Abstract
We used epidemiologic data for human West Nile virus (WNV) disease in Colorado from 2003 and 2007 to determine 1) the degree to which estimates of vector-borne disease occurrence is influenced by spatial scale of data aggregation (county versus census tract), and 2) the extent of concordance between spatial risk patterns based on case counts versus incidence. Statistical analyses showed that county, compared with census tract, accounted for approximately 50% of the overall variance in WNV disease incidence, and approximately 33% for the subset of cases classified as West Nile neuroinvasive disease. These findings indicate that sub-county scale presentation provides valuable risk information for stakeholders. There was high concordance between spatial patterns of WNV disease incidence and case counts for census tract (83%) but not for county (50%) or zip code (31%). We discuss how these findings impact on practices to develop spatial epidemiologic data for vector-borne diseases and present data to stakeholders.
Introduction
In the past two decades, technological capacity to map and model spatial patterns of risk for exposure to arthropod vectors and vector-borne pathogens has progressed rapidly.1–4 Geographic information system (GIS) and remote sensing software have become more user-friendly and are now complemented by easy-to-use tools to assess spatial and space-time clustering, such as SaTScan™ and the DYCAST system.5,6 New mapping software, such as Google Earth™ and MS Virtual Earth™,7,8 provides basic and easy-to-use capacity to generate not only spatial data overlaid on pre-existing satellite imagery or map representations, but also dynamic illustrations of space-time patterns that can be played as movie clips.9–11 These developments provide extended capacity to determine and present spatial patterns of disease incidence, or of occurrence of vectors or vector-borne pathogens. Geographic information system and other mapping technologies are now routinely used in academic institutions and by public health agencies at national, state, county, and city levels in the United States.
We also have seen explosive development in the field of web-based information delivery, which now provides an effective medium to distribute maps to a wide range of stakeholders including the medical community, vector control practitioners, policy makers, and the public at large.12,13 Using West Nile virus (WNV) disease as an example, maps showing spatial distributions of cases or incidence of West Nile fever (WNF) or West Nile neuroinvasive disease (WNND) are readily available from the Centers for Disease Control and Prevention website (http://www.cdc.gov/ncidod/dvbid/westnile/index.htm), the U.S. Geological Survey Disease Maps website (http://diseasemaps.usgs.gov/index.html), and from many state or local health department websites in WNV disease-endemic areas. Such maps can be used as tools to target limited prevention, surveillance and control resources to high-risk areas for WNV exposure, and to inform the public about local risk levels.
However, with this new technological capacity to determine and present spatial risk patterns comes a series of questions regarding how it should be used responsibly in public health. Benefits and drawbacks of using entomological versus epidemiologic data in spatial risk assessments, and the important issue of uncertainty in pathogen exposure locations for patients afflicted with common vector-borne diseases such as WNV disease and Lyme disease, were discussed previously for important vector-borne diseases in North America.9,14,15 We apply quantitative statistical methods to explore two questions that have not been adequately addressed in the United States for vector-borne diseases: 1) How are estimates of vector-borne disease occurrence influenced by spatial scale of data aggregation (i.e., county versus census tract)? and 2) What is the extent of concordance among spatial risk patterns based on disease case counts versus disease incidence for commonly used spatial boundary units such as county, census tract, and zip code? As a case study to address these questions, we use epidemiologic data for WNV disease in Colorado during the outbreak years of 2003 (total = 2,947 WNV disease cases reported from the state) and 2007 (578 reported WNV disease cases).
Materials and Methods
Epidemiologic and GIS-based data.
The study was based on WNV disease cases reported to the Colorado Department of Public Health and Environment during 2003 and 2007, which represent two outbreak years in Colorado with 2,947 WNV disease cases, including 622 WNND cases, reported in 2003 and 578 WNV disease cases, including 100 WNND cases, reported in 2007.16 The ratios of WNF to WNND cases were 3.7:1 in 2003 and 4.8:1 in 2007. The epidemiologic database provided by the Colorado Department of Public Health and Environment included for each case information for county, zip code, and census tract of residence, date of onset of symptoms, and whether the case was classified as WNF or WNND. No personal identifiers were included in the database. The epidemiologic database was complemented with GIS-derived data for geographic boundaries (county, zip code, and census tract; Environmental Systems Research Institute, Redlands, CA) and 2004 human population (data from the U.S. Census Bureau provided by the Environmental Systems Research Institute).
Cases were aggregated to census tract, zip code and county units, and cumulative disease incidence (hereinafter referred to as incidence per 100,000 person-years) was calculated for 2003 and 2007 combined. Combining cases from 2003 and 2007 was justified because WNV disease incidences in Colorado were significantly correlated in these outbreak years for the county scale (Spearman rank correlation, ρs = 0.737, n = 64, P < 0.001), the census tract scale (ρs = 0.434, n = 1,075, P < 0.001), and the zip code scale (ρs = 0.459, n = 443, P < 0.001). Aggregating cases to county, zip code, and census tract units was based on the assumption that the likely WNV exposure site was located within the specific boundary unit where the residence was located. This assumption undoubtedly introduces some degree of error because of occupational, recreational, or travel exposures where persons are exposed outside their resident zip code, census tract, or county. However, in the absence of reliable information for probable WNV exposure sites in WNV disease patient case files, use of residence as the assumed exposure location is the best available solution.
