Abstract
Ca2+ signals are core transducers and regulators in many adaptation and developmental processes of plants. Ca2+ signals are represented by stimulus-specific signatures that result from the concerted action of channels, pumps, and carriers that shape temporally and spatially defined Ca2+ elevations. Cellular Ca2+ signals are decoded and transmitted by a toolkit of Ca2+ binding proteins that relay this information into downstream responses. Major transduction routes of Ca2+ signaling involve Ca2+-regulated kinases mediating phosphorylation events that orchestrate downstream responses or comprise regulation of gene expression via Ca2+-regulated transcription factors and Ca2+-responsive promoter elements. Here, we review some of the remarkable progress that has been made in recent years, especially in identifying critical components functioning in Ca2+ signal transduction, both at the single-cell and multicellular level. Despite impressive progress in our understanding of the processing of Ca2+ signals during the past years, the elucidation of the exact mechanistic principles that underlie the specific recognition and conversion of the cellular Ca2+ currency into defined changes in protein–protein interaction, protein phosphorylation, and gene expression and thereby establish the specificity in stimulus response coupling remain to be explored.
INTRODUCTION
Calcium (Ca2+) likely represents the most versatile ion in eukaryotic organisms. It is involved in nearly all aspects of plant development and participates in many regulatory processes. Because of its flexibility in exhibiting different coordination numbers and complex geometries, Ca2+ can easily form complexes with proteins, membranes, and organic acids. On the one hand, this feature renders Ca2+ a toxic cellular compound at higher concentrations because it would readily form insoluble complexes with phosphate (as present in ATP), but on the other hand, the required tight spatial and temporal control of cellular Ca2+ concentration may have paved the way for the evolutionary emergence of Ca2+ signaling.
Considerable interest and research on this ion has been sparked by the apparent antagonism between the obvious cellular abundance of Ca2+ in certain organelles and cell structures and its required rareness in the cytoplasm. Since the first report in the green algae Chara that changes of cytosolic Ca2+ indicate a function of Ca2+ as a second messenger in plants (Williamson and Ashley, 1982), transient elevations in cytosolic Ca2+ concentration have been documented to be involved in a multitude of physiological processes, including responses to abiotic stresses, hormones, and pathogens. During the last two decades of the 20th century, advances in Ca2+ monitoring techniques have allowed detailed analyses of cellular Ca2+ dynamics. Several groups reported that defined changes of cytosolic Ca2+ concentration are triggered by cellular second messengers, such as NAADP, IP3, IP6, Sphingosine-1-Phospate, and cADPR (Drøbak and Ferguson, 1985; Schumaker and Sze, 1987; Blatt et al., 1990; Gilroy et al., 1990; Allen and Sanders, 1995; Navazio et al., 2000; Lemtiri-Chlieh et al., 2003), and it became evident that the identity and intensity of a specific stimulus impulse results in stimulus-specific and dynamic alterations of cytosolic Ca2+ concentration (Allen et al., 1995; McAinsh et al., 1995). This heterogeneity of increases in cytosolic-free Ca2+ ion concentration in terms of duration, amplitude, frequency, and spatial distribution lead A.M. Hetherington and coworkers to formulate the concept of “Ca2+ signatures” (Webb et al., 1996). Herein, signal information would be encoded by a specific Ca2+ signature that is defined by precise control of spatial, temporal, and concentration parameters of alterations in cytosolic Ca2+ concentration. The spectrum of stimuli that evoke such Ca2+ elevations and their stimulus-specific characteristics has been cataloged and critically discussed in a number of informative reviews (Rudd and Franklin-Tong, 1999; Sanders et al., 1999; Knight and Knight, 2001; Sanders et al., 2002; Scrase-Field and Knight, 2003). Subsequent research suggested that while the shape and spatio-temporal distribution of Ca2+ elevations could be of critical importance for stimulus response coupling (Allen et al., 2001), an additional level of regulation and specificity is achieved by Ca2+ binding proteins that function as signal sensor proteins (Batistič and Kudla, 2004). These proteins decode and relay the information encoded by Ca2+ signatures into specific protein–protein interactions, defined phosphorylation cascades, or transcriptional responses (Luan et al., 2002; Sanders et al., 2002; Finkler et al., 2007a). Consequently, the dynamic interplay between Ca2+ signatures and Ca2+ sensing proteins contributes to generating stimulus specificity of Ca2+ signaling. Since the principles and cellular tool kits of Ca2+ signaling were last reviewed in this journal (Luan et al., 2002; Sanders et al., 2002), remarkable progress has been achieved especially in elucidating the mechanisms that contribute to decoding of Ca2+ signals, and complete Ca2+-triggered regulatory modules have been identified. In this review, we will focus on the description of scientific insights and the discussion of emerging concepts that have been arising over the past few years.
FUNCTIONS OF Ca2+ SIGNALING
Ca2+ is involved in various responses to abiotic and biotic stimuli, including light, high and low temperature, touch, salt and drought, osmotic stress, plant hormones, fungal elicitors, and nodulation factors (Sanders et al., 1999). These stimuli induce a distinct spatio-temporal pattern of changes in cytosolic-free Ca2+ concentration ([Ca2+]cyt). Single-cell systems, such as guard cells, growing pollen tubes, or root hairs, represent excellent models to investigate primary and autonomous Ca2+ responses. However, the final response of the plant to external stimuli is manifested by regulation of complex growth processes in distinct tissues and organs. Concurrently to the diversity of stimulus-specific Ca2+ signatures at the single-cell level, differentiation gives rise to another layer of cell type–specific Ca2+ responses in tissues or organs. This additional level of complexity may contribute to more diversity in local or systemic responses. Therefore, research on plant Ca2+signaling has taken advantage of single-cell model systems but in parallel moves forward to elucidate Ca2+ dynamics in the tissue context and in the whole organism. Consequently, here, we review and discuss how Ca2+ signatures contribute to signaling processes at the single-cell level and the multicellular level.
Ca2+ Signaling at the Single Cell Level
Stomatal Closure and Opening
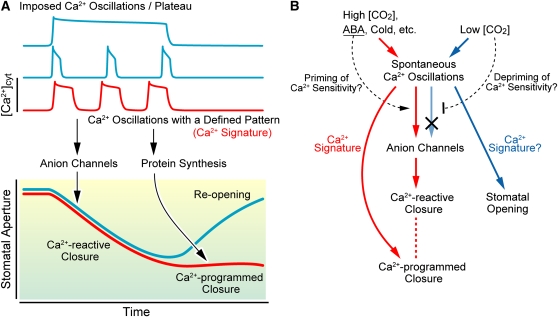
In Arabidopsis thaliana guard cells, stimuli for stomatal closure, including abscisic acid (ABA), hydrogen peroxide, cold, elevation of external Ca2+, and atmospheric CO2, induce cytosolic Ca2+ oscillations (Allen et al., 2001; Young et al., 2006). Artificially imposed Ca2+ oscillations have revealed that cytosolic Ca2+ regulates stomatal closure by two mechanisms: short-term Ca2+-reactive closure and long-term Ca2+-programmed closure (Figure 1A) (Allen et al., 2001; Sanders et al., 2002). The short-term Ca2+-reactive closure is a rapid reaction to exceeding a certain level of [Ca2+]cyt elevation and is irrespective of the elevation pattern (Ca2+ functions likely in a threshold manner). Meanwhile, long-term Ca2+-programmed closure (i.e., prevention of stomatal reopening) is controlled by Ca2+ oscillations within a defined range of amplitude, frequency, duration, and overall transient number (Ca2+ functions as a signature). Recent studies have revealed several elements involved in these mechanisms. The Arabidopsis Ca2+-dependent protein kinase (CDPK) double mutant cpk3 cpk6 exhibits impaired activation of S-type anion channels in response to cytosolic Ca2+. Guard cells in the double mutant are defective in short-term closure but not in long-term closure (Mori et al., 2006). Mutation of the SLOW ANION CHANNEL-ASSOCIATED1 guard cell anion efflux channel abrogates Ca2+-reactive stomatal closure and impairs stomatal responses to CO2, ABA, ozone, light/dark transitions, humidity change, calcium ions, hydrogen peroxide, and nitric oxide (Negi et al., 2008; Vahisalu et al., 2008). By contrast, overexpression of At GLR3.1, a glutamate (Glu) receptor homolog, causes a defect in long-term closure but not in short-term closure (Cho et al., 2009). These findings indicate that the short-term and long-term responses are functionally separable. Interestingly, the translational inhibitor cycloheximide partially inhibits long-term closure but not short-term closure, suggesting no requirement of de novo protein synthesis in the short-term response (Cho et al., 2009). Taken together, these findings imply that certain threshold levels of Ca2+ elevations activate the preexisting cellular machinery, including Ca2+ sensor proteins and ion channels, to regulate rapid stomatal closure. Moreover, defined patterns of Ca2+ oscillations likely activate preexisting proteins and also induce the expression of required genes, resulting in inhibition of stomatal reopening (Figure 1A).
Figure 1.
Ca2+ Signaling in Guard Cell Regulation.
(A) Schematic representation of artificially imposed Ca2+ oscillations/plateau and the corresponding temporal changes in stomatal aperture. The long-term Ca2+-programmed closure is caused by Ca2+ oscillations with a defined pattern (Ca2+ signature), whereas the short-term Ca2+-reactive closure is induced by [Ca2+]cyt elevation, regardless of the pattern.
(B) Simplified model for Ca2+ controlled stomatal closure and opening. Stomatal closing stimuli, such as ABA, high CO2, and cold, induce spontaneous Ca2+ oscillations, resulting in Ca2+-reactive closure and subsequent Ca2+-programmed closure. On the other hand, low CO2-induced Ca2+ oscillations result not in Ca2+-reactive closure but stomatal opening, implying that the stimulus may desensitize the signaling pathways of the Ca2+-reactive closure (Ca2+ sensitivity priming hypothesis). It is unknown whether low CO2-induced Ca2+ oscillation itself encrypts specific information of stomatal opening.
However, it is important to note that nonoscillatory increases in cytosolic Ca2+ are also important features of stomatal responses that result in stomatal closure and that the actual pattern of oscillations evoked by a specific stimulus exhibits some variation and is not “tight” (Hetherington and Brownlee, 2004). Consequently, the Ca2+ decoding machinery that translates these Ca2+ elevations and oscillations into defined downstream responses must be able to decode these variable oscillations (Hetherington and Brownlee, 2004).
Surprisingly, recent studies have shown that Ca2+ SENSING-RECEPTOR (CAS), a thylakoid membrane-localized protein, is crucial for the Ca2+ response, suggesting involvement of chloroplasts in [Ca2+]cyt-mediated stomatal closure (Nomura et al., 2008; Weinl et al., 2008). The elucidation of the functional interplay between Ca2+ releases from different intra- and extracellular sources certainly represents an important goal to further our understanding of guard cell signaling.
Ca2+ signaling likely also contributes to the regulation of stomatal opening (Irving et al., 1992; Schroeder et al., 2001). For example, low atmospheric CO2, which mediates stomatal opening, induces high-frequency Ca2+ oscillation. Treatment with the cytosolic Ca2+ chelator BAPTA-AM abolishes the oscillation and attenuates the stomatal opening in response to low CO2 (Young et al., 2006). It would be interesting to investigate whether the rapid oscillation pattern encrypts specific information. As described above, artificially imposed Ca2+ oscillations induce the short-term Ca2+-reactive closure regardless of their pattern. However, the rapid Ca2+ oscillation induced by low CO2 does not cause such reaction (Young et al., 2006). A model was derived in which low CO2 stimuli may negatively modulate Ca2+ sensitivities of signaling components, which regulate the short-term closure, such as CDPKs. On the other hand, during the stomatal closure response, preexposure to ABA enhances the response of S-type anion channels to [Ca2+]cyt elevation, implying that ABA may positively modulate sensitivities of Ca2+ sensor proteins (Siegel et al., 2009). A Ca2+ sensitivity priming model has been proposed as a hypothesis to explain these observations in the context of mechanisms that contribute to the generation of specificity in Ca2+ signaling (Figure 1B) (Young et al., 2006).
The Establishment of Symbiosis in Root Hairs
In legume root hair cells, exposure to rhizobial-derived nodulation (Nod) factors induces biphasic changes [Ca2+]cyt that comprise an initial Ca2+ influx and a subsequent (10 to 20 min later) long-term Ca2+ oscillation (also designated as Ca2+ spiking) in the perinucleus (Shaw and Long, 2003). The Medicago truncatula mutants does not make infection1 (dmi1) and dmi2 are defective in the Ca2+ spiking but retain the initial Ca2+ influx. Furthermore, low concentration (10−11 to 10−12 M) of Nod factor induces Ca2+ spiking but not Ca2+ influx, suggesting that they are separable responses (Shaw and Long, 2003). Induction of Early Nodulation 11 (ENOD11) is impaired in the Ca2+ spiking-defective mutants dmi1 and dmi2, as well as in dmi3, which is defective in a gene encoding Ca2+ calmodulin-dependent kinase (CCaMK). Remarkably, specific removal of the autoinhibitory domain of this kinase leads to autoactivation of the downstream nodulation signaling pathway with the resultant induction of nodules and nodulation gene expression in the absence of bacterial elicitation (Gleason et al., 2006). This demonstrates not only the essential function of this CCaMK in the regulation of nodule development but also indicates that the activation of this kinase through the oscillatory Ca2+ signal is necessary and sufficient to elicit this process (Gleason et al., 2006). Although the mechanism of generation of Ca2+ spikes is not well understood, some blockers for Ca2+ channels and Ca2+ pumps have been shown to inhibit both Ca2+ spiking (Engstrom et al., 2002) and ENOD11 expression (Charron et al., 2004). This evidence suggests that Ca2+ spiking is essential for regulation of nodulation, raising the question of how the spiking could encode specific information. Miwa et al. (2006) proposed a correlation between the number of Ca2+ spikes and ENOD11 expression levels. In this work, ENOD11 inductions were observed only when the Ca2+ spiking lasted for at least 60 min. The average period between each spike was ∼100 s; therefore, the authors estimated that a minimum of ∼36 spikes is required for ENOD11 induction. Jasmonic acid treatment lengthens the period between each Ca2+ spike but does not affect the required number of spikes for ENOD11 expression, further supporting the notion that the number of Ca2+ spikes functions to encode information (Miwa et al., 2006).
Ca2+ also plays important roles in the symbiosis of legumes with arbuscular micorrhizal fungi. Incubation with micorrhizae induces Ca2+ spiking in legume root hair cells. Ca2+ spiking is abolished in M. truncatula dmi1 and dmi2, suggesting that nodulation and mycorrhizal infection involve common signaling components. Despite the overlap, micorrhizal-induced Ca2+ spiking exhibits a shorter period and smaller amplitudes compared with that in the Nod factor response (Kosuta et al., 2008).
Remarkably, bacterial elicitors of plant pathogens also induce a Ca2+ release in the cytosol and nucleus. Here, the nitric oxide signal following elicitor application is important for the Ca2+ release in the cytosol, but it does not trigger a nuclear Ca2+ response (Lamotte et al., 2004). The nucleus also exhibits specific calcium signals in response to different elicitors. Harpin and flagellin resulted in a different Ca2+ release than observed in response to carbohydrate elicitors such as oligogalacturonides. Remarkably, the cytosolic Ca2+ response to these elicitors was comparable (Lecourieux et al., 2005). These observations suggest that the nucleus does indeed harbor an independent Ca2+ machinery that may involve P-ATPases and nucleotide gated channels located at the inner membrane of the nucleus to regulate the nuclear Ca2+ reservoir (Mazars et al., 2009).
Tip Growth in Pollen Tubes and Root Hair Cells
Pollen tubes are one of the most extensively studied tip-growing model systems. In pollen tubes, [Ca2+]cyt is highly concentrated at the apex by an extracellular influx (Sanders et al., 1999; Hepler et al., 2001). In the Ca2+ gradient, [Ca2+]cyt oscillates with a lag phase behind the corresponding growth rate oscillation, suggesting that stretch-activated Ca2+ channels may be involved in the maintenance of the Ca2+ gradient (Dutta and Robinson, 2004). In accordance with an essential function of stretch-activated channels, pharmacological inhibition of channel activity disrupts the Ca2+ influx at the apex and terminates pollen tube elongation (Picton and Steer, 1985). Moreover, Petunia inflata and M. truncatula mutant lines that are defective in CDPKs exhibit loss of polarity (Ivashuta et al., 2005; Yoon et al., 2006). Therefore, the formation of a Ca2+ gradient as well as the decoding of Ca2+ signals by Ca2+ sensing proteins appear to play important roles in generating spatial determinants for pollen tube growth. Moreover, several studies have suggested a close interaction of intracellular Ca2+ and the cytoskeleton in that the growth regulatory effect of this ion is exerted by Ca2+-dependent regulation of the structure and activity of F-actin (Snowman et al., 2002; Cardenas et al., 2008).
Recent studies have also revealed the importance of Ca2+ signaling in the root hair single-cell model system. Similar to growing pollen, root hair cells exhibit a tip-focused Ca2+ gradient with Ca2+ oscillation lagging behind growth oscillation (Monshausen et al., 2008). Moreover, the Ca2+ oscillation is followed by oscillation of apoplastic reactive oxygen species (ROS) production (Monshausen et al., 2007, 2008). Intriguingly, the RHD2 NADPH oxidase (also known as RBOH C) is localized to the plasma membrane of the tip-growing site and is required for appropriate growth of root hair cells (Takeda et al., 2008). RHD2 contains an EF hand-like Ca2+ binding domain and phosphorylation sites that appear to be targets of calcium-regulated protein kinases (Takeda et al., 2008). Therefore, the Ca2+ oscillation may regulate RHD2 activity. Inhibition of the Ca2+ oscillation by La3+ causes bursting growth, while increasing the Ca2+ gradient by Ca2+ ionophore arrests growth of root hairs (Monshausen et al., 2008). Consequently, it has been proposed that Ca2+ and ROS compose a positive feedback loop to sustain tip growth of root hair cells (Takeda et al., 2008).
