Abstract
The type III secretion system (T3SS) is a macromolecular ‘injectisome’, that allows bacterial pathogens to transport virulence proteins into the eukaryotic host cell. This macromolecular complex is constituted by connected ring-like structures that span both bacterial membranes. The crystal structures of the periplasmic domain of the outer membrane (OM) secretin EscC and the inner membrane (IM) protein PrgH reveal the conservation of a modular fold among the three proteins which form the OM and IM rings of the T3SS. This leads to the hypothesis that this conserved fold provides a common ring-building motif that allows for the assembly of the variably sized OM and IM rings characteristic of the T3SS. Utilizing an integrated structural and experimental approach, ring-models for the periplasmic domain of EscC were generated and placed in the context of the assembled T3SS, providing evidence for direct interaction between the OM and IM ring components and an unprecedented span of the OM secretin.
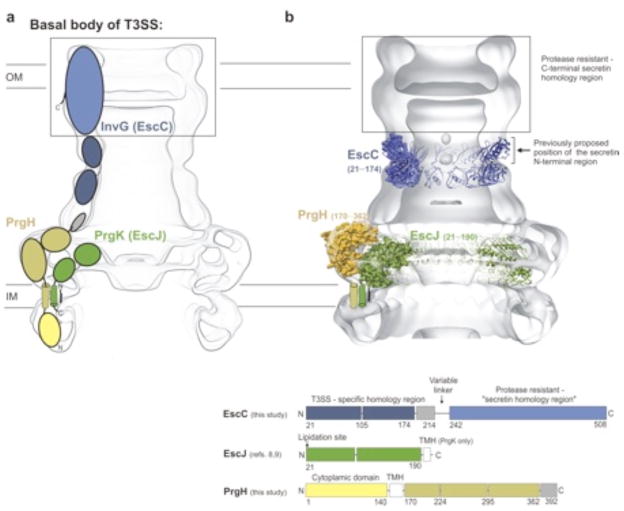
The T3SS is a multi-component macromolecular assembly essential for the virulence of many important Gram-negative bacterial pathogens1. More than 20 conserved proteins constitute the T3SS apparatus which allows for the delivery of bacterial virulence proteins directly into host cells. The structural foundation of this apparatus is composed of a connected series of variably sized ring structures and a protruding “needle” 2–7. Three conserved proteins form these homooligomeric ring assemblies (Fig. 1a): A highly conserved EscJ/PrgK/YscJ family member (from enteropathogenic E. coli (EPEC), S. typhimurium and Yersinia sp. respectively) forms the prominent IM ring 2–7. Recent studies on EscJ/PrgK including the crystal structure of oligomeric EscJ and supporting biochemical data, established a 170 Å 24-subunit ring structure positioned on the periplasmic face of the IM and anchored by an N-terminal lipidation and a C-terminal transmembrane helix (Fig. 1a) 8–10. The EscD/PrgH/YscD family of bitopic membrane proteins, most well-characterized in S. typhimurium (PrgH) consists of an N-terminal cytoplasmic domain and a larger C-terminal periplasmic domain (Fig. 1a) 1–7, 11. The C-terminal domain has been proposed to form a second distinct ring structure (of 18–24 subunits) that encompasses the adjacent EscJ/PrgK ring 2–6, 8, 10. Finally, the highly conserved EscC/InvG/YscC T3SS component, which belongs to the secretin family of proteins, forms the sole OM ring (Fig. 1a) 5–7, 12.
Figure 1.
Basal body of the T3SS. (a) Representation of the basal body of the T3SS and the components that assemble the complex (the basal body contains only the integral T3SS membrane proteins and lacks the internal stalk and extending needle of the assembled T3SS needle complex (NC)) 1–7. InvG/EscC and PrgK/EscJ are the respective orthologs from S. typhimurium and EPEC 2–9. A third prominent component of the basal body has been most well characterized in S. typhimurium (PrgH) and is thought to interact with the inner membrane ring PrgK 1–11. The coloring corresponds to the schematic in the bottom panel, illustrating the domain organization of EscC (InvG), EscJ (PrgK) and PrgH. (b) Crystal structures of T3SS basal body components EscC (21–174), PrgH (170–362) and EscJ (21–190) and their proposed position in the S. typhimurium T3SS EM map, as described here (EM accession codes emd_1224 and emd_1214 5, 21). The box in black indicates the approximate dimensions of the C-terminal protease resistant - “secretin homology region” as determined by EM reconstructions of the secretin PulD and the black arrow below highlights the previously proposed position for the secretin N-terminal region and the secretin N-termus 15, 18. The docked models are equivalent to the models presented in Figures 2 and 4.
Secretins are integral membrane proteins that function as the OM portal common to several bacterial export pathways including type II protein secretion, type III protein secretion, type IV pilus biogenesis and filamentous phage release 12–18. Despite the diversity of these systems, secretins share a remarkably similar overall structure and organization with 12–14 subunits forming a ring structure of ~110 Å in diameter 13–19. The secretin ring not only provides a channel for extrusion of bacterial proteins across the OM, but also presents a critical structural interface with other components of the various export apparatus at its periplasmic face. As such, all secretins consist of two major regions as defined by sequence analysis: an N-terminal periplasmic region and a highly conserved, protease-resistant C-terminal region (often termed the “secretin homology region”) that is embedded in the outer membrane (Fig. 1a) 12. Comparison of EM maps of the full length and protease digested PulD secretin oligomer and nano-gold labeling EM studies on the pIV secretin identified the localization and dimension of a protease-resistant C-terminal domain embedded within the outer membrane (indicated by the grey box in Figs. 1, 2 and 3) and furthermore defined the region on the periplasmic face of this protease resistant core to be occupied by the N-terminal region including the nano-gold labeled secretin N-terminus (as indicated by the black arrow in Figs. 1b and 3b) 15, 18.
Figure 2.
