Abstract
C1q is a subunit of the C1 complex, a key player in innate immunity that triggers activation of the classical complement pathway. Featuring a unique structural organization and comprising a collagen-like domain with a high level of post-translational modifications, C1q represents a challenging protein assembly for structural biology. We report for the first time a comprehensive proteomics study of C1q combining bottom-up and top-down analyses. C1q was submitted to proteolytic digestion by a combination of collagenase and trypsin for bottom-up analyses. In addition to classical LC-MS/MS analyses, which provided reliable identification of hydroxylated proline and lysine residues, sugar loss-triggered MS3 scans were acquired on an LTQ-Orbitrap (Linear Quadrupole Ion Trap-Orbitrap) instrument to strengthen the localization of glucosylgalactosyl disaccharide moieties on hydroxylysine residues. Top-down analyses performed on the same instrument allowed high accuracy and high resolution mass measurements of the intact full-length C1q polypeptide chains and the iterative fragmentation of the proteins in the MSn mode. This study illustrates the usefulness of combining the two complementary analytical approaches to obtain a detailed characterization of the post-translational modification pattern of the collagen-like domain of C1q and highlights the structural heterogeneity of individual molecules. Most importantly, three lysine residues of the collagen-like domain, namely Lys59 (A chain), Lys61 (B chain), and Lys58 (C chain), were unambiguously shown to be completely unmodified. These lysine residues are located about halfway along the collagen-like fibers. They are thus fully available and in an appropriate position to interact with the C1r and C1s protease partners of C1q and are therefore likely to play an essential role in C1 assembly.
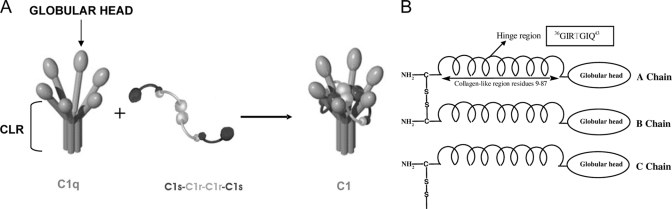
The multiprotein complex C1 (790,000 g·mol−1), the first component of the classical complement pathway, is a key effector in host defense, leading to the elimination of pathogens and altered host cells (1). C1 comprises a subunit named C1q, which insures its recognition and binding functions, and a protease subunit consisting of the Ca2+-dependent tetramer C1s-C1r-C1r-C1s (2). The enzyme subunit is required to be properly folded within C1q to undergo activation upon binding of C1q to a target and to trigger the complement cascade. These requirements for appropriate folding and activation are fulfilled by the C1q subunit through mechanisms remaining to be deciphered. The interface between C1q and the tetramer is not fully identified (3–5). Although the C1q binding sites of C1r and C1s have been recently determined by site-directed mutagenesis (6), much less is known about the interaction sites on C1q. C1q is a multimeric glycoprotein comprising 18 chains of three different types, A, B, and C (217–226 residues). These form three C-C and six A-B disulfide-bonded dimers, which assemble into six heterotrimeric (A-B)(C) subunits through non-covalent association (A-B)(C)-(C)(B-A). This hexameric structure appears in electron microscopy as a bouquet of six flowers and exhibits unique structural features. It consists of a short N-terminal region (3–9 residues) containing the interchain disulfide bonds that is prolonged by a triple helical collagen-like region (CLR;1 about 81 residues) that forms a “stalk” and then diverges into six individual “stems,” each terminating in a C-terminal heterotrimeric globular “head” (or globular region; about 135 residues) (Fig. 1) (7, 8). Several lines of evidence indicate that the tetramer binds to the collagen-like region of C1q. The intact CLR produced by pepsin digestion of C1q competes with C1q for association with the tetramer (9), and a monoclonal antibody to CLR prevents the interaction between C1q and the tetramer (10). Indeed, the effective association of CLR with the tetramer has been observed by analytical ultracentrifugation, providing further indication that the interaction sites between C1q and the tetramer are confined to the collagen region (11). Electron microscopy studies of the cross-linked C1 complex showed that the tetramer is located in the cone formed by the six collagen-like stems between the globular heads and the stalk (12, 13). Electron microscopy and neutron scattering studies of isolated C1s-C1r-C1r-C1s indicated an extended conformation of the tetramer in solution (14, 15). However, the volume inside the C1q cone is too small to contain the entire elongated tetramer. Based on these considerations, on neutron scattering studies of C1 (16), and on the C1s CUB1-EGF homodimer x-ray structure, a structural model of C1 has been proposed in which the tetramer undergoes a large conformational change into a compact “eight-shaped” structure wound in and out of the C1q stems (17). This model has been recently refined based on the identification of the C1q binding sites of C1r and C1s (6). It has been proposed that both C1r/C1s CUB1-EGF-CUB2 heterodimers are located inside the C1q cone and mediate ionic interactions through acidic residues contributed by the C1r and C1s CUB modules. Such ionic interactions at the C1q/C1s-C1r-C1r-C1s interface are expected to involve lysine residues of the collagen-like stems of C1q as suggested by the observation that chemical modification of C1q with lysine-specific reagents inhibits C1 assembly and C1q hemolytic activity (4, 18).
Fig. 1.
Schematic representation of structural organization of human C1q and of C1 assembly. A, model showing the CLR and globular heads of C1q and the association with the C1r/C1s tetramer forming the C1 complex. B, representation of the association between the A, B, and C chains leading to formation of the six ABC heterotrimeric triple helices of C1q CLR. The two following interruptions in the Gly-Y-Z triplet repeats are involved in the kink of the collagen-like region, i.e. the insertion of a threonine at position 39 in the A chain and the replacement of a glycine by an alanine at position 36 in the C chain (only the interruption in the A chain is shown).
Being collagen-like, the sequences of the CLR chains contain the repeating Gly-X-Z triplet where X is often a proline and Z is frequently a hydroxylysine or a hydroxyproline. C1q contains 8.3% carbohydrate (7), most of it being present in the CLR as glucosylgalactosyl disaccharide units linked to hydroxylysine residues (19). Because of the steric hindrance arising from the disaccharide moieties, non-glycosylated lysine residues are more plausible candidates for ionic interactions at the C1q/C1s-C1r-C1r-C1s interface. It was previously reported that 82.6% of the hydroxylysines are glycosylated (19) so that few unmodified lysine residues are available for this purpose. The primary structures initially reported in the 1970s did not allow the post-translational modifications (PTMs; hydroxylations and glycosylations) to be fully accurately identified along the CLR chains (20–25). Furthermore, the full-length C1q protein could not be analyzed to date by nuclear magnetic resonance or x-ray crystallography so the basic residues of the C1q stems involved in the C1q/tetramer interface remain to be identified. We have previously reported the MALDI-TOF mass spectrometry analysis of the entire C1q protein (26). The aim of the present study was to identify the C1q PTMs by MS and locate them along its chains to obtain further insights into the lysine residues likely involved in the interaction with the C1s-C1r-C1r-C1s tetramer. For this purpose, we performed for the first time a comprehensive proteomics study of C1q by using two very complementary approaches. On the one hand, C1q and CLR samples were digested by a combination of collagenase and trypsin, and the resulting peptides were analyzed by capillary LC-MS/MS (nano-LC-MS/MS). On the other hand, the bottom-up approach was completed by top-down analyses consisting of the direct infusion into the LTQ-Orbitrap instrument of the intact CLR domain and of the full-length A, B, and C chains of C1q. Spectra were successively acquired in the MS mode in the Orbitrap analyzer to obtain high accuracy and high resolution measurements of the intact protein masses and in the MSn mode to yield iterative fragmentation spectra of the proteins. The combination of bottom-up and top-down data proved most successful in yielding a significant coverage of the three chain sequences and in deciphering the heterogeneity of the C1q covalent modifications both in terms of proline hydroxylation and lysine glycosylation, thus revealing an unexpected level of complexity of the post-translational modifications.
EXPERIMENTAL PROCEDURES
Reagents and Materials
DTT, iodoacetamide (IAA), and NH4HCO3 were purchased from Sigma-Aldrich. Tris(2-carboxyethyl)phosphine was purchased from Sigma (reference number C-4706), and methyl methane thiosulfonate (MMTS) was obtained from Fluka (reference number 64306). HPLC gradient grade acetonitrile and Normapur grade formic acid were purchased from VWR (West Chester, PA). All buffers and solutions were prepared using ultrapure water (Milli-Q, Millipore, Bedford, MA). Type VII collagenase from Clostridium histolyticum (EC 3.4.24.3) was obtained from Sigma-Aldrich. Sequencing grade modified porcine trypsin (EC 3.4.21.4) was purchased from Promega (Madison, WI).
Fused silica PicoTips (360-μm outer diameter, 20-μm inner diameter) with a nominal tip end inner diameter of 10 ± 1.0 μm used for LC-nano-ESI-LTQ-Orbitrap coupling were obtained from New Objective (Woburn, MA; reference number FS360-20-10-N-20-C10.5). For the analysis of whole proteins by direct nano-ESI infusion, metallized tips (reference number BG12-69-2-CE, New Objective) were used.
Preparation of C1q and Its CLR
Human C1q was purified as described previously (27). For comparison, a commercial preparation of human C1q was purchased from Calbiochem. The CLR of C1q was prepared as described previously (28, 29).
Protein Reduction and Alkylation
A first series of protein samples was prepared as follows. Twenty microliters of commercial and non-commercial C1q as well as 10 μl of CLR (20, 17.8, and 32 μg of protein, respectively) were reduced by incubation for 1 h at 60 °C in the presence of 4.5 mm DTT. Samples were then alkylated by addition of 22 mm IAA and incubation for 45 min at room temperature in the dark. In view of the previously reported overalkylation of proteins by IAA (30), a second series of C1q samples was prepared by reduction with 5 mm tris(2-carboxyethyl)phosphine at 37 °C for 45 min and alkylation with 10 mm MMTS at room temperature for 15 min.