Descriptive mapping combined all reported WNV disease cases (WNF and WNND cases) because our main interest was in risk of exposure to WNV rather than disease manifestations. Furthermore, analysis of data from 2003 and 2007 showed significant correlations between numbers of WNF and WNND cases for the county scale (excluding counties reporting no WNV disease cases; 2003, ρs = 0.851, n = 47, P < 0.001; 2007, ρs = 0.684, n = 34, P < 0.001) and the census tract scale (excluding census tracts reporting no WNV disease cases; 2003, ρs = 0.128, n = 737, P < 0.001; 2007, ρs = 0.161, n = 312, P < 0.005). Data for true infection rates for WNV in Colorado are not available because many infections are inapparent or cause only mild symptoms that are unlikely to result in visits to physicians, laboratory confirmation of WNV exposure, and case reporting. Studies from the United States indicate that approximately 80% of human WNV infections are asymptomatic.17
The statistical analysis of spatial disease case patterns by county versus census tract described below was conducted for 1) all WNV disease cases combined and 2) WNND cases. The rationales for conducting a separate analysis for WNND are that 1) WNV disease cases manifesting in the less severe WNF form are far more likely to go undetected or unreported compared with WNND cases, which often require hospitalization, and 2) WNF is potentially subject to greater awareness and testing bias compared with WNND. The side-by-side analyses for WNND cases and all WNV disease cases combined helped us determine if using only WNND cases would result in different spatial patterns compared with combining WNF and WNND cases.
Spatial model to partition variance of disease incidence at county and census tract scales.
The WNV disease and WNND incidence data for the census tract and county spatial boundary units in Colorado were used to explore the degree to which representations of spatial variability in risk are influenced when data are aggregated to county versus census tract. Zip codes were not used in this analysis because, unlike census tracts, they do not always nest within counties. To identify how variance of WNV disease or WNND incidence was partitioned across counties and within counties, a generalized linear mixed-effects (GLME) model was fitted to the data with the response variable being incidence per 100,000 person-years (2003 and 2007 combined) reported to the specific spatial unit (census tract or county). The population of each spatial unit was assumed to be fixed and the model assumed a binomial distribution for the responses. The GLME model specification is
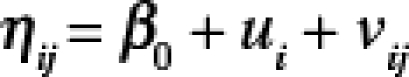 |
where ηij is the linear predictor for the jth census tract in county i, β0 is the intercept, ui, and vij are random effects for county i and census tract j in county i. Case counts were modeled according to the logistic model18 and ui and vij were distributed as a conditionally autoregressive (CAR) model19,20 because exploratory data analysis of the residuals showed spatial dependence among data for WNV disease and WNND (based on Moran's I statistic and neighbor and distance weights matrices). Models were fitted using only the distance spatial weights matrix (assuming that the strength of the correlation between spatial units is inversely proportional to the distances between their centroids) because previous studies21 indicated that unintended correlation results can occur when a neighbor spatial weights matrix (equal correlation among adjacent spatial units) is used in a CAR model.
Hot-spot analysis.
For WNV disease, we also conducted hot-spot analyses based on the Getis-Ord Gi* statistic (using the Spatial Analyst in ArcGIS 9.2; Environmental Systems Research Institute) to determine presence of local clustering of census tracts, within a given county, with either high or low WNV disease incidence based on Z-score values.22 A separate analysis was conducted for each county. A high and positive Z score value indicates that a census tract is surrounded by other census tracts reporting high WNV disease incidence (hot-spot). A high but negative Z-score value indicates that a census tract is surrounded by census tracts reporting low WNV disease incidence (cool-spot).
Disease case count versus incidence.
Zip code, census tract, and county were used to determine the extent of concordance for spatial patterns of areas characterized by high risk of exposure to WNV based on WNV disease case counts versus WNV disease incidence. As before, this analysis was conducted using combined WNV disease data for 2003 and 2007. For each spatial unit (zip code, census tract, county) and disease risk estimate (case count, case incidence), we systematically categorized risk by quartiles. The WNV disease case counts and WNV disease incidences falling within the fourth quartile were considered high risk and used to determine the degree of concordance for spatial patterns of high-risk areas for case count versus incidence for each of the three spatial boundary units examined. This determination was achieved by contingency table analysis. In addition, we determined the overall degree of correlation between case counts and incidence for the three spatial scales using Spearman's rank correlation.
Statistical analyses and map development.
Statistical analyses were conducted using the S-PLUS® version 8.0 (TIBCO Software Inc., Palo Alto, CA) and JMP® 7.0.1 (SAS Institute Inc., Cary, NC) statistical packages. Maps were developed using ArcGIS 9.2 (Environmental Systems Research Institute).
Results
Basic description of spatial patterns for WNV disease case counts and WNV disease incidence.
During 2003 and 2007, a total of 3,525 human WNV disease cases were reported to the Colorado Department of Public Health and Environment. Larimer, Boulder, and Weld counties in north central Colorado accounted for the largest numbers of WNV disease cases during these two years with totals of 650, 537 and 507, respectively. In contrast, the highest WNV disease incidences occurred in the northeastern part of the state, with Logan, Sedgwick, and Phillips counties reporting 293, 273 and 228 cases, respectively, per 100,000 person-years compared with 92–116 cases per 100,000 person-years for Boulder, Larimer, and Weld counties (Figure 1). In the western, mountainous part of Colorado, Mesa and Delta counties reported the highest incidence rates (16–27 cases per 100,000 person-years) (Figure 1). Spatial patterns for WNV disease case counts and WNV disease incidence are displayed visually by county, census tract, and zip code in Figure 2.
Figure 1.