EscC, PrgH and EscJ share a common fold for the assembly of the T3SS multi-ring basal body. (a) Ribbon representation of the EscC (21–174) crystal structure, highlighting the two domain fold and the connecting hinge region. (b) Ribbon representation of the EscJ crystal structure (pdb accession code 1yj7 8). The top shows monomeric EscJ and the ring structure on the bottom illustrates the assembled 24mer oligomeric structure of EscJ, which has been previously described 8, 9. (c) Ribbon representation of the crystal structure of monomeric PrgH (170–362). (d) Superposition of both the outer membrane secretin EscC (21–174) (left panel) and the inner membrane protein PrgH (170–362) (right panel) to EscJ (pdb accession code 1yj7). Highlighted in orange and green are the conserved folds shared among the three proteins. The coloring scheme corresponds to the structures of EscC, EscJ and PrgH in panels (a) to (c). The schematic illustrates the integrated approach, based on the crystal structures and the observed conservation, of molecular modeling, experimental validation and docking into the EM density maps of the S. typhimurium basal body (EM accession codes emd_1224 and emd_1100) 5, 21. PrgH (170–362) was manually docked into the EM maps. The previously published EscJ structure and the EscC (21–174) models are automatically docked as described in the Methods.
Figure 3.
EscC (21–174) and the T3SS specific N-terminal secretin region within the T3SS needle complex (NC). (a) Negative Stain EM of isolated InvG complexes from S. typhimurium. A raw image and representative class averages showing the top and side view of the complex (scale bar represents 50 nm). (b) Comparison of the side view of isolated InvG to the docked EscC ring-models and the previously published EM reconstruction of the T3SS NC 5. The docked EscC (21–174) ring-models represent the ensemble of final models, which fulfill all structural and biochemical constraints and show the highest correlation to the periplasmic region of the EM density maps (EM accession codes emd_1224 and emd_1100). The docked models in orange and blue (both 12mers) are localized directly adjacent to the C-terminal protease resistant core with a notable match to the bilobal structure observed in the EM map and which also encompasses the periplasmic region previously suggested to contain the N-terminal domain as described above. Also shown is a slightly larger ring-model in green with 14fold symmetry that localizes more towards the IM rings, but for which less biochemical support currently exists. The orange and blue models are favored by existing literature and have thus been used as the representative docked EscC model shown in Figs. 1, 2 and 4.
Electron Microscopy on the assembled T3SS has considerably advanced our understanding of the overall architecture of the T3SS, but the complexity and membrane spanning nature of this macromolecular assembly has hampered structural characterization of the molecular details, essential to further investigate this clinically important apparatus. The integrated approach described here, of combining X-ray crystallography, electron microscopy, molecular modeling, in vivo analysis and site-directed mutagenesis allowed us to begin a process of piecing together a structural picture of the T3SS basal body.
Results
Crystal structures of EscC (21–174) and PrgH (170–362)
In the first stage of our analysis we have determined the crystal structures of the periplasmic domains of two integral T3SS basal body components: (1) the OM secretin EscC from EPEC and (2) the IM protein PrgH from S. typhimurium (Fig. 1b). The crystal structure of the OM secretin EscC (21–174) monomer reveals a modular fold of two small domains (domain 1 (blue): residues 21–104 and domain 2 (orange): 106–174) that are connected by a linker centered at residue 105 (Fig. 2a). The two domains are tilted towards each other and form a prominent hydrophobic interface, comprised of a number of aromatic residues. Structure-based sequence analysis of EscC (21–174) and other T3SS species variants reveals a conservation of residues within the core of the two domains as well as a conservation of both the linker and the interface of the two small domains, implying that the fold of the N-terminal region of T3SS-specific secretins is highly analogous (Supplementary Fig. 1). The structure of the IM PrgH (170–362) monomer, illustrated in Figure 2c, also exhibits a modular architecture of 3 topologically similar α/β domains (domain 1 (yellow): residues 170–224, domain 2 (green): 225–295 and domain 3 (orange): 296–362) that are connected by short linkers. These modular domains together fold into a “boot”-shaped structure (Fig. 2c).
EscC, PrgH and EscJ share a common fold
Despite a complete lack of detectable sequence identity, a comparative structural analysis of the IM PrgH (170–362) reveals striking similarity to the other IM ring component EscJ/PrgK as well the OM ring component EscC (21–174) (Fig. 2) 8. Superposition of EscC (21–174), PrgH (170–362) and EscJ shows that the three proteins that constitute the T3SS basal body rings share a similar modular domain architecture with particular resemblance in the domains shown in orange and green in Figure 2 (Supplementary Fig. 2). The domains of EscC and EscJ depicted in orange superimpose with a Dali Z-score of 4.8; the domains of PrgH and EscJ illustrated in green superimpose with a Dali Z-score of 4.9; the orange colored domain of PrgH superimposes with both EscC and EscJ with lower structural homology (Z-scores of 3.9 and 3.2 respectively). These particular domains exhibit an analogous wedge-shaped structure with two α-helices folding against a β-sheet. In EscJ these domains mediate inter-subunit interactions, assembling into a 24-subunit homo-oligomeric symmetrical structure (Figs. 1b and 2b) 8, 9. This striking similarity in fold and architecture between the three proteins suggests the conservation of a wedge-shaped fold that potentially provides a common ring-building motif for the assembly of the symmetrical ring structures that constitute the T3SS basal body. At the same time, the sequence variation between the 3 individually conserved ring family proteins allows for unique differences that mediate formation of rings with varying oligomerization number, overall size and distinct surface features necessary for T3SS assembly and function.
Prediction of EscC (21–174) ring structures
To investigate this appealing hypothesis we have used a new approach for the modeling of symmetrical assemblies to predict the ring structure of the OM secretin EscC (21–174) (see Methods for details) 9. Even though EscC (21–174) was, unlike the IM EscJ, not captured in an oligomerized state within our crystals it can be assumed to adopt a symmetrical ring structure in the assembled secretin, since nano-gold labeling and EM visualization of the homologous pIV secretin revealed a symmetrical ring-like arrangement of the pIV N-terminus at the periplasmic face of the secretin oligomer 18. It is also not surprising that the isolated EscC (21–174) N-terminal fragment does not oligomerize in the absence of the C-terminal transmembrane region as this latter domain of the secretin, as with many outer membrane proteins, has been shown to drive the oligomerization appropriately within the membrane milieu. Our molecular modeling is based on the assumption of 12–14 secretin subunits constituting the symmetrical ring structure. Although the relative stoichiometry of the different T3SS components (InvG/EscC: PrgK/EscJ: PrgH/EscD) is controversial in the field, the stoichiometry of the OM secretin is well established and 12 to 14 subunit arrangements have been consistently observed in a number of studies on various secretins 13–19.
The short linker connecting the two EscC domains (centered around residue 105) was treated as flexible in the modeling approach to account for possible conformational variability of the two domains towards each other. This molecular modeling approach led to a set of energetically and structurally plausible ring-models with an overall bilobal appearance (see Methods for details).