Ca2+ Signaling at the Multicellular Level
Abiotic Stress Responses
In the endodermis and the pericycle of Arabidopsis roots, salt stress causes a biphasic Ca2+ response consisting of an initial transient elevation and a subsequent oscillation that differs in phase and/or period in individual cells (Kiegle et al., 2000). By contrast, only a monophasic [Ca2+]cyt elevation is induced in the epidermis and the cortex (Kiegle et al., 2000). Interestingly, the initial Ca2+ response in the pericycle occurs a few seconds later than that in the other cell types. Moreover, the magnitude in the pericycle is significantly lower. This is likely because the endodermis limits the flow of water and ions to the xylem. Accordingly, whole-plant [Ca2+]cyt measurements have suggested a direct correlation between the strength of NaCl stress and the magnitude of [Ca2+]cyt elevation (Tracy et al., 2008). The function of the subsequent Ca2+ oscillation in the endodermis and the pericycle remains to be elucidated.
In contrast with salt stress, cold stress causes only monophasic [Ca2+]cyt elevation in the four types of Arabidopsis root cells without significant temporal difference, suggesting that all cells sense temperature changes simultaneously (Kiegle et al., 2000). The magnitude of cold-induced Ca2+ responses is dependent on the cooling rate (ΔT/Δt) (Plieth et al., 1999). However, the pericycle exhibits a more pronounced Ca2+ response to cold stress (Kiegle et al., 2000), suggesting that different cell types harbor different Ca2+ homeostasis and signaling components. Interestingly, when cold-induced Ca2+ responses were analyzed by specific expression of the reporter protein Aequorin in guard cells, Dodd et al. (2006) observed circadian modulations of the cold-induced Ca2+ elevation that were significantly more pronounced during the mid-photoperiod than at the beginning or the end of the day. Therefore, it would be interesting to determine if circadian modulation of Ca2+ responses also occurs in root tissues and during responses to other environmental cues.
Dodd et al. (2006) also performed mathematical modeling analysis of calcium signatures to investigate the relationship between single-cell calcium analyses and whole population (or whole plant) calcium monitoring. This study revealed that the population Ca2+ signature may be diagnostic for underlying single-cell Ca2+ oscillations, but it is unlikely to be representative of single-cell Ca2+ signatures. This is a very important finding that needs to be considered when interpreting work of Ca2+ dynamics that has used Aequorin as a (whole plant) reporter protein. For example, the second sustained Ca2+ elevation of biphasic Ca2+ responses that has been observed in such studies may in fact represent an amalgamation of asynchronous Ca2+ oscillations that occur in individual cells of the plant after the initial synchronous Ca2+ transient (Dodd et al., 2006).
Recently, it has been demonstrated that heat shock induces prolonged [Ca2+]cyt elevation (∼20 min duration) via putative heat-sensitive plasma membrane Ca2+-permeable channels in the moss Physcomitrella patens (Saidi et al., 2009). It appears that a larger increase in temperature induces a more intense [Ca2+]cyt elevation and results in a stronger response (e.g., expression of heat shock proteins) compared with a less pronounced temperature elevation shift. In Arabidopsis, Ca2+ and calmodulin proteins have been shown to be involved in the heat shock response (Zhang et al., 2009). It is likely that flowering plants also sense temperature increments with heat-sensitive channels. It would be interesting to elucidate whether heat shock induces cell type–specific [Ca2+]cyt elevations in Arabidopsis and to compare the Ca2+-related mechanisms of heat stress responses with that of the cold response.
Response to Mechanical Stimuli
There is ample evidence that mechanical stimuli, such as touch or wind, induce [Ca2+]cyt elevations in plant cells (Braam, 2005). Recently, Monshausen et al. (2009) reported that different types of mechanical stimuli, such as touch and bending, induce distinct patterns of Ca2+ responses in the Arabidopsis root. Whereas touch stimuli induce monophasic [Ca2+]cyt elevations in the cells at the touch site, bending elicits biphasic transient elevations in the cells on the convex (stretching) side. These Ca2+ responses are essential for apoplastic alkalinization as well as RBOH C-dependent apoplastic ROS production that may contribute to resistance of plants to mechanical stresses (Monshausen et al., 2009). Bending of the root recruits pericycle cells on the convex side to become founder cells of a new lateral root (LR) primordium. Interestingly, blocking the bending-induced [Ca2+]cyt elevation inhibits the recruitment of new LRs (Richter et al., 2009), indicating an essential role of Ca2+ signals in this process. However, it remains to be elucidated how the biphasic pattern of Ca2+ elevation encrypts specific information for bending-induced LR production.
GENERATION OF Ca2+ SIGNALS
A Ca2+ signal is defined by the balanced activation of Ca2+ channels at different cellular membranes, which is followed by the subsequent inactivation of channels and activation of efflux transporters to terminate Ca2+ influx and to rebalance the cellular Ca2+ homeostasis. Both processes are strictly regulated and define the physiological outcome of Ca2+ signaling.
Influx of Ca2+
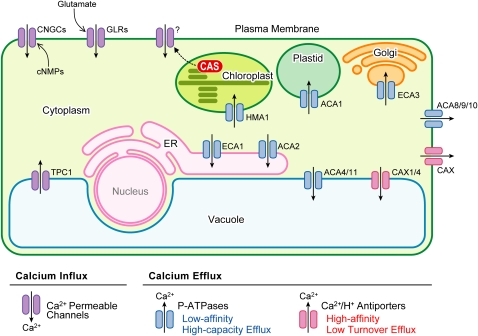
Distinct ion channel types capable of mediating Ca2+ fluxes coexist in different cell types and tissues. According to their activation mechanism, these channels can be classified as voltage-dependent, voltage-independent/ligand-dependent, and stretch-activated Ca2+ channels (Cosgrove and Hedrich, 1991; White et al., 2002; White and Broadley, 2003; Dutta and Robinson, 2004; Nakagawa et al., 2007). Depending on their specific activation properties, Ca2+ channels can shape the parameters of Ca2+ influx and the resulting Ca2+ signature (White et al., 2002; Demidchik and Maathuis, 2007). This enables the plant to translate a wide range of different signals into distinct Ca2+ signatures (Miedema et al., 2001, 2008). Moreover, variability in the specific abundance of the different channel types likely reflects the special needs of a cell type or tissue (Demidchik et al., 2002). A schematic summary of the channels and transporters that are discussed in this review is illustrated in Figure 2.
Figure 2.
Overview of Ca2+ Transport Systems in an Arabidopsis Cell.
Shown are Ca2+ influx/efflux pathways that have been identified at the molecular level. See text for further details. CNGC, cyclic nucleotide channel; GLR, glutamate receptor; TPC1, two pore channel 1; CAS, Ca2+-sensing receptor; ACA, autoinhibited calcium ATPase; ECA, ER type calcium ATPase; HMA1, heavy metal ATPase1; CAX, cation exchanger.
Voltage-Dependent Channels at the Plasma Membrane
Voltage-dependent Ca2+-permeable channels have been classified as depolarization-activated Ca2+-permeable channels (DACCs) and hyperpolarization-activated Ca2+-permeable channels (HACCs) (White et al., 2002). Although the properties of DACCs and HACCs are well studied electrophysiologically (Grabov and Blatt, 1998; Thion et al., 1998; Thuleau et al., 1998; Hamilton et al., 2000; Pei et al., 2000; Klusener et al., 2002), the molecular identity of these channels is still unknown (White et al., 2002). It is assumed that DACCs contribute to the short transient influx of Ca2+ in response to various stimuli, including chilling and microbe interaction (Thion et al., 1998). HACCs contribute to a sustained Ca2+ influx in response to ABA (Hamilton et al., 2000; Pei et al., 2000), blue light (Harada and Shimazaki, 2009), and Ca2+ nutrition (Miedema et al., 2001, 2008).
Plant annexins are ubiquitous, soluble proteins capable of Ca2+-dependent and Ca2+-independent binding to endomembranes and the plasma membrane (Demidchik and Maathuis, 2007; Mortimer et al., 2008). Surprisingly, a recent study indicated that cytosolic annexins create a Ca2+ influx pathway directly, particularly during stress responses involving acidosis in Zea mays (Laohavisit et al., 2009). This suggests that annexins modulate or even represent a part of voltage-gated Ca2+-permeable channels. However, further characterization of plant annexins and especially the molecular identification of voltage-gated Ca2+ channels are required to enable more insights of the contribution of annexins and voltage-gated channels to Ca2+ signaling (Cheong et al., 2007).
Ligand-Gated Channels at the Plasma Membrane
Cyclic nucleotide-gated channels (CNGCs) are ligand-gated channels that are important for cellular homeostasis of cations and can mediate fluxes of Ca2+ ions (Hua et al., 2003a; Ali et al., 2006). Twenty CNGC genes have been identified in Arabidopsis (White et al., 2002). CNGCs are activated by binding of the cyclic nucleotides cAMP and cGMP and harbor a binding site for calmodulin that partially overlaps with the binding domain for cyclic nucleotides (cNMPs). Consequently, binding of Ca2+/calmodulin results in inactivation of CNGCs due to blocking of the cNMP binding domain (Hua et al., 2003b; Ali et al., 2006). Thereby, Ca2+ itself, by binding to calmodulins/calmodulin-like proteins (CMLs), can modulate the influx of Ca2+ mediated by CNGCs. Several CNGCs, including CNGC4, CNGC11, and CNGC12, have been implicated in response reactions to pathogens (Balague et al., 2003; Yoshioka et al., 2006; Urquhart et al., 2007). CNGC2 represents an extensively characterized channel that was originally identified by cloning of the gene responsible for the defense no death1 (dnd1) mutant phenotype. Mutant plants fail to induce the Ca2+-mediated hypersensitive response to an avirulent strain of the pathogen Pseudomonas syringae and exhibit enhanced resistance to pathogens (Yu et al., 1998; Clough et al., 2000). Specifically, cngc2 mutants are impaired in nitric oxide production that is required for the occurrence of a hypersensitive response (Ali et al., 2007) and that was previously reported to depend specifically on Ca2+ influx from extracellular stores (Lamotte et al., 2004).
The function of CNGCs appears not to be restricted to plant pathogen interactions. CNGC3 and CNGC10 are implicated in regulating ion homeostasis especially for establishing the proper Na+/K+ ratio during salt stress adaptation (Gobert et al., 2006; Guo et al., 2008). CNGC18 is asymmetrically localized to the tip region of growing pollen tubes and has been shown to be indispensable for proper tip growth of pollen, suggesting that this channel is important to establish the tip-focused gradient of Ca2+ (Frietsch et al., 2007). Consequently, CNGC18 provides a mechanism for direct transduction of a cNMP signal into an ion flux that can produce a localized signal capable of regulating the pollen tip growth machinery. Considering the mechanistic similarities in the signal transduction processes governing tip growth in pollen tubes and root hairs, it would be most interesting to address the function of CNGCs in root hairs. Moreover, the investigation of Ca2+ dynamics in cngc mutant lines that would express the Ca2+ reporter protein cameleon would be most helpful for further elucidating the contribution of CNGC toward the generation of cellular Ca2+ signals.
Similar to CNGCs, glutamate receptors (GLRs) are nonselective cation channels that have attracted considerable attention as important regulators of Ca2+ influx. In Arabidopsis, 20 genes encode for GLRs that are differentially activated by Glu and Gly, as well as by other amino acids, and mediate an increase of cytosolic Ca2+ (Chiu et al., 2002; Qi et al., 2006; Stephens et al., 2008). It is assumed that GLRs are important for plant Ca2+ nutrition (Kim et al., 2001; Demidchik and Maathuis, 2007) but also in mediating Ca2+ responses upon cold stress (Meyerhoff et al., 2005) or excess aluminum (Sivaguru et al., 2003). At GLR1.1 regulates the expression of enzymes involved in carbon and nitrogen metabolism and regulates ABA biosynthesis (Kang and Turano, 2003; Kang et al., 2004). At GLR1.1 antisense lines contain elevated ABA levels and are hypersensitive to ABA or glucose (Kang and Turano, 2003; Kang et al., 2004). Overexpression of another GLR, At GLR3.1, impaired long-term stomatal closure but did not affect the short-term stomatal closure response or the kinetics of Ca2+ oscillations that were imposed by extracellular Ca2+ (Cho et al., 2009). In rice (Oryza sativa), disruption of the Os GLR3.1 gene results in reduced growth of the primary root and of adventious roots especially in the early seedling stage due to reduced mitotic activity of the root apical meristem (Li et al., 2006a). GLRs also have been implicated in root development in Arabidopsis since Glu can affect root architecture (Walch-Liu et al., 2006). Additionally, GLRs might have a role in Ca2+-dependent generation of electrical potentials in the root (Masi et al., 2009). Moreover, plants that have been treated with antagonists of GLRs display impaired light-induced signal transduction, harbor enlarged hypocotyls, and accumulate less chlorophyll when grown in light, suggesting that GLRs may also function in photomorphogenesis (Lam et al., 1998; Brenner et al., 2000).
These findings suggest that extracellular Glu not only provides a signal for Ca2+ nutrition but in addition regulates different developmental aspects, electrical transmission, supply of nutrients, and responses to abiotic stress. Therefore, it would be of considerable interest to investigate the functional interrelation of plant CNGCs and GLRs and to investigate whether they act in concert to mediate Ca2+ signaling.
Voltage-Dependent Channels at Other Cellular Membranes
The coexistence of voltage-dependent channels and ligand-gated channels appears not to be restricted to the plasma membrane since compelling evidence points to a function of both channel types also in the vacuolar membrane. An important voltage-dependent channel activity was identified as a slow vacuolar type (SV) channel under depolarized conditions (Johannes et al., 1992; Allen and Sanders, 1994). This SV channel originally was described as voltage-dependent channel that is regulated by cytoplasmic Ca2+ concentrations (Hedrich and Neher, 1987), and subsequent work reported the Ca2+ release capability of this channel (Allen and Sanders, 1994). In addition, this channel seems to be regulated by Ca2+ in two opposing ways. While elevation of cytoplasmic Ca2+ can activate the channel, an increase in vacuolar luminal Ca2+ inactivates the channel (Pottosin and Schonknecht, 2007). This electrophysiologically characterized channel activity was recently shown to be conferred by the protein Two-pore channel1 (TPC1) (Peiter et al., 2005). Although the SV channel is the most abundant vacuolar channel and TPC1 represents a single gene in the Arabidopsis genome, loss of SV channel function in Arabidopsis does not result in a lethal phenotype or severe growth defects. However, loss of TPC1 function has a significant effect on germination inhibition by ABA, and while there is no effect on guard cell ABA signaling, there is a highly significant effect on stomatal responses to external Ca2+ (Peiter et al., 2005; Ranf et al., 2008). Although the SV channels appear to make only a minor contribution to a global Ca2+ release into the cytosol by Ca2+ influx from the vacuole, it was proposed that TPC1 channels could form clusters within the tonoplast to form local Ca2+ microdomains (Perez et al., 2008). In contrast with the situation in Arabidopsis, knockout mutation of TPC1 in rice results in reduced plant growth on soil (Kadota et al., 2004). Moreover, overexpression of Os TPC1 resulted in prolonged activation of the mitogen-activated protein kinase (MAPK) pathway, indicating that in rice (Kurusu et al., 2005) and also in tobacco (Nicotiana tabacum; Kadota et al., 2004), channel function could have a role in pathogen response. Similarly, knockout of TPC1 in Arabidopsis results in weaker induction of defense genes after pathogen infection but does not significantly affect pathogen resistance (Bonaventure et al., 2007b). By contrast, a hyperactive gain-of-function allele of TPC1, designated as fou2, leads to enhanced defense gene expression and resistance to pathogens (Bonaventure et al., 2007a, 2007b). Although the exact physiological role of TPC1 in Arabidopsis is not fully understood, the fact that TPC1 mediates the flux of K+ and other cations through the vacuolar membrane (Bihler et al., 2005; Ivashikina and Hedrich, 2005) may point to an important role of TPC1 in regulating cytosolic K+ homeostasis in a Ca2+-dependent manner (Pottosin and Schonknecht, 2007). In agreement with such a function, SV channel activity is downregulated at low vacuolar potassium concentrations (Pottosin et al., 2005), and the fou2 transcriptome resembles the transcript profile of plants after potassium starvation (Bonaventure et al., 2007b). Moreover, fou2 mutant plants contain less K+ and more Ca2+ within the vacuole (Beyhl et al., 2009). Several recent investigations reported that NAADP binds to and confers the activation of animal TPC channels that results in Ca2+ release from lysosomal and endosomal compartments (Brailoiu et al., 2009; Calcraft et al., 2009). Since these animal TPC channels are structurally closely related to plant TPC1 but lack the EF hand motif in their central domain (Ishibashi et al., 2000), it will be most interesting to investigate if the respective plant channels are also responsive to this second messenger.
In addition to TPC1, several other less well characterized voltage-dependent channels appear to exist at the vacuolar membrane (Allen and Sanders, 1994), which were characterized as a fast vacuolar channel (Hedrich and Neher, 1987) and as a Ca2+-insensitive vacuolar channel (Ranf et al., 2008).
Ligand-Gated Channels at the Vacuolar and Endoplasmic Reticulum Membranes
Several electrophysiological studies supported the existence of ligand-gated channels at the tonoplast and suggested regulation of these channels by IP3/IP6 and cADPR (Schumaker and Sze, 1987; Allen et al., 1995; Muir and Sanders, 1996; Martinec et al., 2000; Navazio et al., 2000; Lemtiri-Chlieh et al., 2003). However, despite the existence of multiple full-genome sequences for several higher plant species, no genes encoding for an ADP ribosyl cyclase that would be required for cADPR production have been identified. In addition, ryanodine receptors that are the targets of cADPR in animal cells appear to be missing in higher plants. In this regard, experimental determination and direct proof of cADPR abundance by gas chromatography–mass spectrometry techniques are urgently needed.