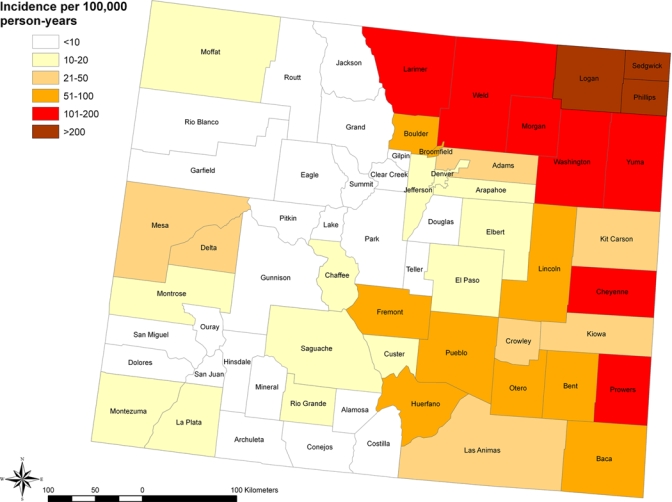
West Nile virus disease incidence per 100,000 person-years by county in Colorado based on combined data for the 2003 and 2007 outbreak years. This figure appears in color at www.ajtmh.org.
Figure 2.
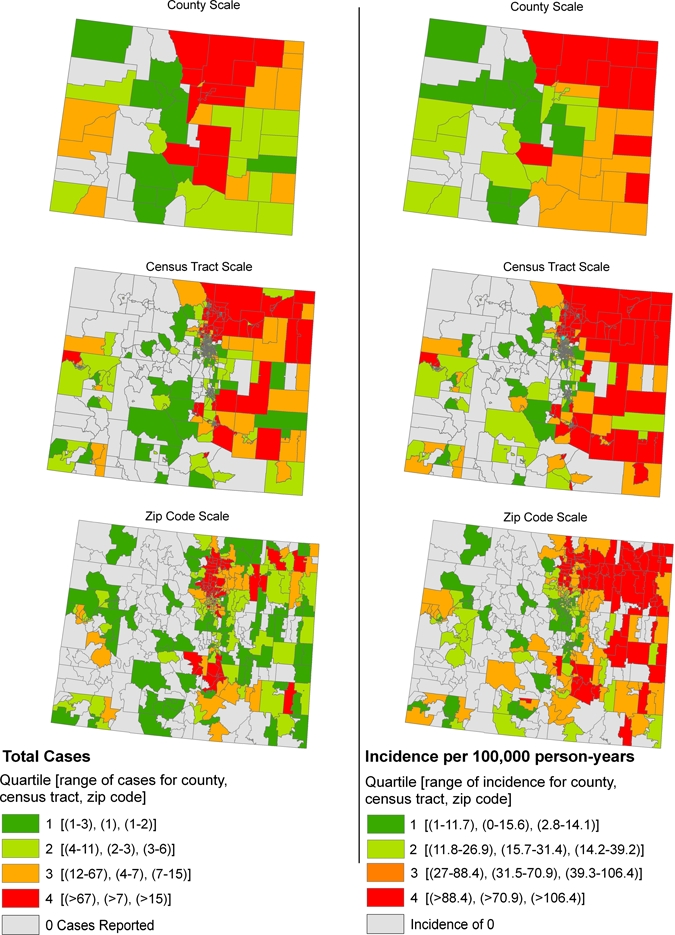
West Nile virus disease case counts and incidences per 100,000 person-years in Colorado by county, census tract, and zip code based on combined data for 2003 and 2007 and classified as no cases reported or by quartile for the spatial boundary units reporting cases. This figure appears in color at www.ajtmh.org.
Spatial model to partition variance of WNV disease and WNND incidence at county and census tract scales.
Significant spatial autocorrelation was detected from the residuals of the GLME model for WNV disease for county (neighbor weights matrix, Moran's I = 0.63, P < 0.01; distance weights matrix, Moran's I = 0.48, P < 0.01) and census tract (neighbor weights matrix, Moran's I = 0.22, P < 0.01; distance weights matrix, Moran's I = 0.08, P = 0.03). The analysis for WNND provided similar results, with spatial autocorrelation occurring for county (neighbor weights matrix, Moran's I = 0.28, P < 0.01; distance weights matrix, Moran's I = 0.25, P < 0.01) and census tract (neighbor weights matrix, Moran's I = 0.15, P < 0.01; distance weights matrix, Moran's I = 0.11, P < 0.01).
To model the observed spatial dependence, CAR models were fit to the WNV disease and WNND data.20 Positive spatial correlation among census tracts occurred for counties located along the Front Range in north central Colorado (WNV disease and WNND), in the southeastern part of the state (WNV disease), and to the southwest (WNV disease and WNND). Negative spatial correlation among census tracts occurred in counties dispersed throughout the state for WNV disease and WNND. The pattern for spatial correlation for WNV disease is shown in Figure 3.
Figure 3.
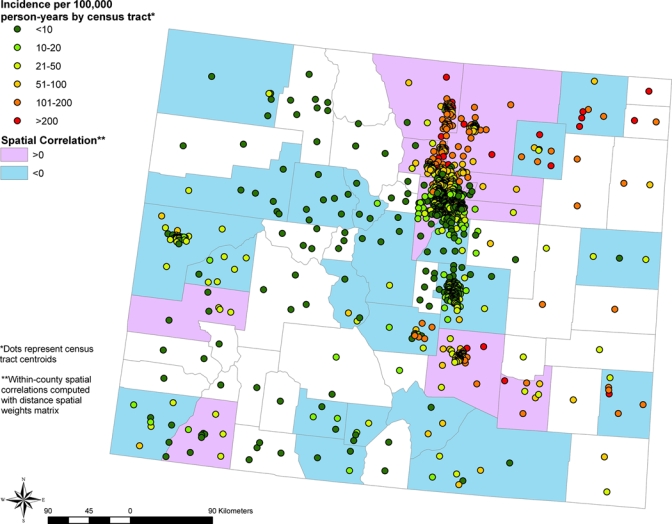
Spatial correlation of West Nile virus disease incidence among census tracts within a given county based on combined data for 2003 and 2007. Colorado counties with positive spatial correlation among census tracts are shown in purple, counties with negative spatial correlation among census tracts in light blue, and counties containing too few census tracts to calculate spatial correlation in white. Points indicate locations of census tract centroids and are color coded by West Nile virus disease incidence. This figure appears in color at www.ajtmh.org.
