Abstract
Background
Memory is the ability to store, retain, and later retrieve learned information. Long-term memory (LTM) formation requires: DNA transcription, RNA translation, and the trafficking of newly synthesized proteins. Several components of these processes have already been identified. However, due to the complexity of the memory formation process, there likely remain many yet to be identified proteins involved in memory formation and persistence.
Results
Here we use a quantitative proteomic method to identify novel memory-associated proteins in neural tissue taken from animals that were trained in vivo to form a long-term memory. We identified 8 proteins that were significantly up-regulated, and 13 that were significantly down-regulated in the LTM trained animals as compared to two different control groups. In addition we found 19 proteins unique to the trained animals, and 12 unique proteins found only in the control animals.
Conclusions
These results both confirm the involvement of previously identified memory proteins such as: protein kinase C (PKC), adenylate cyclase (AC), and proteins in the mitogen-activated protein kinase (MAPK) pathway. In addition these results provide novel protein candidates (e.g. UHRF1 binding protein) on which to base future studies.
Background
Learning and memory are separate but related processes each with its own 'rules and regulations'. Learning is the acquisition of a new behaviour, and memory the ability to store and recall this new information [1]. Memory formation is a complex process involving several necessary biomolecular steps. Long-term memory (LTM), which persists for days or even decades has been shown to require both transcription and translation (i.e. new protein expression) in order to occur [2-4]. In addition to transcription and translation LTM also typically requires the activation of second messengers by an appropriate signalling event, the amplification and translocation of this signal to the nucleus, and the transport of the newly synthesized proteins to the appropriate location in the cell [5,6]. It is apparent that memory formation and its maintenance is a process that requires the involvement of numerous proteins in order to occur, and the identification of the proteins involved is likely far from complete [7]. Owing to the complexity of memory systems, identifying those proteins that are causal to memory formation is no simple task.
In an attempt to work around the problem of the complexity of most memory systems (as based on sheer neuron numbers) we have chosen to use a reductionist model system of memory formation in Lymnaea. The fresh water snail Lymnaea stagnalis has been used in many studies of learning and memory. Owing to its relatively simple nervous system with large identifiable neurons, much progress has been made in characterizing the underlying neuronal circuitry that drives various behaviours [1,8,9]. One such relatively simple behaviour that has been studied in depth is aerial respiration. This breathing behaviour is driven by a 3-neuron central pattern generator (CPG), whose necessity and sufficiency have been shown [10,11]. This aerial respiratory behaviour can be operantly conditioned to form memories of varying duration, including long-term memories that require both the transcription and translation of new proteins to occur [12]. Additionally as this is a non-declarative form of memory, the site of physical storage of the memory should reside within the neurons that mediate the motor behaviour, i.e. the respiratory CPG. Right pedal dorsal 1 (RPeD1), a neuron in the respiratory CPG has been previously shown to be required for long-term memory, extinction, reconsolidation and forgetting [13,14]. This detailed knowledge of the neurobiology controlling respiration presents a unique opportunity to study LTM formation on the cellular level.
In this study we employ a quantitative proteomics technique to tissue taken from both control and animals trained to form a long-term memory. Using this protocol we wish to test whether it is possible to: 1) detect proteins previously associated with memory, and 2) detect any novel proteins not previously associated with the memory formation/storage process.
Methods
Animals
The fresh water snail Lymnaea stagnalis were used in the following experiments. Animals were bred and maintained at the facilities in the University of Calgary, from a colony initially set up at Vrije University in The Netherlands. Adult animals with a shell size larger than 20 mm were used in all experiments. Animals were maintained, and all experiments were performed at room temperature (~20-21°C).
Training Protocols
Three sets of animals were used for this study, an operantly conditioned (LTM) group, a yoked control group, and a naïve group. Lymnaea are bi-modal breathers able to obtain their oxygen requirements cutaneously via diffusion across the skin, or aerially via the pneumostome (an opening into the lung). This allows for the aerial respiratory behaviour to be conditioned without compromising the animals survival [15]. This is done by placing individually labelled animals in water made hypoxic by bubbling N2 through for 20 minutes prior to training, in order to increase the aerial respiratory drive. Upon commencement of training the snails receive a gentle tactile stimulus to the pneumostome area each time aerial respiration is attempted. With training the animals learn to withhold this behaviour thus decreasing the number of attempted pneumostome openings. The LTM animals received three 45 min reinforcement sessions. Two 45 min training sessions on the first day separated by an hour, and a third 45 min training/memory test session on the second day. The yoked animals received the same hypoxia exposure and number of stimuli as the LTM group, except in a non-contingent manner (animals only receive stimulation if pneumostome is closed). Naïve animals received no treatment.
