Abstract
Backcrossed inbred lines (BILs) and a set of reciprocal chromosome segment substitution lines (CSSLs) derived from crosses between japonica rice cultivars Nipponbare and Koshihikari were used to detect quantitative trait loci (QTLs) for pre-harvest sprouting resistance. In the BILs, we detected one QTL on chromosome 3 and one QTL on chromosome 12. The QTL on the short arm of chromosome 3 accounted for 45.0% of the phenotypic variance and the Nipponbare allele of the QTL increased germination percentage by 21.3%. In the CSSLs, we detected seven QTLs, which were located on chromosomes 2, 3 (two), 5, 8 and 11 (two). All Nipponbare alleles of the QTLs were associated with an increased rate of germination. The major QTL for pre-harvest sprouting resistance on the short arm of chromosome 3 was localized to a 474-kbp region in the Nipponbare genome by the SSR markers RM14240 and RM14275 by using 11 substitution lines to replace the different short chromosome segments on chromosome 3. This QTL co-localized with the low-temperature germinability gene qLTG3-1. The level of germinability under low temperature strongly correlated with the level of pre-harvest sprouting resistance in the substitution lines. Sequence analyses revealed a novel functional allele of qLTG3-1 in Nipponbare and a loss-of-function allele in Koshihikari. The allelic difference in qLTG3-1 between Nipponbare and Koshihikari is likely to be associated with differences in both pre-harvest sprouting resistance and low-temperature germinability.
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-010-1275-z) contains supplementary material, which is available to authorized users.
Introduction
Seed dormancy is an important trait in cereal crops because lack of seed dormancy causes pre-harvest sprouting (Harlan et al. 1973). Pre-harvest sprouting often occurs in hot, humid conditions at maturity and results in reduced grain quality. The degree of seed dormancy is affected by genetic factors controlling germination including the mechanical hardness of seed coats, the morphological or physiological immaturity of embryos, and the activity of specific metabolic systems (Bewley 1997; Bentsink et al. 2007). Phytohormones are important signaling molecules for germinability and expression of pre-harvest sprouting (McCarty 1995; Gubler et al. 2005). Gibberellin promotes germination, whereas abscisic acid is involved in the onset and maintenance of dormancy. Environmental conditions such as the temperature during the ripening period and the degree of maturity also control seed dormancy and pre-harvest sprouting (Roberts 1962; Anderson et al. 1993; Li and Foley 1997).
A wide range of variation in the level of seed dormancy or resistance to pre-harvest sprouting has been observed in rice as well as in other crop species (Foley 2001; Mori et al. 2005; Hori et al. 2007). Inheritance of these variations is complex (Foley 2001). Extensive genetic studies have revealed more than 40 quantitative trait loci (QTLs) associated with seed dormancy or pre-harvest sprouting resistance for cultivated rice (Oryza sativa L.) (Lin et al. 1998; Dong et al. 2003; Guo et al. 2004; Wan et al. 2005, 2006; Gao et al. 2008), wild relatives O. rufipogon and O. nivara (Cai and Morishima 2000; Thomson et al. 2003; Lee et al. 2005; Li et al. 2006) and weedy rice (Gu et al. 2004).
For verification and precise mapping of Sdr1, a QTL with a major effect on seed dormancy located on the short arm of chromosome 3, genetic analyses were carried out using backcrossed populations derived from a cross between Nipponbare (japonica cultivar) and Kasalath (indica cultivar) (Takeuchi et al. 2003). This study demonstrated that Sdr1 and the heading date QTL Hd8 were not identical but were closely linked with each other, suggesting that seed dormancy and heading date are independently regulated. Gu et al. (2008) performed fine linkage mapping of qSD12, a QTL with a major effect on seed dormancy detected in weedy rice. This QTL was narrowed down to a ~600 kbp genomic region located on the long arm of chromosome 12 and was suggested to function in the offspring tissue (embryo or endosperm).
Only indica cultivars have been used as dormancy parents in all previous QTL analyses using cultivated rice. Despite a wide variation in the degree of seed dormancy among japonica rice cultivars (Ikeda 1963), there are no reported genetic analyses of seed dormancy in japonica rice cultivars. This is most likely attributable to the extremely low frequency of DNA polymorphisms among japonica rice cultivars preventing genetic analyses, including QTL analysis, using progeny from japonica rice cultivars. The completion of the International Rice Genome Sequencing Project (IRGSP 2005) has allowed effective large-scale screening for polymorphic simple sequence repeats (SSRs) between japonica cultivars and has made it possible to conduct QTL analysis of their progeny. Recent QTL analyses using japonica cultivars have identified several QTLs with major effects on grain quality (Kobayashi et al. 2007; Tabata et al. 2007), heading date (Matsubara et al. 2008), culm length (Monden et al. 2009; Hori et al. 2009), and eating quality (Kobayashi and Tomita 2008; Takeuchi et al. 2008; Wada et al. 2008).
Here we aim to identify QTLs for pre-harvest sprouting resistance in the Japanese japonica rice cultivars Nipponbare and Koshihikari. Koshihikari showed strong resistance to pre-harvest sprouting, whereas Nipponbare showed moderate pre-harvest sprouting resistance. For QTL detection, we developed reciprocal chromosome segment substitution lines (CSSLs) derived from crosses between Nipponbare and Koshihikari. We identified a QTL on the short arm of chromosome 3 with a major effect on pre-harvest sprouting resistance. Substitution mapping narrowed down the position of this QTL to a small genomic region of 474 kbp. Our results suggest that this QTL is located in the same chromosomal region as qLTG3-1, which is a low-temperature germinability gene isolated by Fujino et al. (2008). We investigated seed germinability under low-temperature conditions in the substitution lines and discussed the physiological basis of the QTL for pre-harvest sprouting resistance.
Materials and methods
Plant materials
For primary QTL analysis, we used 79 backcrossed inbred lines (BILs) of Nipponbare/Koshihikari//Nipponbare (N-BILs), which had been developed by Matsubara et al. (2008).
