Abstract
Cutaneous melanoma is an aggressive form of human skin cancer characterized by high metastatic potential and poor prognosis. To better understand the role of microRNAs (miRNAs) in melanoma, the expression of 470 miRNAs was profiled in tissue samples from benign nevi and metastatic melanomas. We identified 31 miRNAs that were differentially expressed (13 up-regulated and 18 down-regulated) in metastatic melanomas relative to benign nevi. Notably, miR-193b was significantly down-regulated in the melanoma tissues examined. To understand the role of miR-193b in melanoma, functional studies were undertaken. Overexpression of miR-193b in melanoma cell lines repressed cell proliferation. Gene expression profiling identified 314 genes down-regulated by overexpression of miR-193b in Malme-3M cells. Eighteen of these down-regulated genes, including cyclin D1 (CCND1), were also identified as putative miR-193b targets by TargetScan. Overexpression of miR-193b in Malme-3M cells down-regulated CCND1 mRNA and protein by ≥50%. A luciferase reporter assay confirmed that miR-193b directly regulates CCND1 by binding to the 3′untranslated region of CCND1 mRNA. These studies indicate that miR-193b represses cell proliferation and regulates CCND1 expression and suggest that dysregulation of miR-193b may play an important role in melanoma development.
Cutaneous melanoma is a form of skin cancer characterized by aggressive metastatic growth and poor prognosis.1 The incidence of melanoma continues to increase in many parts of the world.2 The median survival time of patients with metastatic melanoma is 6 months, and the 5-year survival rate is less than 5%.3 Genetic factors and exposure to ultraviolet radiation are risk factors for melanoma pathogenesis.4
MicroRNAs (miRNAs) are a class of small (≈22 nucleotides) noncoding regulatory RNAs found in animals, plants, and viruses.5 miRNAs regulate gene expression through imperfect or perfect base pairing with the 3′ untranslated region (3′ UTR) of targeted mRNA, resulting in translational repression or mRNA destabilization and degradation.5 The exact mechanisms by which miRNAs recognize and regulate target genes are not well understood.6 Experimental data demonstrated that a 6-nucleotide seed sequence, from nucleotides 2 to 7 at the 5′ end of the miRNA, called miRNA “seed,” is involved in target recognition.7 Genes targeted by a miRNA often contain seed-matched sites at their 3′ UTR. Nevertheless, a single miRNA can regulate expression of hundreds of genes,8,9 and at least one third of human protein-coding genes are thought to be regulated by miRNAs.10 Importantly, miRNA target sites are often evolutionarily conserved in the genomes of many species, suggesting that miRNA:mRNA interactions are functionally significant and biologically important. Consistent with this, functional studies have implicated miRNAs in many biological processes, including development, differentiation, apoptosis, and cell proliferation.5
Recent studies have characterized diverse miRNA regulatory networks and suggested that miRNA dysregulation plays an important role in human cancer. Cancer-specific miRNA expression profiles have been identified in a variety of human malignancies.11 Depending on the specific miRNA and the cellular context, miRNAs are reported to act as either tumor suppressors or oncogenes.12 Differential expression of miRNAs in cancer cells may in part reflect the fact that many miRNA genes are located in cancer-associated genomic regions.13 Abnormal epigenetic regulation and transcriptional factor deregulation may also contribute to distinct patterns of miRNA expression in cancer cells.14,15
Several previous studies have examined possible roles of miRNA dysregulation in melanoma. One study examined the relative expression of 157 miRNAs in primary melanomas and benign nevi using quantitative real-time PCR.16 Schultz et al showed that let-7 family miRNAs were significantly down-regulated in primary melanomas, and let-7b inhibited melanoma cell cycle progression.16 Another study examined miRNA expression profiles in melanocytes and cell lines derived from primary or metastatic melanoma and identified large numbers of miRNAs associated with melanoma progression and metastatic colonization.17 However, it has been suggested that global miRNA abundance is generally higher in tissues compared with cell lines,18 and we believe a study using tissues can add important information to this field. In this study, we profiled 470 miRNAs in metastatic melanomas and benign nevi using a microarray platform and identified 31 miRNAs that were differentially expressed in melanomas compared with nevi. One candidate miRNA identified in this study is miR-193b. We demonstrated miR-193b represses melanoma cell proliferation and that CCND1 is a direct target of miR-193b.
Materials and Methods
Cell Culture and Tissue Samples
Metastatic melanoma cell lines Malme-3M, SKMEL-28, and SKMEL-5 were grown in RPMI medium 1640 (Hyclone, Logan, UT) supplemented with 10% fetal bovine serum (Hyclone) at 37°C and 5% CO2.
Formalin-fixed paraffin-embedded (FFPE) tissues from eight benign nevi, and eight metastatic melanomas were obtained from the Department of Pathology and Molecular Medicine, Kingston General Hospital. All cases were diagnosed and classified by a Dermatopathologist (V.A.T.). Tissue samples used in this study were all within 3 years of formalin fixation/embedding. Ethics approval was obtained from the Faculty of Health Sciences Ethics Board at Queen’s University.
MicroRNA Microarrays
Total RNA was isolated from FFPE samples using the RecoverAll Total RNA Isolation kit (Ambion, Austin, TX) according to the manufacturer’s instructions. For each sample, three 20-μm sections were used for RNA isolation. Agilent MicroRNA V1 arrays, which detect 470 human miRNAs, were used for profiling as described previously.19 Briefly, 100 ng of total RNA from each sample was dephosphorylated and ligated with pCp-Cy3 (Agilent, Santa Clara, CA). Labeled RNA was purified and hybridized in a rotating oven at 55°C for 20 hours. The chips were scanned with the Agilent DNA Microarray scanner, and signals were quantified using the Agilent Feature Extraction 9.5.3.1 software. Raw data can be accessed via the National Center for Biotechnology Information Gene Expression Omnibus website (http://www.ncbi.nlm.nih.gov/geo/accession number GSE18512, release date Dec 20, 2009).
