Abstract
Purpose
Identifying sources of variation in expression microarray data and the effect of variance in gene expression measurements on complex predictive and diagnostic models is essential when translating microarray-based experimental approaches into clinical assays. The technical reproducibility of microarray platforms is well established. Here, we investigate the additional impact of intratumor heterogeneity, a largely unstudied component of variance, on the performance of several microarray-based assays in breast cancer.
Patients and Methods
Genome-wide expression profiling was performed on 50 core needle biopsies from 18 breast cancer patients using Affymetrix GeneChip Human Genome U133 Plus 2.0 arrays. Global profiles of expression were characterized using unsupervised clustering methods and variance components models. Array-based measures of estrogen receptor (ER) and progesterone receptor (PR) status were compared with immunohistochemistry. The precision of genomic predictors of ER pathway status, recurrence risk, and sensitivity to chemotherapeutics was evaluated by interclass correlation.
Results
Global patterns of gene expression demonstrated that intratumor variation was substantially less than the total variation observed across the patient population. Nevertheless, a fraction of genes exhibited significant intratumor heterogeneity in expression. A high degree of reproducibility was observed in single-gene predictors of ER (intraclass correlation coefficient [ICC] = 0.94) and PR expression (ICC = 0.90), and in a multigene predictor of ER pathway activation (ICC = 0.98) with high concordance with immunohistochemistry. Substantial agreement was also observed for multigene signatures of cancer recurrence (ICC = 0.71) and chemotherapeutic sensitivity (ICC = 0.72 and 0.64).
Conclusion
Intratumor heterogeneity, although present at the level of individual gene expression, does not preclude precise microarray-based predictions of tumor behavior or clinical outcome in breast cancer patients.
INTRODUCTION
Since its inception, microarray technology has provided a powerful tool to the research community because of its ability to simultaneously measure the expression of tens of thousands of genes. In particular, breast cancer research has seen great benefits from this technology, with many studies describing multigene expression patterns associated with diagnostic and prognostic subclasses among otherwise indistinguishable tumors.1–3 These studies have also established the ability to predict a cancer patient's treatment response based on gene expression patterns.4–7 With the clear potential of microarray-based assays to guide clinical decisions, translating these assays to the clinical laboratory is imperative.
Clinical translation requires an understanding of factors that influence the precision and accuracy of microarray-based assays. Chief among these factors is the variability of gene expression measurements, which can be divided into technical (intrinsic to the platform) and preanalytic (intrinsic to the sample) sources of variation. The Affymetrix GeneChip Human Genome U133 Plus 2.0 array platform (Affymetrix, Santa Clara, CA), investigated herein, has a high degree of reproducibility and thus little technical variance, as established by several groups including the MicroArray Quality Control (MAQC) project.8–12 Variance intrinsic to a sample is more difficult to control, particularly for solid tumor specimens where intratumor heterogeneity could result in significant sampling bias. Small sampling, such as needle core biopsies, can yield samples from the same tumor with different histologic and biologic features. The effect of tumor heterogeneity on microarray-based assays has been evaluated in some cancers, although breast cancer is surprisingly understudied in this regard.13–17
Here, we investigate the impact of tumor heterogeneity on several microarray-based predictors of biologic behavior and clinical outcome in breast cancer patients. Multiple core biopsies from individual patients were evaluated by routine histology and tested using single-gene measurements and multigene signatures that would be integral to the routine care of the breast cancer patient, including estrogen receptor (ER) status, progesterone receptor (PR) status, risk of cancer recurrence, and chemotherapeutic sensitivity. Precision for each of these predictors was measured and evaluated in the context of performance expectations for clinical assays.
PATIENTS AND METHODS
Tumor Sample Collection and Histologic Analysis
Following patient consent, samples were obtained from breast cancer excisions as part of a Duke University Health System (DUHS) institutional review board–approved tissue banking and research protocol for the Duke University Medical Center (DUMC) Breast Cancer Specialized Program of Research Excellence (SPORE). Immediately after surgical excision, lumpectomy specimens were sampled by 14-gauge needle core biopsy using an imaging device, as previously described.18 The core biopsies were embedded in Tissue-Tek OCT (Qiagen, Valencia, CA) and frozen in liquid nitrogen. One 5-μm frozen section was prepared from each sample, stained with hematoxylin and eosin, and evaluated by an expert breast pathologist (J.G.). Routine pathologic evaluation of the corresponding clinical specimens by the DUHS laboratories included determination of ER and PR status by immunohistochemistry (IHC). Samples with an Allred score of 0 or 2 were classified as negative. Human epidermal growth factor receptor 2 (HER2) status was determined by IHC and fluorescent in situ hybridization. This study included only HER2-negative patients with at least two frozen core biopsies containing neoplastic cells.
RNA Purification and Microarray Hybridization
Total RNA was extracted using a kit-based method (RNeasy, Qiagen, Valencia, CA). RNA quality was assessed using an Agilent bioanalyzer (Agilent Technologies, Santa Clara, CA). Hybridization targets were prepared from 2 μg of total RNA and hybridized according to standard Affymetrix protocols using Affymetrix GeneChip Human Genome U133 Plus 2.0 arrays. Arrays were scanned on the Affymetrix GeneChip scanner and probe set expression values, percent present, and 3′/5′ probe set ratios for actin and glyceraldehyde-3-phosphate dehydrogenase were calculated using the Affymetrix Microarray Analysis Suite v5.0.
