Abstract
Genetic and environmental factors influence brain structure and function profoundly The search for heritable anatomical features and their influencing genes would be accelerated with detailed 3D maps showing the degree to which brain morphometry is genetically determined. As part of an MRI study that will scan 1150 twins, we applied Tensor-Based Morphometry to compute morphometric differences in 23 pairs of identical twins and 23 pairs of same-sex fraternal twins (mean age: 23.8 ± 1.8 SD years). All 92 twins’ 3D brain MRI scans were nonlinearly registered to a common space using a Riemannian fluid-based warping approach to compute volumetric differences across subjects. A multi-template method was used to improve volume quantification. Vector fields driving each subject’s anatomy onto the common template were analyzed to create maps of local volumetric excesses and deficits relative to the standard template. Using a new structural equation modeling method, we computed the voxelwise proportion of variance in volumes attributable to additive (A) or dominant (D) genetic factors versus shared environmental (C) or unique environmental factors (E). The method was also applied to various anatomical regions of interest (ROIs). As hypothesized, the overall volumes of the brain, basal ganglia, thalamus, and each lobe were under strong genetic control; local white matter volumes were mostly controlled by common environment. After adjusting for individual differences in overall brain scale, genetic influences were still relatively high in the corpus callosum and in early-maturing brain regions such as the occipital lobes, while environmental influences were greater in frontal brain regions which have a more protracted maturational time-course.
1 Introduction
3D maps showing the relative contribution of genetic, shared and unique environmental factors to brain structure can facilitate the understanding of the influence of genetics on anatomical variability. Twins have been studied with quantitative genetic models to estimate these different factors. This approach has detected highly heritable (i.e., genetically influenced) brain features, such as the whole brain volume and total gray and white matter volumes (Posthuma et al., 2002).
Identifying genetically-influenced features is important, as genes at least partially mediate many psychiatric disorders (Van’t Ent et al., 2007). In addition, many cognitive or behavioral measures in normal individuals, such as full-scale IQ, are highly genetically influenced (Gray and Thompson, 2004) and are correlated with measures of brain structure (Reiss et al., 1996; Thompson et al., 2001; Haier et al., 2004). These image-derived measures (such as gray matter volume) are often called intermediate phenotypes if they are associated with an illness and are more amenable to quantitative genetic analysis (see Glahn al. (2007a) for a review of the endophenotype concept). Using this approach, researchers have identified and confirmed specific genes that are associated with structural brain deficits in schizophrenia patients (Cannon et al., 2002, 2005; Pietiläinen et al., 2008; Narr et al., 2008). Association studies (Sullivan et al., 2007; Hattersley and McCarthy, 2005) and twin studies using a cross-twin cross-trait design (bivariate genetic models) (Posthuma et al., 2002) have also found specific genes or common sets of genes influencing brain morphology and cognitive performance.
Environmental factors (e.g., cardiovascular health, nutrition, exercise, and education) may also exert protective or harmful effects on the structural integrity of the brain (Raji et al., 2009). In epidemiological studies and drug trials, it may be important to account for genetic and environmental influence on disease progression (e.g., the ApoE4 risk allele in Alzheimer’s disease; Hua et al. (2008b)), to adjust for confounds in the analysis of treatment effects (Jack et al., 2008). Twin studies can reveal whether specific neuroanatomical measures are predominantly influenced by genetics or shared or individual environments (see Peper et al. (2007) for a review), by comparing twin pairs with different degrees of genetic affinity. Identical (or monozygotic, MZ) twins share the same genetic material, whereas fraternal (or dizygotic, DZ) twins share, on average, only half of their genetic polymorphisms (random DNA sequence variations that occur among normal individuals). DZ twins are commonly studied in lieu of other siblings because they are the same age, preventing any age-related confounds. Identical and fraternal twin pairs are compared to ensure, to the greatest possible extent, comparable upbringings and family environments despite varying degrees of genetic resemblance.
The earliest neuroanatomical genetic studies used traditional volumetric measures and region of interest analyses to quantify similarity between MZ and DZ twins. Whole brain and hemispheric volumes were found to be highly heritable (> 80% for the whole brain in Pfefferbaum et al. (2000); Sullivan et al. (2001) and > 94% for the hemispheric volume in Bartley et al. (1997)). Gray matter and white matter volumes were shown to be 82% and 88% genetically determined (Baaré et al., 2001), respectively. Oppenheim et al. (1989); Pfefferbaum et al. (2000); Scamvougeras et al. (2003); Hulshoff Pol et al. (2006a) showed that the corpus callosum is mostly controlled by genes, which was verified at different stages in life. Findings were less consistent for ventricular volume and shape. Reveley et al. (1982); Pfefferbaum et al. (2000) and Styner et al. (2005) showed these structures to be highly heritable, whereas other studies (Baaré et al., 2001; Wright et al., 2002) determined that ventricular volumes are equally influenced by genetics (58%) and environment (42%). Gyral and sulcal patterns were shown to be widely variable in MZ twins (Weinberger et al., 1992; Bartley et al., 1997), suggesting strong environmental influences independent of genetics (Steinmetz et al., 1994).
Computational mapping methods allow the mapping of genetic influences on structure volumes throughout the brain, without requiring a priori specification of regions of interest. Among them, voxel-based methods, such as voxel-based morphometry (VBM) (Ashburner and Friston, 2000), have revealed genetically-mediated deficits in attention deficit hyperactivity disorder (Van’t Ent et al., 2007), anxiety disorders (de Geus et al., 2006) and schizophrenia (Hulshoff Pol et al., 2006b). Tensor-based morphometry (TBM) is another voxel-based method that has been used successfully to detect morphometric differences associated with aging and Alzheimers disease (Hua et al., 2008a,b), HIV/AIDS (Brun et al., 2007; Chiang et al., 2006; Leporé et al., 2008a), Williams sydrome (Chiang et al., 2007), Fragile X syndrome (Lee et al., 2007), schizophrenia (Gogtay al., 2008), and normal brain development (Hua et al., 2007).
As TBM has been extensively used in past studies, we chose this method to analyze our data set of 23 pairs of MZ and 23 pairs of same-sex DZ twins. TBM combines a warping step and a statistical step to determine local volume changes. Here, we detected local similarities between MZ twins, and DZ twins, and compared these two groups, to determine the genetic and environmental effects on brain structures. We first hypothesized that brain structure volumes would be more genetically influenced when the data is not adjusted for the overall brain size. We also predicted that the volumes of brain regions that mature the earliest in infancy (e.g., occipital lobes) would be the most highly heritable, while environmental effects would be more readily detected in structures that have a more protracted maturational time course, such as the frontal lobes.
2 Methods
2.1 Overview
In TBM, a population of images is linearly aligned to a common space, then non-rigidly registered (i.e., warped) to a common target brain, chosen either as one of the subjects in the study or as a specially constructed template with the mean geometry for the group of subjects being studied. The local expansion or compression factor applied during the warping process (also called the Jacobian determinant) is a useful index of volumetric differences between each subject and the template. Morphometric differences are assessed by performing a statistical analysis of the volumetric differences at each location in the brain. Here, we performed the registration using a fluid registration algorithm first proposed in Brun et al. (2007) and further developed in Brun et al. (2008). For more precise quantification of volumes, we used a multi-template scheme described in Leporé et al. (2008c) rather than using one single target brain image. We previously studied whether TBM results depend on the choice of template for normalization (Leporé et al., 2007, 2008c,d). In general, we found that the findings regarding genetic influences are highly consistent, regardless of which template is used for spatial normalization. This is encouraging, because the power to resolve morphometric variation in TBM depends on the ability to align each image accurately to the chosen template. In addition, we also found that it makes very little difference to the results, if the warping is performed on images that have already been segmented into gray and white matter before spatial normalization (Chou et al., 2008). In (Leporé et al., 2007), we built a mean brain template based on averaging a set of deformation fields, such that the mean deformation tensor of the mappings from each individual to the template was minimized in a log-Euclidean metric that measured deviations from the identity tensor. This involved iterative perturbation of the template until the criterion for the mean deformation was optimized. Even though the results with this template were very similar to those achieved with an individual brain as the target, it can be used to rule out a possible source of bias that may arise due to template selection. In complementary work, we also investigated whether multi-template normalization can provide greater power for TBM studies (Leporé et al., 2008d). In that approach, instead of aligning images to a single brain, we aligned all images to a set of different templates, and the results were averaged (Chou et al., 2008, 2009a,b); this approach was originally termed targetless spatial normalization (Kochunov et al., 2002) and is related to multi-atlas segmentation methods (Heckemann et al., 2006). This multi-template method can increase the statistical power to detect disease effects and also leads to more accurate segmentations, as it reduces the overall impact of errors in individual registrations by fusing information from a large number of registrations among different pairs of brains in the study. We assessed anatomical resemblance within twin pairs by computing intraclass correlation coefficient (ICC) maps for the MZ and DZ twins. Falconer’s heritability statistics (Falconer, 1989), which estimate the proportion of variance due to genetic differences, were computed at each voxel and displayed as a map (as in Thompson et al. (2001)). Using a more advanced modeling approach developed in Chiang et al. (2008); Lee et al. (2009), and based on Neale et al. (1999), we fitted a structural equation model (SEM) to estimate the proportion of local volumetric variability attributable to genetics, shared and unique environmental factors across the whole brain both before and after adjusting the data for individual differences in overall brain scale. The same technique was applied for various lobar and subcortical regions of interest (ROIs) traced on the brain template.
2.2 Participants
We scanned 23 pairs of monozygotic (MZ) (11 male and 12 female pairs) and 23 pairs of same-sex dizygotic (DZ) twins (10 male and 13 female pairs), recruited as part of a 5-year research project that will eventually evaluate 1150 twins. As explained below, to eliminate bias and improve registration accuracy, we used a multi-template approach in which we aligned all the scans to a set of 5 target brains randomly selected from our database (Leporé et al. (2008d)). These scans were performed with the same protocol on 5 healthy subjects not part of the genetic analysis (one of them was a monozygotic twin, 2 of them were members of two different dizygotic twin pairs, and 2 did not have twin siblings). These scans were used as templates for the fluid registration step (see Section 2.3), and all the other images were fluidly registered to each of them. The age range for the subjects was 22 – 25 years old for all the subjects, including the templates (mean age: 23.8 ± 1.8 SD years). Each subject was informed of the goals of the study and signed a formal consent. The study was approved by the appropriate Institutional Review and Research Ethics Boards.