MicroRNA Real-Time PCR
To measure specific miRNA expression patterns in FFPE tissues, the same total RNA described above was used for real-time PCR assays. As well, total RNA from transfected cell lines was isolated using the miRNeasy Mini kit (Qiaqen, Valencia, CA) according to the manufacturer’s protocol. miRNA levels were determined using the TaqMan MicroRNA Assays (Applied Biosystems, Foster City, CA) according to the manufacturer’s protocol. Briefly, miRNAs were reverse transcribed using miRNA specific stem-loop RT primers purchased from Applied Biosystems. Subsequent real-time PCR reactions were performed using the Eppendorf Realplex system (Eppendorf, Hamburg, Germany). PCR reactions were incubated in a 96-well plate at 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. All miRNAs were assayed in triplicate and data were normalized to endogenous RNU6B. The relative levels were calculated using the ΔΔCt method.
Reagents
The Pre-miR hsa-miR-193b miRNA Precursor and Pre-miR Negative Control used for miR-193b overexpression studies were purchased from Ambion. The miRNA precursors are modified double-stranded RNA molecules designed to mimic endogenous mature miRNAs. The CCND1 siRNA and the negative control siRNA were purchased from Applied Biosystems. Transfection reagent was Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Dicer-substrate Mcl-1 siRNA was obtained from Integrated DNA Technologies (Coralville, IA). The transfection efficiency was >95% (data not shown). miR-193b overexpression after transfection was confirmed by real-time PCR (see supplemental Figure S1 at http://ajp.amjpathol.org).
Proliferation, Apoptosis, and Cell Cycle Assays
Cells were seeded in 6-well plates at 100,000 cells per well the day before transfection. Malme-3M cells were transfected with 5 nmol/L miRNA precursors (either miR-193b or negative control); SKMEL-28 and SKMEL-5 were transfected with 50 nmol/L miRNA precursors (either miR-193b or negative control).
Proliferation was measured using the Cell Proliferation ELISA, BrdU (chemiluminescence) by Roche Applied Biosciences (Laval, Quebec, Canada) following the manufacturer’s instructions. Briefly, cells were harvested 72 hours posttransfection, counted, and reseeded in 96-well black plates (Corning®, Corning, NY) at 3500 cells per well. Cells were incubated for 24 hours in the presence of BrdU before fixing and labeling with anti-BrdU antibody. Chemiluminescence signal was measured using a EG&G Berthold microplate luminometer. Results are presented as relative level of proliferation and are the mean of three independent experiments ± SEM.
For apoptosis analysis, the proportion of cells undergoing apoptosis and necrosis was measured using the Annexin V FITC Apoptosis Detection kit (Calbiochem, San Diego, CA). In brief, cells were collected 72 hours posttransfection, washed twice with cold PBS, then resuspended in annexin V binding buffer. FITC-conjugated annexin V (1.25 μl/sample) was added and cells incubated for 30 minutes at room temperature in the dark. Cells were centrifuged, resuspended in binding buffer, and propidium iodide was added (10 μl per sample). Samples were kept on ice and analyzed immediately by flow cytometry, using a Beckman Coulter EPICS Altra HSS flow cytometer. For positive control, Malme-3M cells were treated with 10 nmol/L Mcl-1 DsiRNA and 10 μmol/L ABT-737 as described previously.20 Representative data from one of three independent experiments are shown.
For cell cycle analysis, samples were harvested 72 hours posttransfection, fixed overnight in ice-cold ethanol (70% v/v), and stained for 3 hours at 4°C with 50 μg ml−1 propidium iodide (PI, Calbiochem) in PBS containing 3.8 mmol/L sodium citrate and 0.5 μg ml−1 RNase A (Sigma-Aldrich, Oakville, ON, Canada) in 10 mmol/L Tris pH 7.5, 15 mmol/L NaCl. DNA content was determined using a Beckman Coulter EPICS Altra HSS flow cytometer. Results are presented as % of cell population in each cell cycle phase. Data are the mean of three independent experiments ± SEM. P value was calculated using G1 population by independent samples t test.
Gene Expression Microarray
Malme-3M cells were seeded 100,000 cells per well in six-well plate and transfected the following day with 5 nmol/L miRNA precursors (either miR-193b or negative control). Total RNA was harvested 24 hours after transfection using miRNeasy Mini kit (Qiagen). The Agilent Microarray Platform for One-Color Analysis of Gene Expression was used for profiling (Agilent). Briefly, 500 ng total RNA of each sample was mixed with 5 μl of a 5000-fold dilution of Agilent One-Color Spike-in RNA control. The mixture was amplified and labeled using the One color, Quick Amp Labeling kit (Agilent). mRNA was primed with an oligo (dT) primer containing a T7 RNA polymerase promoter to synthesize double-stranded cDNA as a template for in vitro transcription to generate Cy3-labeled cRNA. After amplification and labeling, cRNA yield and specific activity were assessed using Nanodrop ND-1000 (Nanodrop, Wilmington, DE). Only samples with cRNA yields >1.65 μg and specific activities >9.0 pmol Cy3/μg cRNA were processed further. Successfully amplified and labeled samples were fragmented at 60°C for 30 minutes and hybridized to Agilent Human 4X44K Whole genome microarrays in a rotating oven at 65°C for 17 hours. Microarray chips were scanned with the Agilent DNA Microarray scanner and quantified as described above for miRNA chips. Raw data can be accessed via National Center for Biotechnology Information Gene Expression Omnibus website (http://www.ncbi.nlm.nih.gov/geo/accession number GSE18512, release date Dec 20, 2009).
mRNA Real-Time PCR
Total RNA from transfected cell cultures was isolated using miRNeasy Mini kit (Qiaqen), and then reverse transcribed to cDNA using TaqMan Reverse Transcription Reagents (Applied Biosystems). 300 ng total RNA was random primed and reverse transcribed on a thermal cycler. CCND1 quantification was then performed using CCND1 TaqMan Gene Expression Assay (Applied Biosystems) on an Eppendorf Realplex platform (Eppendorf). The PCR mixture was incubated at 50°C for 2 minutes, 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 s, 60°C for 60 s. β-actin was used as housekeeping gene for normalization. The relative levels were calculated using the ΔΔCt method.
