The mechanism of vacuole inheritance is poorly understood. This work makes use of the model organism C. merolae, which has a minimum set of organelles that are systematically inherited, and identifies vig1 as being essential for vacuole inheritance.
Abstract
Vacuoles/lysosomes function in endocytosis and in storage and digestion of metabolites. These organelles are inherited by the daughter cells in eukaryotes. However, the mechanisms of this inheritance are poorly understood because the cells contain multiple vacuoles that behave randomly. The primitive red alga Cyanidioschyzon merolae has a minimum set of organelles. Here, we show that C. merolae contains about four vacuoles that are distributed equally between the daughter cells by binding to dividing mitochondria. Binding is mediated by VIG1, a 30-kD coiled-coil protein identified by microarray analyses and immunological assays. VIG1 appears on the surface of free vacuoles in the cytosol and then tethers the vacuoles to the mitochondria. The vacuoles are released from the mitochondrion in the daughter cells following VIG1 digestion. Suppression of VIG1 by antisense RNA disrupted the migration of vacuoles. Thus, VIG1 is essential for tethering vacuoles to mitochondria during vacuole inheritance in C. merolae.
INTRODUCTION
The inheritance of single membrane–bound and DNA-less organelles, such as vacuoles/lysosomes, Golgi bodies, endoplasmic reticulum, and microbodies, as well as double membrane–bound organelles, such as mitochondria and plastids, is an essential feature of eukaryotic cell division. Vacuoles are lytic compartments, function as reservoirs for ions and metabolites, including pigments, and are crucial for detoxification processes and general cell homeostasis (Marty, 1999). The vacuoles of algae, plants, and yeasts share some of their basic properties with the lysosomes of animal cells (Marty, 1999). In humans, loss of lysosome function causes lysosomal storage disorders, such as Fabry disease and GM1 gangliosidosis (Futerman and van Meer, 2004).
In mammals, plants, and yeasts, lysosomes/vacuoles are inherited by the daughter cells (Bergeland et al., 2001; Han et al., 2003; Kutsuna et al., 2003). In Madin-Darby canine kidney cells, video and confocal microscopy analyses have shown that lysosomes accumulate in the vicinity of the microtubule organization center during cytokinesis and then separate into daughter cells (Bergeland et al., 2001). In plants, the dynamic changes in vacuoles during mitosis were examined by monitoring the tubular structures of the vacuolar membrane (TVMs) in living transgenic tobacco (Nicotiana tabacum) cultivar Bright Yellow-2 cells stably expressing green fluorescent protein (GFP)-AtVam3p, a fusion protein of GFP with AtVam3p/SYP22, which belongs to the syntaxin family of Arabidopsis thaliana proteins (Kutsuna et al., 2003). The TVMs, which were initially derived from large vacuoles, elongated to encircle the spindle at metaphase. Subsequently, the TVMs invaded the equatorial region from anaphase to telophase and were divided between the two daughter cells by the cell plate at cytokinesis. Furthermore, actin filaments were indispensable for the development and maintenance of TVMs. However, the underlying molecular mechanisms were not established.
During vacuole inheritance in Saccharomyces cerevisiae, the vacuole forms vesicular-tubular projections known as segregation structures. The segregation structures originate from the vacuole membrane and extend to the daughter bud. This inheritance is based on an actin cable and myosin-V motor protein Myo2 with Vac8 and Vac17 (Han et al., 2003; Weisman, 2006). The Myo2-Vac8-Vac17 complex drives the segregation structures of the vacuole to the bud along the actin cable. Vac8 is a myristoylated and palmitoylated armadillo repeat-containing protein that binds to the vacuole membrane. Vac17 has a Vac8 binding domain and a Myo2 binding domain and links Vac8-associated vacuoles to Myo2 (Weisman, 2006).
In the primitive red alga Cyanidioschyzon merolae, the vacuoles are inherited by daughter cells through binding to the dividing mitochondrion regardless of the absence of actin filaments and myosin (Takahashi et al., 1995; Matsuzaki et al., 2004; Yagisawa et al., 2007). In C. merolae, no functional actin filaments were detected by immunological methods or staining with rhodamine-conjugated phalloidin (Takahashi et al., 1995). One actin gene is encoded in the C. merolae genome, but not expressed, whereas myosin genes are absent altogether (Matsuzaki et al., 2004). Therefore, a currently unknown mechanism that does not depend on actin filaments must be involved in vacuole inheritance. C. merolae cells offer many advantages for studies of vacuole inheritance. Each C. merolae cell has a minimum set of organelles comprising one plastid, one mitochondrion, and a few vacuoles (Kuroiwa, 1998; Misumi et al., 2005; Yagisawa et al., 2007), and organelle divisions can be highly synchronized by a light/dark cycle (Suzuki et al., 1994). Furthermore, the C. merolae genome has been completely sequenced and is one of the smallest (16.5 Mb) among free-living eukaryotes, containing the fewest number of genes (Ohta et al., 1998, 2003; Matsuzaki et al., 2004; Nozaki et al., 2007). Because most of the genes are present in low copy numbers and lack introns, designing oligonucleotide probes for microarrays is simple and easy. Microarray analyses of C. merolae have provided clues to solve biological problems. For example, genome-wide microarray analysis of C. merolae cells in a nitrogen-depleted condition identified the key nitrogen assimilation genes (Imamura et al., 2009) and the mitochondrial division gene ZED (Yoshida et al., 2009) and was useful for finding the candidate genes for salt tolerance (Sakajiri et al., 2008). Therefore, we expected that C. merolae might provide a unique tool for resolving the issue of vacuole inheritance. Previously, we conducted microarray analysis to obtain a genome-wide gene expression profile during organelle division and inheritance (Fujiwara et al., 2009). In this study, we used the expression profiling data of Fujiwara et al. (2009) together with domain analysis to identify candidate genes with a role in vacuole migration during inheritance and show that one such gene, vacuole inheritance gene1 (vig1), is essential for this process.