Zygosity was established objectively by typing nine independent DNA microsatellite polymorphisms (Polymorphism Information Content > 0.7), using standard polymerase chain reaction (PCR) methods and genotyping. These results were cross-checked with blood group (ABO, MNS and Rh), and phenotypic data (hair, skin and eye color), giving an overall probability of correct zygosity assignment of greater than 99.99%. All subjects underwent physical and psychological screening to exclude cases of pathology known to affect brain structure. None of the twin subjects reported a history of significant head injury, a neurological or psychiatric illness, substance abuse or dependence, or had a first-degree relative with a psychiatric disorder.
2.3 Image Acquisition and preprocessing
All MR images were collected using a 4 Tesla Bruker Medspec whole body scanner (Bruker Medical, Ettingen, Germany) at the Center for Magnetic Resonance (University of Queensland, Australia). Three-dimensional T1-weighted images were acquired with a magnetization prepared rapid gradient echo (MP-RAGE) sequence to resolve anatomy at high resolution. Acquisition parameters were: inversion time (TI) /repetition time (TR) /echo time (TE) = 1500/2500/3.83 msec; flip angle = 15°; slice thickness = 0.9 mm with a 256×256×256 acquisition matrix.
Extracerebral (non-brain) tissues were manually deleted from the MRI images using the 3D interactive program, Display (Montreal Neurological Institute, McGill University, Canada). All scans were then aligned to the ICBM53 template using 9-parameter registration (i.e., translational and rotational alignment, allowing scaling in 3 independent directions) found in the FMRIB’s Linear Image Registration Toolbox, FLIRT (Jenkinson et al., 2002). The ICBM53 template is one of several standardized adult brain templates, and was generated by nonlinearly registering and averaging 53 high-resolution brain MRI scans in the ICBM standard space to improve the signal-to-noise ratio (Collins et al. (1995); Montreal Neurological Institute, McGill University, Canada).
2.4 Fluid Registration
Non-rigid warping is usually performed by registering each subject’s image S to a common target image, T. Most commonly, this target is an image selected from the data set or a specially constructed Mean Deformation Template (MDT), which has the average geometry (shape) and intensity for the group of subjects studied (Kochunov et al., 2001). Sometimes, a single image is used rather than group average, because it has sharper features and greater anatomical detail. The choice of an MDT can affect the precision of the registration (Kochunov et al., 2001; Leporé et al., 2008b). To avoid any bias in the registration, we did not use one single target image. Instead we used the multi-template method described in Leporé et al. (2008d), which is a variant of previously proposed multi-template registration approaches; we registered all the subjects to four different templates. The use of 4 templates offers a reasonable trade-off between using more templates, which can reduce registration errors but is very time-consuming, and preserving computational efficiency, which requires keeping the number of templates reasonably low (see Chou et al. (2008), for an optimization study).
These 4 templates were fluidly registered to a common 5th template (see Section 2.3 for a description of these subjects). These registration vector fields were concatenated with the previous registrations, so that all the obtained vector fields (4 per subject), sit in a common space.
For each subject, Jacobian matrices J were derived from these 4 vector fields (see Section 2.5.1). As all the defined mathematical entities (4 vector fields and their 4 corresponding Jacobian matrices per subject) were computed in a common space, we computed Javerage from the 4 Jacobian matrices existing at each voxel. We used the mean defined in Arsigny et al. (2006) (this mean is defined in a Riemannian space).
The first deformable registration methods were inspired by continuum mechanics. In these elastic or fluid registration approaches, the image is considered to be embedded in a 3D elastic or fluid medium and its deformation at each voxel is governed by mechanical equilibrium equations. A similarity measure is defined between the deforming image and the target image (simple squared-intensity difference, cross-correlation or information-theoretic measures, such as normalized mutual information or Jensen-Rényi divergence (Chiang et al., 2006)) and this similarity term is included as a force term in a partial differential equation (PDE), that regularizes the deformation to avoid any folds or tears in the image (see Christensen et al. (1996); Bro-Nielsen et al. (1996); Gramkow (1996); Leporé et al. (2008b)).
Fluid registration methods have been advocated as they guarantee diffeomorphisms (i.e., smooth invertible, one-to-one mappings) even when the magnitude of the deformations is large (Christensen et al., 1996), whereas elastic registration methods are typically simpler to implement but only generate diffeomorphic mappings when the deformations are small in magnitude. Our study used a fluid registration algorithm built upon an elastic registration method that was developed in a Riemannian framework by Pennec et al. (2005). This method ensures diffeomorphic maps when matching anatomy across subjects (for details, see Brun et al. (2007, 2008)).
It is worth noting that the twins’ images are not registered by aligning one twin’s image directly to the other twin’s image. Instead they are each individually aligned to a common target image from a randomly selected control subject who is not a member or the twin pair. As such, because the MZ twins vary as much, as a group, as the DZ group (and the general population) what matters most for the registration is how much they deviate from the target image. As the target image was not the other member of a twin pair used in the analysis, and was an unrelated individual, a zygosity x registration error interaction is unlikely. Consequently, statistical measures assessing resemblance between members of twin pairs can be computed from registration-derived measures without any bias introduced by the registration.
2.5 Statistical analysis
2.5.1 Measuring Volumetric Differences
For each registration, there is a corresponding displacement vector field. At each voxel, each of the registrations (4 per subject) gives a 3-component vector (noted ux, uy, uz), that describes the displacement of a particle from the template to the initial image (defined in the template coordinate system). From those vectors, a Jacobian matrix is computed, defined as
Scalar values such as the trace, geodesic anisotropy or the determinant of the matrices may be analyzed (Leporé et al., 2008a). Here, we chose to focus on the analysis of det(J), the expansion factor that detects local volume changes; this is the most common form of TBM analysis. detJ(u⃗) > 1 indicates a local volumetric expansion of the image being studied in comparison to the template and detJ(u⃗) < 1 indicates a local volume shrinkage.
2.5.2 Measuring the genetic influence on brain structures
Intraclass correlation and Falconer’s heritability statistic
To measure the resemblance between twin pairs, we first computed the intraclass correlation coefficient (ICC) for both the MZ and the DZ groups, according to the formula
| (1) |
Here, is the pooled variance between pairs and is the variance within pairs (Scout and Fleiss, 1979).
Heritability is the proportion of the variation in a measurement that is attributable to genetic factors. We first computed Falconer’s heritability statistic, h2, defined as twice the difference in correlation between MZ and DZ pairs
where r(MZ) and r(DZ) are the ICC measures for the MZ and DZ groups, respectively. This statistic has been debated in the twin literature as it is based on the equal environments assumption (that MZ and DZ twins have comparable rearing environments; see Thompson et al. (2002a) for a discussion). Even so, it is perhaps the most widely-reported index of genetic effects on a trait (e.g., in the twin studies reviewed by Peper et al., 2007), so we report it first before going on to a more sophisticated analysis of genetic effects, based on structural equation models.
We did not want to assume that the data on volumetric differences (here det(J)) were normally distributed, so we computed p values at each voxel with a voxelwise permutation test, to establish a null distribution for the ICC statistics at each voxel (Nichols and Holmes, 2002). The null hypothesis for the intraclass correlation was ICC = 0 (no correlation). At each permutation, a given subject’s scalar map det(J) was randomly assigned to another subject, and a null distribution was computed at each voxel; the r-values computed from this randomly-generated distribution were compared to the r-values for the true assignment to give statistical maps of the significance of the ICCs. All voxels from the same subject were permuted in the same way at each randomization, as is required to maintain spatial continuity of the null distribution. The resulting permutation-based (non-parametric) p-value is defined as the quantile of the empirical null distribution where the real data falls. To control the standard error of p, we performed 5000 permutations at each voxel (Edgington, 1995).
To control for multiple spatial comparisons implicit in computing maps of statistics, we computed the omnibus probability, which we call pcorrected, using the suprathreshold volume, for maps thresholded at p = 0.05. This value assesses the overall significance of the observed pattern of effects in the statistical maps (Nichols and Holmes, 2002). These values were computed for both MZ and DZ probability maps.
Structural equation modeling
Eight regions of interest (ROI) were manually traced on the 5th (common) template, using BrainSuite (Shattuck and Leahy, 2002). These ROIs consisted of the five lobes, the thalamus, the lateral ventricles and the basal ganglia. For each individual, the volume of each ROI was computed using the determinant of the Jacobian matrix, by integrating its value over the appropriate region in the template (see Section 2.5.1). This allows the volume of the ROI in each subject to be estimated.
A variance component analysis was performed at each voxel in addition to the analysis of the lobar ROI volumes by fitting structural equation models (SEMs) at each voxel in the images. This method was first implemented and promoted by Neale et al. (1999) with the genetic modeling program, Mx. Here, we used a more computationally efficient version that we recently implemented (Chiang et al., 2008; Lee et al., 2009), and this permitted a voxel-wise analysis. To analyze genetic and environmental correlations in twins, structural equation models can compute the relative contributions of additive genetic (A), shared environmental (C) and random environmental (E) components to the variance in the observed local and ROI volume, y. For each twin subject, we can model the value of y as the combination of 3 latent factors, y = aA + cC + eE; by standard analysis (Neale et al., 1999), the covariance matrix Σ for the vector (y1, y2)T, where 1 or 2 stands for the first or second twin in the same pair, can be modeled by:
| (2) |
where coυ(u, υ) means covariance between u and υ. γ = 1 for MZ twins, and for DZ twins, as MZ twins share all and DZ twins share on average half of their genetic polymorphisms. Since A, C, and E are unobservable variables, the path coefficients Θ = (a, c, e) are estimated by comparing the covariance matrix implied by the model, Σ(Θ), with the sample covariance matrix (S) of the observed variables (y1 and y2), using maximum-likelihood fitting (Fornell and Larcker, 1981).
| (3) |
where p = 2 is the number of observed variables. Under the null hypothesis that S = Σ(Θ), minus twice the logarithm of the likelihood ratio was simplified to TLM,Θ = (nMZ − 1)FML,Θ,MZ + (nDZ − 1)FML,Θ,DZ, which follows a χ2 distribution with p(p + 1)t degrees of freedom, where t = 3 is the number of free model parameters, and nMZ and nDZ are the number of MZ and DZ twin pairs. Acceptance of the null hypothesis (p > 0.05) indicates a good fit for the model. The maximum likelihood computation was performed to find the model parameters and expected covariance for both MZ and DZ pairs.
