Summary
Both natural and newly synthesized allopolyploids display nonadditive gene expression changes through genetic and epigenetic mechanisms. The nonadditively expressed genes include many microRNA (miRNA) targets, suggesting a role for miRNAs and their targets in morphological variation in the allopolyploids and their progenitors.
We produced dominant-negative transgenic allotetraploid plants in Arabidopsis using RNA interference (RNAi) that downregulates the expression of miRNA biogenesis genes, including DCL1 and AGO1.
RNAi of DCL1 and AGO1 led to dominant negative phenotypes and decreased accumulation of several miRNAs and a tasiRNA tested in the transgenic resynthesized allotetraploids.
The results demonstrated that miRNA biogenesis genes are effectively downregulated in the resynthesized allotetraploids containing redundant homoeologous genes that are difficult to be manipulated by conventional mutation screens. These lines will be useful for studying the effects of miRNA biogenesis genes on growth and developmental variation in the allopolyploids.
Keywords: AGO1, DCL1, polyploidy, hybrid vigor, microRNA, RNAi, small RNA
Introduction
Polyploids can be formed by whole-genome duplication within a species (autopolyploids) or by hybridization between related species followed by chromosomes doubling (allopolyploids) (Chen, 2007; Doyle et al., 2008). Estimates indicate that over 75% of flowering plants are polyploids (Masterson, 1994), and many important crops such as wheat, cotton, and canola are allopolyploids. Arabidopsis allotetraploids are readily formed by hybridizing two autotetraploids, Arabidopsis thaliana and Arabidopsis arenosa (Comai et al., 2000; Wang et al., 2004). Genome-wide analysis of gene expression indicates c. 15% (up to c. 35%) of the transcriptome divergence between A. thaliana and A. arenosa (Wang et al., 2006b). The majority of differentially expressed genes (c. 68%) between the parents are nonadditively expressed in two independently derived allotetraploids. Nonadditive expression refers to that the expression level of a gene (two homoeologous loci) in the allotetraploid is not equal to the sum of the two parental loci. It is considered to be a major source of nonadditive or novel phenotypic variation in the allotetraploids. For example, nonadditive regulation of FRI and FLC loci mediates flowering-time variation in Arabidopsis resynthesized and natural allopolyploids (Wang et al., 2006a). A recent study showed that nonadditive expression of the genes involved in the circadian clock regulation leads to vigorous growth through altered expression of downstream metabolic genes in the allotetraploids (Ni et al., 2009). Many microRNA (miRNA) target genes encode transcription factors and proteins that are important to plant growth and development (Carrington & Ambros, 2003; Kidner & Martienssen, 2005; Chuck et al., 2009). Interestingly, many miRNAs and their targets are nonadditively expressed in the allotetraploids (Wang et al., 2006b; Ha et al., 2009), suggesting their role in morphological and phenotypic variation in the allopolyploids.
One of the difficulties in studying gene function and epigenetic regulation in allotetraploids is genetic redundancy, because two or more homoeologous loci of the progenitors’ genes are present in an allopolyploid. Although Transfer DNA (T-DNA) insertion and Ethane Methyl Sulfonate (EMS) mutagenesis have been widely used to create gene knockout or knockdown mutants for genetic studies in plants, including Arabidopsis (Alonso et al., 2003; Maple & Moller, 2007), these conventional genetic screening strategies do not work well in polyploids. The dominant negative strategy using RNA interference (RNAi) or hairpin RNA (hp-RNA) is an effective approach to overcome genetic redundancy and has been used to downregulate the expression of MET1, DDM1 and HDT1 in natural allotetraploid, Arabidopsis suecica (Lawrence & Pikaard, 2003; Lawrence et al., 2004; Wang et al., 2004). Resynthesized Arabidopsis allotetraploids have not been transformed, and miRNA biogenesis genes have not been subjected to RNAi in allotetraploids. It is likely that RNAi for miRNA biogenesis genes may not work well in diploids because the small RNA biogenesis genes such as DCL1 are essential for growth and development and can only be maintained in heterozygous states (Gasciolli et al., 2005). In Arabidopsis, DICER-LIKE 1 (DCL1) is an RNaseIII-type enzyme and functions in the sequential processing of primary miRNAs (pri-miRNAs) to pre-mature miRNAs (pre-miRNAs) and finally mature miRNAs. The guide strand of a miRNA is then incorporated into the RNA-induced silencing complex (RISC) containing ARGONAUTE 1 (AGO1) for target gene downregulation (Chapman & Carrington, 2007b). In a previous study we showed that DCL1 and AGO1 are nonadditively expressed in allotetraploids, suggesting that miRNA biogenesis genes have a role in nonadditive accumulation of miRNAs in allopolyploids (Ha et al., 2009b). In this study, we used gene-specific hp-RNAs to downregulate DCL1 or AGO1 expression in Arabidopsis resynthesized allotetraploids, namely, Allo733 (Wang et al., 2006b). The RNAi of DCL1 and AGO1 allotetraploids displayed dominant negative phenotypes that are characteristic of these miRNA biogenesis genes in diploids. These lines are valuable materials for testing the effects of miRNAs and their targets on growth and developmental variation in resynthesized and natural allotetraploids.