To evaluate the biological relevance of each of the potential ring-models, we performed limited biotinylation experiments on extracted S. typhimurium T3SS needle complexes (NCs, which contain the basal rings of the inner and outer membrane as well as the connecting rod and protruding needle filaments) to specifically identify accessible and buried lysine residues of the assembled secretin within the intact NC (Supplementary Fig. 3). This information was used in conjunction with our additional site directed mutagenesis data to evaluate the generated ring-models (see Methods and Supplementary Figs. 4–6 for detailed information on the evaluation and filtering process). This procedure led to an ensemble of ring-models for EscC (21–174) that satisfy all structural and experimental constraints, demonstrating the observed ring-building motif has the potential to mediate ring formation.
The N-terminal secretin domain in the context of the T3SS
To understand the relevance of these models, we first attempted to better delineate the boundaries of the N- and C-terminal secretin regions within the context of the published cryo-EM maps of the S. typhimurium T3SS NC. Our analysis was based on analogy with the previously proposed position for the C-terminal protease resistant core and the secretin N-terminus, deduced from a combination of proteolysis, EM and nano-gold labeling on the PulD and pIV secretin, as described above (Fig. 1). Importantly, our tryptic digestion of both the isolated secretin InvG and fully assembled T3SS NCs from S. typhimurium followed by mass spectrometry, identified an equivalent C-terminal domain fragment of InvG with residue K269 demarking the boundary of the protease resistant C-terminal secretin homology region (data not shown), showing the comparability of the domain organization and exposure of a T3SS secretin in its isolated and NC assembled forms. Sequence comparison shows, that this position directly aligns with the trypsin cleavage site of PulD, thereby confirming the equivalence of the C-terminal protease resistant core of PulD and InvG (Supplementary Fig. 7). Furthermore, our averaged negative stain EM analysis of purified InvG complexes from S. typhimurium emphasizes the very similar overall structure of the T3SS secretin InvG to PulD and pIV (Fig. 3 and Supplementary Fig. 8) 15, 18. Collectively, these results indicate the relative boundaries of the N- and C-terminal regions of the secretin within the assembled T3SS EM mask as shown in Figure 1.
Docking of EscC (21–174) ring-models into the T3SS NC
Guided by this information, the ensemble of experimentally validated EscC (21–174) ring-models were then docked to the cryo-EM maps of the S. typhimurium T3SS basal body and NC using the automated EM map fitting algorithm COLORES of the SITUS program package and analyzed for overall complimentarity to the periplasmic density regions between the defined C-terminal secretin core and the IM rings (Fig. 3b) 5, 20. Figure 3b illustrates a representative subset of the validated final models resulting from this procedure (Supplementary Fig. 8). The docked models (both 12mers) shown in orange and blue in Figure 3b are localized directly adjacent to the C-terminal protease resistant core with a notable match to the bilobal structure observed in the EM map and which also encompasses the periplasmic region previously suggested to contain the N-terminal domain as described above (the model in orange was chosen to represent the subset of potential ring-models in Figs. 1, 2 and 4) 15, 18. Although we favor this localization of the docked ring model because of its compatibility with existing data from Type III and other secretin families, we acknowledge that the limited effective resolution of the molecular modeling approach and the EM maps to which we are fitting the models limits definitive conclusions based on orientation, atomic details and localization. For example, we note in Figure 3b an alternative generated model, albeit of slightly higher oligomerization number and size (14mer), localized optimally to a distinct region of the periplasm (in an unassigned region of EM density towards the IM components PrgH and PrgK). What is unequivocal from our data however, is that regardless of the predicted possible localizations, our experimental structures and accompanying biochemical data clearly defines that the span of the N-terminal secretin region must be substantially further into the periplasm then previously understood and which suggests a close interaction of the OM secretin and the IM components (Fig. 3 and Supplementary Fig. 8).
Figure 4.
EscC (21–174) and the T3SS specific N-terminal secretin region within the T3SS needle complex (NC). (a) Representative EscC model in the EM density maps of the S. typhimurium basal body and the fully assembled T3SS NC containing in addition the stalk and needle, which are built up by the proteins InvJ, PrgJ, PrgI (EM accession codes emd_1224; emd_1100 5, 21; the docked EscC model is equivalent to the model shown in orange in Figures 3b). The blue bar highlights the important region that undergoes considerable structural rearrangement during the NC assembly process 5, 21. (b) Effects of EscC mutations on type III secretion in EPEC. Shown are the Tir/EspB secretion profiles in secretion assays of escC-deficient EPEC strains complemented with the EscC mutants. The proteins were detected with respective monoclonal antibodies. (c) Effects of InvG mutation on type III secretion in Salmonella. Secreted proteins were separated by SDS-PAGE and stained with Coomassie Blue from wildtype and chromosomal InvG (E106P) S. typhimurium strains. Western-blots of secreted proteins were performed with respective monoclonal antibodies. Needle Complexes (NC) were isolated, separated on SDS-PAGE and stained with Coomassie (molecular weight reference shown in kDa). Levels of InvG in NC were detected with a polyclonal antibody.
Importance of the linker in EscC (21–174) and InvG (21–175)
Specifically, the EscC ring-models are localized to a region of the NC EM maps, which in S. typhimurium has been implicated to accommodate factors that modulate assembly and regulation of the needle filament 5, 21. During the assembly process of the T3SS NC and the insertion of the needle components, considerable structural rearrangement in this region have been observed by EM (Fig. 4a shows a comparison of the EM densities of the T3SS basal body and the fully assembled NC containing the needle and the connecting stalk) 5, 21. We propose these rearrangements are mediated by changes in relative disposition between the two flexibly linked domains we observe in the EscC (21–174) structure. Interestingly, the recently published crystal structure of the octameric outer membrane export protein Wza shows a similar modular architecture of flexibly linked symmetrical rings that constitute a large periplasmic pore structure (Supplementary Fig. 9) 22. This modular architecture of ring structures connected by conserved linker regions implies that both of these elements are important requirements for the function of the assembled oligomeric complex and it is tempting to speculate that this architecture might facilitate structural rearrangements in the fully assembled complex through variations in the connecting linker and resulting conformational changes between the rings.