Similarly, no sequences with significant similarity to animal IP3 receptors have been identified in genomes of higher plants. Interestingly, while higher plants lack classical endoplasmic reticulum (ER)-localized IP3 receptors, which are well known and characterized in animals (Berridge, 2009), several algae species, such as Volvox and Chlamydomonas, appear to harbor these receptor channels, suggesting that this channel type was present in the ancient plant progenitor of the evolutionary plant lineage and may have been lost during the further evolution of the plant lineage (Wheeler and Brownlee, 2008). This raises the obvious question of how IP3 can function in plant Ca2+ signaling in the absence of canonical IP3 receptors. The absence of IP3 receptors in higher plants in combination with the extremely low levels of phosphatidyl-inositol-4,5-bisphosphate in plant tissues, that in animal cells is converted by PLC into IP3, has raised concerns about the simple assignability of mechanistic principles of IP3-induced Ca2+ release from animal systems to the situation in plant cells (Munnik and Testerink, 2009). Therefore, future research in this field of plant Ca2+ signaling may unravel unexpected mechanisms that interconnect Ca2+ and IP3.
In addition to the previously mentioned ligand gated channels, a unique ligand-gated channel that appears to reside in the ER membrane is activated by NAADP (Navazio et al., 2000). However, since the molecular identity of all these channels is still unknown, it remains to be established if these ligands directly activate channels or, alternatively, function via receptors that indirectly modulate the activity of channels.
Efflux of Ca2+
Much of the research on Ca2+ signaling has focused on advancing our understanding of the generation of Ca2+ elevations by Ca2+-releasing channels. However, to represent a distinct signal, the regulation of Ca2+ efflux that not only terminates but also shapes the Ca2+ signature is as important as the Ca2+-releasing events. During the past years, we have witnessed significant insights into the regulation of Ca2+ extrusion. However, in the future, it will be most important to reveal the interconnected regulation of Ca2+ release and extrusion that is required to generate defined signals.
Extrusion of Ca2+ from the cytosol is achieved by P-type Ca2+-ATPases and by the Ca2+/proton antiporter systems. While antiporters mediate a high-affinity low turnover efflux, ATPases mediate a low-affinity high-capacity efflux. Therefore, it is assumed that antiporters reduce the Ca2+ concentration back to a few micromolar after signal mediated influx, while Ca2+-ATPases are important to maintain the low resting concentration of Ca2+ (Hirschi, 1999).
The coordinative regulation of the cellular extrusion systems still is not fully understood. Transport activity is clearly activated after influx of Ca2+, as in response to different hormones (Erdei et al., 1979; Bush et al., 1993; Zocchi and Rabotti, 1993), salt stress (Gao et al., 2004), or mechanical stimulation (Bourgeade et al., 1991). Enhanced Ca2+ extrusion was reported to occur in senescent potato (Solanum tuberosum; Fakhrai and Hall, 1984). Moreover, enhanced transcription of ATPases was observed in response to ABA (Cerana et al., 2006), sugar (Mito et al., 1996), or by sodium (Wimmers et al., 1992).
Hormones can also differentially activate transporter systems of different cellular membranes. Ca2+ released by GA seems to be transported mainly via ER transporters out of the cytoplasm, while ABA activates transport activity at the ER as well as at the tonoplast (Bush and Sze, 1986; Bush et al., 1989a, 1993). Interestingly, overall, the Ca2+ extrusion system seems to be less effective in protoplasts or cells from epidermal strips than in intact leaves (Levchenko et al., 2008). This important observation should be considered in the design of future experiments addressing the regulation of Ca2+ extrusion.
Ca2+-Proton Antiporter
In the Arabidopsis genome, six genes encode for putative Ca2+-proton antiporters, also named cation exchangers (CAX) (Maser et al., 2001; Shigaki et al., 2006), which regulate the homeostasis of Ca2+ and other divalent cations, such as cadmium (Catala et al., 2003; Cheng et al., 2003; Koren'kov et al., 2007; Zhao et al., 2008). Five additional antiporters that were previously designated as CAX7-11 resemble potassium (Na2+)-dependent Na2+/Ca2+ antiporters and were therefore renamed cation Ca2+ exchanger proteins (Shigaki et al., 2006). Additionally, four putative antiporters encoded in the Arabidopsis genome contain EF hand Ca2+ binding motifs, implicating that these transporters are directly regulated by Ca2+ (Shigaki et al., 2006). It is unknown to which extent the latter two types of antiporters are important for Ca2+ extrusion.
The antiporters CAX1 to CAX4 are localized to the vacuole (Hirschi et al., 2000; Cheng et al., 2002, 2003, 2005), but antiporter activity was also reported to reside at the plasma membrane (Kasai and Muto, 1990; Luo et al., 2005). CAX proteins harbor an N-terminal regulatory/autoinhibitory domain, which binds to an adjacent region within the N terminus (Pittman et al., 2002; Mei et al., 2007). The exact mechanistic regulation by the N terminus is not well understood, and differential N-terminal–dependent regulation of CAX protein activity may occur. Autoinhibition can be relieved by formation of heteromers of, for example, CAX1 and CAX3 (Zhao et al., 2009), and different regulatory proteins could additionally interact with CAX proteins to regulate transport activity (Cheng and Hirschi, 2003; Cheng et al., 2004a, 2004b). The importance of specific transport activity regulation by the N-terminal region was demonstrated by overexpressing an N-terminal truncated, deregulated version of the vacuolar Ca2+/proton antiporter CAX1 from Arabidopsis in tobacco. Although tobacco plants contained more total Ca2+, these plants showed Ca2+ deficiency symptoms and displayed hypersensitivity to magnesium, sodium, and cold shock (Hirschi, 1999). It was assumed that overexpression of At CAX1 leads to overloading of Ca2+ into the vacuole and to a severe reduction of cytosolic Ca2+ concentration, which caused the observed deficiency symptoms.
The formation of functional heteromers does not exclude the possibility that single CAX proteins are important for responses to specific signals. While both cax1 and cax3 mutant plants are hypersensitive to ABA during germination (Zhao et al., 2008), cax3 mutants exhibited increased sensitivity toward NaCl or LiCl and low pH levels, while cax1 mutants displayed normal wild-type responses (Catala et al., 2003; Zhao et al., 2008). This difference can be partially explained by the observation that CAX3 is prominently expressed in roots, whereas CAX1 is predominantly expressed in leaves (Zhao et al., 2008).
P-ATPases
Classical Ca2+ P-ATPases belong to the second subclass (II) of phosphorylated (P)-type ATPases. They are classified as PIIA- or ER-type Ca2+ ATPases (ECAs; which include four members in Arabidopsis) and PIIB ATPases (10 family members in Arabidopsis, all of which contain an autoinhibtory N-terminal region; Sze et al., 2000). Therefore, the latter ATPases also have been named autoinhibited Ca2+ ATPases (ACAs) (Sze et al., 2000). The autoinhibitory domain in PIIB-type proteins can be relieved by binding of calmodulin to the regulatory domain, which results in activation of the pump (Harper et al., 1998). On the other hand, the activity of the PIIB-type Ca2+ ATPase ACA2 can be inhibited by phosphorylation within the N-terminal regulatory domain by other Ca2+-regulated proteins, such as CDPKs (Hwang et al., 2000).
PIIA-type ATPases are localized at the ER (ECA1) (Liang et al., 1997), the Golgi (ECA3) (Mills et al., 2008), and endosomes (also ECA 3) (Li et al., 2008), implicating that the latter organelles can function as mobile Ca2+ stores that could contribute to the spatial specificity of Ca2+ signaling (Menteyne et al., 2006).
PIIB types are localized at the ER (ACA2) (Harper et al., 1998), vacuole (ACA4 and ACA11) (Geisler et al., 2000; Lee et al., 2007b), plasma membrane (ACA8, ACA9, and ACA10) (Bonza et al., 2000; Schiott et al., 2004; George et al., 2008), and at the plastid envelope (ACA1) (Huang et al., 1993). The importance of a PIIa-type Ca2+-ATPase activity regulating the cytoplasmic Ca2+ dynamics was recently impressively exemplified by the analysis of a Ca2+-ATPase loss-of-function mutant in the moss P. patens (Qudeimat et al., 2008). Whereas wild-type plants exhibit a transient cytosolic Ca2+ signature after applying sodium stress, loss-of-function mutant lines displayed a sustained elevation of Ca2+ (Qudeimat et al., 2008). Interestingly, the sustained level of Ca2+ leads to a reduced upregulation of salt stress–induced genes and renders mutant plants specifically less tolerant to sodium stress (Qudeimat et al., 2008). These findings not only establish the importance of regulated Ca2+ extrusion for appropriate formation of Ca2+ signatures, they also implicate a direct interaction between the shape of a Ca2+ signature and proper stress responsiveness. In Arabidopsis, analysis of loss-of-function mutants of ACA9 and ACA10 indicated that the pumps specifically function in pollen tube growth and in inflorescence development of plants, respectively (Schiott et al., 2004; George et al., 2008)
Besides ECAs and ACAs, PI-type ATPases, which are known as heavy metal transporters, were recently implicated in Ca2+ transport. At HMA1, a heavy metal transporter involved in detoxification processes for heavy metals, is a PI-ATPase that localizes to the chloroplast envelope. At HMA1 transports Ca2+ and heavy metals, such as copper, with high affinity and, similar to Ca2+ pumps from animals, is specifically inhibited by thapsigargin (Seigneurin-Berny et al., 2006; Moreno et al., 2008).
All of these observations support the notion that finely regulated Ca2+ extrusion is as important as the influx of Ca2+, as the deregulated hyperextrusion of Ca2+ as well as downregulated extrusion result in deregulated Ca2+ dynamics and homeostasis. Certainly, a minimal concentration of the toxic cation Ca2+ is required to sustain correct signaling and metabolic functions, while decelerated extrusion of Ca2+ disturbs the formation of defined Ca2+ signatures and impairs the capabilities of the plants to correctly respond to a signal.
Calcium Signal Modulation by Organelles
Plastids can accumulate high levels of Ca2+ (Nobel et al., 1966) in the milimolar range and thereby can contribute to the homeostasis of cellular Ca2+ and other ions (Portis and Heldt, 1976). Ca2+ within plastids, especially the correct distribution between stroma and thylakoid lumen, is important for the regulation of plastidial enzymes (Brand and Becker, 1984; Kreimer et al., 1988). The level of Ca2+ within plastids is regulated and can raise upon illumination (Muto et al., 1982; Kreimer et al., 1988) or during transition to dark (Sai and Johnson, 2002).
A protein identified as the Ca2+ binding protein CAS may influence the loading capacity of chloroplasts or may be important for sensing the loading status of Ca2+ within the chloroplasts, thereby modulating cytoplasmic Ca2+ signaling (Figure 2) (Han et al., 2003; Nomura et al., 2008; Weinl et al., 2008). CAS was first reported as an extracellular Ca2+-sensing receptor, exhibiting a high capacity to bind Ca2+ (10 to 12 Ca2+ ions per molecule) (Han et al., 2003). However, the protein contains an N-terminal chloroplast targeting signal peptide, and subsequent reports identified CAS as a chloroplast-localized protein regulating cytoplasmic Ca2+ levels (Nomura et al., 2008; Vainonen et al., 2008; Weinl et al., 2008). Within the chloroplast, CAS is targeted to the thylakoid membrane (Vainonen et al., 2008; Weinl et al., 2008). CAS can be phosphorylated in a light-dependent manner depending on the activity of the light-regulated kinase STN8 (Vainonen et al., 2005, 2008). Although the activity of the photosystem is unaltered, cas knockout plants show retarded growth. Especially when grown under low Ca2+ conditions, plants show delayed bolting and impaired induction of flowering (Han et al., 2003). Mutant plants also show a strong deficit in regulating cytoplasmic Ca2+ levels (Vainonen et al., 2008) and are not able to induce stomatal closure provoked by extracellular Ca2+ (Han et al., 2003; Nomura et al., 2008; Weinl et al., 2008). However, cas knockout plants can respond to externally imposed Ca2+ oscillations and then display normal stomatal closure compared with the wild type, indicating a defect in the generation of Ca2+ transients that are required for stomatal closure, rather than the response machinery per se (Weinl et al., 2008). This indicates that the chloroplast-targeted Ca2+ sensor protein somehow connects cytoplasmic and chloroplast Ca2+ levels. This function resembles that of the Ca2+ buffer protein CRT within the ER lumen (Persson et al., 2001; Jia et al., 2009). Similarly, loss of CAS may lead to a reduced buffer capacity of the chloroplasts, suggesting that less Ca2+ can be allocated from the chloroplasts to the transient cytoplasmic increase of Ca2+. Therefore, it will be of interest to analyze the concentration and dynamics of Ca2+ in chloroplasts of cas mutant lines.
DECODING AND RELAY OF Ca2+ SIGNALS
Ca2+ signals evoked by a specific stimulus are presented as defined Ca2+ signatures on the cellular level (as for example in guard cells) as well as in distinct cell types and tissue regions (for example, in roots after mechanical stimulation). It is apparent that the ability of a given cell or tissue to translate these Ca2+ signals into defined molecular and biochemical responses primarily depends on the presence, concentration, cellular localization, and Ca2+ binding affinity of signaling components that can sense such Ca2+ signatures and convey specific output reactions for further information processing (McAinsh and Hetherington, 1998). In plants, a diverse and extensive set of Ca2+ binding proteins that function as cellular Ca2+ sensors represent these first information translation points (Luan et al., 2002; Batistič and Kudla, 2004; McCormack et al., 2005; Kim et al., 2009).
Plant Ca2+ sensor proteins have been classified conceptually into sensor relays and sensor responders (Sanders et al., 2002). Sensor responder proteins, for example, CDPKs, combine within one protein a sensing function (Ca2+ binding and Ca2+-induced conformational changes) with a response activity (e.g., protein kinase activity). By contrast, sensor relay proteins, like calmodulin, also effectively bind Ca2+ ions and usually undergo Ca2+-induced conformational changes but lack other effector domains. To transmit the Ca2+ signal, sensor relay proteins interact with target proteins and regulate their activity (Luan et al., 2002; Sanders et al., 2002).
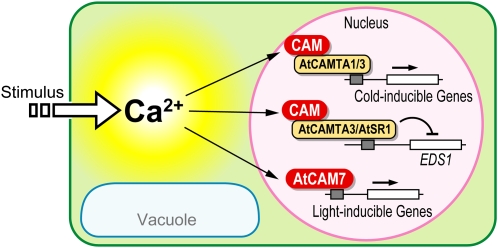
Initially, this concept of functional classification for Ca2+ sensing proteins was applied to the conversion of Ca2+ signals into phosphorylation responses (Sanders et al., 2002). However, considering new insights into the function of Ca2+-sensing proteins, we propose extending this concept to the conversion of Ca2+ signals into transcriptional responses. Whereas Ca2+ binding to calmodulin 7 (CAM7) appears to result in direct promoter interaction and regulation, other calmodulins are likely to mediate gene regulation by interacting with calmodulin binding transcription factors (CAMTAs) that function as transcriptional coregulators (see below). Consequently, CAM7 would represent a bona fide sensor responder directly contributing to activating gene expression, while other calmodulins function as sensor relays modulating gene expression via their interaction with proteins that function as transcription factors (Kushwaha et al., 2008).
In addition to calmodulins, CMLs, which are represented by >50 diverse calcium sensor proteins in Arabidopsis, appear to fulfill important functions in plant development and responses to environmental cues (McCormack et al., 2005). Loss of CML42 function leads to aberrant trichomes with increased branching, while mutation of CML24 causes alterations in flowering time (Delk et al., 2005; Tsai et al., 2007; Dobney et al., 2009). Moreover, CML24 functions in responses to ABA and ionic stress, and mutation of CML9 alters plant responses to ABA and abiotic stresses (Delk et al., 2005; Magnan et al., 2008). Exploring the function of the other members of this diverse protein family and especially identifying targets of these proteins therefore represents a promising area of plant signaling research.
Many biological processes, such as metabolic starch degradation by α-amylase (Bush et al., 1989b); biosynthetic processes, such as brassinosteroid synthesis (Du and Poovaiah, 2005); or even the mechanical occlusion of sieve tube elements (Furch et al., 2009) are likely important targets of direct Ca2+-dependent modulation. However, with respect to the signaling function of Ca2+ ions, Ca2+-dependent phosphorylation events and Ca2+-dependent gene regulation represent the major cellular currencies for converting defined Ca2+ signatures into specific downstream reactions. Recent advances in these two facets of Ca2+ signaling will be discussed in detail here.
Translating Ca2+ Signatures into Protein Phosphorylation
Signaling requires messengers whose concentration varies in time and space. Ca2+ ions and phosphate ions have come to dominate cellular signaling. Ca2+ binding triggers changes in protein shape and charge. Similarly, phosphorylation imparts a negative charge, modulating protein conformations and protein interactions (Hunter, 1995; Soderling, 1999; Clapham, 2007). Ca2+-dependent kinases and protein kinases regulated by interaction with Ca2+ binding proteins functionally combine these two major cellular currencies of signal transduction and allow for the perception and transmission of Ca2+ signatures directly into phosphorylation events that orchestrate downstream signaling responses (Harmon et al., 2000; Batistič and Kudla, 2004). Plants possess an extensive repertoire of calcium-regulated protein kinases, which are represented by the three families of CCaMKs, CDPKs, and CBL-interacting protein kinases (CIPKs) (Sanders et al., 2002; Batistič and Kudla, 2004; Gleason et al., 2006). While CDPKs and CCaMKs (the later appear not to exist in the Arabidopsis genome) represent typical sensor responders, the CIPKs are targets of Calcineurin B-like (CBL) sensor relay proteins. These kinases and Ca2+ binding proteins form an intricate cellular network for decoding Ca2+ signals in diverse cellular processes (Batistič and Kudla, 2009; Weinl and Kudla, 2009).
