for peptide protein molar ratios of 0.07 (black) and 3.4 (open). Lys15 and His18 are broadened out and are indicated by an asterisk.
Figure 3.
A molecular plot of the IL-8monomer (PDB id 1IKM(7)) showing the N-loop residues involved in CXCR1 N-domain binding (panel A). The solvent-exposed residues, Ile10, Lys15, and Lys20, are highlighted in red, and partially buried residues, Thr12, Try13, His18, and Phe21, are highlighted in green. The plot was rendered using PyMOL(52). Chemical shift changes and ASA (panel B). A plot of the chemical shift change (top y-axis) and the side chain ASA (bottom y-axis) of the IL-8 monomer N-loop residues shows that they are not necessarily correlated. Chemical shift changes for Lys15 and His18 are not available due to line broadening. The ASA of the IL-8 monomer (PDB id 1IKM) was calculated using the program VADAR (53).
Ile10, Ser14, Lys15, and Lys20 are solvent exposed (ASA ~ 0.8) and so exert their influence by making direct contact with the receptor residues. On the other hand, residues Thr12, Tyr13, His18, and Phe21 are partially buried in the structure (ASA ~ 0.3–0.6); therefore, describing their role in binding on the basis of a static structure is less straightforward (Figure 3B). For instance, the structure reveals that one face of the aromatic ring side chain of His18 and Phe21 is solvent exposed, while the other face is buried in the structure and is probably involved in some form of favorable packing/aromatic interactions (Figure 3A). However, as the N-loop is conformationally flexible, the side chain could become completely exposed and engage in binding. It is also possible that these residues indirectly influence the binding of Ile10, Lys15, and Lys20 through packing interactions. NMR studies of binding-induced structural changes of these N-loop mutants are essential to provide amore definite answer as to how these residues mediate binding affinity.
Our NMR data showing that the entire N-loop is involved in receptor binding is inconsistent with a structural model proposed by Williams et al. and Hammond et al. (14, 33). On the basis of mutagenesis, these authors propose a binding site as that formed by residues Phe17, Phe21, and Ile22 (14, 33). Our data show only minimal perturbation for residues Phe17 and Ile22, discounting any major role for these residues in direct binding. Phe17 is largely buried (ASA ~ 0.2) and Ile22 is completely buried, and they are involved in extensive packing interactions with each other and with residues Phe21 and Leu43. Further, mutation of Phe17 to Ala resulted in an unfolded protein, indicating that Phe17 is essential for maintaining the integrity of the structural scaffold and so is involved in the presentation of the N-loop residues for receptor binding (14). Similarly, we propose that Ile22 is part of the structural scaffold and plays a role in function by indirectly influencing the binding of N-loop residues.
The solution structure of a CXCR1 N-domain peptidomimetic bound to dimeric IL-8 has been solved using NMR spectroscopy (18). The structure was determined on the basis of intermolecular NOEs between 13C- and 15N-labeled IL-8 and unlabeled CXCR1 peptide and reveals that the binding interface predominantly involves the N-loop residues, including hydrophobic residues Ile10, Tyr13, Phe17, and Phe21 and charged residues Lys11 and Lys15. Our data are largely consistent with the structure, though it is very unlikely that Phe17 is involved in direct binding interactions (vide supra).
Other studies have shown that mutating His18, Lys15, and Lys20 results in significant loss of binding, whereas mutating residues Thr12 and Ser14 had only a modest effect (11–13, 33, 34). Our NMR data show that Lys15 and His18 broaden out, whereas Thr12 shows large binding-induced perturbation and remains significantly weak, indicating that these residues are conformationally flexible in the bound form (Figures 1 and 2). The structure of the complex also fails to reveal specific interactions for residues Thr12 and Ser14, suggesting a more subtle role for these residues (18). The dynamic nature of these residues in the receptor-bound form suggests that such a property could play a more prominent role for receptor activities such as chemotaxis and Ca2+ release than for binding affinity.
Role of the Third β-Strand Residues
The third β-strand residues that are significantly perturbed are Glu48, Leu49, Cys50, Leu51, and Asp52 (Figure 1C). The ASA indicates that Glu48 alone is exposed while others are largely buried. Interestingly, Asp52 shows the largest perturbation of all amino acids, which is unexpected considering that the CXCR1 N-domain is highly negatively charged. The CXCR1 N-domain sequence has only one Lys, and it can be argued that Asp52 interacts with this Lys. However, IL-8 also binds CXCR2 receptor with similar high affinity, and the corresponding residue in the CXCR2N-domain is a Leu. Asp52 is also significantly perturbed on binding to the CXCR2 N-domain (Ravindran et al., unpublished results), indicating that the perturbation of Asp52 should be due to indirect interactions.
The NMR structure of the monomer reveals that both Leu51 andAsp52 interact with the N-loop residuesThr12 and Tyr13 (7), and so binding-induced changes in Thr12 and Tyr13 are most likely propagated to Leu51 and Asp52 through packing interactions (Figure 4A). The structure also reveals that Leu49 is largely buried but nevertheless shows side chain packing interactions with Tyr13, suggesting binding-induced conformational changes in Tyr13 could be propagated through packing interactions to Leu49 (Figure 4A). Another mechanism by which conformational changes could be propagated is through the Cys9–Cys50 disulfide bridge, which tethers the conformationally flexible N-loop to the third β-strand. Chemical shift data show that both Cys9 and Cys50 are significantly perturbed and also broaden out during the titration (Figures 1 and 2). These data together indicate that the changes observed in the third β-strand are not due to direct binding but due to indirect coupling interactions propagated through the N-loop.