Microarray Preprocessing and Statistical Analysis
Expression estimates for the 50 DUMC tumor biopsies and for the publicly available breast cancer microarray data sets (GEO# GSE349419 and GSE145620) were obtained by robust multiarray average21 and log2 transformed. The ratio of intratumor variance to total variance among the DUMC biopsy specimens was calculated for all probe sets. The total variance was calculated as the sum of squared differences from the mean expression for all samples. Intratumor variance was calculated as the sum of squared differences from the mean expression within individual tumors.14 Global patterns of expression were evaluated by principal component analysis (PCA) and hierarchical clustering using average linkage of the Pearson correlation coefficient.
Predictors of ER pathway activation and breast cancer recurrence were generated from published breast cancer microarray data sets (GEO# GSE349419 and GSE1456,20 respectively), using established methodologies.22,23 Briefly, tests of differential expression were used to select gene sets strongly correlated to phenotype. Expression values were summarized by the top principal components and fitted to a Bayesian probit regression model. Predicted probabilities were generated for these predictors, and for previously identified signatures of sensitivity to chemotherapeutic agents doxorubicin and docetaxel.23 Binary classifications were made using thresholds defined a priori for each signature.
The precision of all signatures was evaluated using fixed-effects analysis of variance (ANOVA) models and the intraclass correlation coefficient (ICC [1,1]).24 ICC values ranging from 0 to 1 have been characterized by Landis and Koch25 as indicating moderate (0.41-0.60), substantial (0.61-0.80), and almost perfect agreement (0.81-1.00). Accuracy of the single-gene ER and PR predictors and the multigene ER pathway predictor are reported with 95% CIs. The influence of clinical and technical covariates on precision and accuracy are assessed using analysis of covariance (ANCOVA) models. All microarray preprocessing and analysis were performed in R/Bioconductor and Matlab (The Mathworks, Natick, MA) with graphics generated using Graphpad Prism (GraphPad Software, La Jolla, CA) and Cluster/Treeview software (Eisen Lab, Berkeley, CA).26
RESULTS
Characterization of Morphologic Heterogeneity in Discrete Samplings of Individual Breast Tumors
Fifty samples were obtained from 18 patients as part of a tissue banking and research protocol for the DUMC Breast Cancer SPORE. One patient had four replicate samples, 12 patients had triplicate samples, and five patients had duplicate samples (Table 1). Analysis was performed on frozen sections stained with hematoxylin and eosin for each of these core biopsies. The sample set contained a mixture of ER- and PR-positive and ER- and PR-negative cases (11 ER-positive/PR-positive, two ER-positive/PR-negative, one ER-negative/PR-positive, four ER-negative/PR-negative) using the diagnostic core biopsy IHC as the standard. Invasive carcinoma was present in 49 samples, while one biopsy contained ductal carcinoma in situ only. One set of biopsies (patient B) contained lobular carcinoma; the remaining cases were ductal-type carcinomas. Invasive tumor cellularity varied from 10% to 90%. RNA and microarray quality control metrics are provided in Table 1.
Table 1.
Histology, RNA, and Microarray Quality Control Measures
| Patient Sample | Histology QC |
RNA QC |
Microarray QC |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Histology | Size | Grade | ER | PR | HER2 | CA (%) | Other Features | Concentration (μg/μL) | 260/280 | 260/230 | % P | Actin | GAPDH | |
| A1 | Ductal | 2.3 | 3 | + | + | − | 90 | 0.339 | 2.08 | 2.03 | 54.2 | 1.49 | 1.44 | |
| A2 | 80 | 0.097 | 2.09 | 2.37 | 49.7 | 2.09 | 2.09 | |||||||
| A3 | 65 | 0.162 | 2.09 | 2.08 | 53.9 | 2.04 | 1.77 | |||||||