The CDPK Signaling System
The Arabidopsis genome encodes 34 CDPKs and eight additional CDPK-related kinases (Hrabak et al., 2003). The biochemistry and regulation of CDPKs have been comprehensively reviewed (Harper et al., 2004; Ludwig et al., 2004; Harper and Harmon, 2005). Binding of Ca2+ to the C-terminal EF hand–containing regulatory domain leads to conformational changes relieving the active site of the kinase domain from masking by an autoinhibitory domain, resulting in activation of the respective CDPK. This process is enhanced by autophosphorylation of the CDPKs that contribute to full activation of the kinases (Ludwig et al., 2004).
The first conclusive evidence that linked the activation of CDPKs with the induction of signaling responses in vivo was performed on the Cf9-avr9 disease response of tomato (Solanum lycopersicum) and was obtained by suppression of Nt CDPK2 by viral-induced gene silencing in Nicotiana benthamiana (Romeis et al., 2001). CDPK-silenced plants displayed a reduced and delayed hypersensitive response after race-specific Avr9 elicitation in a gene-for-gene interaction and lacked an accompanying wilting phenotype. Subsequent further analysis of Nt CDPK2 function by T. Romeis and coworkers additionally indicated that elevated CDPK signaling compromises stress-induced MAPK activation and that this inhibition requires ethylene synthesis and perception (Ludwig et al., 2005). These important findings suggest that CDPK and MAPK pathways do not function independently and that a concerted regulation of both pathways controls response specificity to biotic and abiotic stress. Clearly, such aspects of interconnection and interplay between different plant signaling systems will need to be investigated in more depth in the future.
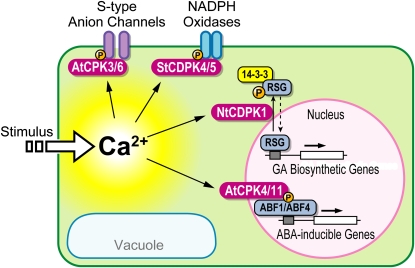
The application of reverse genetics techniques has recently extended our knowledge of the physiological function of several members of the CDPK protein family (see Figure 3 for an illustrated summary). Arabidopsis CPK3 and CPK6 function synergistically in the regulation of stomatal closure in response to ABA and external Ca2+ elevation (Mori et al., 2006). Moreover, in independent alleles of single and double mutants of cpk3 and cpk6, ABA and Ca2+ activation of slow-type anion channels and ABA activation of plasma membrane Ca2+-permeable channels were impaired in guard cells (Mori et al., 2006). In addition to CPK3 and CPK6, Arabidopsis CPK4 and CPK11 are critical for proper ABA responsiveness of guard cells and have been shown to phosphorylate the ABA-responsive transcription factors ABF1 and ABF4 in vitro (Zhu et al., 2007). Research in tobacco revealed that CDPK1 regulates the transcription factor RSG in response to gibberellins (Ishida et al., 2008). Phosphorylation of Ser-114 in RSG by CDPK1 promotes interaction with 14-3-3 proteins that appears to regulate the intracellular localization of this transcription factor. Work in potato suggests that two CDPKs (St CDPK4 and St CDPK5) phosphorylate NADPH oxidases and thereby positively regulate the production of ROS (Kobayashi et al., 2007). Accordingly, ectopic expression of a constitutively active mutant of St CDPK5 provoked ROS production in N. benthamiana leaves, and this CDPK-mediated ROS production was disrupted by knockdown of St CDPK5. Together, these findings point to a critical role of CDPK-mediated Ca2+ signaling in a diverse set of biological processes. The identification of additional CDPK target proteins and CDPK-regulated process should in the near future allow us to address more precisely how specific CDPKs mechanistically contribute to a specific decoding of distinct Ca2+ signatures.
Figure 3.
The CDPK Signaling System for Translating Ca2+ Signatures into Protein Phosphorylation.
Shown are characterized components of the CDPK system. See text for further details. RSG, repression of shoot growth; ABF, ABA response element-binding factor.
The CBL/CIPK Signaling System
CBL proteins and their interacting protein kinases (CIPKs) were originally identified in Arabidopsis (Kudla et al., 1999; Shi et al., 1999). Subsequent comprehensive bioinformatics analyses have identified a complement of 10 CBLs and 26 CIPKs in Arabidopsis and 10 CBLs and 30 CIPKs in the genome of rice, respectively (Kolukisaoglu et al., 2004; Weinl and Kudla, 2009). In contrast with the CDPK sensor responders, the families of CBL proteins and their interacting CIPKs separate Ca2+ binding function (sensor relay function) and kinase activity (response activity) into two flexible combinable modules. In addition, preferential complex formation of individual CBLs with defined subsets of CIPKs appears to be one of the mechanisms generating the temporal and spatial specificity of Ca2+ signals in plant cells (Albrecht et al., 2001). Together, these features allow for the formation of a complex and dynamic Ca2+-decoding signaling network. Since its initial discovery, this signaling system has been subject to intensive research. Advances in our understanding of the physiological function and structural features of these proteins and functional principles of this Ca2+-decoding system have been discussed in several reviews (Luan et al., 2002; Batistič and Kudla, 2004, 2009; Luan, 2009; Luan et al., 2009; Weinl and Kudla, 2009).
CBL proteins exhibit significant similarity to the regulatory B subunit of calcineurin and Neuronal Ca2+ Sensor proteins from animals and yeast (Kudla et al., 1999). All CBL proteins share a rather conserved core region consisting of four EF hand Ca2+ binding sites that are arranged in completely invariant spacing within the protein (Kolukisaoglu et al., 2004). All CIPK-type kinases are composed of a conserved N-terminal kinase domain and a C-terminal regulatory domain, which are separated by a variable junction domain. Within the rather divergent regulatory domain, the conserved NAF domain has been identified as required and sufficient for mediating CBL interaction (Albrecht et al., 2001). It is assumed that binding of CBL proteins to the NAF domain of CIPKs releases the C-terminal (autoinhibitory) domain from the kinase domain, thereby transforming the kinase into an active state (Guo et al., 2001; Gong et al., 2002). Additional phosphorylation of the activation loop within the kinase domain by an unidentified kinase further contributes to the activation of CIPKs (Gong et al., 2002). An interesting new facet of CIPK–CBL interaction is provided by recent reports of phosphorylation of CBL proteins by their interacting CIPKs that appears to enhance the CBL–CIPK interaction (Mahajan et al., 2006; Lin et al., 2009). Lin et al. (2009) reported that the kinase CIPK24/SOS2 specifically phosphorylates CBL10 (which these authors renamed SCaBP8), but not any other investigated CBL protein, at position Ser-237, a finding that is rather surprising considering that this amino acid position and the surrounding sequence motif is conserved in eight out of the 10 CBL proteins in Arabidopsis (Lin et al., 2009). In addition, a protein-phosphatase interaction (PPI) domain mediating CIPK interaction with phosphatases of the PP2C group has been identified within the C terminus of these kinases (Ohta et al., 2003). Unfortunately, the functional implications of this interaction are currently unknown. It appears conceivable that CIPKs may phosphorylate PP2Cs or that PP2Cs dephosphorylate CIPKs in vivo, as it has been shown for sucrose nonfermenting-related kinases (SnRKs) of the SnRK2 family that are dephosphorylated by a subgroup of PP2Cs (Umezawa et al., 2009). Alternatively, CIPK/PP2C complexes could serve as signaling kinase/phosphatase modules allowing for rapid alternating phosphorylation/dephosphorylation of target proteins. However, crystallization studies of CBL4 (SOS3) in complex with the regulatory domain of CIPK24 (SOS2) suggest that either CBLs or PP2Cs may interact in a mutually exclusive manner with the regulatory domain of CIPKs, thereby preventing the formation of trimeric complexes (Sanchez-Barrena et al., 2007). Consequently, PP2C interaction with the PPI domain of CIPKs would lead to replacement of the CBL protein, which binds to the NAF and partly to the PPI domain, and would allow for competitive formation of either CBL/CIPK or CIPK/PP2C complexes. Considering the recent identification of PP2C phosphatases as direct targets of the PYR/RCAR ABA receptors (Ma et al., 2009; Park et al., 2009), it is tempting to speculate that PP2C-bound CIPKs contribute to early signaling steps after ABA perception, an assumption that would be in agreement with the observation of ABA response phenotypes in many analyzed CIPK mutants (see below).
Evolution and Functional Diversification of the CBL/CIPK Signaling System
The increasing number of available full-genome sequences has facilitated the study of the evolution of the CBL/CIPK signaling system. Single CBL and CIPK genes have been identified in green alga species, such as Ostreococcus tauri and Chlorella sp, whereas the moss P. patens contains four CBLs and seven CIPKs, and the genome of the fern Selaginella moellendorfii possesses a complement of five CBL and five CIPK genes (Batistič and Kudla, 2009; Weinl and Kudla, 2009; Batistič et al., 2010). These observations suggest that the complexity of the CBL/CIPK system evolved concurrently with the increasing morphological and developmental sophistication of land plants that enabled the colonization of ecologically diverse and environmentally fluctuating habitats. Surprisingly, CBLs and CIPKs also were recently identified in protozoan eukaryotic species, such asTrichomonas vaginalis and Naegleria gruberi, raising the question about the function of this Ca2+ decoding components in nonplant species (Batistič and Kudla, 2009). Remarkably, the occurrence and function of plant-specific Ca2+ signaling components in human pathogens appears not to be restricted to CBLs and CIPKs, as CDPKs have been identified in Plasmodium falciparum and Plasmodium berghei, where Pb CDPK4 fulfills a critical function during the life cycle of this human pathogen (Billker et al., 2004; Harper and Harmon, 2005). Considering the absence of related proteins in their human host, further advancements of our understanding of the regulation of plant Ca2+ sensor proteins may facilitate the identification of therapeutic inhibitors specifically affecting the plant-like proteins of these protozoan species.
Local Ca2+ signals at specific microdomains are assumed to be the basis for differential Ca2+ signaling (Berridge, 2006). Consequently, Ca2+-decoding systems would be expected to reflect this spatial specification of Ca2+ signaling. The CBL/CIPK network appears to meet this demand. Localization studies of all 10 Arabidopsis CBL proteins revealed the importance of their variable N-terminal extensions for specific subcellular targeting (Batistič et al., 2010). Four CBL proteins are localized to the plasma membrane, while another four CBLs are localized to vacuolar membrane and two CBLs are detected in the cytoplasm and nucleus (Batistič et al., 2010). These distinct subcellular localizations suggest that CBL Ca2+ sensors might function as fast relays of local Ca2+ release events from internal and external stores and that the spatial separation of distinct CBL/CIPK complexes contributes to spatial specificity in Ca2+ signaling. Dual lipid modification by myristoylation and S-acylation are required for CBL1 function and for localization of this Ca2+ sensor at the plasma membrane. This localization is achieved by a two-step targeting process in which initial myristoylation results in localization at the ER, and subsequent S-acylation is crucial for ER–to–plasma membrane trafficking (Batistič et al., 2008). As CBL4, CBL5, and CBL9 have been shown to undergo myristoylation and share the adjacent palmitoylation motif, a similar targeting mechanism can be predicted for these proteins (Batistič et al., 2008).
In contrast with CBL proteins, most CIPKs when expressed as green fluorescent protein fusions exhibit a cytoplasmic and nucleoplasmic localization (D'Angelo et al., 2006; Batistič et al., 2010). However, CBL–CIPK interaction analyses using bimolecular fluorescence complementation revealed that the subcellular localization of CIPKs incorporated into distinct CBL/CIPK complexes is determined by the identity of their CBL moiety (D'Angelo et al., 2006; Cheong et al., 2007; Batistič et al., 2008, 2010; Waadt et al., 2008). For example, CIPK1 is targeted to the plasma membrane by CBL1 or CBL9 (Cheong et al., 2007; Waadt et al., 2008). However, upon interaction with CBL2, the resulting CBL2/CIPK1 complexes are localized exclusively to the tonoplast (Batistič et al., 2008). Similarly, CIPK14/CBL2 complexes have been detected at the tonoplast, while the same kinase is targeted to the plasma membrane upon interaction with CBL8 (Batistič et al., 2010). Remarkably, the cellular targeting of plasma membrane– or tonoplast-localized CBL/CIPK complexes appears not to involve the conventional protein trafficking pathway because inhibition experiments using a dominant-negative form of the SAR1 protein (that interferes with COPII vesicle formation) or brefeldin A (that impedes COPI vesicle formation) did not affect the cellular targeting of singular CBL proteins or CBL/CIPK complexes (Batistič et al., 2008, 2010).
Physiological Functions of CBLs and CIPKs
Forward genetic screens aiming to identify critical components of plant salt tolerance have provided insights into the physiological function of CBLs and CIPKs. The CBL Ca2+ sensor SOS3 (At CBL4) and the CIPK-type kinase SOS2 (At CIPK24) appear to be part of a Ca2+-regulated signaling pathway that specifically mediates salt stress adaptation by regulating the Na+/H+ antiporter SOS1 (Liu and Zhu, 1998; Halfter et al., 2000; Qiu et al., 2002). Recent studies revealed that mutation of CBL10 also renders plants salt sensitive and that CBL10 is also able to interact with the kinase CIPK24 (Kim et al., 2007; Quan et al., 2007). In vivo analyses revealed a tonoplast localization of the CBL10/CIPK24 complex, thereby supporting a functional model wherein alternative complex formation of CIPK24 kinases with either CBL4 or CBL10 creates a dual functioning kinase. While CBL4/CIPK24 complexes mediate Na+ extrusion via the regulation of the H+/Na+ antiporter SOS1 at the plasma membrane, formation of CBL10/CIPK24 likely results in Na+ sequestration into the vacuole by regulating unknown targets (Kim et al., 2007; Weinl and Kudla, 2009). In addition, recent studies have suggested that CBL/CIPK24 complexes at the tonoplast membrane activate and regulate the vacuolar Ca2+/H+ antiporter CAX1 independently from CBL4 (Cheng et al., 2004b) and have identified subunits of the V-ATPase complex as interacting proteins of CIPK24 (Batelli et al., 2007).
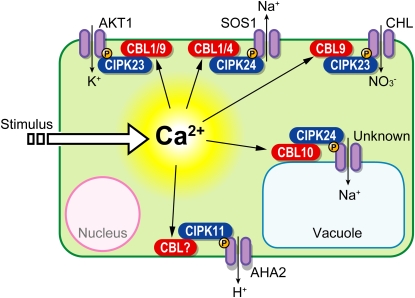
Reverse genetics analyses have greatly advanced our understanding of CBLs and CIPKs and have uncovered crucial functions of these proteins for plant mineral nutrition and for responses to abiotic stresses and hormones, such as ABA (see Figure 4 for an illustrated summary). Characterization of cbl1 loss-of-function mutant lines revealed a function of CBL1 as a central integrator of responses to abiotic stresses, including drought, cold, and salt (Albrecht et al., 2003; Cheong et al., 2003). While the mutant studies of CBL1 revealed an ABA-independent function of this protein in several abiotic stress responses, loss of function of the closely related Ca2+ sensor CBL9 renders plants hypersensitive to ABA (Pandey et al., 2004). Alternative complex formation of the kinase CIPK1 with either CBL1 or CBL9 mediates ABA-dependent and ABA-independent signaling responses (D'Angelo et al., 2006). In addition, CBL9 appears to form a complex with CIPK3 for modulating ABA responses (Pandey et al., 2008).
Figure 4.
The CBL/CIPK Signaling System for Translating Ca2+ Signatures into Protein Phosphorylation.
Shown are characterized complexes consisting of CBL proteins and their interacting CIPKs. AKT1, Arabidopsis K+ transporter 1; SOS1, salt overly sensitive 1; CHL, a nitrate transporter; AHA2, Arabidopsis H+ ATPase 2.
A breakthrough study in 2006 by Wei-Hua Wu and colleagues uncovered a role of the CBL/CIPK system in regulating K+ homeostasis and provided the first molecular insights into how plant ion channels might be regulated by phosphorylation (Xu et al., 2006). CIPK23 is targeted to the plasma membrane and activated by the two highly related Ca2+ sensors CBL1 and CBL9 (Xu et al., 2006; Cheong et al., 2007). These complexes then regulate the activity of the shaker-like potassium channel AKT1 (Li et al., 2006b; Xu et al., 2006). CIPK23 interacts exclusively with AKT1 but not with other K+ transporters from Arabidopsis (Hedrich and Kudla, 2006; Lee et al., 2007a; Geiger et al., 2009). Interaction analysis in yeast in combination with electrophysiological studies indentified the 2C-type protein phosphatase AIP1 as a negative regulator of AKT1 that counteracts activation by CIPK23 (Lee et al., 2007a). Considering that AIP1 can interact with both CIPK23 and AKT1, it will be most interesting to distinguish if this activation/deactivation switch is brought about by phosphorylation/dephosphorylation or, alternatively, results from competitive binding of either the phosphatase or the kinase to the potassium channel. Besides the regulation of K+ uptake in roots, the Ca2+-decoding CBL1/CBL9/CIPK23 module appears to be involved in stomata regulation under dehydrating conditions (Cheong et al., 2007).
An exciting novel twist in our understanding of CIPK23 function results from the recent report that this kinase also phosphorylates the nitrate transporter CHL1 (also called NRT1.1). Taking advantage of a novel CHL1 mutant allele (chl1-9), Yi-Fang Tsay and colleagues provided compelling evidence that this mutation impairs the nitrate uptake function of CHL1 without affecting the signaling response to nitrate as analyzed by the transcriptional response of the NRT2.1 gene (Ho et al., 2009). Importantly, biochemical and reverse genetics analyses identified CIPK23 as a critical regulator mediating the switch between low- and high-affinity nitrate transport modes by phosphorylating residue Thr-101 of CHL1. Specifically, at low external nitrate concentrations, CIPK23-mediated phosphorylation results in low-level nitrate signaling (Ho et al., 2009). Moreover, Ho et al. reported that CIPK23 is independently involved in potassium and nitrate responses, indicating a lack of crosstalk between both ions. Considering the critical involvement of the same CBL1 (and CBL9) Ca2+ sensor proteins and CIPK23-dependent phosphorylation in both processes, this puzzling observation underscores the urgent need for further investigation of the primary ion-sensing mechanism(s) that confer this remarkable specificity in plant signaling.