Dissection
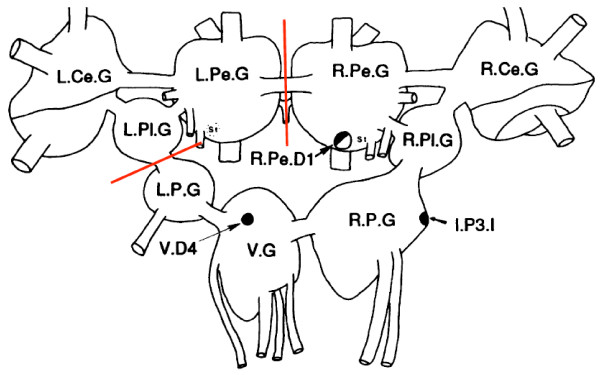
Animals were dissected one hour after the completion of the final training or control session. The animals were incapacitated by submersion in ice-cold saline, with the dissections also carried out in ice-cold saline. The central ganglia of the animals were quickly dissected out. Next the buccal ganglia were dissected away from the central ganglia followed by the removal of the left pedal (L.Pe.G), cerebral (L.Ce.G), and pleural ganglia (L.Pl.G) (Figure 2). This was performed in order to better isolate the brain regions containing the central pattern generator which controls aerial respiration [10], in an attempt to enhance the signal to noise ratio (cells where memory is stored to non-memory cells) of the samples. Upon completion of dissection the ganglia were placed in eppendorf tubes, and immediately snap frozen and stored at -80°C until use.
Figure 2.
Dissection of the Lymnaea CNS. After excising the CNS from the animal, it was then further dissected by removing the left pedal, pleural and cerebral ganglia (L.Pe.G, L.Pl.G, and L.Ce.G respectively) via cutting at the indicated areas (red lines). The remaining ganglia of the CNS containing the neurons of the aerial respiratory CPG right pedal dorsal one (R.Pe.D1), input 3 interneuron (I.P3.I), and visceral dorsal 4 (V.D4) were processed for proteomic analysis. Figure adapted from Syed et al., 1990 [10].
Sample Preparation
0.5 mL of solubilization buffer (Urea-Thiourea-CHAPS buffer containing, 40 mM Tris, 5 M Urea, 2 M thiourea, 4% CHAPS, 10 mM DTT, 1 mM EDTA, 1 mM PMSF, and a Protein inhibitor cocktail) was added to each sample, and the samples were homogenised on ice using the loose pestle of a Dounce homogeniser. The samples were then mixed on a rotary mixer for 30 minutes and then centrifuged at 10 000 g for 10 minutes. The supernatant was transferred to a new tube and this was centrifuged for 45 minutes at 150 000 g to sediment any un-dissolved material. The supernatant was transferred to a new tube, and protein concentration was determined using the EZQ assay (Invitrogen). The samples were stored at -20 until used. The samples were then processed as follows for mass spectrometry analysis.
Nano UPLC MS/MS
Protein samples were analyzed via an ultra-performance liquid chromatography tandem mass spectrometry (nano UPLC-MS/MS) protocol as previously described [16,17]. Briefly, the methanol-chloroform precipitated samples were subjected to in-solution tryptic digestion. Samples were then desalted using Sepak C18 columns (Waters, Milford, MA, USA). Next lyophilized peptides were resuspended (97% H2O, 3% acetonitrile, 0.1% formic acid) and subjected to the nano UPLC-MS/MS analysis using a Waters Nano-Acquity UPLC system with a 75-um inner diameter × 25 cm UPLC column and a 90 min gradient of 2-45% solvent B (solvent A: 99.9% H2O, 0.1% formic acid; solvent B: 99.9% acetonitrile, 0.1% formic acid; final flow rate 250 nl/min, 7000 psi), coupled to a Waters QTOF-premier tandem mass spectrometer (Waters, Milford, MA, USA). Data was acquired in high-definition MSE mode (low collision energy 4 eV, high collision energy ramping from 15 to 40 eV, switching every 1.5 seconds) and processed with ProteinLynx Global Server (PLGS version 2.2.5, Waters, Milford, MA, USA) to reconstruct MS/MS spectra by combining all masses with identical retention times. MS/MS spectra (peaklists) were searched against the Swiss Prot database using PLGS. Each sample was analysed in triplicate runs and were spiked with an alpha-enolase tryptic digest (125 fmol) as an internal standard. Quantitative changes in protein abundance, based on peptide ion peak intensities were analyzed using the Waters Expression Analysis Software (WEPS), which is part of the PLGS package.