Protein Digestion by Collagenase and Trypsin
Reduced and alkylated samples were supplemented with collagenase to obtain an enzyme to substrate ratio of 0.5 (w/w) and incubated for 3 h at 37 °C. Before performing trypsin digestion of the mixture containing the C1q globular regions and collagenase, 50 mm NH4HCO3, pH 8.0, was added to the samples to reach a final concentration of 20 mm to obtain pH conditions more suitable to the serine protease activity (pH >7.5). Two and 3 μl of trypsin at 0.4 μg/μl were then added to each C1q and CLR sample, respectively, before incubation for 18 h at 37 °C.
Reversed Phase LC-MS/MS Analysis on QqTOF Instrument
A first series of LC-MS/MS analyses was performed using a Famos-Switchos-UltiMate chromatographic system (LC Packings/Dionex) coupled to a hybrid quadrupole QqTOF mass spectrometer QSTAR Pulsar i (Applied Biosystems/MDS Sciex) equipped with a nanoelectrospray source Protana XYZ manipulator (Protana, Odense, Denmark). Protein digests (1 pmol) were loaded onto a C18 precolumn (PepMap100 C18, 300-μm inner diameter, 5-mm length, 5-μm particle size, 100-Å porosity; Dionex) for desalting and concentrating at a flow rate of 30 μl/min in solvent A (water/acetonitrile/formic acid, 98:2:0.1, v/v/v). Peptides were then eluted from the precolumn and separated on a capillary column (PepMap C18, 75-μm inner diameter, 150-mm length, 3-μm particle size, 100-Å porosity; Dionex) at 200 nl/min using a gradient as follows: solvent A for 5 min, linear increase to 60% solvent B (water/acetonitrile/formic acid, 10:90:0.1, v/v/v) in 50 min, then ramp to 90% B in 5 min (held 10 min), and return to 100% A in 5 min for a 15-min-long re-equilibration of the columns. The autosampler was kept at 10 °C. Peptides eluting from the column were analyzed using the information-dependent data acquisition feature in the Analyst QS software v1.1: species ionized in the nano-ESI source were detected for 1 s in the MS mode, and the three most intense signals associated to either doubly or triply charged species were subsequently selected to be fragmented in the MS/MS mode for 3 s each. MS detection and MS/MS acquisitions were performed over the m/z ranges 400–1400 and 100–2000, respectively. Analyses were carried out with the dynamic exclusion of already fragmented m/z values for 3 min.
Reversed Phase LC-MS/MS Analysis on LTQ-Orbitrap Instrument
A second series of LC-MS/MS analyses was realized using an UltiMate 3000 chromatographic system (Dionex) coupled to a hybrid LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific). Typically, 200–300 fmol of digested C1q or CLR were injected onto a C18 precolumn (Dionex). After desalting for 5 min with buffer A (0.1% formic acid in water), peptides were separated on a capillary column (same reference as the one used on the QqTOF instrument) using a gradient from 100% solvent A to 60% solvent B (water/acetonitrile/formic acid, 10:90:0.1, v/v/v) in 60 min. The column was then further washed with 95% solvent B for 10 min. One series of MS analyses (ITMS2 analyses) consisted of acquiring cycles composed of one MS scan in the Orbitrap analyzer (profile mode; resolution, 15,000; m/z range, 400–2000) followed by three MS/MS scans (CID fragmentation and detection in the linear ion trap analyzer; centroid mode; isolation width, 2 Da) triggered on the three most intense species detected in the preceding MS scan. Singly charged species were excluded from fragmentation; dynamic exclusion of already fragmented ions was applied for 90 s with a repeat count of 1, a repeat duration of 20 s, and an exclusion mass width of ±5 ppm. Automatic gain control allowed accumulation of up to 5·105 ions for FTMS scans, 105 ions for FTMSn scans, and 104 ions for ITMSn scans. The maximum injection time was 100 ms for acquiring FTMS and ITMSn scans. Only one microscan was acquired for each scan type, although three were accumulated in FTMS mode in initial experiments. When testing acquisitions consisting of CID fragmentation in the ion trap analyzer and detection of the resulting fragments in the Orbitrap (OTMS2 analyses), the maximum injection time was 200 ms. In experiments combining MS2 and neutral loss-triggered MS3 scans (MS2/MS3 analyses), MS2 spectra were only acquired in the ion trap on the two precursor ions giving the most intense signals in MS, and MS3 was launched whenever a neutral loss of m/z 108.035, 162.053, 216.070, 324.106, and 486.158 was detected with a ±0.5-Da tolerance among the eight most intense MS2 fragments. MS3 was oriented toward the MS2 fragment corresponding to the biggest neutral loss. For these MS2/MS3 analyses, a precursor selection window of 2 and 5 Da was used for MS2 and MS3 scans, respectively. In addition, three microscans were accumulated to build an MS3 scan. Acquired raw data were processed by the software Bioworks to create Mascot-compatible .MGF files (no grouping of MS/MS scans was allowed).
Protein Identification Using Nano-ESI Direct Infusion on LTQ-Orbitrap
Five microliters of reduced non-commercial C1q and of CLR were desalted on a ZipTipC4 (Millipore), eluted in (water/acetonitrile/formic acid, 50:50:0.5, v/v/v) to 5 pmol/μl, and loaded into a metallized nanoelectrospray needle (PicoTip emitters, reference number BG12-69-2-CE-20, New Objective). A spray was obtained while working at a capillary temperature of 240 °C and adjusting the voltage applied to the nano-ESI tip between 1.4 and 2.4 kV. Automatic gain control parameters were set to 2·106 for FTMS, 2·105 for FTMS2, and 104 for ITMSn scans. Target resolution was 60,000 for FTMS and FTMS2 analyses. The maximum injection time was set to 500 ms in FTMS and FTMS2 and to 100 ms in ITMSn scans. The precursor selection width for FTMS2 fragmentation was around 3 Da and always adjusted to find a compromise between sensitivity and the clean selection of a single isotopic distribution. The selection window for ITMSn fragmentation (n ≥ 3) was 5 Da. The spectra shown in this study correspond to the accumulation of scans over approximately 1 min, yet good signal to noise ratios could be obtained within less time.
Database Searches
LC-MS/MS data (.MGF files obtained from .WIFF or .RAW data) were searched using the Mascot software (Matrix Science). Acquired data were compared with a homemade database named “mature C1q” consisting of only five sequences extracted from Swiss-Prot database release 54.7: porcine trypsin (accession number P00761), collagenase (accession number Q9X721), and the three mature protein sequences, C1qA, C1qB, and C1qC (accession numbers P02745, P02746, and P02747; the signal peptides 1–22, 1–25, and 1–28, respectively, were removed from the sequences stored in Swiss-Prot to obtain the mature sequences). Searches were first performed while considering that the analyzed peptides resulted from the combined use of collagenase and trypsin (collagenase + trypsin search). The sequential use of collagenase and trypsin to digest C1q was considered to produce peptides resulting from cleavages N-terminally of glycine residues and C-terminally of lysine and arginine residues. Collagenase is indeed known to specifically degrade collagen by cleaving N-terminally of GPX triplets. In a second step, searches were run by considering that trypsin may have produced half-tryptic peptides (collagenase + semitrypsin search). In both searches, nine missed cleavages were allowed. Finally, an error-tolerant search was performed after the two previous search steps to allow in particular identification of the N-terminal region of chain C1qB, which is known to be cyclized into pyrrolidone carboxylic acid. For data obtained on the QqTOF instrument, tolerances on mass measurement of precursors and fragments were set to 50 ppm and 0.4 Da, respectively. For Orbitrap data, tolerances on mass measurements were 5 ppm (FTMS) and 0.8 Da (ITMSn). Cysteine residues were considered to be fully alkylated either by IAA or MMTS; hydroxylation of lysine and proline residues, oxidation of methionine residues, and glucosylgalactosyl modification on hydroxylysines were included as potential modifications. We modified the definition of the modification “glucosylgalactosyl (Lys)” initially present in Mascot for neutral losses of 162.0528 and 324.1056 mass units to be taken into account for scoring.
RESULTS
Bottom-up Analysis of C1q and Its CLRs
Three types of samples were analyzed to determine the PTMs of C1q: laboratory-purified C1q, commercial C1q, and CLR. Each of them was digested using successively collagenase and trypsin and analyzed by LC-MS/MS. To facilitate reading, hydroxylated proline and lysine residues are noted P* and K*, respectively, and a lysine residue bearing a glucosylgalactosyl (Glc-Gal) moiety is noted K# in sequences.
Overall Sequence Coverage of Chains A, B, and C
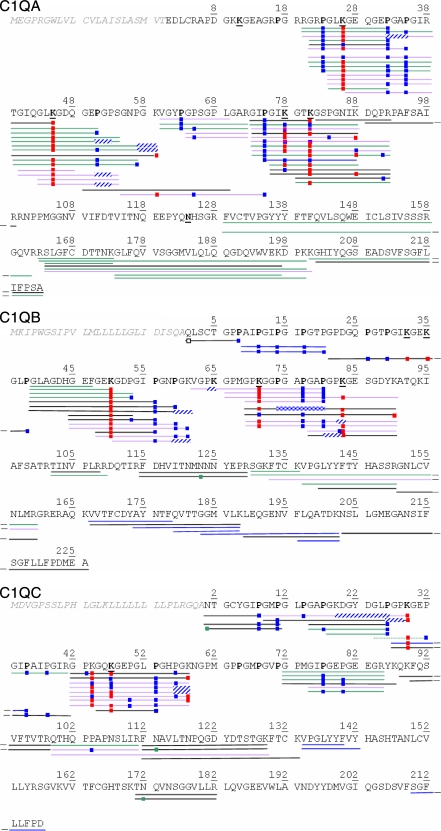
The digested samples were first characterized by triplicate LC-MS/MS analyses on QqTOF and LTQ-Orbitrap instruments (MS2 analyses). In the latter case, fragmentation and detection of the fragments were carried out in the linear ion trap of the hybrid instrument (ITMS2 analyses). The peptide sequences identified in the three chains constituting C1q are shown in Fig. 2. Database searches were performed assuming that collagenase would cleave N-terminally of glycine residues within the CLR regions (see “Experimental Procedures”). Indeed, nearly all identified sequences resulted from cleavages N-terminally of glycine residues and/or C-terminally of Lys/Arg residues. It is worth mentioning that, except for glycosylated peptides (see below), the identification of a peptide was routinely validated only if a minimal continuous sequence stretch of five amino acids was identified due to y-type or b-type fragment ions. Nonetheless, the frequent occurrence of proline residues at the Z position within GXZ triplets led us to be more tolerant: some peptide identifications were accepted when series of y ions, separated by three residues, corresponded to fragments containing an N-terminal proline (31, 32).