The plasma membrane H+-ATPase AHA2 has been identified as an additional target of CBL/CIPK signaling complexes (Fuglsang et al., 2007). Phosphorylation of a Ser residue within the C-terminal domain of AHA2 by CIPK11 (designated as PKS5 by these authors) prevents binding of 14-3-3 proteins to this domain and leads to downregulation of AHA2 proton transport activity. Moreover, yeast two-hybrid interaction studies suggested a potential function of CBL2 in the CIPK11-dependent regulation of AHA2 (Fuglsang et al., 2007). However, since CBL2 has so far only been detected at the tonoplast in plants cells (Batistič et al., 2008, 2010), unambiguously determining the identity of the CBL protein mediating Ca2+-dependent regulation of AHA2 in plants may require additional in planta or reverse genetics analyses.
Finally, recent work performed in rice further extended the identified physiological functions of CIPKs (Lee et al., 2009). The results from this study indicate that protein kinase Os CIPK15 plays a key role in O2 deficiency tolerance in rice and is required for growth of rice under flooded conditions. Moreover, CIPK15 regulates the plant global energy and stress sensor SnRK1A, thereby integrating responses to O2 deficiency with sugar signaling and enabling rice growth under floodwater. The latter finding may point to a general role of the CBL/CIPK system in fine-tuning plant metabolism in response to adverse environmental conditions.
The results of all these studies suggest that the CBL/CIPK network represents a central and critical signaling system for decoding Ca2+ signals in response to a wide range of stimuli. So far, research on CBLs and CIPKs has been focused mainly on abiotic stress responses and ion uptake. However, it is safe to predict that future studies will most likely uncover a role of this Ca2+ decoding system in additional processes, such as plant pathogen interactions. It is now well established that each CBL and each CIPK represents a multifunctional signaling component that can undergo alternative protein interactions determining the flow of information processing through this signaling system. Therefore, furthering our understanding of this signaling system will require the combined application of state-of-the-art cell biological techniques that enable the monitoring of dynamic protein–protein interactions and protein targeting. Such approaches should also allow the elucidation of mechanistic factors that determine the decision making in this flexible interaction network and will be of primary importance to advance our understanding of Ca2+ decoding mechanisms.
Converting Ca2+ Signals into Transcriptional Responses
Transcriptional regulation is coupled with numerous signaling processes that can be conveyed by several second messengers and the function of Ca2+ binding proteins (Ikura et al., 2002). Despite the obvious importance of defined Ca2+ signatures in eliciting specific transcriptional response reactions in plants to stimuli, such as cold, drought, and salt stress, the molecular mechanisms mediating Ca2+-responsive gene expression have long remained little understood (Scrase-Field and Knight, 2003). This is due in part to the difficulty in discriminating between stress-dependent Ca2+ responses per se and stress-dependent but Ca2+-independent transcriptional reactions (Finkler et al., 2007b). Recently, the induction of defined artificial Ca2+ transients in response to calmodulin antagonists like WP7 and SKF-7171 has allowed the identification of 230 Ca2+-responsive genes that were differentially expressed 1 h after stimulus in Arabidopsis (Kaplan et al., 2006). Since the microarrays used in this study covered only 25% of the known Arabidopsis genes, this finding suggests that ∼3% of the protein-coding genes in the Arabidopsis genome are subject to regulation by Ca2+. Remarkably, many of the genes identified in this study as being Ca2+ regulated were previously characterized as early stress-induced genes.
Within the promoter regions of Ca2+-regulated genes, this study identified abscisic acid–responsive element (ABRE)-related cis-elements as being sufficient to confer transcriptional regulation in response to cytosolic Ca2+ signatures. This finding is significant because such ABREs have been identified in the promoter of CBF/DREB1 transcription factors that function as master regulators of abiotic stress responses (Finkler et al., 2007b). Therefore, these results point to a direct interconnection of Ca2+-regulated gene transcription and abiotic stress responses. Such an immediate conversion of Ca2+ signatures into transcriptional regulation may be in part achieved by the function of calmodulin binding transcription factors (CAMTA proteins).
Ca2+-Regulated Transcription Factors
CAMTAs are Ca2+-dependent CaM binding transcription factors forming a family of six members in Arabidopsis (Finkler et al., 2007a). They share a conserved domain structure including an N-terminal CG-1 domain mediating binding to DNA cis-elements (CAMTA binding sites), including ABREs (Finkler et al., 2007a). The C-terminal CaM binding domain of CAMTAs mediates interactions with calmodulin. First insights into the physiological function of plant CAMTA proteins came from a reverse genetic analysis of Arabidopsis CAMTA3 function that revealed a critical role of this protein in suppressing plant responses to pathogens such as P. syringae and Botrytis cinerea (Galon et al., 2008). Most recent work on CAMTA3 (designated At SR1) uncovered that CAMTA3 directly interacts with the promoter of the EDS1 gene, an established regulator of salicylic acid levels, and represses its expression (Figure 5) (Du et al., 2009). Moreover, Ca2+/calmodulin binding to CAMTA3 was required for the suppression of plant defense, indicating a direct role of Ca2+/calmodulin in regulating the function of CAMTA3 (Du et al., 2009). Important evidence for a direct link between Ca2+ signaling via CAMTA1 and CAMTA3 and the regulation of cold tolerance in plants was provided by the discovery that these CAMTA proteins bind to regulatory elements in the promoter region of the DREB1c/CBF2 gene (Doherty et al., 2009). A single camta3 mutant exhibited a significant reduction in cold induction of a number of known cold-induced genes, including CBF2. Moreover, camta1 camta3 double mutant plants were impaired in their cold acclimation to freezing tolerance (Doherty et al., 2009). These findings establish a crucial role of Ca2+/calmodulin-regulated CAMTA transcription factors in controlling CBF-regulated cold-responsive gene expression in plants (Figure 5). Direct decoding of cold-induced Ca2+ signatures into the regulation of gene expression may occur through direct and Ca2+-dependent interaction of CAMTAs with members of the Arabidopsis calmodulin Ca2+ sensors family (McCormack et al., 2005). The function of calmodulin in the regulation of gene expression by direct interaction with transcription factors appears not to be restricted to their interaction with CAMTAs because interactions of calmodulin with MYB and WRKY transcription factors has also been reported (Park et al., 2005; Yoo et al., 2005). In this regard, it will be important to investigate exactly how the stimulus-induced changes in Ca2+ concentration that occur in the cytoplasm are connected into transcriptional regulation that proceeds in the nucleus.
Figure 5.
Converting Ca2+ Signals into Transcriptional Responses.
Recent studies have unveiled that Ca2+/CAMs (calmodulins) regulate CAMTAs that interact with promoters of abiotic stress-responsive genes. Interestingly, Arabidopsis CAM7 is likely a direct converter of cytoplasmic Ca2+ signatures into regulation of gene expression. EDS1, enhanced disease susceptibility 1.
Transcriptional Regulation by Calmodulin
Calmodulins are conserved eukaryotic Ca2+ sensor relay proteins that upon Ca2+ binding undergo extensive conformational changes that result in protein interactions with their target proteins (Luan et al., 2002; McCormack et al., 2005). In Arabidopsis, seven genes encode four CAM isoforms, of which CAM1/CAM4 differ by four amino acid substitutions from CAM7, whereas CAM2/3/5 and CAM6 differ in one amino acid position from CAM7 (McCormack et al., 2005). Classical microinjection experiments into hypocotyl cells of the phytochrome deficient tomato aurea mutant suggested a function of Ca2+ and calmodulin in light signal transduction that controls regulation of light-responsive gene expression (Neuhaus et al., 1993; Bowler et al., 1994). A recent study of CAM7 from Arabidopsis revealed surprising insights into the regulation of light-induced gene expression by calmodulin (Kushwaha et al., 2008). The authors identified specifically CAM7 but not CAM2/3/5 as a transcriptional regulator that directly interacts with promoters of several light-inducible genes (Figure 5). In accordance with such a function of CAM7, cam7 mutants exhibited a reduced expression of light-inducible genes. Conversely, overexpression of CAM7 resulted in an increase in the expression of light-inducible genes. The findings of this study suggest a direct translation of cytoplasmic Ca2+ signatures by the Ca2+ sensor responder protein CAM7 into regulation of gene expression by DNA binding. To further elucidate details of this regulatory circuit, it will be most interesting to investigate if this function requires the translocation of CAM7 from the cytoplasm to the nucleus and/or the interaction of CAM7 with additional (transcription factor) proteins.
Conclusions and Future Perspectives
During the past years we have witnessed tremendous progress in our understanding of Ca2+ signaling processes in plants. When comparing research on Ca2+ signaling in animals and plants during the past decade, remarkably different developments become apparent. Whereas in animals the components and mechanisms that exercise primary Ca2+-releasing events and shape Ca2+ signatures have seen the most significant advances, in plant research, most progress has been achieved in identifying and characterizing components that decode Ca2+ signals.
A central question around which Ca2+ signaling research in plants is centered is: How can one ion specifically control so many different processes and events? Current research is beginning to provide answers. Ca2+ signaling in plants involves many facets that can define and adjust responses in both time and space. The unequal distribution of this ion in the cell provides the basis for rapid Ca2+ fluxes and the resulting concentration changes. Many energized cation transporters and channels that were previously assumed to merely be regulators of Ca2+ homeostasis actually appear to play a role in Ca2+ signaling. However, a major bottleneck in our understanding of the generation of Ca2+ signals is the paucity of molecular information about true Ca2+ channels. It may require novel genetic screens or the application of chemical genetic screens (that have been successful, for example, in identifying plant ABA receptors) to successfully tackle this serious limitation of plant Ca2+ signaling research.
It is now widely accepted that complex families of Ca2+ binding proteins that function as calcium sensors provide the toolkit for deciphering various Ca2+ signatures. Calmodulins, CMLs, CDPKs, and CBL/CIPK complexes form intricate signaling networks for translating these signatures into downstream phosphorylation events and transcriptional responses. An important role of Ca2+-regulated transcription factors like CAMTAs in these processes is emerging. It is very likely the network-like character of these signaling systems that warrants flexible but robust information processing. Accumulating evidence also points to two levels at which Ca2+ signaling is operative. Within specific cells, such as guard cell or root hairs, intracellular Ca2+ signaling processes occur at high spatial and temporal specificity. This layer of Ca2+ signaling appears to be superimposed on the tissue or organismic level where specific tissue layers or cell types exhibit defined and prominent changes in Ca2+ concentration.
It has become evident that parameters such as the specific Ca2+ binding affinity, the specific cellular concentration and subcellular localization, and the specific interaction affinities of Ca2+ decoders are critical components that contribute to generating specificity in signal-to-response coupling. However, a major challenge that remains is to understand mechanistically how exactly calcium signatures or variable oscillations are sensed and transduced by calcium sensor proteins. Tackling this important task will require serious investments in structural analyses of Ca2+ sensors, NMR-based studies of their structural dynamics, extensive investigation of Ca2+ binding and Ca2+-dissociation parameters of Ca2+ sensors, and novel cell biological approaches to visualize the dynamics of protein interactions and protein complex formation in planta. The integration of these data with advanced fluorescence resonance energy transfer-based monitoring of cellular Ca2+ dynamics and the mathematical modeling of such processes represent the route to be pursued in plant Ca2+ signaling research in the near future. Currently, despite all the exciting progress described in this review article, we are still not close to finally cracking the calcium code in plants.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft, the Alexander von Humboldt Foundation, the Deutscher Akademischer Austauschdienst, and the Human Frontier of Science Program.
References
- Albrecht V., Ritz O., Linder S., Harter K., Kudla J. (2001). The NAF domain defines a novel protein-protein interaction module conserved in Ca2+-regulated kinases. EMBO J. 20: 1051–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht V., Weinl S., Blazevic D., D'Angelo C., Batistič O., Kolukisaoglu U., Bock R., Schulz B., Harter K., Kudla J. (2003). The calcium sensor CBL1 integrates plant responses to abiotic stresses. Plant J. 36: 457–470 [DOI] [PubMed] [Google Scholar]
- Ali R., Ma W., Lemtiri-Chlieh F., Tsaltas D., Leng Q., von Bodman S., Berkowitz G.A. (2007). Death don't have no mercy and neither does calcium: Arabidopsis CYCLIC NUCLEOTIDE GATED CHANNEL2 and innate immunity. Plant Cell 19: 1081–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali R., Zielinski R.E., Berkowitz G.A. (2006). Expression of plant cyclic nucleotide-gated cation channels in yeast. J. Exp. Bot. 57: 125–138 [DOI] [PubMed] [Google Scholar]
- Allen G.J., Chu S.P., Harrington C.L., Schumacher K., Hoffmann T., Tang Y.Y., Grill E., Schroeder J.I. (2001). A defined range of guard cell calcium oscillation parameters encodes stomatal movements. Nature 411: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Allen G.J., Muir S.R., Sanders D. (1995). Release of Ca2+ from individual plant vacuoles by both InsP3 and cyclic ADP-ribose. Science 268: 735–737 [DOI] [PubMed] [Google Scholar]
- Allen G.J., Sanders D. (1994). Two voltage-gated, calcium release channels coreside in the vacuolar membrane of broad bean guard cells. Plant Cell 6: 685–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen G.J., Sanders D. (1995). Calcineurin, a Type 2B protein phosphatase, modulates the Ca2+-permeable slow vacuolar ion channel of stomatal guard cells. Plant Cell 7: 1473–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balague C., Lin B., Alcon C., Flottes G., Malmstrom S., Kohler C., Neuhaus G., Pelletier G., Gaymard F., Roby D. (2003). HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide-gated channel ion channel family. Plant Cell 15: 365–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batelli G., Verslues P.E., Agius F., Qiu Q., Fujii H., Pan S., Schumaker K.S., Grillo S., Zhu J.K. (2007). SOS2 promotes salt tolerance in part by interacting with the vacuolar H+-ATPase and upregulating its transport activity. Mol. Cell. Biol. 27: 7781–7790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistič O., Kudla J. (2004). Integration and channeling of calcium signaling through the CBL calcium sensor/CIPK protein kinase network. Planta 219: 915–924 [DOI] [PubMed] [Google Scholar]
- Batistič O., Kudla J. (2009). Plant calcineurin B-like proteins and their interacting protein kinases. Biochim. Biophys. Acta 1793: 985–992 [DOI] [PubMed] [Google Scholar]
- Batistič O., Sorek N., Schultke S., Yalovsky S., Kudla J. (2008). Dual fatty acyl modification determines the localization and plasma membrane targeting of CBL/CIPK Ca2+ signaling complexes in Arabidopsis. Plant Cell 20: 1346–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistič O., Waadt R., Steinhorst L., Held K., Kudla J. (2010). CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J. 61: 211–222 [DOI] [PubMed] [Google Scholar]
- Berridge M.J. (2006). Calcium microdomains: Organization and function. Cell Calcium 40: 405–412 [DOI] [PubMed] [Google Scholar]
- Berridge M.J. (2009). Inositol trisphosphate and calcium signalling mechanisms. Biochim. Biophys. Acta 1793: 933–940 [DOI] [PubMed] [Google Scholar]
- Beyhl D., Hortensteiner S., Martinoia E., Farmer E.E., Fromm J., Marten I., Hedrich R. (2009). The fou2 mutation in the major vacuolar cation channel TPC1 confers tolerance to inhibitory luminal calcium. Plant J. 58: 715–723 [DOI] [PubMed] [Google Scholar]
- Bihler H., Eing C., Hebeisen S., Roller A., Czempinski K., Bertl A. (2005). TPK1 is a vacuolar ion channel different from the slow-vacuolar cation channel. Plant Physiol. 139: 417–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billker O., Dechamps S., Tewari R., Wenig G., Franke-Fayard B., Brinkmann V. (2004). Calcium and a calcium-dependent protein kinase regulate gamete formation and mosquito transmission in a malaria parasite. Cell 117: 503–514 [DOI] [PubMed] [Google Scholar]