| B1 | Lobular | 3.6 | 2 | + | + | − | 50 | 0.068 | 2.02 | 2.24 | 53.4 | 1.34 | 1.26 | |
| B2 | 85 | 0.08 | 2.09 | 2.44 | 50.6 | 1.21 | 1.12 | |||||||
| B3 | 70 | 0.068 | 2.11 | 1.86 | 53 | 1.81 | 1.36 | |||||||
| C1 | Ductal | 2 | 2 | + | + | − | 10 | Biopsy site | 0.042 | 1.98 | 2.39 | 54.3 | 1.49 | 1.14 |
| C2 | 80 | Biopsy site | 0.093 | 2.06 | 1.63 | 56 | 1.37 | 1.01 | ||||||
| C3 | 40 | Biopsy site | 0.07 | 2.04 | 0.84 | 57 | 1.24 | 1 | ||||||
| D1 | Ductal | 2.8 | 2 | + | + | − | 30 | 0.158 | 2.08 | 2.27 | 55.3 | 1.54 | 1.31 | |
| D2 | 50 | 0.147 | 2.08 | 1.59 | 50.4 | 1.35 | 1.28 | |||||||
| D3 | 70 | 0.182 | 2.1 | 2.22 | 52 | 1.21 | 1.23 | |||||||
| E1 | Ductal | 2.1 | 3 | − | − | − | 25 | 0.269 | 2.08 | 1.82 | 52.7 | 1.17 | 1.03 | |
| E2 | 75 | 0.324 | 2.08 | 2.27 | 50.9 | 1.14 | 1.05 | |||||||
| E3 | 50 | 0.26 | 2.08 | 2.2 | 53.5 | 1.08 | 1.09 | |||||||
| F1 | Ductal/micropapillary | Multi-focal | 2 | + | − | − | 25 | 0.06 | 2.11 | 1.8 | 54.1 | 1.89 | 1.17 | |
| F2 | 60 | 0.292 | 2.06 | 2.19 | 54 | 1.19 | 1.13 | |||||||
| F3 | 25 | 0.079 | 2.04 | 2.13 | 54.2 | 1.18 | 0.99 | |||||||
| G1 | Ductal | 3.4 | 3 | − | − | − | 70 | 0.515 | 2.08 | 2.19 | 51.3 | 3.31 | 1.31 | |
| G2 | 30 | 0.076 | 2.03 | 2.39 | 56.1 | 1.13 | 0.98 | |||||||
| G3 | 70 | 0.331 | 2.07 | 2.24 | 54.4 | 1.21 | 1.04 | |||||||
| H1 | Ductal | 1.6 | 3 | − | − | − | 30 | 0.112 | 2.11 | 1.59 | 50.3 | 2.94 | 1.12 | |
| H2 | 30 | 0.096 | 1.92 | 0.92 | 54.1 | 2.21 | 1.09 | |||||||
| H3 | 30 | 0.072 | 2.12 | 1.29 | 49.7 | 1.49 | 1.09 | |||||||
| H4 | 20 | 0.285 | 2.09 | 1.98 | 49.6 | 6.66 | 1.95 | |||||||
| I1 | Ductal/lobular | > 2 | 2 | + | + | − | 70 | 0.237 | 2.11 | 1.96 | 52.5 | 2.35 | 1.19 | |
| I2 | 70 | 0.331 | 2.09 | 2.03 | 51 | 2.17 | 1.15 | |||||||
| I3 | 70 | 0.173 | 2.12 | 2.07 | 51.5 | 1.79 | 1.1 | |||||||
| J1 | Ductal | 3.5 | 3 | + | + | − | 80 | 0.682 | 2.08 | 1.93 | 49.8 | 3.85 | 1.47 | |
| J2 | 80 | 0.941 | 2.08 | 2.11 | 51.3 | 2.91 | 1.42 | |||||||
| J3 | 80 | 0.561 | 2.07 | 2.17 | 50.8 | 3.49 | 1.57 | |||||||
| K1 | Ductal | 2.4 | 2 | + | + | − | 15 | DCIS | 0.068 | 2.12 | 1.66 | 51.4 | 1.88 | 1.22 |
| K2 | 75 | DCIS | 0.044 | 2.06 | 1.85 | 55 | 1.85 | 1.04 | ||||||
| L1 | Ductal | 1.3 | 3 | + | + | − | 0 | DCIS | 0.055 | 2.1 | 1.31 | 51.2 | 1.55 | 1.17 |
| L2 | 15 | DCIS | 0.083 | 2.11 | 1.84 | 50.7 | 1.39 | 0.92 | ||||||
| L3 | 15 | DCIS | 0.099 | 2.09 | 1.21 | 51 | 1.69 | 1.06 | ||||||
| M1 | Ductal | 1.9 | 2 | + | + | − | 50 | 0.116 | 2.1 | 1.74 | 50.3 | 2.49 | 1.75 | |
| M2 | 50 | Necrosis | 0.09 | 2.08 | 1.74 | 49.1 | 1.58 | 1.1 | ||||||
| M3 | 30 | 0.084 | 2.12 | 1.65 | 45.5 | 2.06 | 1.35 | |||||||
| N1 | Ductal | 1.8 | 2 | + | + | − | 20 | 0.077 | 2.14 | 1.8 | 50.5 | 1.57 | 1.19 | |
| N2 | 70 | 0.068 | 2.06 | 1.96 | 52.9 | 1.28 | 1.09 | |||||||
| N3 | 70 | 0.069 | 2.13 | 0.67 | 51.6 | 1.74 | 1.33 | |||||||
| O1 | Ductal | 3 | 1 | + | + | − | 20 | Carcinoma + papilloma | 0.261 | 2.11 | 2.2 | 53.7 | 2 | 0.95 |
| O2 | 50 | Carcinoma + papilloma | 0.143 | 2.13 | 1.24 | 55.5 | 2.06 | 1.13 | ||||||
| P1 | Ductal | 0.9 | 3 | − | − | − | 60 | 0.05 | 2.11 | 1.89 | 54.4 | 1.34 | 1.06 | |
| P2 | 10 | 0.044 | 2.15 | 1.61 | 56.6 | 1.34 | 1.05 | |||||||
| Q1 | Ductal | 1.8 | 3 | − | + | − | 70 | Biopsy site | 0.474 | 2.07 | 2.03 | 51.9 | 2.13 | 1.08 |
| Q2 | 30 | Biopsy site | 0.226 | 2.11 | 2.03 | 55.1 | 2.02 | 1.02 | ||||||
| R1 | Ductal | 2.8 | 3 | + | − | − | 70 | Biopsy site | 0.156 | 2.07 | 2.1 | 52.8 | 1.66 | 1.12 |
| R2 | 20 | Biopsy site | 0.132 | 2.1 | 2.14 | 52.8 | 2.33 | 1.26 | ||||||
NOTE. 260/280 and 260/230 are optical density ratios; CA is the tumor content in percent; % P is the percent of probe sets present; actin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) are the 3′ to 5′ probe set ratios for the actin and GAPDH probe sets, respectively.
Abbreviations: QC, quality control; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; DCIS, ductal carcinoma-in-situ.