The scale factors 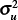 and
and 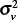 had a ratio for WNV disease of
had a ratio for WNV disease of
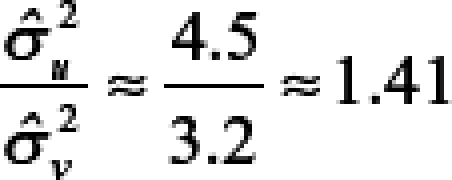 |
with a 95% confidence interval of 0.91–2.26, indicating no statistical difference from a ratio of 1 and suggesting that the spatial model partitions the total variability between counties and census tracts similarly.
The scale factors 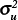 and
and 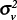 for WNND had a ratio of
for WNND had a ratio of
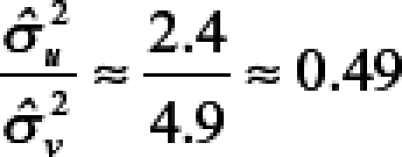 |
with a 95% confidence interval of 0.27–0.80, indicating the ratio is statistically less than 1 and suggesting that the spatial model attributes more variability to the census tracts than to the counties. After accounting for spatial dependence, the results thus indicate that 1) variability in disease incidence within counties is approximately the same as the variability between counties for WNV disease and approximately twice the variability between counties for WNND, and 2) county-scale determination of spatial disease incidence patterns account for only approximately 50% of the variance in WNV disease incidence and approximately 33% for WNND incidence at the finer census tract scale.
Hot-spot analysis for WNV disease.
The Getis-Ord Gi* statistic identified hot-spot census tracts for WNV disease, within a given county, that were surrounded by other census tracts reporting high WNV disease incidence and cool-spot census tracts surrounded by other low WNV disease incidence census tracts. We found numerous instances where hot-spots or cool-spots within counties are obscured when WNV disease incidence is displayed at the county scale. Denver County provided an example of census tract hot-spots occurring within a county reporting overall low incidence (17.09 cases per 100,000 person-years). As shown in Figure 4A, eight statistically significant (P < 0.05) census tract hot-spots were identified within this county. Conversely, cool-spots occurred in several counties in north central Colorado that reported high overall WNV disease incidence (Larimer, Weld, and Morgan counties; range = 109.03–134.48 cases per 100,000 person-years). Within these counties, 14 census tracts were statistically significant cool-spots (Figure 4B).
Figure 4.
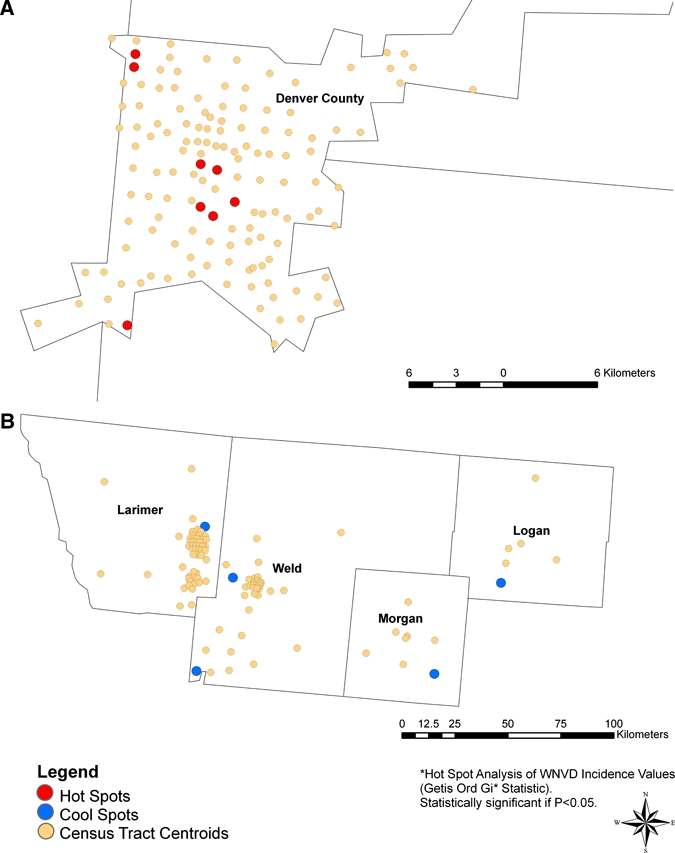
Hot spots and cool spots for West Nile virus disease incidence based on combined data for 2003 and 2007 for census tracts located within selected counties in Colorado. A, Denver County with a low overall incidence (17.09 cases per 100,000 person-years) and B, Larimer, Weld, Morgan, and Logan counties with high overall incidences (range = 109.03–134.48 cases per 100,000 person-years). Points indicate locations of census tract centroids and are color coded to indicate presence of statistically significant (P < 0.05) hot or cool spots of West Nile virus disease incidence. A separate analysis was conducted for each county. This figure appears in color at www.ajtmh.org.
Concordance between spatial patterns based on WNV disease case count versus incidence.