- Blatt M.R., Thiel G., Trentham D.R. (1990). Reversible inactivation of K+ channels of Vicia stomatal guard cells following the photolysis of caged inositol 1,4,5-trisphosphate. Nature 346: 766–769 [DOI] [PubMed] [Google Scholar]
- Bonaventure G., Gfeller A., Proebsting W.M., Hortensteiner S., Chetelat A., Martinoia E., Farmer E.E. (2007a). A gain-of-function allele of TPC1 activates oxylipin biogenesis after leaf wounding in Arabidopsis. Plant J. 49: 889–898 [DOI] [PubMed] [Google Scholar]
- Bonaventure G., Gfeller A., Rodriguez V.M., Armand F., Farmer E.E. (2007b). The fou2 gain-of-function allele and the wild-type allele of Two Pore Channel 1 contribute to different extents or by different mechanisms to defense gene expression in Arabidopsis. Plant Cell Physiol. 48: 1775–1789 [DOI] [PubMed] [Google Scholar]
- Bonza M.C., Morandini P., Luoni L., Geisler M., Palmgren M.G., De Michelis M.I. (2000). At-ACA8 encodes a plasma membrane-localized calcium-ATPase of Arabidopsis with a calmodulin-binding domain at the N terminus. Plant Physiol. 123: 1495–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeade P., de Jaegher G., Boyer N. (1991). Microsomal ATP-dependent Ca2+ transport as affected by environmental stress in Bryonia dioica internodes. Plant Sci. 79: 23–30 [Google Scholar]
- Bowler C., Neuhaus G., Yamagata H., Chua N.H. (1994). Cyclic GMP and calcium mediate phytochrome phototransduction. Cell 77: 73–81 [DOI] [PubMed] [Google Scholar]
- Braam J. (2005). In touch: plant responses to mechanical stimuli. New Phytol. 165: 373–389 [DOI] [PubMed] [Google Scholar]
- Brand J.J., Becker D.W. (1984). Evidence for direct roles of calcium in photosynthesis. J. Bioenerg. Biomembr. 16: 239–249 [DOI] [PubMed] [Google Scholar]
- Brailoiu E., Churamani D., Cai X., Schrlau M.G., Brailoiu G.C., Gao X., Hooper R., Boulware M.J., Dun N.J., Marchant J.S., Patel S. (2009). Essential requirement for two-pore channel 1 in NAADP-mediated calcium signaling. J. Cell Biol. 186: 201–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner E.D., Martinez-Barboza N., Clark A.P., Liang Q.S., Stevenson D.W., Coruzzi G.M. (2000). Arabidopsis mutants resistant to S+-beta-methyl-alpha, beta-diaminopropionic acid, a cycad-derived glutamate receptor agonist. Plant Physiol. 124: 1615–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush D.R., Sze H. (1986). Calcium transport in tonoplast and endoplasmic reticulum vesicles isolated from cultured carrot cells. Plant Physiol. 80: 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush D.S., Biswas A.K., Jones R.L. (1989a). Gibberellic-acid-stimulated Ca2+ accumulation in endoplasmic reticulum of barley aleurone: Ca2+ transport and steady-state levels. Planta 178: 411–420 [DOI] [PubMed] [Google Scholar]
- Bush D.S., Biswas A.K., Jones R.L. (1993). Hormonal regulation of Ca2+ transport in the endomembrane system of the barley aleurone. Planta 189: 507–515 [Google Scholar]
- Bush D.S., Sticher L., van Huystee R., Wagner D., Jones R.L. (1989b). The calcium requirement for stability and enzymatic activity of two isoforms of barley aleurone alpha-amylase. J. Biol. Chem. 264: 19392–19398 [PubMed] [Google Scholar]
- Calcraft P.J., et al. , (2009). NAADP mobilizes calcium from acidic organelles through two-pore channels. Nature 459: 596–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas L., Lovy-Wheeler A., Kunkel J.G., Hepler P.K. (2008). Pollen tube growth oscillations and intracellular calcium levels are reversibly modulated by actin polymerization. Plant Physiol. 146: 1611–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala R., Santos E., Alonso J.M., Ecker J.R., Martinez-Zapater J.M., Salinas J. (2003). Mutations in the Ca2+/H+ transporter CAX1 increase CBF/DREB1 expression and the cold-acclimation response in Arabidopsis. Plant Cell 15: 2940–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerana M., Bonza M.C., Harris R., Sanders D., De Michelis M.I. (2006). Abscisic acid stimulates the expression of two isoforms of plasma membrane Ca2+-ATPase in Arabidopsis thaliana seedlings. Plant Biol. 8: 572–578 [DOI] [PubMed] [Google Scholar]
- Charron D., Pingret J.L., Chabaud M., Journet E.P., Barker D.G. (2004). Pharmacological evidence that multiple phospholipid signaling pathways link Rhizobium nodulation factor perception in Medicago truncatula root hairs to intracellular responses, including Ca2+ spiking and specific ENOD gene expression. Plant Physiol. 136: 3582–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng N.H., Hirschi K.D. (2003). Cloning and characterization of CXIP1, a novel PICOT domain-containing Arabidopsis protein that associates with CAX1. J. Biol. Chem. 278: 6503–6509 [DOI] [PubMed] [Google Scholar]
- Cheng N.H., Liu J.Z., Nelson R.S., Hirschi K.D. (2004a). Characterization of CXIP4, a novel Arabidopsis protein that activates the H+/Ca2+ antiporter, CAX1. FEBS Lett. 559: 99–106 [DOI] [PubMed] [Google Scholar]
- Cheng N.H., Pittman J.K., Barkla B.J., Shigaki T., Hirschi K.D. (2003). The Arabidopsis cax1 mutant exhibits impaired ion homeostasis, development, and hormonal responses and reveals interplay among vacuolar transporters. Plant Cell 15: 347–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng N.H., Pittman J.K., Shigaki T., Hirschi K.D. (2002). Characterization of CAX4, an Arabidopsis H+/cation antiporter. Plant Physiol. 128: 1245–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng N.H., Pittman J.K., Shigaki T., Lachmansingh J., LeClere S., Lahner B., Salt D.E., Hirschi K.D. (2005). Functional association of Arabidopsis CAX1 and CAX3 is required for normal growth and ion homeostasis. Plant Physiol. 138: 2048–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng N.H., Pittman J.K., Zhu J.K., Hirschi K.D. (2004b). The protein kinase SOS2 activates the Arabidopsis H+/Ca2+ antiporter CAX1 to integrate calcium transport and salt tolerance. J. Biol. Chem. 279: 2922–2926 [DOI] [PubMed] [Google Scholar]
- Cheong Y.H., Kim K.N., Pandey G.K., Gupta R., Grant J.J., Luan S. (2003). CBL1, a calcium sensor that differentially regulates salt, drought, and cold responses in Arabidopsis. Plant Cell 15: 1833–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong Y.H., Pandey G.K., Grant J.J., Batistič O., Li L., Kim B.G., Lee S.C., Kudla J., Luan S. (2007). Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J. 52: 223–239 [DOI] [PubMed] [Google Scholar]
- Chiu J.C., Brenner E.D., DeSalle R., Nitabach M.N., Holmes T.C., Coruzzi G.M. (2002). Phylogenetic and expression analysis of the glutamate-receptor-like gene family in Arabidopsis thaliana. Mol. Biol. Evol. 19: 1066–1082 [DOI] [PubMed] [Google Scholar]
- Cho D., Kim S.A., Murata Y., Lee S., Jae S.K., Nam H.G., Kwak J.M. (2009). De-regulated expression of the plant glutamate receptor homolog AtGLR3.1 impairs long-term Ca2+-programmed stomatal closure. Plant J. 58: 437–449 [DOI] [PubMed] [Google Scholar]
- Clapham D.E. (2007). Calcium signaling. Cell 131: 1047–1058 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Fengler K.A., Yu I.C., Lippok B., Smith R.K., Jr, Bent A.F. (2000). The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc. Natl. Acad. Sci. USA 97: 9323–9328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D.J., Hedrich R. (1991). Stretch-activated chloride, potassium, and calcium channels coexisting in plasma membranes of guard cells of Vicia faba L. Planta 186: 143–153 [DOI] [PubMed] [Google Scholar]
- D'Angelo C., et al. , (2006). Alternative complex formation of the Ca2+-regulated protein kinase CIPK1 controls abscisic acid-dependent and independent stress responses in Arabidopsis. Plant J. 48: 857–872 [DOI] [PubMed] [Google Scholar]
- Delk N.A., Johnson K.A., Chowdhury N.I., Braam J. (2005). CML24, regulated in expression by diverse stimuli, encodes a potential Ca2+ sensor that functions in responses to abscisic acid, daylength, and ion stress. Plant Physiol. 139: 240–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidchik V., Bowen H.C., Maathuis F.J., Shabala S.N., Tester M.A., White P.J., Davies J.M. (2002). Arabidopsis thaliana root non-selective cation channels mediate calcium uptake and are involved in growth. Plant J. 32: 799–808 [DOI] [PubMed] [Google Scholar]
- Demidchik V., Maathuis F.J. (2007). Physiological roles of nonselective cation channels in plants: From salt stress to signalling and development. New Phytol. 175: 387–404 [DOI] [PubMed] [Google Scholar]
- Dobney S., Chiasson D., Lam P., Smith S.P., Snedden W.A. (2009). The calmodulin-related calcium sensor CML42 plays a role in trichome branching. J. Biol. Chem. 284: 31647–31657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd A.N., Jakobsen M.K., Baker A.J., Telzerow A., Hou S.W., Laplaze L., Barrot L., Poethig R.S., Haseloff J., Webb A.A. (2006). Time of day modulates low-temperature Ca signals in Arabidopsis. Plant J. 48: 962–973 [DOI] [PubMed] [Google Scholar]
- Doherty C.J., Van Buskirk H.A., Myers S.J., Thomashow M.F. (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21: 972–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drøbak B.K., Ferguson I.B. (1985). Release of Ca2+ from plant hypocotyl microsomes by inositol-1,4,5-trisphosphate. Biochem. Biophys. Res. Commun. 130: 1241–1246 [DOI] [PubMed] [Google Scholar]
- Du L., Ali G.S., Simons K.A., Hou J., Yang T., Reddy A.S., Poovaiah B.W. (2009). Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457: 1154–1158 [DOI] [PubMed] [Google Scholar]
- Du L., Poovaiah B.W. (2005). Ca2+/calmodulin is critical for brassinosteroid biosynthesis and plant growth. Nature 437: 741–745 [DOI] [PubMed] [Google Scholar]
- Dutta R., Robinson K.R. (2004). Identification and characterization of stretch-activated ion channels in pollen protoplasts. Plant Physiol. 135: 1398–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom E.M., Ehrhardt D.W., Mitra R.M., Long S.R. (2002). Pharmacological analysis of nod factor-induced calcium spiking in Medicago truncatula. Evidence for the requirement of type IIA calcium pumps and phosphoinositide signaling. Plant Physiol. 128: 1390–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdei L., Toth I., Zsoldos F. (1979). Hormonal regulation of Ca2+-stimulated K+ influx and Ca2+, K+-ATPase in rice roots: in vivo and in vitro effects of auxins and reconstitution of the ATPase. Physiol. Plant. 45: 448–452 [Google Scholar]
- Fakhrai H., Hall J.L. (1984). Changes in calcium-ATPase activity associated with the washing (ageing) of potato tuber discs. J. Plant Physiol. 117: 69–79 [DOI] [PubMed] [Google Scholar]
- Finkler A., Ashery-Padan R., Fromm H. (2007a). CAMTAs: Calmodulin-binding transcription activators from plants to human. FEBS Lett. 581: 3893–3898 [DOI] [PubMed] [Google Scholar]
- Finkler A., Kaplan B., Fromm H. (2007b). Ca2+-responsive cis-elements in plants. Plant Signal. Behav. 2: 17–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frietsch S., Wang Y.F., Sladek C., Poulsen L.R., Romanowsky S.M., Schroeder J.I., Harper J.F. (2007). A cyclic nucleotide-gated channel is essential for polarized tip growth of pollen. Proc. Natl. Acad. Sci. USA 104: 14531–14536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglsang A.T., Guo Y., Cuin T.A., Qiu Q., Song C., Kristiansen K.A., Bych K., Schulz A., Shabala S., Schumaker K.S., Palmgren M.G., Zhu J.K. (2007). Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. Plant Cell 19: 1617–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furch A.C., van Bel A.J., Fricker M.D., Felle H.H., Fuchs M., Hafke J.B. (2009). Sieve element Ca2+ channels as relay stations between remote stimuli and sieve tube occlusion in Vicia faba. Plant Cell 21: 2118–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galon Y., Nave R., Boyce J.M., Nachmias D., Knight M.R., Fromm H. (2008). Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Lett. 582: 943–948 [DOI] [PubMed] [Google Scholar]
- Gao D., Knight M.R., Trewavas A.J., Sattelmacher B., Plieth C. (2004). Self-reporting Arabidopsis expressing pH and [Ca2+] indicators unveil ion dynamics in the cytoplasm and in the apoplast under abiotic stress. Plant Physiol. 134: 898–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D., Becker D., Vosloh D., Gambale F., Palme K., Rehers M., Anschuetz U., Dreyer I., Kudla J., Hedrich R. (2009). Heteromeric AtKC1/AKT1 channels in Arabidopsis roots facilitate growth under K+-limiting conditions. J. Biol. Chem. 284: 21288–21295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M., Frangne N., Gomes E., Martinoia E., Palmgren M.G. (2000). The ACA4 gene of Arabidopsis encodes a vacuolar membrane calcium pump that improves salt tolerance in yeast. Plant Physiol. 124: 1814–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George L., Romanowsky S.M., Harper J.F., Sharrock R.A. (2008). The ACA10 Ca2+-ATPase regulates adult vegetative development and inflorescence architecture in Arabidopsis. Plant Physiol. 146: 716–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy S., Read N.D., Trewavas A.J. (1990). Elevation of cytoplasmic calcium by caged calcium or caged inositol triphosphate initiates stomatal closure. Nature 346: 769–771 [DOI] [PubMed] [Google Scholar]
- Gleason C., Chaudhuri S., Yang T., Munoz A., Poovaiah B.W., Oldroyd G.E. (2006). Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441: 1149–1152 [DOI] [PubMed] [Google Scholar]
- Gobert A., Park G., Amtmann A., Sanders D., Maathuis F.J. (2006). Arabidopsis thaliana cyclic nucleotide gated channel 3 forms a non-selective ion transporter involved in germination and cation transport. J. Exp. Bot. 57: 791–800 [DOI] [PubMed] [Google Scholar]
- Gong D., Guo Y., Jagendorf A.T., Zhu J.K. (2002). Biochemical characterization of the Arabidopsis protein kinase SOS2 that functions in salt tolerance. Plant Physiol. 130: 256–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabov A., Blatt M.R. (1998). Membrane voltage initiates Ca2+ waves and potentiates Ca2+ increases with abscisic acid in stomatal guard cells. Proc. Natl. Acad. Sci. USA 95: 4778–4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo K.M., Babourina O., Christopher D.A., Borsics T., Rengel Z. (2008). The cyclic nucleotide-gated channel, AtCNGC10, influences salt tolerance in Arabidopsis. Physiol. Plant. 134: 499–507 [DOI] [PubMed] [Google Scholar]
- Guo Y., Halfter U., Ishitani M., Zhu J.K. (2001). Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13: 1383–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfter U., Ishitani M., Zhu J.K. (2000). The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA 97: 3735–3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton D.W., Hills A., Kohler B., Blatt M.R. (2000). Ca2+ channels at the plasma membrane of stomatal guard cells are activated by hyperpolarization and abscisic acid. Proc. Natl. Acad. Sci. USA 97: 4967–4972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Tang R., Anderson L.K., Woerner T.E., Pei Z.M. (2003). A cell surface receptor mediates extracellular Ca2+ sensing in guard cells. Nature 425: 196–200 [DOI] [PubMed] [Google Scholar]
- Harada A., Shimazaki K. (2009). Measurement of changes in cytosolic Ca2+ in Arabidopsis guard cells and mesophyll cells in response to blue light. Plant Cell Physiol. 50: 360–373 [DOI] [PubMed] [Google Scholar]
- Harmon A.C., Gribskov M., Harper J.F. (2000). CDPKs: A kinase for every Ca2+ signal? Trends Plant Sci. 5: 154–159 [DOI] [PubMed] [Google Scholar]
- Harper J.F., Breton G., Harmon A. (2004). Decoding Ca2+ signals through plant protein kinases. Annu. Rev. Plant Biol. 55: 263–288 [DOI] [PubMed] [Google Scholar]
- Harper J.F., Harmon A. (2005). Plants, symbiosis and parasites: A calcium signalling connection. Nat. Rev. Mol. Cell Biol. 6: 555–566 [DOI] [PubMed] [Google Scholar]
- Harper J.F., Hong B., Hwang I., Guo H.Q., Stoddard R., Huang J.F., Palmgren M.G., Sze H. (1998). A novel calmodulin-regulated Ca2+-ATPase (ACA2) from Arabidopsis with an N-terminal autoinhibitory domain. J. Biol. Chem. 273: 1099–1106 [DOI] [PubMed] [Google Scholar]
- Hedrich R., Kudla J. (2006). Calcium signaling networks channel plant K+ uptake. Cell 125: 1221–1223 [DOI] [PubMed] [Google Scholar]
- Hedrich R., Neher E. (1987). Cytoplasmic calcium regulates voltage-dependent ion channels in plant vacuoles. Nature 329: 833–836 [Google Scholar]
- Hepler P.K., Vidali L., Cheung A.Y. (2001). Polarized cell growth in higher plants. Annu. Rev. Cell Dev. Biol. 17: 159–187 [DOI] [PubMed] [Google Scholar]
- Hetherington A.M., Brownlee C. (2004). The generation of Ca2+ signals in plants. Annu. Rev. Plant Biol. 55: 401–427 [DOI] [PubMed] [Google Scholar]
- Hirschi K.D. (1999). Expression of Arabidopsis CAX1 in tobacco: Altered calcium homeostasis and increased stress sensitivity. Plant Cell 11: 2113–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschi K.D., Korenkov V.D., Wilganowski N.L., Wagner G.J. (2000). Expression of Arabidopsis CAX2 in tobacco. Altered metal accumulation and increased manganese tolerance. Plant Physiol. 124: 125–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C.H., Lin S.H., Hu H.C., Tsay Y.F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194 [DOI] [PubMed] [Google Scholar]
- Hrabak E.M., et al. , (2003). The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 132: 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua B.G., Mercier R.W., Leng Q., Berkowitz G.A. (2003a). Plants do it differently. A new basis for potassium/sodium selectivity in the pore of an ion channel. Plant Physiol. 132: 1353–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua B.G., Mercier R.W., Zielinski R.E., Berkowitz G.A. (2003b). Functional interaction of calmodulin with a plant cyclic nucleotide gated cation channel. Plant Physiol. Biochem. 41: 945–954 [Google Scholar]