Western Blot and analysis
For the western blot analysis, the CNS of another set of animals were dissected out and snap frozen after treatment as described above (naïve n = 8, yoked n = 8, and trained n = 8). 0.5 mL of Nonidet-P40 (NP-40) buffer (containing, 20 mM Tris HCl pH 8, 137 mM NaCl, 10% glycerol, 1% NP-40, 2 mM EDTA, 1 mM PMSF, and a Pearce Halt protease inhibitor cocktail) was added to each sample, and the samples were homogenised on ice using the loose pestle of the glass homogeniser. The samples were then mixed on a rotary mixer for 30 minutes and then centrifuged at 10 000 g for 10 minutes. The supernatant was transferred to a new tube and this was centrifuged for 45 minutes at 150 000 g to sediment any insoluble material. Protein levels were standardised and samples were resolved using 10% SDS-PAGE gel. Proteins were transferred overnight at 4°C onto PVDF membranes. Membranes were blocked in 5% non-fat milk/TBST (20 mM Tris-HCl, 0.15 M NaCl, 01% TWEEN 20), and incubated with polyclonal anti-ADCY8 (1:5000) or anti-β-actin (1:10000) (Abcam, Cambridge, MA, USA). Membranes were then washed and incubated with an HRP-conjugated secondary antibody (1:10000). Western blots were visualized using ECL plus (GE healthcare, Piscataway, NJ, USA). Membranes were scanned using a Molecular Dynamics Storm 860 phosphoimager, and densiometric analysis was completed using ImageJ software.
Results
LTM training and control animals
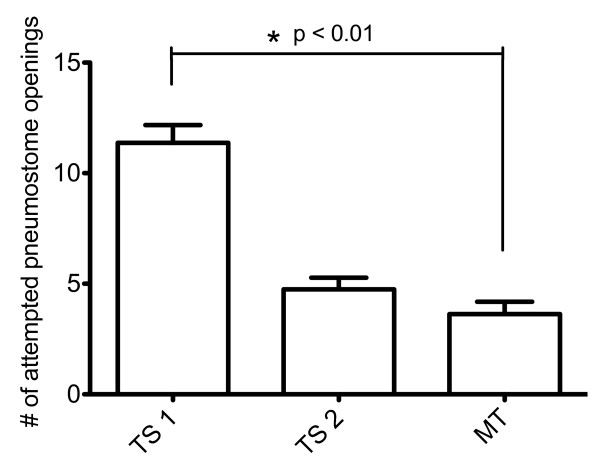
Three groups of animals were used in this study; an operantly conditioned group (n = 8), a yoked control group (n = 8), and a naïve group (n = 8). The operantly conditioned animals received two 45-minute training sessions (TS1 and TS2) separated by an hour on day 1. We tested for memory 24 h later (MT). We define memory to be present if the number of attempted pneuomostome openings in the MT session is significantly less than that in TS1, and not significantly greater than that of TS2. As expected the operantly trained animals showed a significant reduction in the number of attempted pneumostome openings in the last session (MT) as compared to the first (TS1) (ANOVA F (2,7) = 45.24, p < 0.001), and thus we conclude that they successfully formed LTM (Figure 1). This data is consistent with previously published findings using this training regime [18]. Yoked control snails did not exhibit LTM (data not shown).
Figure 1.
LTM formation. Animals given two 45 min training sessions separated by an hour (TS1 and TS2) have long-term memory when assayed 24 hours later (MT), as observed by a significant reduction in the number of attempted pneumostome openings (TS1 vs MT ANOVA F (2,7) = 45.24, p < 0.001).
Characterization of altered protein expression as a result of Long-term memory training
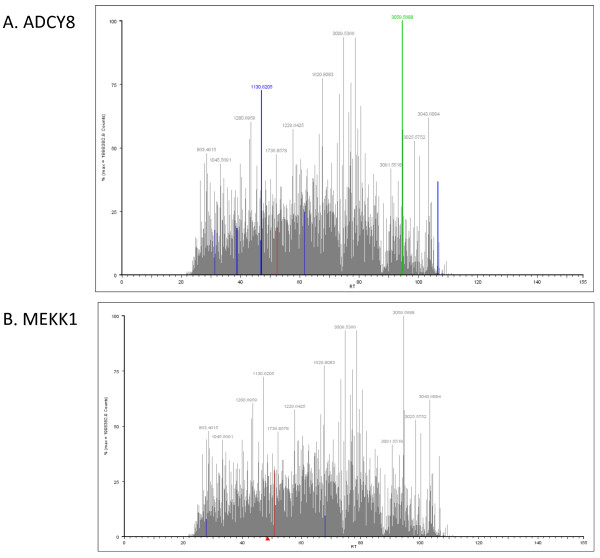
Following behavioural testing for LTM, all snails (including the two control groups) had their nervous systems dissected out (Figure 2) and processed for proteomic screening as described in the methods. Briefly the snail brains were homogenized, proteins were extracted, subjected to a trypsin digestion and analyzed by inline liquid chromatography and mass spectrometry. Hundreds of proteins were positively identified in each sample (LTM = 503, Naïve = 506, and Yoked = 518). From these identified proteins there were found to be a large number that were significantly different between the LTM group and the controls (yoked and naïve). To better focus the presentation of the data we are reporting only those proteins that were found to be significantly different (increased/decreased, present/absent) in the LTM group as compared to both the naïve and yoked control groups. That is any protein found to be significantly different in the LTM group, showed a significant difference from both the naïve and the yoked groups for the identical protein, and in the same direction. Using these criteria there were 8 proteins that were found to be significantly more abundant in the LTM trained animal sample as compared to the controls, and 13 proteins which were significantly less abundant (Table 1). 19 proteins were found to be unique to the LTM trained animal sample, while 12 proteins were detected in both control samples but not in the LTM sample (Table 2). Included in the list of proteins that were up regulated in LTM animals were adenylate cyclase type 8 (ADCY8) and mitogen activated protein kinase kinase kinase 1 (MEKK1) whose chromatograms are shown in figure 3. These two proteins have previously been associated with the memory formation process. ADCY8 is membrane bound enzyme that catalyses the formation of cyclic AMP (cAMP) in response to stimulus evoked entry of calcium into neurons. ADCY8 knockout mice show deficits for memory retention in object recognition and passive avoidance tasks [19], as well as deficits in the formation of mossy fiber long-term potentiation (LTP) in the hippocampus [20]. MEKK1 is a serine/threonine kinase, and a member of the MAPK signal transduction pathway. MEKK1 has been shown to be an activator of both the extracellular signal-regulated kinases 1 and 2 (ERK1/2) and the c-Jun N-terminal kinase 2 (JNK 2) pathways [21], with ERK activity being necessary for the induction of LTP [22], and the formation of various kinds of memories [23-25].