Western Blotting
30 μg cell lysates were separated on 15% SDS-polyacrylamide gels, and then were transferred to PVDF membrane (Millipore, Bedford, MA). The CCND1 antibody used for blotting was purchased from BD Biosciences (San Jose, CA), and gamma tubulin was purchased from Sigma-Aldrich. Densitometry was performed using Quantity One software (Bio-Rad, Mississauga, ON, Canada).
Vector Construction and Luciferase Assay
We generated pGL3-CCND1 by amplifying a 512 bp 3′ UTR fragment of CCND1 gene harboring the miR-193b binding site predicted by the TargetScan (http://www.targetscan.org/, accession date Oct 27, 09) and subsequently cloning it into the pGL3 control vector (Promega, Madison, WI) at the XbaI site immediately downstream of firefly luciferase. The primer sequences used for amplification were (XbaI sites are in bold): sense 5′-GCTCTAGAGGAGGCTGCGTGCCAGTCAAGAAG-3′ and antisense 5′-GCTCTAGACCTTGCACCCATGCCTGTCCAATC-3′. pGL3-CCND1 was subsequently used as a template to generate the control vector pGL3-CCND1 MM, which has two mismatch mutations in the miR-193b seed complementary site. Overlapping PCR was used to introduce mutations into the control vector. The following primers were used for PCR amplification: sense 5′-GCAGAGGATGTTCATAAGGGCAGAATGATTTATAAATGCAATCTCC-3′ and antisense 5′-GGAGATTGCATTTATAAATCATTCTGCCCTTATGAACATCCTCTGC-3′ (mismatch nucleotides are in bold).
Malme-3M cells were seeded at 75,000 cells per well in a 12-well plate the day before transfection. The cells were cotransfected with 5 nmol/L miRNA precursor (either miR-193b or negative control), 400 ng of firefly luciferase vector (either pGL3-CCND1 or pGL3-CCND1 MM), and 50 ng of Renilla luciferase vector (pRL-TK). Luciferase activity was measured 24 hours after transfection using the Dual-Glo Luciferase Assay System (Promega). The Renilla luciferase activity served as internal control.
Immunohistochemistry and Image Analysis
Immunohistochemistry was performed in the Department of Pathology and Molecular Medicine at the Kingston General Hospital using a standard protocol on tissue microarray slides. Anti-Cyclin D1 antibody (NeoMarkers, clone SP4) was used at a 1/100 dilution and incubated for 32 minutes at 37C. Stained slides were scanned on an Aperio scanner (Vista, CA), and the percentage of CCND1-positive nuclei in cell population was quantified using immunohistochemistry Image Analysis software.
Statistical Analysis
In microRNA microarray study, unsupervised hierarchical clustering was performed on log2 transformed data (signal values less 1 were reassigned to 1) with Cluster 3.0 using uncentered correlation metric and average linkage methods.21 The results were visualized by Java Treeview 1.1.1 (http://jtreeview.sourceforge.net/, accession date Oct 27, 09). To identify differentially expressed miRNAs, significance analysis of microarray (SAM) was performed on the array data without log transformation.22,23 miRNAs were excluded after SAM analysis when the absolute mean difference of their expression between melanomas and nevi (numerator r ) was <15. The heatmap, presenting the log2-transformed fold changes of miRNAs, was generated using Cluster 3.0 and Java Treeview as described above. In gene expression microarray study, raw data were normalized and analyzed using GeneSpring GX 10.0.2 (Agilent). For quality control, probe sets were filtered to include only genes that were described as “Present” or “Marginal” by GeneSpring in all 4 samples, and genes with raw signals <25.0 were removed from subsequent analyses. P values were calculated for two pair-wise comparisons between Malme-3M cells with or without miR-193b overexpression using the paired t test.
In the bar graphs, data were analyzed by t test using SPSS Statistics 17.0 (SPSS Inc., Chicago, IL). A P value <0.05 was considered statistically significant.
Results
Distinct miRNA Signatures in Metastatic Melanoma and Benign Nevus
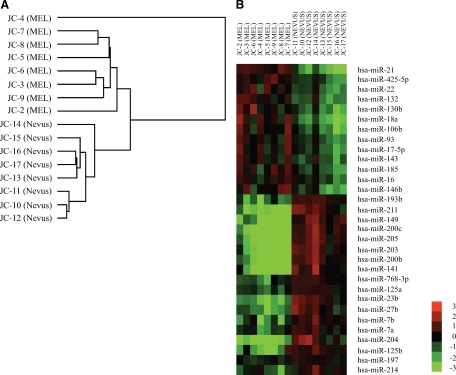
Our initial studies examined the expression of 470 miRNAs in FFPE samples from eight benign nevi and eight metastatic melanomas using the Agilent miRNA microarray platform (version 1.5) to identify possible miRNA candidates for functional studies. We used these two groupings because they represent two biologically and clinically distinct melanocytic tissues. In our opinion, nevus is the best surrogate for benign melanocytes, whereas metastatic melanoma, by virtue of the fact that it has spread, is clearly malignant. The validity of using this method with FFPE tissue samples was confirmed previously by us and others.19,24 Data were analyzed by unsupervised hierarchical clustering, and the results were summarized in a dendrogram (Figure 1A), which clearly shows that benign nevi cluster separately from metastatic melanomas.
Figure 1.
Unsupervised hierarchical clustering of miRNA expression data and heatmap of differentially expressed miRNAs. A: The dendrogram was generated by comparing miRNA expression profiles of metastatic melanomas (MEL) and benign nevi (NEVUS) using Cluster 3.0. Average linkage clustering was performed using uncentered correlation metric and visualized by Java Treeview. B: miRNAs were identified by SAM after comparing microarray results between metastatic melanomas and benign nevi (false discovery rate q < 0.001). The heatmap displays the log2 transformed fold changes. Red indicates overexpression, and green indicates down-regulation.
