Abstract
The E. coli single stranded DNA binding (SSB) protein plays a central role in DNA metabolism through its high affinity interactions with ssDNA, as well as its interactions with numerous other proteins via its unstructured C-termini. Although SSB interacts with at least 14 other proteins, it is not understood how SSB might recruit one protein over another for a particular metabolic role. To probe the specificity of these interactions we have used isothermal titration calorimetry to examine the thermodynamics of binding of SSB to two E. coli proteins important for DNA replication, the χ subunit of DNA polymerase III holoenzyme and the PriA helicase. We find that an SSB tetramer can bind up to four molecules of either protein primarily via interactions with the last ~ 9 amino acids in the conserved SSB C-terminal tails (SSB-Ct). We observe intrinsic specificity for the binding of an isolated SSB-Ct peptide to PriA over χ due primarily to a more favorable enthalpic component. PriA and χ also bind with weaker affinity to SSB (in the absence of ssDNA) than to isolated SSB-Ct peptides, indicating an inhibitory effect of the SSB protein core. Although the binding affinity of SSB for both χ and PriA is enhanced if SSB is prebound to ssDNA, this effect is larger with PriA indicating a further enhancement of SSB specificity for PriA. These results also suggest that DNA binding proteins such as PriA, which also interact with SSB, could use this interaction to gain access to ssDNA by first interacting with the SSB C-termini.
A major role of single stranded DNA (SSB) binding proteins is to coat extended regions of single stranded (ss) DNA formed transiently during DNA metabolic processes (1, 2). However, in recent years, it has become clear that SSB plays the additional more complex role of interacting with a variety of other proteins and enzymes to organize and facilitate their functions in DNA metabolism (3). To date, at least 14 other proteins have been shown to interact with the E. coli SSB protein during DNA replication (DNA pol III holoenzyme (4–7), primase (6, 8)), recombination (RecQ (9–11), RecO (12–15), RecJ (16), RecG (17, 18)), repair (ExoI (19–21), uracil DNA glycosylase (22, 23), DNA pol II (24, 25), DNA pol V (26)) and replication restart (PriA (27, 28), PriB (29)). Hence, rather than providing solely an inert protective function, E. coli SSB protein also serves as a central scaffolding protein to recruit other proteins to their sites of function on DNA. All of these multiple interacting partners appear to bind via the unstructured C-termini of SSB.
The homotetrameric E. coli SSB protein (30) binds with high affinity, but little sequence specificity to ssDNA (1, 31, 32). Even ssDNA binding is complex in that SSB can bind ssDNA in multiple modes differing in the number of subunits used to contact the DNA. Two major ssDNA binding modes, (SSB)35 and (SSB)65, have been identified where the subscripts denote the average number of nucleotides occluded by an SSB tetramer (33). The relative stabilities of these binding modes depend on salt concentration and type (33–35) as well as protein to DNA ratio (36–39). In the (SSB)65 mode, favored at [NaCl]>0.2M, ~65 nucleotides of ssDNA wrap around all four subunits of the tetramer, while displaying only “limited” cooperativity between adjacent tetramers. SSB is also able to readily diffuse along ssDNA in its (SSB)65 binding mode, thus making it easy to be moved without fully dissociating from the ssDNA (40, 41). In the (SSB)35 mode, favored at [NaCl]<0.02M and high SSB to DNA ratios, ~35 nucleotides interacts with an average of only two subunits of the tetramer interact with ssDNA, and SSB binding to ssDNA displays high cooperativity and an ability to form protein clusters (31, 33, 34, 37, 39, 42, 43). As such, the fully wrapped (SSB)65 binding mode can be populated even at low salt concentrations at low binding density (low protein to ssDNA ratio) (39, 43).
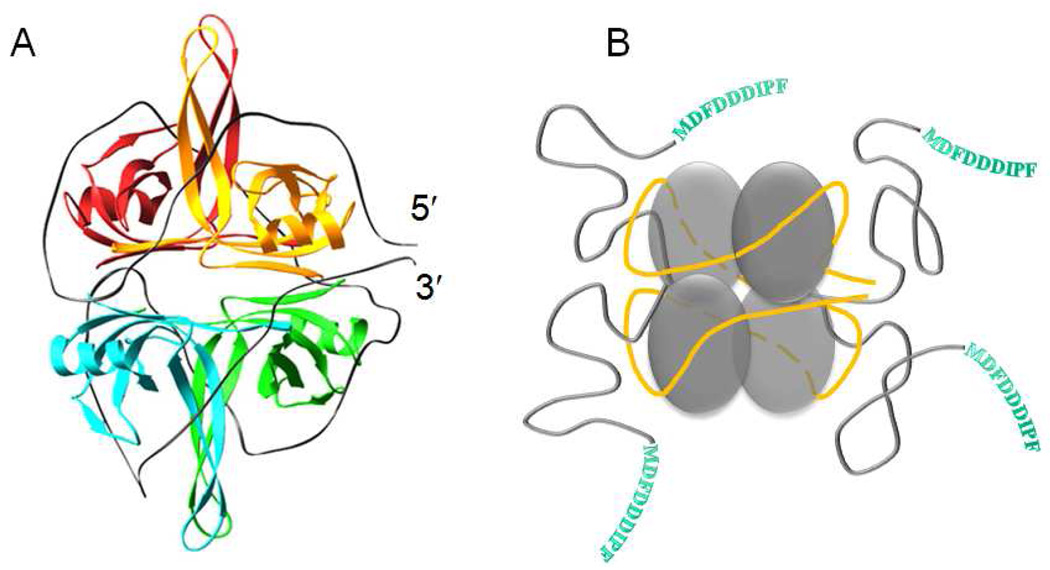
Models have been proposed for both the (SSB)35 and (SSB)65 binding modes based on x-ray crystal structures of a C-terminal truncation of SSB (missing residues 135–177) bound to two molecules of dC35 (44). Figure 1a shows the homotetrameric structure of the core DNA binding domains (residues 1–112) each of which forms an OB-fold, as well as the proposed topology of ssDNA wrapping in the (SSB)65 binding mode, where ~65 nucleotides of ssDNA enter and exit in close proximity. The electron densities for the four C-terminal tails (residues 113–177) are not observable in the structure, even when SSB is bound to ssDNA (45), suggesting as depicted in Figure 1b, that the SSB C-terminal tails are disordered, consistent with early proteolysis studies (46). In fact, the C-termini of E. coli SSB possess the characteristics of intrinsically disordered proteins (IDP) (47). Its sequence is significantly enriched with E, P, Q, S, R, M and D (51% of all residues), the amino acids which are believed to hinder protein folding, and A and G (35% of residues), considered as neutral (48).
Figure 1.
SSB homotetramer complexed with ssDNA in its (SSB)65 binding mode. (A) - model depicting 70 nucleotides of ssDNA (grey ribbon) wrapped around the four SSB subunits, consisting of four OB folds, based on the X-ray crystallographic structure of the SSBc tetramer bound to two molecules of (dC)35 (44). (B) – a cartoon representing ssDNA (yellow ribbon) wrapped around the SSB core in its (SSB)65 binding mode, corresponding to the structural model in panel A, with the addition of the unstructured C-terminal tails (shown in grey) that are not observed in the crystal structure. The 9 amino acids end sequence of each C-terminal, responsible for the interaction of SSB with other metabolic proteins, is shown in single letter amino acids codes.
Previous studies of the interaction of E. coli SSB with its binding partners suggest that the major point of interaction is with its unstructured C-termini, in particular the last nine amino acids, which is highly acidic (3). Isothermal titration calorimetry (ITC) was used to characterize interactions of SSB and its terminal peptide with RecQ helicase (10). Binding of SSB to the χ subunit of DNA pol III holoenzyme in the presence or absence of ssDNA was investigated using Surface Plasmon Resonance (SPR) (4, 5, 7), analytical ultracentrifugation (AU) (7) and equilibrium gel filtration (4). Limited SPR data are also available for PriA helicase interaction with SSB C-terminal peptide (28), E. coli UDG (23) and RecO (12) with SSB, and AU has been used to study exo I interaction with SSB and its C terminus (20). However, since all previous studies investigated the binding of a single protein to SSB, it is not clear whether SSB shows any specificity for its binding partners. In addition, since the homotetrameric SSB protein possesses four C-termini, it is also possible that the stoichiometry of binding might differ depending on its binding partner or by the presence of bound DNA. In the current study we have used isothermal titration calorimetry (ITC) to examine questions of specificity and stoichiometry for the interaction of SSB and its ssDNA complexes with two of its binding partners, the χ subunit of DNA pol III and the PriA helicase. Many IDPs involved in signaling also interact with multiple binding partners (49), similar to SSB (3). Therefore, our studies of the binding specificity of the SSB-Ct with its many binding partners will also contribute to our understanding of the mechanisms of IDP function.
E. coli DNA polymerase III holoenzyme is a multisubunit replicase consisting of three subassemblies: (a) DNA pol III core - (comprised of the α polymerase, ε exonuclease and θ stability factor; (b) the β clamp processivity factor, and (c) the γ clamp loader complex (comprised of the γ,δ, δ’, ψ and χ subunits) (50–52). The latter subcomplex loads the processivity factor onto DNA in an ATP dependent manner and helps tie the holoenzyme together through a network of protein-protein interactions. It has been shown that the χ subunit of the γ complex stimulates β clamp loading and helps to localize the holoenzyme to ssDNA coated with SSB via an interaction between the χ subunit and the C-terminal unstructured tail of the SSB protein (4, 5).
E. coli PriA is a key replication protein that plays a critical role in assembly of the primosome, a multiprotein complex involved in restarting stalled replication forks (53–55). PriA is an SF2 superfamily helicase which unwinds dsDNA in 3’ to 5’ direction. The helicase activity of PriA on branched DNA substrates is stimulated specifically by SSB (27, 28), and this stimulation appears to be dependent upon an interaction with the C-termini of SSB.
Materials and Methods
Reagents and Buffers
All buffer solutions were prepared with reagent grade chemicals and distilled water that was subsequently treated with a Milli Q (Millipore, Bedford, MA) water purification system. Buffer T is 10 mM Tris , pH 7.5, 10% (v/v) glycerol, buffer H is 10 mM Hepes, pH 8.1, and buffer C is 10 mM Cacodylate, pH 7.0, 25% glycerol. All buffers contained 0.1 mM Na3EDTA and either 1mM BME or 0.5 mM TCEP. The concentrations of NaCl were 20 or 200 mM for “low” and “moderate” salt conditions, respectively, and are specified in the text.
E. coli SSB, PriA helicase and χ subunit of DNA Polymerase III Holoenzyme
SSB protein was purified as described (56) with the addition of a double stranded DNA cellulose column to remove a minor exonuclease contaminant (57). PriA helicase was expressed and purified as described (58, 59). The E. coli chi (χ) subunit was overexpressed from E. coli strain BL21(DE3)PlysS transformed with plasmid pET3c-χ , which was a generous gift from Dr. M. O’Donnell. Cells were grown in 4 Liters of LB broth at 37°C to OD600=1.0 and then induced by adding 0.4 mM IPTG. Lysis and further purification of the χ protein was performed following a modified protocol (60). The ammonium sulfate precipitation and ATP agarose column steps were skipped and an additional purification step using a Mono-Q column (1ml, Amersham Bioscience, 0–0.5 M NaCl gradient) was added after the Q-sepharose Fast Flow and Heparin agarose columns. The purified protein (>98% pure, judged by SDS-PAGE) was dialyzed versus purification buffer (20 mM Tris, pH 7.5, 10% glycerol, 2mM EDTA, 2 mM DTT), concentrated and stored at −80°C. The purified protein has no detectable exonuclease activity on 5’ 32P labeled dT20 (61) even after 2 hours of incubation. Sedimentation equilibrium analysis of the protein in Tris and Hepes buffers (pH 8.1, 0.02 and 0.2M NaCl) in the range of concentrations from 2 to 13 µM indicated a monomeric species (MW ≈ 17 kDa) with no detectable higher order oligomeric species. SSB protein concentrations were determined spectrophotometrically in Tris buffer (pH 8.1, 0.2 M NaCl) using an extinction coefficient of ε280=1.13×105 M−1 (tetramer) cm−1 (34). The concentrations of PriA and χ were determined using extinction coefficients of ε280=1.06×105 M−1 cm−1 (59) and ε280=2.92×104 M−1 cm−1 (60), respectively.