Table 1.
Proteins significantly up/down regulated in LTM vs. controls
| Protein description | Accession number | PLGS Score | Mass kDa. | No. of MS/MS Spectra | Seq. Coverage % | Fold change LTM vs. Naive | p-value | Fold change LTM vs. Yoked | p-value |
|---|---|---|---|---|---|---|---|---|---|
| Kinesin like protein KIF14 | Q15058 | 109.68 | 186.49 | 21 | 13 | 1.12 | < 0.01 | 1.19 | < 0.01 |
| Canalicular multispecific organic anion transporter | O15438 | 74.4 | 169.34 | 24 | 23 | 1.11 | 0.01 | 1.18 | < 0.01 |
| UHRF1 binding protein | Q6BDS2 | 65.7 | 159.49 | 10 | 8 | 1.16 | < 0.01 | 1.25 | < 0.01 |
| Putative hexokinase HKDC1 | Q2TB90 | 53.72 | 102.52 | 2 | 2 | 1.19 | 0.01 | 1.39 | < 0.01 |
| Uncharacterized protein C2orf54 | Q08AI8 | 49.33 | 49.57 | 11 | 31 | 1.12 | 0.05 | 1.16 | 0.01 |
| Adenylate cyclase type 8 | P40145 | 87.68 | 140.12 | 35 | 36 | 1.22 | < 0.01 | 1.11 | 0.03 |
| Bile salt export pump | O95342 | 43.39 | 146.41 | 1 | 1 | 1.12 | 0.04 | 1.11 | 0.03 |
| Mitogen activated protein kinase kinase kinase 1 | Q13233 | 59.93 | 164.47 | 13 | 12 | 1.11 | 0.01 | 1.11 | 0.04 |
| 5 azacytidine induced protein 1 | Q9UPN4 | 91.48 | 122.06 | 18 | 17 | 0.93 | < 0.01 | 0.93 | < 0.01 |
| Zinc finger protein 28 homolog | Q8NHY6 | 79.58 | 98.71 | 13 | 14 | 0.93 | 0.03 | 0.8 | < 0.01 |
| Kelch repeat and BTB domain containing protein 5 | Q2TBA0 | 37.8 | 69.26 | 9 | 16 | 0.85 | 0.01 | 0.78 | < 0.01 |
| Diacylglycerol kinase kappa | Q5KSL6 | 36.22 | 141.83 | 14 | 15 | 0.88 | < 0.01 | 0.88 | < 0.01 |
| Phosphatidylinositol 4 phosphate 3 kinase C2 domain containing alpha polypeptide | O00443 | 90.69 | 190.68 | 24 | 17 | 0.92 | 0.04 | 0.88 | 0.01 |
| Acyl CoA dehydrogenase family member 10 | Q6JQN1 | 67.81 | 118.83 | 11 | 11 | 0.93 | 0.03 | 0.92 | 0.01 |
| Arf-GAP with SH3 domain, ANK repeat and PH domain-containing protein 1 | Q9ULH1 | 60.8 | 125.47 | 15 | 15 | 0.88 | 0.03 | 0.88 | 0.01 |
| ATP binding cassette sub family A member 3 | Q99758 | 47.19 | 191.36 | 16 | 13 | 0.88 | 0.01 | 0.92 | 0.01 |
| Cytochrome P450 26A1 | O43174 | 41.39 | 56.2 | 6 | 14 | 0.87 | 0.03 | 0.89 | 0.02 |
| Tyrosine protein kinase JAK2 | O60674 | 56.35 | 130.67 | 10 | 9 | 0.92 | 0.03 | 0.9 | 0.03 |
| NTPase KAP family P loop domain containing protein 1 | Q17RQ9 | 52.69 | 67.78 | 13 | 26 | 0.83 | 0.01 | 0.88 | 0.03 |
| Ectonucleotide pyrophosphatase phosphodiesterase family member 2 | Q13822 | 47.65 | 99 | 9 | 14 | 0.83 | 0.05 | 0.83 | 0.03 |
| Multidrug resistance associated protein 9 | Q96J65 | 59.07 | 152.3 | 1 | 1 | 0.86 | 0.02 | 0.94 | 0.05 |
Accession number for Swiss Prot.