Figure 4.
Binding-induced indirect coupled interactions in third β-strand residues (panel A). A molecular plot of the IL-8 monomer showing packing interactions between the N-loop residues Thr12 and Tyr13 (blue) and the third β-strand residues Asp52 and Leu49 (pink). Spheres are shown to highlight the side chain packing. One of the methyl groups of Leu49 is packed against the aromatic ring of Tyr13, and the side chain carboxylate of Asp52 is in contact with the hydroxyl side chain of Thr12. Coupled interactions in the C-terminal helix (panel B). A molecular plot of the IL-8 monomer showing the packing interactions between the C-terminal helical residues, Val58, Val61, Val62, and Phe65 (red), and the β-strand residues, Ile22, Leu25, and Leu51 (blue). Spheres are shown to highlight the side chain packing. Solvent-exposed residues Arg60 and Lys64 are shown in black. The plots were rendered using PyMOL (52).
Structure–function studies of conservatively mutating Asp52 to Asn had no effect on binding affinity, indicating that the carboxylate side chain is not important for function (33). This is consistent with our conclusion that Asp52 is involved in packing interactions and not in direct binding. Mutating Leu49 and Leu51 to Ala results in reduced binding, and Glu48 to Lys had no effect on binding (12, 14, 33). We have previously shown that it is possible to substitute cysteines with a non-natural cysteine analogue in a disulfide without breaking the disulfide and that Cys50 plays a role in receptor binding (35). Cys50 like Leu49 and Leu51 is largely buried, and we propose that these residues form a part of the hydrophobic core, which functions as a scaffold, and indirectly influence the binding of the N-loop residues by coupled interactions.
Role of the C-Terminal Helix Residues
Significant chemical shift perturbations and line broadening are observed for the C-terminal helical residues Val58, Arg60, Val61, Val62, and Phe65 (Figures 1 and 2). ASA data indicate that except Arg60 all other residues are buried (ASA < 0.2). We observe a periodicity in chemical shift perturbation with residues Val58, Val61, and Phe65 showing the largest change (Figure 1). The side chains of these residues are oriented in the same direction and are packed against hydrophobic residues of the β-strand (Figure 4B). Except for Arg60, the remaining solvent-exposed helical residues Gln59, Glu63, and Lys64 are relatively less perturbed (Figure 1). The side chains of Arg60 and Lys64 are located on the surface of the helix and pointed away from the N-loop (Figure 4B). Therefore, the perturbation of the C-terminal helical residues cannot be due to direct interaction with the N-domain.
The role of C-terminal helix for receptor binding and function has been investigated using C-terminal deletion mutants and IL-8 chimera containing helices of IP-10 or PF-4 (10, 11). The chimeras are as active as the WT IL-8, but successive deletion of helical residues in WT IL-8 results in progressive loss of binding affinity (9). On the other hand, mutagenesis studies have shown that positively charged residues Arg60 and Lys64 bind to negatively charged glycosaminoglycans, and such binding has been shown to be essential in the neutrophil recruitment process (36). Our NMR studies and the structure–function data together indicate that the helical residues are part of the structural scaffold and are not involved in direct receptor binding but indirectly influence the binding of the N-loop residues through long-range coupling interactions.
Role of the First and Second β-Strand Residues
Chemical shift perturbations are also observed for residues in the first and second β-strand and in the 40s turn which link the second and third β-strands. The extent of perturbation is lower compared to that observed for the N-loop, third β-strand, and the C-terminal helix (Figure 1). However, perturbation is differential, involving relatively large as well as small chemical shift changes. For instance, relatively large perturbations are observed for Lys23 and Leu25 in the first β-strand, Ile39 and Val41 in the second β-strand, and Leu43 and Ser44 in the 40s turn, and in contrast, minimal perturbations are observed for Arg26 in the first β-strand and Ile40 in the second β-strand. The monomer structure reveals packing interactions between Lys23 and Ser44, between Leu25 and Val41, and between Ile39 and Leu51, suggesting indirect coupling interactions as the cause for the observed perturbations (7).
Hammond et al. have proposed, on the basis of mutagenesis, that Val41 and Leu43 play a direct role in receptor binding and that these and N-loop residues Tyr13, Phe17, and Ile22 together form a receptor-binding site (33). Williams et al. have characterized the function of a wide variety of mutants including those of the 40s residues and also studied the consequence of mutation on the tertiary structure using NMR spectroscopy. On the basis of their results, the authors propose that N-loop residues Phe17, Phe21, Ile22, and Leu43 form the receptor-binding pocket (14). Skelton et al. also propose that Leu43 is involved in binding on the basis of the structure of the complex (18).