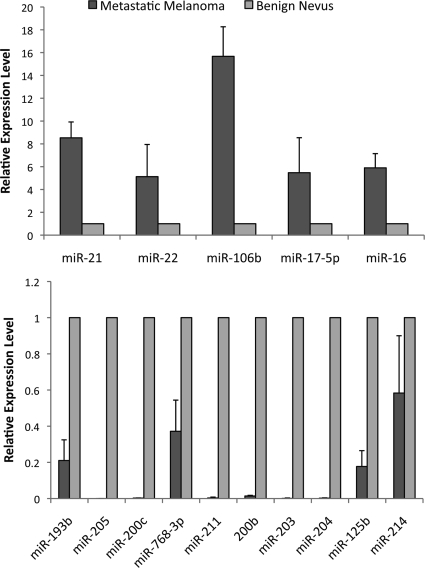
We compared the expression of miRNAs between metastatic melanomas and benign nevi, and identified differentially expressed miRNAs by SAM. By using 1000 permutations and selecting a false discovery rate q < 0.001, we identified 31 differentially expressed miRNAs including 13 up-regulated and 18 down-regulated in metastatic melanomas compared with benign nevi (Figure 1B). To provide a technical validation, 5 significantly up-regulated miRNAs and 10 significantly down-regulated miRNAs were quantified in four metastatic melanoma samples and four benign nevus samples by real-time PCR using the same RNA samples isolated for the microarray studies. The levels of miRNAs detected by real-time PCR agreed with the microarray results (Figure 2). On average, the five up-regulated miRNAs were expressed at a fourfold higher level in metastatic melanomas than in nevi, and the 10 down-regulated miRNAs, with the exception of miR-214, were expressed on average at twofold lower level in metastatic melanoma samples.
Figure 2.
Real-time PCR analysis of miRNAs in melanoma. Candidates were selected from dysregulated miRNAs identified in the microarray study: up-regulated miRNAs in metastatic melanomas (upper) and down-regulated miRNAs in metastatic melanomas (lower). Their expression levels were quantified by the TaqMan microRNA assays. RNU6B was used as an internal control. Each sample was measured in triplicate. Data are the mean ± SEM of four tissue samples.
miR-193b Suppresses Melanoma Cell Proliferation
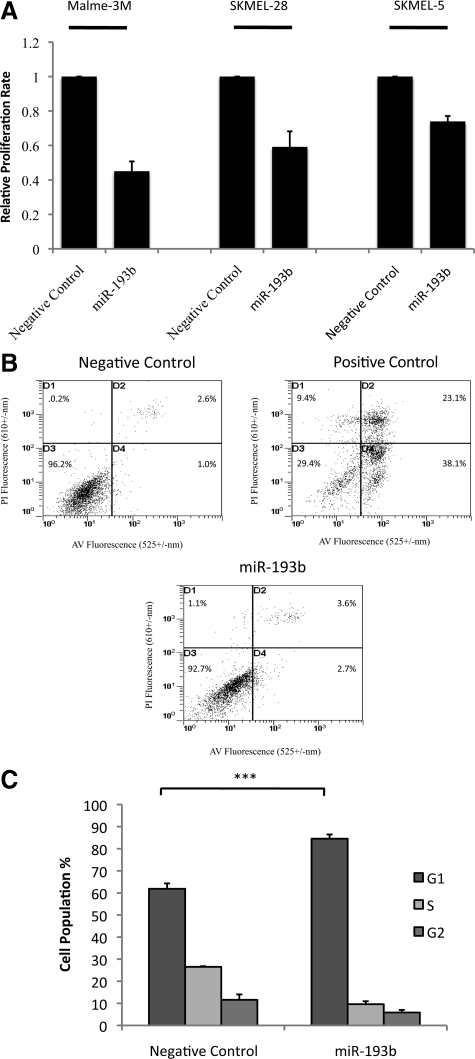
In the SAM analysis,22 miR-193b had the largest relative difference (score d = −6.9) among the down-regulated miRNAs, and its expression was 3.4-fold lower in metastatic melanomas compared with benign nevi. The down-regulation of miR-193b was also confirmed by real-time PCR (Figure 2). Its function in melanoma is unknown. To investigate the functional roles of miR-193b, three metastatic melanoma cell lines, Malme-3M, SKMEL-28, and SKMEL-5 were transfected with miRNA precursors (either miR-193b or negative control). Cell proliferation was measured using the BrdU incorporation assay (Roche). Overexpression of miR-193b repressed cell proliferation in all three melanoma cell lines (Figure 3A). Among them, Malme-3M cells were the most responsive, showing a nearly 60% reduction in proliferation after miR-193b overexpression. Thus, additional functional studies on miR-193b were conducted in Malme-3M cells.
Figure 3.
Effect of miR-193b overexpression on proliferation and apoptosis in melanoma cells. Malme-3M, SKMEL-28, and SKMEL-5 cells were transfected with miRNA precursors (either miR-193b or negative control) for 72 hours. A: Transfected cells, incubated with BrdU, were seeded at 3500 cells per well into 96-well black plate. After 24 hours, cell proliferation rate was assessed using the cell proliferation ELISA. The data are mean ± SEM from three independent experiments, each performed in octuplicate. B: Cells were fixed, stained with Annexin V FITC and PI, and analyzed by flow cytometry. Necrotic cells in D1, necrotic and late apoptotic cells in D2, viable cells in D3, and early apoptotic cells in D4. As a positive control, Malme-3M cells were treated with Mcl-1 DsiRNA and ABT-737 as described previously.20 Graphs show representative results from one of three independent experiments. C: Malme-3M cells were fixed and treated with RNase. After PI staining, cells were analyzed by flow cytometry. Data are the mean of four independent experiments, presented as mean ± SEM. ***P < 0.001 was calculated using G1 population by independent samples t test.
To determine whether reduced cell proliferation was apoptosis-independent, we performed Annexin V–FITC staining to detect apoptosis. Annexin V–FITC data showed that overexpression of miR-193b did not significantly alter the fraction of necrotic or apoptotic Malme-3M cells (Figure 3B). Cell cycle analysis by PI staining was performed. Overexpression of miR-193b increased the fraction of cells in the G1 phase from 62% to 85% while decreasing the fraction of cells in the S phase and G2 phase from 27% and 12% to 10% and 6%, respectively (Figure 3C). This result suggests that miR-193b represses cell proliferation by arresting cells in the G1 phase, without inducing cell death.