PLGS Score is calculated by the Protein Lynx Global Server (PLGS 2.2.5) software using a Monte Carlo algorithm to analyse all available mass spec. data and is a statistical measure of accuracy of assignation. A higher score implies greater confidence of protein identity [16].
Table 2.
Proteins unique to LTM and control conditions
| Protein description | Mass kDa | Accession number | PLGS Score | No. of MS/MS spectra | Seq. coverage % | Unique to (LTM or yoked/naïve) |
|---|---|---|---|---|---|---|
| Protein kinase C epsilon | 83.67 | Q02156 | 53.24 | 10 | 16 | LTM |
| Centrosomal protein of 63 kDa | 81.34 | Q96MT8 | 47.33 | 11 | 18 | LTM |
| Ankyrin repeat and IBR domain containing protein 1 | 122 | Q9P2G1 | 40.79 | 8 | 9 | LTM |
| Bcl 2 related proline rich protein | 36.82 | Q9HB09 | 40.7 | 8 | 21 | LTM |
| Olfactomedin like protein 1 | 45.92 | Q6UWY5 | 40.69 | 4 | 9 | LTM |
| Zinc finger protein 271 | 75.54 | Q14591 | 40.68 | 6 | 10 | LTM |
| Importin 9 | 115.96 | Q96P70 | 40.27 | 14 | 20 | LTM |
| Uncharacterized aarF domain containing protein kinase 5 | 65.9 | Q3MIX3 | 39.92 | 11 | 24 | LTM |
| Gamma parvin | 37.49 | Q9HBI0 | 38.99 | 6 | 20 | LTM |
| Nucleolar protein 11 | 81.12 | Q9H8H0 | 38.88 | 10 | 18 | LTM |
| 39S ribosomal protein L17 mitochondrial | 20.05 | Q9NRX2 | 38.59 | 8 | 42 | LTM |
| Chromobox protein homolog 6 | 43.9 | O95503 | 37.55 | 7 | 18 | LTM |
| Zinc finger protein 101 | 50.34 | Q8IZC7 | 37.42 | 3 | 7 | LTM |
| Transmembrane protein 68 | 37.43 | Q96MH6 | 36.96 | 8 | 28 | LTM |
| Ras association domain containing protein 5 | 47.09 | Q8WWW0 | 36.83 | 6 | 20 | LTM |
| Peroxiredoxin 4 | 30.54 | Q13162 | 36.11 | 6 | 26 | LTM |
| Solute carrier family 25 member 35 | 32.44 | Q3KQZ1 | 35.33 | 7 | 29 | LTM |
| Golgin subfamily A member 5 | 82.99 | Q8TBA6 | 35.2 | 9 | 15 | LTM |
| Putative uncharacterized protein CXorf55 | 114.94 | Q8N7X1 | 35 | 7 | 8 | LTM |
| Ras GTPase activating protein 1 | 116.4 | P20936 | 60.25 | 17 | 18 | N/Y |
| Elongator complex protein 1 | 150.25 | O95163 | 52.49 | 11 | 12 | N/Y |
| Alpha type platelet derived growth factor receptor precursor | 122.67 | P16234 | 49.83 | 12 | 10 | N/Y |
| Evolutionarily conserved signaling intermediate in Toll pathway | 49.15 | Q9BQ95 | 48.69 | 3 | 8 | N/Y |
| Vacuolar proton translocating ATPase 116 kDa subunit a isoform 4 | 96.36 | Q9HBG4 | 48.44 | 10 | 16 | N/Y |
| Exportin T | 109.96 | O43592 | 45.66 | 11 | 16 | N/Y |
| C type lectin domain family 4 member F | 65.52 | Q8N1N0 | 44.32 | 8 | 15 | N/Y |
| Transmembrane protein 16B | 113.97 | Q9NQ90 | 43.27 | 8 | 9 | N/Y |
| Hepatocellular carcinoma down regulated mitochondrial carrier protein | 33.44 | Q6Q0C1 | 41.69 | 7 | 20 | N/Y |
| Cell division cycle 7 related protein kinase | 63.89 | O00311 | 41.36 | 6 | 13 | N/Y |
| Myotubularin | 69.93 | Q13496 | 39.76 | 7 | 13 | N/Y |
| Aladin Adracalin | 59.57 | Q9NRG9 | 39.32 | 6 | 15 | N/Y |
Accession number for Swiss Prot.