Intertumor Variance Exceeds Intratumor Variance at the Level of Gene Expression
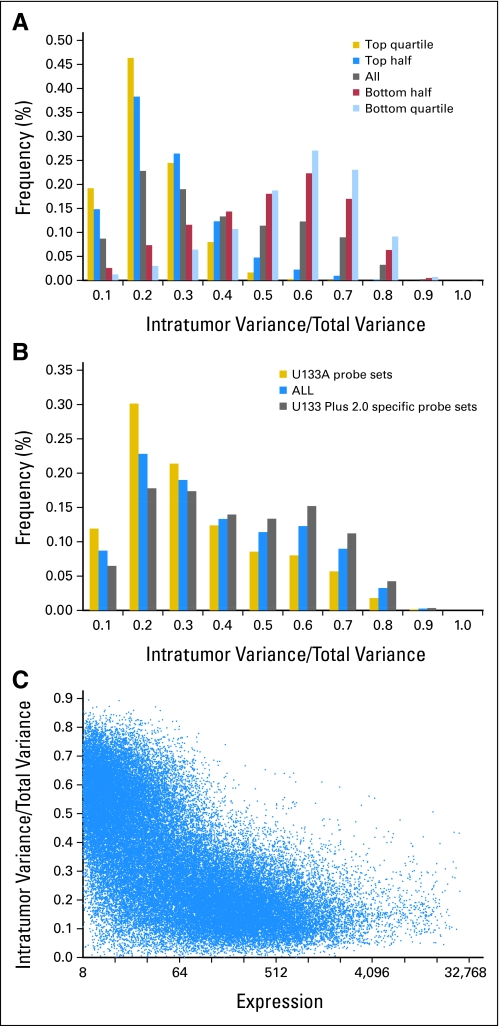
To evaluate the performance of individual features on the Affymetrix array, we calculated the ratio between intratumor variance and total variance for all probe sets. As shown in Figure 1A to 1C, the majority of samples had an intratumor/total variance ratio below 0.5. A distinct inverse relationship exists between expression intensity and the proportion of intratumor variance. Specifically, within the top quartile of probe sets of highly expressed genes (Fig 1A) more than 90% had an intratumor variance ratio less than one third of the total variance. Conversely, only 11% of genes expressed at comparatively low levels (bottom quartile, Fig 1A) had an intratumor/total variance ratio less than one third. We also found that probe sets corresponding to the U133B platform generally had higher intratumor variability when compared with those on the original U133A array (P < .001; Fig 1B), and a similar pattern was noted for probe sets annotated to known genes compared with unannotated probe sets (P < .001).
Fig 1.
For Affymetrix probe sets, intratumor variance is generally a small component of total variance. Histograms stratified by (A) expression level demonstrate that profiles differ sharply between highly and lowly expressed genes, and (B) source demonstrate that U133A probe sets are generally more reproducible. (C) Scatter plot demonstrates an inverse relationship between variance ratios and average expression.
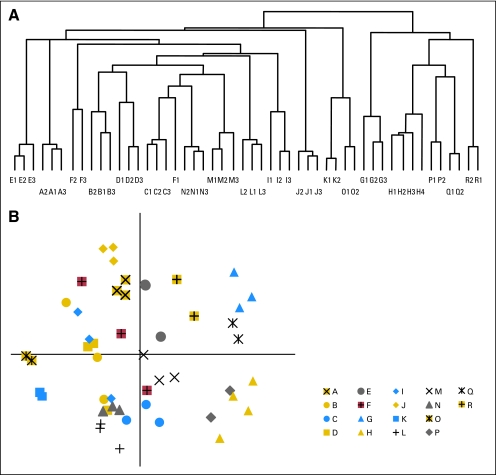
To evaluate the global patterns of expression, we performed PCA and hierarchical clustering on robust multiarray average–normalized data. Hierarchical clustering of all samples using probe sets with log2 expression values > 5 demonstrated that replicate samples from a single tumor tended to group together in a robust and statistically significant fashion (Fig 2A and Data Supplement, online only). One tumor showed imperfect clustering (patient F). There were no histology, RNA, or array quality metrics that could account for discordance between samples F1, F2, and F3. A PCA of the filtered expression data (Fig 2B) demonstrates that patient replicates largely cluster together in the top two principal components (capturing 28% of the total variance in global expression), indicating that more heterogeneity is seen across patients than within replicates. In summary, while most genes exhibit a low degree of intratumor variability and most replicate samples demonstrate similar global expression patterns, poor performing probe sets exist and could potentially have an impact on the precision of array-based predictors of tumor biology or behavior.
Fig 2.
Unsupervised clustering demonstrates that replicate samples have similar global patterns of expression. (A) Pearson Correlation Coefficient–based hierarchical clustering shows complete segregation of replicates in 17 of 18 patients and is confirmed by (B) a scatter plot of the top two principal components of all expressed genes. The discordant patient, F, is highlighted in red. Principal component 1 captures estrogen receptor status.