For the verification and fine-mapping of QTLs detected in BILs, we developed a set of reciprocal chromosome segment substitution lines (CSSLs). The CSSLs were developed essentially as described by Ebitani et al. (2005) and this procedure is outlined in Supplemental Fig S1. Nipponbare was crossed with Koshihikari to produce F1 plants. The F1 plants were backcrossed three or four times with Nipponbare or Koshihikari to obtain reciprocal BC3F1 and BC4F1 plants. In every backcrossed generation, heterozygous plants at the target region to be substituted were selected by using DNA markers for further backcrossing or self-pollination. BC3F1 and BC4F1 plants were consecutively self-pollinated five and four times to substitute the target regions homozygous for the donor cultivar allele and those homozygous for the recurrent cultivar at other regions. Finally, the genotypes of the CSSLs were again examined by using the DNA markers. Sets of CSSLs were designated as N-CSSLs (Koshihikari segments in Nipponbare genetic background) and K-CSSLs (Nipponbare segments in Koshihikari genetic background), respectively.
To delimit the genomic region of the QTL, we developed 11 advanced substitution lines including nine lines with smaller Koshihikari segments of the short arm of chromosome 3 in a Nipponbare genetic background and 2 lines with smaller Nipponbare segments of the short arm of chromosome 3 in a Koshihikari genetic background. Two plants in which the target region was heterozygous and the other regions were homozygous for the Nipponbare or Koshihikari allele were selected from BC4F4 families, which were in the process of developing the N-CSSLs or K-CSSLs. Selected plants were consecutively self-pollinated one or two times to obtain the 11 advanced substitution lines in which the target region was homozygous for the Nipponbare or Koshihikari allele.
Genotyping of CSSLs and substitution lines by DNA markers
For genotyping the backcrossed plants during development of CSSLs, we used 104 SSR and 4 SNP markers that covered the 12 chromosomes from the lists provided by McCouch et al. (2002), IRGSP (2005), and Matsubara et al. (2008). Procedures of DNA extraction and polymorphism detection for the SSR markers are described by Matsubara et al. (2008) and Hori et al. (2009). An additional 24 SSR markers (McCouch et al. 2002; IRGSP 2005) that showed polymorphisms between Nipponbare and Koshihikari on the short arm of chromosome 3 were used to develop the 11 advanced substitution lines.
Evaluation for pre-harvest sprouting resistance
Nipponbare and Koshihikari were cultivated in an experimental field at the National Institute of Agrobiological Sciences (NIAS) (Kannondai, Tsukuba, Ibaraki, Japan) in 2007 and 2008. N-BILs, N-CSSLs, K-CSSLs, and advanced substitution lines were cultivated at the NIAS field in 2008. Seeds were sown in mid April, and seedlings were transplanted into the paddy field in mid May in double-row plots with a distance of 18 cm between plants and 30 cm between rows. Germination percentage (GP) per panicle was used to score the level of resistance to pre-harvest sprouting. Ten panicles of Nipponbare and Koshihikari were harvested at 4–8 weeks (28–56 days) after heading in 2007 and at 4–10 weeks (28–70 days) after heading in 2008. Five panicles of each line in N-BILs, N-CSSLs, K-CSSLs, and advanced substitution lines were harvested at 7 weeks (49 days) after heading in 2008. Harvested panicles were immediately placed under 30°C and 100% humidity. After 7 days incubation, the number of germinated seeds per panicle was counted. Average GPs were calculated for each line.
QTL detection and substitution mapping
In N-BILs, linkage mapping was performed using version 3.0 of MAPMAKER/EXP (Lander et al. 1987). The Kosambi map function was used to calculate genetic distances (Kosambi 1944). QTL analyses were performed using composite interval mapping as implemented by the Zmapqtl program (model 6) provided by version 2.5 of the software package QTL Cartographer (Basten et al. 2005; http://statgen.ncsu.edu/qtlcart/WQTLCart.htm). Genome-wide threshold values (α = 0.05) were used to detect putative QTLs based on the results of 1,000 permutations.
In N-CSSLs, K-CSSLs, and advanced substitution lines, GPs were compared with each recurrent parent, Nipponbare or Koshihikari, by using the Dunnett’s multiple comparison procedure provided by the JMP 6.0 software. QTLs were declared present when GPs were significantly different between each line and the recurrent parents.
Sequence analysis of the low-temperature germinability gene qLTG3-1
To clarify sequence differences within the low-temperature germinability gene qLTG3-1 that are located within the same genomic region as the target QTL on chromosome 3, we amplified genomic fragments of Nipponbare and Koshihikari by PCR using primers specific for genomic sequences of the qLTG3-1 gene (Fujino et al. 2008). We directly sequenced the amplified fragments (0.5–1.0 kbp) from both the 3′ and 5′ ends with appropriate primers using an ABI3100 Genetic Analyzer (Applied Biosystems, Foster City, CA).
Germinability under low-temperature conditions
For evaluation of low-temperature germinability (LTG), mature seeds of each line were harvested in the field at an appropriate time and hand-threshed after drying 2 days at 30°C. Harvested seeds were then stored at 4°C for over the following 6 months before testing for germination to break seed dormancy. For each line 30 seeds were placed on a filter paper in a 9-cm Petri dish, and 10 mL of distilled water was added. The dishes were placed in an incubator at 15°C and 11°C. The number of germinated seeds was counted daily. GPs at the 18th day after incubation were used for genetic analysis.
GPs for germinability at low temperature for N-CSSLs, K-CSSLs, and advanced substitution lines were compared with those for each recurrent parent, Nipponbare or Koshihikari, by using the Dunnett’s multiple comparison procedure provided by the software package JMP 6.0. Pre-harvest sprouting resistance and germinability under low-temperature conditions were compared among the N-CSSLs, K-CSSLs, and advanced substitution lines to delimit chromosome regions associated with each trait.