If the variance in the data at each voxel is assumed to be normal, χ2 values could be used to measure the agreement between the observed and expected covariance matrices, as χ2 is essentially a goodness-of-fit index. To free our fitted SEMs from any assumption that the data were normally distributed, we used a permutation-based method to determine the goodness of fit (Lee et al., 2009; Chiang et al., 2008; Bollen and Stine, 1992). FML,Θ,DZ was minimized using an optimization method in the original and in the 5000 permuted samples in which the twin pair’s MZ and DZ labels were randomly shuffled. At each permutation, three hypotheses with different Θ were evaluated, in order to fit the ACE, AE or CE models and the pvalues pace, pce, pae were determined separately by comparing the value of TLM,Θ for the real distribution to the one of the permuted distribution. For each null hypothesis, the sample data was rescaled since the permutation distribution of χ2 statistics, TLM,Θ can differ from its original distribution. At each voxel and for each region, the three probabilities p-values pace, pce and pae were compared and the A, C and E values resulting from the best model were retained. The best model was defined by the p-value that was greater than the two others and also greater than 0.05 (a p-value < 0.05 indicates a lack of fit to the data and rejection of the model - note that this is analogous to the convention in standard multivariate regression, where a p-value < 0.05 denotes rejection of the null hypothesis, but in our case the genetic model, consisting of the ACE factors, is rejected when p < 0.05. The model is therefore considered to fit when p-value > 0.05. Even so, the significance of a specific term in a model (such as the A term) can be assessed by comparing the χ2 goodness-of-fit values of the two competing models also has a χ2 distribution, and its significance can be reported in the conventional way, with p < 0.05 denoting that the added term provides a better overall fit to the data).
A similar path analysis and structural equation model can be used to estimate the A, D and E parameters (relative contributions of additive genetic factors, genetic dominance effects and random environmental components, respectively on the observed volumes). The ADE model differentiates two genetic effects: the sum of all the effects of individual genetic loci (A) and the result of any interactions between alleles at the same genetic locus (D) (Maes, 2005). When the genetic dominance term fits, the DZ correlation tends to be less than half of the MZ correlation (here we take to fit D and to fit A)
2.5.3 Estimating phenotypic variance in the population
To examine the variability of the measured phenotypes (Jacobian determinants, det(J), at each voxel, and ROIs volumes), we computed the mean and variance of each volumetric across the sample. To avoid any bias in estimating variance, we chose a group composed of one subject per pair in the MZ and the DZ groups, taken randomly.
3 Results
3.1 Intraclass Correlations and Falconer’s heritability estimates
Figure 1 shows the intraclass correlation computed for local volumes in both identical and fraternal twins (top left: rMZ and top right: rDZ). Red colors indicate a high correlation (r close to 1), whereas blue colors indicate no detectable correlation (r = 0). The significance of the intraclass correlations was assessed by computing p-values corrected for multiple comparisons (bottom left: pICC(MZ), pcorrected = 0.034; bottom right: pICC(DZ), pcorrected = 0.025). A comparison of these two intraclass correlations is given by the maps of Falconer’s heritability statistics (h2, middle right panel). Red colors indicate greater heritability. The left panel in the middle row shows the common anatomical template and the colored lines indicate the different sections exhibited in the color maps. Three features are evident: first, for most subcortical regions, MZ twin volumes are correlated between members of the pair at around r = 0.5, with values much closer to zero for DZ twins. Second, a correction for multiple comparisons reveals that the overall pattern of correlations in the DZ twins is significantly greater than zero; strictly speaking, a higher proportion of the brain has correlations exceeding the p = 0.05 threshold than would be expected by chance if the null hypothesis of no correlation were true. Third, the high values of heritability (right panel in the middle row), with values over 0.5 for the majority of the subcortical regions, are based on twice the difference in the intraclass correlations for the MZ and DZ twins. These give an estimate of the proportion of the variance in those regions that is genetically mediated. As is also implied by the structural equation models below, the heritability maps suggest that the anatomy of the MZ twins resembles each other to a greater degree than the anatomy of the DZ twins in ventricular, callosal, limbic (cingulate gyrus), occipital and anterior temporal regions, while DZ twins resemble each other to a greater degree than randomly chosen individuals of the same age and sex.
Fig. 1.
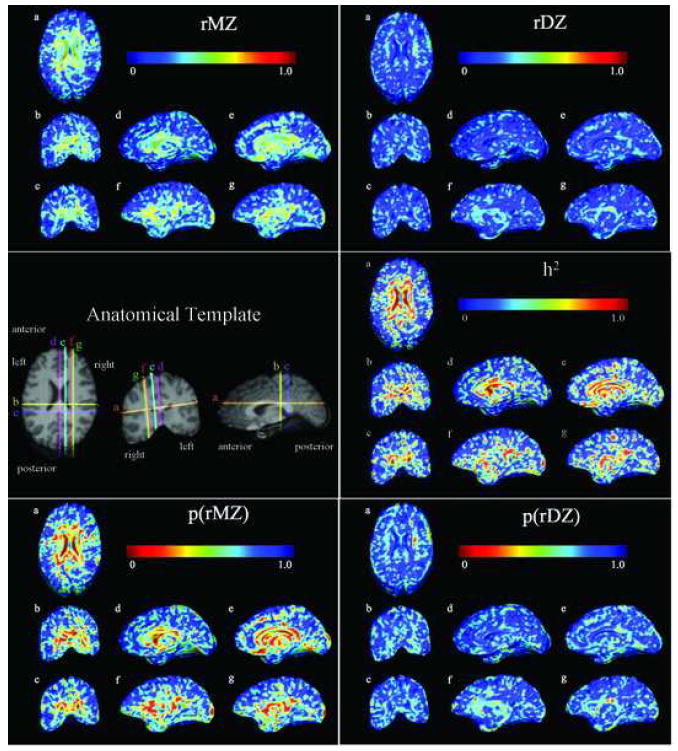
top row: Intraclass correlation maps are shown for the monozygotic twins (rMZ; left panel) and for the dizygotic twins (rDZ; right panel); middle row: An anatomical image (left) shows the sections for which statistics are displayed; maps of Falconer’s heritability estimate (h2) show high heritability in subcortical regions; bottom row: Maps show the p-values (significance) of the intraclass correlation in monozygotic twins (icc MZ) and dizygotic twins (icc DZ).
3.2 Genetic and Environmental influence on brain structure variability
The influence of additive genetic (A), as well as shared (C) and unique (E) environmental factors on brain structure volumes are mapped in Figure 2 and Figure 3 for the unscaled and scaled data, respectively. The corresponding values are also reported for eight ROIs in Table 1 (unscaled data) and Table 2 (scaled data).
Fig. 2.
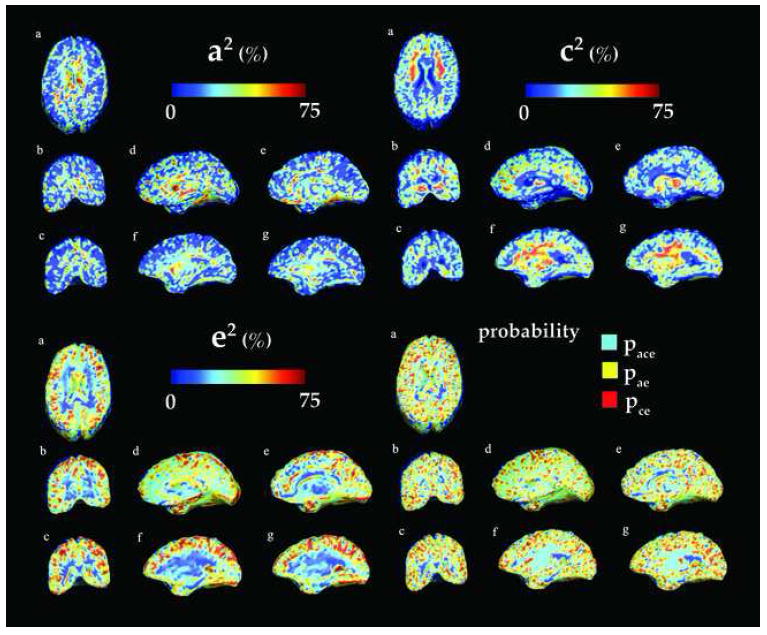
Variance component maps for additive genetic (a2 - top left), common (c2 - top right), and unique environmental (e2 - bottom left) factors for the unscaled data. Bottom right: Color-coded maps representing the model choice at each voxel- Light blue (yellow and red, respectively) indicates that the best fitting model is obtained with ACE (AE and CE, respectively). The corresponding anatomical sections (a-g) are shown in Figure 5 - middle left.
Fig. 3.
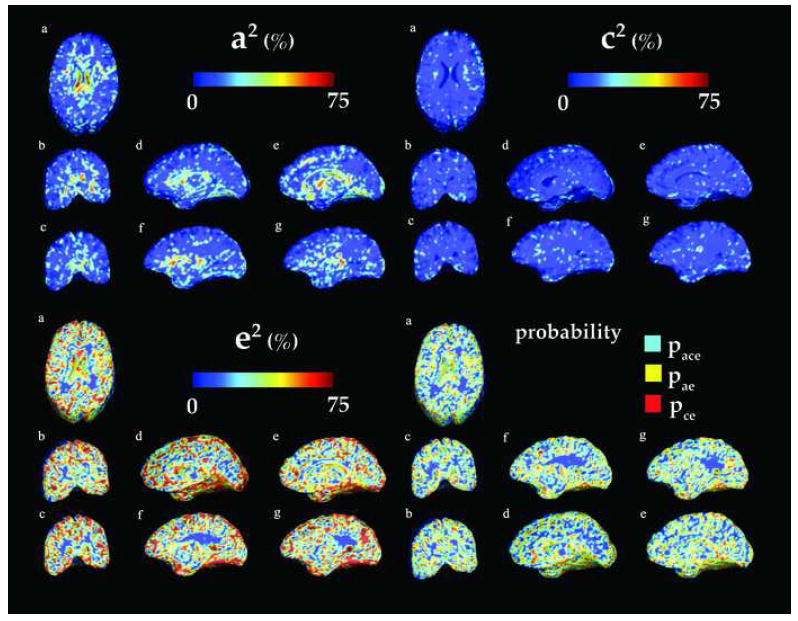
Variance component maps for additive genetic (a2 - top left), common (c2 - top right), and unique environmental (e2 - bottom left) factors for the scaled data. Bottom right: Color-coded maps representing the model choice at each voxel- Light blue (yellow and red, respectively) indicates that the best fitting model fit is obtained with ACE (AE and CE, respectively). The corresponding anatomical sections (a-g) are shown in Figure 5 - middle left.