In order to address this hypothesis and its functional implications on T3SS we performed site directed mutagenesis in EPEC and S. typhimurium on the residue E105 and E106, respectively, which is positioned in the linker connecting the two small N-terminal domains (21–104 and 106–174) and is furthermore conserved in all T3SS specific secretins (Fig. 4a). Analysis of escC-deficient EPEC complemented with either wild type EscC or mutant EscC showed that a proline mutation (E105P) completely abolished type III secretion while the corresponding alanine mutation (E105A) did not cause substantial secretion defects, suggesting that a conformational rather than electrostatic requirement is critical for linker function (Fig. 4b). The equivalent chromosomal mutant of InvG (E106P) in S. typhimurium also abolished type III secretion, confirming the importance of the conformation of the N-terminal domains in T3SS function (Fig. 4c). To investigate this further, NCs were prepared from this mutant and analyzed. The results revealed that oligomeric InvG was present at a slightly reduced level in NC preparations and the amounts of the core inner membrane ring components PrgK and PrgH were severely depleted (Fig. 4c). To test whether the decrease in inner membrane components resulted from changes in amounts and/or localization of InvG, we assessed the cellular and membrane levels of the linker mutant secretin. InvG (E106P) showed similar cellular and membrane levels to wildtype InvG, indicating that abundance and localization remained unaltered (data not shown). Additionally, protein levels and localization of the IM PrgH also remained similar (data not shown), indicating that abundance and localization of PrgH is not affected by InvG (E106P). Cumulatively, these results indicate that the proline mutation prevents proper T3SS assembly at the stage of basal body formation. This observation highlights the importance of the conformation of the T3SS N-terminal secretin domains for its function within the T3SS and implies a potential role of the secretin and its N-terminal region in stabilizing the precursor inner membrane ring structures during T3SS assembly.
Structure and localization of PrgH (170–362)
The predicted span and disposition of the secretin EscC N-terminal region within the T3SS is furthermore in agreement with the high resolution structure of the periplasmic domain of PrgH, determined here: The bitopic inner membrane protein PrgH is localized to the inner membrane by a single transmembrane helix (residues 141–165) and the C-terminal periplamic domain of PrgH (residues 166–392) has been shown to form a complex with PrgK (Fig. 1a) 4, 8. Our structure of PrgH (170–362) from S. typhimurium exhibits a distinctive “boot”-shape which was consistently observed in 12 individual copies of the protein in the asymmetric units of our crystal structures. The “boot”-shape shows a compelling complementarity to the expected localization in the EM map and the orientation of the docked structure accommodates the predicted position of the N-terminal transmembrane helix (residues 141–165) (Fig. 1 and Fig. 2c). The docked model furthermore explains the previously observed solvent inaccessibility of both PrgK and PrgH in the assembled T3SS NC, with PrgH encompassing PrgK 4, 8. The structures of PrgH (170–362), EscJ (24–190) and EscC (21–174) thus account for the majority of the observed periplasmic EM density and suggest a close disposition of the outer and inner membrane T3SS rings (Fig. 1).
Discussion
The crystal structures of the essential T3SS proteins EscC (21–174) and PrgH (170–362) reveal an unpredicted conservation of a wedge-shaped motif between the outer and inner membrane components of the multi-ring T3SS basal body (Fig. 2). For the IM protein EscJ this conserved fold has previously been shown to assemble into a symmetrical ring structure that constitutes part of the IM T3SS rings. The striking conservation of this modular fold between all three proteins that form the T3SS basal body suggests that the topologically similar, but sequence-diverse folds provide a common ring-building motif that allows for the assembly of the variably sized OM and IM rings characteristic of the T3SS.
Earlier nano-gold labeling studies of the intact pIV secretin labeled at its N-terminus revealed a ring-like arrangement 18. Although, like other outer membrane proteins, the driving force for oligomerization is derived from the membrane spanning C-terminal secretin homology domain 19, 23, the nanogold study clearly implies an ordered ring structure for the N-terminal domain as well in the context of full length secretin. Using molecular modeling, limited biotinylation, protease digestion, electron microscopy and site-directed mutagenesis we have generated experimentally validated ring-models for the secretin N-terminal domain and demonstrated that the conserved motif has the potential to mediate ring formation. The ring-models have in a subsequent analysis been placed in the context of electron microscopy maps of the T3SS needle complex, resulting in an ensemble of experimentally validated final ring-models within the assembled T3SS (Fig. 3).
Despite the limitations, our integrated approach allows for the prediction of the highly conserved and functionally essential N-terminal secretin domain within the assembled export system, a domain which has previously been shown to be difficult to study by conventional means (Supplementary Fig. 8) 24. Furthermore it reveals the relative span of the T3SS secretins within the NC and implies direct interactions of the OM secretin and the IM membrane components. Together with the crystal structure of the complete periplasmic domain of PrgH, determined here, our analysis provides a new structural framework for functional studies on the T3SS.
Comparison to the octameric transport protein Wza, which forms the OM portal of the Wza-Wzc capsular polysaccharide secretion system, supports the relative periplasmic location and span of the EscC models in the T3SS and moreover emphasizes a similar modular architecture of small wedge-shaped periplasmic domains facilitating the assembly of connected symmetrical rings that constitute large pore structures (Supplementary Fig. 9) 22. The observed modular architecture of ring structures connected by conserved linker regions implies that both of these elements are important requirements for the function of the assembled oligomeric complex. It is tempting to speculate that this conserved architecture might facilitate structural rearrangements in the fully assembled complex through variations in the connecting linker and resulting conformational changes between the rings. In support of this hypothesis introduction of a proline residue into the connecting linker markedly reduced assembled NCs while essentially maintaining oligomeric InvG. Since no inner membrane components associated with the mutant InvG oligomer, this data supports the hypothesis that binding of other components of the export apparatus to the secretin, like the inner membrane base components PrgH/PrgK or the regulatory factors InvJ/PrgJ/PrgI, is required for assembly and oligomerization of inner membrane needle complex components and subsequently the intact T3SS. Therefore, rings linked by flexible elements may be a common theme in providing the necessary regulation to perform complex assemblies of apparati that deliver substrates to the extracellular space.
During the final stages of this manuscript the crystal structure of the periplasmic domain of the type II secretion system secretin GspD from enterotoxigenic E.coli (ETEC) was published 25. Although the N-terminal regions of secretins from different export systems are only distantly related the structure and architecture of EscC and GspD are surprisingly similar and seem to be conserved for all secretins. The GspD structure resembles the modular architecture of conserved α/β folds connected by flexible linkers and moreover reveals the same conserved structural motif, which we have identified here as a common ring-building domain for the assembly of modular ring structures. This striking similarity in fold and architecture of the distantly related N-terminal secretin region of type II and type III secretins supports our hypothesis of a conserved motif mediating the formation of the secretin rings and flexibly linked ring structures as a potentially common theme.