Intratumor Variance in Gene Expression Does Not Preclude Precise Predictions of Tumor Biology
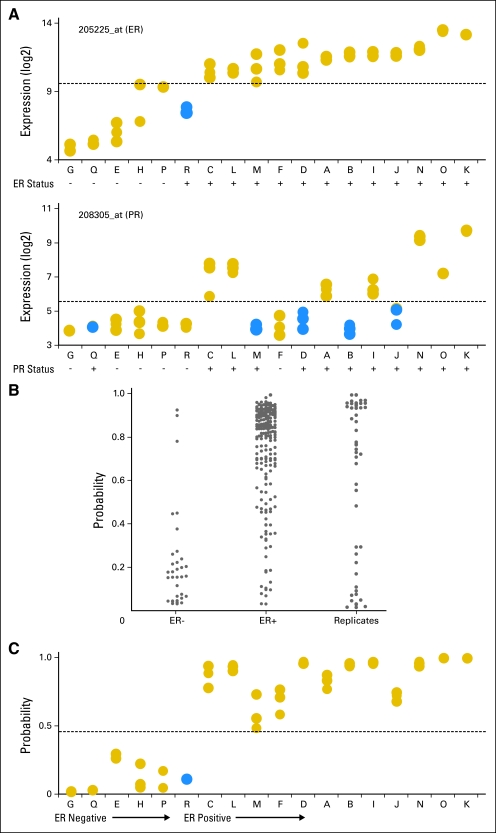
ER and PR status are two critical characteristics of breast cancer that define biologically distinct subgroups of disease. ER status and PR status are prognostic of clinical outcome and often determine the course of treatment. To determine the effect of intratumor variance on array-based assessments of hormone receptor status, we began by evaluating single-gene predictors of ER (probe set 205225_at)27 and PR expression (probe set 208305_at). Expression levels of 205225_at demonstrated almost perfect agreement among the replicates samples (ICC, 0.94; 95% CI, 0.86 to 0.97; Fig 3A). Further, using an optimal binary classification (threshold of log2 = 9.6 to maximize accuracy), only one patient showed discordance with IHC (sensitivity, 1.0; 95% CI, 0.90 to 1.0; specificity, 0.88; 95% CI, 0.62 to 0.98). A single probe set for the PR gene, 208305_at, also showed near perfect agreement among replicates (ICC, 0.95; 95% CI, 0.89 to 0.98), but with an optimal binary classifier (threshold of log2 = 5.2 chosen to maximize accuracy), greater disagreement was noted between PR expression and IHC results. While all samples from the six PR-negative patients were classified correctly (specificity, 1.0; 95% CI, 0.80 to 1.0), samples from five of the 12 PR-positive patients were discordant with IHC (sensitivity, 0.64; 95% CI, 0.45 to 0.80).
Fig 3.
(A) Single-gene predictors of estrogen receptor (ER) and progesterone receptor (PR) status demonstrate high precision among individual patients and agreement with immunohistochemistry. (B) A multigene predictor of ER pathway activation generated from an independent data set of 247 patients (C) demonstrates high precision and agreement with immunohistochemistry in the breast replicate data set. Blue solid circles, discordant or inaccurate samples; gold solid circles, complete concordance. Capital letters are patient designations.
We next created a multigene predictor of ER pathway activation from a previously published breast cancer Affymetrix U133A microarray data set (GEO# GSE3494). This data set was filtered to retain probe sets with mean log2 expression values > 5.0 for the 247 ER-annotated patient samples.19 The ER predictor was based on 1,022 probe sets identified by a Wilcoxon rank sum test with Bonferroni correction (adjusted P < .05), with intentional exclusion of probe sets for ER itself (Data Supplement). The large number of differentially expressed genes highlights the distinct biologic characteristics of these different tumor types. A predictor of ER pathway status was generated by applying the top principal component of expression from the 1,022 probe set list to a Bayesian probit regression model. Under leave-one-out cross validation and a threshold of 0.5, the model classified 91% of the ER-negative samples and 85% of the ER-positive samples by IHC correctly (Fig 3B). Applied prospectively to the breast cancer replicate data set with an optimal threshold of 0.45 (Fig 3C), the model showed near perfect precision (ICC, 0.98; 95% CI, 0.95 to 0.99) and 96% accuracy identical to the single-gene model. Taken together, these data show that when assaying a robust biologic property of breast cancer such as ER status, intratumor variance does not preclude precise predictions from microarray data.
Intratumor Heterogeneity Does Not Preclude Precise Predictions of Clinical Outcome
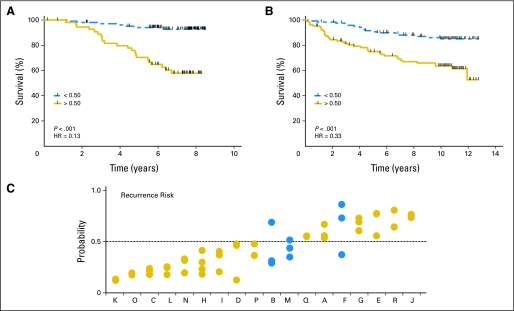
A prognostic model for death attributed to breast cancer was generated from expression data from a previously published data set (GEO# GSE1456; N = 159).20 Probe sets associated with survival were identified using a Cox proportional hazard model and the Benjamini-Hochberg method for controlling false discovery rates.28 A total of 205 probe sets (representing 184 genes) were identified with a false discovery rate < 0.01. These probe sets contain an over-representation of genes involved in cell cycles (20 genes; P < .001), cytokinesis (seven genes; P = .004), and cellular metabolism (84 genes; P = .004; Data Supplement). A binary classifier of survival was created using the first two principal components. The fitted model accurately stratified patients into high and low risk for death from disease with a threshold of 0.50 (Fig 4A). When this genomic signature was next applied to an independent validation data set (GSE3494; N = 315),19 it maintained the ability to identify a high-risk cohort for survival (P = .0069; Fig 4B). The precision of the prognostic genomic signature was found to be substantial (ICC, 0.71; 95% CI, 0.48 to 0.87), when applied to the replicate samples (Fig 4C). The patient with the most variance in recurrence risk predictions (patient F) also showed the poorest internal concordance by hierarchical clustering and PCA analysis, suggesting that the global variance in expression within this tumor may have affected the precision of the risk predictor.