Results
Changes in germination percentages after heading
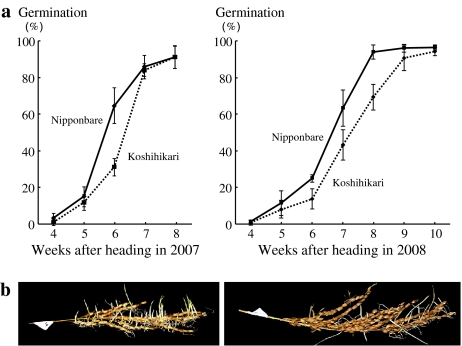
GPs of Nipponbare and Koshihikari were investigated from 4 to 8 weeks after heading in 2007 and from 4 to 10 weeks after heading in 2008 to determine appropriate evaluation points in their progenies (Fig. 1). In 2007 and 2008, the GPs for Nipponbare were increased during the early weeks after heading compared with those for Koshihikari. Differences in GP between Nipponbare and Koshihikari were greatest at 6 weeks in 2007 and at 7 weeks in 2008. The GP of Nipponbare was 64.0 ± 9.6% at 6 weeks in 2007 and 63.3 ± 9.9% at 7 weeks in 2008. The GP of Koshihikari was 31.2 ± 4.7% at 6 weeks in 2007 and 43.3 ± 8.2% at 7 weeks in 2008.
Fig. 1.
a Changes in germination percentages in the japonica rice cultivars Nipponbare and Koshihikari from 4 to 8 weeks after heading in 2007 and from 4 to 10 weeks after heading in 2008. Solid and broken lines indicate germination percentages for the Nipponbare and Koshihikari, respectively. Error bars indicate the standard deviations. b Panicles of Nipponbare (left) and Koshihikari (right) at 6 weeks after heading in 2007 following incubation at 30°C for 7 days
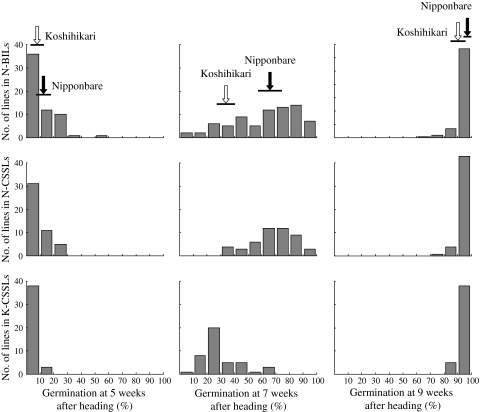
In N-BILs, N-CSSLs, and K-CSSLs over 80% of the lines showed a GP of less than 20% at 5 weeks and almost all lines showed a GP of more than 90% at 9 weeks in 2008 (Fig. 2). At 7 weeks, these populations showed a wide range of distribution in GPs and the BILs showed continuous and transgressive segregation (Fig. 2). Thus, we used the GPs at 7 weeks after heading in the following genetic analyses.
Fig. 2.
Frequency distributions for germination percentages at 5, 7 and 9 weeks after heading in 2008 in 79 of the Nipponbare/Koshihikari//Nipponbare backcrossed inbred lines (N-BILs), 48 of the Nipponbare/Koshihikari/////Nipponbare chromosome segment substitution lines (N-CSSLs) and 41 of the Nipponbare/Koshihikari/////Koshihikari chromosome segment substitution lines (K-CSSLs). Black and white arrows indicate the mean values for Nipponbare and Koshihikari, respectively. Horizontal lines under the arrows indicate the standard deviations
QTL detection in N-BILs
A QTL on chromosome 3 with a major effect on pre-harvest sprouting resistance was detected at the marker interval RM4108-RM5849 with an LOD score of 12.5 and explained variance of 45.0% (Table 1). Another QTL was detected on the short arm of chromosome 12. The Nipponbare alleles of the QTLs on chromosomes 3 and 12 increased the GP by 21.3 and 9.9%, respectively. The digenic interaction between these QTLs was not significant based on a two-way ANOVA using the marker locus closest to both the QTLs (data not shown). Some of the minor LOD peaks with lower statistical significance were found on the short arm of chromosome 5 and on the long arm of chromosome 11 (Suppl Fig. S2). No significant QTLs were detected at 5 and 9 weeks after heading when differences in GPs between the parents were small (data not shown).
Table 1.
QTL for resistance to pre-harvest sprouting at 7 weeks after heading in N-BILs
| Chr. | Marker interval | Nearest marker | LOD | PVE | AE (%) |
|---|---|---|---|---|---|
| 3 | RM4108-RM5849 | RM4108 | 12.5 | 45.0 | 21.3 |
| 12 | RM3455-RM6905 | RM3455 | 3.8 | 10.8 | 9.9 |
All genetic parameters were calculated by QTL Cartographer ver.2.5 (Basten et al. 2005). The LOD threshold for detection of QTL in the composite interval mapping procedure was 3.5
LOD Log-likelihood value, PVE percentage of total phenotypic variance explained by the QTL, AE additive effect of Nipponbare allele for germination percentage
QTL detection in N-CSSLs and K-CSSLs
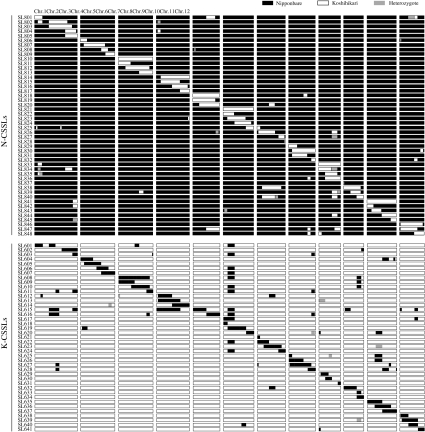
We developed a set of reciprocal CSSLs carrying Koshihikari chromosome segments in the Nipponbare genetic background (N-CSSLs) and carrying Nipponbare chromosome segments in the Koshihikari genetic background (K-CSSLs). Graphical representations of the genotypes of 48 N-CSSLs and 41 K-CSSLs using 104 SSR and 4 SNP markers are shown in Fig. 3. Each line contained a substituted segment of a particular chromosomal region and additional small segments in non-target regions. The substituted segments covered most of the rice genome, with the exception of four small regions on chromosomes 1 (defined by RM10695) and 4 (defined by RM16316) in N-CSSLs and on chromosome 1 (defined by RM10695 and RM1349) in K-CSSLs. Although we could not rule out the possibility that tiny chromosomal segments of donor lines might not be substituted (particularly at the distal end of each chromosome), the CSSLs developed can be used for the detection of QTLs with minor effects and verification of effects of the QTLs detected in BILs.