C-terminal SSB peptides and DNA
Synthetic peptides, MDFDDDIPF (P9) and, PSNEPPMDFDDDIPF (P15), corresponding to the sequences of the last 9 and 15 amino acids of the SSB C-terminus were obtained from Celtek Peptides ( Celtek Bioscience, LLC, TN). In addition, and to allow for better quantification of the peptide concentration, a modified P9 peptide containing a Trp residue on the N terminal side, designated WP9 (Celtek Peptides), was also used. In order to examine the effect of sequence specificity on peptide binding we also synthesized a peptide with the randomly generated sequence, WDFMDDPFID (WP9r) (obtained from GenScript Corp., NJ) and a peptide containing a Pro to Ser mutation, WMDFDDDISF (WP9-113) (Celtek Peptides), corresponding to the SSB-113 mutation, which is known to disrupt a number of protein interactions with the SSB C-terminus (1, 32). For peptides that do not contain Trp, their concentration was measured based on an extinction coefficient calculated for the two Phe residues, ε258=195×2=390 M−1 cm−1, otherwise the extinction coefficient corresponding to that of a single Trp residue was used ε280=5500 M−1 cm−1.
The oligodeoxynucleotides, (dT)20, (dT)35 and (dT)70, were synthesized and purified as described (43) and were ≥98% pure as judged by denaturing gel electrophoresis and autoradiography of a sample that was 5′ end-labeled with 32P using polynucleotide kinase. All oligo(dT) concentrations were determined spectrophotometrically in buffer T (pH 8.1), 100 mM NaCl using the extinction coefficient ε260=8.1×103 M−1 (nucleotide) cm−1 (62).
Isothermal Titration Calorimetry (ITC)
ITC experiments were performed using a VP-ITC titration microcalorimeter (MicroCal Inc., Northhampton, MA) (63). Generally, experiments were carried out by titrating χ or PriA solutions (1–5 µM ) with the peptides (generally stock concentrations ranging from 60 to 180 µM)) or with SSB or SSB-ssDNA complexes (reverse titrations) with the concentrations ranging from 5 to 9 µM (tetramer). Some experiments with χ protein were performed with SSB in the cell (2–4 µM tetramer) and titrating with χ protein (20–40 µM stock).
The heats of dilution were usually obtained by a reference titration in which the species in the syringe is titrated into the cell containing buffer solution. All corrections for heats of dilution were applied as described (64). Oligo(dT) and protein samples were dialyzed extensively vs. each particular buffer at the indicated salt concentration used in the ITC experiments.
The stoichiometry of binding and the values of Kobs and ΔHobs were obtained by fitting the ITC titration curves to a model of ligand (X= χ or PriA) binding to n identical and independent sites on the macromolecule (M=SSB) using eq. 1a:
| (1a) |
where is the total heat after the i-th injection and V0 is the volume of the calorimetric cell. The concentration of the free ligand (X) was obtained by solving eq. 1b:
| (1b) |
In eqs 1a and 1b, Xtot and Mtot are the total concentrations of the ligand and macromolecule, respectively, in the calorimetric cell after i-th injection. The same model was applied for analysis of the interaction of the SSB-Ct peptides (X) with χ or PriA (M). Non-linear least squares fitting of the data was performed using the “ITC Data Analysis in Origin” software provided by the manufacturer. The details of the conversion of integral heats to differential heats (heats per injection observed in the experiment) and the fitting routine including corrections for heat displacement effects and ligand and macromolecule dilutions in the calorimetric cell are as described previously (64) and in the ITC Data Analysis in Origin Tutorial Guide (MicroCal Inc.). When necessary additional fitting and simulation of the data were performed using the nonlinear regression package in Scientist (MicroMath Scientist Software, St. Louis, MO).
Data in figure 3A and 3B were fit globally to Eqs. 2,3:
| (2) |
| (3) |
Where R is the gas constant, ΔKobs,ref and ΔHobs,ref are the association equilibrium binding constant and enthalpy change at Tref=25°C and ΔCp is the heat capacity change, assumed to be independent on temperature.
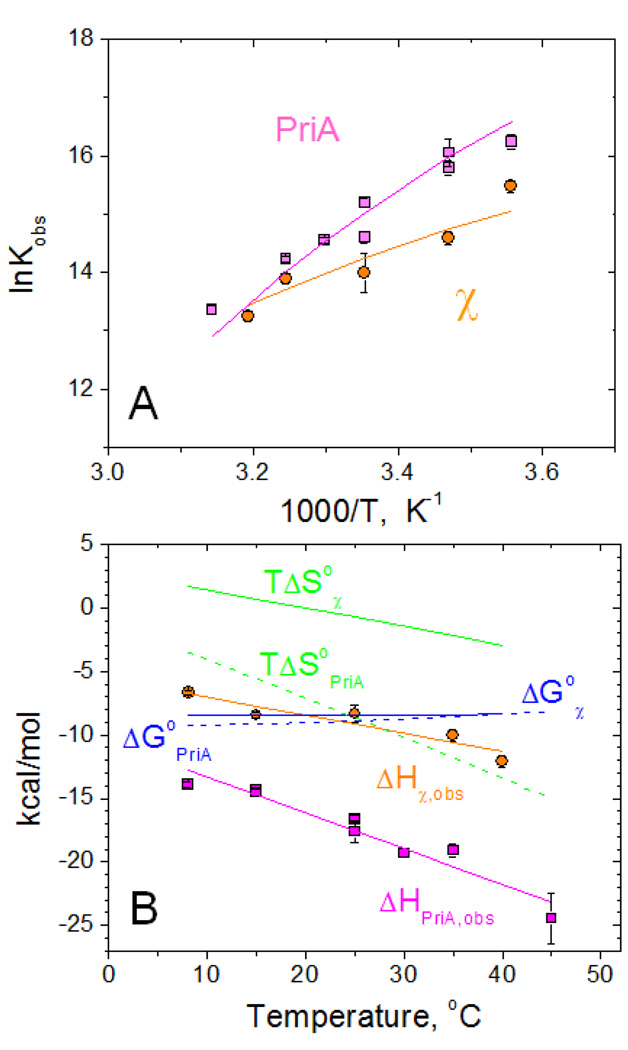
Figure 3.
Temperature dependence of the interaction of PriA and χ with SSB-Ct peptide (WP9) performed in low salt conditions (20 mM NaCl) in buffer C (10mM Cacodylate, pH 7.0, 25% glycerol)
(A) – van't Hoff plots of the dependences of Kobs on temperature for PriA (magenta squares) and χ (orange cirles).
(B) - Temperature dependences of thermodynamic parameters: ΔHobs (PriA – magenta squares and χ - orange circles), ΔG°obs (blue dashed and solid lines for PriA and χ, respectively) and TΔS°obs (green dashed and solid lines for PriA and χ, respectively)
Solid lines (magenta for PriA and orange for χ ) through the experimental points in panels A and B represent global fits of the data to eqs 2–3 (see Materials and Methods) with the following parameters: Kobs,25°C=(3.4±0.6)×106 M−1, ΔHobs,25°C = −17.6±0.2 kcal/mol and ΔCpobs= −281±21 cal/mol deg for PriA and Kobs,25°C =(1.6±0.4)×106 M−1, ΔHobs,25°C = −9.2±0.3 kcal/mol and ΔCpobs= −143±21 cal/mol deg for χ. The dependences of ΔG°obs and TΔS°obs shown in panel B were simulated using ΔG°obs=−RTlnKobs and TΔS°obs=ΔHobs − ΔG°obs.
RESULTS
Interactions of χ and PriA with SSB C-terminal peptides and ssDNA
Using ITC we first investigated the binding of short synthetic peptides containing the SSB C-terminal tail sequence to χ and PriA. We examined the effects of peptide length and amino acid sequence as well as the effects of solution conditions.
Binding of PriA and χ to SSB C-terminal peptides in the absence of ssDNA
Functional interactions of SSB with other proteins appear to require a minimum of the last 9–10 amino acids of the SSB C-terminus since deletion of these amino acids renders inviable E. coli cells expressing the truncated protein (65). Figure 2 shows typical ITC titration results (buffer T, 20 mM NaCl, 25°C) for the binding of χ to peptides, P9 and P15, corresponding to the last 9 and 15 amino acids of the SSB C-terminus. The titrations were analyzed using an n-independent and identical sites model (see eq.1 in Materials and Methods), providing estimates of the binding stoichiometry (n), equilibrium association constant (Kobs) and binding enthalpy (ΔHobs). The results indicate little quantitative difference in the binding of these peptides to the χ subunit (see Table 1). Similar equilibrium binding parameters were obtained by ITC for the interaction of these peptides with PriA under identical solution conditions (Table 1). To attain more accurate quantification of the peptide concentration we introduced an additional Trp residue at the N -terminal position of the P9 peptide (WP9 peptide in Table 1). No differences in binding parameters were found compared to the peptide without Trp, and thus we used the WP9 peptide for the remaining studies.
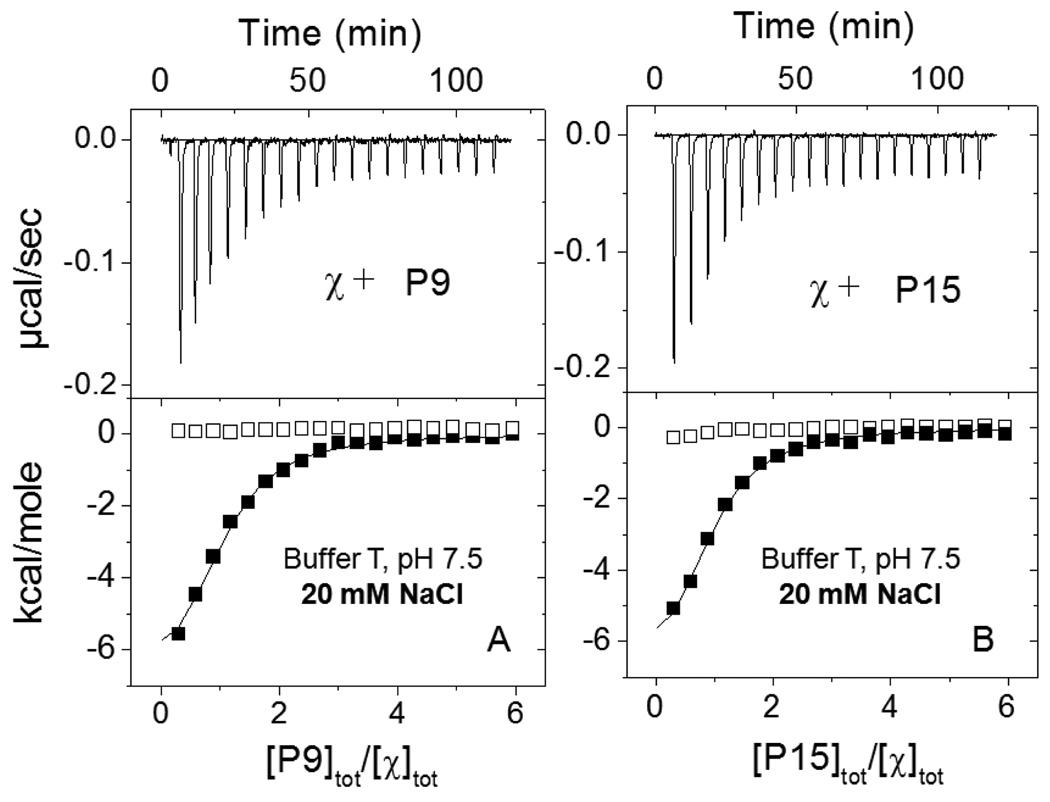
Figure 2.