RESULTS
Vacuole Inheritance during the Cell Cycle of C. merolae
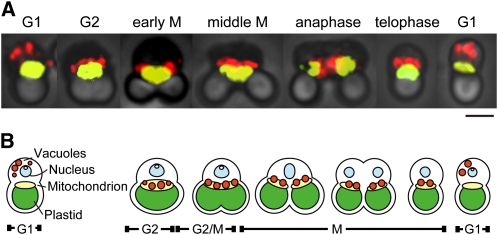
The cell cycle in C. merolae was synchronized by a 12-h light/dark cycle, and cells were harvested and observed mainly in the G2 and M phases. To examine the behavior and inheritance of vacuoles during the cell cycle, the vacuole marker V-ATPase (vacuole H+-ATPase) and mitochondrial marker Porin/CMO111C (a mitochondrial outer membrane channel [POR]) of C. merolae were used to generate antibodies. Antisera against POR specifically recognized the ∼30-kD band that was the predicted molecular mass of POR (see Supplemental Figure 1 online). Immunofluorescence microscopy using the antibodies against V-ATPase along with the antibodies against POR revealed that vacuoles were distributed in the cytosol during the G1 phase. Vacuoles approached the mitochondrial surface from the cytosol during the S phase (see Supplemental Figure 2 online) and then became bound to the dividing mitochondrion by G2. During the M phase, the vacuoles were inherited by the daughter cells via the binding to the dividing mitochondria. In the early G1 phase, the vacuoles were released from the mitochondrion and returned to the cytosol in the daughter cells (Figures 1A and 1B).
Figure 1.
Behavior of Vacuoles during the Cell Cycle of C. merolae.
(A) Phase-contrast immunofluorescence images showing behavior of vacuoles during the cell cycle. Vacuoles (red) and mitochondria (yellow) were immunostained by antibodies against V-ATPase and mitochondrial porin, respectively. Cell cycle phases are indicated on images. Bar = 2 μm.
(B) Diagrammatic summary of vacuole inheritance in C. merolae. Vacuoles were located around the nucleus in the G1 phase but moved to the mitochondrial surface by G2 and continued to localize there during the M phase. After cytokinesis, vacuoles were released from the mitochondrial surface in the G1 phase of daughter cells. Vacuoles, orange; nucleus, light blue; mitochondria, yellow; plastid, green. Cell cycle phases are indicated under the diagrams.
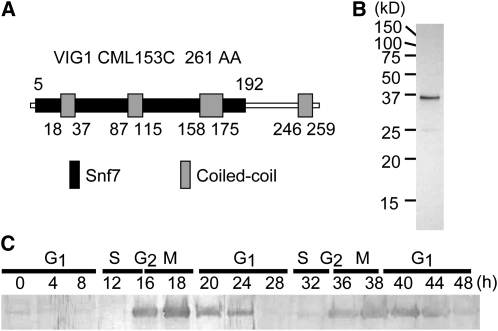
Therefore, genes related to vacuole inheritance must be strongly expressed during the G2 and M phases. In C. merolae, the transcriptional and translational levels of ftsZ2 and dnm2, which are required for plastid division (Takahara et al., 2000; Miyagishima et al., 2003; Yoshida et al., 2006), were reported to peak during plastid division in the predicted S and G2 phases (Takahara et al., 2000; Miyagishima et al., 2003). Therefore, gene expression profiling during the cell cycle is a useful tool to search for genes related to organelle division and inheritance that depend on cell cycle progression. We used microarray data in a previous study (Fujiwara et al., 2009) and performed a preliminary screen to identify candidate genes involved in vacuole inheritance. A previous microarray study analyzed gene expression in the cell cycle of C. merolae and selected 158 genes induced during S and G2-M phases (Fujiwara et al., 2009). Within these 158 genes, we searched for candidate genes functioning on the vesicle surface and related to vesicle trafficking by a conserved domain search because filamentous structures linking the vacuolar surface to the mitochondrial surface have been recorded (Yagisawa et al., 2007). Only one gene, CML153C, was found. CML153C has an Snf7 domain, with an expected value (Ev) of 2e-8 in a conserved domain search (Figure 2A). The Snf7 superfamily protein CHMP5/MOS10/VPS60 is localized on the endosome membrane (Kranz et al., 2001) and is suggested to function in protein sorting and transport from the endosomes to the vacuoles/lysosomes (Kranz et al., 2001). Therefore, it seemed possible that CML153C had a function related to the migration of the vacuole to the mitochondrial surface. The gene expression profiling showed that the level of CML153C transcripts began to increase in the S phase and peaked mainly in the M phase. ftsZ2 and dnm2 were also periodically expressed during the S and G2 phases, respectively (see Supplemental Figure 3A online). Real-time RT-PCR analysis also supported that CML153C transcripts accumulated during the S and M phases (see Supplemental Figure 3B online). The oscillation of the CML153C transcript level corresponded to the migration of vacuoles to the mitochondria. Therefore, we analyzed the function of CML153C, hereafter named vig1.
Figure 2.
Domain Structure and Expression of VIG1 Protein.
(A) Predicted domain structure of VIG1/CML153C. VIG1 has aN Snf7 domain (black box) and four coiled-coil regions (gray boxes).
(B) Characterization of the affinity-purified antibody against VIG1. Total protein from mitotic cells was separated and immunoblotted using antibody raised against the recombinant VIG1.
(C) Protein expression throughout the cell cycle detected with an immunoblot for VIG1. VIG1 showed cell cycle–specific expression and accumulated during the G2 and M phases. Total proteins from cells were harvested at the indicated time after synchronization.
VIG1 Is a Coiled-Coil Protein and Is Regulated in Cell Cycle Progression
VIG1 is a predicted coiled-coil protein with no transmembrane domain. Three coiled-coil regions are contained within the Snf7 domain, and one coiled-coil region is found outside the Snf7 domain in the C terminus (Figure 2A). Immunoblotting analysis showed that anti-VIG1 antibodies, generated against recombinant VIG1 protein, specifically detected a 35-kD band from the total cell lysate of mitotic cells (Figure 2B) and that VIG1 was expressed mainly during the G2 and M phases (Figure 2C).