Fig. 3.
Graphical representation of genotypes of 48 N-CSSLs and 41 K-CSSLs by the 104 simple sequence repeat (SSR) and four single-nucleotide polymorphism (SNP) markers. Black, white, and shaded bars indicate genotypes homozygous for Nipponbare alleles, those homozygous for Koshihikari alleles, and those heterozygous for these alleles, respectively. The graphical genotypes shown here are based on the physical distance included in Build 4.0 Pseudomolecules of the Rice Genome (IRGSP 2005). Genotype data can be obtained on the website of the Rice Genome Resource Center at NIAS (http://www.rgrc.dna.affrc.go.jp/index.html)
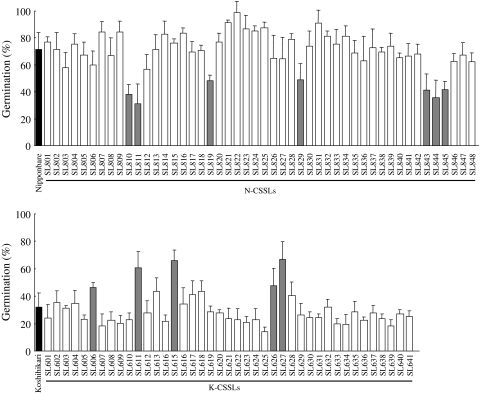
Accordingly, the GP at 7 weeks after heading was measured in the N-CSSLs and K-CSSLs (Fig. 4; Suppl Table 1). Among the N-CSSLs, seven lines including SL810 (substitution of short arm of chr. 3), SL811 (chr. 3), SL819 (short arm of chr. 5), SL829 (short arm of chr. 8), SL843 (long arm of chr. 11), SL844 (long arm of chr. 11) and SL845 (long arm of chr. 11), exhibited significantly lower GPs than those of Nipponbare. Among the K-CSSLs, five lines including SL606 (long arm of chr. 2), SL611 (long arm of chr. 3), SL615 (short arm of chr. 5), SL626 (short arm of chr. 8) and SL627 (short arm of chr. 8), exhibited significantly higher GPs than of Koshihikari. These results indicated that putative QTLs were located in the respective chromosome substitution segments. In all cases, the Koshihikari segments decreased the GP.
Fig. 4.
Germination percentages and standard deviations of Nipponbare, Koshihikari, 48 N-CSSLs and 41 K-CSSLs at 7 weeks after heading in 2008. Black bars indicate germination of the recurrent parents Nipponbare and Koshihikari. Shaded bars indicate significant difference (P < 0.05) between CSSLs and each recurrent parent by Dunnett’s test
Substitution mapping of the QTL on chromosome 3
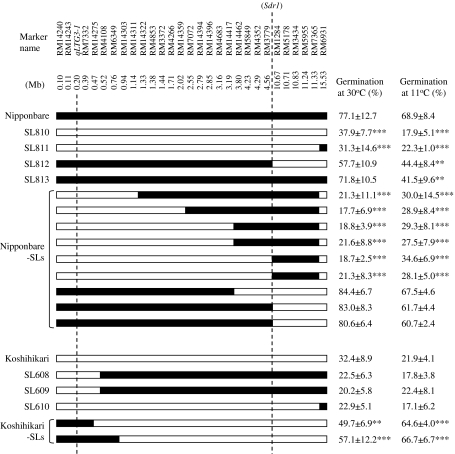
We performed substitution mapping to delimit the candidate genomic regions of the QTL on the short arm of chromosome 3. By means of marker-assisted selection using an additional 24 SSR markers, we developed nine advanced substitution lines harboring different Koshihikari segments and two advanced substitution lines harboring different Nipponbare segments on the short arm of chromosome 3 (Fig. 5). For six substitution lines with Koshihikari alleles at the distal region of the short arm of chromosome 3 in a Nipponbare genetic background, the GPs were significantly lower than those calculated for the Nipponbare. Two substitution lines with Nipponbare alleles on a Koshihikari genetic background substituted at marker intervals RM14240-RM7332 and RM14240-RM6349. The GPs of these two substitution lines were significantly greater than those calculated for the Koshihikari. Genotyping by using additional SSR markers revealed that the chromosome segments of Nipponbare in the two K-CSSLs SL608 and SL609 were not substituted at the marker interval RM14240-RM14275 (Fig. 3). The GPs of the three K-CSSLs were not significantly different to those calculated for Koshihikari. Taken together, these results reveal that the QTL was localized to a 474-kbp region in the distal region of short arm of chromosome 3 (Fig. 5).
Fig. 5.