Materials and Methods
Plant materials and growth conditions
The resynthesized allotetraploids were derived from a cross of A. thaliana (L.) Heynh. tetraploid pollinated with pollen from A. arenosa (L.) Lawalrée tetraploid and used for gene expression analysis in the F6 generation (Comai et al., 2000; Wang et al., 2004). One of the stable allotetraploids was self-pollinated to the F8 generation and used to generate transgenic plants. dcl4-2 T-DNA mutant (CS6954) was obtained from the Arabidopsis Biological Resource Center (ABRC). All plants were grown at 22°C under a 16 h light and 8 h dark cycle. All samples were collected between 10 o’clock in the morning and noon. for gene expression analyses.
RNAi constructs and plant transformation
Partial cDNA fragment of DCL1 (436-bp; At1g01040) and AGO1 (470-bp; At1g48410) were amplified from A. thaliana cDNA using the primer pairs with flanking 5′ attB1 and 3′ attB2 sites (Supporting Information Table S1). Individually amplified PCR product was cloned into pDONR221 using Gateway technology (Invitrogen) to generate an entry construct for subsequent recombination into the destination vector, pAGRIKOLA (Hilson et al., 2004). The resulting hp-RNA construct contained the partial cDNA fragment that was cloned into sense and antisense orientation flanking the Pdk and Cat introns. Individual RNAi construct was co-transformed with the helper plasmid, pSoup, into Agrobacterium tumefaciens GV3101 by electroporation. For plant transformation, Allo733 plants with many flower buds (c. 10 wk old) were dipped twice (with 1 wk apart) in Agrobacterium cultures containing the DCL1 or AGO1 RNAi construct using floral dip method (Clough & Bent, 1998). The plants were grown four more weeks after transformation, and the seeds were harvested for screening transgenics using herbicides. Primary transformants were selected on Gamborg’s B-5 Basal Medium (G5893; Sigma) containing 20 mg l−1 glufosinate-ammonium (Sigma). Genomic DNA was extracted from 4-wk-old leaves of selected transformants for PCR genotyping using a 5′ gene-specific primer (with the attB2 site) and a 3′-primer target the OCS terminator (Table S1). Here T1 seeds were referred to a mixture of transformed and untransformed seeds from floral dipped plants (T0). The transformant plants were selected from T1 seeds, and T2 plants were obtained from progenies (T2 seeds) of a T1 plant.
Total RNA isolation and qRT-PCR
Total RNA was extracted from leaves of 7–8 wk old plants (c. 100 mg tissues) using Trizol Reagent (Invitrogen) and treated with Turbo DNase (Ambion) according to the manufacturers’ instructions. An aliquot (1 μg) of DNase-treated RNAs was prepared for cDNA synthesis using Superscript III reverse transcriptase (Invitrogen) in the presence of 25 ng μl−1 oligo dT (12–18) primer. The synthesized cDNAs were diluted to 100 μl, and 1 μl of the diluted cDNA was used for quantitative (q)PCR using power SYBR green PCR mix (Applied Biosystems, Foster City, CA, USA) in the presence of 0.5 μm gene specific primers (Table S1). The qPCR was performed using 7500 Real Time PCR thermal cycler (Applied Biosystems). Expression levels of the endogenous target genes in RNAi lines were normalized against the Ubiquitin10 transcript levels. Relative expression levels of the target transcripts were derived by comparisons between an RNAi line and control transgenic lines that contain a plasmid vector only. A Student t-test was used to determine the significance of target gene downregulation in the RNAi lines relative to the control transgenic plants in three biological replications.
Small RNA detection
Total RNA (10 μg) was separated in a 15% polyacrylamide gel and transferred to a Hybond N+ membrane (GE Healthcare, Piscataway, NJ, USA) using a Mini-Protean II wet-transfer cell (BioRad). After ultraviolet (UV) crosslinking, the membrane was prehybridized in Church buffer (Church & Gilbert, 1984) at 37°C for 1 h. An oligonucleotide with the sequence complementary to the target mature miRNA was end-labeled with γ-P32 ATP (6000 Ci mmol−1) and purified using a G-25 Sephadex column (Pharmacia Fine Chemicals Inc., Piscataway, NJ, USA). The antisense U6 oligonucleotide was also end-labeled using 2 mCi ml−1 γ-P32. For hybridization, the small RNA blot was incubated overnight at 40°C in the presence of radioactive labeled probes. After hybridization, the blot was rinsed with 2× standard saline citrate (SSC) for 5 min, then washed twice with 2× SSC and 0.1% sodium dodecyl sulfate (SDS) for 15 min at 40 °C. Small RNA hybridization signals were detected by exposing the blot to a Phosphor imaging plate from 4 h to overnight and quantified using imagequant (Bio-Rad). Each hybridization experiment was replicated once. For detection of other miRNAs, the same blot was stripped in 0.1× SSC and 0.1% SDS solution at 95°C for 1 h and reprobed with an antisense oligonucleotide probe of another miRNA, as previously described, following a 1-h prehybridization in Church buffer at 37°C.