Spatial patterns for WNV disease case counts and WNV disease incidence are shown by county, census tract, and zip code in Figure 2. Positive correlations between case counts and disease incidence occurred for all three spatial units but was much stronger for census tract (ρs = 0.877, n = 1,075, P < 0.001) than for zip code (ρs = 0.238, n = 443, P < 0.001) or county (ρs = 0.558, n = 64, P < 0.001). A similar pattern among the spatial units was detected for high-risk areas falling in the fourth quartile for each disease measure. High-risk counties based on WNV disease case counts were distributed throughout the Front Range in the central part of the state, whereas high-risk counties based on WNV disease incidence more commonly were located in far eastern Colorado (Figure 2). Of the 12 counties classified as high risk based on WNV disease incidence, 6 were also classified as high risk based on WNV disease case counts (50% concordance between spatial patterns for high-risk counties based on WNV disease case count versus WNV disease incidence; Table 1).
Table 1.
Concordance between spatial patterns for high-risk areas based on WNV disease incidence versus WNV disease case counts for the county, zip code, and census tract scales, Colorado*
| WNV disease incidence | WNV disease case count | ||
|---|---|---|---|
| High risk† | Other‡ | % Concordance | |
| County scale | |||
| High risk† | 6 | 6 | 50 |
| Other‡ | 6 | 46 | 88 |
| Zip code scale | |||
| High risk† | 23 | 51 | 31 |
| Other‡ | 51 | 318 | 86 |
| Census tract scale | |||
| High risk† | 158 | 33 | 83 |
| Other‡ | 33 | 851 | 96 |
WNV = West Nile virus.
Spatial units with WNV disease case counts or WNV disease incidences falling within the fourth quartile for each spatial scale were considered high risk.
Spatial units either lacking cases and with incidence of 0, or falling within the first to third quartile.
High-risk zip codes based on WNV disease case counts occurred in three distinct clusters in the north central, northeastern, and south central parts of Colorado, whereas high-risk zip codes based on WNV disease incidence were shifted to the far eastern parts of the state (Figure 2). Of the 74 zip codes classified as high risk based on WNV disease incidence, 23 were also classified as high risk based on WNV disease case counts (31% concordance; Table 1). In contrast, we found far higher concordance (83%) between spatial patterns for high risk based on WNV disease case count versus WNV disease incidence for the census tract scale (Table 1). High-risk census tracts were ubiquitous in northeastern Colorado, occurred commonly to the southeast, and were found only sporadically in the western, mountainous part of the state (Figure 2).
Discussion
We used WNV disease in Colorado as a case study to quantitatively examine 1) the degree to which estimates of vector-borne disease incidence is influenced by spatial scale of data aggregation (i.e., county versus census tract), and 2) the extent of concordance between spatial risk patterns based on disease case counts versus disease incidence for commonly used spatial boundary units. The analyses showed that variability in WNV disease incidence within counties is approximately the same as the variability between counties, and that county-scale determinations of spatial WNV disease incidence patterns therefore account for only approximately 50% of the variance in WNV disease incidence that is shown at the census tract scale. This pattern was even stronger for WNND, with variability in incidence within counties approximately twice the variability between counties and the county scale accounting for only approximately 33% of the variability evident at the census tract scale. Use of the county scale was also found to mask hot-spots for WNV disease evident at finer scale (census tract or zip code) in counties with low overall WNV disease incidence. Furthermore, there was high concordance between spatial patterns of areas with high risk for exposure to WNV based on WNV disease incidence and WNV disease case counts for the census tract scale but not for the county or zip code scales. The primary weakness of the study, which needs to be addressed in prospective follow-up studies, is the lack of reliable information for WNV exposure sites for patients. Developing a more detailed understanding of the spatial dimensions of WNV transmission to humans in different environments, for example in urban versus rural areas, is an important next step to provide additional data to guide the public health community in the choice of appropriate spatial boundary units for presentation of aggregated vector-borne disease data.
There is a diverse stakeholder community with an interest in spatial patterns of risk for contracting diseases caused by vector-borne pathogens. In the specific case of WNV disease, stakeholders include federal, state, and local public health agencies, mosquito control programs, health care providers, purveyors of disease prevention products, and the general public. These stakeholders have needs for spatial information that differ not only in terms of scale but also in type of information. For example, a mosquito control program aiming to implement control activities to suppress vector mosquitoes and reduce the burden of WNV disease likely will be most interested in finding out where high numbers of WNV disease cases occur at sub-county scales to focus expensive prevention efforts. Conversely, a member of the public seeking information to help determine his/her personal risk of exposure to WNV, and the need for use of personal protective measures such as repellents, will be more interested in a spatial risk estimate based on WNV disease incidence (which accounts for population size) in the area of interest. The challenge presented to public health map-makers is to present stakeholders with a package of suitable and easy-to-understand information for spatial risk patterns in electronic map formats while at the same time protecting patient privacy and carefully considering benefits and drawbacks to determination and presentation of risk assessments at different spatial scales.23
Basic options to present information for spatial risk of vector-borne diseases in map formats include point locations for disease cases or aggregation of disease case counts or disease incidence to administrative boundary units (summarized in Table 2). A map showing individual case point locations is obviously the most precise way to present spatial disease data. However, this has distinct disadvantages including 1) the possibility that the address of residence is not the site of pathogen exposure, 2) a lack of accounting for population size, and 3) in some countries, including the United States, strict regulations to guide the use of patient health information.24–26 The latter issue can be addressed by random offsets from the actual location of the patient's residence but this essentially means that an inaccurate disease case location map is presented.
Table 2.