Graphical genotype of the short arm of chromosome 3 in Nipponbare, N-CSSLs, Nipponbare substitution lines, Koshihikari, K-CSSLs and Koshihikari substitution lines. Positions of the SSR markers, the low-temperature germinability gene qLTG3-1, and the pre-harvest sprouting resistance QTL Sdr1 in the rice genome sequence (Build 4.0 Pseudomolecules of the Rice Genome; IRGSP 2005) are oriented from the short arm (left) to the long arm (right). Black and white bars indicate homozygous for the Nipponbare and Koshihikari alleles, respectively. ** and *** indicate germination percentages in pre-harvest sprouting test (30°C) and low-temperature germinability test (11°C) between CSSLs or substitution lines and their recurrent parents are significantly different at P < 0.01 and P < 0.001, respectively
Low-temperature germinability of CSSLs and advanced substitution lines
The qLTG3-1 gene is located at 198 kbp from the distal end of the short arm of chromosome 3, and a rice cultivar Italica Livorno has a functional allele whereas Hayamasari has a loss-of-function allele of qLTG3-1 (Fujino et al. 2008). It was interesting that qLTG3-1 is located within the 474-kbp candidate genomic region of the major QTL for pre-harvest sprouting resistance detected in the present study. To reveal the relationship between pre-harvest sprouting resistance and LTG, we determined the genomic sequences of about 1.5 kbp of the qLTG3-1 gene in Nipponbare and Koshihikari. We found a 71-bp deletion in the qLTG3-1 gene in Koshihikari, which is identical to the allele from Hayamasari (DDBJ accession No. AB510407) (Suppl Fig. S3). We found a nonsynonymous substitution in the first exon of qLTG3-1 in Nipponbare when we compared with the sequence of Italica Livorno [adenine in Nipponbare/thymine in Italica Livorno (DDBJ accession No. AB510406)] (Suppl Fig. S3). This substitution resulted in a leucine (Italica Livorno) to histidine (Nipponbare) substitution at 17th amino acid based on the genomic sequences of qLTG3-1.
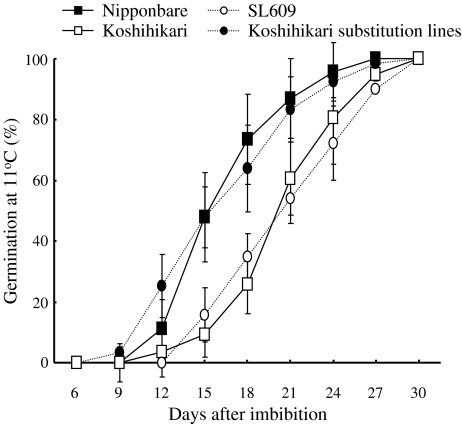
Nipponbare showed higher GPs than Koshihikari at 11°C from the 12th to the 27th day after seed imbibition (Fig. 6). At 11°C at the 18th day after seed imbibition, the average GP of Nipponbare was 63.8 ± 4.6%, whereas the average GP of Koshihikari was 26.3 ± 10.8% (Figs. 5, 6). All CSSLs and SLs having Nipponbare segments at the marker interval RM14240-qLTG3-1-RM14275 on the distal region of chromosome 3 showed high GPs at 11°C at the 18th day after seed imbibition (Fig. 5). These results indicate that the Nipponbare segments increased germinability at both 30 and 11°C.
Fig. 6.
Changes in germination percentages in Nipponbare, Koshihikari, SL609, and one advanced substitution line of Koshihikari under low-temperature conditions at 11°C. Germination percentages and standard deviations are shown as indicated
Discussion
Despite a wide variation in the degree of seed dormancy among japonica rice cultivars (Ikeda 1963), there are no reported genetic analyses of seed dormancy in japonica rice cultivars. Even though Koshihikari showed a relatively high level of pre-harvest sprouting resistance under field evaluation, this level was not comparable with that of indica cultivar and wild relatives, which exhibited deep seed dormancy. One of the reasons why no genetic analysis on the seed dormancy in japonica cultivars has been performed so far is the relatively small range of difference. In general, seed dormancy is deeply affected by the seed developmental stage and environmental conditions, particularly temperature and precipitation during ripening stage (Roberts 1962; Anderson et al. 1993; Li and Foley 1997). Therefore, evaluation of the level of seed dormancy under natural field conditions may often not be reliable. To consider this risk, we carefully characterized the changes in the level of seed dormancy (GP) at different stages of ripening for 2 years. In fact, the pattern of changes in the GP during ripening stage is slightly different in 2007 and 2008. In 2007, increasing of GPs of Nipponbare and Koshihikari was observed at the early weeks in comparison to 2008. This might be caused by the difference in temperature during ripening stage between 2007 and 2008. However, the general tendency of the changes in GP seems to be similar between 2007 and 2008. In 2008, differences in GPs between Nipponbare and Koshihikari were not observed early after heading at 5 weeks or late after heading at 9 weeks (Fig. 1). Thus, evaluations of backcrossed populations must be conducted at a particular stage of maturity around 7 weeks after heading in 2008. Careful monitoring of sampling stage made it possible to perform reliable genetic analyses of pre-harvest sprouting resistance between Nipponbare and Koshihikari.
A QTL on the short arm of chromosome 3 with a major effect on pre-harvest sprouting resistance was confirmed in N-CSSLs, but was not detected in K-CSSLs because of the lack of a line harboring Nipponbare segments at the distal end of the short arm of chromosome 3. Seven QTLs on pre-harvest sprouting resistance on chromosomes 2, 3 (two), 5, 8, and 11 (two) were detected by analysis of advanced mapping populations of CSSLs. We demonstrated that CSSLs, which carry a particular chromosome segment from a donor line in the genetic background of another line, is superior for detection of QTLs with small additive effects that are masked by QTLs with larger effects in primary populations including F2 and BILs (Ebitani et al. 2005; Takai et al. 2007; Ando et al. 2008). The large allelic difference at the QTL on chromosome 3 might make it difficult to show statistical significance for other QTLs with minor effects in the BILs. Use of CSSLs allowed verification of QTLs and demonstration of the potential of this approach for identifying QTLs with minor effects. The putative QTL on chromosome 12 in the BILs could not be verified in both N-CSSLs and K-CSSLs. We could not clearly explain this discrepancy. One possible explanation is that putative QTL may be a false positive (type I error). Another explanation is that multigenic interaction may be involved in the expression of the QTL on chromosome 12, although we could not detect any digenic interaction between the two QTLs on chromosomes 3 and 12 by ANOVA. Additional trials to detect QTL for pre-harvest sprouting under different environmental conditions might be necessary to clear this discrepancy.