Leu43 is largely buried (ASA ~0.2), and the monomer structure reveals that the side chain is packed against residues Phe17, Tyr21, and Ile22 (7). Though mutating Leu43 to Ala resulted in ~15-fold lower affinity, mutating to Asp had no effect, indicating that the large hydrophobic side chain is not critical for binding but important for packing interactions (14). Val41 is completely buried and is packed against Leu25 and Phe17, and mutating it to Ala results only in marginal loss of binding (2-fold), suggesting that it cannot be involved in direct binding interactions (14). Though chemical shifts of Ile40 are only minimally perturbed, mutating Ile40 to Ala results in substantial loss of binding and also in perturbation of the structure, indicating that loss of function is due to indirect interactions (14). Our NMR and the mutagenesis data together indicate that the 40s residues, if any, play a minimal role in direct binding and, like the C-terminal helical and third β-strand residues, are part of the structural scaffold and indirectly influence the binding of the N-loop residues by coupling interactions.
Role of the N-Terminal and 30s Loop Residues
Our NMR data show minimal chemical shift and intensity changes in the N-terminal and the 30s loop regions (Figure 1). The 30s loop residues link the first and second β-strands. The N-terminal region binds to the receptor extracellular residues (site II) and is linked to the 30s loop region via the Cys7–Cys34 disulfide bridge. Mutating the 30s loop residues and the cysteines of the 7–34 disulfide results in significant loss of binding, indicating a role for coupled interactions in site II binding (11). Minimal chemical shift changes indicate that these regions are not directly involved in site I binding. Perturbation of these residues nevertheless indicates that site I binding could influence site II binding.
Binding of the IL-8 WT and Trapped R26C Dimer
WT IL-8 dissociates at micromolar concentrations (Kd ~10 μM) and therefore is predominantly dimeric at the NMR concentrations used in this study. Most interestingly, we observe a set of new peaks emerging during the latter titration points (>1:1 molar ratio), whose chemical shifts correspond to those of the monomer- bound N-domain complex (Figure 5). These observations indicate receptor N-domain binding induces dissociation of the dimer–receptor complex to the monomer–receptor complex. Previous titration studies did not report similar binding-induced dissociation (18, 37) and can be mainly attributed to the fact that in their studies the final molar ratio was ~1:1, whereas we carried out titration up to >10-fold excess of the receptor N-domain.
Figure 5.
Binding-induced dissociation of the WT IL-8 dimer. A section of the 15N–1H HSQC spectra showing binding-induced chemical shift changes in the WT dimer and the appearance of bound monomer (labeled as M in red). The peaks are color coded for peptide:protein molar ratios of 0 (black), 2.7 (green), and 11.2 (red).
In order to better understand the structural basis of dimer binding and binding-induced dissociation of the dimer-bound complex, we also carried out NMR studies of the disulfide-trapped R26C IL-8 dimer. The trapped R26C dimer has been shown to behave like the WT dimer but does not dissociate (8). We observe that the extent of overall chemical shift perturbation in the trapped dimer is substantially lower compared to the monomer. In addition to perturbation of the N-loop, third β-strand, and the C-terminal helical residues, perturbation of the second β-strand residues is also observed (Figure 6). Two previous NMR titration studies of WT IL-8 binding to the CXCR1 N-domain showed a similar chemical shift perturbation profile, including perturbation of the 40s residues (18, 37). On the basis of monomer NMR data (vide supra), only N-loop residues are involved in direct binding, and any chemical shift perturbation for other regions, such as the 40s residues, should be due to indirect coupling interactions. Therefore, 40s residues in the dimer cannot be involved in direct binding, suggesting mechanism(s) other than binding should be contributing to the chemical shift change.
Figure 6.
Binding-induced chemical shift changes in the trapped IL-8 dimer (panel A). The regions showing significant perturbations are the N-loop, second β-strand, third β-strand, and the C-terminal helix. For residues that completely broaden out in the bound form (Gln8, Thr12, Lys15, Lys20, Leu25, Arg26, Glu48, and Asp52), shift changes are calculated from the last observable titration point, and data do not exist for four prolines (residues 16, 19, 32, and 53). A section of the 15N–1H HSQC titration spectra showing binding-induced chemical shift changes in the IL-8 trapped dimer (panel B). The peaks are color coded for peptide:protein molar ratios of 0 (black), 1.4 (green), and 2.6 (red). Data for higher molar ratios are not shown as peaks either disappear or are significantly weak.
We observe binding-induced NMR line broadening in the trapped dimer to be distinctly different from the monomer. In the case of the monomer, significant line broadening was observed only for those residues that are involved in binding either by direct or by coupled interactions and could be described as due to exchange between free and receptor-bound form. On initial binding, these peaks in the monomer become weak and disappear and then reappear, becoming stronger at later titration points as the population of the bound form dominates; these residues also show the largest binding-induced chemical shift perturbations (Figure 1). However, in the case of trapped dimer, resonances of almost all residues become weak or completely disappear at later points (Figures 6B and 7). Most importantly, we observe no correlation between the extent of peak intensity change and chemical shift perturbation, indicating that the line broadening in the dimer is caused by mechanisms other than exchange between the free and bound forms. For instance, in the trapped dimer, Arg47 shows a small binding-induced chemical shift change, but its line broadening is similar to Ser44 that shows the largest chemical shift change (Figures 6 and 7). However, in the monomer, Arg47 shows minimal chemical shift change and minimal line broadening, and Ser44 shows significant chemical shift change and significant line broadening (Figures 1 and 7).