Figure 3.
Representative LC-MS/MS chromatograms. LC-MS/MS chromatograms for ADCY8 (A) and MEKK1 (B).
Western blot
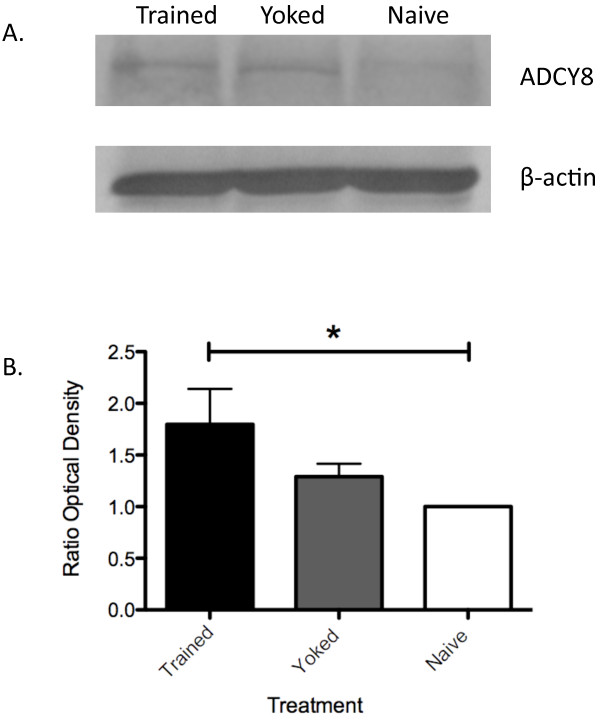
To validate the results of the quantitative MS experiments, we performed a set of western-blot experiments on one of the proteins found to be up-regulated in the LTM sample; ADCY8. For this experiment three new sets of samples were prepared, as previously described, and four separate gels and blots were done to allow for quantification. As shown in figure 4, animals trained to form a LTM showed a significant increase in the amount of ADCY8 as compared to naïve animals. While the western blot results show a similar pattern of results when compared to the UPLC-MS/MS data, they differed slightly as the change in ADCY8 expression between yoked and trained animals did not reach statistical significance by western blot analysis. This difference may be attributed to the much higher precision and sensitivity of measurement made by the UPLC MS/MS method.
Figure 4.
ADCY8 is significantly up-regulated in trained animals. A) Animals trained to form a LTM have increased levels of ADCY8 as compared to Naïve animals by western blot analysis B) Average change in ADCY8 expression (n = 4, LTM vs. p < 0.05).
Discussion
In this study we used quantitative proteomic methods to identify changes in protein expression in Lymnaea stagnalis trained to form a long-term memory as compared to controls. At the time point measured (1 hr after the final training session), we observed that the abundance in 21 proteins was significantly altered by the training procedure when compared against both control groups. In addition to changes found in those 21 proteins, we also identified 19 proteins that were found to be uniquely detected in the LTM group and not in either control, and conversely we found 12 proteins that were expressed in both the control groups but not found in the LTM group.
Several of the proteins identified have already been implicated in various learning/memory paradigms. For example in the present experiment adenylate cyclase type 8 (AC8) was found to have increased expression in LTM animals as compared to the controls. AC8 is stimulated by Ca2+/calmodulin and acts to increase the production of cyclic adenosine monophosphate (cAMP), which in turn activates protein kinase A (PKA) that can phosphorylate and bind to promotor regions of DNA. Previously it has been shown that AC1/AC8 double knock out mice display hippocampal dependent LTM deficits [26], and a more recent study has shown AC8 to be required for working/episodic memory as well as the acquisition of newer spatial information in mice [19].
We also found that protein kinase C-epsilon (PKC-e) was uniquely expressed in LTM trained animals but not detected in either of the controls. PKC enzymes operate by phosphorylating various target proteins and therefore can modulate their actions. PKC epsilon has been shown to modulate Na+ channel activity and neurotransmitter release in hippocapal neurons [27]. In numerous studies the PKC family of enzymes have been shown to be necessary in various forms of synaptic plasticity, and memory formation, with the epsilon isoform required for long-term potentiation (LTP) in the mossy fiber CA3 hippocampal pathway [28]. Additionally in Lymnaea, the PKC agonist bryostatin will allow a LTM to form with a reduced training paradigm, and increase the duration of the memory formed [29].
In addition to the proteins for which a role in the memory process has already been established, we identified some whose roles are less than clear. One example is the ubiquitin like containing PHD and RING finger domains 1 binding protein (UHRF1 binding protein). UHRF1 is a protein that is involved in the maintenance and replication of global and local DNA methylation in vivo [30]. DNA methylation has been shown to be an important step in memory formation [31], and thus UHRF1 may be one of the proteins involved in mediating this alteration in the histone code that takes place when a memory is formed.