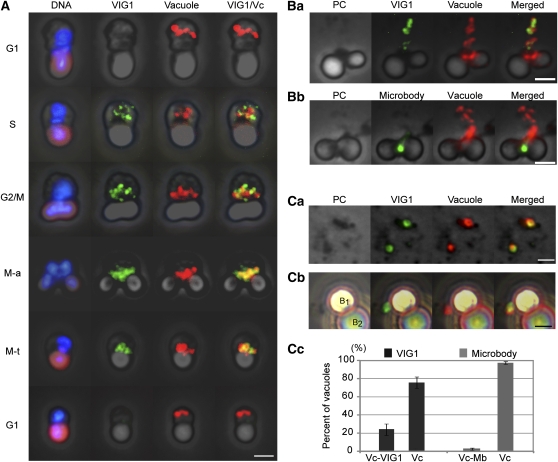
VIG1 Is Localized on the Surface of Vacuoles during Vacuole Inheritance
We used immunofluorescence microscopy with antibodies against VIG1 and V-ATPase to reveal the dynamics of VIG1 during the cell cycle. VIG1 was undetectable in the cell during G1 but began to accumulate at the vacuoles in the cytosol during late S phase. VIG1 and vacuoles migrated to the mitochondrial regions by G2. The localization of VIG1 at the vacuoles on the mitochondria continued during the M phase. In the G1 phase, VIG1 was not detected in the cytosol or around the vacuoles as they released from the mitochondrial region in early G1 (Figure 3A).
Figure 3.
Immunofluorescence for VIG1 and V-ATPase on the Vacuoles.
(A) Immunofluorescence showing localization of VIG1 (green) and behavior of vacuoles (red) during the cell cycle. DNA was stained with DAPI (blue). The plastid emitted autofluorescence (purple-red) under UV excitation. M-a, M-anaphase; M-t, M-telophase. Bar = 2 μm.
(B) Localization of VIG1, microbodies, and vacuoles in smeared cells.
(Ba) Together with VIG1, the vacuoles were separated from the mitochondria. VIG1 colocalized with the vacuoles. Bar = 2 μm.
(Bb). Localization of a microbody (green) and vacuoles in a smeared mitotic cell. The microbody did not localize near the separated vacuoles. Bar = 2 μm.
(C) IP of vacuoles.
(Ca) VIG1 localized at vacuoles in homogenates. Bar = 2 μm.
(Cb) Magnetic IP of vacuoles. Vacuole was caught by secondary antibodies conjugated magnetic beads. B1 and B2 show magnetic bead1 and bead2, respectively. VIG1 is attached to the vacuole binding B1. Bar = 2 μm.
(Cc) Percentage of vacuoles on the beads colocalized with VIG1 or microbodies. Localization of vacuoles with VIG1 or microbodies was individually examined. n = 40, number of vacuoles examined. Bars indicate sd. Vc, vacuoles to which magnetic beads were bound; Vc-VIG1, vacuoles attached to VIG1 on the beads; Vc-Mb, vacuoles attached to microbodies on the beads. All images were merged with phase-contrast (PC) images.
To detect the relationship between VIG1 and the vacuoles among dispersed organelles, we examined smeared cells using immunofluorescence microscopy (Figure 3B). A microbody served as a negative control. In C. merolae, a microbody is a single membrane–bound organelle that binds to a mitochondrion during inheritance in a similar way to the vacuoles (Miyagishima et al., 1999a). Together with the vacuoles, VIG1 mostly peeled from the mitochondrial region in smeared cells (Figure 3Ba), while the microbody remained at the mitochondrial region (Figure 3Bb). Whereas most of the vacuoles overlapped with VIG1 signal, only a small number of the vacuoles overlapped with and/or touched the microbodies (Table 1). The results showed that VIG1 was colocalized with the vacuoles.
Table 1.
Percentage of Vacuoles Overlapping with and Touching VIG1 or Microbodies in Smeared Cells
| Vacuoles with VIG1 | Vacuoles with Microbodies |
| 88.6 ± 9.9 (%) | 2.9 ± 1.4 (%) |
Count for percentage of vacuoles was performed in double immunofluorescence assays for vacuoles with VIG1 or vacuoles with microbodies. They were counted if any part of their signals overlapped. Values are means and sd. n = 150, number of vacuoles examined. Count was performed in two technical repeats.
To confirm that VIG1 was localized at the juxta-vacuolar region, we immunostained VIG1 in homogenates and isolated vacuoles by immunoprecipitation (IP). The result confirmed that VIG1 was colocalized with the vacuoles (Figure 3Ca). IP showed that 25% of vacuoles were observed with VIG1, and 3% of vacuoles were with microbodies (Figures 3Cb and 3Cc).
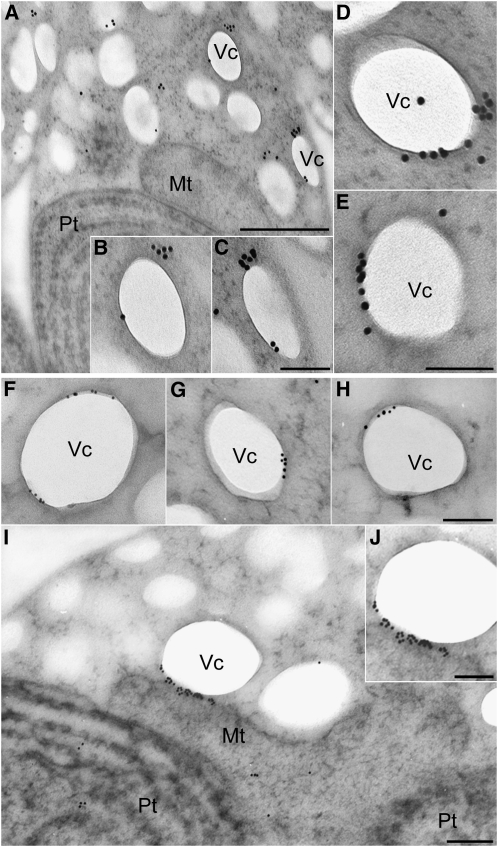
To test the hypothesis that VIG1 was associated with the vacuoles, immunogold labeling and transmission electron microscopy was used. To avoid the artifacts resulting from the size of gold particles, localization of VIG1 was examined using immunogold labeling with 15- and 10-nm gold particles. The vacuolar membrane protein metallopeptidase, which is localized at the vacuolar boundary, was used as the control (Yagisawa et al., 2009). The distribution of immunogold particles was quantified.