Options for presentation of spatial patterns of vector-borne diseases
| Method | Advantages | Disadvantages |
|---|---|---|
| Disease case point locations | Fine-scale information allowing for precise communication of disease risk. Facilitates development of predictive spatial risk models from knowledge of case point locations (based on kriging interpolation, associations of case locations with environmental factors, etc). Not subject to modifiable areal unit problem. | Privacy issues associated with dissemination of information for disease case locations (can be addressed by random offsets from actual case locations). Address of residence may not be the site of pathogen exposure. May simply reflect population density. |
| Aggregate of disease cases | Useful for targeting vector control resources to the areas with the highest case loads. Improved likelihood that the probable site of pathogen exposure is included within the boundary unit associated with the address of residence. | Less precise compared with disease case point locations. Subject to modifiable areal unit problem. May simply reflect population density. |
| Aggregate of disease incidence | Disease risk measure that accounts for population and thus reflects level of personal risk. Improved likelihood that the probable site of pathogen exposure is included within the boundary unit associated with the address of residence. | Less precise compared with disease case point locations. Under-represents total variance. Subject to modifiable areal unit problem. Problematic for boundary units with low population bases. |
A commonly used approach to avoid privacy issues is to aggregate disease case counts or disease incidence to administrative boundaries. This approach in turn raises the issue of the modifiable areal unit problem,27 which occurs when numerical results vary when the same set of data is grouped at different levels of spatial resolution, and raises the question of which boundary unit best captures the variability of spatial vector-borne disease data without compromising data quality.23 Another issue to consider is that data collection practices for patients afflicted with common and less severe vector-borne diseases, such as WNF and Lyme disease, often do not enable reliable determination of probable pathogen exposure sites.15 This issue introduces uncertainty for pathogen exposure sites related to recognized disease cases and places restrictions on the use of fine spatial boundary units such as census blocks. In the United States, the Centers for Disease Control and Prevention and nearly all individual state health agencies provide spatial WNV disease information to the public at the county scale. One exception is the Colorado Department of Public Health and Environment, which in addition to county-based information, also provides maps for WNV disease incidence by census tract.
Although our results provide a compelling argument for display of risk patterns for exposure to vector-borne pathogens at sub-county scales, there are several problems that need to be considered before sub-county information is presented to end-users. There is no question that sub-county variability exists for risk of exposure to mosquito and tick vectors of human pathogens such as WNV and the Lyme disease spirochete, Borrelia burgdorferi, in the United States.28–32 The basic problem when working with sub-county spatial risk patterns developed based on epidemiologic data is to determine which of the resulting patterns are real and which are likely to be analysis artifacts. Such artifacts may occur for several reasons including that 1) case files for common vector-borne diseases, such as WNV disease and Lyme disease, often lack information for likely site of vector and pathogen exposure and thus the address of residence may not be the exposure location; 2) information that a case has occurred may result in other nearby cases being detected through increased risk perception and health care seeking; and 3) lack of access to health care among lower income zip codes or census tracts may prevent reporting and thus mask the presence of disease in those areas. These problems also occur at the county scale but can be assumed to have greater impact at sub-county scales.
One way to evaluate the accuracy of sub-county scale risk patterns that are based on epidemiologic data is to develop complementary spatial models based on entomological risk measures such as abundance of vectors or pathogen-infected vectors and compare the spatial patterns based on epidemiologic versus entomological data.14,32,33 Concordance between epidemiologic and entomological risk measures can validate sub-county scale risk patterns, whereas discordance indicates the need for additional investigations. For example, ground-based entomological surveillance in areas with high projected epidemiologic risk but low projected entomological risk can be used to assess whether the observed epidemiologic pattern represents real risk or more likely is a data artifact.
When choosing the most appropriate spatial scale to use for presentation of epidemiologic data for vector-borne diseases to stakeholder communities, we are faced with a situation where use of the county scale obscures variability in spatial risk patterns evident at sub-county scales. However, use of sub-county scales introduces more potential error in terms of actual pathogen exposure location not falling within the spatial boundary unit containing the case's residence. Prospective studies are urgently needed to determine the extent of this error for county versus sub-county scales for various vector-borne diseases. Use of sub-county units with small population sizes may also present the problem of unstable incidence rates.23 Numerous spatial statistical smoothing methods exist to deal with the problem of rate instability including local-area averaging or geostatistical smoothing such as kriging.34,35 Finally, our findings also highlight the need to present maps of vector-borne disease incidence at either county or sub-county scales together with information on the limitations for the scale at which data are presented.
Figure 2 provides a powerful visual example of the value of side-by-side presentations of spatial disease patterns based on case counts versus incidence. At the county scale, there was low overall correlation between WNV disease incidence and case counts and poor concordance (50%) for counties categorized as high risk for WNV exposure based on case counts versus incidence. Because some stakeholders are better served knowing disease case counts (e.g., mosquito control programs) whereas other stakeholders need information based on disease incidence (e.g., general public), our findings argue for presentations of WNV disease data at the county scale that include maps showing WNV disease case counts and WNV disease incidence. Concordance between high-risk areas determined by case counts versus incidence was also poor for the zip code scale (31%) but much higher for the census tract scale (83%). This pattern of higher concordance for census tracts than for either zip codes or counties in Colorado likely results, in part, from census tracts having a more uniform population size (mean population = 4,427, SD = 2,321) than either zip codes (mean population = 10,742, SD = 13,584) or counties (mean population = 74,355, SD = 148,158).