Results of ITC titrations of χ protein with the P9 and P15 peptides corresponding to the SSB C-terminal sequences of 9 (A) and 15 (B) amino acids under low salt conditions (buffer T: 10 mM Tris, pH 7.5, 10% glycerol, 20 mM NaCl, 25°C). Upper panels show the raw titration data, plotted as the heat signal (microcalories per second) versus time (minutes), obtained for 19 injections (15 µl each) of the peptide (100µM) into a solution containing χ protein (3.7 µM). Lower panels show the integrated heat responses per injection, normalized to the moles of injected peptide, after subtraction of the heats of dilution obtained from the blank titration of peptide into buffer. The smooth curves represent the best fit of the data to an n - independent and identical sites model (eq. 1 in Materials and Methods) with n=0.92±0.04, Kobs=(0.80±0.08)×106 M−1 ΔH= −8.2±0.4 kcal/mol for P9 (A) and n=0.88±0.04, Kobs=(0.83±0.09)×106 M−1 ΔH= −7.7±0.4 kcal/mol for P15 (B).
Table 1.
ITC binding data for the interaction of χ and PriA with E.coli SSB C-terminal peptides in different solution conditions*.
| χ P15 |
χ P9 |
χ WP9 |
PriA P15 |
PriA P9 |
PriA WP9 |
|
|---|---|---|---|---|---|---|
| Buffer T, pH 7.5, 25°C 20 mM NaCl |
n=0.88±0.04 K=(0.8±0.1)×106 ΔH= −7.7±0.4 |
n=0.92±0.04 K=(0.8±0.1)×106 ΔH= −8.2±0.4 |
_ | n=1.03±0.06 K=(1.6±0.4)×106 ΔH= −7.0±0.5 |
n=0.89±0.05 K=(2.0±0.3)×106 ΔH= −6.7±0.3 |
_ |
| Buffer C, pH 7.0, 25°C 20 mM NaCl |
n=0.91±0.02 K=(1.5±0.2)×106 ΔH= −9.5±0.3 |
n=1.0±0.1 K=(1.6±0.4)×106 ΔH= −7.9±0.4 |
n=1.02±0.03 K=(1.2±0.1)×106 ΔH= −8.0±0.3 |
n=0.90±0.02 K=(2.7±0.2)×106 ΔH= −20.7±0.8 |
_ | n=0.95±0.02 K=(1.6±0.2)×106 ΔH= −14.5±0.5 |
| Buffer C, pH 7.0, 25°C 200 mM NaCl |
_ | _ | n=0.96±0.03 K=(3.0±0.2)×105 ΔH= −9.2±0.4 |
n=0.92±0.03 K=(3.6±0.2)×105 ΔH= −14.0±0.6 |
n=0.81±0.12 K=(1.8±0.6)×105 ΔH= −17.3.0±0.6 |
n=1.1±0.1 K=(2.3±0.3)×105 ΔH= −13.0±1.3 |
binding parameters: n – stoichiometry of binding, K (M−1) – observed association equilibrium constant, ΔH (kcal/mol) – enthalpy change
We next examined the effect of solution conditions, changing buffer type, pH and glycerol concentration, but maintaining the same salt concentration (20mM NaCl) and temperature (25°C). The results of ITC experiments performed in buffer C (pH 7.0, 25% glycerol) (see Table 1) are similar to those obtained in buffer T, except that the ΔHobs is larger in magnitude for the interaction with PriA. Therefore, at low salt (20mM NaCl) both χ and PriA interact with the SSB-Ct peptides with similar affinity but with different binding enthalpies that depend on buffer conditions in the case of PriA. The following summary of the binding parameters is based on an average of the data in Table 1: for χ (in both buffer T and buffer C), n=0.95±0.06, Kobs=(1.2±0.4)×106 M−1, ΔHobs= −8.3±0.8 kcal/mol; for PriA (in buffer T), n=0.96±0.08, Kobs=(1.8±0.5)×106 M−1, ΔHobs= −6.9±0.6 kcal/mol and for PriA (in buffer C), n=0.93±0.03, Kobs=(2.2±0.2)×106 M−1, ΔHobs= −17.6±3.1 kcal/mol.
The approximately two-fold increase in the magnitude of the PriA binding enthalpy in buffer C (Cacodylate), which has a much lower ionization enthalpy (ΔHion= −0.47 kcal/mol (66)) compared to buffer T (Tris, ΔHion= 11.34 kcal/mol (67)), may indicate that the binding of PriA is accompanied by some linked protonation (68). If we ascribe the observed difference in ΔHobs to only protonation effects and neglect the slight difference in pH values we obtain a rough estimate that ~one proton is absorbed upon complex formation (see Supplementary information). Although more comprehensive studies performed in different buffers at multiple pH values (68) are required to quantify this effect, these are beyond the scope of this investigation. Nonetheless, the observed difference in ΔHobs for PriA and χ indicates that the dependences of Kobs on temperature should differ for the binding of the SSB peptide to χ vs. PriA. We therefore performed ITC titrations of both proteins with WP9 over a wide temperature range from 8°C to 45°C. These were performed in buffer C to minimize the possible contribution to ΔHobs due to ionization of the buffer if any protonation effects are linked to binding. The results summarized in Fig. 3 clearly indicate a very different thermodynamic profile for binding of WP9 to PriA vs. χ. In the higher temperature range, the affinities of WP9 for the two proteins are comparable. However, as the temperature is lowered, differences in the binding affinities become evident, suggesting a larger and more negative ΔHobs for the PriA interaction as compared to χ. Indeed, the data in Fig. 3B show that this is the case. Moreover, it appears that the dependence of ΔHobs on temperature for PriA binding is larger indicating a larger negative heat capacity change, ΔCp, for PriA binding than for χ binding. Global fitting to Eqs. 2–3 (see Materials and Methods) of the data in Fig. 3A and 3B yields the following parameters for WP9 binding at 25°C: Kobs,25=(3.4±0.6)×106 M−1, ΔHobs,25= −17.6±0.2 kcal/mol and ΔCpobs= −281±21 cal/mol deg for PriA and Kobs,25=(1.6±0.4)×106 M−1, ΔHobs,25= −9.2±0.3 kcal/mol and ΔCpobs= −143±21 cal/mol deg for χ.
We also examined the interaction of SSB-Ct peptides with PriA and χ at moderate salt (0.2M NaCl, buffer C, 25°C) and the results are shown in Fig. 4 and Table 1. As at lower salt, both proteins also bind the C-terminal peptides with similar affinity, although the values of Kobs are lower by approximately a factor of ten at the higher [NaCl]. The one consistent difference is that the binding of WP9 to PriA (ΔHobs ≈ -(13–17 kcal/mol) is more exothermic than to χ (ΔHobs ≈ −9 kcal/mol). Hence, although the values of ΔGobs (Kobs) for WP9 binding to χ and PriA are similar at 25°C, the overall thermodynamics of binding are significantly different.
Figure 4.
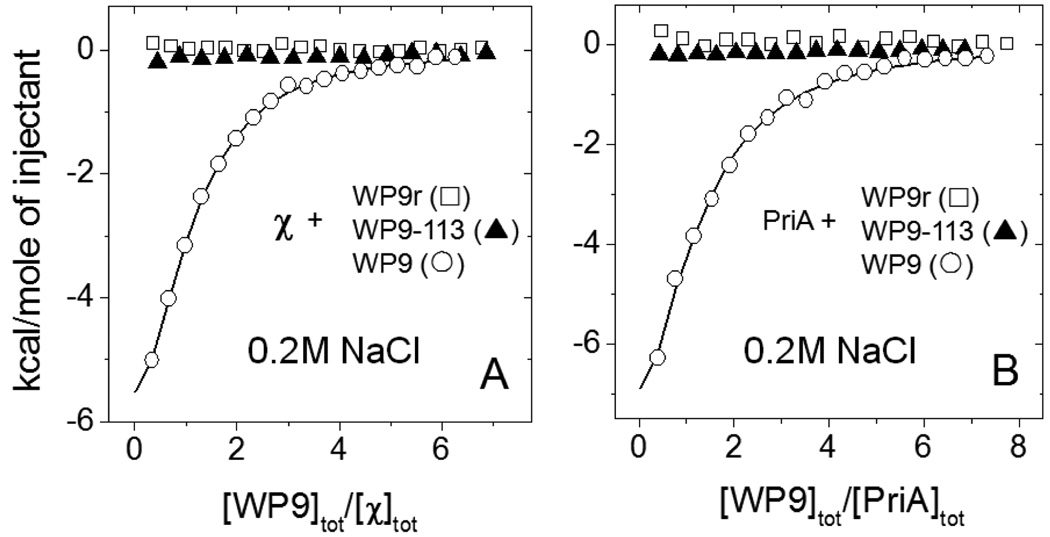
Results of ITC experiments performed at moderate salt conditions (0.2M NaCl, buffer C, 25°C) for the interaction of the χ (A) and PriA (B) with different peptides: WP9 (−) - wild type peptide containing the last 9 amino acid sequence of SSB C-terminus; WP9–113 (7) – last 9 amino acid peptide sequence containing Pro to Ser replacement corresponding to the sequence of the SSB-113 mutation P176S and WP9r (∀) - a peptide containing the same amino acids as in wild type, but randomized. Throughout all the titrations the concentrations of the proteins in the cell were within 4–5µM and the concentrations of the peptides in the syringe were 150–200 µM. The smooth curves represent the best fit of the data to an n - independent and identical sites model (eq. 1 in Materials and Methods) with n=0.96±0.03, Kobs=(3.04±0.17)×105 M−1 ΔH= −9.2±0.4 kcal/mol for χ-WP9 titration (A) and n=1.01±0.10, Kobs=(2.25±0.24)×105 M−1, ΔH= −13.7±0.2 kcal/mol for PriA-WP9 titration (B).
We next performed ITC titrations of χ and PriA with two peptides, one having a single Pro to Ser substitution at the penultimate position, which corresponds to the P176S mutation found in the SSB-113 mutant (designated WP9-113), and another peptide containing the same amino acids as in WP9, but with a randomized sequence (designated WP9r) (see Fig. 4). For comparative purposes these ITC titrations were performed using the same concentrations of proteins and peptides as was used in the experiments with the wild type peptide. No binding can be detected for either peptide indicating that peptide binding is sequence specific.
PriA vs. χ binding to ssDNA
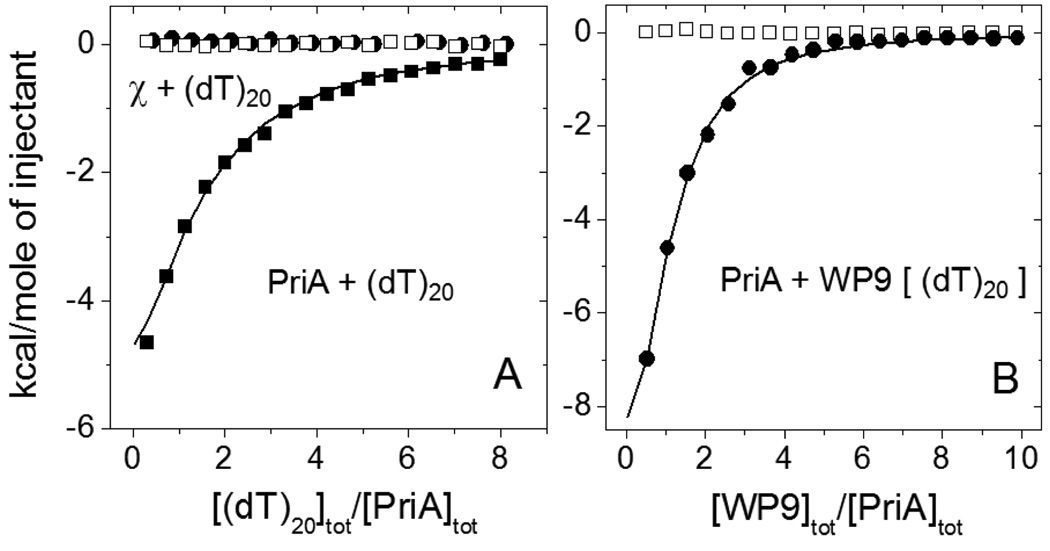
PriA binds to ssDNA in the absence of nucleotide cofactors with an occluded site size of ~20 nucleotides and with some specificity for pyrimidines (58, 59). However, there is little information on the binding of χ to ssDNA. As necessary background for our SSB studies, we therefore compared the binding of both PriA and χ to the oligodeoxythymidylates, dT20 and dT70, under both moderate and low salt conditions (buffer C, 25°C). These direct binding studies showed no detectable binding of χ to ssDNA under these conditions (open squares in Fig. 5A and 6C, respectively). In contrast, PriA shows significant binding to dT20 (Fig. 5A) and dT70 (data not shown) with stoichiometries of one and three PriA molecules per oligo(dT), respectively, and with affinities similar to those determined for PriA binding to the C terminal SSB peptides (see Table 2). We note that these PriA-(dT)L binding data were obtained at moderate salt conditions (0.2 M NaCl). When experiments were performed at lower NaCl concentration (20 mM) PriA-ssDNA complex formation was accompanied by significant aggregation, hence we could not obtain quantitative binding information under these conditions.