The results using 15-nm gold particles showed that VIG1 was located on the vacuole membrane in the cytosol during S phase as shown in Figure 3A but had an asymmetric distribution (Figures 4A to 4E, Table 2). The results using 10-nm gold particles clearly supported the results with the 15-nm gold particles (Figures 4F to 4H). Moreover, VIG1 was mostly localized between the vacuoles and the mitochondria when vacuoles were tethered to mitochondria (Figures 4I and 4J, Table 3; see Supplemental Figure 4 online). Metallopeptidase was localized uniformly around the vacuolar membrane (see Supplemental Figure 5 online). To examine localization of VIG1 on the entire surface of the vacuole, negative staining of an isolated complex of a plastid accompanied by vacuoles was performed. The association between vacuoles and the mitochondrial region became loose, as in the smeared cells (see Supplemental Figure 6A online). Immunogold-labeled VIG1 was localized at patch structures binding to the vacuolar surface (see Supplemental Figure 6B online). These structures localized on one part of the vacuole membrane. This observation corresponded with the transmission electron microscopy observations of the vacuolar surface.
Figure 4.
Immunoelectron Microscopy for VIG1.
(A) to (E) Sections of cell were immunolabeled with anti-VIG1 antibodies and 15-nm gold particle–conjugated secondary antibodies. This assay was performed in three independent replicates.
(A) Section showing a cell detected with anti-VIG1antibodies.
(B) and (C) Magnified images of vacuoles in (A).
(D) and (E) Localization of VIG1 on vacuoles in the cytosol from other sections.
(F) to (J) Sections of cell were immunolabeled with anti-VIG1 antibodies and 10-nm gold particle–conjugated secondary antibodies. This assay was performed in three independent replicates.
(F) to (H) Localization of VIG1 on vacuoles in the cytosol.
(I) Section showing VIG1 localization when the vacuole was bound to the mitochondria. VIG1 was localized between the vacuoles and mitochondria.
(J) A magnified image of the vacuole binding to the mitochondrial membrane in (I).
Mt, mitochondrion; Pt, plastid; Vc, vacuole. Bars = 500 nm in (A), 100 nm in (B) to (H) and (J), and 200 nm in (I).
Table 2.
Density of Labeling by Antibodies against VIG1 among Organelles
| Number of Gold Particles (100 nm−2) |
||
| Organelle | Membranea | Inner Regionb |
| Vacuoles | 22.0 ± 3.0 | 0.0 ± 0.0 |
| Nuclei | 1.9 ± 0.5 | 1.1 ± 0.3 |
| Mitochondria | 2.1 ± 0.7 | 0.3 ± 0.1 |
| Plastids | 0.2 ± 0.0 | 0.7 ± 0.1 |
| Cytosol | 0.0 ± 0.0 | 0.3 ± 0.2 |
Numbers of immunogold particles labeling VIG1 on each organelle of sections were counted.
Membrane region was defined as the line of membrane in the image and 50 nm outside of it.
Inner region includes the inside of the organelle membrane or cytoplasmic membrane. Immunogold particles of VIG1 selectively localized on the vacuolar membrane. Values are means ± se. n = 50, number of cell sections examined.
Table 3.
Density of Labeling by Antibodies against VIG1 on Membranes of Vacuoles Tethered to Mitochondria
| Organelles | Number of Gold Particles (100 nm−2) | P Value in Welch's t Test |
| Vacuolar membrane | ||
| Cytosol sidea | 2.0 ± 0.7 | 6.2 × 10−5 d |
| Mitochondrial sideb | 27.8 ± 1.9 | – |
| Mitochondrial membranec | 1.7 ± 0.4 | 5.8 × 10−5 e |
Numbers of immunogold particles on the parts of the vacuoles bound to the mitochondria were counted. The area of the vacuole was divided into a and b. a, The two-thirds of the vacuolar membrane toward the cytosol; b, the one-third of the vacuolar membrane facing the mitochondria. Mitochondrial membrane (c) was used as a control. Divided areas of the vacuole membrane are shown in a schematic diagram (see Supplemental Figure 4 online). Welch's t test showed a significant difference of density of immunogold particles of VIG1 among the mitochondrial side on the vacuole (b) and the other area (a or c). d, Welch's t test for b versus a. e, Welch's t test for b versus c. VIG1 accumulated on the mitochondrial side of the vacuole membrane. Values are means ± se. n = 50, number of cell sections examined.
In sum, the immunofluorescence and electron microscopy show that VIG1 is localized at the surface of vacuoles during S and M phases and is mostly localized between the vacuoles and mitochondria when the vacuoles are tethered to mitochondria.
Analysis of Morphological Effects of Downregulation of VIG1 by Antisense RNA
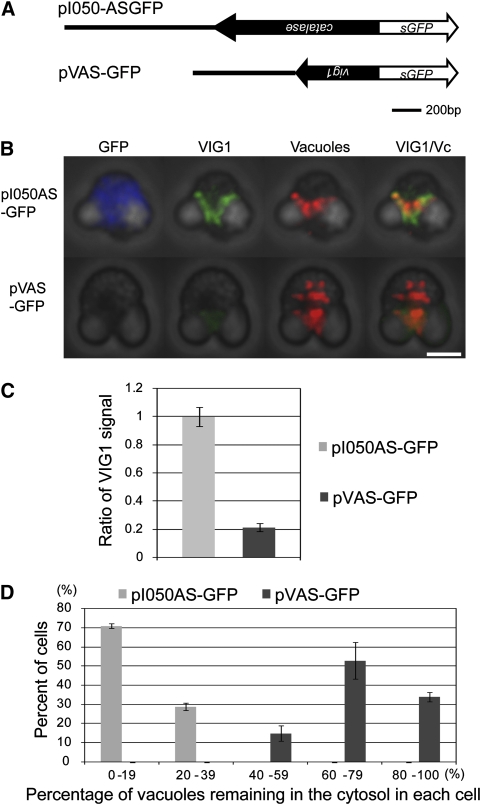
We performed downregulation of VIG1 using antisense RNA (AS RNA) methods to suppress VIG1 selectively. AS RNA hybridizes to mRNA of the target gene and can repress translation (Yin and Ji, 2002). Transient AS RNA expression is a powerful method to downregulate gene expression in C. merolae (Ohnuma et al., 2009). The vig1 promoter was added upstream of the full-length vig1 AS sequence. Synthetic GFP (sGFP) was added downstream of the AS sequence as a marker for the transcription of exogenous genes. Since no paralog of vig1 was found in the C. merolae genome, vig1 AS should inhibit the translation of vig1 only. This plasmid was named pVAS-GFP (Figure 5A). A catalase AS construct was used as a control experiment for the vig1 AS experiment (pI050AS-GFP) (Ohnuma et al., 2009) (Figure 5A). The cells were harvested at 24 h after transformation. RT-PCR showed that the exogenous AS RNA of vig1 was transcribed (see Supplemental Figure 7 online).