Table 1.
Measures of the intraclass correlation coefficients (ICC) for the MZ and DZ groups, Falconer’s heritability estimate (h2), the additive genetic (a2), dominant genetic (d2), shared (c2) and unique environmental (e2) variance components, their confidence interval, the probability p (computed from the structural equation models; these exceed 0.05 when the model fits) and the model that provided the best fit when explaining variance in the volumes of these brain regions across subjects for the unscaled data. ROIs: WB- whole brain ; Fr -Frontal lobes ; Par - Parietal lobes ; Temp - Temporal lobes ; Occ - Occipital lobes ; Lmbc - Limbic lobes ; Vent - Ventricles ; BG - Basal Ganglia ; Th - Thalamus
| Regions | WB | Fr | Par | Temp | Occ | Lmbc | Vent | BG | Th |
|---|---|---|---|---|---|---|---|---|---|
| ICCMZ | 0.87 | 0.86 | 0.87 | 0.88 | 0.87 | 0.85 | 0.71 | 0.86 | 0.84 |
| ICCDZ | 0.74 | 0.74 | 0.74 | 0.74 | 0.75 | 0.77 | 0 | 0.71 | 0.76 |
| h2 | 0.26 | 0.25 | 0.27 | 0.29 | 0.24 | 0.17 | 1.42 | 0.30 | 0.17 |
| a2(%) | 34.82 (0.47 - 65.66) | 35.28 (0.32 - 54.18) | 33.18 (3.07 - 55.81) | 36.58 (31.89 - 96.97) | 33.26 (30.69 - 91.19) | 27.81 (1.85 - 36.86) | 0 (0 - 49.64) | 39.58 (0.22 - 42.57) | 24.08 (0.05 - 68.59) |
| c2/d2(%) | 55.03 (1.52 - 56.59) | 54.44 (28.16 - 73.67) | 56.57 (0 - 57.26) | 54.98 (0.15 - 55.90) | 56.71 (4.9 - 56.93) | 62.03 (30.17 - 78.73) | 63.51 (0.79 - 63.91) | 49.53 (17.02 - 95.13) | 63.67 (9.43 - 98.97) |
| e2(%) | 10.14 (7.87 - 53.03) | 10.28 (7.51 - 41.24) | 10.25 (5.65 - 85.93) | 8.44 (1.90 - 38.22) | 10.03 (3.23 - 40.38) | 10.15 (10.13 - 51.53) | 36.49 (0.80 - 63.91) | 10.99 (0.35 - 70.10) | 12.26 (0.26 - 41.98) |
| probability | 0.74 | 0.74 | 0.77 | 0.58 | 0.56 | 0.41 | 0.09 | 0.91 | 0.74 |
| Best fitting model | ACE | ACE | ACE | ACE | ACE | ACE | ACE | ACE | ACE |
Table 2.
Measures of the intraclass correlation coefficients (ICC) for the MZ and DZ groups, Falconer’s heritability estimate (h2), the additive genetic (a2), dominant genetic (d2), shared (c2) and unique environmental (e2) variance components, their confidence interval, the probability p (computed from the structural equation models; these exceed 0.05 when the model fits) and the model that provided the best fit when explaining variance in the volumes of these brain regions across subjects for the scaled data. ROIs: WB- whole brain ; Fr - Frontal lobes ; Par - Parietal lobes ; Temp - Temporal lobes ; Occ - Occipital lobes ; Lmbc - Limbic lobes ; Vent - Ventricles ; BG - Basal Ganglia ; Th - Thalamus
| Regions | WB | Fr | Par | Temp | Occ | Lmbc | Vent | BG | Th |
|---|---|---|---|---|---|---|---|---|---|
| ICCMZ | 0.21 | 0.38 | 0.09 | 0.12 | 0.55 | 0.57 | 0.59 | 0.49 | 0.61 |
| ICCDZ | 0.33 | 0.46 | 0.26 | 0.21 | 0.26 | 0.12 | 0 | 0.35 | 0 |
| h2 | 0 | 0 | 0 | 0 | 0.58 | 0.91 | 1.17 | 0.27 | 1.21 |
| a2(%) | 0 (0 - 25.23) |
0 (0-34.76) |
- - |
14.91 (0-38.41) |
46.84 (1.46 - 65.11) |
17.27 (0.02 - 79.76) |
0 (0-34.92) |
56.97 (0 - 99.93) |
0 (0 - 68.10) |
| c2/d2(%) | 27.13 (3.27 - 61.47) |
42.29 (0 - 45.53) |
- - |
0 0 |
7.01 (5.72 - 54.36) |
34.54 (0.01 - 41.67) |
53.12 (9.40 - 53.62) |
0 0 |
58.32 (1.66 - 58.67) |
| e2(%) | 73.87 (24.43 - 89.48) |
57.05 (19.70 - 83.03) |
- - |
85.08 (61.58 - 100) |
46.13 (28.53 - 72.13) |
48.18 (28.75 - 73.95) |
46.88 (26.14 - 73.95) |
43.03 (0 - 99.99) |
41.68 (0 - 79.60) |
| probability | 0.26 | 0.54 | - | 0.55 | 0.38 | 0.06 | 0.23 | 0.61 | 0.25 |
| Best fitting model | ACE | ACE | no fit | AE | ADE | ADE | ADE | AE | ADE |
Figure 2 and Figure 3 display voxelwise maps of the ACE variance components. In each map, the proportion of the overall variance is expressed on a scale of 0 (dark blue) to 75% (red). The variance components, a2, c2, e2 are proportions, and vary from 0 to 1, but their contribution to the overall variance is often stated as a percentage). In the bottom left panel, color-coded maps are presented that show the model that provided the best fit. Light blue corresponds to the ACE model, yellow to the AE model and red to the CE model
If we assume that all the regions of the brain have a partly shared genetic influence related to overall scale of the brain, then after adjusting for individual differences in brain scale across subjects, a lesser residual effect of genetic factors should remain (this is based on the fact that overall brain volume is heritable). Therefore, we hypothesized that all brain regions would show a higher heritability prior to the adjustment and we also expected the proportion of variance due to environmental factors to be greater afterwards.
Figure 3 shows that the influence of genetic factors is detectable throughout the brain in the unscaled data (top left): from 20% in the white matter to 75% in subcortical structures such as the corpus callosum and the ventricles, 20–40% in the basal ganglia and the thalamus and 50% in the occipital lobes (d) The effects of the shared environment, as shown by the c2 values (Figure 2, top right panel), are more prominent than their genetic counterparts in the white matter, such as the internal capsule, the uncinate fasciculus and the superior longitudinal fasciculus and mostly located in the frontal lobes. The unique environment variance (e2) maps demonstrate high variance in the gray matter. As this term not only accounts for the individual environment influence but also measurement errors from all sources, it is not possible to distinguish unique environmental effects from sources of measurement errors that are uncorrelated between the twins. These maps should therefore be interpreted cautiously.
As hypothesized, the scaled maps showed less genetic effects throughout the brain (see Figure 3 - top right). While scaling the data depleted the effects of the common environment on brain structure (only the white matter partly exhibits c2 values equal to 20 – 30%), the genetics influence is still very strong in the limbic lobe, and the subcorticular structures, in particular in the ventricles (60%). Effects are also noticeable in the occipital lobes (20 – 30% - top left - d) The comparison of the top right panels in Figure 2 and 3 shows that overall, genetic influences (a2) are relatively high in the subcortical areas, as well as in the occipital areas, which are the earliest to mature in infancy.
To summarize the effect of the three factors on global structure volumes, ICC, h2 and proportion of variance factors were computed for the five lobes, the ventricles, the thalamus, basal ganglia and the whole brain (Table 1 and Table 2). In the unscaled data, the proportion of genetic, shared and unique environmental variance was approximately the same for all the lobes (see Table 1), with around 30 – 40% of the variance being attributable to genetic differences in the cohort. The shared environment also accounted for around half of the variance in these volumes, with the rest of the differences being attributable either to unique environment or measurement errors. Volumes for the basal ganglia and the thalamus were shown to be influenced by genetic factors (A), as well as shared (C) and unique (E) environmental factors (a2 = 40%, and c2 = 50%, for the basal ganglia - a2 = 25% and c2 = 63% for the thalamus). Between 60% and 70% of the variance in ventricular volumes was attributable to dominant genetic factors (the ADE model resulted in a better fit than the ACE model, which may be related to the undetectable DZ correlation (ICC = 0) and to the increased difference between MZ and DZ correlations .
This value was still high (50% to 70%) in the scaled data (Table 2), where the ADE model was also proved to be the best fit. The strong influence of genetics was also seen in the thalamus and basal ganglia (where a2 = 58% and d2 = 57%, respectively). Even so, the genetics influence on the whole brain volume was considerably smaller after scaling, whereas the effect of the common environment decreased from 10%. This trend was found for all lobar structures except for the occipital lobes and the temporal lobes scaled .
In the scaled and unscaled data, the A, C, E or A, D, E terms fitted in all cases except for the parietal lobes (where the best p-value = 0.01 was found for the ACE model in the scaled data, and indicates a lack of fit). The full ACE model gave the best fit for all the structures except for the lateral ventricles (where pace = 0.048 and pade = 0.091) in the unscaled data. In comparison, the best fit with ACE was found in the scaled whole brain and frontal lobes only (p = 0.26 and p = 0.54, respectively). The ADE model was the best fit for the most genetically influenced structures, such as the occipital lobes, the ventricles, and the thalamus, whereas the AE model performed better in the basal ganglia (pace = 0.44, pade = 0.40 and pae = 0.61) and in the temporal lobes (pace = 0.41, pade = 0.36 and pae = 0.55).