Figure 5.
ITC data for the interaction of χ and PriA with (dT)20 and PriA with WP9 in the presence of saturating amount of (dT)20.
(A) – Titration of PriA (4.1µM) with (dT)20 (161µM) (!) in buffer C, 0.2M NaCl, 25°C. The smooth curve represents the best fit of the data to an n - independent and identical sites model (eq. 1 in Materials and Methods) with n=1.0±0.2, Kobs=(1.49±0.25)×105 M−1, ΔH= −12.3±3.1 kcal/mol. The injection heats for the titration of χ (5.0 µM) with (dT)20 (195 µM) (,) are indistinguishable from reference titrations of the proteins into the buffer (∀).
(B) - Titration of PriA (3.8 µM) with WP9 (180 µM) (,) in buffer C, 0.2M NaCl, 25°C, in the presence of saturating amount of (dT)20 (16µM, ~80% of PriA estimated to be in the complex with (dT)20 based on the data presented in panel (A)). The smooth curve represents the best fit of the data to an n - independent and identical sites model with n=0.90±0.08, Kobs=(4.04±0.45)×105 M−1, ΔH= −14.4±1.4 kcal/mol.
Figure 6.
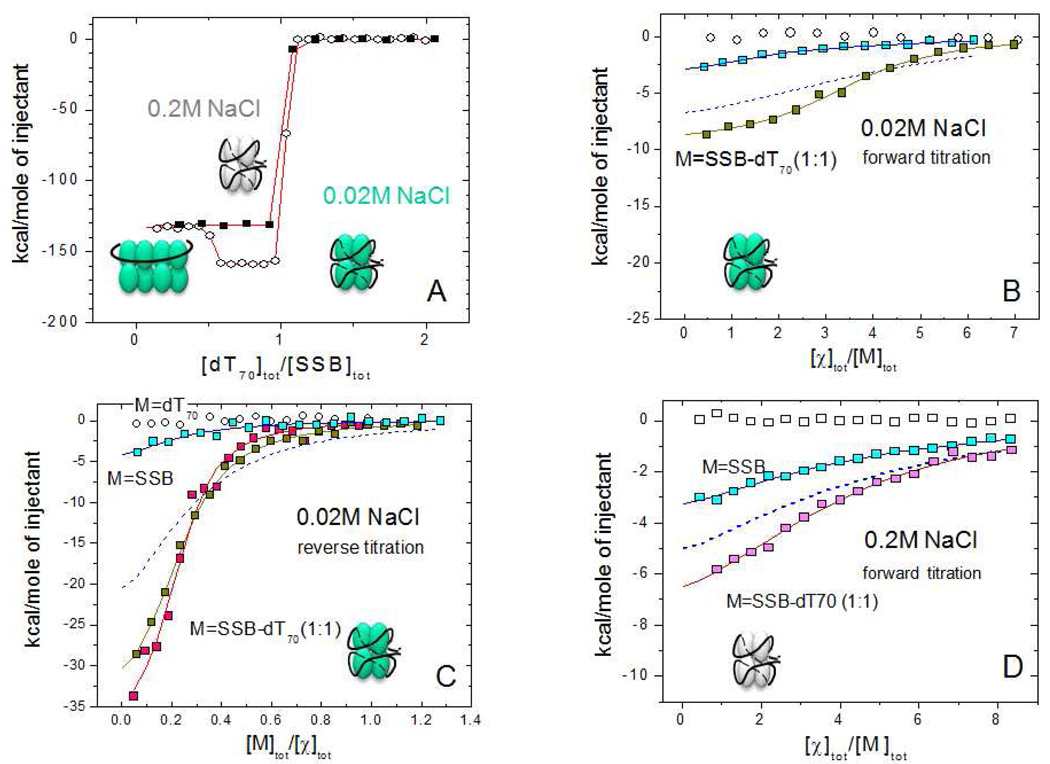
Results of ITC experiments for the binding of χ to SSB and SSB-dT70 at low (0.02M) and moderate (0.2M) NaCl concentrations.
(A) – ITC titration of SSB (1 µM) with (dT)70 at 0.2 M NaCl (!) and 0.02 M NaCl (−) in buffer C, 25°C (concentrations of dT70 are 10 and 14 µM, respectively). At low salt the formation of a stoichiometric (SSB)35 complex (with two SSB tetramers bound per (dT)70 ) occurs at [dT70]tot/[SSB]tot ≤ 0.5, which then rearranges to a 1:1 (SSB)65 complex when [dT70]tot/[SSB]tot=1.0.
(B) – Forward titrations of SSB (0.6 µM) and an SSB-dT70 (1:1) complex (0.53 µM ) with χ protein (17 µM) (blue and yellow squares, respectively) in buffer H, low salt (0.02M NaCl). The smooth curve through SSB-dT70 data points represents the best fit to an n - independent and identical sites model (eq. 1 in Materials and Methods) with n=3.5±0.1, Kobs=(5.5±0.8)×106 M−1, ΔH= −9.5±0.7 kcal/mol. The isotherm shown with a blue dashed curve was simulated using n=4, Kobs=1.2×106 M−1, ΔH= −8.3 kcal/mol (based on the χ-SSB-Ct binding parameters).
(C) - Reverse titrations of χ protein (0.9–1.1 µM) with (dT)70 (open squares), SSB (6.4 µM , blue squares) and SSB-(dT)70 (1:1) complex in buffer H (5.4 µM complex, yellow squares) and buffer C (4 µM complex, magenta squares) at low salt ( 0.02M NaCl). The smooth curves through SSB-(dT)70 data points represent the best fits to an n -independent and identical sites model with n=4.3±0.2, Kobs=(3.8±0.4)×106 M−1, ΔH= −8.9±0.2 kcal/mol (buffer H) and n=4.5±0.2, Kobs=(7.6±1.6)×106 M−1, ΔH= −8.7±0.3 kcal/mol (buffer C). The isotherm shown as a blue dashed curve was simulated using n=4, Kobs=1.2×106 M−1, ΔH= −8.3 kcal/mol (based on the χ-SSB-Ct binding parameters)
(D) – Forward titration of SSB (1µM) (blue squares) and SSB-(dT)70 (1:1) complex (1.1 µM) (magenta squares) with χ protein ( 40 µM ) in buffer C (0.2M NaCl). The smooth lines represent the best fit of the data to an n - independent and identical sites model with n=3.9±0.4, Kobs=(3.0±0.7)×105 M−1, ΔH= −6.1±1.1 kcal/mol (SSB) and n=3.9±0.3, Kobs=(5.4±1.0)×105 M−1, ΔH= −9.5±1.0 kcal/mol (SSB-dT70 complex). The isotherm shown as a blue dashed curve is a simulation using n=4, Kobs=3×105 M−1, ΔH= −9.2 kcal/mol (based on the χ-SSB-Ct binding parameters)
Table 2.
Comparison of the binding parameters for the interaction of χ and PriA with SSB-Ct peptides, SSB, SSB-dT70 (1:1) complexes and oligo(dT)*
| SSB-Cta | SSBb | SSB-dT70 (1:1)b | dT20 | dT70 | |
|---|---|---|---|---|---|
|
0.2 M NaCl χ |
n=0.96±0.03 K=(3.0±0.2)×105 ΔH= −9.2±0.4 |
n=3.9±0.4 K=(3.0±0.7)×105 ΔH= −6.1±1.1 |
n=3.9±0.3 K=(5.4±1.0)×105 ΔH= −9.5±1.0 |
Not detectable | Not detectable |
|
0.2 M NaCl PriA |
n=0.94±0.15 K=(2.6±0.9)×105 ΔH= −14.8±2.2 |
n=4.0 (fixed) K=(4.5±0.5)×105 ΔH= −7.5±0.5 |
n=3.0±0.3 K=(4.0±0.5)×106 ΔH= −18.3±2.0 |
n=1.1±0.2 K=(1.5±0.3)×105 ΔH= −12.3±3.1 |
n=3.1±1.0 K=(1.0±0.7)×105 ΔH= −12.9±0.6 |
|
0.02 M NaCl χ |
n=0.95±0.06 K=(1.2±0.4)×106 ΔH= −8.3±0.8 |
Weak (see text) |
n=4.1±0.5 K=(5.6±1.9)×106 ΔH= −9.0±0.4 |
Not detectable | Not detectable |
|
0.02 M NaCl PriA |
n=0.93±0.02 K=(2.2±0.2)×106 ΔH= −17.6±3.1 |
Precip. | Precip. | Precip. | Precip. |
binding parameters: n – stoichiometry of binding, K (M−1) – observed association equilibrium constant, ΔH (kcal/mol) – enthalpy change
the results for the interaction of χ and PriA with SSB-Ct peptides are averaged based on the data in Table 1 for experiments performed in buffer C
the results represent an average of 2–4 experiments performed in buffer C
To determine whether prebinding of ssDNA to PriA affects PriA binding to the SSB-Ct tail, we performed titrations of PriA with the C-terminal peptide WP9 in the presence of nearly saturating dT20 (~80% of PriA is complexed with dT20). The results shown in Fig. 5B, and the averaged values from two titrations (n=0.91±0.11, Kobs=(4.1±0.8)×105 M−1, ΔHobs= −13.9±2.0 kcal/mol) indicate that there is little or no effect of prebound ssDNA on the binding of PriA to the C-terminal peptide (compare with the titration of PriA with WP9 in the absence of ssDNA (n=1.1±0.1, Kobs=(2.3±0.3)×105 M−1, ΔHobs= −13.0±1.3 kcal/mol, averaged based on two titrations, see Table 1)).
PriA and χ binding to SSB and SSB-oligo(dT) complexes
We next examined the extent to which full length SSB or SSB-ssDNA complexes show any binding specificity for χ vs. PriA. We also examined whether the stoichiometry of χ or PriA binding to SSB is influenced by the different modes of ssDNA binding to the SSB tetramer. Since one of the main factors affecting the formation of the different SSB binding modes on long ssDNA is the monovalent salt concentration (33, 34, 42) we also probed the interactions of χ and PriA with SSB-ssDNA complexes at low (20mM NaCl) and moderate (0.2M NaCl) salt conditions, which favor the (SSB)35 and (SSB)65 binding modes, respectively. Figure 6A shows that for both NaCl concentrations, an SSB tetramer forms a high affinity (stoichiometric) 1:1 molar complex with (dT)70 (Kobs >1010 M−1 ) (Buffer C, pH 7.0, 25% glycerol, 25°C). In this complex (dT)70 interacts with all four subunits of the SSB tetramer to form a fully wrapped (SSB)65 complex (32, 44). As demonstrated previously by Roy et al (39) and by sedimentation equilibrium (Kozlov and Lohman, unpublished data) and as is evident in Figure 6A, an intermediate 2:1 molar complex of SSB tetramer bound to (dT)70 can also form at the lower [NaCl] (20 mM) when SSB is in molar excess over (dT)70. Due to the very high affinity of the 1:1 SSB-(dT)70 complex, no appreciable dissociation of this complex occurs in the ITC experiments even under conditions of a reverse titration (Fig. 6C and Fig. 7) when an SSB-(dT)70 complex (5–9µM) is diluted ~ 100-fold upon injection into the calorimetric cell. This was confirmed by performing control titrations of the 1:1 molar SSB-(dT)70 complex into the buffer solution for every experiment. In all cases the resulting reference heats of dilution are comparable to those obtained for reference titrations of buffer into buffer (data not shown).