Results
RNAi-mediated downregulation of DCL1 and AGO1 in allotetraploids
Nonadditive expression of miRNAs in the allotetraploids may be caused by transcriptional regulation of homoeologous miRNA precursor loci and/or posttranscriptional regulation of miRNA biogenesis genes (Ha et al., 2009). In A. thaliana, DCL1 and AGO1 are two key components of the miRNA biogenesis pathway that play a critical role in growth and development (Meins et al., 2005; Ramachandran & Chen, 2008). Interestingly, several miRNA biogenesis genes, including DCL1 and AGO1, are nonadditively expressed in the allotetraploids, which may lead to differential accumulation of miRNAs and growth and developmental variation in resynthesized and natural allotetraploids (Ha et al., 2009). To test the role of miRNA biogenesis genes in miRNA production in allotetraploids, we produced dominant negative transgenic plants using RNAi of DCL1 and AGO1. The transgenic allotetraploid plants were generated using two hp-RNA constructs targeting A. thaliana DCL1 and AGO1, respectively, with pAGRIKOLA (Hilson et al., 2004) as the destination vector (Fig. 1a). To target DCL1, a 436-bp AtDCL1 cDNA region corresponding to amino acid residues 1–146 of AtDCL1 (At1g01040) (Fig. 1b) was used for the hp-RNA construct. A 470-bp cDNA encoding amino acid residues 675–831 of AtAGO1 (At1g48410) (Fig. 1c) was used as the hp-RNA construct to target AGO1. The nucleotide sequence alignment between AtDCL1 and AtAGO1 cDNA fragments and the corresponding A. arenosa homoeologous BAC sequences (Fig. 1b,c) showed 97.3% and 97% identities, respectively. With the hairpin construct, a 21- to 24-nt RNA predicted by computational analysis showed a perfect match with both AtDCL1 and AaDCL1. By contrast, the AtDCL1 fragment in the hp-RNA construct had no homology with other members (AtDCL2, AtDCL3 and AtDCL4) of the gene family in A. thaliana (data not shown). Among the 10 AtAGO genes, AGO1, AGO5 and AGO10 are in the same clade. The AtAGO1 sequence used in the hairpin construct is 64.9% identical to AGO10 and 64.5% identical to AGO5 (Fig. S1). Computational analysis did not find a region with continuous 21- to 24-bp that could target a sequence region other than AGO1. Outside the clade, the AtAGO1 fragment has a low sequence identity with other AGO family members (e.g. AGO4 and AGO6).
Fig. 1.
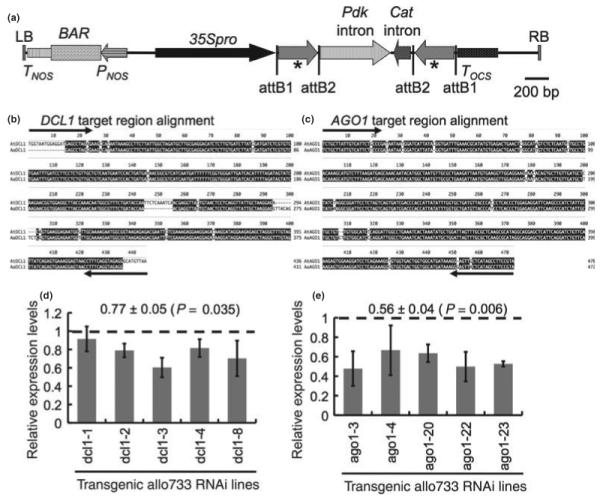
Hairpin RNA constructs for DCL1 and AGO1 and downregulation of DCL1 and AGO1 in transgenic plants. (a) Schematic diagram of T-DNA region of the hairpin RNA construct in pAGRIKOLA (Hilson et al., 2004). The hairpin RNA cassette was produced using partial cDNA fragments of DCL1 (436 bp) or AGO1 (470 bp) that was cloned into the T-DNA region flanking the Pdk and Cat introns using Gateway cloning vector. 35Spro, cauliflower mosaic virus 35S promoter; attB1 and attB2, recombination attachment sites from Gateway cloning vector; BAR, bialaphos resistance gene; Cat, intron of the castor bean Cat gene; LB, left border; Pdk, 2nd intron of the Flaveria Pdk gene; PNOS, nopaline synthase promoter; RB, right border; TNOS, nopaline synthase terminator; TOCS, octopine synthase terminator. Asterisks indicate the DCL1 or AGO1 partial cDNA fragments in the sense and antisense orientation. (b,c) clustal w alignment of target AtDCL1 (b) or AtAGO1 (c) fragment used in the RNAi construct with the corresponding bacterial artificial chromosome (BAC) sequence from Arabidopsis arenosa. Arrows show the primer binding sites in generating the hairpin constructs. (d,e) quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) showing downregulation of DCL1 and AGO1 transcripts in dcl1(Allo):RNAi lines (d) and ago1(Allo):RNAi lines (e) compared with the average expression level of the corresponding transcripts in two control transgenic lines containing a plasmid vector only (dashed line). Relative expression levels were estimated using Ubiquitin10 gene as an internal control. Values at top indicate the mean ± SE and P-value of target transcript reduction in the RNAi lines compared with the control transgenic lines. Error bars indicate standard deviation from replicated experiments.