Figure 7.
Binding-induced line broadening in dimer vs monomer. Binding-induced intensity changes for the trapped dimer (○) and the monomer (●) are shown for residues Ser44 and Arg47. Line broadening and chemical shift changes are correlated for the monomer but not for the trapped dimer.
In order to better understand the dynamic characteristics of the trapped dimer, we also carried out 15N–T2 relaxation measurements of the free and receptor-bound form. We could characterize the relaxation properties of all of the residues of the trapped dimer except for the overlapped resonances (Figure 8). In stark contrast, we could measure relaxation values of only ~25% of the residues in the bound form due to significant line broadening. Peaks corresponding to the dimer interface β1 as well as the β2 and β3 residues broaden out, and only peaks corresponding to the N-terminal, C-terminal helix, and loop residues are observed (Figure 8). Indeed, previous relaxation measurements of the free WT dimer show residues of the β-strand, including those at the dimer interface, exhibit chemical exchange line broadening (38). Consistent with our observations from the trapped dimer, the authors conclude that these residues undergo concerted rather than independent motions due to extensive H-bonding and packing interactions.
Figure 8.
Relaxation properties of free and bound trapped IL-8 dimer. Overlay of a section of the 15N-T2 relaxation data when there is no relaxation delay (τ=0; black) and at T2 delay of τ=70 ms (red) of the free (A) and the receptor peptide-bound (B) trapped dimer. The bound spectrum is shown at a level where most of the broadened peaks are visible and is not to scale with respect to the unbound spectrum. In contrast to the free monomer where most peaks are still visible at a T2 delay of τ=70 ms, additional peaks have disappeared in the bound form. The dimer interface residues and the characteristic upfield shifted peaks are labeled. (C) Plot of T2 values for the free and bound trapped dimer. Due to poor s/n ratio in the bound form, T2 values could be fitted only for 18 residues.
Global line broadening is evidence of increased dynamics in the bound form, providing a snapshot of events that precede dissociation of the bound complex. We propose that binding induced conformational changes disrupt dimer–interface interactions resulting in the dissociation of the complex, and that the receptor-bound dimer is thermodynamically unfavored and the receptor-bound monomer is the favored state.
Binding Affinities of the Monomer and Dimer
The apparent binding constants for the IL-8(1–66) monomer and the trapped R26C dimer were obtained by fitting binding induced chemical shift changes from the titration experiments. We observe that the monomer binds the CXCR1 N-domain with an affinity of ~10 μM, and the trapped R26C dimer binds with an affinity of ~340 μM. Our measured binding constant for the IL- 8(1–66) monomer is similar to the previously measured value of ~8 μM using ITC (39). Previous studies have shown that the trapped dimer binds the intact CXCR1 receptor with ~70-fold lower affinity than the trapped monomer (8). Our observation that the trapped dimer binds the isolated CXCR1 N-domain with ~35-fold lower affinity indicates that most of the reduced binding is due to reduced affinity to the receptor N-domain site. All of these observations together emphasize that the dimer is the low-affinity ligand and the monomer is the high-affinity ligand for the CXCR1 receptor.
Linkage Scheme
Results from our titration studies allow us to discuss the linkage scheme of the binding of the monomer and dimer to the N-domain (Figure 9). The individual steps involve binding of the monomer to the receptor N-domain (ΔG1), binding of the dimer to the receptor N-domain (ΔG2), dissociation of the dimer to monomer (ΔG3), and dissociation of the N-domain-bound dimer to the N-domain-bound monomer (ΔG4). On the basis of calculated binding constants (Kd) from our NMR titration studies, free energies of monomer and dimer binding to the CXCR1 N-domain are ΔG1=−13.9 kcal mol−1 K−1 and ΔG2 is −5.0 kcal mol−1 K−1. The free energy of dimer dissociation has been measured from both ITC and ultracentrifugation methods, ΔG3=7.4 kcal mol−1 K−1 (21). Therefore, free energy for dissociation of the receptor-bound dimer (ΔG4) is calculated to be −1.5 kcal mol−1 K−1. Our observation that binding-induced dissociation of the dimer–receptor N-domain complex is thermodynamically favorable suggests that such dissociation also occurs on dimer binding to the intact CXCR1 receptor.
Figure 9.

A thermodynamic linkage scheme representing the binding of the monomer and trapped dimer to the CXCR1 N-domain.
DISCUSSION
IL-8 belongs to the subclass of CXC chemokines that have the characteristic N-terminal “ELR” motif. All ELR chemokines recruit and activate neutrophils by binding two receptors, CXCR1 and CXCR2. Only IL-8 is a high-affinity ligand for CXCR1, whereas all neutrophil-activating chemokines including IL-8 are high-affinity ligands for CXCR2. We have shown previously that our trapped IL-8 dimer binds CXCR1 compared to CXCR2 with much lower affinity and that dissociation of the WT dimer is essential for binding CXCR1 with high affinity (8, 40). Structure–function studies have shown that mutating IL-8 N-loop residues results in greater loss for CXCR1 binding, indicating that properties of N-loop residues are more stringent for CXCR1 binding (12, 13, 33).