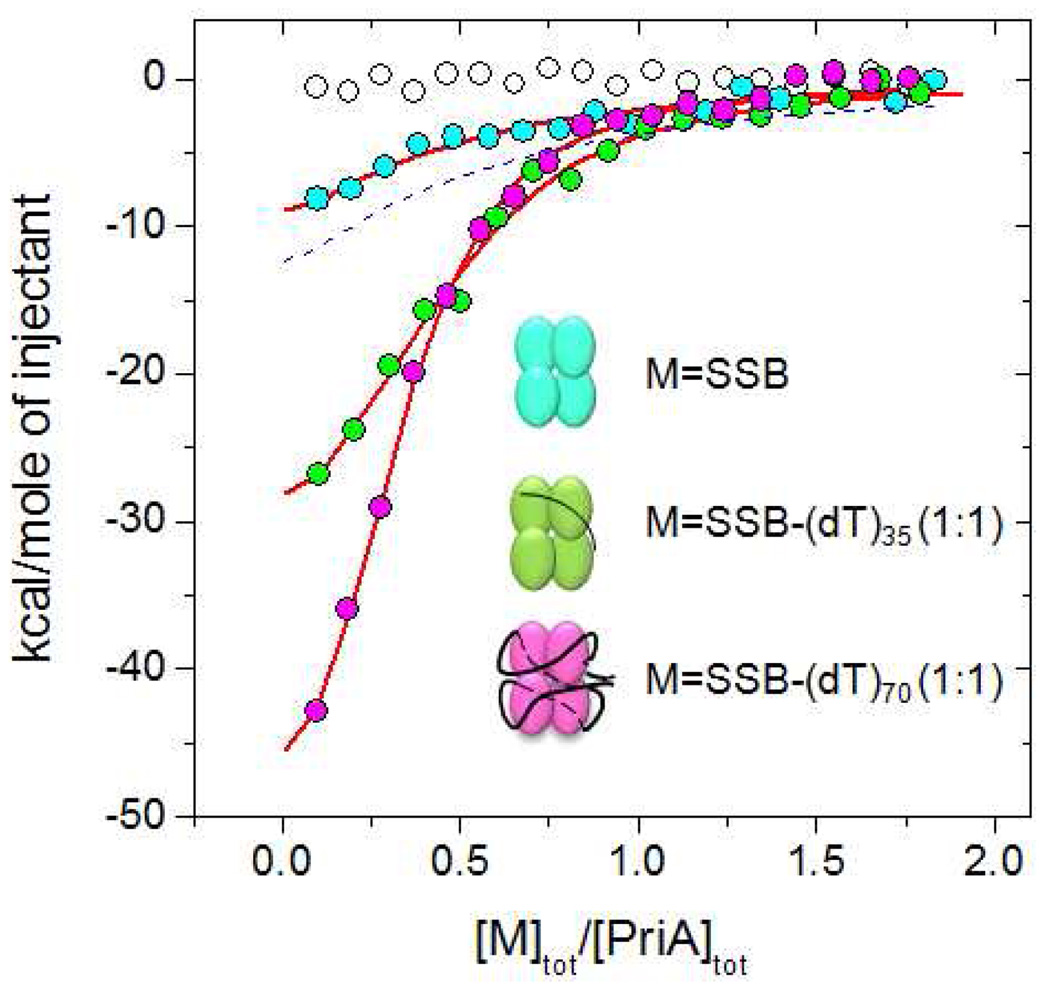
Figure 7.
Comparison of PriA binding to SSB and SSB-(dT)35 and SSB-(dT)70 (1:1) complexes at moderate salt (0.2M NaCl, buffer C, 25°C). The blue, green and magenta circles represent ITC titrations with SSB and SSB-(dT)35 and SSB-(dT)70, respectively. The experiments were performed by titrating 1µM of PriA with 9µM of SSB tetramer. The smooth curves represent the best fits of the data to an n – independent and identical sites model ( eq. 1 in Materials and Methods) with the following parameters: Kobs=(3.9±0.9)×105 M−1, ΔH= −8.1±1.0 kcal/mol n=4 (fixed) for SSB; n=2.2±0.1, Kobs=(3.2±0.7)×106 M−1, ΔH= −16.6±0.6 kcal/mol for the SSB-dT35 (1:1) complex and n=3.2±1.0, Kobs=(3.7±0.5)×106 M−1, ΔH= −18.3±0.4 kcal/mol for the SSB-dT70 (1:1) complex. The blue dashed curve is a simulation using n=4, Kobs=2.6×105 M−1, ΔH= −14.8 kcal/mol (based on the PriA-SSB-Ct binding parameters)
Interaction of χ with SSB and SSB-oligo(dT) complexes at low and moderate salt conditions
We examined χ binding to SSB and SSB-(dT)70 (1:1 molar) complexes at low salt concentrations (Buffer H, pH 8.1, 20mM NaCl, 25°C) by titrating χ into the SSB solutions (forward titration) (Fig. 6B) as well as by titrating either SSB or SSB-(dT)70 complexes into a χ protein solution (reverse titration) (Fig. 6C). Both experiments indicate that χ binds with much higher affinity to SSB when SSB is prebound to (dT)70. The titrations in Fig. 6 were analyzed using an n-independent and identical sites model (see eq. 1 in Materials and Methods), providing estimates of the binding stoichiometry (n), equilibrium association binding constant (Kobs) and binding enthalpy (ΔHobs). For both forward and reverse titrations of SSB alone (blue squares), the observed heats differ only slightly from those obtained in the reference titrations (χ or SSB into buffer - open circles), hence we could not obtain accurate binding parameters from a nonlinear least squares analysis. However, if we simulate an isotherm (dashed blue lines in Figures 6B and 6C) by assuming a stoichiometery of four χ per SSB tetramer using the binding parameters obtained from the χ-SSB-Ct peptide experiments ( Kobs = 1.2 × 106 M−1 and ΔHobs = −8.3 kcal/mol, see Table 2), we find that the experimental χ-SSB interaction is weaker than predicted from the simulation (solid vs. dashed blue lines, respectively). We hypothesize that the weaker interaction of SSB with χ may be due to an inhibitory effect of the SSB core, which may interact with the SSB-Ct in a nonspecific manner and therefore inhibit χ binding (see Discussion).
ITC titrations of χ with SSB-(dT)70 complexes (yellow squares, Fig. 6B and 6C) demonstrate that χ binds with higher affinity to an SSB-ssDNA complex, as shown previously at low salt (7). Both forward and reverse titrations fit well to an n-independent and identical site model with similar binding parameters: n=3.5±0.1, Kobs=(5.5±0.8)×106 M−1, ΔHobs = −9.5±0.7 kcal/mol., and n=4.3±0.2, Kobs=(3.8±0.4)×106 M−1, ΔHobs = −8.9±0.2 kcal/mol, respectively. It appears that there is at least a 3-fold increase in the observed affinity of χ for the SSB-ssDNA complex compared to the C-terminal peptides, while no significant change in ΔHobs is observed. Titrations performed at the same salt concentration but in buffer C (see Fig. 6C, magenta squares) indicate that there is little effect of pH, glycerol concentration or buffer type (n=4.5±0.2, Kobs=(7.6±1.6)×106 M−1, ΔHobs = −8.7±0.3 kcal/mol). This observation is important for our comparisons of these results with those for PriA, which could only be obtained in buffer C due to solubility problems in other conditions.
Due to the decreased affinity at moderate salt concentrations (0.2M NaCl) we could only obtain reliable binding data for the interaction of χ with SSB and SSB-(dT)70 complexes using forward titrations. The data shown in Fig. 6D were fit to an n-independent and identical sites model, yielding a similar stiochiometry (~4), but a somewhat lower affinity and less favorable binding enthalpy for the binding to SSB alone (see Figure 6 legend and Table 2). Generally, the results show at least a 10-fold decrease in affinity due to the increase in NaCl concentration, similar to what we observed for the SSB-Ct peptide experiments (see Table 2). However, χ binds stronger to the SSB-(dT)70 complex than to SSB alone. On the other hand, it appears that in the absence of ssDNA the interaction of χ with the C-terminal tails is weaker when the tails are part of the SSB protein. To emphasize this point, Figure 6D shows a simulated isotherm (dashed blue curve) for χ binding to four isolated SSB C-terminal tails, based on the parameters for χ binding to the WP9 peptide (see Table 2) (n=4, Kobs=3×105 M−1, ΔH= −9.2 kcal/mol). This comparison indicates a lower binding affinity of χ for the Ct tail when it is attached to the core SSB protein than for the Ct peptide alone, again suggesting an inhibitory effect of the SSB core on binding to the Ct tail, as observed at the lower [NaCl].
PriA binding to SSB and SSB-oligo(dT) complexes at moderate NaCl concentrations
In order to compare PriA binding to SSB and SSB-oligo(dT) complexes with that of χ, we first tested low salt conditions but found that PriA binding is accompanied by partial aggregation. This was also the case in three other buffers (buffers H, T and C, see Materials and Methods), which differ in both pH and glycerol concentration. Although we find that PriA is more soluble in buffer C (10mM Cacodylate, pH 7.0, 25% glycerol), at low [NaCl] (20 mM) we still observe partial aggregation when PriA binds to DNA alone, SSB alone or an SSB-oligo(dT) complex. These problems were eliminated by increasing the [NaCl] to 200 mM (25°C).
Typical results of titrations of PriA with SSB alone or an SSB-(dT)70 (1:1 molar) complex are shown in Fig. 7, from which it is clear that PriA affinity for the SSB-(dT)70 complex is much higher than for SSB alone. For PriA binding to the SSB-(dT)70 complex, we find a stoichiometry of ~ four PriA per SSB tetramer with Kobs=(3.7±0.5)×106 M−1, and ΔHobs = −18.3±0.4 kcal/mol. Unfortunately, the binding of PriA to SSB alone is sufficiently weak that we were unable to obtain reliable binding parameters without constraining the stoichiometry. However, we can compare this isotherm to that predicted from the averaged values for PriA binding to the SSB-Ct peptides alone (n=4, K=2.6×105 M−1 and ΔHobs = −14.8 kcal/mol), shown as a dashed blue curve in Fig. 7. Although the difference is small, this simulated isotherm differs from the isotherm for PriA binding to SSB alone (blue squares) and again suggests that the SSB C-terminal tails bind PriA weaker when they are attached to the SSB protein. Thus, the binding of both PriA and χ to the SSB Ct tails appear to be inhibited when the Ct tail is part of the SSB DNA binding core. Importantly, the data in Fig. 7 also show that PriA binds to the SSB-(dT)70 complex with more than 10-fold higher affinity than does χ (Table 2) indicating a clear and enhanced binding specificity.
We also performed experiments in 0.2M NaCl in buffer T (10mM Tris, pH 7.5, 10% glycerol) since Tris buffer has a higher enthalpy of ionization than does cacodylate buffer. We found that the stoichiometry and affinity of PriA binding were unchanged at 25°C (n=3.8±0.4, Kobs=(1.9±0.5)×106 M−1 and ΔHobs= −8.6±0.3 kcal/mol, data not shown), although a ~two-fold decrease in the magnitude of the binding enthalpy is observed, suggesting that some protonation (68) might be linked to complex formation. In contrast, we note that similar comparative experiments with χ performed in Hepes vs. cacodylate buffer, which also have quite different ionization enthalpies, show no evidence of an effect of buffer type (see Fig. 6C). Therefore, the intrinsic specificity of the free SSB-Ct peptides for binding of PriA vs. χ (see Table 1), is enhanced for the SSB-oligo(dT) complexes.