Methods
Protein expression, purification and crystallization
Cloning, protein expression and purification was carried out using standard techniques as described previously 8. EscC (21–174) crystallized in microbatch under the conditions 2.0 M Ammonium sulfate + 0.1 M sodium acetate pH 4.5 or Bis-Tris pH 6.5. PrgH (170–392) crystals were obtained in 16% (w/v) PEG 3350 + 0.2 M tri-ammonium citrate + 0.1M Tris pH 8.0. and PrgH (170–362) crystallized in an orthorhombic (2.8M NaCl + 0.1M Tris pH 8.5 + 0.01M cupric chloride) and a trigonal crystal form (30% (w/v) PEG 1500 + 0.1M lithium sulfate + 0.1M Tris 8.5 + 0.109M octanoyl-n-hydroxyethylglucamide).
Structure determination of EscC (21–174), PrgH (170–362) and PrgH (170–392)
The crystal structures of EscC (21–174) and PrgH (170–362) were determined by single anomalous dispersion (SAD) at the selenium edge using crystals of the selenomethionine incorporated proteins. The structure of PrgH (170–392) was subsequently solved by molecular replacement. Data collection and refinement statistics are summarized in Table 1 and detailed in the accompanying Supplementary methods.
Table 1.
Data collection, phasing and refinement statistics for EscC (21–174), PrgH (170–362) and PrgH (170–392)
| EscC (21–174) | PrgH (170–362) | PrgH (170–362) | PrgH (170–392) | |
|---|---|---|---|---|
| SeMet (peak) | SeMet (peak) | native | native | |
| Data collection | ||||
| Space group | P6522 | P21212 | P31 | C2221 |
| Cell dimensions | ||||
| a, b, c (Å) | 90.6, 90.6, 133.4 | 87.0, 106.6, 55.5 | 53.5, 53.5, 282.5 | 79.6, 188.7, 305.6 |
| Resolution (Å) | 2.05 (2.05–2.1) * | 2.8 (2.8–2.87) | 2.3 (2.3–2.36) | 2.8 (2.8–2.87) |
| Rmerge | 0.072 (0.501) | 0.074 (0.275) | 0.037 (0.137) | 0.069 (0.293) |
| I/σI | 31.9 (5.9) | 29.4 (9.9) | 20.6 (5.5) | 27.5 (4.2) |
| Completeness (%) | 99.9 (99.9) | 100 (100) | 94.1 (71.0) | 96.8 (81.4) |
| Redundancy | 20.9 (19.6) | 13.9 (14.4) | 3.1 (2.0) | 11.5 (5.2) |
| Refinement | ||||
| Resolution (Å) | 2.05 | 2.3 | 2.8 | |
| No. reflections | 20941 | 39588 | 52376 | |
| Rwork/Rfree | 0.215/0.251 | 0.206/0.255 | 0.249/0.295 | |
| No. atoms | ||||
| Protein | 1237 | 6081 | 12624 | |
| Water | 77 | 231 | - | |
| B-factors (Å2) | ||||
| Protein | 41.6 | 23.2 | 54.16 | |
| Water | 43.3 | 33.4 | - | |
| R.m.s deviations | ||||
| Bond lengths (Å) | 0.032 | 0.013 | 0.014 | |
| Bond angles (°) | 2.10 | 1.33 | 1.55 | |
Values in parentheses are for highest-resolution shell.
Computational methodology
The prediction of the oligomeric structure of EscC and homology modeling of InvG was performed using the computational modeling software rosetta 26. It was assumed that EscC adopts a ring structure with cyclical symmetry consisting of 12–14 subunits. The protocol for symmetrical protein-protein docking using rosetta has been previously described 9, 27. The described protocol was modified to allow backbone flexibility in the hinge region of EscC. Two variants were used, one aggressive and one conservative. In the aggressive variant the conformational space of the linker is explored using fragment insertion in the linker region (residue 102–108) where backbone conformations are generated with a simulated annealing protocol as used in ab-initio structure prediction with rosetta 26. The monomeric structure generated by the fragment insertion is used as a starting point for the oligomer-assembly protocol described above. In the coarse-grained search part not only the rigid-body orientation but also the linker backbone is optimized. This is achieved by adding torsion angle perturbations (small moves) in the Monte Carlo search. A torsion angle perturbation consists of φ and ϕ changes of up to 1.8° in the hinge region. Before the high-resolution refinement step the residues in the linker region are repacked. The high-resolution Monte Carlo minimization consists of three perturbations: small, rigid- body and shear moves. Shear moves consists of random φ and ϕ perturbations of up to 2° in helix or strand regions or 3° in loop regions and the preceding ϕ is perturbed by the same amount of degrees in the opposite direction to produce a compensatory shear motion in the peptide plane. The conservative involves the same steps as the aggressive protocol but excludes the fragment insertion. In all simulations described so far symmetry is maintained for rigid body, backbone and side-chain degrees of freedom throughout at all stages of the run.
Several types of simulations with different protein constructs were used to create models of the oligomeric structure of EscC. In the most conservative approach the backbone conformation of EscC was kept in the crystallographically determined state. Backbone flexibility was introduced by allowing the backbone to adjust during the simulation using the hinge refinement protocol described above. When large-scale conformational changes in the hinge region were sampled a limited number of initial binding modes were explored. These were selected by running rigid-body docking simulations of the N-terminal domain (residue 21–101) and clustering the structures generated by this procedure. The clusters with largest number of members were selected and simple filters were used to reduce the number of starting binding modes. These filters require for example that the hinge region does not point into the binding interface region of the next subunit. Three different starting orientations of the domain 1 was sampled in the fragment insertion protocol (described above) devised to sample large-scale conformational changes in the linker. Models generated starting from a random rigid-body conformation protocols was further refined by the hinge refinement protocol. Models were selected first on the number of contacts in the interface to identify models that are well packed and this subset was ranked based on Rosetta full-atom energy. For each run, the oligomerization state was fixed in the range 12–14 and 5×103–3×104 structures were generated. The model identified as the most probable based on the fit with experimental data has a monomeric structure that is very similar to the crystal structure (0.4 Å backbone RMSD) and was generated after a hinge refinement run with relatively small conformational changes relative the native structure. Figure 5 illustrates the conformational search space generated by hinge refinement protocol and the effect of selection of models with large number of interface contacts and low energy. Figure 5a was generated by superposition of single subunits extracted from the docking models superimposed onto a specific orientation of the domain 1 and illustrate how the conformational space is reduced by adding more energetic and experimental constraints. Figure 5b shows the result of symmetrical docking of domain 1 as 12mer rings. The plot shows the energy vs. backbone RMSD for three consecutive subunits relative the model selected as best fitting with the experimental data. The result shows that a large number of conformations are sampled during docking of the N-terminal ring and illustrates that the docking of the N- and C-terminal domains cannot be treated as independent problems in order to generate the model that best fits the experimental data since the very low rmsd values (below 2 Å) are not sampled by this procedure. Although the orientation found in the model for the full protein construct that best fits with the experimental data is not the lowest energy conformation, the lowest energy models are found in the vicinity of this model.