Gene Expression Profiling Reveals CCND1 as One of miR-193b Potential Targets
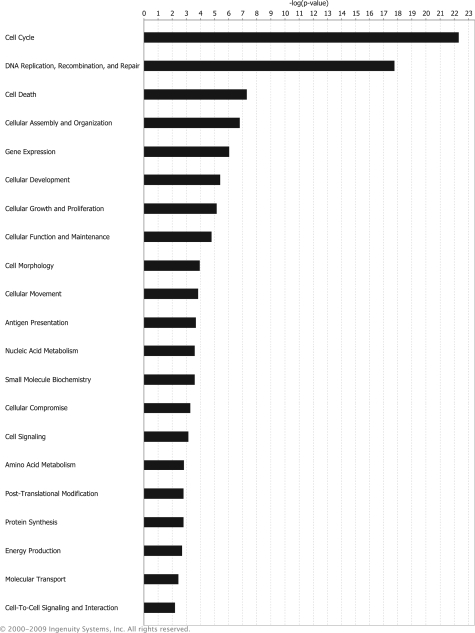
It is crucial to identify functionally important miRNA targets to understand how a specific miRNA functions. Recent studies showed that mRNA destabilization is the major component of the miRNA repression mechanism in targets with robust protein reduction.9 Thus, we performed gene expression analysis to globally screen for miR-193b targets that may affect cell proliferation. By comparing mRNA expression levels in Malme-3M cells transfected with miRNA precursors (either miR-193b or negative control) using the whole human genome microarray (Agilent), we identified 314 genes that were down-regulated more than 1.5 fold (P < 0.05) by miR-193b overexpression (see supplemental Table S1 at http://ajp.amjpathol.org). Ingenuity pathways analysis functional analysis was performed to examine the molecular and cellular functions of those genes. Ingenuity pathways analysis revealed that the most significantly enriched gene ontology category among down-regulated genes was cell cycle, indicating miR-193b, directly or indirectly, regulates genes involved in cell cycle progression (Figure 4).
Figure 4.
Ingenuity pathways analysis. Core analysis was performed on 314 genes down-regulated in Malme-3M cells after overexpressing miR-193b. Molecular and cellular functions associated with down-regulated genes were determined and analyzed by Fisher exact test. The threshold P value was 0.05. These data were generated through the use of Ingenuity Pathways Analysis online (Ingenuity Systems, Redwood City, CA).
Interestingly, 18 of the genes down-regulated after miR-193b overexpression were also the targets of miR-193b predicted by TargetScan (Human 5.1, http://www.targetscan.org/, accession date Oct 27, 09; Table 1). We reasoned that genes predicted by TargetScan are more likely to be the direct targets of miR-193b because they contain the predicted evolutionary conserved miR-193b seed binding sites. Of those 18 genes, CCND1 was of particular interest. CCND1 plays an important role in regulating the G1/S transition during cell cycle progression. This is consistent with the observation that overexpression of miR-193b inhibited cell growth and increased G1 cell cycle arrest, effects that could be mediated by repression of CCND1.
Table 1.
Genes Down-regulated by miR-193b That Are Predicted by TargetScan
| Down-regulated genes | Fold change |
|---|---|
| Homo sapiens | |
| Abl interactor 2 (ABI2) | 3 |
| Adenylate cyclase 9 (ADCY9) | 1.94 |
| Rho GTPase activating protein 19 (ARHGAP19) | 2.38 |
| Atonal homolog 8 (Drosophila) (ATOH8) | 1.54 |
| Cyclin D1 (CCND1) | 2.02 |
| Calsyntenin 1 (CLSTN1) | 1.81 |
| CCR4-NOT transcription complex, subunit 6 (CNOT6) | 2.17 |
| E2F transcription factor 6 (E2F6) | 1.54 |
| v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS) | 2.63 |
| Leucine rich repeat containing 8 family, member A (LRRC8A) | 1.77 |
| Myeloid cell leukemia sequence 1 (BCL2-related) (MCL1) | 1.64 |
| RNA binding motif protein 8A (RBM8A) | 1.72 |
| Remodeling and spacing factor 1 (RSF1) | 1.67 |
| Selenoprotein N, 1 (SEPN1) | 2.05 |
| Stathmin 1/oncoprotein 18 (STMN1) | 4.47 |
| Tumor necrosis factor receptor superfamily, member 21 (TNFRSF21) | 2.16 |
| WD repeat domain 68 (WDR68) | 2.89 |
| Zinc finger protein 365 (ZNF365) | 1.74 |
Gene expression microarray analysis was performed on Malme-3M cells 24 hours post-transfection with miR-193b.