The Nipponbare alleles of the all QTLs detected in the present study were associated with increased GPs. This result contradicts our observation of transgressive segregation for the GPs at 7 weeks after heading in N-BILs (Fig. 2). We monitored the GPs at several points after heading in parental lines, CSSLs, and advanced substitution lines, and showed that GPs dramatically changed within 1 week (Fig. 1). Although we tried to synchronize the developmental stage of the BILs for the evaluation of GPs, it is likely that the sampling stage was not the same for all lines. This resulted in a wider range of GPs in the BILs, in particular towards high GPs, suggesting that there might not be transgressive segregation.
More than 40 QTLs for pre-harvest sprouting resistance have been previously reported for cultivated, wild, and weedy rice (Lin et al. 1998; Cai and Morishima 2000; Dong et al. 2003; Thomson et al. 2003; Gu et al. 2004; Guo et al. 2004; Lee et al. 2005; Wan et al. 2005, 2006; Li et al. 2006; Gao et al. 2008). Among them, several QTLs for seed dormancy or pre-harvest sprouting were detected on the short arm of chromosome 3 (Lin et al. 1998; Guo et al. 2004; Lee et al. 2005; Wan et al. 2005, 2006). Takeuchi et al. (2003) have carried out fine-mapping of Sdr1, a QTL with a major effect on seed dormancy located on the short arm of chromosome 3, using a Nipponbare/Kasalath population. Sdr1 was mapped to the marker interval R10942-C2045 around 10.2 Mbp from the distal end of the short arm of chromosome 3. Here, we narrowed down the candidate genomic region of a major QTL for pre-harvest sprouting resistance to a 474-kbp region within the marker interval RM14240-RM14275 on the short arm of chromosome 3 (Fig. 5). This finding suggests that the major QTL for pre-harvest sprouting resistance detected in the present study is not Sdr1. In addition, based on comparisons of genomic positions of DNA markers, two QTLs detected previously near the Sdr1 locus (Guo et al. 2004; Wan et al. 2005) were also different from the QTLs detected here. We cannot, however, rule out the possibility that the QTL detected in present study is same as those reported by Lee et al. (2005) and Wan et al. (2006). To our knowledge, qSD12 is the only QTL to be identified on the long arm of chromosome 12 (Gu et al. 2004). Here, we identified a novel QTL on the short arm of chromosome 12 in the BILs, suggesting that this QTL was clearly different from the QTL reported by Gu et al. (2004). The QTLs on chromosomes 2 and 11 reported by Cai and Morishima (2000) are located near those identified in the present study. Lee et al. (2005) detected one QTL, sd3.2, located on the long arm of chromosome 3. On the short arm of chromosome 5, Lin et al. (1998), Dong et al. (2003), and Lee et al. (2005) identified the QTLs Sdr2, qPHS-5, and sd5, respectively. On the short arm of chromosome 8, Lin et al. (1998) detected the QTL Sdr5. These QTL regions overlapped with the QTLs detected in the present study. Based on the current information, we could not clarify whether the QTLs detected in the present study were the same as those previously identified. Further analysis is required for verification of these QTLs.
Several seed dormancy mutants were reported in Arabidopsis, maize and barley, and the responsible genes were involved in the abscisic acid and gibberellin biosynthesis or signaling pathways (McCarty 1995; Gubler et al. 2005). However, we could not find any homologous genes in rice for the mutant genes at the marker intervals of the six QTLs detected in the present study.
Ikehashi (1973) and Seshu and Sorrells (1986) reported that seed dormancy or pre-harvest sprouting resistance is correlated with germinability under various conditions including low temperature and osmotic pressure. It is necessary to clarify the relationships between pre-harvest sprouting resistance and germinability under different conditions to gain a deeper understanding of pre-harvest sprouting resistance. In the present study, we demonstrated a possible close relationship between LTG and resistance to pre-harvest sprouting. Although germinability of Nipponbare was significantly higher than that of Koshihikari at 11°C (Fig. 6), no difference in germinability could be observed between Nipponbare and Koshihikari at 15°C (Suppl Fig. S4). These results suggest that the phenotypic difference in LTG due to qLTG3-1 between Nipponbare and Koshihikari is smaller than that between Italica Livorno and Hayamasari. This notion could be supported by sequence analysis of qLTG3-1 in both Nipponbare and Koshihikari. The Nipponbare allele of qLTG3-1 having one nonsynonymous substitution may be a novel functional allele that is less functional than the Italica Livorno allele. Further genetic and physiological analyses are necessary to clarify the relationship between allelic difference in qLTG3-1 and variations in pre-harvest sprouting resistance.
Expression of qLTG3-1 is associated with vacuolation of the tissues covering the embryo and reduction of the mechanical resistance to the growth potential of the coleoptile (Fujino et al. 2008). The protein encoded by qLTG3-1 controls germinability under various stress conditions including low and high temperature, high salinity, and high osmotic pressure. Seedling emergence after direct seeding in submerged soil was a target trait in recent breeding programs (Ismail et al. 2009). Gao et al. (2008) suggested that resistance to pre-harvest sprouting might result in low germinability and insufficient seedling establishment. The results of our current study suggest that the inferior characteristic of low-temperature germinability might be attributable to the QTL associated with pre-harvest sprouting resistance. Regardless of whether the cause is pleiotropy or tight linkage, this association between positive and negative characteristics must be dissolved in order to breed improved cultivars. To this end, positional cloning or further delimitation of the candidate genomic regions of the QTLs for resistance to pre-harvest sprouting is required.
In general, map-based cloning of QTLs in populations from crosses between japonica rice cultivars is often difficult due to the low frequency of DNA polymorphisms. However, once we delimit the candidate genomic region of the target gene, a low level of polymorphism become advantageous. In the present study, the candidate genomic region of the QTL on chromosome 3 was delimited to less than 474 kbp by using SSR markers. Sequencing of the candidate genomic region containing this QTL is now feasible with the recent establishment of large-scale sequencing technologies (Blow 2007; Hutchison 2007). Sequence comparisons between the two parental lines will allow us to identify DNA polymorphisms that are associated with the QTL and have functional significance in regulating resistance to pre-harvest sprouting.