Figure 5.
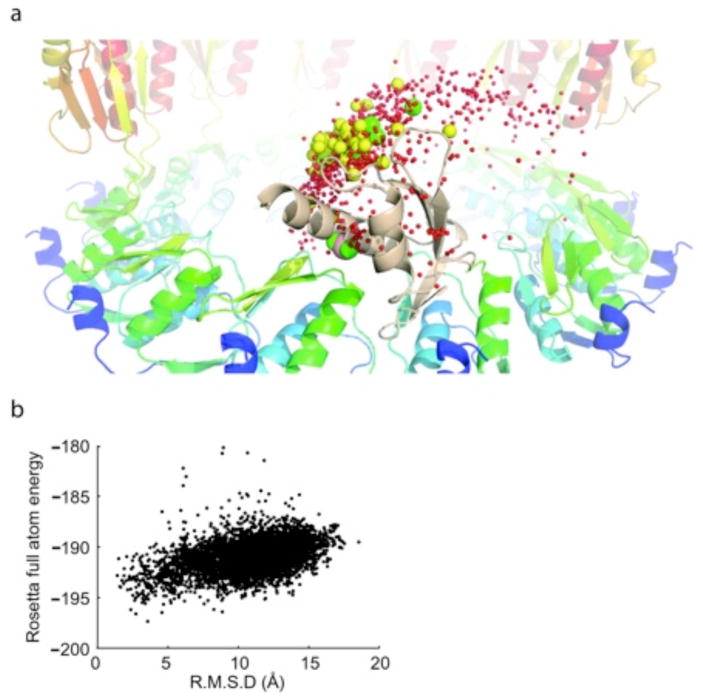
Illustration of conformational sampling during protein-protein docking simulations. (a) Conformational sampling of the hinge region (residue 102–108) of EscC. The position of the α-carbon of residue 162 in a random selection of docking models after a hinge perturbation protocol is shown in red spheres. The yellow spheres indicate a subset of docking models with more than 300 contacts across an interface and green the 10% lowest energy subset of the yellow spheres. The wheat colored domain 2 correspond to the orientation of the model best fitting the experimental data and the magenta sphere is the α-carbon of residue 162 of this model. When a smaller sized hinge-perturbation is used spheres clusters around the magenta sphere, which roughly coincides with the position of the residue 162 in the crystal structure. In order to pick out the best models out of the green ensemble (and correspondingly in other simulations), geometric information and biotinylation data was used as a filter, see Figure 6. (b) Symmetrical docking of the N-terminal EscC domain (21–101) into 12mer ring models. The plot shows the Rosetta full atom energy vs backbone rmsd for three consecutive subunits relative the model selected as best fitting with the experimental data.
The crystallized portion of EscC (residue 21–174) does not oligomerize on its own in solution and is expected to be weakly interacting in isolation. In addition, it is likely that the crystal structure will adopt a slightly different backbone conformation in the context of the intact macromolecular complex and we have explored such conformational flexibility by allowing the linker to adjust. Due to the relatively weak interactions and conformational flexibility a strong energy signal is not expected for a near native conformation. To identify an ensemble of feasible models a set of evaluation criteria was used: The N-and C-termini must point to solution and be able to connect to the rest of the EscC structure and the dimensions of the ring must fit at least one portion of the region of the cryo-EM map that can be attributed to the secretin. Finally, the models were filtered by how well they match experimentally determined biotinylation data reporting on the exposure of lysine residues on the assembled secretin within the NC (Fig. 6).
Figure 6.
Illustration of the evaluation process of the ring-models. The predicted models (Fig. 5) are further discriminated by experimentally derived data of accessible and buried lysine residues, identified by limited biotinylation (Supplementary Fig. 3). The models in panels a – f show a representative subset of models to illustrate the process in detail. Solvent accessible residues are rendered in green and buried in yellow spheres. In addition we utilized site-directed mutagenesis data on two negatively charged patches of EscC that show defects in type III secretion upon mutation (residues in orange spheres) (Supplementary Fig. 4). The illustrated process identifies a subset of models that all satisfy both the modeling/structural constraints and the experimental data (the models a, b, c, d and e are representative for the identified subset; the model f can be excluded because it does not meet the experimental data). This ensemble of validated models was further evaluated by docking to the EM density maps of the T3SS basal body and NC from S. typhimurium 5, 21. This process led to a set of final models that fulfil all structural/modelling, experimental and docking constrains. In the representative subset above only models a and b show considerable complementarity to the periplasmic regions of the EM density maps and satisfy all imposed criteria (Supplementary Fig. 8). These two models and their docking positions are equivalent to the models described in Figure 3b.
The accuracy of the proposed models is dependent on the resolution of the experimental data and the correctness of the assumptions about the degree of flexibility which must be incorporated to go from the crystallographically determined conformation to the conformation in the intact NC. Given the assumptions about flexibility described above, the experimental data pick out a limited ensemble of low energy models, which are the solution to the modeling problem as posed. It must be recognized that if our assumptions about flexibility are incorrect and there are much larger conformational changes in forming the intact NC, these models could be considerably in error.
Since the experimental biotinylation was performed on the T3SS NCs from S. typhimurium and therefore the secretin ortholog InvG, a homology model of InvG was generated from the structure of EscC in order to interpret the biotinylation data (Supplementarty Fig. 5). The general protocol has been previously described 28. The sequences of EscC and InvG were aligned using clustalW and only regions with loop secondary structure in EscC were rebuilt 29.