Fig. 2.
Peptide identified by LC-MS/MS analyses of C1q successively digested with collagenase and trypsin using QqTOF and LTQ-Orbitrap instruments. Green lines, peptides identified in both laboratory-purified and commercial C1q; black and violet lines, peptides only identified in either laboratory-purified or commercial C1q, respectively; blue lines, peptides specifically identified from the “collagenase + semitrypsin” database search; dotted line (in C1q C), sequence GLPGPK#GEP* identified from the “collagenase + trypsin” search was finally rejected in favor of sequence GPK#GEPGIP* (collagenase + semitrypsin) because the latter better matched the experimental spectrum. Blue squares, hydroxylation; red squares, Glc-Gal modification; green squares, deamidation; empty square, pyrrolidone cyclization. When precise localization of one (two) hydroxylation(s) could not be established from MS/MS spectra, an extended striped blue/white rectangle overlapping the possibly modified residues (Pro or Lys) is shown.
The C1q A chain, its CLR (residues 9–87), and its globular head region (residues 88–223) were covered at 77, 78, and 76%, respectively. The sequence 100–128 of the C1q A globular head could not be detected likely because of the presence of an N-linked oligosaccharide on residue Asn124 (33). The C1q B chain, its CLR (residues 6–89), and its globular head region (residues 90–226) were covered at 90, 100, and 83%, respectively. The “error-tolerant” search provided the identification of the N-terminal peptide of C1q B, 1%QLSCTGPP*8 (% indicates loss of NH3 by the N-terminal glutamine) in which Gln1 was detected as a pyroglutamic acid (Fig. 2, white square) in agreement with previous data (22). Finally, the C1q C chain, its CLR (residues 3–86), and its globular head region (residues 87–217) were covered at 68, 84, and 57%, respectively. These analyses resolved reported conflicts (22, 23) and confirmed the identity of residues deduced from the cDNA sequence (34): Pro75, Lys81, Cys150, Ser156, and the sequence LIFP (218–221) in the C1q A chain; Asn58 and Gly73 in the C1q B chain; and Lys29, Pro38, Lys44, and Pro56 in the C1q C chain.
Identification of Hydroxylated Sequences
The CLR of C1q contains the repeating triplet Gly-X-Z where Z is frequently a proline or a hydroxyproline residue. In most cases, peptides containing hydroxylated residues (but no glycosylation) fragmented in a manner similar to non-modified species, allowing confident determination of their sequence; yet, as stated above, the frequent occurrence of proline residues was detrimental to the detection of continuous y/b fragment series. As presented in Fig. 2, several proline residues of the three C1q chains were detected in both a non-modified and a hydroxylated form, indicating incomplete modification. For instance, peptide 72GPMGIPGEPGEEGR85 of C1q C was confidently identified in a non-modified form as well as in singly and doubly hydroxylated forms with modifications observed on residue Pro77 alone and additionally on Pro80. Taking into account the possible oxidation of methionine residues, this peptide was finally identified as doubly hydroxylated (on both Pro77 and Pro80) and oxidized on Met74. Although oxidation and hydroxylation yield equal mass increments, the detected MS2 fragments ruled out the possible hydroxylation of Pro73.
Several peptides containing one or two glycosylated lysine residues and fully or partially hydroxylated proline residues were also identified. Although identification of glycosylated sequences was often tedious (see section below), Pro23 and Pro35 from C1q A were clearly identified as being fully modified, whereas Pro32 was confidently determined to be partially modified due to the detection of several overlapping peptides.
Identification of Glycosylated Sequences
Composition analysis has indicated that C1q contains 8.3% carbohydrate (7), most of it being present in CLR as Glc-Gal disaccharide units linked to 82.6% of the hydroxylysine residues (19). In contrast to peptides containing only hydroxylated residues, glycosylated sequences systematically provided MS2 spectra dominated by sequential neutral losses of saccharide moieties (162.05 mass units), whereas fragmentation along the peptide backbone produced peaks of minor intensity, often precluding robust sequence determination and localization of the glycosylation site. We therefore tested different fragmentation/detection schemes on the LTQ-Orbitrap instrument to obtain complementary and redundant identification of glycosylated sequences and thus increase confidence in the identification of lysine residues bearing a Glc-Gal motif.
The MS2 data acquired on the QqTOF and LTQ-Orbitrap instruments allowed identification of the glycosylated sequences shown in Fig. 2. All sequences identified by the Mascot software are shown provided that all fragment peaks of major intensities matched the theoretical fragments expected for the proposed sequence. We checked the relevance of hydroxylation positioning and reintroduced uncertainty when the detected fragments were insufficient to definitely localize this modification on a specific proline or lysine residue (see for example peptide 39–59 of C1q A). Some peptides bearing one glycosylation and containing one lysine residue could be readily identified; supplemental Fig. S1 shows an MS2 spectrum obtained on the QqTOF instrument and unambiguously identifying peptide 39TGIQGLK#GDQGEP51 from C1q A. Determination was more challenging when a glycosylation site(s) occurred within sequences containing more lysine residues than the number of disaccharide moieties. This is exemplified in sequences 73–92 from C1q A, 69–90 from C1q B, and 42–58 from C1q C, which all contain three lysine residues and were identified as being doubly modified by Glc-Gal motifs. The Mascot search software then often provided identification of sequences corresponding to the different possible combinations of free and glycosylated lysines. For example, peptide 69–90 from C1q B was proposed to be modified either on Lys71 and Lys83 or on Lys83 and Lys90. Interestingly, glycosylated lysine residues were sometimes detected as still bearing one sugar moiety (the lysine was modified by 178.0477 mass units) during fragmentation in the linear ion trap of the LTQ-Orbitrap instrument; this behavior was never observed using the QqTOF instrument with which initially glycosylated lysine residues were systematically detected in a simply hydroxylated form in MS2 spectra. Two spectra showing lysine residues carrying a single sugar unit during MS/MS fragmentation are provided in supplemental Fig. S2, A and B; such fragmentation patterns helped us confirm the glycosylated nature of some lysine residues. More generally, to discriminate which lysine residues were most probably modified by Glc-Gal, we determined the best consensus resulting from the different proposed glycosylated sequences. Taking again the doubly glycosylated sequence 69–90 from C1q B as an example, the identification of peptide 69GPK#GGPGAP*GAP*80 unambiguously pointed to Lys71 as being glycosylated (Fig. 2); additionally, several MS2 spectra identifying sequence 69GPK#GGPGAP*GAP*GP(KGESGDYK)90# (the glycosylation is located in the partial sequence in parentheses) allowed detection of the y2 ion at m/z 310.176 indicating that the C-terminal lysine was free. We therefore concluded on the consensus sequence 69GPK#GGPGAP*GAP*GPK#GESGDYK90. This reasoning aiming to deduce the most probable modified residues in the three C1q chains from the largest number of converging sequences identified was applied systematically. The conclusions derived from MS2 analyses are collected in Fig. 3. In addition, identification of glycosylated sequences was further confirmed by performing LC-MS/MS analyses of digested CLR and detecting both the precursor and the fragment ions in the Orbitrap cell (OTMS2 analyses) (35). Visual inspection of the fragmentation data allowed us to select MS2 spectra from glycosylated sequences by pointing to multiple losses of 162.0528 mass units. Then, by simply combining the knowledge of the accurate mass of the precursor and of the number of sugar losses, we could match certain experimental spectra to theoretical sequences of C1q obtained by in silico digestion of the three chains (considering cleavages N-terminally of Gly and C-terminally of Lys/Arg residues). The glycosylated sequences thus identified are listed in supplemental Table S1. This highlighted the fact that the knowledge of the precursor mass at 5-ppm accuracy and of its glycosylated nature was often sufficient to unequivocally identify sequences from C1q; this observation supports the identification of glycosylated sequences obtained from the previous ITMS2 data acquired on the LTQ-Orbitrap instrument.
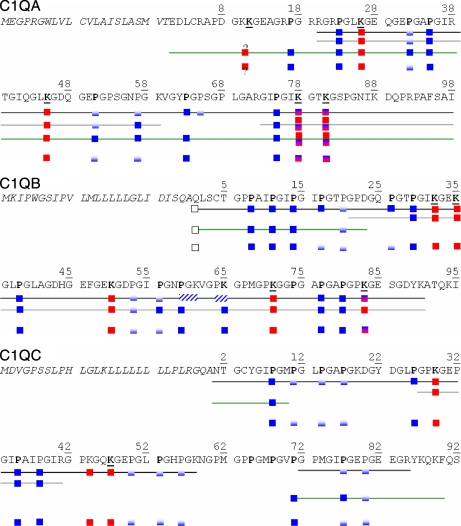
Fig. 3.
Summary of post-translational modifications determined by bottom-up and top-down analyses of CLR and C1q samples. Black line, identifications from MS2 data; gray line, identification from MS3 data; green line, identification from top-down data; no line, merged information on PTMs from bottom-up and top-down analyses. Fully blue squares, residue always detected as hydroxylated; blue/white squares, residue detected either as unmodified or hydroxylated, indicating partial modification; red squares, lysine always detected in a glycosylated form; red/blue squares, lysine detected either as glycosylated or hydroxylated. ? indicates that ambiguity remains as to whether Lys10 or Lys11 of C1q A is glycosylated.