miR-193b Suppresses Cell Proliferation by Directly Down-Regulating CCND1
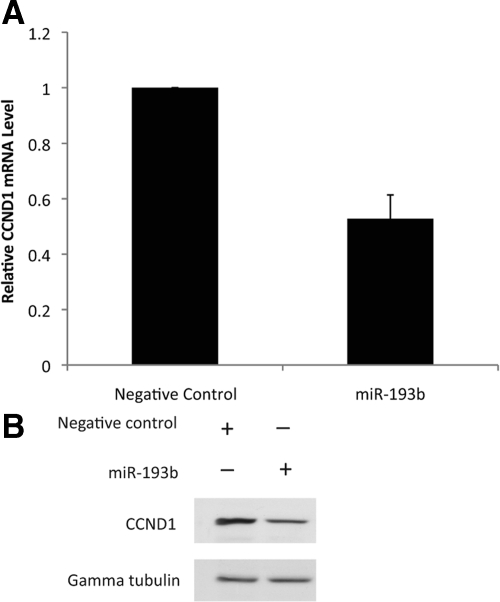
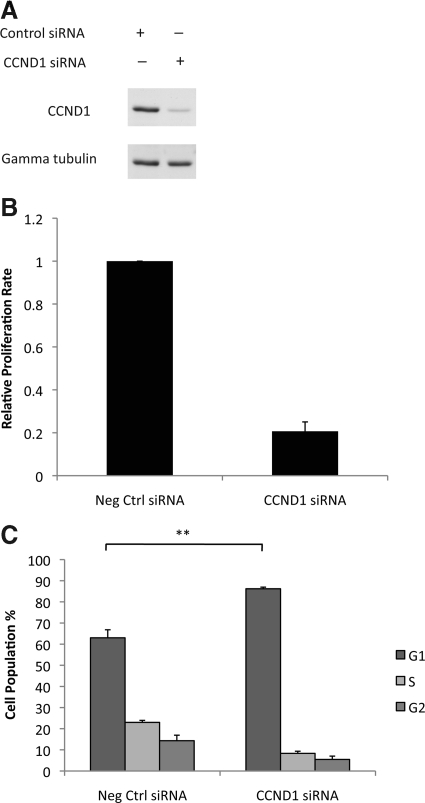
To validate CCND1 as a target of miR-193b, CCND1 mRNA and protein were quantified in Malme-3M cells with or without overexpression of miR-193b. The results showed that CCND1 mRNA and protein levels were approximately 50% lower in cells overexpressing miR-193b than in control cells (Figures 5, A and B). It is predicted that lower expression of CCND1 will inhibit cell proliferation. We examined this prediction by knocking down CCND1 expression in Malme-3M cells using siRNA and measuring cell proliferation and cell cycle progression (Figure 6, A–C). The results confirmed that CCND1 knockdown inhibits cell proliferation and causes G1 phase arrest in Malme-3M cells (Figure 6, B and C). Therefore, CCND1 may be an important miR-193b target gene and may play a direct role in mediating the effects of miR-193b overexpression on proliferation of melanoma cells.
Figure 5.
miR-193b represses CCND1 expression. Malme-3M cells were transfected with miRNA precursors (either miR-193b or negative control) for 72 hours. A: Real-time PCR was used to quantify CCND1 mRNA; β-actin was used as the internal control. Data are mean ± SEM of three independent experiments, each performed in triplicate. B: CCND1 protein was quantified by Western blotting. Gamma tubulin was used as the loading control. Representative data from one of three independent experiments are shown.
Figure 6.
CCND1 knockdown suppresses cell proliferation. Malme-3M cells were transfected with either CCND1 siRNA or negative control (Neg Ctrl) siRNA for 72 hours. A: CCND1 protein was quantified by Western blotting. Gamma tubulin was used as the loading control. Representative data from one of three independent experiments are shown. B: Cell proliferation was measured as described in the legend to Figure 3A. The data are mean ± SEM from three independent experiments, each performed in octuplicate. C: Cell cycle progression was analyzed as described in the legend to Figure 3C. **P < 0.004 was calculated using G1 population by independent samples t test.
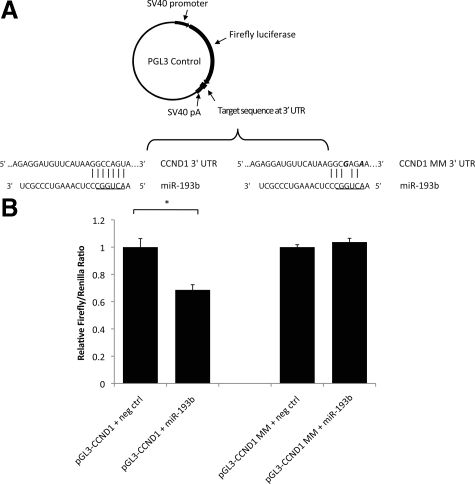
As mentioned above, TargetScan identified an evolutionarily conserved binding site for miR-193b in the 3′ UTR of CCND1. To investigate whether miR-193b directly binds to the predicted binding site, we cloned a 512-bp fragment of the CCND1 3′ UTR harboring the potential binding site into a pGL3 control plasmid and downstream of a luciferase reporter gene (Figure 7A). Similar sequence containing two mismatch mutations at the conserved miRNA seed binding site was cloned into the luciferase reporter plasmid (Figure 7A). These luciferase reporter constructs were cotransfected with miRNA precursors (either miR-193b or negative control). The results showed that activity of the luciferase reporter gene carrying the wild-type CCND1 3′ UTR on average was 30% lower in cells overexpressing miR-193b, while no repression of luciferase activity was observed in the reporter construct carrying the mutant CCND1 3′ UTR (Figure 7B). These results indicate miR-193b can repress CCND1 expression by directly binding to a specific 3′ UTR site of CCND1.
Figure 7.
CCND1 is a direct target of miR-193b. A: Luciferase reporter vectors. A fragment of the CCND1 3′ UTR harboring the predicted binding site of miR-193b was cloned into a pGL3 control plasmid, downstream of a firefly luciferase reporter gene (pGL3-CCND1). The control vector (pGL3-CCND1 MM) included two mismatch mutations at the conserved binding site. The underlined 6-nucleotide indicates miR-193b seed sequence. Mismatch mutations are in bold and italic. B: Cells were cotransfected with 5 nmol/L miRNA precursors (either miR-193b or negative control), 400 ng firefly luciferase report plasmids (either pGL3-CCND1 or pGL3-CCND1 MM), and 50 ng renilla plasmids (pRL-TK). Cells were harvested 24 hours after transfection. Protein extracts were prepared and assayed for firefly and renilla luciferase activity. Firefly luciferase activity was normalized to renilla luciferase activity. Data are shown as the mean ± SEM of three replicates and are representative of three independent experiments. *P < 0.013 was calculated using independent samples t test.
CCND1 Level Is Elevated in Melanoma
CCND1 has been reported as a potential oncogene in melanoma.25 To confirm that our melanoma samples demonstrated increased CCND1 levels as previously described,25 we created a tissue microarray that included our benign nevi and melanoma tumors. Immunohistochemistry was performed using a rabbit monoclonal antibody directed against CCND1. Image analysis confirmed that the average cell population with CCND1 positively stained nuclei increased from 9.5% in nevi to 37.9% in melanomas (see supplemental Figure S2 at http://ajp.amjpathol.org).