The SSB tetramer can bind to ssDNA in a number of binding modes that differ in the average number of subunits that interact with the ssDNA (32). The two major binding modes are referred to as the (SSB)35 mode and the (SSB)65 mode. In the (SSB)35 mode, an average of two SSB subunits interact with 35 nucleotides of ss-DNA, whereas in the (SSB)65 mode, all four subunits interact with ~ 65 nucleotides of ss-DNA and form a fully wrapped structure as shown in Fig.1. Since ssDNA binding enhances the affinity of PriA and χ for the SSB Ct tails, we examined whether the mode of SSB binding to ssDNA might influence the binding (stoichiometry, as well as energetics) of PriA or χ to SSB. For this reason we also examined the binding of PriA to an SSB-(dT)35 (1:1 molar) complex (Fig. 7). This complex is believed to mimic aspects of the (SSB)35 binding mode (44), although without the inter-tetramer positive cooperativity. We have previously shown that two molecules of (dT)35 can bind per SSB tetramer, occupying all four subunits, although the second molecule binds with a salt-dependent negative cooperativity (64, 69, 70). At 0.2 M NaCl the first molecule of (dT)35 binds to the SSB tetramer with high affinity such that binding is stoichiometric and interacts with an average of two SSB subunits (44, 69, 70). We compared the binding of PriA to a 1:1 molar SSB-(dT)35 complex as a mimic for the (SSB) 35 mode with PriA binding to the SSB-(dT)70 complex as a mimic for the fully wrapped (SSB)65 mode.
The titrations of PriA with the SSB-(dT)35 and SSB-(dT)70 complexes are compared directly in Fig. 7. At the beginning of the titration when a small amount of either SSB-DNA complex is titrated into a large excess of PriA in the cell, the normalized heats are almost twice as large for the SSB-(dT)70 titration, which either indicates a higher affinity or higher stoichiometry for the SSB-dT70 complex. Direct fitting of the data shown in Fig. 7 provides parameters (see Fig. 6 legend) suggesting that the difference between the titrations reflects a higher stoichiometry for PriA binding to the SSB-(dT)70 complex. However, since the parameters obtained from these fits are highly correlated, this conclusion must be viewed with caution.
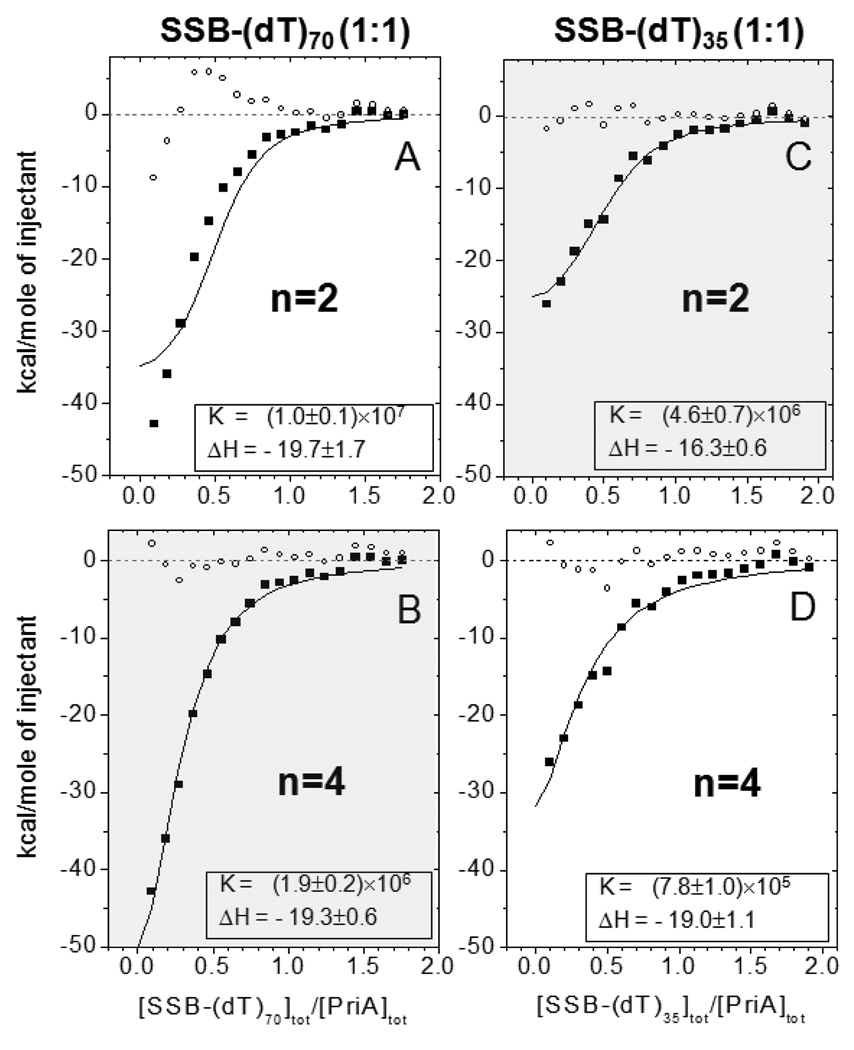
In an attempt to address this concern we also fit these data by constraining the stoichiometry of binding to n=2 or n=4 for both sets of titrations and allowing both Kobs and ΔHobs to float in a NLLS analysis. The fits to these data are shown in Fig. 8 and indicate that the isotherm for PriA binding to the SSB-(dT)70 complex is better described by a stoichiometry of n=4, whereas the isotherm for PriA binding to the SSB-(dT)35 complex is better described by a stoichiometry of n=2 (Fig. 8B and 8C, respectively). In both cases the affinities are similar and in the range (2–5)×106 M−1 . Further analysis of additional experiments (data not shown) showed similar results with comparable binding parameters. We note that if we constrain n=3 for both titrations, reasonable fits are obtained for both, although the affinity and magnitude of the enthalpy change for PriA binding to the fully wrapped SSB-(dT)70 complex are higher, (Kobs =(3.7±1.0)×106 M−1; ΔHobs = −18.1±2.6 kcal/mol (5 titrations)), than for the SSB-(dT)35 complex (Kobs=(1.1±0.4)×106 M−1 and ΔHobs = −16.9±1.4 kcal/mol (2 titrations)). We also examined the binding of PriA to the SSB-(dT)70 (SSB:dT70=1:1) complex using sedimentation equilibrium. Those experiments (data not shown) were performed using a large molar excess of PriA (6 µM) over the SSB-dT70 (1:1) complex (0.4 µM) and indicated a minimum stoichiometry of three PriA per SSB:dT70 (1:1) complex. Although we are unable to definitively assign the differences in PriA binding to SSB:(dT)70 and SSB:(dT)35 complexes to differences in stoichiometry rather than affinity, it is clear that PriA interacts with SSB with significantly higher affinity if SSB is pre-bound to ssDNA and secondly, the effectiveness of the PriA-SSB interaction increases as the length of ssDNA involved in the complex increases.
Figure 8.
The stoichiometry of PriA binding to to an SSB-ssDNA complexis increased as the number of SSB subunits contacting the DNA is increased. Comparison of different fits for the data presented in Fig. 7 for the interaction of PriA with the SSB-(dT)70 (1:1) complex, for fixed values of n=2 (A) and n=4 (B), and the SSB-dT35 (1:1) complex for fixed values of n=2 (C) and n=4 (D). The better fitting is achieved when the stoichiometry is n=4 for the SSB-dT70 (1:1) complex (B) and n=2 for the SSB-dT35 (1:1) complex (C). The residuals of the fits are shown as small open circles.
DISCUSSION
Intrinsically disordered proteins (IDP) have become a subject of a great interest (47) since the recognition that many proteins and regions of proteins involved in signaling and regulation use unstructured amino acid tethers as interaction sites. It has been suggested that the advantages of using intrinsically disordered regions for these interactions include: high specificity/low affinity binding (49, 71, 72), and the ability to bind multiple targets (49, 71, 73).
The E. coli SSB tetramer, which possesses four intrinsically disordered C-termini (residues 113–177), even when SSB is tightly bound to ssDNA (45), represents another example of a protein that uses intrinsically disordered regions as sites of interaction. Through its C-termini, SSB interacts with at least 14 other proteins involved in DNA replication, recombination or repair and sequesters these proteins to their sites of function on the DNA (3). These interactions occur primarily with the last ~9 amino acid residues at the end of the SSB-Ct sequence which is a highly negatively charged region (MDFDDDIPF) (3)(see Table 1). In fact, SSB proteins with 8–10 residues deleted from the C-terminus show no interaction with accessory proteins (10, 18, 20) and do not support their functions (10, 11, 15, 28). Moreover, even the single Pro to Ser mutation, corresponding to the P176S mutation in SSB-113, dramatically diminishes Ct binding to both χ and PriA proteins, as has also been reported for RecQ helicase (10).
Although interactions of SSB with its partner proteins have been documented previously (3), little was known about whether these interactions showed any specificity. Binding specificity must exist at some level, otherwise SSB would be unable to discriminate among its many binding partners and thus control which proteins are recruited to particular sites on the DNA. Our study indicates that SSB, through its C-termini, possesses intrinsic specificity towards at least two of its binding partners, PriA and χ, and that this specificity is further enhanced by ssDNA binding. Besides the ability to interact with a variety of metabolic proteins involved in genome maintenance (3), the SSB C-termini can also affect the ways in which SSB binds to ssDNA. Deletion of the SSB-Ct tails can influence the relative stabilities of the (SSB)35 and (SSB)65 binding modes (39). Our results also indicate that the SSB core can inhibit binding of the Ct tails to its binding partners and that ssDNA binding can relieve this inhibition. This inhibition may be caused by the acidic end of the SSB C-terminal tails interacting with the DNA binding sites within the SSB core and may explain partially why ssDNA binding enhances these protein interactions with the SSB tails. In fact, deletion of the acidic C-terminal tail of SSB has been shown to enhance its affinity for the RNA, poly(U) (65). Similar effects have also been shown for the phage T4 SSB (gene 32 protein) (74) and the phage T7 SSB (gene 2.5 protein) (75) and may play a regulatory role.
SSB C-terminal peptides show specificity for binding to PriA vs. χ
At 25°C both χ and PriA interact with the isolated SSB C-terminal peptides with similar affinities. However, although there is little difference in affinities at 25°C, we observed clear differences in binding enthalpy such that binding of PriA is much more enthalpically favorable at both low and moderate salt conditions (see Table 1). Although this enthalpic difference disappears for experiments performed in Tris (see Table 1), this is likely due to a fortuitous balance of protonation effects. The larger magnitudes of ΔHobs,PriA obtained in Cacodylate buffer, which has a significantly lower ionization enthalpy, suggest that PriA binding to the Ct peptide may be linked to protonation events (68).
This difference in binding enthalpy indicates that binding specificity should be greater at lower temperatures and this was observed for χ and PriA interaction with the SSB-Ct peptide (WP9) in buffer C (pH 7.0, 0.02M NaCl) (see Fig. 3B). Both interactions are enthalpy driven, with ΔHobs,PriA twice as large in magnitude than ΔHobs,χ and PriA binding showing a larger temperature dependence indicating a larger negative heat capacity change (ΔCpobs = −281±21 for PriA vs. −143±21 cal/mol deg for χ). On the other hand, the interactions with PriA are more costly entropically than with χ. These results underscore the fact that measurements of only binding constants made at a single temperature may not detect important thermodynamic differences that become apparent only at different temperatures or solution conditions.
The binding site on RecQ helicase for the C-termini of SSB was identified recently (76). Comparison with the SSB-Ct binding site on exo I (21) reveals a number of structural similarities, including a hydrophobic pocket lined by a “basic lip” and adjoining “basic ridge” (21, 76). A highly conserved basic region within χ was identified in the crystal structure of the χψ heterodimer as a potential binding site for the C-terminal tail of SSB (77). These structural similarities suggest the importance of both electrostatic and hydrophobic components of the interactions with highly conserved acidic (170-Asp-Phe-Asp-Asp-Asp-174) and hydrophobic (175-Ile-Pro-Phe-177) parts of the SSB-Ct. We show that the binding of both χ and PriA with the isolated SSB C-terminal peptides is modulated by [NaCl] (see Table 1), also suggesting a strong electrostatic component to these interactions likely due to the acidic nature of the Ct peptides.