Figure 5.
Downregulation of VIG1 by AS RNA of vig1.
(A) Plasmids used in study. Black line represents the native promoter region of catalase and vig1. Closed arrow represents the antisense strand of catalase and vig1.Open arrow represents sGFP.
(B) Phase-contrast immunofluorescent image of transformants. Immunostained GFP, VIG1, and vacuoles are shown in blue, green, and red, respectively. In the pI050AS-GFP cell, VIG1 localized around vacuoles located in the mitochondrial region. In the pVAS-GFP cell, VIG1 signal decreased and vacuoles failed to localize in the mitochondrial region. Vc, vacuole. Bar = 2 μm.
(C) Ratio of VIG1 expression level between transformants. Signal intensity of immunostained VIG1 in the cytosol was measured. The signal intensity of each transformant was normalized by that in cells that were not transformed in each experiment. n = 20, number of cells examined. Bars indicate se.
(D) Index of the migration of vacuoles in transformants. The x axis shows categories based on the percentage of vacuoles remaining in the cytosol in each cell. The y axis shows percentage of cells falling in each category. In pI050AS-GFP cells (gray bars), most vacuoles did not remain in the cytosol and localized around the mitochondrial region. In pVAS-GFP cells (black bars), most vacuoles remained in the cytosol. pI050C: n = 30; pVAS-GFP: n = 25, number of mitotic cells examined. Bars indicate sd. This assay was performed in two biological repeats.
In cells that were transformed with pI050AS-GFP cells, GFP and VIG1 signals were detected, and the vacuole moved to the mitochondrial region (Figure 5B, pI050AS-GFP). By contrast, in the pVAS-GFP cell, the immunostained signal of VIG1 decreased and vacuole migration failed (Figure 5B). Since the sGFP sequence linked to the AS of vig1 was not translated to a functional GFP-fused protein, GFP was not immunostained in the pVAS-GFP cell (Figure 5B, pVAS-GFP). The immunostained-VIG1 signal in pVAS-GFP cells was decreased to 20% of that in pI050AS-GFP cells (Figure 5C). In the pVAS-GFP cells, 60 to 79% of vacuoles did not migrate to the mitochondrial region (Figure 5D). Therefore, we suggest that VIG1 is essential for tethering vacuoles to mitochondria.
DISCUSSION
Vacuole Inheritance by Tethering Vacuoles to Mitochondria
In this study, vacuole movement during inheritance was visualized by immunofluorescence against V-ATPase and POR. The results were consistent with those of Yagisawa et al. (2007), who used fluorescent probes (e.g., LysoTracker and 4',6-diamidino-2-phenylindole [DAPI]) and electron microscopy. This work provides evidence that vacuoles are inherited by binding to mitochondria in C. merolae. Since C. merolae cells have only a few vacuoles and one mitochondrion, vacuole interaction with the mitochondrion during inheritance was easily observed. It is possible that some of the numerous vacuoles/lysosomes in higher-plant and mammalian cells also interact with mitochondria. If they exist, these interactions would be easy to overlook because these organisms have large or numerous vacuoles/lysosomes and mitochondria that exhibit random behavior. The fact that lysosomes tend to lie close to mitochondria (Beard and Novikoff, 1969) suggests that vacuoles/lysosomes and mitochondria in higher eukaryotes may also bind to and separate from each other.
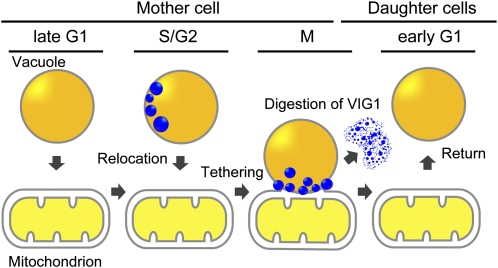
Model for VIG1's Role in Vacuole Inheritance in C. merolae
Vacuole migration during inheritance does not dependent on actin in C. merolae (Yagisawa et al., 2007), and our study shows that VIG1 is essential for vacuole migration. The following model is proposed to explain the role of VIG1 in vacuole inheritance (Figure 6). Immunofluorescent microscopy showed that VIG1 localized at vacuoles before migration to a mitochondrion (Figure 3A, S phase). Immunoelectron microscopy suggested that VIG1 accumulates in patch structures at the vacuolar surface in the cytosol (Figures 4A to 4H; see Supplemental Figure 6 online). Furthermore, the AS experiment showed that vacuoles failed to migrate to mitochondria when VIG1 levels decreased (Figure 5B, pVAS-GFP). These results suggested that VIG1 is needed for transport of the vacuoles (Figure 6, S/G2). Subsequently, vacuoles were tethered to mitochondria during the M phase by the structure including VIG1 (Figure 1A, G2 to telophase; Figure 6, M). Electron microscopy showed that VIG1 accumulated between the vacuolar surface and the mitochondrial surface (Figures 4I and 4J). An examination of thin sections of mitotic cells of C. merolae revealed filamentous structures between the vacuoles and mitochondria (Yagisawa et al., 2007). VIG1 between the vacuoles and mitochondria may correspond to these filamentous structures. In the daughter cell, VIG1 was not detected in the cytosol or at vacuoles released from the mitochondrion (Figure 3A, lower G1). This suggests that the vacuoles return to the cytosol by the digestion of VIG1 (Figure 6, early G1). Therefore, VIG1 is a previously undescribed protein that enables vacuoles to be transported by dividing mitochondria during inheritance in C. merolae. Since VIG1 is a predicted soluble protein and does not have a transmembrane region (see Supplemental Figure 8B online), VIG1 probably requires receptors to localize on the vacuolar surface. The asymmetrical pattern of VIG1 localization might be established by an asymmetrical distribution of preexisting receptors localized on the vacuolar surface.