Overall, when the scaling effect was removed, the explanatory value of the genetic term (A) decreased in all lobar regions. This is in line with expectation, because the variance in substructure volumes obeys an approximate power law relative to the overall size of the brain (Thompson et al., 2003); in other words, the logarithms of the substructure volumes and overall brain volumes are tightly correlated in normal populations. Because of this dependency, some of the variance in substructure volumes is correlated with variations in overall brain volume, which is also highly heritable (see Table 1 and Introduction). If some of the same genes influence substructure volumes as influence the overall brain volume (which is likely), then adjusting for overall brain volume is likely to decrease the remaining genetic proportion of variation in substructure volumes; however, if different genes mediate overall brain volume and substructure volumes, adjusting for overall brain volume may (at least in theory) increase the proportion of the remaining variation in sub-structure volumes that is genetically mediated. In our data, even after adjusting for brain volume (see Figure 2), the adjusted occipital, limbic lobar volumes were still genetically influenced. Temporal lobes volumes were also controlled by genetics before and after adjustment, but to a lesser extend, which may be explained by a high c2 value in the inferior temporal lobes, and a high a2 value in the anterior temporal area, that persisted after scaling (see Figure 2 - top right and left - f and g and Figure 3 - Top left - f and g). The environmental (C) component remained high in frontal regions. This effect was not seen for subcortical structures, where the genetic term was still dominant in the lateral ventricles, basal ganglia and thalamus after adjusting for brain scale.
3.3 Phenotypic variability in the population
Figure 4 shows the voxelwise variance of the phenotype in our sample. Whether the brains are scaled or unscaled, variance remains relatively high in the subcorticular regions (around 5%), where the genetic influence is the highest (see the two previous paragraphs). Second, although not so clearly evident, values measured in the lobar white matter (Figure 4 f and g) show a difference in variance between the unscaled and the scaled data (1% and 0.3%, respectively). This effect is also shown in Table 3, where a similar measurement is presented for the ROIs. While the variance is higher in the unscaled data (8% versus 0.6% for the scaled data) for the whole brain volume, frontal, parietal, temporal lobar and basal ganglia volumes, the scaling has little influence on the other structures that are the most genetically determined.
Fig. 4.
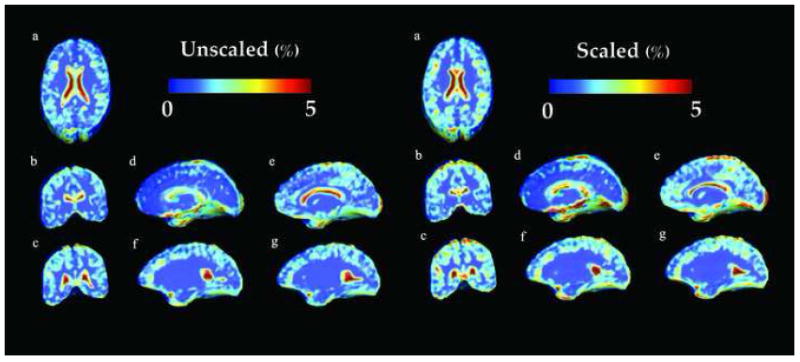
Variance of the Jacobian across the population for the unscaled (left) and the scaled data (right) displayed as the percentage of the mean at each voxel. Here, blue indicates a small variance in the trait (0%), whereas red indicates a higher variance (5%). It is worth noting that values in the unscaled white matter are higher than values in the scaled white matter except for the subcortical regions (1% versus 0.3%).
Table 3.
Standard deviation of the ROI volumes across the population reported as a percentage of the mean of the whole region. ROIs: WB- whole brain ; Fr - Frontal lobes ; Par - Parietal lobes ; Temp - Temporal lobes ; Occ - Occipital lobes ; Lmbc - Limbic lobes ; Vent - Ventricles ; BG - Basal Ganglia ; Th – Thalamus
| Regions | WB | Fr | Par | Temp | Occ | Lmbc | Vent | BG | Th |
|---|---|---|---|---|---|---|---|---|---|
| unscaled | 8.99 | 8.94 | 8.93 | 8.71 | 8.81 | 8.69 | 17.31 | 8.93 | 8.91 |
| scaled | 0.41 | 0.40 | 0.67 | 0.71 | 10.37 | 17.11 | 11.84 | 0.77 | 11.41 |
4 Discussion
4.1 Findings
In this study, we combined Tensor-Based Morphometry, a method that analyzes morphological brain differences, with models traditionally used in genetic studies, including structural equation models, which were computed using a new and efficient method (Chiang et al., 2008; Lee et al., 2009). The study had three main findings. First, we computed correlation maps to visualize the level of anatomical similarity for identical and fraternal twin groups, from which we derived a commonly used measure of heritability. This voxel-wise method indicated a genetically mediated component of variance in subcortical regions (≈ 50%). The maps revealed the expected pattern of genetic influence for a heritable trait, in which identical twins resembled each other the most, and fraternal twins less so. The resemblance for each type of twin was significant overall and was statistically confirmed to be greater than that likely to be observed by chance in random pairings of individuals of the same age and sex.
Subcortical regions, in particular the lateral ventricles and corpus callosum, and occipital lobes that mature the earliest in infancy, showed strong evidence of hereditary influences even after adjustment for whole brain scale. Our voxel-based maps of the heritability indicated genetically influenced regions previously implicated in independent studies by Hulshoff Pol et al. (2006a), such as limbic regions (such as the cingulate gyrus), anterior temporal and occipital lobe regions. In Hulshoff Pol et al. (2006a), the authors used VBM and structural equation models. The additive genetic components of variance, a2, were slightly higher in their study than the ones in our study for similar regions (e.g., ≈ 80% versus ≈ 40% for unscaled data). This may be attributable to a larger e2 component in our study (which contains any measurement errors). Differences may be due to differences in the definitions of regions of interest. In additional, in sample sizes around 100 subjects (as were used in this study and the one by Hulshoff Pol et al. (2006a), there is a certain margin of error in estimating a2, and these confidence intervals have been reported to allow comparisons with other reports and with future studies.
Also, as hypothesized, when unscaled data was assessed (i.e., before accounting for differences in overall brain scale), the proportion of genetic variance was higher for all the unscaled lobar volumes than after adjusting for brain volume. This is reasonable given that brain volume is itself heritable and is correlated with regional volumes (Giedd al., 2007). This finding is also supported by prior twin studies (Pfefferbaum et al., 2000; Bartley et al., 1997).
Third, our ACE variance component models largely confirmed the results obtained with the statistical maps, namely that genetic influences were relatively strong subcortically. Very similar results were found for the voxel-wise maps as for the ROIs. Even so, voxel-wise maps, derived from the SEMs, may be used to define a more detailed pattern for the different influences. For example, the superior/anterior regions of the temporal lobes were found to be under genetic control, whereas their inferior parts were more influenced by common environment, at least in the unscaled data. We also observed strong shared environmental effects on all the unscaled structure volumes but the ventricles, which was not observed in other studies. This finding may be due to the mean age (and small variance) of the sample population (young adults). As the brain changes throughout life, one might propose that the common environmental effects are at their strongest immediately after teenage years (the age range studied here) and that this effect may decrease with age, as the twin subjects no longer live in the same environment. A similar explanation has been proposed in prior studies of heritability of IQ, in which effects of the childhood rearing environment were detected early in life but dissipated later in life when the twin subjects lived apart. After scaling effects were removed, subcortical structures were generally the most highly genetically influenced, while frontal lobe volumes were mostly influenced by shared and unique environment. We were unable to detect any environmental effect on lateral ventricular volume. Reveley et al. (1982); Pfefferbaum et al. (2000) and Styner et al. (2005) showed that ventricular volume is genetically influenced, and others (Baaré et al., 2001; Wright et al., 2002) found that ventricular volumes are equally influenced by genetics (58%) and environment (42%). Inspection of our data revealed that the ventricular volume correlation for DZ twins was relatively low. Choosing the ADE model rather than the ACE helped explaining this findings: the genetics effect on ventricles before and after scaling was shown to be dominant more than additive, leading to a correlation in MZ more than twice higher than the correlation in DZ twins.
Besides, generally speaking the more genetically influenced traits were also the more variable phenotypes in the sample.
4.2 Genetics and the developmental sequence
The extent to which different brain structures are genetically influenced is likely to vary spatially across the brain. One plausible developmental hypothesis, based on the developmental sequence during childhood, is that the earliest-maturing brain regions have structural volumes that are more genetically influenced. Environmental influences may be greater for the later-developing brain regions (e.g., the frontal lobes). As we found in our cortical mapping study of brain development from childhood to early adulthood, (Gogtay al., 2004), brain regions that mature first in childhood tend to support more basic cognitive processes, such as movement (motor and primary sensory cortices), taste (insula), vision (occipital lobes), and hearing (anterior temporal lobes). The regions with the most protracted maturational time-course include the prefrontal lobes (involved in complex reasoning), temporal lobes and hippocampus (Gogtay al., 2004). White matter continues to mature throughout life (Bartzokis et al., 2008), but most of the white matter myelination that affects regional brain volumes is completed by adolescence, with heavy myelination of the corticospinal tracts and occipital lobes occurring in early infancy (Aubert-Broche et al., 2008). This developmental sequence may relate to the profile of genetic influences on brain structure; our initial data are consistent with the hypothesis that structures subserving basic processes may be under stronger genetic control than those that mature over a more extended period.