Identification of glycosylated sequences was hampered by the preferential loss of the labile disaccharide moieties upon CID fragmentation. This is usually also observed with peptides phosphorylated on Ser/Thr residues of which a better characterization has been largely described using an additional fragmentation step (MS3) triggered on the MS2 fragment corresponding to the loss of phosphoric acid (36). To more confidently identify glycosylated peptides from the C1q chains, we therefore analyzed digested C1q and CLR samples while specifying the acquisition of MS3 spectra on the MS2 fragments corresponding to the heaviest detected sugar loss from 2+ and 3+ precursor species. MS3 spectra were then interpreted by considering lysines as possibly hydroxylated. The glycosylated sequences identified from these ITMS3 analyses are represented in detail in Fig. 4 and merged in Fig. 3. Interestingly, overlapping peptides containing Lys78 and Lys81 in C1q A mostly exhibited a loss of two disaccharide moieties between MS2 and MS3 but also happened to only lose one such modification. This confirmed that these two lysines were mostly doubly glycosylated but also occurred as a glycosylated/hydroxylated pair as observed previously in MS2 analyses (Fig. 2).
Fig. 4.
Sequences identified from MS3 scans acquired during LC-MS/MS analyses including sugar loss-triggered MS3 scans performed on both C1q and CLR samples. Dotted line, one disaccharide lost between consecutive MS2 and MS3 scans; full line, two disaccharides lost. The color code is the same as in Fig. 2; additionally, orange lines, identifications from the CLR sample. Circled in red, lysines determined to be glycosylated from these MS2/MS3 analyses; circled in black, unmodified lysines.
Outcome of Bottom-up Analyses
The analysis of digested C1q and CLR samples provided significant sequence coverage of the three C1q chains. The majority of proline residues considered to be hydroxyproline in previous reports (Fig. 3, bold) were confirmed to be fully modified (22). Nonetheless, our study revealed a more subtle modification pattern in which some prolines (e.g. Pro31, Pro51, and Pro57 from C1q A and Pro20, Pro53, and Pro56 from C1q B), recorded as either modified or free, were in fact shown to be partially hydroxylated. In addition, C1q A, B, and C were determined to bear (minimally) four, five, and three glycosylations, respectively, on 5-hydroxylysine residues. In particular, three previously unknown glycosylation sites were identified on Lys50 of C1q B and Lys29 and Lys44 of C1q C. Besides, peptides containing Lys78 and Lys81 of C1q A were observed as either doubly or singly glycosylated; similarly, Lys83 from C1q B was mostly observed in a glycosylated form but was also found in a hydroxylated form in one peptide. Nevertheless, several sequence stretches with potential PTMs remained uncovered by this bottom-up approach, such as in the N-terminal end and the globular region of C1q A. Furthermore, these data revealed an additional, unanticipated level of post-translational heterogeneity. To increase the sequence coverage and address the question of the PTM heterogeneity at the whole protein level, we next completed the bottom-up analyses by acquiring top-down data.
Top-down Analysis of C1q and Its Collagen-like Regions
LTQ-Orbitrap instruments have already been demonstrated to allow analysis of intact proteins up to 50 kDa, to determine their molecular mass with ppm accuracy, and to yield sequence information due to iterative MSn level analyses (37). C1q was therefore subjected to top-down characterization at the C1q A, B, and C chain levels (<26 kDa); similarly, the CLR polypeptides were analyzed by iterative MSn experiments (up to MS4). The latter sample was obtained from C1q using pepsin, a nonspecific enzyme that preferentially cleaves on the C-terminal side of aromatic residues (Phe, Trp, and Tyr) as well as Leu, Ala, Glu, and Gln (proteolysis at pH > 2), generating several possible sequences. The challenge in this study was to handle the simultaneous analysis, without prior separation by LC, of a mixture consisting of different protein chains exhibiting variable levels of hydroxylation and glycosylation.
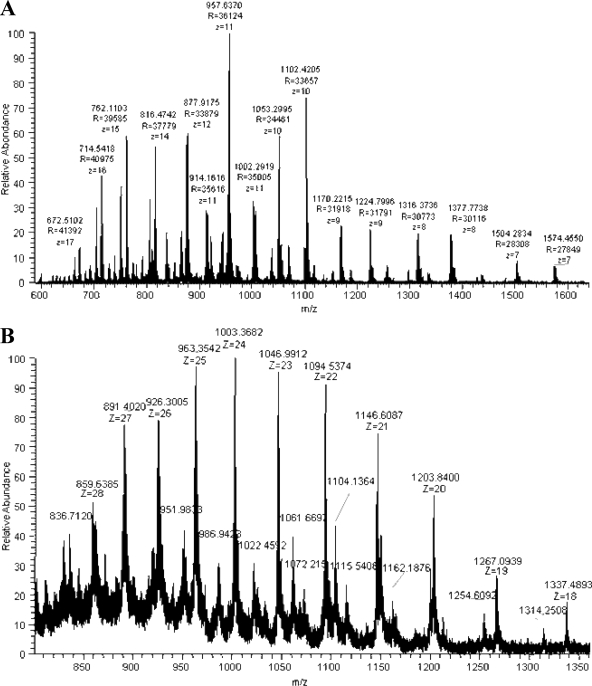
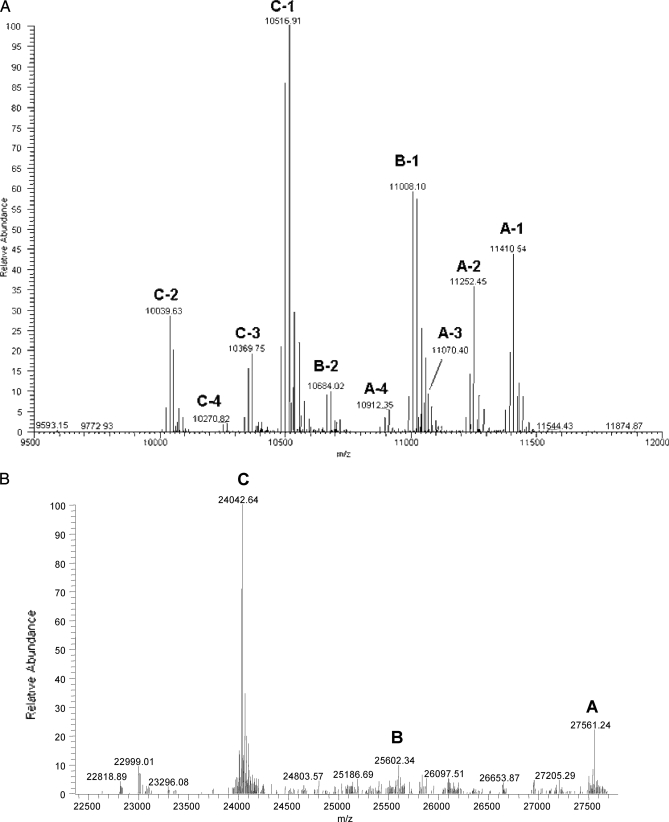
This complexity is illustrated by the mass spectra of reduced CLR and C1q acquired by direct nano-ESI infusion of the proteins on the LTQ-Orbitrap analyzer. Ionic species with charge states ranging from 7 to 17+ and from 18 to 28+ were detected during MS analysis of CLR (Fig. 5A) and reduced C1q (Fig. 5B), respectively. The corresponding deconvoluted spectra are provided in Fig. 6, A and B. The sequences that could be attributed to the signals detected during top-down analysis of C1q and CLR are listed in Table I. More details on the MSn data that allowed establishing these matches are provided in the next section.
Fig. 5.
Mass spectra of reduced CLR and C1q acquired by direct nano-ESI infusion of the proteins on LTQ-Orbitrap instrument. A, MS spectrum obtained on reduced CLR analyzed at a protein concentration of 5 pmol/μl. B, MS spectrum obtained on reduced C1q analyzed at a protein concentration of 5 pmol/μl.
Fig. 6.
Deconvoluted MS spectra obtained by top-down MS analysis of reduced CLR (A) and reduced C1q (B).
Table I. Sequences and PTMs attributed to ionic signals detected during top-down analyses of reduced C1q and CLR chains.
? indicates that the C1q B chain was only detected by a weak signal at a single mass. # represents a Glc-Gal moiety, and * represents a hydroxylation. exp, experimental; th, theoretical.