On the basis of our current data, we propose that the protein core of IL-8 functions as a structural scaffold that is not rigid but dynamic and that a network of direct and indirect backbone–backbone, backbone–side chain, and side chain–side chain interactions are involved in the binding of IL-8 to the CXCR1 N-domain. Residues that span the entire N-loop participate in binding to the N-domain, and a combination of polar, electrostatic, and hydrophobic/packing interactions mediate this event. Sequence analysis reveals that the receptor N-domain is highly negatively charged and shows characteristics of an unstructured or minimally structured domain, and so it can be envisioned that the receptor N-domain binds IL-8 by the fly-casting mechanism (41). The structure of the complex also reveals aromatic/hydrophobic/packing interactions between IL-8 aromatic and receptor Pro/aromatic residues and electrostatic interactions between IL-8 Lys and receptor N-domain Asp/Glu residues. Our NMR data show that the receptor N-domain engages the entire N-loop for the binding, which contrasts previous models, where a subset of highly clustered hydrophobic N-loop residues was defined as the binding site (14, 33).
We propose that binding engagement by multiple N-loop residues triggers propagated conformational changes throughout the protein and that these changes play a critical role in mediating binding affinity and function. Considering that binding-induced structural and dynamic changes are extensive, we propose that this binding event as active and not passive. It is possible that site I binding results in conformational changes in the receptor triggering downstream interactions and/or triggers changes that are essential for site II binding of IL-8 N-terminal ELR residues to the receptor extracellular domain which triggers downstream signaling events for function.
Binding of several CXC (SDF-1α and IP-10) and CC (RANTES, eotaxin, eotaxin-2) chemokines and CX3C chemokine fractalkine to their receptor N-domains has been studied using NMR spectroscopy (42–47). These studies show not only that binding predominantly perturbs chemokine N-loop residues but also chemokine-dependent perturbation of other regions of the protein. For instance, fractalkine binding to the CX3CR1 N-domain results in significant perturbation of N-terminal and 30s loop residues, whereas these residues showed negligible chemical shift changes in IL-8 (44). These observations together suggest that indirect coupling interactions play a critical role for binding of most chemokines and that these interactions play a variety of functional roles from contributing to binding affinity, receptor selectivity, and/or receptor activation.
IL-8 function involves binding to receptors on neutrophils and to glycosaminoglycans (GAG) on the endothelial cell surface. Under transient conditions of injury or infection, IL-8 levels are upregulated and are translocated from the injury site to the bloodstream, and so its concentration will vary continuously as a function of space and time. Indeed, using a trapped monomer, trapped dimer, and the WT, we observe that the WT exists as both monomers and dimers in vivo and that the IL-8 dimer is extremely potent in its ability to recruit neutrophils in a mouse model (Das et al., manuscript in preparation). Whereas mice express only the CXCR2 receptor on neutrophils, humans express both CXCR1 and CXCR2 receptors, suggesting that the two receptors play distinctly different roles in humans. IL-8 plays the dual role of recruiting neutrophils to the site of infection and eliciting cytotoxic activities at the site of infection. These events have to be coordinated as the initial receptor engagement should lead to recruitment and chemotaxis, and receptor activation leading to cytotoxic event should not be triggered until late at the site of infection. It is believed in humans that CXCR2 plays a more active role for trafficking neutrophils into the tissue and CXCR1 is more active for eliciting cytotoxic events (48). Consistent with our observations, the IL-8 dimer is less active for CXCR1-related functions (Wassim et al., manuscript in press), suggesting in vivo IL-8 dimerization negatively regulates CXCR1 function.
Interestingly, the role of dimerization seems to vary from chemokine to chemokine. CXC chemokine SDF-1α dimerizes like IL-8, but receptor-binding properties of the SDF-1α dimer are distinctly different from the IL-8 dimer (47, 49). Whereas the structure of the IL-8 dimer–receptor complex reveals binding interactions to only one subunit, the structure of the trapped SDF-1α–receptor complex reveals that the binding involves residues from both subunits of the dimer (18, 47). Whereas binding of the IL-8 dimer to the CXCR1 N-domain results in the dissociation of the complex, binding of the SDF-1α monomer to the CXCR4 N-domain actually results in ligand dimerization (49). Though CXC and CC monomer structures are similar, they dimerize using different regions of the protein. CXC chemokine dimerization involves the third β-strand and α-helical residues, and CC chemokine dimerization involves the N-loop residues. The disulfide-linked CC dimer MIP-1β was shown to be incapable of binding its receptor, and under conditions where both monomer and dimer exist, only the RANTES monomer was shown to bind the CCR5 N-domain (42, 50). Nevertheless, CC chemokine monomer mutants are inactive in vivo, indicating that their dimerization is essential for functions other than for binding their GPCR receptors (51).
In summary, our results together indicate that the two fundamental properties shared by chemokines, the ability to exist as monomers and dimers and the interaction of ligand N-loop residues with receptor N-domain residues for binding and function, are not independent but coupled. We propose that such coupling interactions play a critical role in regulating in vivo function.