4.3 TBM: advantages and limitations
Prior research has found that key measures of cognitive performance, such as IQ, are highly heritable and correlate with frontal lobe gray and white matter volumes (Peper et al., 2007). Here we found moderate genetic influences on frontal lobe volumes, in the range 30 – 40% before scaling; values that are comparable with, but not quite as high as prior studies. The method used to measure structures is likely to affect the values for the genetic variance components, because all methods are likely to vary in their measurement error variance, which is part of the e2 component of variance, and all variances components are by definition proportions of the overall variance and sum to one. For more specific analysis of cortical gray matter variations, for example, specialized processing methods may be used to analyze the cortical surface (see Thompson et al. (2004) for a review), as in our prior genetic studies (Thompson et al., 2001) and in the cortical surface modeling studies by Schmitt et al. (2008). Although these analyses are more time consuming to perform, a precise estimate of cortical thickness or gray matter density (and other features such as cortical curvature and complexity) can be derived for each subject. In particular, registration methods developed for more precise cortical pattern matching (e.g., Thompson et al. (2004)) can be used to integrate and compare data from subjects whose gyral patterns are different, by fluidly transforming entire networks of gyral landmarks into correspondence. Registration error is reduced, and measurement precision is increased. In general, one would expect the genetic and common environmental components to be higher, as a proportion of the overall variance for methods that further reduce the measurement error variance, even if these methods are more labor-intensive to perform. As such, heritability estimates may be slightly lower for high-throughput voxel-based methods such as TBM or VBM (Hulshoff Pol et al., 2006a) than for methods that explicitly model and register cortical anatomy (Thompson et al., 2001; Schmitt et al., 2008). As twin studies begin to routinely assess hundreds or even thousands of subjects, there will ultimately be a trade-off between methods that are most efficient to use on large cohorts versus those are offer the greatest power to detect gene effects per subject analyzed, given the resources available. A related problem pertains to the subcortical structures as registration accuracy could potentially be influenced by the degree of spatial regularization involved in the warping approach. A highly regularized method, in which the deformation fields are extremely smooth, may increase the genetic proportion of variance (a2) locally if a2 was high for global measures. This issue can be further investigated in future by studying how the genetic effects on brain structures depend on the smoothness and acscuracy of the registration vector fields. In the future, we plan to further examine genetic influences on cortical gray matter differences using surface-based cortical thickness maps Thompson et al. (2004), which will take advantage of the cortical features and explicit surface models to allow higher order spatial normalization and matching of cortical anatomy.
A second more conceptual issue relates to the disentanglement of genetic effects on overall brain volume from regional heritability estimates. In imaging studies designed to identify genes that regulate overall brain volume, there may be no need to adjust the heritability maps for global volume effects, as potentially important genetic effects would be discounted. In the future, however, there will be a growing interest in genes that regulate specific functional systems and not others, or distributed cortical networks that operate as a functional unit. A first step towards understanding regionally-specific genetic influences has been pioneered by Wright et al. (2002) and Schmitt et al. (2007), who proposed a multivariate spatial decomposition of the genetic variance in a dataset. These methods hold promise for identifying genetic effects that affect the development of an entire system, or overlapping sets of genes involved in the maturation of different brain regions. These methods often require very large cohorts to rigorously establish these subtle, second-order effects on heritability. These include the examination of differential heritability between different measures or different regions and the detection of common versus specific genetic influences on different measures, which will be a key target of future studies.
Third, there may be age effects on heritability, which means that any study of a specific cohort must be interpreted as applying to the age range specifically examined (here, early adulthood). In Lenroot (2007), who studied children, the authors argued that there is an age-related difference in the heritability of cortical thickness. The primary sensory and motor cortex were found to be predominantly determined by genes in childhood, while later developing regions, such as the dorsolateral prefrontal cortex and the temporal lobes may experience greater genetic influence later in life.
Lenroot et al. (2007) suggested that brain regions related to high cognitive functions may have morphometric variance that is more heritable in adolescence than childhood, as that is when they are most rapidly developing. Gene-environment interaction effects may also complicate and further enhance this increase in heritability during the most rapid period of development. As such, longitudinal studies will be needed on large datasets (Wright et al., 2001) to account for age effects, or even temporary effects on heritability that may occur during an active growth spurt and may not persist later when growth rates are lower and population variance is lower (Giedd al., 2007).
An additional fruitful direction involves the genetics of diffusion tensor imaging (DTI). Hulshoff Pol et al. (2006a) showed that the cross-sectional area and fiber integrity (fractional anisotropy) of the corpus callosum was mostly determined by genes, a finding that has been verified at different ages across the human lifespan. More recent bivariate genetic modeling studies using high-angular resolution diffusion imaging (HARDI) in twins suggest that DTI-derived measures of fiber architecture and IQ may be both highly heritable and influenced by overlapping sets of genes (Chiang et al., 2008; Lee et al., 2009). Future studies combining DTI, functional, and cognitive measurements with neuroanatomy (Glahn al., 2007b) are needed to better establish links between genetics, functional MRI signals and cognition (see Blokland et al. (2008) and de Zubicaray et al. (2008)).
Acknowledgments
This work was generously supported by NIH grant R01 HD050735 and National Health and Medical Research Council, Australia grant 496682.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arsigny V, Fillard P, Pennec X, Ayache N. Log-Euclidean metrics for fast and simple calculus on diffusion tensors. Mag Res Med. 2006;56(2):411–421. doi: 10.1002/mrm.20965. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Voxel-based Morphometry: the methods. NeuroImage. 2000;11(6):805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Aubert-Broche B, Fonov V, Leppert I, Pike GB, Collins LD. Human Brain Myelination from birth to 4.5 years. International conference on Medical Image Computing and Computer Assisted Intervention (MICCAI); New-York, USA. September 7-9 (2008); [DOI] [PubMed] [Google Scholar]
- Baaré WF, Hulshoff Pol HE, Boomsma DI, Posthuma D, de Geus EJ, Schnack HG, van Haren NE, van Oel CJ, Kahn RS. Quantitative genetic modeling of variation in human brain morphology. Cereb Cortex. 2001;11(9):816–824. doi: 10.1093/cercor/11.9.816. [DOI] [PubMed] [Google Scholar]
- Bartley AJ, Jones DW, Weinberger DR. Genetic variability of human brain size and cortical gyral patterns. Brain. 1997;120(2):257–269. doi: 10.1093/brain/120.2.257. [DOI] [PubMed] [Google Scholar]
- Bartzokis G, Lu PH, Tingus K, Mendez MF, Huang D, Thompson PM, Mintz J. Lifespan trajectories of myelin integrity and tapping speed in healthy males: implications for aging and degenerative brain diseases. Neurobiol Aging. 2008 October; [Google Scholar]
- Blokland GA, McMahon KL, Hoffman J, Zhu G, Meredith M, Martin NG, Thompson PM, de Zubicaray GI, Wright MJ. Quantifying the heritability of task-related brain activation and performance during the N-back working memory task: a twin fMRI study. Biol Psychol. 2008;79(1):70–79. doi: 10.1016/j.biopsycho.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollen KA, Stein RA. Bootstrapping goodness-of-fit measures in structural equation models. Sociol Methods Res, (1992) 1992;21(2):205–229. [Google Scholar]
- Bro-Nielsen M, Gramkow C. Fast fluid registration of medical images. Proceedings of the 4th International conference on Visualization in Biomedical Computing; Hamburg, Germany. September 22-25 (1996); pp. 272–276. [Google Scholar]
- Brun C, Leporé N, Pennec X, Chou Y-Y, Lopez OL, Aizenstein OJ, Becker JT, Toga AW, Thompson PM. Comparison of Standard and Riemannian Fluid Registration for Tensor-Based Morphometry in HIV/AIDS. Workshop on Statistical Registration: Pair-wise and Group-wise Alignment and Atlas Formation, 10th International conference on Medical Image Computing and Computer Assisted Intervention (MICCAI); Brisbane, Australia. October 29- November 2 (2007). [Google Scholar]