Fig 4.
Novel multigene signatures for breast cancer survival demonstrate prognostic value by Kaplan-Meier plot and log-rank tests under (A) cross-validation in the training data (GSE1456; N = 159), (B) independent validation set in a second data set (GSE3494; N = 315), and (C) substantial agreement when applied to the replicate data set. Blue solid circles, discordant or inaccurate samples; gold solid circles, complete concordance. Capital letters are patient designations. HR, hazard ratio.
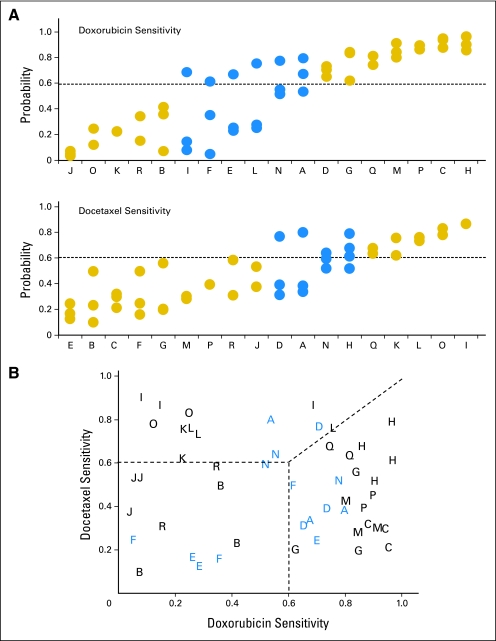
Finally, we established the precision of previously published chemotherapeutic sensitivity predictors for doxorubicin and docetaxel (Fig 5A and Data Supplement), as implemented in a randomized phase II trial to direct neoadjuvant therapy.23 A substantial level of agreement is observed for the doxorubicin sensitivity predictor (ICC, 0.72; 95% CI, 0.49 to 0.88). Further, 12 of 18 patients showed complete concordance when applying an a priori threshold of 0.6 used in the clinical trial. The docetaxel sensitivity predictions showed a slightly lower level of precision (ICC, 0.64; 95% CI, 0.38 to 0.83), with 14 of 18 patients showing complete concordance. Under a multilevel classification of higher sensitivity to one agent or of double resistance (Fig 5B), 13 of 18 patients showed complete concordance. By resampling from a binomial mixture distribution, the observed agreement in the three-level classification was highly significant (P < .001). Variation in tumor content and quality control measures for RNA and Affymetrix arrays was found not to be associated with the discordance in the multigene predictors of clinical outcome (all ANCOVA P > .05).
Fig 5.
(A) Multigene signatures of chemosensitivity to doxorubicin and docetaxel demonstrate substantial agreement when applied prospectively to the replicate samples (A-R) breast cancer data set. Capital letters are patient designations. Gold solid circles, complete concordance; blue solid circles, discordant samples. (B) Under a multilevel classification of higher sensitivity to one agent or of double resistance, 13 of 18 patients show complete concordance.
DISCUSSION
Here, we demonstrate that intratumor variance at the level of gene expression does not preclude the development of precise microarray-based clinical prediction models in breast cancer. We show intertumor heterogeneity is greater than intratumor heterogeneity at the level of global gene expression for this breast tumor data set. While a small group of genes exhibits a significant level of intratumor variation, many of these genes are expressed at relatively low levels and can be filtered as background noise when creating predictive algorithms. Finally, we demonstrate that a high degree of precision was seen among replicate samples when assayed using single-gene predictors of ER and PR status and PCA-based predictors of ER pathway activation, cancer recurrence, and chemotherapeutic sensitivity.
The heterogeneous nature of solid neoplasms has been recognized and studied by pathologists for decades. In the context of breast cancer, the varying presence of normal breast tissue, inflammatory cells, vessels, necrosis, and neoplastic epithelium gives rise to a variably mixed population of cells with unique or distinct biologic makeup in any given tumor sampling. Thus, the question has been raised of how this cellular heterogeneity may affect assays typically performed for diagnostic and prognostic purposes in breast cancer patients, including ER, PR, and HER2.
The largest study of the reproducibility of HER2 testing was performed on patients enrolled in the North Central Cancer Treatment Group (NCCTG) N9831 adjuvant trial of trastuzumab. In this study, only 85.8% of the 2,535 patients registered in the trial had concordant results for HER2 positivity between the local and central performing laboratories (88.1% concordance by fluorescent in situ hybridization and 81.6% concordance by IHC).29 A similar study of ER and PR IHC measurements in 776 patients enrolled on Eastern Cooperative Oncology Group (ECOG) 2197 showed 90% and 84% concordance between local and central laboratory studies for ER and PR status, respectively.30 These studies, however, are strictly a measure of assay reproducibility; a measure of technical variance rather than tumor heterogeneity.
Discordance attributable solely to tumor heterogeneity between breast core biopsy and resection specimens ranges from 1.2% for ER status on the low end31 to 14% for ER status and 20% for PR status on the high end.32 Discordance rates attributable to technique and tumor heterogeneity that develops over time was determined to be even higher (18.4%, 40.3%, and 13.6% for ER, PR, and HER2, respectively) when comparing primary tumors and their corresponding metastatic lesions.33 The low discordance rates (0% in this study) of our array-based predictors of ER and PR status are at least equivalent to the good performance of these traditional techniques.