Acknowledgments
We thank Dr. Shanmin Zhang and Dr. Sean Moran for help with NMR instrumentation and the John S. Dunn Gulf Course Consortium for Magnetic Resonance for access to the 800MHz spectrometer. We also thank Dr. Gregg Nagle for protein purification, Mr. Christopher Chin for assistance with mass spectrometry, and Dr. Junji Iwahara for useful discussions.
Footnotes
Abbreviations: IL-8, interleukin-8; CXCR, CXC chemokine receptor; GPCR, G protein coupled receptor; N-domain, N-terminal domain; NMR, nuclear magnetic resonance; WT, wild type; HSQC, heteronuclear single-quantum coherence; ASA, accessible surface area; ITC, isothermal titration calorimetry.
This work was supported in part by grants from the National Institutes of Health (R01AI069152) and the John Sealy Endowment Fund (to K.R.) and a predoctoral fellowship from the American Heart Association (0815354F to A.R.)
References
- 1.Moser B, Wolf M, Walz A, Loetscher P. Chemokines: multiple levels of leukocyte migration control. Trends Immunol. 2004;25:75–84. doi: 10.1016/j.it.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 2.Luster AD. The role of chemokines in linking innate and adaptive immunity. Curr Opin Immunol. 2002;14:129–135. doi: 10.1016/s0952-7915(01)00308-9. [DOI] [PubMed] [Google Scholar]
- 3.Gerard C, Rollins BJ. Chemokines and disease. Nat Immunol. 2001;2:108–115. doi: 10.1038/84209. [DOI] [PubMed] [Google Scholar]
- 4.Rajarathnam K. Designing decoys for chemokine-chemokine receptor interaction. Curr Pharm Des. 2002;8:2159–2169. doi: 10.2174/1381612023393233. [DOI] [PubMed] [Google Scholar]
- 5.Luster AD. Chemokines—chemotactic cytokines that mediate inflammation. N Engl J Med. 1998;338:436–445. doi: 10.1056/NEJM199802123380706. [DOI] [PubMed] [Google Scholar]
- 6.Clore GM, Appella E, Yamada M, Matsushima K, Gronenborn AM. Three-dimensional structure of interleukin 8 in solution. Biochemistry. 1990;29:1689–1696. doi: 10.1021/bi00459a004. [DOI] [PubMed] [Google Scholar]
- 7.Rajarathnam K, Clark-Lewis I, Sykes BD. 1H NMR solution structure of an active monomeric interleukin-8. Biochemistry. 1995;34:12983–12990. doi: 10.1021/bi00040a008. [DOI] [PubMed] [Google Scholar]
- 8.Rajarathnam K, Prado GN, Fernando H, Clark-Lewis I, Navarro J. Probing receptor binding activity of interleukin-8 dimer using a disulfide trap. Biochemistry. 2006;45:7882–7888. doi: 10.1021/bi0605944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark-Lewis I, Schumacher C, Baggiolini M, Moser B. Structure-activity relationships of interleukin-8 determined using chemically synthesized analogs. Critical role of NH2-terminal residues and evidence for uncoupling of neutrophil chemotaxis, exocytosis, and receptor binding activities. J Biol Chem. 1991;266:23128–23134. [PubMed] [Google Scholar]
- 10.Clark-Lewis I, Dewald B, Geiser T, Moser B, Baggiolini M. Platelet factor 4 binds to interleukin 8 receptors and activates neutrophils when its N terminus is modified with Glu-Leu-Arg. Proc Natl Acad Sci USA. 1993;90:3574–3577. doi: 10.1073/pnas.90.8.3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark-Lewis I, Dewald B, Loetscher M, Moser B, Baggiolini M. Structural requirements for interleukin-8 function identified by design of analogs and CXC chemokine hybrids. J Biol Chem. 1994;269:16075–16081. [PubMed] [Google Scholar]
- 12.Lowman HB, Slagle PH, DeForge LE, Wirth CM, Gillece-Castro BL, Bourell JH, Fairbrother WJ. Exchanging interleukin-8 and melanoma growth-stimulating activity receptor binding specificities. J Biol Chem. 1996;271:14344–14352. doi: 10.1074/jbc.271.24.14344. [DOI] [PubMed] [Google Scholar]
- 13.Suetomi K, Lu Z, Heck T, Wood TG, Prusak DJ, Dunn KJ, Navarro J. Differential mechanisms of recognition and activation of interleukin-8 receptor subtypes. J Biol Chem. 1999;274:11768–11772. doi: 10.1074/jbc.274.17.11768. [DOI] [PubMed] [Google Scholar]
- 14.Williams G, Borkakoti N, Bottomley GA, Cowan I, Fallowfield AG, Jones PS, Kirtland SJ, Price GJ, Price L. Mutagenesis studies of interleukin-8. Identification of a second epitope involved in receptor binding. J Biol Chem. 1996;271:9579–9586. doi: 10.1074/jbc.271.16.9579. [DOI] [PubMed] [Google Scholar]
- 15.Gayle RB, III, Sleath PR, Srinivason S, Birks CW, Weerawarna KS, Cerretti DP, Kozlosky CJ, Nelson N, Vanden Bos T, Beckmann MP. Importance of the amino terminus of the interleukin-8 receptor in ligand interactions. J Biol Chem. 1993;268:7283–7289. [PubMed] [Google Scholar]