Figure 6.
Model for Vacuole Inheritance in C. merolae.
VIG1 accumulates at patch structures on the vacuolar surface in the cytosol. The patch structures have an asymmetric distribution during S phase. During M phase, the VIG1 patch localizes between the vacuole membrane and mitochondrial surface and tethers the vacuole to the mitochondria. After cytokinesis, digestion of VIG1 allows the vacuole to return to the cytosol.
[See online article for color version of this figure.]
Conservation of VIG1 in Eukaryotes
We searched for a homolog of VIG1 in eukaryotes and found that the CHMP5 proteins have some coiled-coil structures and a similar charge distribution (see Supplemental Figures 8A and 8B online). VIG1 has a weak similarity to CHMP5 of Homo sapiens (Ev, 3e-9; identities, 25%), CHMP5/VPS60/MOP10 of S. cerevisiae (Ev, 4e-7; identities, 25%), the Snf7 family protein of Dictyostelium discoideum (Ev, 9e-7; identities, 24%), and VPS60.2 of Arabidopsis (Ev, 7e-16; identities, 28%) (see Supplemental Figure 8C online). CHMP5 is a subunit of Endosomal Sorting Complex Required for Transport III (Bowers et al., 2004) and functions in multivesicular bodies (MVBs)/endosome sorting to the lysosome/vacuole in the endocytotic pathway (Kranz et al., 2001; Shim et al., 2006). In yeasts, CHMP5 was detected in a fraction with the endosomal protein Pep12 following fractionation of CHMP5 by density gradient centrifugation (Kranz et al., 2001). In mice, CHMP5 is essential in the final step of MVB sorting (Shim et al., 2006). These previous studies suggested that CHMP5 functions on the MVBs/endosome membrane. It has not been reported that CHMP5 is involved in the vacuolar/lysosomal transport to mitochondria. However, similarity of VIG1 to CHMP5 offers suggestions for the design of future studies for vacuole inheritance and the association between vacuoles and mitochondria in higher organisms.
METHODS
Synchronous Culture
Cyanidioschyzon merolae 10D-14 was synchronized according to the method of Suzuki et al. (1994). Cells were cultured in 2× Allen's medium at pH 2.3 (Allen, 1959). Flasks were shaken under continuous light (40 W/m2) at 42°C. The cells were subcultured to 107 cells/mL and then synchronized by subjecting them to a 12-h-light/12-h-dark cycle at 42°C while the medium was aerated.
Amino Acid Sequence Analysis
The amino acid sequences of VIG1 and CHMP5 were obtained from the National Center for Biotechnology Information (NCBI). Coiled-coil regions in protein were predicted by the COILS program (http://www.ch.embnet.org/software/COILS_form.html) (Lupas et al., 1991). Hydrophobicity (Hopps and Woods program) and charge distribution were analyzed by Genetyx version 6. The amino acid sequences were aligned using the default settings by ClustalX v1.81 (Thompson et al., 1997).
Antibody Generation and Immunoblotting
To generate an anti-VIG1 antiserum, the coding region for the amino acid sequence of VIG1/CML153C protein was amplified by PCR with the following primers: 5′-CGCCGCCGCGGATCCATGTTCTGGAAAAAGAAGGCACC-3′ (BamHI site underlined) and 5′-CCCCCCCCCAAGCTTGACCGCCATGCTGCGTCTC-3′ (HindIII site underlined). To generate an antimitochondrial porin (POR) antiserum, the coding region for the amino acid sequence of POR/CMO111C was amplified by PCR with the following primers: 5′- ACATACATGCATGCATGGTGATCAAGCTATTCTCAAAGC-3′ (SphI underlined) and 5′- ACGCACGCGTCGACGGCATCGTATGTCAAAGACAACC-3′ (SalI site underlined). After digestion by restriction enzymes, the DNA fragment was cloned into the pQE80 vector (Qiagen) and transformed into the Escherichia coli strain XL1Blue. The recombinant protein was produced as a fusion protein with a 6-his tag at the N terminus and purified using a nickel affinity column. The purified proteins were injected into guinea pigs. The anti-VIG1 antiserum was purified with a 6-his tag fused, VIG1-conjugated, NHS-activated sepharose column (GE Healthcare). Immunoblotting for VIG1 and POR was performed by conventional methods. For immunoblots, C. merolae cells were harvested at each time from initiation of the synchronized culture and lysed in conventional SDS containing sample buffer. Thirty micrograms of proteins of each sample were separated by SDS-PAGE and then were immunoblotted. The primary antisera or antibodies against VIG1 and POR were each used at a dilution of 1:1000. The secondary antibodies and alkaline phosphatase–conjugated goat anti-guinea pig (Kirkegaad and Perry Laboratories) were used at a dilution of 1:3000. The signal was detected by AP conjugate substrate kit (Bio-Rad) and the Printgraph AE-6905H (ATTO).
Real-Time RT-PCR
The details of the RNA isolation procedure, reverse transcription, and real-time PCR are described in the Supplemental Methods online. Expression of vig1 transcripts was normalized by that of ef-1a. This assay was performed in three technical repeats. The primers used in this assay for vig1 were 5′-TGCGCCTTGGCTTGGT-3′ and 5′-GCGTGTTGCCAGTTCCATT-3′, and for ef-1a were 5′-CCTGGGCATCGTGACTTTATC-3′ and 5′-GGAGACGCAACCACCAAGAT-3′.