Docking of oligomeric EscC models and EscJ into EM density maps
The ensemble of validated symmetrical ring-models of EscC (21–174) were docked into the published EM density maps of the S. typhimurium T3SS needle complex (NC) and basal body (accession codes emd_1100, emd_1224 and emd_1214 respectively), using the automated FFT Accelerated 6D Exhaustive Search protocol implemented in the program COLORES of the SITUS package 5, 20. The docked models were analyzed for overall complimentarity to the periplasmic density regions between the defined C-terminal secretin core and the IM rings. Figure 3b illustrates a representative subset of the validated final models resulting from this procedure. We acknowledge that the 20 fold symmetry imposed on the only available cryoEM map of the S. typhimurium NC may alter the observed surface features in the docking process. However, the overall dimensions and shape of this region, the main structural constraints for the docking procedure in addition to the experimentally derived constraints, will not be substantially altered. Rotational differences of the map were not considered in the docking procedure.
The previously published EscJ ring model was docked in the same manner (pdb accession code 1yj7). The structure of monomeric PrgH (170–362) was manually docked into the EM maps (Figs. 1 and 2). The docked models were analysed in Pymol and Chimera 30, 31.
Purification of InvG complexes from S. typhimurium
InvG was expressed in a S. typhimurium prgH deletion strain under conditions previously described 2. The cells were lysed in the presence of 20% (w/v) sucrose, 1mM EDTA, lysozyme and 0.4% (w/v) CHAPS. Oligomeric InvG was extracted from the collected membranes in 1% (w/v) n-dodecyl-β-D-maltoside (DDM) buffer and further purified in DDM by Anion- and Cation- exchange chromatography. The purifed complex was finally subjected to gelfiltration in either DDM or n-octyl-β-D-glucoside (βOG).
Electron Microscopy and Image Processing
Samples were prepared by conventional negative staining with 0.75% (w/v) uranyl formate as described previously 32. Using BOXER, the display program associated with the EMAN software package 33, 4,591 particles were interactively selected from 88 images of InvG. Individual particles were windowed into 80 × 80 pixel images. Using the SPIDER software package 34, the particles were rotationally and translationally aligned and subjected to 10 cycles of multireference alignment. Each round of multireference alignment was followed by k means classification specifying 50 output classes. The references used for the first multireference alignment were randomly chosen from the raw images.
Trypsin proteolysis of purified S. typhimurium NCs
Purified NC was incubated with sequencing grade modified trypsin (Sigma) at 100:1 mass:mass for 16h at 22°C. The reaction was stopped with SDS sample buffer. The reaction mixture was run on a 12% (w/v) SDS PAGE gel and unique bands were excised and incubated with trypsin. The peptides were analysed by LC MS on a QTOF instrument.
Surface accessibility analysis of S. typhimurium InvG secretin component of NC with limited biotinylation and matrix assisted laser desorption ionization-time offlight (MALDI-TOF) MS
Purified NC were biotinylated and analyzed as described elsewhere 8.
Mutagenesis, complementation, and secretion assays in EPEC
Secretion assays were performed on these complemented strains and wild type EPEC as previously described 35.
Strain construction and bacterial growth conditions for S. typhimurium
S. typhimurium strains ATCC 14028s and NC overexpressing strain TK385 were used for secreted protein analysis and NC purification, respectively 2. Salmonella strains were grown in Luria-Bertani (LB) medium and LB supplemented with 300 mM sodium chloride for NC purification. Bacterial strains were constructed using the λ-RED system 36.
Secreted protein and S. typhimurium NC protein component analysis
Secreted proteins were prepared for S. typhimurium as described previously 2. NC’s were purified as described previously 2. Protein samples were run on SDS-PAGE and Western blotted as described 37.
Supplementary Material
Acknowledgments
We thank the staff at the Advanced Light Source beamlines 8.2.2 and 8.3.1 for data collection time and assistance. C.K.Y. thanks the Natural Science and Engineering Research Council of Canada and the Michael Smith Foundation of Health Research. I.A. is supported by the Knut and Alice Wallenberg foundation. S.S. thanks the Natural Sciences and Engineering Research Council of Canada for postdoctoral funding. N.C.J.S. and B.B.F. thank the Howard Hughes International Scholar program and the CIHR for funding. N.C.J.S. also thanks the MSFHR and CFI for infrastructure funding support. N.C.J.S. is a MSFHR Senior Scholar and CIHR Investigator and B.B.F is the Peter Wall Distinguished Professor. D.B. is an HHMI investigator. This work was supported by the NHI grant P20 GM076222 to D.B and by grants from the NIH U54 AI05741 and RO1 AI030479 to S.I.M. The authors would also like to thank Tom Walz (Harvard Medical School, USA) for generously providing resources for the electron microscopy measurements. E.E. Galyov (University of Leicester, UK) generously provided the SipA, SipB and SipC antibodies while V. Koronakis (University of Cambridge, UK) generously provided the InvG antibody. The authors also thank Heather Felise, Scott Schafer, and Dave Goodlett for technical advice and assistance.
Footnotes
Author Contributions
C.Y., M.V. and A.Y. cloned, purified and crystallized the EscC and PrgH constructs and C.Y. determined the crystal structures. The molecular modeling was carried out by I.A. and the models were analyzed, interpreted and docked into the EM maps by T.S. and I.A. S.S. did the biochemical characterization in S. typhimurium and W.D. performed the experiments in EPEC. T.S. established the purification protocol for InvG. T.S. and N.C.J.S. wrote the paper, I.A. prepared Fig. 5 and the computational methods, S.S. prepared Fig. 4c and Supplementary Fig. 3 and W.D. Fig. 4b and Supplementary Fig. 4. All authors discussed the results and commented on the manuscript. D.B. supervised all computational analysis. S.I.M. and B.B.F. provided resources for the biochemical characterizations in S. typhimurium and EPEC respectively. N.C.J.S. provided resources for all data other than Figs. 4b,c and 5 and Supplementary Figs. 3,4.
Accession codes
The structure factors and atomic coordinates for EscC (21–174), PrgH (170–362) and PrgH (170–392) have been deposited in the Protein Data Bank with accession codes 3GR5, 3GR0 and 3GR1 respectively.