- Huang L., Berkelman T., Franklin A.E., Hoffman N.E. (1993). Characterization of a gene encoding a Ca2+-ATPase-like protein in the plastid envelope. Proc. Natl. Acad. Sci. USA 90: 10066–10070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. (1995). Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell 80: 225–236 [DOI] [PubMed] [Google Scholar]
- Hwang I., Sze H., Harper J.F. (2000). A calcium-dependent protein kinase can inhibit a calmodulin-stimulated Ca2+ pump (ACA2) located in the endoplasmic reticulum of Arabidopsis. Proc. Natl. Acad. Sci. USA 97: 6224–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura M., Osawa M., Ames J.B. (2002). The role of calcium-binding proteins in the control of transcription: Structure to function. Bioessays 24: 625–636 [DOI] [PubMed] [Google Scholar]
- Irving H.R., Gehring C.A., Parish R.W. (1992). Changes in cytosolic pH and calcium of guard cells precede stomatal movements. Proc. Natl. Acad. Sci. USA 89: 1790–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi K., Suzuki M., Imai M. (2000). Molecular cloning of a novel form (two-repeat) protein related to voltage-gated sodium and calcium channels. Biochem. Biophys. Res. Commun. 270: 370–376 [DOI] [PubMed] [Google Scholar]
- Ishida S., Yuasa T., Nakata M., Takahashi Y. (2008). A tobacco calcium-dependent protein kinase, CDPK1, regulates the transcription factor REPRESSION OF SHOOT GROWTH in response to gibberellins. Plant Cell 20: 3273–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashikina N., Hedrich R. (2005). K+ currents through SV-type vacuolar channels are sensitive to elevated luminal sodium levels. Plant J. 41: 606–614 [DOI] [PubMed] [Google Scholar]
- Ivashuta S., Liu J., Lohar D.P., Haridas S., Bucciarelli B., VandenBosch K.A., Vance C.P., Harrison M.J., Gantt J.S. (2005). RNA interference identifies a calcium-dependent protein kinase involved in Medicago truncatula root development. Plant Cell 17: 2911–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X.-Y., He L.-H., Jing R.-L., Li R.-Z. (2009). Calreticulin: Conserved protein and diverse functions in plants. Physiol. Plant. 136: 127–138 [DOI] [PubMed] [Google Scholar]
- Johannes E., Brosnan J.M., Sanders D. (1992). Parallel pathways for intracellular Ca2+ release from the vacuole of higher plants. Plant J. 2: 97–102 [Google Scholar]
- Kadota Y., Furuichi T., Ogasawara Y., Goh T., Higashi K., Muto S., Kuchitsu K. (2004). Identification of putative voltage-dependent Ca2+-permeable channels involved in cryptogein-induced Ca2+ transients and defense responses in tobacco BY-2 cells. Biochem. Biophys. Res. Commun. 317: 823–830 [DOI] [PubMed] [Google Scholar]
- Kang J., Mehta S., Turano F.J. (2004). The putative glutamate receptor 1.1 (AtGLR1.1) in Arabidopsis thaliana regulates abscisic acid biosynthesis and signaling to control development and water loss. Plant Cell Physiol. 45: 1380–1389 [DOI] [PubMed] [Google Scholar]
- Kang J., Turano F.J. (2003). The putative glutamate receptor 1.1 (AtGLR1.1) functions as a regulator of carbon and nitrogen metabolism in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 100: 6872–6877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan B., Davydov O., Knight H., Galon Y., Knight M.R., Fluhr R., Fromm H. (2006). Rapid transcriptome changes induced by cytosolic Ca2+ transients reveal ABRE-related sequences as Ca2+-responsive cis elements in Arabidopsis. Plant Cell 18: 2733–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai M., Muto S. (1990). Ca2+ pump and Ca2+/H+ antiporter in plasma membrane vesicles isolated by aqueous two-phase partitioning from corn leaves. J. Membr. Biol. 114: 133–142 [DOI] [PubMed] [Google Scholar]
- Kiegle E., Moore C.A., Haseloff J., Tester M.A., Knight M.R. (2000). Cell-type-specific calcium responses to drought, salt and cold in the Arabidopsis root. Plant J. 23: 267–278 [DOI] [PubMed] [Google Scholar]
- Kim B.G., Waadt R., Cheong Y.H., Pandey G.K., Dominguez-Solis J.R., Schultke S., Lee S.C., Kudla J., Luan S. (2007). The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J. 52: 473–484 [DOI] [PubMed] [Google Scholar]
- Kim S.A., Kwak J.M., Jae S.K., Wang M.H., Nam H.G. (2001). Overexpression of the AtGluR2 gene encoding an Arabidopsis homolog of mammalian glutamate receptors impairs calcium utilization and sensitivity to ionic stress in transgenic plants. Plant Cell Physiol. 42: 74–84 [DOI] [PubMed] [Google Scholar]
- Kim M.C., Chung W.S., Yun D.J., Cho M.J. (2009). Calcium and calmodulin-mediated regulation of gene expression in plants. Mol. Plant 2: 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klusener B., Young J.J., Murata Y., Allen G.J., Mori I.C., Hugouvieux V., Schroeder J.I. (2002). Convergence of calcium signaling pathways of pathogenic elicitors and abscisic acid in Arabidopsis guard cells. Plant Physiol. 130: 2152–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H., Knight M.R. (2001). Abiotic stress signalling pathways: Specificity and cross-talk. Trends Plant Sci. 6: 262–267 [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Ohura I., Kawakita K., Yokota N., Fujiwara M., Shimamoto K., Doke N., Yoshioka H. (2007). Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell 19: 1065–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolukisaoglu U., Weinl S., Blazevic D., Batistič O., Kudla J. (2004). Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 134: 43–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren'kov V., Park S., Cheng N.H., Sreevidya C., Lachmansingh J., Morris J., Hirschi K., Wagner G.J. (2007). Enhanced Cd2+-selective root-tonoplast-transport in tobaccos expressing Arabidopsis cation exchangers. Planta 225: 403–411 [DOI] [PubMed] [Google Scholar]
- Kosuta S., Hazledine S., Sun J., Miwa H., Morris R.J., Downie J.A., Oldroyd G.E. (2008). Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc. Natl. Acad. Sci. USA 105: 9823–9828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreimer G., Melkonian M., Holtum J.A., Latzko E. (1988). Stromal free calcium concentration and light-mediated activation of chloroplast fructose-1,6-bisphosphatase. Plant Physiol. 86: 423–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J., Xu Q., Harter K., Gruissem W., Luan S. (1999). Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc. Natl. Acad. Sci. USA 96: 4718–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurusu T., Yagala T., Miyao A., Hirochika H., Kuchitsu K. (2005). Identification of a putative voltage-gated Ca2+ channel as a key regulator of elicitor-induced hypersensitive cell death and mitogen-activated protein kinase activation in rice. Plant J. 42: 798–809 [DOI] [PubMed] [Google Scholar]
- Kushwaha R., Singh A., Chattopadhyay S. (2008). Calmodulin7 plays an important role as transcriptional regulator in Arabidopsis seedling development. Plant Cell 20: 1747–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H.M., Chiu J., Hsieh M.H., Meisel L., Oliveira I.C., Shin M., Coruzzi G. (1998). Glutamate-receptor genes in plants. Nature 396: 125–126 [DOI] [PubMed] [Google Scholar]
- Lamotte O., Gould K., Lecourieux D., Sequeira-Legrand A., Lebrun-Garcia A., Durner J., Pugin A., Wendehenne D. (2004). Analysis of nitric oxide signaling functions in tobacco cells challenged by the elicitor cryptogein. Plant Physiol. 135: 516–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laohavisit A., Mortimer J.C., Demidchik V., Coxon K.M., Stancombe M.A., Macpherson N., Brownlee C., Hofmann A., Webb A.A., Miedema H., Battey N.H., Davies J.M. (2009). Zea mays annexins modulate cytosolic free Ca2+ and generate a Ca2+-permeable conductance. Plant Cell 21: 479–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecourieux D., Lamotte O., Bourque S., Wendehenne D., Mazars C., Ranjeva R., Pugin A. (2005). Proteinaceous and oligosaccharidic elicitors induce different calcium signatures in the nucleus of tobacco cells. Cell Calcium 38: 527–538 [DOI] [PubMed] [Google Scholar]
- Lee K.W., Chen P.W., Lu C.A., Chen S., Ho T.H., Yu S.M. (2009). Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Sci. Signal. 2: ra61. [DOI] [PubMed] [Google Scholar]
- Lee S.C., Lan W.Z., Kim B.G., Li L., Cheong Y.H., Pandey G.K., Lu G., Buchanan B.B., Luan S. (2007a). A protein phosphorylation/dephosphorylation network regulates a plant potassium channel. Proc. Natl. Acad. Sci. USA 104: 15959–15964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.M., Kim H.S., Han H.J., Moon B.C., Kim C.Y., Harper J.F., Chung W.S. (2007b). Identification of a calmodulin-regulated autoinhibited Ca2+-ATPase (ACA11) that is localized to vacuole membranes in Arabidopsis. FEBS Lett. 581: 3943–3949 [DOI] [PubMed] [Google Scholar]
- Lemtiri-Chlieh F., MacRobbie E.A., Webb A.A., Manison N.F., Brownlee C., Skepper J.N., Chen J., Prestwich G.D., Brearley C.A. (2003). Inositol hexakisphosphate mobilizes an endomembrane store of calcium in guard cells. Proc. Natl. Acad. Sci. USA 100: 10091–10095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko V., Guinot D.R., Klein M., Roelfsema M.R., Hedrich R., Dietrich P. (2008). Stringent control of cytoplasmic Ca2+ in guard cells of intact plants compared to their counterparts in epidermal strips or guard cell protoplasts. Protoplasma 233: 61–72 [DOI] [PubMed] [Google Scholar]
- Li J., Zhu S., Song X., Shen Y., Chen H., Yu J., Yi K., Liu Y., Karplus V.J., Wu P., Deng X.W. (2006a). A rice glutamate receptor-like gene is critical for the division and survival of individual cells in the root apical meristem. Plant Cell 18: 340–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Kim B.G., Cheong Y.H., Pandey G.K., Luan S. (2006b). A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 12625–12630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Chanroj S., Wu Z., Romanowsky S.M., Harper J.F., Sze H. (2008). A distinct endosomal Ca2+/Mn2+ pump affects root growth through the secretory process. Plant Physiol. 147: 1675–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F., Cunningham K.W., Harper J.F., Sze H. (1997). ECA1 complements yeast mutants defective in Ca2+ pumps and encodes an endoplasmic reticulum-type Ca2+-ATPase in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94: 8579–8584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Yang Y., Quan R., Mendoza I., Wu Y., Du W., Zhao S., Schumaker K.S., Pardo J.M., Guo Y. (2009). Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell 21: 1607–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Zhu J.K. (1998). A calcium sensor homolog required for plant salt tolerance. Science 280: 1943–1945 [DOI] [PubMed] [Google Scholar]
- Luan S. (2009). The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 14: 37–42 [DOI] [PubMed] [Google Scholar]
- Luan S., Kudla J., Rodriguez-Concepcion M., Yalovsky S., Gruissem W. (2002). Calmodulins and calcineurin B-like proteins: Calcium sensors for specific signal response coupling in plants. Plant Cell 14 (suppl.): S389–S400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S., Lan W., Chul Lee S. (2009). Potassium nutrition, sodium toxicity, and calcium signaling: connections through the CBL-CIPK network. Curr. Opin. Plant Biol. 12: 339–346 [DOI] [PubMed] [Google Scholar]
- Ludwig A.A., Romeis T., Jones J.D. (2004). CDPK-mediated signalling pathways: Specificity and cross-talk. J. Exp. Bot. 55: 181–188 [DOI] [PubMed] [Google Scholar]
- Ludwig A.A., Saitoh H., Felix G., Freymark G., Miersch O., Wasternack C., Boller T., Jones J.D., Romeis T. (2005). Ethylene-mediated cross-talk between calcium-dependent protein kinase and MAPK signaling controls stress responses in plants. Proc. Natl. Acad. Sci. USA 102: 10736–10741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G.Z., Wang H.W., Huang J., Tian A.G., Wang Y.J., Zhang J.S., Chen S.Y. (2005). A putative plasma membrane cation/proton antiporter from soybean confers salt tolerance in Arabidopsis. Plant Mol. Biol. 59: 809–820 [DOI] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Magnan F., Ranty B., Charpenteau M., Sotta B., Galaud J.P., Aldon D. (2008). Mutations in AtCML9, a calmodulin-like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid. Plant J. 56: 575–589 [DOI] [PubMed] [Google Scholar]
- Mahajan S., Sopory S.K., Tuteja N. (2006). Cloning and characterization of CBL-CIPK signalling components from a legume (Pisum sativum). FEBS J. 273: 907–925 [DOI] [PubMed] [Google Scholar]
- Martinec J., Feltl T., Scanlon C.H., Lumsden P.J., Machackova I. (2000). Subcellular localization of a high affinity binding site for D-myo-inositol 1,4,5-trisphosphate from Chenopodium rubrum. Plant Physiol. 124: 475–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser P., et al. (2001). Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 126: 1646–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masi E., Ciszak M., Stefano G., Renna L., Azzarello E., Pandolfi C., Mugnai S., Baluska F., Arecchi F.T., Mancuso S. (2009). Spatiotemporal dynamics of the electrical network activity in the root apex. Proc. Natl. Acad. Sci. USA 106: 4048–4053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazars C., Bourque S., Mithofer A., Pugin A., Ranjeva R. (2009). Calcium homeostasis in plant cell nuclei. New Phytol. 181: 261–274 [DOI] [PubMed] [Google Scholar]
- McAinsh M.R., Hetherington A.M. (1998). Encoding specificity in Ca2+ signalling systems. Trends Plant Sci. 3: 32–36 [Google Scholar]
- McAinsh M.R., Webb A., Taylor J.E., Hetherington A.M. (1995). Stimulus-induced oscillations in guard cell cytosolic free calcium. Plant Cell 7: 1207–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack E., Tsai Y.C., Braam J. (2005). Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 10: 383–389 [DOI] [PubMed] [Google Scholar]
- Mei H., Zhao J., Pittman J.K., Lachmansingh J., Park S., Hirschi K.D. (2007). In planta regulation of the Arabidopsis Ca2+/H+ antiporter CAX1. J. Exp. Bot. 58: 3419–3427 [DOI] [PubMed] [Google Scholar]
- Menteyne A., Burdakov A., Charpentier G., Petersen O.H., Cancela J.M. (2006). Generation of specific Ca2+ signals from Ca2+ stores and endocytosis by differential coupling to messengers. Curr. Biol. 16: 1931–1937 [DOI] [PubMed] [Google Scholar]
- Meyerhoff O., Muller K., Roelfsema M.R., Latz A., Lacombe B., Hedrich R., Dietrich P., Becker D. (2005). AtGLR3.4, a glutamate receptor channel-like gene is sensitive to touch and cold. Planta 222: 418–427 [DOI] [PubMed] [Google Scholar]
- Miedema H., Bothwell J.H., Brownlee C., Davies J.M. (2001). Calcium uptake by plant cells-Channels and pumps acting in concert. Trends Plant Sci. 6: 514–519 [DOI] [PubMed] [Google Scholar]
- Miedema H., Demidchik V., Very A.A., Bothwell J.H., Brownlee C., Davies J.M. (2008). Two voltage-dependent calcium channels co-exist in the apical plasma membrane of Arabidopsis thaliana root hairs. New Phytol. 179: 378–385 [DOI] [PubMed] [Google Scholar]
- Mills R.F., Doherty M.L., Lopez-Marques R.L., Weimar T., Dupree P., Palmgren M.G., Pittman J.K., Williams L.E. (2008). ECA3, a Golgi-localized P2A-type ATPase, plays a crucial role in manganese nutrition in Arabidopsis. Plant Physiol. 146: 116–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito N., Wimmers L.E., Bennett A.B. (1996). Sugar regulates mRNA abundance of H+-ATPase gene family members in tomato. Plant Physiol. 112: 1229–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa H., Sun J., Oldroyd G.E., Downie J.A. (2006). Analysis of calcium spiking using a cameleon calcium sensor reveals that nodulation gene expression is regulated by calcium spike number and the developmental status of the cell. Plant J. 48: 883–894 [DOI] [PubMed] [Google Scholar]
- Monshausen G.B., Bibikova T.N., Messerli M.A., Shi C., Gilroy S. (2007). Oscillations in extracellular pH and reactive oxygen species modulate tip growth of Arabidopsis root hairs. Proc. Natl. Acad. Sci. USA 104: 20996–21001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monshausen G.B., Bibikova T.N., Weisenseel M.H., Gilroy S. (2009). Ca2+ regulates reactive oxygen species production and pH during mechanosensing in Arabidopsis roots. Plant Cell 21: 2341–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monshausen G.B., Messerli M.A., Gilroy S. (2008). Imaging of the Yellow Cameleon 3.6 indicator reveals that elevations in cytosolic Ca2+ follow oscillating increases in growth in root hairs of Arabidopsis. Plant Physiol. 147: 1690–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno I., Norambuena L., Maturana D., Toro M., Vergara C., Orellana A., Zurita-Silva A., Ordenes V.R. (2008). AtHMA1 is a thapsigargin-sensitive Ca2+/heavy metal pump. J. Biol. Chem. 283: 9633–9641 [DOI] [PubMed] [Google Scholar]
- Mori I.C., Murata Y., Yang Y., Munemasa S., Wang Y.-F., Andreoli S., Tiriac H., Alonso J.M., Harper J.F., Ecker J.R., Kwak J.M., Schroeder J.I. (2006). CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca2+-permeable channels and stomatal closure. PLoS Biol. 4: 1749–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer J.C., Laohavisit A., Macpherson N., Webb A., Brownlee C., Battey N.H., Davies J.M. (2008). Annexins: Multifunctional components of growth and adaptation. J. Exp. Bot. 59: 533–544 [DOI] [PubMed] [Google Scholar]