Discussion
This study analyzed expression of 470 miRNAs in eight benign nevi and eight metastatic melanomas, and identified 31 differentially expressed miRNAs, which may contribute to melanoma development. Furthermore, a potentially important role in melanoma progression was identified for miR-193b. miR-193b overexpression correlated with decreased expression of >300 genes in Malme-3M cells, a number of which are involved in cell cycle progression, including CCND1, E2F1, cyclin A2, and cell division cycle 2 (CDC2; see supplemental Table S1 at http://ajp.amjpathol.org). In this study, we focused on a well-known cell cycle regulatory gene, CCND1, and validated it as a direct target of miR-193b. Although we did not explore the potential role of all of other putative targets on cell cycle progression, we acknowledge that many of them may be important targets to validate. A previous study reported that expression of miR-193b was lower in endometrioid adenocarcinoma cells than in adjacent normal endometrium.26 More recently, miR-193b was found to repress the expression of estrogen receptor-α and urokinase-type plasminogen activator in breast cancer cell lines.27,28 This study reports for the first time that miR-193b overexpression reduces cell proliferation in melanoma cell lines and suggests that miR-193b may regulate cell cycle progression by down-regulating expression of CCND1 protein. Although we believe G1 arrest is the most likely explanation for the increased proportion of cells in G1/G0 after miR-193b overexpression, we cannot rule out other possibilities, such as a lengthening of G1 phase or the movement of cells into G0 or along differentiation pathways.
CCND1 plays a key role in controlling the cell cycle progression.29 CCND1 binds and activates CDK4/6, which subsequently phosphorylates tumor suppressor protein Rb and allows the cell cycle to progress through G1 into S. In normal cells, the expression of CCND1 is induced by extracellular growth factors. Overexpression of CCND1 has been found in a variety of human malignancies, including parathyroid adenoma, breast cancer, colon cancer, lymphoma, lung cancer, melanoma, and prostate cancer.29 Although gene amplification is observed in a subset of melanomas, tumors with normal CCND1 gene copy number can also express an elevated level of CCND1.25,30 CCND1 protein levels were also higher in the metastatic melanoma samples compared with benign nevi in our study (see supplemental Figure S2 at http://ajp.amjpathol.org). Therefore, transcriptional or translational mechanisms, possibly involving oncogene induction, may increase expression of CCND1 in melanoma cells that lack CCND1 amplification. For example, the mitogen-activated protein kinase pathway induces CCND1 expression and is often hyperactivated in melanoma.31 Mitogen-activated protein kinase pathway activation may result from Braf mutations, which are associated with 50% to 70% of melanoma cases.4 Interestingly, more than 80% of benign nevi, which rarely progress to melanoma, also harbor Braf mutations.32 It is hypothesized that these melanocytes fail to proliferate and progress because of oncogene-induced cell senescence and that additional mutations are needed for complete cellular transformation.4 The results presented in this study suggest that miR193b is down-regulated several-fold in melanoma cells and that miR-193b represses expression of CCND1. Thus, down-regulation of miR-193b in metastatic melanomas but not in benign nevi may contribute to melanoma progression.
In addition, many of the dysregulated miRNAs identified in our study are dysregulated in other cancers, and several of the miRNAs that were up-regulated in our melanoma samples have oncogenic properties. For example, the most significantly up-regulated miRNAs in metastatic melanomas is miR-21. Since first reported as an antiapoptotic factor in human glioblastoma cells,33 miR-21 has been shown to repress a number of tumor suppressors, including phosphatase and tensin homolog (PTEN) in human hepatocellular cancer cells,34 tromopyosin 1 (TPM1) and programmed cell death 4(PDCD4) in breast cancer cells,35,36,37 as well as reversion-inducing cysteine rich protein with Kazal motifs (RECK) and tissue inhibitor of matrix metalloproteinase 3 (TIMP3) in glioma cells.38 Members of miR-17-92 cluster (miR17-5p and miR-18a) and its paralog miR-106-25 cluster (miR-106b and miR-93) were also found up-regulated in melanoma samples examined in this study. A large body of evidence suggests that the miR-17-92 cluster and its paralogs act as oncogenes by regulating apoptosis and cell proliferation.39 Targets of these miRNAs include transcription factor E2Fs, cyclin-dependent kinase inhibitor CDKN1A, and proapoptotic protein BIM.39 However, there is one exception. miR-16 is a well-known tumor suppressor that targets oncogenes like Bcl-240 and is up-regulated in our melanoma samples. This is consistent with the fact that Bcl-2 was reported to be down-regulated in metastatic melanoma.41
Several of the miRNAs reported in this study to be down-regulated in melanomas act as tumor suppressors. For instance, members of the miR-200 family (miR-200b, miR-200c, and miR-141) and miR-205, down-regulated in melanomas in this study, were reported to be down-regulated in cells that had undergone epithelial to mesenchymal transition.42 These miRNAs regulate expression of E-cadherin transcriptional repressors, and E-cadherin is down-regulated during melanoma progression.43 Let-7a and let-7b are among miRNAs down-regulated in melanomas in this study. Previous studies revealed that Let-7 family miRNAs function as tumor suppressors by targeting RAS.44 More recently, let-7b was found to target cyclins and cyclin-dependent kinase 4 (CDK4) in melanoma cells.16
In summary, this study characterizes distinct miRNA expression patterns in benign nevi and melanoma metastases. More importantly, the functional studies suggest that miR-193b may repress melanoma cell proliferation and regulate CCND1. These results may help explain the abnormal expression of CCND1 protein in melanoma. We have begun to examine miR-193b levels in an independent set of metastatic melanomas to provide a biological validation of our observation in this study. Further study of miR-193b in primary melanomas is necessary to understand whether down-regulation of miR-193b is an early event in melanoma progression. Because miR-193b appears to have antiproliferative effects in melanoma cells, it may have potential as a novel therapeutic agent for melanoma treatment.
Acknowledgments
We thank Dr. Tom Radcliffe for statistical support. We thank members of the Tron lab for their helpful comments. We also thank Matthew Gordon (Cancer Research Laboratories, Queen’s University) for conducting the flow cytometry analysis.
Footnotes
Address reprint requests to Dr. Victor A. Tron, Department of Pathology and Molecular Medicine, Queen’s University, 88 Stuart Street, Richardson Laboratory, Rm 202, Kingston, ON K7L 3N6. E-mail: tronv@kgh.kari.net.