Fluorescence Microscopy
Cells were harvested during the mitotic phase from synchronized cultures. After fixation (Nishida et al., 2004) and blocking of the cells, vacuoles were stained with an anti-V-ATPase primary antibody at a dilution of 1:100 (Yagisawa et al., 2009) and secondary goat anti-rabbit IgG conjugated with Alexa 488 at a dilution of 1:1000 or Alexa 555 at a dilution of 1:100 (both from Molecular Probes). Mitochondria were stained with anti-POR primary antisera at a dilution of 1:200 and secondary goat anti-rat IgG conjugated with Alexa 555 at a dilution of 1:300. VIG1 was stained with an anti-VIG1 primary antibody at a dilution of 1:200 and secondary goat anti-guinea pig IgG conjugated with Alexa 488 at a dilution of 1:100. The microbody was stained with antisera against catalase primary antibody at a dilution of 1:200 and secondary goat anti-rat IgG conjugated with Alexa 488 at a dilution of 1:300 (Ohnuma et al., 2009). For the observation of DNA, the cells stained with 1 mg/mL DAPI.
Cells were observed using a fluorescence microscope (Olympus BX51) with a combination of narrow band-pass filter sets: BP330-385 BA420 (U-MWUA2; Olympus) for DAPI, BP470-490 BA510-550 (U-MNIBA2; Olympus) for Alexa-488, BP541-551 BA565-595 (XF37; Omega) for Alexa-555, and using a Hg arc lamp as a source of excitation light. Images were collected using a 3CCD camera system (C7780-10; Hamamatsu Photonics) and processed using the M8458-03 RCA-3CCD Photoshop plug-in software. The change of color channels and adjustment of fluorescence and merging of images were performed by Photoshop CS2 (Adobe Systems). The distance between vacuoles and mitochondria was measured by Photoshop CS2, after immunostaining of vacuoles and mitochondria and collecting images by C7780-10.
Vacuole Isolation
For preparation of the smeared cells in which vacuoles separated from the mitochondrial region were observed, the cover glass on top of the immunostained cells was slightly pushed and moved. The isolation of vacuoles was demonstrated as follows. The synchronized culture was harvested at 18 h in the M phase and centrifuged at 1500g for 3 min. The pellet was suspended in a vacuole isolation buffer (20 mM Tris-HCl, pH 7.5, 5 mM EGTA, 2 mM MgCl, 5 mM KCl, and 6% [w/v] sucrose) and then incubated at 42°C for 30 min. The cells were mixed with 20% (w/v) maize starch (final concentration) with the protease inhibitor cocktail Complete (Roche, Applied Science), and then the mixture was homogenized by a Dounce homogenizer (Wheaton Industrial). DNase I was added to give a final concentration of 200 g/mL to the lysate, and the mixture was incubated on ice for 30 min. Following centrifugation at 500g for 1 min to remove starch, the lysate was recentrifuged at 500g for 3 min. The supernatant containing vacuoles was centrifuged at 6000g for 3 min. After fixation of the pellet, IP of vacuoles was performed using magnetic Dynabeads M-280 conjugated sheep anti-rabbit IgG (Invitrogen). For IP, the vacuole fraction was mixed with the anti-V-ATPase primary antibody (1:100 dilution) and then with the magnetic bead solution at a dilution of 1:10 for 2 h on ice. Vacuoles were stained with Alexa555-conjugated secondary antibody (1:100 dilutions). VIG1 and microbodies were stained with the anti-VIG1 primary antibody (1:100 dilution) or anticatalase primary antisera (1:200 dilution) and then with Alexa488-conjugated secondary antibody (1:100 dilution).
Electron Microscopy
Transmission electron microscopy was performed as previously described (Miyagishima et al., 2001). Briefly, a cell pellet was rapidly frozen in liquid propane chilled in liquid nitrogen, transferred to dried acetone containing 1% osmium tetroxide (OsO4) at −80°C, embedded in Spurr's resin, and cut into 90-nm-thick serial thin sections. The sections were immunostained with anti-VIG1 antibody and then with 10- or 15-nm gold particle–conjugated goat anti-guinea pig IgG antibody (Aurion) and stained with uranyl acetate and lead citrate. In the experiments using 10-nm gold particle–conjugated secondary antibodies, primary antibodies were used at a dilution of 1:50 and secondary antibodies were diluted 1:60. In the 15-nm gold particle experiments, both primary and secondary antibodies were used at dilutions of 1:20. To determine the number of immunogold particles per unit area, the area containing the membrane and inner regions of organelles and the cell was measured by HC Image (Hamamatsu Photonics). After the F-test, Welch's t test was calculated by Excel 2007 (Microsoft). Antimetallopeptidase antibody was previously prepared (Yagisawa et al., 2009). For immunoelectron microscopy of metallopeptidase, the sections were immunostained with antimetallopeptidase antibody at a dilution of 1:50 and then with 10-nm gold particle–conjugated goat anti-rat IgG antibody at a dilution of 1:60 (Yagisawa et al., 2009).
The isolation of the plastid accompanied with vacuoles was performed according to the methods of plastid isolation described (Miyagishima et al., 1999b). The complex was immunostained with anti-VIG1 antibodies at a dilution of 1:20, followed by 15-nm gold particle–conjugated goat anti-guinea pig IgG at a dilution of 1:20, and then stained with 0.5% phosphotungstic acid, pH 7.0. Images were captured with conventional film or using a super coolscan 9000 ED (Nikon) and digitally processed. Sections and sample were examined with a JEM-1200EX electron microscope (JEOL).