- 16.Crump MP, Gong JH, Loetscher P, Rajarathnam K, Amara A, Arenzana-Seisdedos F, Virelizier JL, Baggiolini M, Sykes BD, Clark-Lewis I. Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1. EMBO J. 1997;16:6996–7007. doi: 10.1093/emboj/16.23.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajagopalan L, Rajarathnam K. Structural basis of chemokine receptor function—a model for binding affinity and ligand selectivity. Biosci Rep. 2006;26:325–339. doi: 10.1007/s10540-006-9025-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skelton NJ, Quan C, Reilly D, Lowman H. Structure of a CXC chemokine-receptor fragment in complex with interleukin-8. Structure. 1999;7:157–168. doi: 10.1016/S0969-2126(99)80022-7. [DOI] [PubMed] [Google Scholar]
- 19.Leong SR, Kabakoff RC, Hebert CA. Complete mutagenesis of the extracellular domain of interleukin-8 (IL-8) type A receptor identifies charged residues mediating IL-8 binding and signal transduction. J Biol Chem. 1994;269:19343–19348. [PubMed] [Google Scholar]
- 20.LaRosa GJ, Thomas KM, Kaufmann ME, Mark R, White M, Taylor L, Gray G, Witt D, Navarro J. Amino terminus of the interleukin-8 receptor is a major determinant of receptor subtype specificity. J Biol Chem. 1992;267:25402–25406. [PubMed] [Google Scholar]
- 21.Burrows SD, Doyle ML, Murphy KP, Franklin SG, White JR, Brooks I, McNulty DE, Scott MO, Knutson JR, Porter D, et al. Determination of the monomer-dimer equilibrium of interleukin-8 reveals it is a monomer at physiological concentrations. Biochemistry. 1994;33:12741–12745. doi: 10.1021/bi00209a002. [DOI] [PubMed] [Google Scholar]
- 22.Rajarathnam K, Kay CM, Clark-Lewis I, Sykes BD. Characterization of quaternary structure of interleukin-8 and functional implications. Methods Enzymol. 1997;287:89–105. doi: 10.1016/s0076-6879(97)87009-7. [DOI] [PubMed] [Google Scholar]
- 23.Lowman HB, Fairbrother WJ, Slagle PH, Kabakoff R, Liu J, Shire S, Hebert CA. Monomeric variants of IL-8: effects of side chain substitutions and solution conditions upon dimer formation. Protein Sci. 1997;6:598–608. doi: 10.1002/pro.5560060309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajagopalan L, Rajarathnam K. Ligand selectivity and affinity of chemokine receptor CXCR1. Role of N-terminal domain. J Biol Chem. 2004;279:30000–30008. doi: 10.1074/jbc.M313883200. [DOI] [PubMed] [Google Scholar]
- 25.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 26.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 27.Johnson BA, Blevins RA. NMR View: a computer program for the visualization and analysis of NMR data. J Biol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 28.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 29.Spyracopoulos L. A suite of Mathematica notebooks for the analysis of protein main chain 15N NMR relaxation data. J Biomol NMR. 2006;36:215–224. doi: 10.1007/s10858-006-9083-0. [DOI] [PubMed] [Google Scholar]
- 30.Lian L, Roberts GCK. Effects of chemical exchange on NMR spectra, in NMR of macromolecules. In: Roberts GCK, editor. A practical approach. Oxford University Press; Oxford, U.K: 1993. pp. 153–182. [Google Scholar]
- 31.Qin J, Vinogradova O, Gronenborn AM. Protein-protein interactions probed by nuclear magnetic resonance spectroscopy. Methods Enzymol. 2001;339:377–389. doi: 10.1016/s0076-6879(01)39323-0. [DOI] [PubMed] [Google Scholar]
- 32.Reuben J, Fiat D. Nuclear magnetic resonance studies of solutions of the rare-earth ions and their complexes. IV. Concentration and temperature dependence of the oxygen-17 transverse relaxation in aqueous solutions. J Chem Phys. 1969;51:10. [Google Scholar]
- 33.Hammond ME, Shyamala V, Siani MA, Gallegos CA, Feucht PH, Abbott J, Lapointe GR, Moghadam M, Khoja H, Zakel J, Tekamp-Olson P. Receptor recognition and specificity of interleukin-8 is determined by residues that cluster near a surface-accessible hydrophobic pocket. J Biol Chem. 1996;271:8228–8235. doi: 10.1074/jbc.271.14.8228. [DOI] [PubMed] [Google Scholar]
- 34.Schraufstatter IU, Ma M, Oades ZG, Barritt DS, Cochrane CG. The role of Tyr13 and Lys15 of interleukin-8 in the high affinity interaction with the interleukin-8 receptor type A. J Biol Chem. 1995;270:10428–10431. doi: 10.1074/jbc.270.18.10428. [DOI] [PubMed] [Google Scholar]