| Species name | Detected masses | Matched sequence stretch | Modifications | Mass error (exp − th)/th | Variable numbers of hydroxylations |
|---|---|---|---|---|---|
| mass units | ppm | ||||
| CLR | |||||
| A-1 | 11,410.54 | 1–97 from C1q A | 8*, 5# | 8.6 | 6* to 9* |
| A-2 | 11,252.45 | 1–95 from C1q A | 8*, 5# | 6.9 | 6* to 9* |
| A-3 | 11,070.40 | 1–97 from C1q A | 8*, 4# | 5.3 | 6* to 9* |
| A-4 | 10,912.35 | 1–95 from C1q A | 8*, 4# | 7.2 | 6* to 9* |
| B-1 | 11,008.10 | 1–97 from C1q B | 12*, 5#, −NH3 | 5.0 | 11* to 14* |
| B-2 | 10,684.02 | 1–97 from C1q B | 13*, 4#, −NH3 | 7.5 | 11* to 14* |
| C-1 | 10,516.91 | 1–94 from C1q C | 14*, 3# | 6.6 | 12* to 15* |
| C-2 | 10,055.62 | 1–90 from C1q C | 14*, 3# | 0 | 12* to 15* |
| C-3 | 10,369.75 | 1–93 from C1q C | 14*, 3# | −2.9 | 12* to 15* |
| C-4 | 10,270.82 | 1–92 from C1q C | 14*, 3# | 10 | 12* to 15* |
| C1q | |||||
| A | 27,561.24 | C1q A | 8*, 5#, 1 N-glycan | 2.5 | 7* to 9* |
| B | 25,602.34 | C1q B | 12*, 5#, −NH3 | 4.1 | ? |
| C | 24,042.64 | C1q C | 14*, 3# | 2.1 | 12* to 15* |
Four signals, designated A-1 to A-4 in Fig. 6A, were detected for the A chain in the CLR sample. A-1 and A-2 were attributed to sequences 1–97 and 1–95 from C1q A, respectively, decorated with eight hydroxylations and five glycosylations. The lower intensity signals A-3 and A-4 corresponded to the same sequence stretches yet bearing only four glycosylations (−340.10 mass units). These top-down data indicated a heterogeneous level of glycosylation for the A species. Because the residue pair (Lys78, Lys81) was detected by bottom-up analysis as being either doubly glycosylated or hydroxylated/glycosylated, one of these residues must be glycosylated in chains A-1 and A-2 and only hydroxylated in A-3 and A-4. Similarly, two signals, B-1 and B-2, could be attributed to sequence 1–97 from C1q B. Whereas B-1 bears 12 hydroxylations and five glycosylations, the minor fraction B-2 only carries four saccharide moieties but 13 hydroxylations (−324.1 mass units). In B-2, Lys83 could lack glycosylation considering that this residue was found by bottom-up analysis to be either glycosylated or hydroxylated. Finally, four signals were assigned to different sequence stretches of C1q C: polypeptides 1–90, 1–92, 1–93, and 1–94 could be detected; each was decorated with three glycosylations and 14 hydroxylations. The most intense species, C-1, i.e. probably the most abundant CLR C1q C polypeptide, corresponded to the cleavage C-terminally of a Phe residue in agreement with the cleavage specificity of pepsin. Each peak pattern detected for the CLR polypeptides additionally consisted of a series of ionic species by increments of 16 mass units, obviously indicating a variable level of hydroxylation (15.9949 mass units). The ranges of hydroxylation motifs present on each C1q chain are indicated in Table I.
Iterative Fragmentation of CLR and C1q Chains
A Chain Characterization
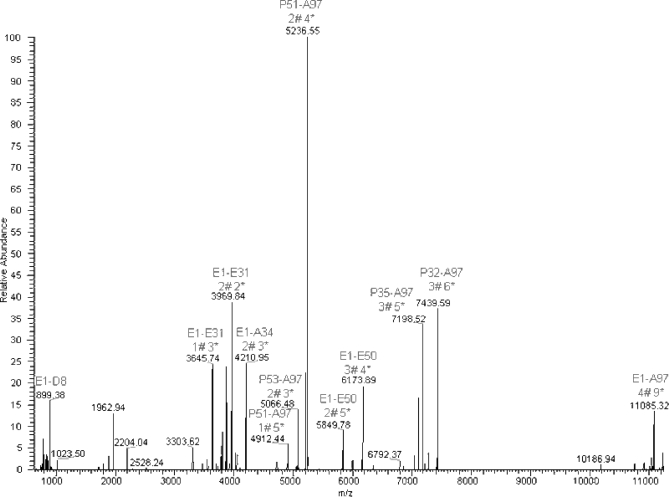
Species A-1 was fragmented by MS/MS, in its 14+ (m/z 817.35) and 16+ (m/z 715.90) charge states, with detection of the resulting fragments in the Orbitrap analyzer. Globally, the same fragment ions were generated in both cases. The deconvoluted MS2 spectrum of the m/z 817.35 ion is provided in Fig. 7. Detailed inspection of MS2 fragments obtained on CLR C1q A and of the matched theoretical sequences allowed us to determine the position of some hydroxylations and glycosylations; this information is represented in Fig. 3. Based on these MS2 data, the calculated monoisotopic mass of 11,410.4432 mass units (species A-1) could be attributed definitely to the CLR sequence 1–97 of C1q A modified with five glycosylations and eight hydroxylations.
Fig. 7.
Deconvoluted and deisotoped MS/MS spectrum obtained from fragmentation of ionic species at about m/z 817.35 (14+) of CLR C1q A. # represents a Glc-Gal moiety, and * represents a hydroxylation. The 33 most intense ions were manually interpreted even though the deconvolution algorithm happened to be unsuccessful in determining the monoisotopic mass (errors of 1 mass unit). The most intense fragment at 5236.55 mass units corresponded to the CLR C1q A Pro51–Ala97 stretch (y47 ion) modified by two glycosylations and four hydroxylations. Its corresponding deglycosylated form was detected at 4912.44 mass units. The complementary sequence Glu1–Glu50 (b50 ion), bearing three glycosylations and four hydroxylations, was detected at 6173.89 mass units. Once again, its deglycosylated form was detected at 5849.78 mass units. The b50 ion revealed glycosylation of either Lys10 or Lys11, a modification that could not be detected by bottom-up analysis. Another couple of fragments was detected at 7439.59 mass units (y65 ion; recalculated monoisotopic mass at 7440.59), corresponding to sequence Pro32–Ala97, bearing three glycosylations and six hydroxylations, and at 3969.84 mass units (b32 ion) corresponding to sequence Glu1–Glu31, bearing two glycosylations and two hydroxylations.
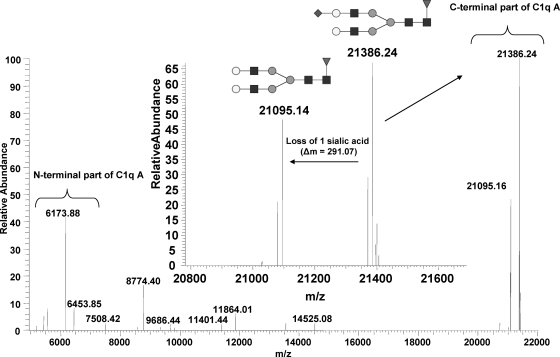
To validate the N-linked glycosylation at Asn124, we acquired an MS/MS spectrum on the ionic cluster centered on m/z 1061.63 (26+), corresponding to the C1q A chain (supplemental Fig. S3). The detected fragments of higher charge states (y173 ion) and highest intensities at m/z 1189.7425 (18+) and m/z 1259.9043 (17+) merged into the same monoisotopic mass of 21,386.24 mass units in the deconvoluted spectrum (Fig. 8). This value matched the C-terminal region of C1q A starting at residue Pro51 and bearing two O-glycosylations, four hydroxylations, and a monosialylated fucosylated biantennary N-glycan with a mass of 2059.74 mass units (theoretical mass, 21,387.28 mass units). In addition, the intense ion signals detected at m/z 1173.6829 (18+) and m/z 1242.6631 (17+), corresponding to a monoisotopic mass of 21,095.16 mass units, were assigned to the same C-terminal sequence, but the terminal sialic acid group was lost (Δm, −291.073 mass units). The detection of sialylated glycan chains is in agreement with a previous analysis indicating that 80% of Asn-linked glycans of C1q contain sialic acid (38). Whereas a minor N-glycan population with two sialic acid residues has been reported (38), we found that these glycans were substituted with a single sialic acid residue. The signal detected at m/z 883.4193 (7+) (b507+ ion) corresponded to the N-terminal sequence 1–50 (6173.879 mass units), observed previously during fragmentation of CLR C1q A and bearing three glycosylations and four hydroxylations, and was complementary to the C-terminal fragment at 21,387.24 mass units. As a whole, five glycosylations and eight hydroxylations, as well as one sialylated fucosylated biantennary N-glycan of 2059.74 mass units, were thus confirmed to be present on C1q A, accounting for a mass of 27,561.29 mass units. The minor fraction possessing four Lys# probably contains one of the two lysines, Lys78 and Lys81, in a hydroxylated form.
Fig. 8.
Deconvoluted and deisotoped MS/MS spectrum acquired on whole C1q A chain. Detection of complementary N- and C-terminal regions from C1q A is indicated. Inset, zoom on the loss of one sialic acid group.
B Chain Characterization
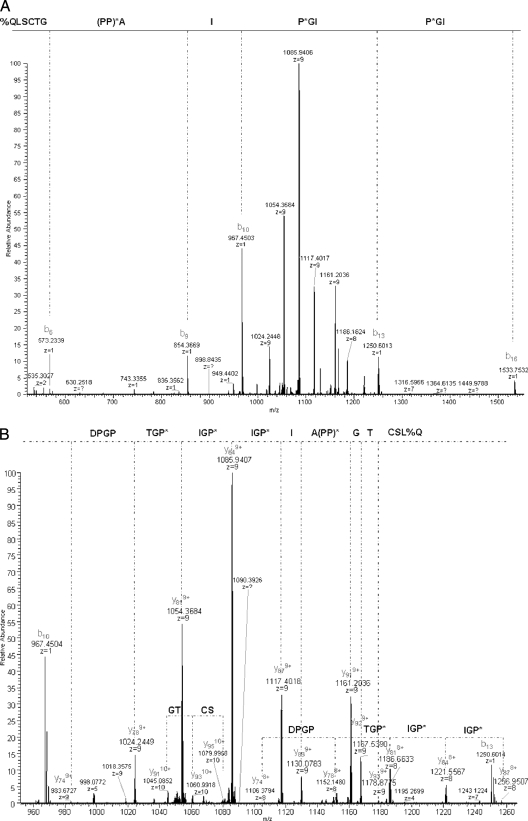
The MS2 spectrum acquired on the ionic species around m/z 1102.41 (10+) is provided in Fig. 9, A and B. As reported above in the bottom-up analysis, preferential fragmentation was observed at the N terminus of proline residues. The sequence 1%QLSCTG(PP)*AIP*GIP*GIP*GTPGPD23 could be confidently determined from the MS2 spectrum, unambiguously identifying CLR C1q B. We could thus precisely match the B-1 polypeptide at mass 11,008.10 mass units, with 5.1-ppm error, to the sequence stretch 1–97 of C1q B (-----KATQKIAF) bearing five glycosylations and 12 hydroxylations and with a loss of NH3 at the protein N terminus due to conversion of the glutamine residue into pyrrolidone carboxylic acid (−17.0265 mass units).