Two hairpin constructs were transformed into the stable resynthesized allotetraploid line (Allo733). We produced 10 AGO1 (ago1(Allo):RNAi) and 11 DCL1 (dcl1(Allo):RNAi) transgenic plants. The transgenic plants were examined for the presence of transgenes using PCR and genomic DNA (data not shown). The T1 transgenic plants were self-pollinated, and one-third of them died or did not produce seeds. The expression levels of endogenous DCL1 and AGO1 were examined using quantitative reverse-transcription (qRT-)PCR analysis (Fig. 1d,e). Among the dcl1(Allo):RNAi lines tested, endogenous DCL1 expression levels were reduced, ranging from c. 8% in the dcl1-1(Allo):RNAi line to c. 40% in the dcl1-3(Allo):RNAi line relative to the control transgenic plants that contained a plasmid vector construct (Fig. 1d). On average, the DCL1 expression levels were reduced by 23% in the DCL1 RNAi lines (0.77 ± 0.05; n = 5; P = 0.035) compared with the DCL1 expression levels in the control transgenic plants. Downregulation of DCL1 expression by RNAi increased the number of leaves in some dcl1(Allo):RNAi lines (Fig. 2b), suggesting a role for DCL1 in cell proliferation and development, as observed in the caf-1 (CARPEL FACTORY, a DCL1-like homolog) mutants (Jacobsen et al., 1999; Park et al., 2002). Similar to homozygous dcl (sin1) mutants in Arabidopsis diploids (Ray et al., 1996), flowering time was delayed by 1–3 wk in the dcl1(Allo):RNAi lines compared with the control plants (flowered in c. 8 wk) in the T1 generation (Fig. 2f). However, flowering time of several dcl1(Allo):RNAi lines in the T2 generation was variable, suggesting that the RNAi effects on DCL1 repression are relatively unstable. Further investigation is required to fully characterize the effects of DCL1 downregulation on phenotypic variation, including flowering time.
Fig. 2.

RNAi-induced phenotypes in dcl1(Allo):RNAi and ago1(Allo):RNAi transgenic allotetraploids (Allo733, 8 wk old). (a) Wild-type control: resynthesized allotetraploids (Allo733) derived from Arabidopsis thaliana and Arabidopsis arenosa tetraploids. (b) dcl1-2(Allo):RNAi transgenic plants showing increased number of rosette leaves. (c) ago1-3(Allo):RNAi transgenic plant displaying an increased number of rosette leaves and an altered pattern of leaf serration. (d) Adaxial and (e) abaxial leaf morphology comparison between ago1(Allo):RNAi transgenic plants and other lines including A. thaliana diploid Col-0 and dcl4-2 mutant, tetraploid A. arenosa (Aa), natural allotetraploid Arabidopsis suecica (As), and resynthesized allotetraploid (Allo733). (f) Late-flowering phenotypes of dcl1-1(Allo) and ago1-1(Allo) RNAi lines compared with the control plant.
In the ago1(Allo):RNAi lines tested, the expression levels of endogenous AGO1 were reduced 33–52% (Fig. 1e). The ago1(Allo):RNAi lines showed an average of 44% reduction in AGO1 expression levels (0.56 ± 0.04; n = 5; P = 0.006) compared with the control transgenic plants. Phenotypically, some ago1(Allo):RNAi lines flowered late (Fig. 2f), and others displayed dramatic changes in leaf morphology with reduced levels of leaf serration and curled leaf phenotypes, which curled toward the abaxial side of the leaves (Fig. 2c,d,e). Interestingly, the curled leaf phenotype in ago1(Allo):RNAi lines resembles that in dcl4-2 mutant in A. thaliana diploids (Fig. 2d,e) (Xie et al., 2005; Yoshikawa et al., 2005). In addition, like dcl1(Allo):RNAi lines, down-regulation of AGO1 in Allo733 led to late flowering in several transgenic lines.