- 35.Rajarathnam K, Sykes BD, Dewald B, Baggiolini M, Clark-Lewis I. Disulfide bridges in interleukin-8 probed using non-natural disulfide analogues: dissociation of roles in structure from function. Biochemistry. 1999;38:7653–7658. doi: 10.1021/bi990033v. [DOI] [PubMed] [Google Scholar]
- 36.Kuschert GS, Hoogewerf AJ, Proudfoot AE, Chung CW, Cooke RM, Hubbard RE, Wells TN, Sanderson PN. Identification of a glycosaminoglycan binding surface on human interleukin-8. Biochemistry. 1998;37:11193–11201. doi: 10.1021/bi972867o. [DOI] [PubMed] [Google Scholar]
- 37.Clubb RT, Omichinski JG, Clore GM, Gronenborn AM. Mapping the binding surface of interleukin-8 complexes with an N-terminal fragment of the type 1 human interleukin-8 receptor. FEBS Lett. 1994;338:93–97. doi: 10.1016/0014-5793(94)80123-1. [DOI] [PubMed] [Google Scholar]
- 38.Grasberger BL, Gronenborn AM, Clore GM. Analysis of the backbone dynamics of interleukin-8 by 15N relaxation measurements. J Mol Biol. 1993;230:364–372. doi: 10.1006/jmbi.1993.1152. [DOI] [PubMed] [Google Scholar]
- 39.Fernando H, Nagle GT, Rajarathnam K. Thermodynamic characterization of interleukin-8 monomer binding to CXCR1 receptor N-terminal domain. FEBS J. 2007;274:241–251. doi: 10.1111/j.1742-4658.2006.05579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fernando H, Chin C, Rosgen J, Rajarathnam K. Dimer dissociation is essential for interleukin-8 (IL-8) binding to CXCR1 receptor. J Biol Chem. 2004;279:36175–36178. doi: 10.1074/jbc.C400283200. [DOI] [PubMed] [Google Scholar]
- 41.Shoemaker BA, Portman JJ, Wolynes PG. Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc Natl Acad Sci USA. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duma L, Haussinger D, Rogowski M, Lusso P, Grzesiek S. Recognition of RANTES by extracellular parts of the CCR5 receptor. J Mol Biol. 2007;365:1063–1075. doi: 10.1016/j.jmb.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 43.Ye J, Kohli LL, Stone MJ. Characterization of binding between the chemokine eotaxin and peptides derived from the chemokine receptor CCR3. J Biol Chem. 2000;275:27250–27257. doi: 10.1074/jbc.M003925200. [DOI] [PubMed] [Google Scholar]
- 44.Mizoue LS, Bazan JF, Johnson EC, Handel TM. Solution structure and dynamics of the CX3C chemokine domain of fractalkine and its interaction with an N-terminal fragment of CX3CR1. Biochemistry. 1999;38:1402–1414. doi: 10.1021/bi9820614. [DOI] [PubMed] [Google Scholar]
- 45.Mayer KL, Stone MJ. NMR solution structure and receptor peptide binding of the CC chemokine eotaxin-2. Biochemistry. 2000;39:8382–8395. doi: 10.1021/bi000523j. [DOI] [PubMed] [Google Scholar]
- 46.Booth V, Keizer DW, Kamphuis MB, Clark-Lewis I, Sykes BD. The CXCR3 binding chemokine IP-10/CXCL10: structure and receptor interactions. Biochemistry. 2002;41:10418–10425. doi: 10.1021/bi026020q. [DOI] [PubMed] [Google Scholar]
- 47.Veldkamp CT, Seibert C, Peterson FC, Sakmar TP, Volkman BF. Recognition of a CXCR4 sulfotyrosine by the chemokine stromal cell-derived factor-1alpha (SDF-1alpha/CXCL12) J Mol Biol. 2006;359:1400–1409. doi: 10.1016/j.jmb.2006.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chuntharapai A, Kim KJ. Regulation of the expression of IL-8 receptor A/B by IL-8: possible functions of each receptor. J Immunol. 1995;155:2587–2594. [PubMed] [Google Scholar]
- 49.Veldkamp CT, Seibert C, Peterson FC, De la Cruz NB, Haugner JC, III, Basnet H, Sakmar TP, Volkman BF. Structural basis of CXCR4 sulfotyrosine recognition by the chemokine SDF-1/CXCL12. Sci Signal. 2008;1:ra4. doi: 10.1126/scisignal.1160755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin H, Shen X, Baggett BR, Kong X, LiWang PJ. The human CC chemokine MIP-1beta dimer is not competent to bind to the CCR5 receptor. J Biol Chem. 2007;282:27976–27983. doi: 10.1074/jbc.M702654200. [DOI] [PubMed] [Google Scholar]
- 51.Proudfoot AE, Handel TM, Johnson Z, Lau EK, LiWang P, Clark-Lewis I, Borlat F, Wells TN, Kosco-Vilbois MH. Glycosaminoglycan binding and oligomerization are essential for the in vivo activity of certain chemokines. Proc Natl Acad Sci U SA. 2003;100:1885–1890. doi: 10.1073/pnas.0334864100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 53.Willard L, Ranjan A, Zhang H, Monzavi H, Boyko RF, Sykes BD, Wishart DS. VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acids Res. 2003;31:3316–3319. doi: 10.1093/nar/gkg565. [DOI] [PMC free article] [PubMed] [Google Scholar]