PriA and χ interactions with SSB are weaker than to the isolated SSB tails indicating an inhibitory effect of the SSB core
Our equilibrium binding studies indicate that the χ-SSB and PriA-SSB interactions are weaker than expected based on the binding parameters determined for PriA and χ interactions with the isolated SSB-Ct peptides. We hypothesize that the lower affinities when the Ct tails are part of the full length SSB protein reflect inhibition, possibly due to non-specific interactions of the C-termini with the SSB core (N-terminal residues 1–112) that forms the four ssDNA binding OB folds (44). Positively charged regions comprising the ssDNA binding site are accessible on the surface of the SSB core and it is possible that the negatively charged C-termini can interact with these regions in nonspecific manner. Consistent with this suggestion, we find that deletion of the C-terminal tails from SSB increases its equilibrium binding affinity for some ss-DNA (A. G. Kozlov and T. M. Lohman, unpublished observations). A similar inhibitory effect on ss-DNA binding has been observed for the C-terminal tail of the T7 gene 2.5 SSB (75).
Pre-binding ssDNA to SSB increases its affinity much more for PriA than for χ
The effects of pre-binding ss-DNA to SSB on its affinity for a partner protein have previously been examined only for the χ protein (4, 7). Little enhancement of affinity was observed in the presence of ssDNA at moderate salt concentrations (0.1–0.3M NaCl), although at low salt (5mM NaCl) a ~10-fold enhancement was reported (7.4±1.0)×106 M−1) (7). At 20 mM NaCl, the affinity of χ for SSB alone is too weak to determine its binding parameters, but we measure a similar affinity (5.6±1.9)×106 M−1) for the interaction of χ with an SSB-(dT)70 complex. This is a ~4-fold enhancement compared to χ binding to a Ct peptide alone at low salt (20 mM NaCl) (see Table 2). Unfortunately, we were unable to make the same comparisons for PriA at these low [NaCl] conditions. However, at higher salt (0.2 M NaCl), we could compare directly the binding of PriA and χ to an isolated SSB-Ct peptide, SSB and a fully wrapped (1:1 molar ratio) SSB-(dT)70 complex. The results (Table 2) indicate that there is little increase in affinity of χ for an SSB-(dT)70 complex compared to its affinity for the Ct peptides and SSB in agreement with previous reports (4, 5, 7).
For PriA, we observe an ~10-fold increase in affinity for binding to an SSB-(dT)70 complex compared to SSB alone or an isolated Ct peptide (0.2 M NaCl) (Table 2). Thus the binding of SSB to ssDNA increases the affinity of SSB for PriA, to a much larger extent than for χ. How this occurs is not completely clear, although it is possible that PriA makes additional contacts with parts of the ssDNA not involved in the interactions with SSB, which is not the case for χ, since χ does not interact with ssDNA. The enhancement may also result if ssDNA binding eliminates the auto-inhibition of the Ct tails. It is also possible that SSB plays a kinetic role in facilitating loading of PriA onto DNA. Our simple competition experiments (Fig.5B) show that the presence of saturating (dT)20 does not affect binding of the SSB-Ct peptide to PriA, indicating that PriA has separate binding sites for the SSB-Ct and ssDNA. This could allow PriA to be recruited by SSB when it is bound to ssDNA through an interaction with the C-terminal tails followed by a transfer to the ssDNA through formation of ternary complex in which both PriA and SSB are bound to the same ssDNA.
In this study we have also observed that an increase in stoichiometry and affinity of PriA binding to the SSB Ct tails occurs when an increasing number of SSB subunits are occupied by ss-DNA. This may result if the acidic region of the C-terminal tails interact with the DNA binding sites within the SSB core and a longer ssDNA that interacts with more SSB subunits (2 vs. 4) would release more of the C-terminal tails for interaction with its protein partner. This also suggests that the (SSB)65 binding mode, in which all four subunits are involved in DNA interactions, should be more effective in providing a larger local concentration of PriA to be loaded onto ssDNA structures where it will function.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. M. O’Donnell for providing the plasmid for overexpression of χ protein and T. Ho for synthesis and purification of the oligodeoxynucleotides.
Abbreviations
- SSB
Single Stranded Binding protein
- SSB-Ct
SSB C-terminal
- ssDNA
single stranded; DNA
- DNA Pol III HE
DNA polymerase III Holoenzyme
- ITC
isothermal titration calorimetry
- Tris
tris(hydroxymethyl)aminomethane
- EDTA
ethylenediaminetetraacetic acid
- DNA Pol III HE
DNA polymerase III Holoenzyme
Footnotes
This research was supported in part by the NIH (GM30498 to TML and GM46679 to WB)
SUPPORTING INFORMATION AVAILABLE
The estimate of protonation effect on PriA interaction with SSB-Ct peptides. This material is available free of charge via the Internet at http://pubs.asc.org.
REFERENCES
- 1.Chase JW, Williams KR. Single-stranded DNA binding proteins required for DNA replication. Annu Rev Biochem. 1986;55:103–136. doi: 10.1146/annurev.bi.55.070186.000535. [DOI] [PubMed] [Google Scholar]
- 2.Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990;54:342–380. doi: 10.1128/mr.54.4.342-380.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an organizer/mobilizer of genome maintenance complexes. Crit Rev Biochem Mol Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelman Z, Yuzhakov A, Andjelkovic J, O'Donnell M. Devoted to the lagging strand-the subunit of DNA polymerase III holoenzyme contacts SSB to promote processive elongation and sliding clamp assembly. Embo J. 1998;17:2436–2449. doi: 10.1093/emboj/17.8.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glover BP, McHenry CS. The chi psi subunits of DNA polymerase III holoenzyme bind to single-stranded DNA-binding protein (SSB) and facilitate replication of an SSB-coated template. J Biol Chem. 1998;273:23476–23484. doi: 10.1074/jbc.273.36.23476. [DOI] [PubMed] [Google Scholar]
- 6.Yuzhakov A, Kelman Z, O'Donnell M. Trading places on DNA--a three-point switch underlies primer handoff from primase to the replicative DNA polymerase. Cell. 1999;96:153–163. doi: 10.1016/s0092-8674(00)80968-x. [DOI] [PubMed] [Google Scholar]
- 7.Witte G, Urbanke C, Curth U. DNA polymerase III chi subunit ties single-stranded DNA binding protein to the bacterial replication machinery. Nucleic Acids Res. 2003;31:4434–4440. doi: 10.1093/nar/gkg498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun W, Godson GN. Structure of the Escherichia coli primase/single-strand DNA-binding protein/phage G4oric complex required for primer RNA synthesis. J Mol Biol. 1998;276:689–703. doi: 10.1006/jmbi.1997.1471. [DOI] [PubMed] [Google Scholar]
- 9.Umezu K, Nakayama H. RecQ DNA helicase of Escherichia coli. Characterization of the helix-unwinding activity with emphasis on the effect of single-stranded DNA-binding protein. J Mol Biol. 1993;230:1145–1150. doi: 10.1006/jmbi.1993.1231. [DOI] [PubMed] [Google Scholar]
- 10.Shereda RD, Bernstein DA, Keck JL. A central role for SSB in Escherichia coli RecQ DNA helicase function. J Biol Chem. 2007;282:19247–19258. doi: 10.1074/jbc.M608011200. [DOI] [PubMed] [Google Scholar]
- 11.Suski C, Marians KJ. Resolution of converging replication forks by RecQ and topoisomerase III. Mol Cell. 2008;30:779–789. doi: 10.1016/j.molcel.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Umezu K, Kolodner RD. Protein interactions in genetic recombination in Escherichia coli. Interactions involving RecO and RecR overcome the inhibition of RecA by single-stranded DNA-binding protein. J Biol Chem. 1994;269:30005–30013. [PubMed] [Google Scholar]
- 13.Hegde SP, Qin MH, Li XH, Atkinson MA, Clark AJ, Rajagopalan M, Madiraju MV. Interactions of RecF protein with RecO, RecR, and single-stranded DNA binding proteins reveal roles for the RecF-RecO-RecR complex in DNA repair and recombination. Proc Natl Acad Sci U S A. 1996;93:14468–14473. doi: 10.1073/pnas.93.25.14468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kantake N, Madiraju MV, Sugiyama T, Kowalczykowski SC. Escherichia coli RecO protein anneals ssDNA complexed with its cognate ssDNA-binding protein: A common step in genetic recombination. Proc Natl Acad Sci U S A. 2002;99:15327–15332. doi: 10.1073/pnas.252633399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hobbs MD, Sakai A, Cox MM. SSB protein limits RecOR binding onto single-stranded DNA. J Biol Chem. 2007;282:11058–11067. doi: 10.1074/jbc.M611007200. [DOI] [PubMed] [Google Scholar]
- 16.Han ES, Cooper DL, Persky NS, Sutera VA, Jr, Whitaker RD, Montello ML, Lovett ST. RecJ exonuclease: substrates, products and interaction with SSB. Nucleic Acids Res. 2006;34:1084–1091. doi: 10.1093/nar/gkj503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lecointe F, Serena C, Velten M, Costes A, McGovern S, Meile JC, Errington J, Ehrlich SD, Noirot P, Polard P. Anticipating chromosomal replication fork arrest: SSB targets repair DNA helicases to active forks. Embo J. 2007;26:4239–4251. doi: 10.1038/sj.emboj.7601848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buss JA, Kimura Y, Bianco PR. RecG interacts directly with SSB: implications for stalled replication fork regression. Nucleic Acids Res. 2008;36:7029–7042. doi: 10.1093/nar/gkn795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sandigursky M, Mendez F, Bases RE, Matsumoto T, Franklin WA. Protein-protein interactions between the Escherichia coli single-stranded DNA-binding protein and exonuclease I. Radiat Res. 1996;145:619–623. [PubMed] [Google Scholar]
- 20.Genschel J, Curth U, Urbanke C. Interaction of E. coli single-stranded DNA binding protein (SSB) with exonuclease I. The carboxy-terminus of SSB is the recognition site for the nuclease. Biol Chem. 2000;381:183–192. doi: 10.1515/BC.2000.025. [DOI] [PubMed] [Google Scholar]
- 21.Lu D, Keck JL. Structural basis of Escherichia coli single-stranded DNA-binding protein stimulation of exonuclease I. Proc Natl Acad Sci U S A. 2008;105:9169–9174. doi: 10.1073/pnas.0800741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Handa P, Acharya N, Varshney U. Chimeras between single-stranded DNA-binding proteins from Escherichia coli and Mycobacterium tuberculosis reveal that their C-terminal domains interact with uracil DNA glycosylases. J Biol Chem. 2001;276:16992–16997. doi: 10.1074/jbc.M100393200. [DOI] [PubMed] [Google Scholar]
- 23.Purnapatre K, Handa P, Venkatesh J, Varshney U. Differential effects of single-stranded DNA binding proteins (SSBs) on uracil DNA glycosylases (UDGs) from Escherichia coli and mycobacteria. Nucleic Acids Res. 1999;27:3487–3492. doi: 10.1093/nar/27.17.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molineux IJ, Gefter ML. Properties of the Escherichia coli in DNA binding (unwinding) protein: interaction with DNA polymerase and DNA. Proc Natl Acad Sci U S A. 1974;71:3858–3862. doi: 10.1073/pnas.71.10.3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonner CA, Stukenberg PT, Rajagopalan M, Eritja R, O'Donnell M, McEntee K, Echols H, Goodman MF. Processive DNA synthesis by DNA polymerase II mediated by DNA polymerase III accessory proteins. J Biol Chem. 1992;267:11431–11438. [PubMed] [Google Scholar]