Fig. 9.
MS2 spectrum acquired on ionic species at about m/z 1102.41 (10+) detected during top-down MS analysis of reduced CLR. Fragmentation was performed in the linear IT, and detection of fragments was carried out in the Orbitrap cell.
The ionic species around m/z 1104.02 (10+) (deconvoluted monoisotopic mass of 11,024.10 mass units) was also fragmented and led to a fragmentation pattern similar to that obtained for the ion at m/z 1102.41 (10+) (supplemental Fig. S4). As deduced from the mass difference between both 10+ species, they only differed by one hydroxylation, which could be placed within the sequence stretch 20–23. Indeed, a common y74 fragment was detected at 983.67 (9+), whereas fragment y78 was either seen at 1024.245 (9+) or at 1026.135 (9+), thus definitely localizing the additional hydroxylation within the sequence 20PGPD23. Based on these results, we concluded that CLR C1q B molecules bear 11–14 hydroxylations, indicating a variable level of hydroxylation, as observed for CLR C1q A.
In addition to 8+ and 9+ fragments, the two fragmented polypeptides at m/z 1102.41 (10+) and m/z 1104.02 (10+) produced common 1+ and 2+ fragments at m/z 535.3027 (2+), m/z 854.3668 (b9+ ion), m/z 967.4524 (b10+ ion), m/z 1250.6036 (b13+ ion), and m/z 1533.75 (b16+ ion). These MS2 fragments were then selected to be fragmented in MS3. Table II lists the peptide sequences determined from MS3 scans, and the manually interpreted spectra are provided as supplemental data. Fragmentation of b ions validated the hydroxylation of Pro8, Pro11, and Pro14. Residue Pro7 appeared as possibly hydroxylated from the MS2 fragmentation spectrum obtained on m/z 1250.6036 (1+); however, this modification would be unusual given its location at position 2 within a GXZ triplet. The doubly charged ion at m/z 535.3027 corresponded to the y92+ ion, validating the C-terminal region of the CLR C1q B B-1 species as follows: 89YKATQKIAF97.
Table II. Peptide sequences identified from iterative MSn analyses of reduced CLR C1q B and CLR C1q C by direct infusion on LTQ-Orbitrap instrument.
% indicates loss of NH3 by the N-terminal glutamine. ND, not determined.
| Chain | Precursor ion for FTMS2m/za | Precursor ion for ITMS3 m/z | Identified sequences | Further ITMS4 |
|---|---|---|---|---|
| B | 1102.41 (10+) | 854.3670 (1+) | 1%QLSCTGPP*A | ND |
| 1104.02 (10+) | 967.4504 (1+) | 1%QLSCTGPP*AI | ND | |
| 1250.6014 (1+) | 1%QLSCTGP*PAIP*GI | ND | ||
| 1533.7533 (1+) | 1%QLSCTG(PP)*AIP*GIP*GI | ND | ||
| 535.3027 (2+) | 89YKATQKIAF97 | ND | ||
| C | 1256.71 (8+) | 1103.5312 (2+) | 71(PGP)*MGIP*GEPGEEGRYKQKF90 | ND |
| 736.0229 (3+) | 71P*GPMGIP*GEPGEEGRYKQKF90 | ND | ||
| 819.3975 (2+) | 77P*GEPGEEGRYKQKF90 | ND | ||
| 1258.71 (8+) | 1111.5269 (2+) | 71P*GPMGIP*GEP*GEEGRYKQKF90 | ND | |
| 741.3538 (3+) | 71P*GPMGIP*GEP*GEEGRYKQKF90 | ND | ||
| 827.3938 (2+) | 77P*GEP*GEEGRYKQKF90 | ND | ||
| 1314.23 (8+) | 1050.0091 (2+) | 77P*GEPGEEGRYKQKFQSVF94 | ND | |
| 889.7638 (3+) | 71P*GPMGIP*GEPGEEGRYKQKFQSVF94 | 399.17 | ||
| 1316.38 (8+) | 1058.0063 (2+) | 77P*GEP*GEEGRYKQKFQSVF94 | ND | |
| 895.0957 (3+) | 71P*GPMGIP*GEP*GEEGRYKQKFQSVF94 | 569.33 |
a The m/z value of the isotope of highest intensity in the pattern selected for fragmentation is provided.
C Chain Characterization
Due to the occurrence of Phe/Gln/Tyr amino acids between residues 86 and 94 of the C1q C chain, several polypeptides could be expected to be generated from cleavage of this chain by pepsin. MS/MS fragmentation performed on the ions around m/z 1314.23 (8+) (monoisotopic mass, 10,516.843 mass units) and m/z 1258.71 (8+) (monoisotopic mass, 10,055.616 mass units) allowed identifying the same sequence, 1NTGCYGIP*GM10, based on 7+ fragments (supplemental Figs. S5 and S6, respectively), thus unambiguously attributing this species to a CLR C1q C polypeptide.
The MS/MS spectra acquired on species around m/z 1314.23 (8+), m/z 1316.38 (8+), m/z 1256.71 (8+), and m/z 1258.71 (8+) allowed us to detect peptides at charge states between 1+ and 3+, which were selected for MS3 fragmentation followed by MS4 when possible (supplemental data). Table II contains the sequences thus identified. These iterative fragmentations validated the C-terminal sequences of C-1 (-----KQKFQSVF) and C-2 (-----KQKF). MS3 fragmentation of the species at m/z 1256.71 and 1314.23 (containing three Lys# and 13 Lys* or Pro*) revealed hydroxylation of Pro71 (left unidentified by bottom-up analysis) and of Pro77. MS3 fragmentation of the species at m/z 1258.71 and 1316.38 (containing three Lys# and 14 Lys* or Pro*) highlighted an additional hydroxylation on Pro80. Very interestingly, MS2 fragmentation of the species at m/z 1316.38 (supplemental Fig. S7) unambiguously revealed the heterogeneous hydroxylation of biomolecules corresponding to that mass. Indeed, this ion fragmented into two y182+ ions at m/z 1058.51 (corresponding to 77P*GEP*GEEGRYKQKFQSVF94) and m/z 1050.01 (corresponding to 77(PGEP)*GEEGRYKQKFQSVF94) as well as two y242+ ions containing three (71P*GPMGIP*GEP*GEEGRYKQKFQSVF94) or only two hydroxylated proline residues. The detection of these two pairs of y ions, with one or two hydroxylations in the sequence 77PGEP80, indicated the presence of counterbalancing proline residues being either free or hydroxylated within the 1–70 stretch of the fragmented CLR C1q C chain.
Outcome of Top-down Analysis
The top-down analysis of the collagen-like regions of C1q indicated two sites of pepsin cleavage in C1q A (after Phe95 and Ala97), one site in C1q B (after Phe97), and four sites in C1q C (after Phe90, Ser92, Val93, and Phe94). Top-down analyses of CLR and intact reduced C1q allowed identification of the PTMs decorating each chain (summarized in Fig. 3), some of which (e.g. the glycosylation of Lys10/Lys11 in C1q A) had not been detected by bottom-up analyses. The complexity of the hydroxylation patterns was further illustrated: in particular, fragmentation of CLR C1q C at m/z 1316.38 (supplemental Fig. S7) showed that a biomolecule at a given mass can actually exhibit different combinations of hydroxylation sites. As a whole, the signals detected on intact CLR chains indicated that C1q A contains five Lys# and 6–9 hydroxylations (for biomolecules at detectable levels), C1q B bears five Lys# and 11–14 hydroxylations, and C1q C contains three Lys# and 12–15 hydroxylations.
DISCUSSION
We report here for the first time the proteomics analysis of the human complement protein C1q, which is composed of 18 chains from three different polypeptides and exhibits a unique structural organization comprising a collagen-like domain with a high level of post-translational modifications: hydroxylations on Lys/Pro residues, Glc-Gal disaccharides on Lys residues, and a branched N-linked sugar moiety on the globular moiety of the A chains.
Clearly, identifying the lysine residues modified by Glc-Gal motifs was not trivial. It is worth mentioning that unlike for the common N-glycosylation no known enzyme allows removal of these disaccharides and that attempts to chemically eliminate these modifications led to disruption of the polypeptide chains (data not shown). The fact that the loss of sugar cycles (162.05 mass units) was favored during MS/MS, together with the inefficient fragmentation of the peptide backbone, rendered sequence determination by the Mascot software difficult. Nonetheless, we could check from OTMS2 analyses that the high mass accuracy (<5 ppm) of the LTQ-Orbitrap provided confident identification of glycosylated sequences from chains A, B, and C. Additionally, we performed LC-MS/MS analyses with MS3 scans triggered on the heaviest neutral loss of sugar moieties (multiples of 324.1 mass units). This method appeared to be efficient in producing pairs of MS2/MS3 spectra whose precursors differed by the total number of glycosylation motifs present on the initial peptide (for usually 1# and 2#). The obtained MS3 scans allowed reading a sequence in amino acids with much more confidence than the corresponding MS2 scans. Such a method programming MS3 scans triggered on the heaviest sugar loss would be of general interest when studying proteins that contain a collagen-like domain exhibiting glycosylated lysine residues. This method may be more generally applicable to the study of labile oligosaccharide-type modifications. We combined bottom-up and top-down analyses of the C1q and CLR samples to obtain complementary information on the PTMs decorating the three C1q chains. Obtaining a really exhaustive characterization of the variable modification level of the individual chains would have required separation of the intact proteins by LC to systematically acquire MSn information from each species by off-line infusion (39). Nevertheless, our combined bottom-up and top-down data were sufficient to identify Pro/Lys residues that are fully hydroxylated (or glycosylated) in all C1q molecules (given their systematic detection in a modified form in LC-MS/MS analyses) and others that are either unmodified (or solely hydroxylated) in different biomolecules. A variable level of hydroxylation at specific Pro/Lys residues was described previously for other collagen-containing proteins (40–43). In our case, top-down analyses revealed that proteins with the same sequence can bear between N and N + 4 hydroxylation motifs (with N being 6, 11, and 12 for the C1q A, B, and C chains, respectively). They also showed that proteins with the same sequence and mass can exhibit different distributions of hydroxylation sites, thus highlighting a further level of PTM pattern complexity.