- Brun C, Leporé N, Pennec X, Chou Y-Y, Lee AD, de Zubicaray G, McMahon K, Wright M, Barysheva M, Toga AW, Thompson PM. A new registration method based on log-euclidean tensor metrics and its application to genetic studies. Proceedings of the 5th IEE International Symposium on Biomedical Imaging (ISBI); Paris, France. May 14-17 (2008); [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon TD, Thompson PM, Van Erp TG, Toga AW, Poutanen VP, Huttunen M, Lonnqvist J, Standerskjold-Nordenstam CG, Narr KL, Khaledy M, Zoumalan CI, Dail R, Kaprio J. Cortex mapping reveals regionally specific patterns of genetic and disease-specific gray-matter deficits in twins discordant for schizophrenia. Proc Natl Acad Sci. 2002;99(5):3228–3233. doi: 10.1073/pnas.052023499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon TD, Hennah W, van Erp TG, Thompson PM, Lönnqvist J, Huttunen M, Gasperoni T, Tuulio-Henriksson A, Pirkola T, Toga AW, Kaprio J, Mazziotta J, Peltonen L. Association of DISC1/TRAX haplotypes with schizophrenia, reduced prefrontal gray matter, and impaired short- and long-term memory. Arch Gen Psychiatry. 2005;62(11):1205–1213. doi: 10.1001/archpsyc.62.11.1205. [DOI] [PubMed] [Google Scholar]
- Chiang M-C, Dutton RA, Hayashi KM, Toga AW, Lopez OL, Aizen-stein HJ, Becker JT, Thompson PM. Fluid registration of medical images using Jensen-Rényi divergence reveals 3D-profile of brain atrophy in HIV/AIDS. Proceedings of the 3rd IEEE International Symposium on Biomedical Imaging; Arlington, Virginia, USA. 6-9 April (2006). [Google Scholar]
- Chiang M-C, Dutton RA, Hayashi KM, Toga AW, Lopez OL, Aizenstein HJ, Becker JT, Thompson PM. 3D pattern of brain abnormalities in Williams syndrome visualized using tensor-based morphometry. Neuroimage. 2007;36(4):1096–1109. doi: 10.1016/j.neuroimage.2007.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang M-C, Barysheva M, Lee AD, Madsen SK, Klunder AD, Toga AW, McMahon KL, de Zubicaray GI, Meredith M, Wright MJ, Srivastava A, Balov N, Thompson PM. Brain Fiber Architecture, Genetics, and Intelligence: A High Angular Resolution Diffusion Imaging (HARDI) Study. Workshop on Computational diffusion MRI, International conference on Medical Image Computing and Computer Assisted Intervention (MICCAI); New-York, USA. September 6 (2008); [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Y-Y, Leporé N, de Zubicaray GI, Carmichael OT, Becker JT, Toga AW, Thompson PM. Automated Ventricular Mapping with Multi-Atlas Fluid Image Alignment Reveals Genetic Effects in Alzheimers Disease. Neuroimage. 2008;40(2):615–630. doi: 10.1016/j.neuroimage.2007.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Y-Y, Leporé N, Chiang M-C, Avedissian C, Barysheva M, McMahon KL, de Zubicaray GI, Meredith M, Wright MJ, Toga AW, Thompson PM. Mapping Genetic Influences on Ventricular Structure in Twins. NeuroImage. 2009;44(4):1312–1323. doi: 10.1016/j.neuroimage.2008.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Y-Y, Leporé N, Brun CC, Barysheva M, McMahon K, de Zubicaray GI, Wright MJ, Toga AW, Thompson PM. an Tissue Segmentation Improve Registration? A study of 92 Twins. Organization for Human Brain Mapping. 2009 [Google Scholar]
- Christensen EG, Rabbitt RD, Miller MI. Deformable templates using large deformation kinematics. IEEE Trans Image Process. 1996;5(10):1435–1447. doi: 10.1109/83.536892. [DOI] [PubMed] [Google Scholar]
- Collins L, Holmes CJ, Peters TM, Evans AC. Automatic 3D model-based neuroanatomical segmentation. Hum Brain Map. 1995;3(3):190–208. [Google Scholar]
- De Geus EJC, Van’t Ent D, Wolfensberger SPA, Heutink P, Hoogendijk WJG, Boomsma DI, Veltman DJ. Intrapair differences in hippocampal volume in monozygotic twins discordant for the risk for anxiety and depression. Biol Psychiatry. 2006;61(9):1062–1071. doi: 10.1016/j.biopsych.2006.07.026. [DOI] [PubMed] [Google Scholar]
- De Zubicaray GI, Chiang M-C, McMahon KL, Shattuck DW, Toga AW, Martin NG, Wright MJ, Thompson PM. Meeting the challenges of neuroimaging genetics. Brain Imaging Behav. 2008;2(4):258–263. doi: 10.1007/s11682-008-9029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgington ES. Randomization Tests. 3. Marcel Dekker; 1995. [Google Scholar]
- Falconer DS. Introduction to Quantitative Genetics. 3. Essex, UK: Longman; 1989. [Google Scholar]
- Fornell C, Larcker DF. Evaluating structural equation models with unobservable variables and measurement error. J Marketing Res. 1981;18(3):375–381. [Google Scholar]
- Giedd JN, Schmitt JE, Neale MC. Structural brain magnetic resonance imaging of pediatric twins. Hum Brain Mapp. 2007;28(6):474–481. doi: 10.1002/hbm.20403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn DC, Thompson PM, Blangero J. Neuroimaging endophenotypes: strategies for finding genes influencing brain structure and function. Hum Brain Mapp. 2007;28(6):488–501. doi: 10.1002/hbm.20401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn DC, Paus T, Thompson PM. Imaging genomics: Mapping the influence of genetics on brain structure and function. Hum Brain Mapp. 2007;28(6):461–463. doi: 10.1002/hbm.20416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogtay N, Giedd JN, Lusk L, Hayashi KM, Greenstein D, Vaituzis AC, Nugent TF, 3rd, Herman DH, Clasen LS, Toga AW, Rapoport JL, Thompson PM. Dynamic mapping of human cortical development during childhood through early adulthood. Proc Natl Acad Sci. 2004;101(21):8174–8179. doi: 10.1073/pnas.0402680101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogtay N*, Lu A*, Leow AD, Klunder AD, Lee AD, Chavez A, Greenstein D, Giedd JN, Toga AW, Rapoport JL, Thompson PM. 3D Growth Pattern Abnormalities Visualized in Childhood-Onset Schizophrenia using Tensor-Based Morphometry. Proc Natl Acad Sci. 2008 doi: 10.1073/pnas.0806485105. * equal contribution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramkow C. Master’s thesis. Danish Technical University; Copenhagen, Denmark: 1996. Registration of 2D and 3D medical images. [Google Scholar]
- Gray JR, Thompson PM. Neurobiology of Intelligence: Science and Ethics. Nat Rev Neurosci. 2004;4(6):471–482. doi: 10.1038/nrn1405. [DOI] [PubMed] [Google Scholar]
- Haier RJ, Jung RE, Yeo RA, Head K, Alkire MT. Structural brain variation and general intelligence. Lancet. 2004;23(1):425–433. doi: 10.1016/j.neuroimage.2004.04.025. [DOI] [PubMed] [Google Scholar]
- Hattersley AT, McCarthy MI. What makes a good genetic association study? Lancet. 2005;366(9493):1315–1323. doi: 10.1016/S0140-6736(05)67531-9. [DOI] [PubMed] [Google Scholar]
- Heckemann RA, Hajnal JV, Aljabar P, Rueckert D, Hammers A. Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. NeuroImage. 2006;33(1):115–126. doi: 10.1016/j.neuroimage.2006.05.061. [DOI] [PubMed] [Google Scholar]
- Hua X, Leow AD, Levitt JG, Caplan R, Thompson PM, Toga AW. Detecting brain growth patterns in normal children using tensor-based morphometry. Hum Brain Map. 2007 Dec 6; doi: 10.1002/hbm.20498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Leow AD, Lee S, Klunder AD, Toga AW, Leporé N, Chou Y-Y, Chiang M-C, Barysheva M, Jack CR, Jr, Bernstein MA, Britson PJ, Gunter JL, Ward CP, Whitwell JL, Borowski B, Fleisher AS, Fox NC, Boyes R, Barnes J, Harvey D, Kornak J, Schuff N, Boreta L, Studholme C, Alexander GE, Weiner MW, Thompson PM. 3D Characterization of Brain Atrophy in Alzheimer’s disease and mild cognitive impairment using Tensor-Based Morphometry. NeuroImage. 2008;41(1):19–34. doi: 10.1016/j.neuroimage.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Leow AD, Parikshak N, Lee S, Chiang MC, Toga AW, Jack CR, Jr, Weiner MW, Thompson PM. Alzheimer’s Disease Neuroimaging Initiative, Tensor-based morphometry as a neuroimaging biomarker for Alzheimer’s disease: an MRI study of 676 AD, MCI, and normal subjects. NeuroImage. 2008;43(3):458–469. doi: 10.1016/j.neuroimage.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulshoff Pol HE, Schnack HG, Posthuma D, Mandl RCW, Baaré WF, Van Oel CJ, Van Haren NE, Collins DL, Evans AC, Amunts K, Bürgel U, Zilles K, de Geus E, Boomsma DI, Kahn RS. Genetic Contributions to Human Brain Morphology and Intelligence. J Neurosci. 2006;26(40):10235–10242. doi: 10.1523/JNEUROSCI.1312-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulshoff Pol HE, Schnack HG, Mandl RCW, Brans RG, Van Haren NE, Baaré WF, Van Oel CJ, Collins DL, Evans AC, Kahns RS. Gray and white matter density changes in monozygotic and same-sex dizygotic twins discordant for schizophrenia using voxel-based morphometry. Neuroimage. 2006;31(2):482–488. doi: 10.1016/j.neuroimage.2005.12.056. [DOI] [PubMed] [Google Scholar]
- Jack CR, Jr, Petersen RC, Grundman M, Jin S, Gamst A, Ward CP, Sencakova D, Doody RS, Thal LJ. Members of the Alzheimer’s Disease Cooperative Study (ADCS). Longitudinal MRI findings from the vitamin E and donepezil treatment study for MCI. Neurobiol Aging. 2008;29(9):1285–1295. doi: 10.1016/j.neurobiolaging.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Bannister PR, Brady JM, Smith SM. Improvised optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17(2):825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Kochunov P, Lancaster JL, Thompson PM, Woods R, Mazziotta J, Hardies J, Fox P. Regional spatial normalization: toward an optimal target. J Comput Assist Tomogr. 2001;25(5):805–816. doi: 10.1097/00004728-200109000-00023. [DOI] [PubMed] [Google Scholar]
- Kochunov P, Lancaster JL, Thompson PM, Toga AW, Brewer P, Hardies J, Fox P. An Optimized Individual Target Brain in the Talairach Coordinate System. NeuroImage. 2002;17(2):922–927. [PubMed] [Google Scholar]