While the effect of tumor heterogeneity and technical variance on ER, PR, and HER2 testing has been well studied, few studies have examined the effect of tumor heterogeneity on multigene predictive algorithms. A study comparable to the one presented here reached similar conclusions using a 48-gene TaqMan-based assay.34 This study showed high concordance among three replicate samplings for 12 breast cancer patients. A similar study examining the contribution of technical variance to the reproducibility of microarray-based assays in breast cancer, demonstrated that gene expression–based signatures developed from replicate experiments among 35 patients resulted in precise predictions of breast cancer chemotherapeutic responsiveness.13
To the best of our knowledge, our work is the first to demonstrate the impact of intratumor heterogeneity on the precision of multigene predictive models focused on tumor behavior. Precision is an integral component of clinical testing that is often overlooked in the early stages of translational research. Accuracy always seems of primary interest at that stage. However, our data suggest that a lack of accuracy in microarray-based assays may in fact be caused by a lack of precision, particularly for more indirect measures of tumor behavior (eg, chemosensitivity or recurrence). For these more abstract measures, a lack of precision may be attributable to true differences in tumor microenvironment. This is supported by our finding that the genes in these complex predictors tend to show higher intratumor variance (Data Supplement). In fact, there is a direct correlation between each predictor's ICC and the proportion of genes with high intratumor variance. Our data also highlight the fact that testing of replicate samples, even early in the development of a microarray-based assay, can clearly differentiate between inaccurate and imprecise.
The importance of assessing performance of multigene microarray-based assays, as described here, bears on the future use of this technology as a single assay for breast cancer patients that can provide not only measures of prognosis or predicted therapeutic response, but can also supplement or replace the standard assays of ER, PR, and HER2. The feasibility of this approach is, in part, demonstrated in this study. This full genome expression profile may find additional uses as assays become algorithms applied to expression data sets. As we move toward this, understanding the role of tumor heterogeneity in measures of tumor behavior and developing approaches and data sets (like the one provided here) to test the precision of these algorithms is essential.
Supplementary Material
Footnotes
Supported by Duke Breast Specialized Program of Research Excellence (SPORE) Grant No. P50CA068438, Congressionally Directed Medical Research Programs Department of Defense Clinical Trial Grant No. W81XWH-07-1-0394 (P.K.M.), and a grant from the V Foundation for Cancer Research.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: John A. Olson, Core Prognostex (U) Consultant or Advisory Role: Joseph R. Nevins, Millennium Pharmaceuticals (C), Johnson & Johnson (C) Stock Ownership: John A. Olson, Core Prognostex; Joseph R. Nevins, Expression Analysis Honoraria: Joseph R. Nevins, Eli Lilly Research Funding: None Expert Testimony: None Other Remuneration: Michael B. Datto, Affymetrix
AUTHOR CONTRIBUTIONS
Conception and design: William T. Barry, John A. Olson, Jeff R. Marks, Geoffrey S. Ginsburg, Paul K. Marcom, Joseph R. Nevins, Joseph Geradts, Michael B. Datto
Financial support: Paul K. Marcom, Joseph R. Nevins
Administrative support: Geoffrey S. Ginsburg, Joseph R. Nevins
Provision of study materials or patients: Paul K. Marcom, Joseph Geradts
Collection and assembly of data: William T. Barry, Holly K. Dressman, Ryan J. Griffis, J'Vonne D. Hunter, Joseph Geradts, Michael B. Datto
Data analysis and interpretation: William T. Barry, Dawn N. Kernagis, Jeff R. Marks, Paul K. Marcom, Joseph R. Nevins, Joseph Geradts, Michael B. Datto
Manuscript writing: William T. Barry, Dawn N. Kernagis, Paul K. Marcom, Joseph R. Nevins, Joseph Geradts, Michael B. Datto
Final approval of manuscript: William T. Barry, Dawn N. Kernagis, Holly K. Dressman, Ryan J. Griffis, J'Vonne D. Hunter, John A. Olson, Jeff R. Marks, Geoffrey S. Ginsburg, Paul K. Marcom, Joseph R. Nevins, Joseph Geradts, Michael B. Datto
REFERENCES
- 1.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Rouzier R, Perou CM, Symmans WF, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005;11:5678–5685. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- 3.van de Vijver MJ, He YD, van't Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 4.Potti A, Nevins JR. Utilization of genomic signatures to direct use of primary chemotherapy. Curr Opin Genet Dev. 2008;18:62–67. doi: 10.1016/j.gde.2008.01.018. [DOI] [PubMed] [Google Scholar]
- 5.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360:790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 6.Bonnefoi H, Potti A, Delorenzi M, et al. Validation of gene signatures that predict the response of breast cancer to neoadjuvant chemotherapy: A substudy of the EORTC 10994/BIG 00-01 clinical trial. Lancet Oncol. 2007;8:1071–1078. doi: 10.1016/S1470-2045(07)70345-5. [DOI] [PubMed] [Google Scholar]