The analytical methods used in our study on WNV disease in Colorado are broadly applicable to vector-borne diseases in North America where humans are incidental pathogen hosts. These include a wide range of diseases caused by pathogens transmitted by fleas (e.g., plague), mosquitoes (e.g., eastern equine encephalitis, La Crosse encephalitis, St. Louis encephalitis, western equine encephalitis and WNV disease) and ticks (e.g., babesiosis, Colorado tick fever, human granulocytic anaplasmosis, human monocytic ehrlichiosis, Lyme disease, Rocky Mountain spotted fever, tick-borne relapsing fever, and tularemia). The same methods may also be applicable to mosquito-borne diseases where humans serve as important or primary pathogen hosts (e.g., dengue and malaria), but this needs to be corroborated in future studies.
Our study demonstrates the potential value of using sub-county scales to determine and present spatial assessments of risk for vector-borne pathogens based on epidemiologic data. It also underscores some problem areas that need to be addressed in future studies including 1) development of a more detailed understanding of the spatial dimensions of WNV transmission to humans in different environments to assess the potential for increases in error of spatial assignation of WNV disease cases by address of residence at census tract or zip code scale, compared with the county scale, related to pathogen exposure occurring outside of the census tract or zip code of residence but within the county of residence, and 2) assessment of how data collection practices could be changed to provide improved information regarding potential pathogen exposure sites without placing undue burdens on the medical community. Other important research needs include 1) development of spatial risk models based on entomological risk measures to complement risk assessments based on epidemiologic data, and 2) assessment of the extent to which model results may differ based on the scale of the data used to develop the model (for example home location versus census tract or county of residence for models based on epidemiologic data). The latter question applies not only to vector-borne diseases but also broadly to other diseases with causes linked to environmental conditions that are spatially heterogeneous.
There also is need for extensive research on delivery mechanisms for spatial risk maps and other risk assessment information to stakeholder communities, especially through web-based information delivery mechanisms. This need includes 1) gaining a better understanding of what type of information different stakeholder groups feel that they require, and 2) determining optimal map and text formats to ensure that the message we aim to transmit is clear to the user. Evaluating the effect of different data presentations for disease risk (e.g., maps of WNV disease case counts versus disease incidence) also merits future research because threat perception is closely linked to use of personal protective measures such as mosquito repellents.
Acknowledgments
We thank Saul Lozano-Fuentes (Colorado State University) for helpful discussions.
Footnotes
Financial support: The study was funded, in part, by a grant from the Centers for Disease Control and Prevention (T01/CCT822307) and a contract from the National Institutes of Allergy and Infectious Diseases (N01-AI-25489).
Authors' addresses: Anna M. Winters, Rebecca J. Eisen, Mark J. Delorey, Marc Fischer, Roger S. Nasci, and Emily Zielinski-Gutierrez, Division of Vector-Borne Infectious Diseases, Centers for Disease Control and Prevention, Fort Collins, CO. Chester G. Moore and Lars Eisen, Department of Microbiology, Immunology and Pathology, Colorado State University, Fort Collins, CO. W. John Pape, Communicable Disease Program, Colorado Department of Public Health and Environment, Denver, CO.
References
- 1.Beck LR, Lobitz BM, Wood BL. Remote sensing and human health: new sensors and new opportunities. Emerg Infect Dis. 2000;6:217–227. doi: 10.3201/eid0603.000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalluri S, Gilruth P, Rogers D, Szczur M. Surveillance of arthropod vector-borne infectious diseases using remote sensing techniques: a review. PLoS Pathog. 2007;3:e116. doi: 10.1371/journal.ppat.0030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kitron U. Landscape ecology and epidemiology of vector-borne diseases: tools for spatial analysis. J Med Entomol. 1998;35:435–445. doi: 10.1093/jmedent/35.4.435. [DOI] [PubMed] [Google Scholar]
- 4.Rogers DJ, Randolph SE. Studying the global distribution of infectious diseases using GIS and RS. Nat Rev Microbiol. 2003;1:231–237. doi: 10.1038/nrmicro776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.SaTScan™ http://www.satscan.org/ Available at.
- 6.Theophilides CN, Ahearn SC, Grady S, Merlino M. Identifying West Nile virus risk areas: the dynamic continuous-area space-time system. Am J Epidemiol. 2003;157:843–854. doi: 10.1093/aje/kwg046. [DOI] [PubMed] [Google Scholar]
- 7.GoogleEarth http://earth.google.com/ Available at.
- 8.MSVirtualEarth http://www.microsoft.com/virtualearth/ Available at.
- 9.Eisen L, Lozano-Fuentes S. Use of mapping and spatial and space-time modeling approaches in operational control of Aedes aegypti and dengue. PLoS Negl Trop Dis. 2009;3:e411. doi: 10.1371/journal.pntd.0000411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamadjeu R. Tracking the polio virus down the Congo River: a case study on the use of Google Earth™ in public health planning and mapping. Int J Health Geogr. 2009;8:4. doi: 10.1186/1476-072X-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lozano-Fuentes S, Elizondo-Quiroga D, Farfan-Ale JA, Loroño-Pino MA, Garcia-Rejon J, Gomez-Carro S, Lira-Zumbardo V, Najera-Vazquez R, Fernandez-Salas I, Calderon-Martinez J, Dominguez-Galera M, Mis-Avila P, Coleman M, Morris N, Moore CG, Beaty BJ, Eisen L. Use of Google Earth to strengthen public health capacity and facilitate management of vector-borne diseases in resource-poor environments. Bull World Health Organ. 2008;86:718–725. doi: 10.2471/BLT.07.045880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bell BS, Hoskins RE, Pickle LW, Wartenberg D. Current practices in spatial analysis of cancer data: mapping health statistics to inform policymakers and the public. Int J Health Geogr. 2006;5:49. doi: 10.1186/1476-072X-5-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao S, Mioc D, Yi X, Anton F, Oldfield E, Coleman D. Towards Web-based representation and processing of health information. Int J Health Geogr. 2009;8:3. doi: 10.1186/1476-072X-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisen RJ, Eisen L. Spatial modeling of human risk of exposure to vector-borne pathogens based on epidemiological versus arthropod vector data. J Med Entomol. 2008;45:181–192. doi: 10.1603/0022-2585(2008)45[181:smohro]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 15.Eisen RJ, Eisen L. Need for improved methods to collect and present spatial epidemiologic data for vectorborne diseases. Emerg Infect Dis. 2007;13:1816–1820. doi: 10.3201/eid1312.070211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colorado Department of Public Health and Environment http://www.cdphe.state.co.us/dc/zoonosis/wnv/index.html Available at.