The determination of the primary structure of C1q initially carried out by proteolytic digestion and Edman sequencing yielded the sequences of the C1q A and B chains and that of the 94 N-terminal residues of the C1q C chain (22) and resulted in incomplete identification of the PTMs in the CLR moieties of the chains. The proteomics study reported here allowed us to cover the sequences of the whole C1q A, B, and C chains and to verify the cDNA-derived sequences, confirming that the few discrepancies noticed with the initial protein-derived sequence do not arise from polymorphism. In addition, we made the most of the analytical potentialities of the LTQ-Orbitrap instrument (acquisition of MS2 and sugar loss-based MS3 scans in bottom-up analyses and of iterative MSn fragmentations on intact proteins in top-down analyses) to confidently and comprehensively identify glycosylated residues. The large majority of the identified Glc-Gal-bearing hydroxylysines appear to be fully modified (Lys10 or Lys11, Lys26, and Lys45 in C1q A; Lys32, Lys35, Lys50, and Lys71 in C1q B; and Lys29, Lys44, and Lys47 in C1q C), yet a few were also detected as being either glycosylated or hydroxylated (Lys78 or Lys81 in C1q A and Lys83 in C1q B). Finally, only three lysine residues within the CLR have been systematically and unambiguously identified in a non-modified form, i.e. residues Lys59 in C1q A, Lys61 in C1q B, and Lys58 in C1q C. This identification is consistent with the early analyses reported by Reid (22) and represents highly meaningful information with respect to the assembly of the C1 complex.
Given the important function of the C1 complex in the immune system and its role in the triggering of complement-mediated inflammation, the understanding of its assembly is key information that, in addition, can be extended to the collectins mannan-binding lectin (MBL) and ficolins, two other important classes of pattern recognition molecules. These collectins, which trigger complement through the lectin pathway, share with C1q the ability to associate in a homologous manner with their partner proteases, MBL-associated serine proteases through their collagen domain (44). Point mutations of recombinant MBL and ficolins have revealed the essential role of a single unmodified lysine residue in their collagen domains for the association with the MBL-associated serine proteases (45, 46). Unlike MBL and the ficolins, which are assembled from a single polypeptide chain and thus have been produced recombinantly, so far C1q could not be studied by site-directed mutagenesis to identify the CLR residues involved in the assembly of the C1 complex. Nevertheless, point mutants of C1r and C1s have been produced, providing evidence for the essential role of Asp and Glu residues within the C1r/C1s CUB modules in the interaction of the C1r/C1s tetramer with C1q likely through ionic bonds. Given the homologous assembly of the collectins MBL/ficolins and C1q, we postulate that the free residues C1q A-Lys59, C1q B-Lys61, and C1q C-Lys58 are fully accessible and available for interaction with acidic residues contributed by the C1r and C1s CUB modules. In support of this proposal, the location of these three unmodified lysine residues about halfway along the CLR is in agreement with the recently proposed refined model of C1 assembly in which the C1r/C1s tetramer is positioned inside the cone defined by the C1q stems (6). According to this model, both C1r/C1s CUB1-EGF-CUB2 heterodimers provide six binding sites distributed radially to make contacts with each of the six C1q collagen-like stems. As they are located halfway along the CLR, these unmodified lysine residues are thus in an appropriate position for making individual contacts with the CUB modules and mediating effective interaction with the C1r/C1s tetramer (Fig. 10).
Fig. 10.
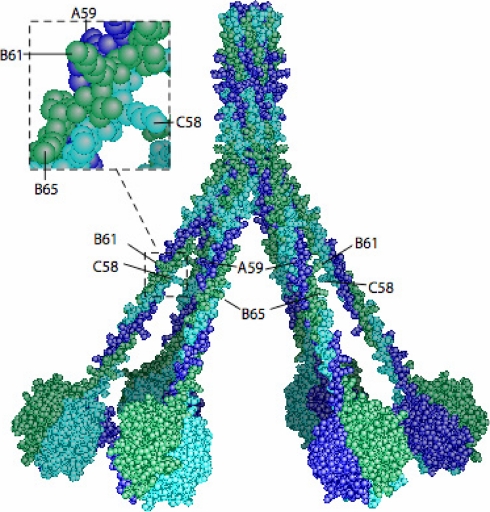
Three-dimensional model of human C1q highlighting positions of unmodified lysine residues. The model (2) was assembled as described previously (50). The C1q chains are colored dark blue (A), green (B), and light blue (C). The positions of the side chains of Lys59 A, Lys61 B, and Lys58 C (unmodified) and of Lys65 B (hydroxylated) are shown.
The MS characterization reported here is therefore fully consistent with the proposed model for the C1q-C1r/C1s tetramer assembly (6), accounting for the architecture and function of the C1 complex. Recent data reveal that the role of C1 extends beyond pathogen recognition to include implications in autoimmune diseases, ischemia-reperfusion injury, organ graft rejection, and neurodegeneration (1, 47). It is therefore generally considered that the early inhibition of the complement cascade at the C1 level would provide a therapeutic benefit (48, 49). In this context, the experimental data reported here on the interface between C1q and its C1r/C1s protease partners provide useful clues for the design of inhibitory molecules aimed at targeting the C1 complex.
Supplementary Material
Acknowledgments
We gratefully acknowledge the CNRS, Genopole-France, Institut National de la Recherche Agronomique, and Région Ile de France, in particular for funding of the LTQ-Orbitrap, and warmly thank C. Gaboriaud for preparing the C1q model displayed in Fig. 10.
* This work was supported by the CNRS, Genopole-France, Institut National de la Recherche Agronomique, and Région Ile de France.
 This article contains supplemental Figs. S1–S7, Table S1, and other data.
This article contains supplemental Figs. S1–S7, Table S1, and other data.
1 The abbreviations used are:
- CLR
- collagen-like region
- EGF
- epidermal growth factor
- PTM
- post-translational modification
- IAA
- iodoacetamide
- MMTS
- methyl methane thiosulfonate
- QqTOF
- quadrupole/quadrupole time-of-flight
- IT
- ion trap
- OT
- Orbitrap
- MBL
- mannan-binding lectin
- LTQ
- linear quadrupole ion trap
- CUB
- Complement C1r/C1s-Uegf-Bone morphogenetic protein 1.
REFERENCES
- 1.Lu J. H., Teh B. K., Wang L., Wang Y. N., Tan Y. S., Lai M. C., Reid K. B. (2008) The classical and regulatory functions of C1q in immunity and autoimmunity. Cell. Mol. Immunol 5, 9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaboriaud C., Thielens N. M., Gregory L. A., Rossi V., Fontecilla-Camps J. C., Arlaud G. J. (2004) Structure and activation of the C1 complex of complement: unraveling the puzzle. Trends Immunol 25, 368–373 [DOI] [PubMed] [Google Scholar]
- 3.Busby T. F., Ingham K. C. (1990) Amino-terminal calcium-binding domain of human complement C1s mediates the interaction of C1r with C1q. Biochemistry 29, 4613–4618 [DOI] [PubMed] [Google Scholar]
- 4.Illy C., Thielens N. M., Arlaud G. J. (1993) Chemical characterization and location of ionic interactions involved in the assembly of the C1 complex of human complement. J. Protein Chem 12, 771–781 [DOI] [PubMed] [Google Scholar]
- 5.Thielens N. M., Illy C., Bally I. M., Arlaud G. J. (1994) Activation of human complement serine proteinase C1r is down-regulated by a Ca2+-dependent intramolecular control that is released in the C1 complex through a signal transmitted by C1q. Biochem. J 301, 509–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bally I., Rossi V., Lunardi T., Thielens N. M., Gaboriaud C., Arlaud G. J. (2009) Identification of the C1q-binding sites of human C1r and C1s: a refined three-dimensional model of the C1 complex of complement. J. Biol. Chem 284, 19340–19348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper N. R. (1985) The classical complement pathway: activation and regulation of the first complement component. Adv. Immunol 37, 151–216 [DOI] [PubMed] [Google Scholar]
- 8.Kishore U., Reid K. B. (2000) C1q: structure, function, and receptors. Immunopharmacology 49, 159–170 [DOI] [PubMed] [Google Scholar]
- 9.Reid K. B., Sim R. B., Faiers A. P. (1977) Inhibition of reconstitution of hemolytic activity of the first component of human complement by a pepsin-derived fragment of subcomponent C1q. Biochem. J 161, 239–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsiung L., Dodds A. W., Mason D. W., Reid K. B. (1988) A monoclonal antibody to Clq which appears to interact with Clr2Cls2-binding site. FEBS Lett 229, 21–24 [DOI] [PubMed] [Google Scholar]
- 11.Siegel R. C., Schumaker V. N. (1983) Measurement of the association constants of the complexes formed between intact Clq or pepsin-treated Clq stalks and the unactivated or activated Clr2cls2 tetramers. Mol. Immunol 20, 53–66 [DOI] [PubMed] [Google Scholar]
- 12.Poon P. H., Schumaker V. N., Phillips M. L., Strang C. J. (1983) Conformation and restricted segmental flexibility of C1, the first component of human complement. J. Mol. Biol 168, 563–577 [DOI] [PubMed] [Google Scholar]