- Munnik T., Testerink C. (2009). Plant phospholipid signaling: In a nutshell. J. Lipid Res. 50 (suppl.): S260–S265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir S.R., Sanders D. (1996). Pharmacology of Ca2+ release from red beet microsomes suggests the presence of ryanodine receptor homologs in higher plants. FEBS Lett. 395: 39–42 [DOI] [PubMed] [Google Scholar]
- Muto S., Izawa S., Miyachi S. (1982). Light-induced Ca2+ uptake by intact chloroplast. FEBS Lett. 139: 250–254 [Google Scholar]
- Nakagawa Y., et al. (2007). Arabidopsis plasma membrane protein crucial for Ca2+ influx and touch sensing in roots. Proc. Natl. Acad. Sci. USA 104: 3639–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navazio L., Bewell M.A., Siddiqua A., Dickinson G.D., Galione A., Sanders D. (2000). Calcium release from the endoplasmic reticulum of higher plants elicited by the NADP metabolite nicotinic acid adenine dinucleotide phosphate. Proc. Natl. Acad. Sci. USA 97: 8693–8698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi J., Matsuda O., Nagasawa T., Oba Y., Takahashi H., Kawai-Yamada M., Uchimiya H., Hashimoto M., Iba K. (2008). CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 452: 483–486 [DOI] [PubMed] [Google Scholar]
- Neuhaus G., Bowler C., Kern R., Chua N.H. (1993). Calcium/calmodulin-dependent and -independent phytochrome signal transduction pathways. Cell 73: 937–952 [DOI] [PubMed] [Google Scholar]
- Nobel P.S., Murakami S., Takamiya A. (1966). Localization of light-induced strontium accumulation in spinach chloroplasts. Plant Cell Physiol. 7: 263–275 [Google Scholar]
- Nomura H., Komori T., Kobori M., Nakahira Y., Shiina T. (2008). Evidence for chloroplast control of external Ca2+-induced cytosolic Ca2+ transients and stomatal closure. Plant J. 53: 988–998 [DOI] [PubMed] [Google Scholar]
- Ohta M., Guo Y., Halfter U., Zhu J.K. (2003). A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc. Natl. Acad. Sci. USA 100: 11771–11776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey G.K., Cheong Y.H., Kim K.N., Grant J.J., Li L., Hung W., D'Angelo C., Weinl S., Kudla J., Luan S. (2004). The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell 16: 1912–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey G.K., Grant J.J., Cheong Y.H., Kim B.G., Li le G., Luan S. (2008). Calcineurin-B-like protein CBL9 interacts with target kinase CIPK3 in the regulation of ABA response in seed germination. Mol. Plant 1: 238–248 [DOI] [PubMed] [Google Scholar]
- Park C.Y., Lee J.H., Yoo J.H., Moon B.C., Choi M.S., Kang Y.H., Lee S.M., Kim H.S., Kang K.Y., Chung W.S., Lim C.O., Cho M.J. (2005). WRKY group IId transcription factors interact with calmodulin. FEBS Lett. 579: 1545–1550 [DOI] [PubMed] [Google Scholar]
- Park S.Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z.M., Murata Y., Benning G., Thomine S., Klusener B., Allen G.J., Grill E., Schroeder J.I. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406: 731–734 [DOI] [PubMed] [Google Scholar]
- Peiter E., Maathuis F.J., Mills L.N., Knight H., Pelloux J., Hetherington A.M., Sanders D. (2005). The vacuolar Ca2+-activated channel TPC1 regulates germination and stomatal movement. Nature 434: 404–408 [DOI] [PubMed] [Google Scholar]
- Perez V., Wherrett T., Shabala S., Muniz J., Dobrovinskaya O., Pottosin I. (2008). Homeostatic control of slow vacuolar channels by luminal cations and evaluation of the channel-mediated tonoplast Ca2+ fluxes in situ. J. Exp. Bot. 59: 3845–3855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S., Wyatt S.E., Love J., Thompson W.F., Robertson D., Boss W.F. (2001). The Ca(2+) status of the endoplasmic reticulum is altered by induction of calreticulin expression in transgenic plants. Plant Physiol. 126: 1092–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picton J.M., Steer M.W. (1985). The effects of ruthenium red, lanthanum, fluorescein isothiocyanate and trifluoperazine on vesicle transport, vesicle fusion and tip extension in pollen tubes. Planta 163: 20–26 [DOI] [PubMed] [Google Scholar]
- Pittman J.K., Shigaki T., Cheng N.H., Hirschi K.D. (2002). Mechanism of N-terminal autoinhibition in the Arabidopsis Ca2+/H+ antiporter CAX1. J. Biol. Chem. 277: 26452–26459 [DOI] [PubMed] [Google Scholar]
- Plieth C., Hansen U.P., Knight H., Knight M.R. (1999). Temperature sensing by plants: The primary characteristics of signal perception and calcium response. Plant J. 18: 491–497 [DOI] [PubMed] [Google Scholar]
- Portis A.R., Jr, Heldt H.W. (1976). Light-dependent changes of the Mg2+ concentration in the stroma in relation to the Mg2+ dependency of CO2 fixation in intact chloroplasts. Biochim. Biophys. Acta 449: 434–436 [DOI] [PubMed] [Google Scholar]
- Pottosin I.I., Martinez-Estevez M., Dobrovinskaya O.R., Muniz J. (2005). Regulation of the slow vacuolar channel by luminal potassium: Role of surface charge. J. Membr. Biol. 205: 103–111 [DOI] [PubMed] [Google Scholar]
- Pottosin I.I., Schonknecht G. (2007). Vacuolar calcium channels. J. Exp. Bot. 58: 1559–1569 [DOI] [PubMed] [Google Scholar]
- Qi Z., Stephens N.R., Spalding E.P. (2006). Calcium entry mediated by GLR3.3, an Arabidopsis glutamate receptor with a broad agonist profile. Plant Physiol. 142: 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Q.S., Guo Y., Dietrich M.A., Schumaker K.S., Zhu J.K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 99: 8436–8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan R., Lin H., Mendoza I., Zhang Y., Cao W., Yang Y., Shang M., Chen S., Pardo J.M., Guo Y. (2007). SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 19: 1415–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qudeimat E., Faltusz A.M., Wheeler G., Lang D., Brownlee C., Reski R., Frank W. (2008). A PIIB-type Ca2+-ATPase is essential for stress adaptation in Physcomitrella patens. Proc. Natl. Acad. Sci. USA 105: 19555–19560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranf S., Wunnenberg P., Lee J., Becker D., Dunkel M., Hedrich R., Scheel D., Dietrich P. (2008). Loss of the vacuolar cation channel, AtTPC1, does not impair Ca2+ signals induced by abiotic and biotic stresses. Plant J. 53: 287–299 [DOI] [PubMed] [Google Scholar]
- Richter G.L., Monshausen G.B., Krol A., Gilroy S. (2009). Mechanical stimuli modulate lateral root organogenesis. Plant Physiol. 151: 1855–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis T., Ludwig A.A., Martin R., Jones J.D. (2001). Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J. 20: 5556–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd J.J., Franklin-Tong V.E. (1999). Calcium signaling in plants. Cell. Mol. Life Sci. 55: 214–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sai J., Johnson C.H. (2002). Dark-stimulated calcium ion fluxes in the chloroplast stroma and cytosol. Plant Cell 14: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saidi Y., Finka A., Muriset M., Bromberg Z., Weiss Y.G., Maathuis F.J., Goloubinoff P. (2009). The heat shock response in moss plants is regulated by specific calcium-permeable channels in the plasma membrane. Plant Cell 21: 2829–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Barrena M.J., Fujii H., Angulo I., Martinez-Ripoll M., Zhu J.K., Albert A. (2007). The structure of the C-terminal domain of the protein kinase AtSOS2 bound to the calcium sensor AtSOS3. Mol. Cell 26: 427–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D., Brownlee C., Harper J.F. (1999). Communicating with calcium. Plant Cell 11: 691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D., Pelloux J., Brownlee C., Harper J.F. (2002). Calcium at the crossroads of signaling. Plant Cell 14 (suppl.): S401–S417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiott M., Romanowsky S.M., Baekgaard L., Jakobsen M.K., Palmgren M.G., Harper J.F. (2004). A plant plasma membrane Ca2+ pump is required for normal pollen tube growth and fertilization. Proc. Natl. Acad. Sci. USA 101: 9502–9507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder J.I., Kwak J.M., Allen G.J. (2001). Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410: 327–330 [DOI] [PubMed] [Google Scholar]
- Schumaker K.S., Sze H. (1987). Inositol 1,4,5-trisphosphate releases Ca2+ from vacuolar membrane vesicles of oat roots. J. Biol. Chem. 262: 3944–3946 [PubMed] [Google Scholar]
- Scrase-Field S.A., Knight M.R. (2003). Calcium: Just a chemical switch? Curr. Opin. Plant Biol. 6: 500–506 [DOI] [PubMed] [Google Scholar]
- Seigneurin-Berny D., Gravot A., Auroy P., Mazard C., Kraut A., Finazzi G., Grunwald D., Rappaport F., Vavasseur A., Joyard J., Richaud P., Rolland N. (2006). HMA1, a new Cu-ATPase of the chloroplast envelope, is essential for growth under adverse light conditions. J. Biol. Chem. 281: 2882–2892 [DOI] [PubMed] [Google Scholar]
- Shaw S.L., Long S.R. (2003). Nod factor elicits two separable calcium responses in Medicago truncatula root hair cells. Plant Physiol. 131: 976–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Kim K.N., Ritz O., Albrecht V., Gupta R., Harter K., Luan S., Kudla J. (1999). Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell 11: 2393–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigaki T., Rees I., Nakhleh L., Hirschi K.D. (2006). Identification of three distinct phylogenetic groups of CAX cation/proton antiporters. J. Mol. Evol. 63: 815–825 [DOI] [PubMed] [Google Scholar]
- Siegel R.S., Xue S., Murata Y., Yang Y., Nishimura N., Wang A., Schroeder J.I. (2009). Calcium elevation-dependent and attenuated resting calcium-dependent abscisic acid induction of stomatal closure and abscisic acid-induced enhancement of calcium sensitivities of S-type anion and inward-rectifying K channels in Arabidopsis guard cells. Plant J. 59: 207–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaguru M., Pike S., Gassmann W., Baskin T.I. (2003). Aluminum rapidly depolymerizes cortical microtubules and depolarizes the plasma membrane: evidence that these responses are mediated by a glutamate receptor. Plant Cell Physiol. 44: 667–675 [DOI] [PubMed] [Google Scholar]
- Snowman B.N., Kovar D.R., Shevchenko G., Franklin-Tong V.E., Staiger C.J. (2002). Signal-mediated depolymerization of actin in pollen during the self-incompatibility response. Plant Cell 14: 2613–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderling T.R. (1999). The Ca2+-calmodulin-dependent protein kinase cascade. Trends Biochem. Sci. 24: 232–236 [DOI] [PubMed] [Google Scholar]
- Stephens N.R., Qi Z., Spalding E.P. (2008). Glutamate receptor subtypes evidenced by differences in desensitization and dependence on the GLR3.3 and GLR3.4 genes. Plant Physiol. 146: 529–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze H., Liang F., Hwang I., Curran A.C., Harper J.F. (2000). Diversity and regulation of plant Ca2+ pumps: Insights from expression in yeast. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51: 433–462 [DOI] [PubMed] [Google Scholar]
- Takeda S., Gapper C., Kaya H., Bell E., Kuchitsu K., Dolan L. (2008). Local positive feedback regulation determines cell shape in root hair cells. Science 319: 1241–1244 [DOI] [PubMed] [Google Scholar]
- Thion L., Mazars C., Nacry P., Bouchez D., Moreau M., Ranjeva R., Thuleau P. (1998). Plasma membrane depolarization-activated calcium channels, stimulated by microtubule-depolymerizing drugs in wild-type Arabidopsis thaliana protoplasts, display constitutively large activities and a longer half-life in ton 2 mutant cells affected in the organization of cortical microtubules. Plant J. 13: 603–610 [DOI] [PubMed] [Google Scholar]
- Thuleau P., Schroeder J.I., Ranjeva R. (1998). Recent advances in the regulation of plant calcium channels: Evidence for regulation by G-proteins, the cytoskeleton and second messengers. Curr. Opin. Plant Biol. 1: 424–427 [DOI] [PubMed] [Google Scholar]
- Tracy F.E., Gilliham M., Dodd A.N., Webb A.A., Tester M. (2008). NaCl-induced changes in cytosolic free Ca2+ in Arabidopsis thaliana are heterogeneous and modified by external ionic composition. Plant Cell Environ. 31: 1063–1073 [DOI] [PubMed] [Google Scholar]
- Tsai Y.C., Delk N.A., Chowdhury N.I., Braam J. (2007). Arabidopsis potential calcium sensors regulate nitric oxide levels and the transition to flowering. Plant Signal. Behav. 2: 446–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T., Sugiyama N., Mizoguchi M., Hayashi S., Myouga F., Yamaguchi-Shinozaki K., Ishihama Y., Hirayama T., Shinozaki K. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urquhart W., Gunawardena A.H., Moeder W., Ali R., Berkowitz G.A., Yoshioka K. (2007). The chimeric cyclic nucleotide-gated ion channel ATCNGC11/12 constitutively induces programmed cell death in a Ca2+ dependent manner. Plant Mol. Biol. 65: 747–761 [DOI] [PubMed] [Google Scholar]
- Vahisalu T., Kollist H., Wang Y.F., Nishimura N., Chan W.Y., Valerio G., Lamminmaki A., Brosche M., Moldau H., Desikan R., Schroeder J.I., Kangasjarvi J. (2008). SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452: 487–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainonen J.P., Hansson M., Vener A.V. (2005). STN8 protein kinase in Arabidopsis thaliana is specific in phosphorylation of photosystem II core proteins. J. Biol. Chem. 280: 33679–33686 [DOI] [PubMed] [Google Scholar]
- Vainonen J.P., Sakuragi Y., Stael S., Tikkanen M., Allahverdiyeva Y., Paakkarinen V., Aro E., Suorsa M., Scheller H.V., Vener A.V., Aro E.M. (2008). Light regulation of CaS, a novel phosphoprotein in the thylakoid membrane of Arabidopsis thaliana. FEBS J. 275: 1767–1777 [DOI] [PubMed] [Google Scholar]
- Waadt R., Schmidt L.K., Lohse M., Hashimoto K., Bock R., Kudla J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Walch-Liu P., Liu L.H., Remans T., Tester M., Forde B.G. (2006). Evidence that L-glutamate can act as an exogenous signal to modulate root growth and branching in Arabidopsis thaliana. Plant Cell Physiol. 47: 1045–1057 [DOI] [PubMed] [Google Scholar]
- Webb A.A.R., McAinsh M.R., Taylor J.E., Hetherington A.M. (1996). Calcium ions as intracellular second messengers in higher plants. Adv. Bot. Res. 22: 45–96 [Google Scholar]
- Weinl S., Held K., Schlucking K., Steinhorst L., Kuhlgert S., Hippler M., Kudla J. (2008). A plastid protein crucial for Ca2+-regulated stomatal responses. New Phytol. 179: 675–686 [DOI] [PubMed] [Google Scholar]
- Weinl S., Kudla J. (2009). The CBL-CIPK Ca2+-decoding signaling network: Function and perspectives. New Phytol. 184: 517–528 [DOI] [PubMed] [Google Scholar]
- Wheeler G.L., Brownlee C. (2008). Ca2+ signalling in plants and green algae-changing channels. Trends Plant Sci. 13: 506–514 [DOI] [PubMed] [Google Scholar]
- White P.J., Bowen H.C., Demidchik V., Nichols C., Davies J.M. (2002). Genes for calcium-permeable channels in the plasma membrane of plant root cells. Biochim. Biophys. Acta 1564: 299–309 [DOI] [PubMed] [Google Scholar]
- White P.J., Broadley M.R. (2003). Calcium in plants. Ann. Bot. (Lond.) 92: 487–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson R.E., Ashley C.C. (1982). Free Ca2+ and cytoplasmic streaming in the alga Chara. Nature 296: 647–650 [DOI] [PubMed] [Google Scholar]
- Wimmers L.E., Ewing N.N., Bennett A.B. (1992). Higher plant Ca2+-ATPase: Primary structure and regulation of mRNA abundance by salt. Proc. Natl. Acad. Sci. USA 89: 9205–9209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Li H.D., Chen L.Q., Wang Y., Liu L.L., He L., Wu W.H. (2006). A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125: 1347–1360 [DOI] [PubMed] [Google Scholar]
- Yoo J.H., et al. (2005). Direct interaction of a divergent CaM isoform and the transcription factor, MYB2, enhances salt tolerance in Arabidopsis. J. Biol. Chem. 280: 3697–3706 [DOI] [PubMed] [Google Scholar]
- Yoon G.M., Dowd P.E., Gilroy S., McCubbin A.G. (2006). Calcium-dependent protein kinase isoforms in Petunia have distinct functions in pollen tube growth, including regulating polarity. Plant Cell 18: 867–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka K., Moeder W., Kang H.G., Kachroo P., Masmoudi K., Berkowitz G., Klessig D.F. (2006). The chimeric Arabidopsis CYCLIC NUCLEOTIDE-GATED ION CHANNEL11/12 activates multiple pathogen resistance responses. Plant Cell 18: 747–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J.J., Mehta S., Israelsson M., Godoski J., Grill E., Schroeder J.I. (2006). CO2 signaling in guard cells: Calcium sensitivity response modulation, a Ca2+-independent phase, and CO2 insensitivity of the gca2 mutant. Proc. Natl. Acad. Sci. USA 103: 7506–7511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I.C., Parker J., Bent A.F. (1998). Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc. Natl. Acad. Sci. USA 95: 7819–7824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Zhou R.G., Gao Y.J., Zheng S.Z., Xu P., Zhang S.Q., Sun D.Y. (2009). Molecular and genetic evidence for the key role of AtCaM3 in heat-shock signal transduction in Arabidopsis. Plant Physiol. 149: 1773–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Barkla B.J., Marshall J., Pittman J.K., Hirschi K.D. (2008). The Arabidopsis cax3 mutants display altered salt tolerance, pH sensitivity and reduced plasma membrane H+-ATPase activity. Planta 227: 659–669 [DOI] [PubMed] [Google Scholar]
- Zhao J., Shigaki T., Mei H., Guo Y.Q., Cheng N.H., Hirschi K.D. (2009). Interaction between Arabidopsis Ca2+/H+ Exchangers CAX1 and CAX3. J. Biol. Chem. 284: 4605–4615 [DOI] [PubMed] [Google Scholar]
- Zhu S.Y., et al. (2007). Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19: 3019–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zocchi G., Rabotti G. (1993). Calcium transport in membrane vesicles isolated from maize coleoptiles (effect of indoleacetic acid and fusicoccin). Plant Physiol. 101: 135–139 [DOI] [PMC free article] [PubMed] [Google Scholar]