- 17.Mostashari F, Bunning ML, Kitsutani PT, Singer DA, Nash D, Cooper MJ, Katz N, Liljebjelke KA, Biggerstaff BJ, Fine AD, Layton MC, Mullin SM, Johnson AJ, Martin DA, Hayes EB, Campbell GL. Epidemic West Nile encephalitis, New York, 1999: results of a household-based seroepidemiological survey. Lancet. 2001;358:261–264. doi: 10.1016/S0140-6736(01)05480-0. [DOI] [PubMed] [Google Scholar]
- 18.Hardin JW, Hilbe JM. Generalized Linear Models and Extensions. 2nd edition. College Station, TX: Stata Press; 2007. [Google Scholar]
- 19.Cressie N. Statistics for Spatial Data. New York: John Wiley & Sons, Inc; 1993. [Google Scholar]
- 20.Fotheringham A, Brunsdon C, Charlton M. Geographically Weighted Regression, the Analysis of Spatially Varying Relationships. West Sussex, United Kingdom: John Wiley and Sons Ltd; 2002. [Google Scholar]
- 21.Wall M. A close look at the spatial structure implied by the CAR and SAR models. J Statist Plann Inference. 2004;121:311–324. [Google Scholar]
- 22.Getis A, Ord J. The analysis of spatial association by use of distance statistics. Geogr Anal. 1992;24:189–206. [Google Scholar]
- 23.Rytkonen MJP. Not all maps are equal: GIS and spatial analysis in epidemiology. Int J Circumpolar Health. 2004;63:9–24. doi: 10.3402/ijch.v63i1.17642. [DOI] [PubMed] [Google Scholar]
- 24.Curtis A, Mills JW, Leitner M. Keeping an eye on privacy issues with geospatial data. Nature. 2006;441:150. doi: 10.1038/441150d. [DOI] [PubMed] [Google Scholar]
- 25.Health Insurance Portability and Accountability Act of 1996. hhs.gov/HIPAAGenInfo/Downloads/HIPAALaw.pdf Available at.
- 26.VanWey LK, Rindfuss RR, Gutmann MP, Entwisle B, Balk DL. Confidentiality and spatially explicit data: concerns and challenges. Proc Natl Acad Sci USA. 2005;102:15337–15342. doi: 10.1073/pnas.0507804102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Openshaw S. The Modifiable Areal Unit Problem. Norwich, UK: Geo Books; 1984. [Google Scholar]
- 28.Diuk-Wasser MA, Brown HE, Andreadis TG, Fish D. Modeling the spatial distribution of mosquito vectors for West Nile virus in Connecticut, USA. Vector Borne Zoonotic Dis. 2006;6:283–295. doi: 10.1089/vbz.2006.6.283. [DOI] [PubMed] [Google Scholar]
- 29.Eisen RJ, Eisen L, Lane RS. Predicting density of Ixodes pacificus nymphs in dense woodlands in Mendocino County, California, based on geographical information systems and remote sensing versus field-derived data. Am J Trop Med Hyg. 2006;74:632–640. [PubMed] [Google Scholar]
- 30.Nielsen CF, Armijos MV, Wheeler S, Carpenter TE, Boyce WM, Kelley K, Brown D, Scott TW, Reisen WK. Risk factors associated with human infection during the 2006 West Nile virus outbreak in Davis, a residential community in northern California. Am J Trop Med Hyg. 2008;78:53–62. [PMC free article] [PubMed] [Google Scholar]
- 31.Rodgers SE, Mather TN. Evaluating satellite sensor-derived indices for Lyme disease risk prediction. J Med Entomol. 2006;43:337–343. doi: 10.1603/0022-2585(2006)043[0337:essifl]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 32.Winters AM, Bolling BG, Beaty BJ, Blair CD, Eisen RJ, Meyer AM, Pape WJ, Moore CG, Eisen L. Combining mosquito vector and human disease data for improved assessment of spatial West Nile virus disease risk. Am J Trop Med Hyg. 2008;78:654–665. [PubMed] [Google Scholar]
- 33.Eisen RJ, Lane RS, Fritz CL, Eisen L. Spatial patterns of Lyme disease risk in California based on disease incidence data and modeling of vector-tick exposure. Am J Trop Med Hyg. 2006;75:669–676. [PubMed] [Google Scholar]
- 34.Elliot P, Wakefield J, Best N, Briggs D. Spatial Epidemiology Methods and Applications. Oxford, United Kingdom: Oxford University Press; 2000. [Google Scholar]
- 35.Waller LA, Gotway CA. Applied Spatial Statistics for Public Health. Hoboken, NJ: John Wiley & Sons, Inc; 2004. [Google Scholar]