- Lee AD, Leow AD, Lu A, Reiss AL, Hall S, Chiang M-C, Toga AW, Thompson PM. 3D Pattern of Brian Abnormalities in Fragile X Syndrome visualized using Tensor-Based Morphometry. Neuroimage. 2007;34(3):924–938. doi: 10.1016/j.neuroimage.2006.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AD, Leporé N, Brun C, Barysheva M, Chou Y-Y, Chiang M-C, Madsen SK, McMahon KL, de Zubicaray GI, Wright M, Toga AW, Thompson PM. The Multivariate A/C/E model and the genetics of Fiber Architecture. Proceedings of the 6th IEEE International Symposium on Biomedical Imaging (ISBI); Boston, MA, USA. June 28- July 1rst (2009); [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenroot RK, Schmitt JE, Ordaz SJ, Wallace GL, Neale MC, Lerch JP, Kendler KS, Evans AC, Giedd JN. Differences in genetic and environmental influences on the human cerebral cortex associated with development during childhood and adolescence. Hum Brain Mapp. 2007 doi: 10.1002/hbm.20494. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leporé N, Brun CC, Pennec X, Chou YY, Lopez OL, Aizenstein HJ, Becker JT, Toga AW, Thompson PM. Mean Template for Tensor-Based Morphometry using Deformation Tensors. Proceedings, 10th International conference on Medical Image Computing and Computer Assisted Intervention (MICCAI); Brisbane, Australia. October 29- November 2 (2007); pp. 826–33. [DOI] [PubMed] [Google Scholar]
- Leporé N, Brun C, Chou Y-Y, Chiang M-C, Dutton RA, Hayashi KM, Lueders E, Lopez OL, Aizenstein HK, Toga AW, Becker JT, Thompson PM. Generalized Tensor-Based Morphometry of HIV/AIDS using multivariate statistics on deformation tensors. IEEE Transactions on Medical Imaging. 2008;27(1):129–141. doi: 10.1109/TMI.2007.906091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leporé N, Chou Y-Y, Lopez OL, Aizenstein HJ, Becker JT, Toga AW, Thompson PM. Fast 3D Fluid Registration of Brain Magnetic Resonance Images. SPIE Medical Imaging conference; San Diego, CA. February 17-21 (2008); [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leporé N, Brun C, Chou Y-Y, Lee AD, Barysheva M, Pennec X, McMahon KL, Meredith M, de Zubicaray GI, Wright M, Toga AW, Thompson PM. Best individual template selection from deformation tensor minimization. Proceedings of the 5th IEEE International Symposium on Biomedical Imaging (ISBI); Paris, France. May 14-17 (2008); [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leporé N, Brun C, Chou Y-Y, Lee AD, Barysheva M, Pennec X, McMahon KL, Meredith M, de Zubicaray GI, Wright M, Toga AW, Thompson PM. Multi-Atlas tensor-based morphometry and its application to a genetic study of 92 twins. Workshop on Mathematical Foundations of Computational Anatomy, International conference on Medical Image Computing and Computer Assisted Intervention (MICCAI); New-York, USA. September 6 (2008). [Google Scholar]
- Maes HH. Encyclopedia of Statistics in Behavioral Science. John Wiley & Sons, LTD; Chichester: 2005. ACE Model. [Google Scholar]
- Narr KL, Szeszko PR, Lencz T, Woods RP, Hamilton LS, Phillips O, Robinson D, Burdick KE, DeRosse P, Kucherlapati R, Thompson PM, Toga AW, Malhotra AK, Bilder RM. DTNBP1 is associated with imaging phenotypes in schizophrenia. Hum Brain Map. 2008 doi: 10.1002/hbm.20806. under revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MC, Boker SM, Xie G, Maes HH. Mx: Statistical modeling. 5. Richmond; 1999. [Google Scholar]
- Nichols TE, Holmes AP. Non parametric permutation tests for functional neuroimaging: a primer with examples. Hum Brain Map. 2002;15(1):1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppeheim JS, Skerry JE, Tramo MJ, Gazzaniga MS. Magnetic resonance imaging morphology of the corpus callosum in monozygotic twins. Ann Neurol. 26(1):100–104. doi: 10.1002/ana.410260117. [DOI] [PubMed] [Google Scholar]
- Pennec X, Stefanescu R, Arsigny V, Fillard P, Ayache N. Riemannian elasticity: A statistical regularization framework for non-linear registration. Medical Image Computing and Computer-Assisted Intervention (MICCAI); Palm Springs, CA, USA: 2005. pp. 943–950. [DOI] [PubMed] [Google Scholar]
- Peper JS, Brouwer RM, Boomsma DI, Kahn RS, Hulshoff Pol HE. Genetic influences on human brain structure: a review of brain imaging studies in twins. Hum Brain Mapp. 2007;28(6):464–473. doi: 10.1002/hbm.20398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferbaum A, Sullivan EV, Swan GE. Brain structure in men remains highly heritable in the seventh and eighth decades of life. Neurobiol Aging. 2000;21(1):63–74. doi: 10.1016/s0197-4580(00)00086-5. [DOI] [PubMed] [Google Scholar]
- Pietiläinen OPH, Paunio T, Loukola A, Tuulio-Henriksson A, Kieseppä T, Thompson PM, Toga AW, van Erp TGM, Soronen P, Hennah W, Turunen JA, Peltonen JO, Palo OM, Silander K, Lönnqvist J, Kaprio J, Cannon TD, Peltonen L. Association of AKT1 with verbal learning, verbal memory and regional cortical grey matter density in twins. Am J Med Genet B Neuropsychiatr Genet. 2008 December; doi: 10.1002/ajmg.b.30890. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posthuma D, De Geus EJ, Baaré WF, Hulshoff Pol HE, Kahn RS, Boomsma DI. The association between brain volume and intelligence is of genetic origin. Nat Neurosci. 2002;5(2):83–84. doi: 10.1038/nn0202-83. [DOI] [PubMed] [Google Scholar]
- Raji CA*, Ho AJ*, Parikshak N, Becker JT, Lopez OL, Kuller LH, Hua X, Leow AD, Toga AW, Thompson PM. Tensor Based Morphometry of Body Mass Index, Insulin, and Type II Diabetes Effects on Brain Structure in the Cardiovascular Health Study Cognition Study, submitted to Hum Brain Mapp. 2009 * equal contribution. [Google Scholar]
- Reiss AL, Abrams MT, Singer HS, Ross JL, Denckla MB. Brain development, gender and IQ in children. A volumetric imaging study. Brain. 1996;119(5):1763–1774. doi: 10.1093/brain/119.5.1763. [DOI] [PubMed] [Google Scholar]
- Reveley AM, Reveley MA, Clifford CA, Murray RM. Cerebral ventricular size in twins discordant for schizophrenia. Lancet. 1982;1(8271):540–541. doi: 10.1016/s0140-6736(82)92047-5. [DOI] [PubMed] [Google Scholar]
- Scamvougeras A, Kigar DL, Jones D, Weinberger DR, Witelson SF. Size of the human corpus callosum is genetically determined: an MRI study in mono and dizygotic twins. Neurosci Lett. 2003;338(2):91–94. doi: 10.1016/s0304-3940(02)01333-2. [DOI] [PubMed] [Google Scholar]
- Shattuck DW, Leahy RM. BrainSuite: an automated cortical surface identification tool. 2002;6(2):129–142. doi: 10.1016/s1361-8415(02)00054-3. [DOI] [PubMed] [Google Scholar]
- Schmitt JE, Wallace GL, Rosenthal MA, Molloy EA, Ordaz S, Lenroot RK, Clasen LS, Blumenthal JD, Kendler KS, Neale MC, Giedd JN. A multivariate analysis of neuroanatomic relationships in a genetically informative pediatric sample. Neuroimage. 2007;35(1):70–82. doi: 10.1016/j.neuroimage.2006.04.232. [DOI] [PubMed] [Google Scholar]
- Schmitt JE, Lenroot RK, Wallace GL, Ordaz S, Taylor KN, Kabani N, Greenstein D, Lerch JP, Neale MC, Giedd JN. Identification of genetically mediated cortical networks: a multivariate study of pediatric twins and siblings. Cereb Cortex. 2008;18(8):1737–1747. doi: 10.1093/cercor/bhm211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scout PE, Fleiss JL. Intraclass correlations: Uses in assessing rater reliability. Psychol Bull. 1979;86(2):420–428. doi: 10.1037//0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- Steinmetz H, Herzog A, Huang Y, Hacklander T. Discordant brain-surface anatomy in monozygotic twin. N Engl J Med. 1994;331(14):952–953. doi: 10.1056/nejm199410063311419. [DOI] [PubMed] [Google Scholar]
- Styner M, Lieberman JA, McClure RK, Weinberger DR, Jones DW, Gerig G. Morphometric analysis of lateral ventricles in schizophrenia and healthy controls regarding genetic and disease-specific factors. Proc Natl Acad Sci USA. 2005;102(13):4872–4877. doi: 10.1073/pnas.0501117102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan EV, Pfefferbaum A, Swan GE, Carmelli D. Heritability of hippocampal size in elderly twin men: Equivalent influence from genes and environment. Hippocampus. 2001;11(6):754–762. doi: 10.1002/hipo.1091. [DOI] [PubMed] [Google Scholar]
- Sullivan PF. Spurious genetic associations. Biol Psychiatry. 2007;61(10):1121–1126. doi: 10.1016/j.biopsych.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Cannon TD, Narr KL, Van Erp T, Poutanen VP, Huttunen M, Lönnqvist J, Standertskjöld-Nordenstam CG, Kaprio J, Khaledy M, Dail R, Zoumalan CI, Toga AW. Genetic influences on brain structure. Nat Neuroscience. 2001;4(12):1253–1258. doi: 10.1038/nn758. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Cannon TD, Toga AW. Mapping genetic influences on human brain structure. Ann Med. 2002;34(7-8):523–536. doi: 10.1080/078538902321117733. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Narr KL, Toga AW. Mapping structural alterations of the corpus callosum during brain development and degeneration, chapter. In: Zaidel E, Iacoboni M, editors. The Parallel Brain: the Cognitive Neuroscience of the Corpus Callosum. MIT press; 2003. [Google Scholar]
- Thompson PM, Hayashi KM, Sowell ER, Gogtay N, Giedd JN, Rapoport JL, de Zubicaray GI, Janke AL, Rose SE, Semple J, Doddrell DM, Wang Y, van Erp TG, Cannon TD, Toga AW. Mapping cortical change in Alzheimer’s disease, brain development, and schizophrenia. Neuroimage. 2004;23(1):2–18. doi: 10.1016/j.neuroimage.2004.07.071. [DOI] [PubMed] [Google Scholar]
- Van’t Ent D, Lehn H, Derks EM, Hudziak JJ, Van Strien NM, Veltman DJ, de Geus EJC, Todd RD, Boomsma DI. A structural MRI study in monozygotic twins concordant or discordant for attention/hyperactivity problems: evidence for genetic and environmental heterogeneity in the developing brain. Neuroimage. 2007;35(3):1004–1020. doi: 10.1016/j.neuroimage.2007.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger DR, Bartley AJ, Jones DW, Zigun JR. Regional variations in human monozygotic twins. Soc Neurosci Abstr. 1992;18:595. [Google Scholar]
- Wright M, de Geus E, Ando J, Luciano M, Posthuma D, Onon Y, Hansell N, Van Baal C, Hiraishi K, Hasegawa T, Smith G, Geffen G, Geffen L, Kanba S, Miyake A, Martin N, Boomsma D. Genetic of cognition: outline of a collaborative twin study. Twin Res. 2001;4(1):48–56. doi: 10.1375/1369052012146. [DOI] [PubMed] [Google Scholar]
- Wright IC, Sham P, Murray RM, Weinberger DR, Bullmore ET. Genetic contributions to regional variability in human brain structure: methods and preliminary results. Neuroimage. 2002;17(1):256–271. doi: 10.1006/nimg.2002.1163. [DOI] [PubMed] [Google Scholar]