The novel CSSLs developed in the present study can be used to enhance the genetic analysis of phenotypic variations in japonica rice cultivars. The seeds and genotype data of these CSSLs are available at the Rice Genome Resource Center of the NIAS (http://www.rgrc.dna.affrc.go.jp/index.html).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors are grateful to Dr. Kenji Fujino for his useful suggestions. This work was supported by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Green Technology Project QT-1005 and Genomics for Agricultural Innovation NVR-0001 and IPG-0010).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Anderson JA, Sorrells ME, Tanksley SD. RFLP analysis of genomic regions associated with resistance to pre-harvest sprouting in wheat. Crop Sci. 1993;33:453–459. [Google Scholar]
- Ando T, Yamamoto T, Shimizu T, Ma XF, Shomura A, et al. Genetic dissection and pyramiding of quantitative traits for panicle architecture by using chromosomal segment substitution lines in rice. Theor Appl Genet. 2008;116:881–890. doi: 10.1007/s00122-008-0722-6. [DOI] [PubMed] [Google Scholar]
- Basten CJ, Weir BS, Zeng ZB. QTL cartographer, version 1.17. Raleigh: Department of Statistics, North Carolina State University; 2005. p. 189. [Google Scholar]
- Bentsink L, Soppe W, Koornneef M. Genetic aspects of seed dormancy. In: Bradford KJ, Nonogaki H, editors. Seed development, dormancy and germination. Oxford: Blackwell; 2007. pp. 264–304. [Google Scholar]
- Bewley JD. Seed germination and dormancy. Plant Cell. 1997;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow N. The personal side of genomics. Nature. 2007;449:627–630. doi: 10.1038/449627a. [DOI] [PubMed] [Google Scholar]
- Cai HW, Morishima H. Genomic regions affecting seed shattering and seed dormancy in rice. Theor Appl Genet. 2000;100:840–846. doi: 10.1007/s001220051360. [DOI] [Google Scholar]
- Dong Y, Tsozuki E, Kamiunten H, Terao H, Lin D, et al. Identification of quantitative trait loci associated with preharvest sprouting resistance in rice (Oryza sativa L.) Field Crops Res. 2003;81:133–139. doi: 10.1016/S0378-4290(02)00217-4. [DOI] [Google Scholar]
- Ebitani T, Takeuchi Y, Nonoue Y, Yamamoto T, Takeuchi K, et al. Construction and evaluation of chromosome segment substitution lines carrying overlapping chromosome segments of indica rice cultivar ‘Kasalath’ in a genetic background of japonica elite cultivar ‘Koshihikari’. Breed Sci. 2005;55:65–73. doi: 10.1270/jsbbs.55.65. [DOI] [Google Scholar]
- Foley ME. Seed dormancy: an update on terminology, physiological genetics, and quantitative trait loci regulating germinability. Weed Sci. 2001;49:305–317. doi: 10.1614/0043-1745(2001)049[0305:SDAUOT]2.0.CO;2. [DOI] [Google Scholar]
- Fujino K, Sekiguchi H, Matsuda Y, Sugimoto K, Ono K, Yano M. Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. Proc Natl Acad Sci USA. 2008;105:12623–12628. doi: 10.1073/pnas.0805303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FY, Ren GJ, Lu XJ, Sun SX, Li HJ, et al. QTL analysis for resistance to preharvest sprouting in rice (Oryza sativa) Plant Breed. 2008;127:268–273. doi: 10.1111/j.1439-0523.2007.01450.x. [DOI] [Google Scholar]
- Gu XY, Kianian SF, Foley ME. Multiple loci and epistases control genetic variation for seed dormancy in weedy rice (Oryza sativa) Genetics. 2004;166:1503–1516. doi: 10.1534/genetics.166.3.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu XY, Turnipseed EB, Foley ME. The qSD12 locus controls offspring tissue-imposed seed dormancy in rice. Genetics. 2008;179:2263–2273. doi: 10.1534/genetics.108.092007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Millar AA, Jacobsen JV. Dormancy release, ABA and preharvest sprouting. Curr Opin Plant Biol. 2005;8:183–187. doi: 10.1016/j.pbi.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Guo L, Zhu L, Xu Y, Zeng D, Wu P, et al. QTL analysis of seed dormancy in rice (Oryza sativa L.) Euphytica. 2004;140:155–162. doi: 10.1007/s10681-004-2293-1. [DOI] [Google Scholar]
- Harlan JR, de Wet JMJ, Price EG. Comparative evolution of cereals. Evolution. 1973;27:311–325. doi: 10.2307/2406971. [DOI] [PubMed] [Google Scholar]
- Hori K, Sato K, Takeda K. Detection of seed dormancy QTL in multiple mapping populations derived from crosses involving novel barley germplasm. Theor Appl Genet. 2007;115:869–876. doi: 10.1007/s00122-007-0620-3. [DOI] [PubMed] [Google Scholar]
- Hori K, Yamamoto T, Ebana K, Takeuchi Y, Yano M. A novel quantitative trait locus on chromosome 1 involved in semi-dwarfism in Japanese rice varieties. Breed Sci. 2009;59:285–295. doi: 10.1270/jsbbs.59.285. [DOI] [Google Scholar]
- Hutchison CA. DNA sequencing: bench to beside and beyond. Nucleic Acids Res. 2007;35:6227–6237. doi: 10.1093/nar/gkm688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M. Studies on the viviparous germination of rice seed. Bull Fac Agric Kagoshima Univ. 1963;13:89–115. [Google Scholar]
- Ikehashi H. Studies on the environmental and varietal differences of germination habitats in rice seeds with special reference to plant breeding. J Cent Agric Exp Stan. 1973;19:1–60. [Google Scholar]