References
- 1.Ghosh P. Process of protein transport by the type III secretion system. Microbiol Mol Biol Rev. 2004;68:771–795. doi: 10.1128/MMBR.68.4.771-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kimbrough TG, Miller SI. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc Natl Acad Sci U S A. 2000;97:11008–11013. doi: 10.1073/pnas.200209497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sukhan A, Kubori T, Wilson J, Galan JE. Genetic analysis of assembly of the Salmonella enterica serovar Typhimurium type III secretion-associated needle complex. J Bacteriol. 2001;183:1159–1167. doi: 10.1128/JB.183.4.1159-1167.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kimbrough TG, Miller SI. Assembly of the type III secretion needle complex of Salmonella typhimurium. Microbes Infect. 2002;4:75–82. doi: 10.1016/s1286-4579(01)01512-x. [DOI] [PubMed] [Google Scholar]
- 5.Marlovits TC, et al. Structural insights into the assembly of the type III secretion needle complex. Science. 2004;306:1040–1042. doi: 10.1126/science.1102610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blocker A, et al. Structure and composition of the Shigella flexneri “needle complex”, a part of its type III secreton. Mol Microbiol. 2001;39:652–663. doi: 10.1046/j.1365-2958.2001.02200.x. [DOI] [PubMed] [Google Scholar]
- 7.Ogino T, et al. Assembly of the type III secretion apparatus of enteropathogenic Escherichia coli. J Bacteriol. 2006;188:2801–2811. doi: 10.1128/JB.188.8.2801-2811.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yip CK, et al. Structural characterization of the molecular platform for type III secretion system assembly. Nature. 2005;435:702–707. doi: 10.1038/nature03554. [DOI] [PubMed] [Google Scholar]
- 9.André I, Bradley P, Wang C, Baker D. Prediction of the structure of symmetrical protein assemblies. Proc Natl Acad Sci U S A. 2007;104:17656–17661. doi: 10.1073/pnas.0702626104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sani M, et al. Structural organization of the needle complex of the type III secretion apparatus of Shigella flexneri. Micron. 2007;38:291–301. doi: 10.1016/j.micron.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 11.Pallen MJ, Beatson SA, Bailey CM. Bioinformatics analysis of the locus for enterocyte effacement provides novel insights into type-III secretion. BMC Microbiol. 2005;5:1–21. doi: 10.1186/1471-2180-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Genin S, Boucher CA. A superfamily of proteins involved in different secretion pathways in gram-negative bacteria: modular structure and specificity of the N-terminal domain. Mol Gen Genet. 1994;243:112–118. doi: 10.1007/BF00283883. [DOI] [PubMed] [Google Scholar]
- 13.Crago AM, Koronakis V. Salmonella InvG forms a ring-like multimer that requires the InvH lipoprotein for outer membrane localization. Mol Microbiol. 1998;30:47–56. doi: 10.1046/j.1365-2958.1998.01036.x. [DOI] [PubMed] [Google Scholar]
- 14.Burghout P, et al. Structure and electrophysiological properties of the YscC secretin from the type III secretion system of Yersinia enterocolitica. J Bacteriol. 2004;186:4645–4654. doi: 10.1128/JB.186.14.4645-4654.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chami M, et al. Structural insights into the secretin PulD and its trypsin-resistant core. J Biol Chem. 2005;280:37732–37741. doi: 10.1074/jbc.M504463200. [DOI] [PubMed] [Google Scholar]
- 16.Collins RF, et al. Structure of the Neisseria meningitidis outer membrane PilQ secretin complex at 12 A resolution. J Biol Chem. 2004;279:39750–39756. doi: 10.1074/jbc.M405971200. [DOI] [PubMed] [Google Scholar]
- 17.Bitter W. Secretins of Pseudomonas aeruginosa: large holes in the outer membrane. Arch Microbiol. 2003;179:307–314. doi: 10.1007/s00203-003-0541-8. [DOI] [PubMed] [Google Scholar]
- 18.Opalka N, et al. Structure of the filamentous phage pIV multimer by cryo-electron microscopy. J Mol Biol. 2003;325:461–470. doi: 10.1016/s0022-2836(02)01246-9. [DOI] [PubMed] [Google Scholar]
- 19.Brok R, et al. The C-terminal domain of the Pseudomonas secretin XcpQ forms oligomeric rings with pore activity. J Mol Biol. 1999;294:1169–1179. doi: 10.1006/jmbi.1999.3340. [DOI] [PubMed] [Google Scholar]
- 20.Chacon P, Wriggers W. Multi-resolution contour-based fitting of macromolecular structures. J Mol Biol. 2002;317:375–384. doi: 10.1006/jmbi.2002.5438. [DOI] [PubMed] [Google Scholar]
- 21.Marlovits TC, et al. Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature. 2006;441:637–640. doi: 10.1038/nature04822. [DOI] [PubMed] [Google Scholar]
- 22.Dong C, et al. Wza the translocon for E. coli capsular polysaccharides defines a new class of membrane protein. Nature. 2006;444:226–229. doi: 10.1038/nature05267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guilvout I, et al. Bacterial outer membrane secretin PulD assembles and inserts into the inner membrane in the absence of its pilotin. EMBO J. 2006;25:5241–5249. doi: 10.1038/sj.emboj.7601402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krehenbrink M, et al. Artificial binding proteins (Affitins) as probes for conformational changes in secretin PulD. J Mol Miol. 2008;383:1058–1068. doi: 10.1016/j.jmb.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 25.Korotkov KV, Pardon E, Steyaert J, Hol WG. Crystal structure of the N-terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody. Structure. 2009;17:255–265. doi: 10.1016/j.str.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rohl CA, Strauss CE, Misura KM, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 27.Kuhlman B, Baker D. Native protein sequences are close to optimal for their structures. Proc Natl Acad Sci U S A. 2000;97:10383–10388. doi: 10.1073/pnas.97.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qian B, et al. High-resolution structure prediction and the crystallographic phase problem. Nature. 2007;450:259–264. doi: 10.1038/nature06249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chenna R, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pettersen EF, et al. UCSF Chimera - A Visualization System for Exploratory Research and Analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 31.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos: 2002. [Google Scholar]
- 32.Ohi M, Li Y, Cheng Y, Walz T. Negative stain and image classification – powerful tools in modeln electron microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated Software for High-Resolution Single-Particle Reconstructions. Journal of Structural Biology. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 34.Frank J, et al. SPIDER and WEB: Processing and Visualization of Images in 3D Electron Microscopy and Related Fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 35.Deng W, et al. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc Natl Acad Sci U S A. 2004;101:3597–3602. doi: 10.1073/pnas.0400326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pegues DA, Hantman MJ, Behlau I, Miller SI. PhoP/PhoQ transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol Microbiol. 1995;17:169–181. doi: 10.1111/j.1365-2958.1995.mmi_17010169.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.