The results of our experiments would indicate that the formation of a long-term memory is a very complex process involving a number of diverse cellular systems. Most research on the subject has historically focused on the various mechanisms of cell signalling, transcriptional regulation, and protein synthesis. Our proteomic data would suggest that memory formation involves changes in a number of cellular systems not limited to; energy/cellular metabolism and ion gradient regulation, cytoskeletal organization, secretion, cell adhesion, and chromatin modification. In future experiments we hope to use this data to design experiments allowing us to test the necessity of these various systems/proteins for the memory formation process. As a brief example the chromatin modifying chromobox protein homolog 6 found to be present in only LTM trained animals, could be inhibited by RNAi prior to training to determine if memory can still be produced.
Conclusions
In this study we were able to use a quantitative proteomic approach to identify changes that occur in protein expression after an in vivo training paradigm that results in long-term memory formation.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
DR designed the study and carried out the behavioural experiments, tissue isolation, western blot experiments, data analysis, and manuscript preparation. CW performed the proteomic analysis and participated in the data interpretation and analysis, and aided in the revision of the manuscript. KL participated in the design and coordination of the project, and manuscript revision. All authors read and approved the final manuscript.
Contributor Information
David Rosenegger, Email: droseneg@ucalgary.ca.
Cynthia Wright, Email: cynthiaw@ccmp.ox.ac.uk.
Ken Lukowiak, Email: lukowiak@ucalgary.ca.
Acknowledgements
The authors would like to thank the lab of Dr. Benedikt M. Kessler (University of Oxford, UK) for their proteomic expertise, and help with this study.
References
- Parvez K, Rosenegger D, Martens K, Orr M, Lukowiak K. Canadian Association of Neurosciences Review: learning at a snail's pace. Can J Neurol Sci. 2006;33:347–356. doi: 10.1017/s0317167100005291. [DOI] [PubMed] [Google Scholar]
- Alberini CM. Transcription factors in long-term memory and synaptic plasticity. Physiol Rev. 2009;89:121–145. doi: 10.1152/physrev.00017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61:10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Bailey CH, Kandel ER, Kaang BK. Transcriptional regulation of long-term memory in the marine snail Aplysia. Mol Brain. 2008;1:3. doi: 10.1186/1756-6606-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CH, Bartsch D, Kandel ER. Toward a molecular definition of long-term memory storage. Proc Natl Acad Sci USA. 1996;93:13445–13452. doi: 10.1073/pnas.93.24.13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel T, Lattal KM. Molecular mechanisms of memory acquisition, consolidation and retrieval. Curr Opin Neurobiol. 2001;11:180–187. doi: 10.1016/S0959-4388(00)00194-X. [DOI] [PubMed] [Google Scholar]
- Cobb SR, Pitt A. Proteomics in the study of hippocampal plasticity. Expert Rev Proteomics. 2008;5:393–404. doi: 10.1586/14789450.5.3.393. [DOI] [PubMed] [Google Scholar]
- Benjamin PR, Staras K, Kemenes G. A systems approach to the cellular analysis of associative learning in the pond snail Lymnaea. Learn Mem. 2000;7:124–131. doi: 10.1101/lm.7.3.124. [DOI] [PubMed] [Google Scholar]
- Lukowiak K, Martens K, Orr M, Parvez K, Rosenegger D, Sangha S. Modulation of aerial respiratory behaviour in a pond snail. Respir Physiol Neurobiol. 2006;154:61–72. doi: 10.1016/j.resp.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Syed NI, Bulloch AG, Lukowiak K. In vitro reconstruction of the respiratory central pattern generator of the mollusk Lymnaea. Science. 1990;250:282–285. doi: 10.1126/science.2218532. [DOI] [PubMed] [Google Scholar]
- Syed NI, Ridgway RL, Lukowiak K, Bulloch AG. Transplantation and functional integration of an identified respiratory interneuron in Lymnaea stagnalis. Neuron. 1992;8:767–774. doi: 10.1016/0896-6273(92)90097-W. [DOI] [PubMed] [Google Scholar]
- Sangha S, Scheibenstock A, McComb C, Lukowiak K. Intermediate and long-term memories of associative learning are differentially affected by transcription versus translation blockers in Lymnaea. J Exp Biol. 2003;206:1605–1613. doi: 10.1242/jeb.00301. [DOI] [PubMed] [Google Scholar]