DCL1 and AGO1 RNAi lines showed decreased accumulation of small RNAs in allotetraploids
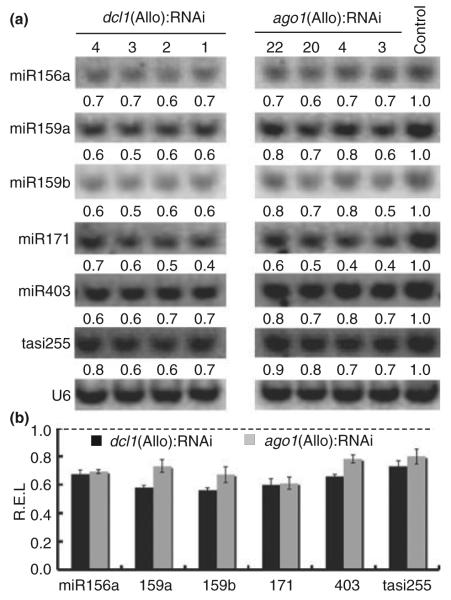
DCL1 and AGO1 are the key components of miRNA biogenesis pathway (Ramachandran & Chen, 2008). Consistent with the reduced expression levels of DCL1 and AGO1, accumulation of selected miRNAs was affected in both dcl1(Allo):RNAi and ago1(Allo):RNAi lines (Fig. 3a). In general, miRNA and tasiRNA levels were downregulated 20–50% in the dcl1(Allo):RNAi and ago1(Allo):RNAi lines. The amounts of miR159a, miR159b and miR403 were reduced more in dcl1(Allo):RNAi lines than in ago1(Allo):RNAi lines (Fig. 3b). The data suggest that RNAi of dcl1 and ago1 correlates with reduced accumulation of RNA and tasiRNA , consistent with the hypomorphic phenotypes observed.
Fig. 3.
Decreased levels of miRNA accumulation in dcl1(Allo) and ago1(Allo) RNAi lines. (a) Representative small RNA blots of selected miRNAs (miR156a, miR159a, miR159b, miR171a, and miR403) and tasiRNA (tasiR255) in dcl1(Allo):RNAi and ago1(Allo):RNAi lines. Averaged numbers (from two independent blots) indicate relative densitometric intensities of miRNA levels between the RNAi lines and a control transgenic plant. The hybridization signals were normalized using the U6 as an endogenous control. Densitometric intensities were obtained using imagequant software (Bio-Rad). (b) Averaged relative expression levels (REL) (mean ± SE) of miRNAs and tasiRNA shown in (a). The REL of each miRNA or tasiRNA was averaged from densitometric intensities in four dcl1(Allo):RNAi lines or ago1(Allo):RNAi lines, which was normalized to the transgenic line containing empty vector construct (dashed line).
Discussion
Many miRNAs and their targets control cell patterning, proliferation, growth and development in plants and animals (Pasquinelli & Ruvkun, 2002; Carrington & Ambros, 2003; Kidner & Martienssen, 2003; Chuck et al., 2009). In resynthesized allotetraploids derived from A. thaliana and its related species A. arenosa, many miRNAs and their targets that are differentially expressed between A. thaliana and A. arenosa are nonadditively expressed, suggesting an important role of miRNAs in morphological and developmental variation between closely related species and allopolyploids (Ha et al., 2009). Both EMS and T-DNA insertional mutants of DCL1 or AGO1 have been identified and characterized in several A. thaliana ecotypes (Col, Ler, Ws and C24) (Bohmert et al., 1998; Jacobsen et al., 1999; Lynn et al., 1999; Golden et al., 2002; Schauer et al., 2002; Baumberger & Baulcombe, 2005; Sorin et al., 2005; Yang et al., 2006; Brodersen et al., 2008). Unlike in A. thaliana diploids, T-DNA insertion and EMS mutagenesis approaches do not work effectively on reducing the expression of target genes in allotetraploids because of genetic redundancy. Therefore, dominant negative strategy has been employed to downregulate the expression of homoeologous genes including several chromatin genes in the allotetraploids. Stable RNAi lines of natural A. suecica have been generated for MET1, DDM1 and HDT1. RNAi of met1 and ddm1 in a natural allotetraploid A. suecica leads to activation of many transposons and heterochromatic genes (Wang et al., 2004; Chen et al., 2008), whereas silenced rDNA was reactivated in RNAi of hdt1 lines (Lawrence & Pikaard, 2003; Lawrence et al., 2004). The phenotypic abnormality of RNAi of met1 and ddm1 was severe in the selfing progeny, consistent with the met1 and ddm1 mutant lines in diploids (Vongs et al., 1993; Kankel et al., 2003). In this study, RNAi of dcl1 and ago1 in the resynthesized allotetraploids (Allo733) results in dramatic changes in leaf morphology and flowering time in the T1 generation. However, almost all dcl1(Allo):RNAi and ago1(Allo):RNAi lines with severe developmental phenotypes were not viable in T2 generation. It is possible that RNAi of dcl1 and ago1 may lead to embryonic lethality as observed in several dcl1 mutants in A. thaliana diploids (Jacobsen et al., 1999; McElver et al., 2001; Schauer et al., 2002). Alternatively, additional mutations may accumulate during selfing in the dcl1(Allo):RNAi lines as observed in the ddm1 mutants (Stokes & Richards, 2002). Only transgenic lines with low RNAi penetrance might have survived through meiotic transmission during selfing. Compared with ddm1 and met1 A. suecica RNAi lines, RNAi of dcl1 and ago1 in resynthesized allotetraploids gives rise to severe effects on growth and development in early generations of polyploidization, suggesting tight regulation of RNAi pathway on repression of DCL1 and AGO1. In addition, RNAi of chromatin genes and small RNA-biogenesis genes might have different effects in resynthesized and natural allotetraploids, and the resynthesized allotetraploids may be more sensitive to epigenetic perturbations than natural allotetraploids.