- IRGSP The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Ismail AM, Ella ES, Vergara GV, Mackill DJ. Mechanisms associated with tolerance to flooding during germination and early seedling growth in rice (Oryza sativa) Ann Bot. 2009;103:197–209. doi: 10.1093/aob/mcn211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Tomita K. QTL detection for stickiness of cooked rice using recombinant inbred lines derived from crosses between japonica rice cultivars. Breed Sci. 2008;58:419–426. doi: 10.1270/jsbbs.58.419. [DOI] [Google Scholar]
- Kobayashi A, Genliang B, Shenghai Y, Tomita K. Detection of quantitative trait loci for white-back and basal-white kernels under high temperature stress in japonica rice varieties. Breed Sci. 2007;57:107–116. doi: 10.1270/jsbbs.57.107. [DOI] [Google Scholar]
- Kosambi DD. The estimation of map distance from recombination values. Ann Eugen. 1944;12:172–175. [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, et al. Mapmaker: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Oh CS, Suh JP, McCouch SR, Ahn SN. Identification of QTL for domestication-related and agronomic traits in an Oryza sativa × O. rufipogon BC1F7 population. Plant Breed. 2005;124:209–219. doi: 10.1111/j.1439-0523.2005.01092.x. [DOI] [Google Scholar]
- Li B, Foley ME. Genetic and molecular control of seed dormancy. Trends Plant Sci. 1997;2:384–389. doi: 10.1016/S1360-1385(97)90053-4. [DOI] [Google Scholar]
- Li C, Zhou A, Sang T. Genetic analysis of rice domestication syndrome with the wild annual species, Oryza nivara. New Phytol. 2006;170:185–193. doi: 10.1111/j.1469-8137.2005.01647.x. [DOI] [PubMed] [Google Scholar]
- Lin SY, Sasaki T, Yano M. Mapping quantitative trait loci controlling seed dormancy and heading date in rice. Oryza sativa L., using backcross inbred lines. Theor Appl Genet. 1998;96:997–1003. doi: 10.1007/s001220050831. [DOI] [Google Scholar]
- Matsubara K, Kono I, Hori K, Nonoue Y, Ono N, et al. Novel QTLs for photoperiodic flowering revealed by using reciprocal backcross inbred lines from crosses between japonica rice cultivars. Theor Appl Genet. 2008;117:935–945. doi: 10.1007/s00122-008-0833-0. [DOI] [PubMed] [Google Scholar]
- McCarty DR. Genetic control and integration of maturation and germination pathways in seed development. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:71–93. doi: 10.1146/annurev.pp.46.060195.000443. [DOI] [Google Scholar]
- McCouch SR, Teytelman L, Xu Y, Lobos KB, Clare K, et al. Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.) DNA Res. 2002;9:257–279. doi: 10.1093/dnares/9.6.257. [DOI] [PubMed] [Google Scholar]
- Monden Y, Naito K, Okumoto Y, Saito H, Oki N, et al. High potential of a transposon mPing as a marker system in japonica × japonica cross in rice. DNA Res. 2009;16:131–140. doi: 10.1093/dnares/dsp004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Uchino N, Chono M, Kato K, Miura H. Mapping QTL for grain dormancy on wheat chromosome 3A and the group 4 chromosomes, and their combined effect. Theor Appl Genet. 2005;110:1315–1323. doi: 10.1007/s00122-005-1972-1. [DOI] [PubMed] [Google Scholar]
- Roberts EH. Dormancy in rice seed. III. The influence of temperature, moisture and gaseous environment. J Exp Bot. 1962;13:75–94. doi: 10.1093/jxb/13.1.75. [DOI] [Google Scholar]
- Seshu DV, Sorrells ME. Genetic studies on seed dormancy in rice. In: International Rice Research Institute, editor. Rice genetics: proceedings of the international rice genetics symposium. Philippines: Manila; 1986. pp. 369–382. [Google Scholar]
- Tabata M, Hirabayashi H, Takeuchi Y, Ando I, Iida Y. Mapping of quantitative trait loci for the occurrence of white-back kernels associated with high temperatures during the ripening period of rice (Oryza sativa L.) Breed Sci. 2007;57:47–52. doi: 10.1270/jsbbs.57.47. [DOI] [Google Scholar]
- Takai T, Nonoue Y, Yamamoto S, Yamanouchi U, Matsubara K, et al. Development of chromosome segment substitution lines derived from backcross between indica donor rice cultivar ‘Nona Bokra’ and japonica recipient cultivar ‘Koshihikari’. Breed Sci. 2007;57:257–261. doi: 10.1270/jsbbs.57.257. [DOI] [Google Scholar]
- Takeuchi Y, Lin SY, Sasaki T, Yano M. Fine linkage mapping enables dissection of closely linked quantitative trait loci for seed dormancy and heading in rice. Theor Appl Genet. 2003;107:1174–1180. doi: 10.1007/s00122-003-1364-3. [DOI] [PubMed] [Google Scholar]
- Takeuchi Y, Hori K, Suzuki K, Nonoue Y, Takemoto-Kuno Y, et al. Major QTLs for eating quality of an elite Japanese rice cultivar, Koshihikari, on the short arm of chromosome 3. Breed Sci. 2008;58:437–445. doi: 10.1270/jsbbs.58.437. [DOI] [Google Scholar]
- Thomson MJ, Tai TH, McClung AM, Lai XH, Hinga ME, et al. Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor Appl Genet. 2003;107:479–493. doi: 10.1007/s00122-003-1270-8. [DOI] [PubMed] [Google Scholar]
- Wada T, Ogata T, Tsubone M, Uchimura Y, Matsue Y. Mapping of QTLs for eating quality and physicochemical properties of the japonica rice ‘Koshihikari’. Breed Sci. 2008;58:427–435. doi: 10.1270/jsbbs.58.427. [DOI] [Google Scholar]
- Wan JM, Cao YJ, Wang CM, Ikehashi H. Quantitative trait loci associated with seed dormancy in rice. Crop Sci. 2005;45:712–716. [Google Scholar]
- Wan JM, Jiang L, Tang JY, Wang CM, Hou MY, et al. Genetic dissection of the seed dormancy trait in cultivated rice (Oryza sativa L.) Plant Sci. 2006;170:786–792. doi: 10.1016/j.plantsci.2005.11.011. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.