- 26.Arad G, Hendel A, Urbanke C, Curth U, Livneh Z. Single-stranded DNA-binding Protein Recruits DNA Polymerase V to Primer Termini on RecA-coated DNA. J Biol Chem. 2008;283:8274–8282. doi: 10.1074/jbc.M710290200. [DOI] [PubMed] [Google Scholar]
- 27.Chen HW, North SH, Nakai H. Properties of the PriA helicase domain and its role in binding PriA to specific DNA structures. J Biol Chem. 2004;279:38503–38512. doi: 10.1074/jbc.M404769200. [DOI] [PubMed] [Google Scholar]
- 28.Cadman CJ, McGlynn P. PriA helicase and SSB interact physically and functionally. Nucleic Acids Res. 2004;32:6378–6387. doi: 10.1093/nar/gkh980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cadman CJ, Lopper M, Moon PB, Keck JL, McGlynn P. PriB stimulates PriA helicase via an interaction with single-stranded DNA. J Biol Chem. 2005;280:39693–39700. doi: 10.1074/jbc.M508521200. [DOI] [PubMed] [Google Scholar]
- 30.Raghunathan S, Ricard CS, Lohman TM, Waksman G. Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength x-ray diffraction on the selenomethionyl protein at 2.9-A resolution. Proc Natl Acad Sci U S A. 1997;94:6652–6657. doi: 10.1073/pnas.94.13.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lohman TM, Bujalowski W. E. coli SSB protein: multiple binding modes and cooperativities. In: Revzin A, editor. The Biology of Nonspecific DNA-Protein Interactions. CRC Press; 1990. pp. 131–170. [Google Scholar]
- 32.Lohman TM, Ferrari ME. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu Rev Biochem. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 33.Bujalowski W, Lohman TM. Escherichia coli single-strand binding protein forms multiple, distinct complexes with single-stranded DNA. Biochemistry. 1986;25:7799–7802. doi: 10.1021/bi00372a003. [DOI] [PubMed] [Google Scholar]
- 34.Lohman TM, Overman LB. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J Biol Chem. 1985;260:3594–3603. [PubMed] [Google Scholar]
- 35.Wei TF, Bujalowski W, Lohman TM. Cooperative binding of polyamines induces the Escherichia coli single-strand binding protein-DNA binding mode transitions. Biochemistry. 1992;31:6166–6174. doi: 10.1021/bi00141a029. [DOI] [PubMed] [Google Scholar]
- 36.Chrysogelos S, Griffith J. Escherichia coli single-strand binding protein organizes single-stranded DNA in nucleosome-like units. Proc Natl Acad Sci U S A. 1982;79:5803–5807. doi: 10.1073/pnas.79.19.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griffith JD, Harris LD, Register J., 3rd Visualization of SSB-ssDNA complexes active in the assembly of stable RecA-DNA filaments. Cold Spring Harb Symp Quant Biol. 1984;49:553–559. doi: 10.1101/sqb.1984.049.01.062. [DOI] [PubMed] [Google Scholar]
- 38.Bujalowski W, Overman LB, Lohman TM. Binding mode transitions of Escherichia coli single strand binding protein-single-stranded DNA complexes. Cation, anion, pH, and binding density effects. J Biol Chem. 1988;263:4629–4640. [PubMed] [Google Scholar]
- 39.Roy R, Kozlov AG, Lohman TM, Ha T. Dynamic structural rearrangements between DNA binding modes of E. coli SSB protein. J Mol Biol. 2007;369:1244–1257. doi: 10.1016/j.jmb.2007.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuznetsov SV, Kozlov AG, Lohman TM, Ansari A. Microsecond dynamics of protein-DNA interactions: Direct observation of the wrapping/unwrapping kinetics of single-stranded DNA around the E. coli SSB tetramer. Journal of Molecular Biology. 2006;359:55–65. doi: 10.1016/j.jmb.2006.02.070. [DOI] [PubMed] [Google Scholar]
- 41.Roy R, Kozlov AG, Lohman TM, Ha T. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature. 2009;461:1092–1097. doi: 10.1038/nature08442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lohman TM, Overman LB, Datta S. Salt-dependent changes in the DNA binding co-operativity of Escherichia coli single strand binding protein. J Mol Biol. 1986;187:603–615. doi: 10.1016/0022-2836(86)90338-4. [DOI] [PubMed] [Google Scholar]
- 43.Ferrari ME, Bujalowski W, Lohman TM. Co-operative binding of Escherichia coli SSB tetramers to single-stranded DNA in the (SSB)35 binding mode. J Mol Biol. 1994;236:106–123. doi: 10.1006/jmbi.1994.1122. [DOI] [PubMed] [Google Scholar]
- 44.Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E-coli SSB bound to ssDNA. Nature Structural Biology. 2000;7:648–652. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 45.Savvides SN, Raghunathan S, Futterer K, Kozlov AG, Lohman TM, Waksman G. The C-terminal domain of full-length E-coli SSB is disordered even when bound to DNA. Protein Science. 2004;13:1942–1947. doi: 10.1110/ps.04661904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams KR, Spicer EK, LoPresti MB, Guggenheimer RA, Chase JW. Limited proteolysis studies on the Escherichia coli single-stranded DNA binding protein. Evidence for a functionally homologous domain in both the Escherichia coli and T4 DNA binding proteins. J Biol Chem. 1983;258:3346–3355. [PubMed] [Google Scholar]
- 47.Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008;18:756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 48.Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic disorder and functional proteomics. Biophys J. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uversky VN, Oldfield CJ, Dunker AK. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J Mol Recognit. 2005;18:343–384. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- 50.Kelman Z, O'Donnell M. DNA polymerase III holoenzyme: structure and function of a chromosomal replicating machine. Annu Rev Biochem. 1995;64:171–200. doi: 10.1146/annurev.bi.64.070195.001131. [DOI] [PubMed] [Google Scholar]
- 51.Johnson A, O'Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 52.O'Donnell M. Replisome architecture and dynamics in Escherichia coli. J Biol Chem. 2006;281:10653–10656. doi: 10.1074/jbc.R500028200. [DOI] [PubMed] [Google Scholar]
- 53.Marians KJ. PriA-directed replication fork restart in Escherichia coli. Trends Biochem Sci. 2000;25:185–189. doi: 10.1016/s0968-0004(00)01565-6. [DOI] [PubMed] [Google Scholar]
- 54.Sandler SJ, Marians KJ. Role of PriA in replication fork reactivation in Escherichia coli. J Bacteriol. 2000;182:9–13. doi: 10.1128/jb.182.1.9-13.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heller RC, Marians KJ. Replisome assembly and the direct restart of stalled replication forks. Nat Rev Mol Cell Biol. 2006;7:932–943. doi: 10.1038/nrm2058. [DOI] [PubMed] [Google Scholar]
- 56.Lohman TM, Green JM, Beyer RS. Large-scale overproduction and rapid purification of the Escherichia coli ssb gene product. Expression of the ssb gene under lambda PL control. Biochemistry. 1986;25:21–25. doi: 10.1021/bi00349a004. [DOI] [PubMed] [Google Scholar]
- 57.Bujalowski W, Lohman TM. Monomer-tetramer equilibrium of the Escherichia coli ssb-1 mutant single strand binding protein. J Biol Chem. 1991;266:1616–1626. [PubMed] [Google Scholar]
- 58.Jezewska MJ, Bujalowski W. Interactions of Escherichia coli replicative helicase PriA protein with single-stranded DNA. Biochemistry. 2000;39:10454–10467. doi: 10.1021/bi001113y. [DOI] [PubMed] [Google Scholar]
- 59.Jezewska MJ, Rajendran S, Bujalowski W. Escherichia coli replicative helicase PriA protein-single-stranded DNA complex. Stoichiometries, free energy of binding, and cooperativities. J Biol Chem. 2000;275:27865–27873. doi: 10.1074/jbc.M004104200. [DOI] [PubMed] [Google Scholar]
- 60.Xiao H, Crombie R, Dong Z, Onrust R, O'Donnell M. DNA polymerase III accessory proteins. III. holC and holD encoding chi and psi. J Biol Chem. 1993;268:11773–11778. [PubMed] [Google Scholar]
- 61.Lohman TM, Chao K, Green JM, Sage S, Runyon GT. Large-scale purification and characterization of the Escherichia coli rep gene product. J Biol Chem. 1989;264:10139–10147. [PubMed] [Google Scholar]
- 62.Kowalczykowski SC, Lonberg N, Newport JW, von Hippel PH. Interactions of bacteriophage T4-coded gene 32 protein with nucleic acids. I. Characterization of the binding interactions. J Mol Biol. 1981;145:75–104. doi: 10.1016/0022-2836(81)90335-1. [DOI] [PubMed] [Google Scholar]
- 63.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 64.Kozlov AG, Lohman TM. Calorimetric studies of E-coli SSB protein single-stranded DNA interactions. Effects of monovalent salts on binding enthalpy. Journal of Molecular Biology. 1998;278:999–1014. doi: 10.1006/jmbi.1998.1738. [DOI] [PubMed] [Google Scholar]
- 65.Curth U, Genschel J, Urbanke C, Greipel J. In vitro and in vivo function of the C-terminus of Escherichia coli single-stranded DNA binding protein. Nucleic Acids Res. 1996;24:2706–2711. doi: 10.1093/nar/24.14.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fukada H, Takahashi K. Enthalpy and heat capacity changes for the proton dissociation of various buffer components in 0.1 M potassium chloride. Proteins. 1998;33:159–166. [PubMed] [Google Scholar]
- 67.Christensen JJ, Hansen LD, Izatt RM. Handbook of proton ionization heats. New York: John Wiley & Sons; 1976. [Google Scholar]
- 68.Kozlov AG, Lohman TM. Large contributions of coupled protonation equilibria to the observed enthalpy and heat capacity changes for ssDNA binding to Escherichia coli SSB protein. Proteins Suppl. 2000;4:8–22. doi: 10.1002/1097-0134(2000)41:4+<8::aid-prot20>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 69.Bujalowski W, Lohman TM. Negative co-operativity in Escherichia coli single strand binding protein-oligonucleotide interactions. I. Evidence and a quantitative model. J Mol Biol. 1989;207:249–268. doi: 10.1016/0022-2836(89)90454-3. [DOI] [PubMed] [Google Scholar]
- 70.Bujalowski W, Lohman TM. Negative co-operativity in Escherichia coli single strand binding protein-oligonucleotide interactions. II. Salt, temperature and oligonucleotide length effects. J Mol Biol. 1989;207:269–288. doi: 10.1016/0022-2836(89)90455-5. [DOI] [PubMed] [Google Scholar]
- 71.Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Intrinsic disorder in transcription factors. Biochemistry. 2006;45:6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu J, Faeder JR, Camacho CJ. Toward a quantitative theory of intrinsically disordered proteins and their function. Proc Natl Acad Sci U S A. 2009;106:19819–19823. doi: 10.1073/pnas.0907710106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wright PE, Dyson HJ. Intrinsically unstructured proteins: reassessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 74.Lonberg N, Kowalczykowski SC, Paul LS, von Hippel PH. Interactions of bacteriophage T4-coded gene 32 protein with nucleic acids. III. Binding properties of two specific proteolytic digestion products of the protein (G32P*I and G32P*III) J Mol Biol. 1981;145:123–138. doi: 10.1016/0022-2836(81)90337-5. [DOI] [PubMed] [Google Scholar]
- 75.Marintcheva B, Hamdan SM, Lee SJ, Richardson CC. Essential residues in the C terminus of the bacteriophage T7 gene 2.5 single-stranded DNA-binding protein. J Biol Chem. 2006;281:25831–25840. doi: 10.1074/jbc.M604601200. [DOI] [PubMed] [Google Scholar]
- 76.Shereda RD, Reiter NJ, Butcher SE, Keck JL. Identification of the SSB binding site on E. coli RecQ reveals a conserved surface for binding SSB's C terminus. J Mol Biol. 2009;386:612–625. doi: 10.1016/j.jmb.2008.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gulbis JM, Kazmirski SL, Finkelstein J, Kelman Z, O'Donnell M, Kuriyan J. Crystal structure of the chi:psi sub-assembly of the Escherichia coli DNA polymerase clamp-loader complex. Eur J Biochem. 2004;271:439–449. doi: 10.1046/j.1432-1033.2003.03944.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.