- 7.Salter KH, Acharya CR, Walters KS, et al. An integrated approach to the prediction of chemotherapeutic response in patients with breast cancer. PLoS ONE. 2008;3:e1908. doi: 10.1371/journal.pone.0001908. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Canales RD, Luo Y, Willey JC, et al. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat Biotechnol. 2006;24:1115–1122. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- 9.Patterson TA, Lobenhofer EK, Fulmer-Smentek SB, et al. Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nat Biotechnol. 2006;24:1140–1150. doi: 10.1038/nbt1242. [DOI] [PubMed] [Google Scholar]
- 10.Shi L, Reid LH, Jones WD, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irizarry RA, Warren D, Spencer F, et al. Multiple-laboratory comparison of microarray platforms. Nat Methods. 2005;2:345–350. doi: 10.1038/nmeth756. [DOI] [PubMed] [Google Scholar]
- 12.Zakharkin SO, Kim K, Mehta T, et al. Sources of variation in Affymetrix microarray experiments. BMC Bioinformatics. 2005;6:214. doi: 10.1186/1471-2105-6-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson K, Hess KR, Kapoor M, et al. Reproducibility of gene expression signature-based predictions in replicate experiments. Clin Cancer Res. 2006;12:1721–1727. doi: 10.1158/1078-0432.CCR-05-1539. [DOI] [PubMed] [Google Scholar]
- 14.Bachtiary B, Boutros PC, Pintilie M, et al. Gene expression profiling in cervical cancer: An exploration of intratumor heterogeneity. Clin Cancer Res. 2006;12:5632–5640. doi: 10.1158/1078-0432.CCR-06-0357. [DOI] [PubMed] [Google Scholar]
- 15.Francis P, Fernebro J, Eden P, et al. Intratumor versus intertumor heterogeneity in gene expression profiles of soft-tissue sarcomas. Genes Chromosomes Cancer. 2005;43:302–308. doi: 10.1002/gcc.20191. [DOI] [PubMed] [Google Scholar]
- 16.Jochumsen KM, Tan Q, Hølund B, et al. Gene expression in epithelial ovarian cancer: A study of intratumor heterogeneity. Int J Gynecol Cancer. 2007;17:979–985. doi: 10.1111/j.1525-1438.2007.00908.x. [DOI] [PubMed] [Google Scholar]
- 17.Trautmann K, Steudel C, Grossmann D, et al. Expression profiling of gastric cancer samples by oligonucleotide microarray analysis reveals low degree of intra-tumor variability. World J Gastroenterol. 2005;11:5993–5996. doi: 10.3748/wjg.v11.i38.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olson JJ, Morris E, Zee KV, et al. Magnetic resonance imaging facilitates breast conservation for occult breast cancer. Ann Surg Oncol. 2000;7:411–415. doi: 10.1007/s10434-000-0411-4. [DOI] [PubMed] [Google Scholar]
- 19.Miller LD, Smeds J, George J, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pawitan Y, Bjöhle J, Amler L, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: Derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7:R953–R964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irizarry R, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 22.West M, Blanchette C, Dressman H, et al. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc Natl Acad Sci U S A. 2001;98:11462–11467. doi: 10.1073/pnas.201162998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potti A, Dressman H, Bild A, et al. Genomic signatures to guide the use of chemotherapeutics. Nat Med. 2006;12:1294–1300. doi: 10.1038/nm1491. [DOI] [PubMed] [Google Scholar]
- 24.Shrout P, Fleiss J. Intraclass correlations: Uses in assessing rater reliability. Psychol Bull. 1979;86:420–428. doi: 10.1037//0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- 25.Landis J, Koch G. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- 26.Eisen MB, Spellman PT, Brown PO, et al. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gong Y, Yan K, Lin F, et al. Determination of oestrogen-receptor status and ERBB2 status of breast carcinoma: A gene-expression profiling study. Lancet Oncol. 2007;8:203–211. doi: 10.1016/S1470-2045(07)70042-6. [DOI] [PubMed] [Google Scholar]
- 28.Benjamini Y, Drai D, Elmer G, et al. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 29.Perez EA, Suman VJ, Davidson NE, et al. HER2 testing by local, central, and reference laboratories in specimens from the North Central Cancer Treatment Group N9831 intergroup adjuvant trial. J Clin Oncol. 2006;24:3032–3038. doi: 10.1200/JCO.2005.03.4744. [DOI] [PubMed] [Google Scholar]
- 30.Badve SS, Baehner FL, Gray RP, et al. Estrogen- and progesterone-receptor status in ECOG 2197: Comparison of immunohistochemistry by local and central laboratories and quantitative reverse transcription polymerase chain reaction by central laboratory. J Clin Oncol. 2008;26:2473–2481. doi: 10.1200/JCO.2007.13.6424. [DOI] [PubMed] [Google Scholar]
- 31.Hodi Z, Chakrabarti J, Lee AH, et al. The reliability of assessment of oestrogen receptor expression on needle core biopsy specimens of invasive carcinomas of the breast. J Clin Pathol. 2007;60:299–302. doi: 10.1136/jcp.2006.036665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taucher S, Rudas M, Gnant M, et al. Sequential steroid hormone receptor measurements in primary breast cancer with and without intervening primary chemotherapy. Endocr Relat Cancer. 2003;10:91–98. doi: 10.1677/erc.0.0100091. [DOI] [PubMed] [Google Scholar]
- 33.Liedtke C, Broglio K, Moulder S, et al. Prognostic impact of discordance between triple-receptor measurements in primary and recurrent breast cancer. Ann Oncol. 2009;20:1953–1958. doi: 10.1093/annonc/mdp263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyng M, Laenkholm A, Pallisgaard N, et al. Intratumor genetic heterogeneity of breast carcinomas as determined by fine needle aspiration and TaqMan low density array. Cell Oncol. 2007;29:361–372. doi: 10.1155/2007/860194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.