Several miRNAs (miR156a, miR159a, miR159b, miR171a and miR403) and a tasiRNA (tasiR255) accumulate in low amounts in both dcl1(Allo):RNAi and ago1(Allo):RNA lines, confirming the functional consequences of DCL1/AGO1 repression by RNAi. In Arabidopsis, small RNAs biogenesis is mediated by four distinct members of the DCL protein family (Henderson et al., 2006). Although four DCL proteins have partially redundant functions (Gasciolli et al., 2005), each DCL protein is associated with the production of a predominant class of small RNAs, including miRNAs by DCL1, siRNAs by DCL2 and DCL3, and tasiRNAs by DCL4 (Vazquez, 2006; Chapman & Carrington, 2007). No recovery of severe dcl1(Allo):RNAi lines is probably the result of specific silencing of DCL1 instead of other DCL genes because DCL1 has very low sequence identity with other DCLs in the RNAi target regions, and AtDCL1 and AaDCL1 are highly conserved in the RNAi target regions. The perfect match of predicted 21- to 24-nt RNA within the hairpin RNA with both AtDCL1 and AaDCL1 suggests that the homoeologous DCL1 loci are downregulated in the allotetraploids.
It is notable that the level of miRNA reduction does not necessarily correspond to the level of DCL1 repression. Several factors may affect the abundance of mature miRNA in allotetraploids. In addition to miRNA biogenesis factors such as DCL1 and AGO1, miRNA loci may be regulated at the transcriptional level (Ha et al., 2009). Homoeologous miRNA precursors in the allotetraploids may also be processed differently, which alters the abundance of mature miRNA.
Specific small RNAs are associated with different AGOs (effector proteins) within the RISC for direct mRNA cleavage, translation repression or transcriptional gene silencing (Chapman & Carrington, 2007; Hutvagner & Simard, 2008). Arabidopsis contains 10 AGO proteins that are classified into three lineages with AGO1, AGO5 and AGO10 within the same clade (Zheng et al., 2007; Hock & Meister, 2008). Both AtAGO5 and AtAGO10 share 65% nucleotide identity with the AtAGO1 hairpin construct. This may cause some complications with RNAi of ago1 in allotetraploids. AGO5 and AGO10 may not be severely repressed by RNAi of ago1. Mild developmental defects observed in ago1(Allo):RNAi lines are probably associated with AGO1, AGO5 and AGO10 redundancy (Lynn et al., 1999). Many genes, including DCL1 and AGO1, are nonadditively expressed in the allotetraploids (Ha et al., 2009), which may alter the effectiveness of RNAi on target (ago1 or dcl1) genes. The high levels of survival RNAi lines in the T2 generation may be related to partial expression of A. arenosa DCL1 and/or AGO1, leading to different levels of phenotypic abnormalities in these RNAi lines. In addition, downregulating AGO1 expression might have disrupted the homoeostasis of this protein and feedback regulation by miR168 (Vaucheret et al., 2006). The effects of AGO1 downregulation on target gene expression in the RNAi lines should be characterized in future studies.
The accumulation levels of miRNAs are more severely affected in dcl1(Allo):RNAi lines than in ago1(Allo):RNAi lines, which is consistent with targeted repression of DCL1 and redundant AGO1, AGO5 and AGO10 that have moderate levels of sequence identities. Moreover, DCL1 plays a major role in miRNA biogenesis, whereas AGO1 is an effector for both miRNA and siRNA pathways (Chapman & Carrington, 2007b). AGO1 functions in mediating miRNA-directed target mRNA cleavage, and incorporation of miRNA into AGO1 may prevent rapid turnover of miRNA. The decrease in miRNA accumulation in ago1(Allo):RNAi lines probably results from indirect effects of AGO1 repression or feedback regulation by miR168 (Vaucheret et al., 2006). The similar curled-leaf phenotypes between diploid dcl4-2 mutant and ago1(Allo):RNAi lines may suggest interactive roles of DCLs and AGOs in miRNA biogenesis and developmental regulation. For example, miRNA-directed cleavage of TAS transcripts (involving DCL1 and AGO1) may lead to DCL4-dependent tasiRNA production (Peragine et al., 2004; Vazquez et al., 2004; Xie et al., 2005). Flowering time variation in the dcl1 and ago1(Allo):RNAi lines can be caused by multiple factors, including environmental conditions and downregulation of miRNAs. For example, the late-flowering phenotype might be related to downregulation of miR172 in the RNAi lines because upregulation of miR172 induces early flowering in diploids (Aukerman & Sakai, 2003). Further investigation is required to fully characterize the effects of DCL1 and AGO1 downregulation on flowering time and other phenotypic changes.