- Scheibenstock A, Krygier D, Haque Z, Syed N, Lukowiak K. The Soma of RPeD1 must be present for long-term memory formation of associative learning in Lymnaea. J Neurophysiol. 2002;88:1584–1591. doi: 10.1152/jn.2002.88.4.1584. [DOI] [PubMed] [Google Scholar]
- Sangha S, Varshney N, Fras M, Smyth K, Rosenegger D, Parvez K, Sadamoto H, Lukowiak K. Memory, reconsolidation and extinction in Lymnaea require the soma of RPeD1. Adv Exp Med Biol. 2004;551:311–318. doi: 10.1007/0-387-27023-x_47. [DOI] [PubMed] [Google Scholar]
- Lukowiak K, Ringseis E, Spencer G, Wildering W, Syed N. Operant conditioning of aerial respiratory behaviour in Lymnaea stagnalis. J Exp Biol. 1996;199:683–691. doi: 10.1242/jeb.199.3.683. [DOI] [PubMed] [Google Scholar]
- Wright CA, Edelmann M, Digleria K, Kollnberger S, Kramer H, McGowan S, McHugh K, Taylor S, Kessler BM, Bowness P. Ankylosing spondylitis monocytes show upregulation of proteins involved in inflammation and the Ubiquitin Proteasome pathway. Ann Rheum Dis. 2008;68(10):1626–32. doi: 10.1136/ard.2008.097204. [DOI] [PubMed] [Google Scholar]
- Xu D, Suenaga N, Edelmann MJ, Fridman R, Muschel RJ, Kessler BM. Novel MMP-9 substrates in cancer cells revealed by a label-free quantitative proteomics approach. Mol Cell Proteomics. 2008;7:2215–2228. doi: 10.1074/mcp.M800095-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowiak K, Sangha S, McComb C, Varshney N, Rosenegger D, Sadamoto H, Scheibenstock A. Associative learning and memory in Lymnaea stagnalis: how well do they remember? J Exp Biol. 2003;206:2097–2103. doi: 10.1242/jeb.00374. [DOI] [PubMed] [Google Scholar]
- Zhang M, Moon C, Chan GC, Yang L, Zheng F, Conti AC, Muglia L, Muglia LJ, Storm DR, Wang H. Ca-stimulated type 8 adenylyl cyclase is required for rapid acquisition of novel spatial information and for working/episodic-like memory. J Neurosci. 2008;28:4736–4744. doi: 10.1523/JNEUROSCI.1177-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Pineda VV, Chan GC, Wong ST, Muglia LJ, Storm DR. Type 8 adenylyl cyclase is targeted to excitatory synapses and required for mossy fiber long-term potentiation. J Neurosci. 2003;23:9710–9718. doi: 10.1523/JNEUROSCI.23-30-09710.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waetzig V, Herdegen T. MEKK1 controls neurite regrowth after experimental injury by balancing ERK1/2 and JNK2 signaling. Mol Cell Neurosci. 2005;30:67–78. doi: 10.1016/j.mcn.2005.06.001. [DOI] [PubMed] [Google Scholar]
- English JD, Sweatt JD. Activation of p42 mitogen-activated protein kinase in hippocampal long term potentiation. J Biol Chem. 1996;271:24329–24332. doi: 10.1074/jbc.271.40.24329. [DOI] [PubMed] [Google Scholar]
- Sweatt JD. Mitogen-activated protein kinases in synaptic plasticity and memory. Curr Opin Neurobiol. 2004;14:311–317. doi: 10.1016/j.conb.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Sharma SK, Carew TJ. The roles of MAPK cascades in synaptic plasticity and memory in Aplysia: facilitatory effects and inhibitory constraints. Learn Mem. 2004;11:373–378. doi: 10.1101/lm.81104. [DOI] [PubMed] [Google Scholar]
- Sweatt JD. The neuronal MAP kinase cascade: a biochemical signal integration system subserving synaptic plasticity and memory. J Neurochem. 2001;76:1–10. doi: 10.1046/j.1471-4159.2001.00054.x. [DOI] [PubMed] [Google Scholar]
- Wong ST, Athos J, Figueroa XA, Pineda VV, Schaefer ML, Chavkin CC, Muglia LJ, Storm DR. Calcium-stimulated adenylyl cyclase activity is critical for hippocampus-dependent long-term memory and late phase LTP. Neuron. 1999;23:787–798. doi: 10.1016/S0896-6273(01)80036-2. [DOI] [PubMed] [Google Scholar]
- Shirai Y, Adachi N, Saito N. Protein kinase Cepsilon: function in neurons. FEBS J. 2008;275:3988–3994. doi: 10.1111/j.1742-4658.2008.06556.x. [DOI] [PubMed] [Google Scholar]
- Hussain RJ, Carpenter DO. A comparison of the roles of protein kinase C in long-term potentiation in rat hippocampal areas CA1 and CA3. Cell Mol Neurobiol. 2005;25:649–661. doi: 10.1007/s10571-005-4045-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenegger D, Parvez K, Lukowiak K. Enhancing memory formation by altering protein phosphorylation balance. Neurobiol Learn Mem. 2008;90:544–552. doi: 10.1016/j.nlm.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–825. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]