Supported by a Canadian Institutes for Health Research grant (to V.A.T. and H.E.F.). J.C. is funded by Queen’s Transdisciplinary Training Program in Cancer Research.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Cummins DL, Cummins JM, Pantle H, Silverman MA, Leonard AL, Chanmugam A. Cutaneous malignant melanoma. Mayo Clin Proc. 2006;81:500–507. doi: 10.4065/81.4.500. [DOI] [PubMed] [Google Scholar]
- Bevona C, Sober AJ. Melanoma incidence trends. Dermatol Clin. 2002;20:589–595, vii.. doi: 10.1016/s0733-8635(02)00037-2. [DOI] [PubMed] [Google Scholar]
- Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445:851–857. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- Miller AJ, Mihm MC., Jr Melanoma. N Engl J Med. 2006;355:51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nat Genet. 2006;38:1452–1456. doi: 10.1038/ng1910. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A, Calin GA, Villanueva A, Ropero S, Sanchez-Cespedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ, Gallagher WM, Eccles SA, Croce CM, Esteller M. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008;18:549–557. doi: 10.1038/cr.2008.45. [DOI] [PubMed] [Google Scholar]
- Mueller DW, Rehli M, Bosserhoff AK. miRNA expression profiling in melanocytes and melanoma cell lines reveals miRNAs associated with formation and progression of malignant melanoma. J Invest Dermatol. 2009;129:1740–1751. doi: 10.1038/jid.2008.452. [DOI] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT. Cell-cell contact globally activates microRNA biogenesis. Proc Natl Acad Sci U S A. 2009;106:7016–7021. doi: 10.1073/pnas.0811523106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chen J, Radcliffe T, Lebrun DP, Tron VA, Feilotter H. An array-based analysis of microRNA expression comparing matched frozen and formalin-fixed paraffin-embedded human tissue samples. J Mol Diagn. 2008;10:513–519. doi: 10.2353/jmoldx.2008.080077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keuling AM, Felton KE, Parker AA, Akbari M, Andrew SE, Tron VA. RNA silencing of Mcl-1 enhances ABT-737-mediated apoptosis in melanoma: role for a caspase-8-dependent pathway. PLoS One. 2009;4:e6651. doi: 10.1371/journal.pone.0006651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu B, Kim DS, Deluca AM, Alani RM. Comprehensive expression profiling of tumor cell lines identifies molecular signatures of melanoma progression. PLoS One. 2007;2:e594. doi: 10.1371/journal.pone.0000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glud M, Klausen M, Gniadecki R, Rossing M, Hastrup N, Nielsen FC, Drzewiecki KT. MicroRNA expression in melanocytic nevi: the usefulness of formalin-fixed, paraffin-embedded material for miRNA microarray profiling. J Invest Dermatol. 2009;129:1219–1224. doi: 10.1038/jid.2008.347. [DOI] [PubMed] [Google Scholar]
- Sauter ER, Yeo UC, von Stemm A, Zhu W, Litwin S, Tichansky DS, Pistritto G, Nesbit M, Pinkel D, Herlyn M, Bastian BC. Cyclin D1 is a candidate oncogene in cutaneous melanoma. Cancer Res. 2002;62:3200–3206. [PubMed] [Google Scholar]
- Wu W, Lin Z, Zhuang Z, Liang X. Expression profile of mammalian microRNAs in endometrioid adenocarcinoma. Eur J Cancer Prev. 2009;18:50–55. doi: 10.1097/CEJ.0b013e328305a07a. [DOI] [PubMed] [Google Scholar]
- Li XF, Yan PJ, Shao ZM. Downregulation of miR-193b contributes to enhance urokinase-type plasminogen activator (uPA) expression and tumor progression and invasion in human breast cancer. Oncogene. 2009;28:3937–3948. doi: 10.1038/onc.2009.245. [DOI] [PubMed] [Google Scholar]
- Leivonen SK, Makela R, Ostling P, Kohonen P, Haapa-Paananen S, Kleivi K, Enerly E, Aakula A, Hellstrom K, Sahlberg N, Kristensen VN, Borresen-Dale AL, Saviranta P, Perala M, Kallioniemi O. Protein lysate microarray analysis to identify microRNAs regulating estrogen receptor signaling in breast cancer cell lines. Oncogene. 2009;28:3926–3936. doi: 10.1038/onc.2009.241. [DOI] [PubMed] [Google Scholar]
- Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: cyclin D1: normal and abnormal functions. Endocrinology. 2004;145:5439–5447. doi: 10.1210/en.2004-0959. [DOI] [PubMed] [Google Scholar]
- Maelandsmo GM, Florenes VA, Hovig E, Oyjord T, Engebraaten O, Holm R, Borresen AL, Fodstad O. Involvement of the pRb/p16/cdk4/cyclin D1 pathway in the tumorigenesis of sporadic malignant melanomas. Br J Cancer. 1996;73:909–916. doi: 10.1038/bjc.1996.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen C, Zavala-Pompa A, Sequeira JH, Shoji M, Sexton DG, Cotsonis G, Cerimele F, Govindarajan B, Macaron N, Arbiser JL. Mitogen-actived protein kinase activation is an early event in melanoma progression. Clin Cancer Res. 2002;8:3728–3733. [PubMed] [Google Scholar]
- Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, Moses TY, Hostetter G, Wagner U, Kakareka J, Salem G, Pohida T, Heenan P, Duray P, Kallioniemi O, Hayward NK, Trent JM, Meltzer PS. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, Krichevsky AM. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369–5380. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Tron VA, Reed JC, Mah KJ, Krajewska M, Li G, Zhou X, Ho VC, Trotter MJ. Expression of apoptosis regulators in cutaneous malignant melanoma. Clin Cancer Res. 1998;4:1865–1871. [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Kuphal S, Poser I, Jobin C, Hellerbrand C, Bosserhoff AK. Loss of E-cadherin leads to up-regulation of NFkappaB activity in malignant melanoma. Oncogene. 2004;23:8509–8519. doi: 10.1038/sj.onc.1207831. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]