In summary, we have produced a set of dominant negative RNAi lines of miRNA biogenesis genes (DCL1 and AGO1) in resynthesized allotetraploids. Although these lines need further characterization and investigation, the dcl1(Allo):RNAi and ago1(Allo):RNAi lines provide useful materials for studying different and specific roles of DCL1 and AGO1 in growth and developmental variation in the allopolyploids and their progenitors.
Supplementary Material
Acknowledgements
We thank Changqing Zhang and other members of the Chen laboratory for helpful discussion, and Mahek Mehta and Ngan Nguyen for plant care and technical assistance. The work was supported by the National Institutes of Health (GM067015) (Z.J.C.).
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC. Arabidopsis argonaute1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proceedings of the National Academy of Sciences, USA. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C. AGO1 defines a novel locus of arabidopsis controlling leaf development. European Molecular Biology Organization Journal. 1998;17:170–180. doi: 10.1093/emboj/17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nature Reviews Genetics. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- Chen ZJ. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annual Review of Plant Biology. 2007;58:377–406. doi: 10.1146/annurev.arplant.58.032806.103835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Ha M, Lackey E, Wang J, Chen ZJ. RNAi of met1 reduces DNA methylation and induces genome-specific changes in gene expression and centromeric small RNA accumulation in arabidopsis allopolyploids. Genetics. 2008;178:1845–1858. doi: 10.1534/genetics.107.086272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Candela H, Hake S. Big impacts by small RNAs in plant development. Current Opinion in Plant Biology. 2009;12:81–86. doi: 10.1016/j.pbi.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proceedings of the National Academy of Sciences, USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Comai L, Tyagi AP, Winter K, Holmes-Davis R, Reynolds SH, Stevens Y, Byers B. Phenotypic instability and rapid gene silencing in newly formed arabidopsis allotetraploids. Plant Cell. 2000;12:1551–1568. doi: 10.1105/tpc.12.9.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Flagel LE, Paterson AH, Rapp RA, Soltis DE, Soltis PS, Wendel JF. Evolutionary genetics of genome merger and doubling in plants. Annual Review of Genetics. 2008;42:443–461. doi: 10.1146/annurev.genet.42.110807.091524. [DOI] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Current Biology. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Golden TA, Schauer SE, Lang JD, Pien S, Mushegian AR, Grossniklaus U, Meinke DW, Ray A. Short integuments1/suspensor1/carpel factory, a dicer homolog, is a maternal effect gene required for embryo development in arabidopsis. Plant Physiology. 2002;130:808–822. doi: 10.1104/pp.003491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Lu J, Tian L, Ramachandran V, Kasschau KD, Chapman EJ, Carrington JC, Chen X, Wang XJ, Chen ZJ. Small RNAs serve as a genetic buffer against genomic shock in arabidopsis interspecific hybrids and allopolyploids. Proceedings of the National Academy of Sciences, USA. 2009;106:17835–17840. doi: 10.1073/pnas.0907003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, Green PJ, Jacobsen SE. Dissecting Arabidopsis thaliana dicer function in small RNA processing, gene silencing and DNA methylation patterning. Nature Genetics. 2006;38:721–725. doi: 10.1038/ng1804. [DOI] [PubMed] [Google Scholar]
- Hilson P, Allemeersch J, Altmann T, Aubourg S, Avon A, Beynon J, Bhalerao RP, Bitton F, Caboche M, Cannoot B, et al. Versatile gene-specific sequence tags for arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Research. 2004;14:2176–2189. doi: 10.1101/gr.2544504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock J, Meister G. The argonaute protein family. Genome Biology. 2008;9:210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nature Reviews Molecular Cell Biology. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Running MP, Meyerowitz EM. Disruption of an RNA helicase/RSAse iii gene in arabidopsis causes unregulated cell division in floral meristems. Development. 1999;126:5231–5243. doi: 10.1242/dev.126.23.5231. [DOI] [PubMed] [Google Scholar]
- Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics. 2003;163:1109–1122. doi: 10.1093/genetics/163.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA. Macro effects of microRNAs in plants. Trends in Genetics. 2003;19:13–16. doi: 10.1016/s0168-9525(02)00011-2. [DOI] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA. The developmental role of microRNA in plants. Current Opinion in Plant Biology. 2005;8:38–44. doi: 10.1016/j.pbi.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Lawrence RJ, Pikaard CS. Transgene-induced RNA interference: a strategy for overcoming gene redundancy in polyploids to generate loss-of-function mutations. Plant Journal. 2003;36:114–121. doi: 10.1046/j.1365-313x.2003.01857.x. [DOI] [PubMed] [Google Scholar]
- Lawrence RJ, Earley K, Pontes O, Silva M, Chen ZJ, Neves N, Viegas W, Pikaard CS. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Molecular Cell. 2004;13:599–609. doi: 10.1016/s1097-2765(04)00064-4. [DOI] [PubMed] [Google Scholar]
- Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton MK. The pinhead/zwille gene acts pleiotropically in arabidopsis development and has overlapping functions with the argonaute1 gene. Development. 1999;126:469–481. doi: 10.1242/dev.126.3.469. [DOI] [PubMed] [Google Scholar]
- Maple J, Moller SG. Mutagenesis in arabidopsis. Methods in Molecular Biology. 2007;362:197–206. doi: 10.1007/978-1-59745-257-1_14. [DOI] [PubMed] [Google Scholar]
- Masterson J. Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science. 1994;264:421–424. doi: 10.1126/science.264.5157.421. [DOI] [PubMed] [Google Scholar]
- McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, Smith K, Thomas C, Schetter A, Zhou Q, Cushman MA, et al. Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics. 2001;159:1751–1763. doi: 10.1093/genetics/159.4.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meins F, Jr, Si-Ammour A, Blevins T. RNA silencing systems and their relevance to plant development. Annual Review of Cell and Developmental Biology. 2005;21:297–318. doi: 10.1146/annurev.cellbio.21.122303.114706. [DOI] [PubMed] [Google Scholar]
- Ni Z, Kim ED, Ha M, Lackey E, Liu J, Zhang Y, Sun Q, Chen ZJ. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature. 2009;457:327–331. doi: 10.1038/nature07523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X. Carpel factory, a dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Current Biology. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Ruvkun GTC. Control of developmental timing by microRNAs and their targets. Annual Review of Cell and Developmental Biology. 2002;18:495–513. doi: 10.1146/annurev.cellbio.18.012502.105832. [DOI] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in arabidopsis. Genes and Development. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran V, Chen X. Small RNA metabolism in arabidopsis. Trends in Plant Science. 2008;13:368–374. doi: 10.1016/j.tplants.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Lang JD, Golden T, Ray S. Short integument (SIN1), a gene required for ovule development in arabidopsis, also controls flowering time. Development. 1996;122:2631–2638. doi: 10.1242/dev.122.9.2631. [DOI] [PubMed] [Google Scholar]
- Schauer SE, Jacobsen SE, Meinke DW, Ray A. DICER-LIKE1: blind men and elephants in arabidopsis development. Trends in Plant Science. 2002;7:487–491. doi: 10.1016/s1360-1385(02)02355-5. [DOI] [PubMed] [Google Scholar]
- Sorin C, Bussell JD, Camus I, Ljung K, Kowalczyk M, Geiss G, McKhann H, Garcion C, Vaucheret H, Sandberg G, et al. Auxin and light control of adventitious rooting in arabidopsis require argonaute1. Plant Cell. 2005;17:1343–1359. doi: 10.1105/tpc.105.031625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes TL, Richards EJ. Induced instability of two arabidopsis constitutive pathogen-response alleles. Proceedings of the National Academy of Sciences, USA. 2002;99:7792–7796. doi: 10.1073/pnas.112040999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Mallory AC, Bartel DP. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of MIR168 by AGO1. Molecular Cell. 2006;22:129–136. doi: 10.1016/j.molcel.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F. arabidopsis endogenous small RNAs: highways and byways. Trends in Plant Science. 2006;11:460–468. doi: 10.1016/j.tplants.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crete P. Endogenous trans-acting siRNAs regulate the accumulation of arabidopsis mRNAs. Molecular Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Vongs A, Kakutani T, Martienssen RA, Richards EJ. Arabidopsis thaliana DNA methylation mutants. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- Wang J, Tian L, Madlung A, Lee HS, Chen M, Lee JJ, Watson B, Kagochi T, Comai L, Chen ZJ. Stochastic and epigenetic changes of gene expression in arabidopsis polyploids. Genetics. 2004;167:1961–1973. doi: 10.1534/genetics.104.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Tian L, Lee HS, Chen ZJ. Non-additive regulation of FRI and FLC loci mediates flowering-time variation in arabidopsis allopolyploids. Genetics. 2006a;173:965–974. doi: 10.1534/genetics.106.056580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Tian L, Lee HS, Wei NE, Jiang H, Watson B, Madlung A, Osborn TC, Doerge RW, Comai L, et al. Genomewide non-additive gene regulation in arabidopsis allotetraploids. Genetics. 2006b;172:507–517. doi: 10.1534/genetics.105.047894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Allen E, Wilken A, Carrington JC. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 2005;102:12984–12989. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Huang W, Wang H, Cai R, Xu Y, Huang H. Characterizations of a hypomorphic argonaute1 mutant reveal novel AGO1 functions in arabidopsis lateral organ development. Plant Molecular Biology. 2006;61:63–78. doi: 10.1007/s11103-005-5992-7. [DOI] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in arabidopsis. Genes and Development. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Zhu J, Kapoor A, Zhu JK. Role of arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. European Molecular Biology Organization Journal. 2007;26:1691–2701. doi: 10.1038/sj.emboj.7601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.