- 13.Strang C. J., Siegel R. C., Phillips M. L., Poon P. H., Schumaker V. N. (1982) Ultrastructure of the first component of human complement: electron microscopy of the crosslinked complex. Proc. Natl. Acad. Sci. U.S.A 79, 586–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tschopp J., Villiger W., Fuchs H., Kilchherr E., Engel J. (1980) Assembly of subcomponents C1r and C1s of first component of complement: electron microscopic and ultracentrifugal studies. Proc. Natl. Acad. Sci. U.S.A 77, 7014–7018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyd J., Burton D. R., Perkins S. J., Villiers C. L., Dwek R. A., Arlaud G. J. (1983) Neutron scattering studies of the isolated C1r2C1s2 subunit of first component of human complement in solution. Proc. Natl. Acad. Sci. U.S.A 80, 3769–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perkins S. J., Villiers C. L., Arlaud G. J., Boyd J., Burton D. R., Colomb M. G., Dwek R. A. (1984) Neutron-scattering studies of subcomponent Clq of first component Cl of human complement and its association with subunit Clr2cls2 within Cl. J. Mol. Biol 179, 547–557 [DOI] [PubMed] [Google Scholar]
- 17.Gregory L. A., Thielens N. M., Arlaud G. J., Fontecilla-Camps J. C., Gaboriaud C. (2003) X-ray structure of the Ca2+-binding interaction domain of C1s. J. Biol. Chem 278, 32157–32164 [DOI] [PubMed] [Google Scholar]
- 18.Tissot B., Montdargent B., Chevolot L., Varenne A., Descroix S., Gareil P., Daniel R. (2003) Interaction of fucoidan with the proteins of the complement classical pathway. Biochim. Biophys. Acta 1651, 5–16 [DOI] [PubMed] [Google Scholar]
- 19.Shinkai H., Yonemasu K. (1979) Hydroxylysine-linked glycosides of human complement subcomponent C1q and various collagens. Biochem. J 177, 847–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reid K. B. (1977) Amino acid sequence of the N-terminal forty-two amino acid residues of the C chain of subcomponent C1q of the first component of human complement. Biochem. J 161, 247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reid K. B. (1974) Collagen-like amino acid sequence in a polypeptide chain of human Clq (a subcomponent of first component of complement). Biochem. J 141, 189–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reid K. B. (1979) Complete amino acid sequences of the three collagen-like regions present in subcomponent C1q of the first component of human complement. Biochem. J 179, 367–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reid K. B., Gagnon J., Frampton J. (1982) Completion of the amino acid sequences of the A and B chains of subcomponent C1q of the first component of human complement. Biochem. J 203, 559–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reid K. B., Porter R. R. (1976) Subunit composition and structure of subcomponent Clq of the first component of human complement. Biochem. J 155, 19–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reid K. B., Thompson E. O. (1978) Amino acid sequence of the N-terminal 108 amino acid residues of the B chain of subcomponent C1q of the first component of human complement. Biochem. J 173, 863–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tissot B., Gonnet F., Iborra A., Berthou C., Thielens N., Arlaud G. J., Daniel R. (2005) Mass spectrometry analysis of the oligomeric C1q protein reveals the B chain as the target of trypsin cleavage and interaction with fucoidan. Biochemistry 44, 2602–2609 [DOI] [PubMed] [Google Scholar]
- 27.Arlaud G. J., Sim R. B., Duplaa A. M., Colomb M. G. (1979) Differential elution of C1q, C1r and C1s from human C1 bound to immune aggregates: use in the rapid purification of C1 subcomponents. Mol. Immunol 16, 445–450 [DOI] [PubMed] [Google Scholar]
- 28.Sasaki T., Yonemasu K. (1983) Chemical studies of the isolated collagen-like and globular fragments of complement component C1q: comparative studies on bovine and human C1q. Biochim. Biophys. Acta 742, 122–128 [DOI] [PubMed] [Google Scholar]
- 29.Tacnet-Delorme P., Chevallier S., Arlaud G. J. (2001) beta-Amyloid fibrils activate the C1 complex of complement under physiological conditions: evidence for a binding site for A beta on the C1q globular regions. J. Immunol 167, 6374–6381 [DOI] [PubMed] [Google Scholar]
- 30.Boja E. S., Fales H. M. (2001) Overalkylation of a protein digest with iodoacetamide. Anal. Chem 73, 3576–3582 [DOI] [PubMed] [Google Scholar]
- 31.Kapp E. A., Schütz F., Reid G. E., Eddes J. S., Moritz R. L., O'Hair R. A., Speed T. P., Simpson R. J. (2003) Mining a tandem mass spectrometry database to determine the trends and global factors influencing peptide fragmentation. Anal. Chem 75, 6251–6264 [DOI] [PubMed] [Google Scholar]
- 32.Tabb D. L., Smith L. L., Breci L. A., Wysocki V. H., Lin D., Yates J. R., 3rd (2003) Statistical characterization of ion trap tandem mass spectra from doubly charged tryptic peptides. Anal. Chem 75, 1155–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu T., Qian W. J., Gritsenko M. A., Camp D. G., 2nd, Monroe M. E., Moore R. J., Smith R. D. (2005) Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J. Proteome Res 4, 2070–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sellar G. C., Blake D. J., Reid K. B. M. (1991) Characterization and organization of the genes encoding the A-, B- and C-chains of human complement subcomponent C1q. The complete derived amino acid sequence of human C1q. Biochem. J 274, 481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scherl A., Shaffer S. A., Taylor G. K., Hernandez P., Appel R. D., Binz P. A., Goodlett D. R. (2008) On the benefits of acquiring peptide fragment ions at high measured mass accuracy. J. Am. Soc. Mass Spectrom 19, 891–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ulintz P. J., Bodenmiller B., Andrews P. C., Aebersold R., Nesvizhskii A. I. (2008) Investigating MS2/MS3 matching statistics: a model for coupling consecutive stage mass spectrometry data for increased peptide identification confidence. Mol. Cell. Proteomics 7, 71–87 [DOI] [PubMed] [Google Scholar]
- 37.Macek B., Waanders L. F., Olsen J. V., Mann M. (2006) Top-down protein sequencing and MS3 on a hybrid linear quadrupole ion trap-orbitrap mass spectrometer. Mol. Cell. Proteomics 5, 949–958 [DOI] [PubMed] [Google Scholar]
- 38.Mizuochi T., Yonemasu K., Yamashita K., Kobata A. (1978) The asparagines-linked sugar chains of subcomponent C1q of the first component of human complement. J. Biol. Chem 253, 7404–7409 [PubMed] [Google Scholar]
- 39.Wu S., Lourette N. M., Toliæ N., Zhao R., Robinson E. W., Tolmachev A. V., Smith R. D., Pasa-Toliæ L. (2009) An integrated top-down and bottom-up strategy for broadly characterizing protein isoforms and modifications. J. Proteome Res 8, 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bos K. J., Rucklidge G. J., Dunbar B., Robins S. P. (1999) Primary structure of the helical domain of porcine collagen X. Matrix Biol 18, 149–153 [DOI] [PubMed] [Google Scholar]
- 41.Rothmann A. B., Mortensen H. D., Holmskov U., Højrup P. (1997) Structural characterization of bovine collectin-43. Eur. J. Biochem 243, 630–635 [DOI] [PubMed] [Google Scholar]
- 42.Leth-Larsen R., Holmskov U., Højrup P. (1999) Structural characterization of human and bovine lung surfactant protein D. Biochem. J 343, 645–652 [PMC free article] [PubMed] [Google Scholar]
- 43.Richards A. A., Stephens T., Charlton H. K., Jones A., Macdonald G. A., Prins J. B., Whitehead J. P. (2006) Adiponectin multimerization is dependent on conserved lysines in the collagenous domain: evidence for regulation of multimerization by alterations in posttranslational modifications. Mol. Endocrinol 20, 1673–1687 [DOI] [PubMed] [Google Scholar]
- 44.Gaboriaud C., Teillet F., Gregory L. A., Thielens N. M., Arlaud G. J. (2007) Assembly of C1 and the MBL- and ficolin-MASP complexes: structural insights. Immunobiology 212, 279–288 [DOI] [PubMed] [Google Scholar]
- 45.Teillet F., Lacroix M., Thiel S., Weilguny D., Agger T., Arlaud G. J., Thielens N. M. (2007) Identification of the site of human mannan-binding lectin involved in the interaction with its partner serine proteases: the essential role of Lys55. J. Immunol 178, 5710–5756 [DOI] [PubMed] [Google Scholar]
- 46.Lacroix M., Dumestre-Pérard C., Schoehn G., Houen G., Cesbron J. Y., Arlaud G. J., Thielens N. M. (2009) Residue Lys57 in the collagen-like region of human L-ficolin and its counterpart Lys47 in H-ficolin play a key role in the interaction with the mannan-binding lectin-associated serine proteases and the collectin receptor calreticulin. J. Immunol 182, 456–465 [DOI] [PubMed] [Google Scholar]
- 47.Bohlson S. S., Fraser D. A., Tenner A. J. (2007) Complement proteins C1q and MBL are pattern recognition molecules that signal immediate and long-term protective immune functions. Mol. Immunol 44, 33–43 [DOI] [PubMed] [Google Scholar]
- 48.Beinrohr L., Dobó J., Závodszky P., Gál P. (2008) C1, MBL-MASPs and C1-inhibitor: novel approaches for targeting complement-mediated inflammation. Trends Mol. Med 14, 511–521 [DOI] [PubMed] [Google Scholar]
- 49.Sjöberg A. P., Trouw L. A., Blom A. M. (2009) Complement activation and inhibition: a delicate balance. Trends Immunol 30, 83–90 [DOI] [PubMed] [Google Scholar]
- 50.Gaboriaud C., Juanhuix J., Gruez A., Lacroix M., Darnault C., Pignol D., Verger D., Fontecilla-Camps J. C., Arlaud G. J. (2003) The crystal structure of the globular head of complement protein C1q provides a basis for its versatile recognition properties. J. Biol. Chem 278, 46974–46982 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.