Transient Gene Expression
To make pVAS-GFP, the 5′-flanking region (970 bp) was amplified with the primer sets 5′-CCCCCCAAGCTTCGAAAACGTTCGTCGACAAG-3′ and 5′-CGCCGCGGATCCATCGAACAATGTTTGTTACCGCC-3′ (HindIII and BamHI sites underlined). The antisense sequence of vig1 was amplified with primer sets 5′-CATGCATGCCATGGCATGTTCTGGAAAAAGAAGGCAC-3′ and 5′-CGCCGCGGATCCGACCGCCATGCTGCGTC-3′ (NcoI and BamHI sites underlined). The 5′-flanking region was cloned into pTH2 (Niwa et al., 1999), followed by cloning of the antisense sequence. pI050AS-GFP was previously prepared (Ohnuma et al., 2009). The transformation of C. merolae cells was performed according to Ohnuma's methods (Ohnuma et al., 2008). After adding 20 μg of plasmid DNA in 200 μL of MA-I (Modified Allen's medium-I) to 50 μL of cell suspension, 250 μL of 60% (w/v) polyethylene glycol solution was added to the mixture to make a 30% polyethylene glycol concentration and then incubated at 40°C. After cultivation for 24 h, the cells were collected for RT-PCR and immunofluorescence microscopy. For the RT-PCR assay, extraction of total RNA and cDNA synthesis was performed by conventional methods (Fujiwara et al., 2009). RT-PCR primers for the antisense regions were previously described in antibody generation of VIG1. The primer sequence for the sGFP region was 5′-TAGGTGGCATCGCCCTCGCG-3′. After fixation, the cells transformed with pI050AS-GFP or pVAS-GFP were immunostained with primary mouse anti-GFP monoclonal antibody (Clontech) at a dilution of 1:100, secondary goat anti-mouse IgG conjugated with Alexa 350 (1:1000 dilution), primary rabbit anti-V-ATPase antibodies (1:200 dilution), secondary goat anti-rabbit IgG conjugated with Alexa 555 (1:200 dilution), primary guinea pig anti-VIG1 antibodies (1:200 dilution), and secondary goat anti-guinea pig IgG conjugated with Alexa 488 (1:300 dilution). Measurement of immunostained VIG1 signal was performed by HCImage (Hamamatsu Photonics). The density of the signal intensity in cytosol area was measured in pI050AS-GFP cells and pVAS-GFP cells. Those intensities were normalized with that of nontransformed cells in each experiment.
Accession Numbers
The VIG1 sequence has been submitted to the DNA Data Bank of Japan (http://www.ddbj.nig.ac.jp/index-j.html) under accession number AB332236. The amino acid sequences and CHMP5 were obtained from the NCBI. The accession numbers are as follows: H. sapiens, AAH21168; S. cerevisiae, AAB64922; D. discoideum, XP636927; and Arabidopsis, AAN13183. The genes of C. merolae in this work can be found in the Cyanidioschyzon merolae Genome Project (http://merolae.biol.s.u-tokyo.ac.jp/) under the following accession numbers: catalase, CMI050C; V-ATPase, CMS342C; POR, CMO111C; metallopeptidase, CMP249C; and EF-1a, CMH226C. The microarray data were from the Gene Expression Omnibus (GPL5399; http://www.ncbi.nlm.nih.gov/geo/).
Author Contributions
T.F., O.M., F.Y., and T.K. designed research. T.F., F.Y., M.O., H.K., and T.K. performed research. T.F., M.O., S.W., and K.T. contributed the development of transient gene expression system. T.F., F.Y., K.N., Y.Y., M.Y., H.K., and T.K. performed discussion. T.F. and T.K. wrote the article.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Characterization of the Antisera against POR/CMO111C.
Supplemental Figure 2. Vacuoles Approach the Mitochondrial Surface According to the Progression of Cell Cycle.
Supplemental Figure 3. Expression Pattern of CML153C/vig1 Transcript during Cell Cycles.
Supplemental Figure 4. Schematic Diagram of the Areas of the Vacuolar Membrane Used to Measure Density of Immunogold Particles.
Supplemental Figure 5. Immunoelectron Microscopy for Metallopeptidase.
Supplemental Figure 6. Localization of VIG1 and Vacuoles in Smeared Cells or Isolated Plastids Accompanied by the Vacuoles.
Supplemental Figure 7. RT-PCR Assay for pVAS-GFP Transformant.
Supplemental Figure 8. The Structure of VIG1 and CHMP5 Orthologs.
Supplemental Methods. Real-Time RT-PCR Assay for vig1 during the First Synchronous Cell Division.
Acknowledgments
We thank Yasuo Niwa (University of Shizuoka, Japan) for providing the pTH-2 vector, Kousuke Tashiro (Kyushu University, Fukuoka, Japan) for preparation of the microarray, and Toshiyuki Mori (RIKEN Frontier Research System, Saitama, Japan) and Yu Kanesaki (Tokyo University of Agriculture) for technical advice. This work was supported by grants from the Japan Society for the Promotion of Science Fellowships (5061 to T.F.) and for Scientific Research on Priority Areas (19207004 to T.K.), the Frontier Project “Adaptation and Evolution of Extremophiles” from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and the program for the Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN to T.K.).
References
- Allen M.B. (1959). Studies with Cyanidium caldarium, an anomalously pigmented chlorophyte. Arch. Microbiol. 32: 270–277 [DOI] [PubMed] [Google Scholar]
- Beard M.E., Novikoff A.B. (1969). Distribution of peroxisomes (microbodies) in the nephron of the rat: A cytochemical study. J. Cell Biol. 42: 501–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeland T., Widerberg J., Bakke O., Nordeng T.W. (2001). Mitotic partitioning of endosomes and vacuoles. Curr. Biol. 11: 644–651 [DOI] [PubMed] [Google Scholar]
- Bowers K., Lottridge J., Helliwell S., Goldthwaite L., Luzio J., Stevens T. (2004). Protein–protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic 5: 194–210 [DOI] [PubMed] [Google Scholar]
- Fujiwara T., Misumi O., Tashiro K., Yoshida Y., Nishida K., Yagisawa F., Imamura S., Yoshida M., Mori T., Tanaka K., Kuroiwa H., Kuroiwa T. (2009). Periodic gene expression patterns during the highly synchronized cell nucleus and organelle division cycles in the unicellular red alga Cyanidioschyzon merolae. DNA Res. 16: 59–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futerman A.H., van Meer G. (2004). The cell biology of lysosomal storage disorders. Nat. Rev. Mol. Cell Biol. 7: 554–565 [DOI] [PubMed] [Google Scholar]
- Han B.K., Aramayo R., Polymenis M. (2003). The G1 cyclin Cln3p controls vacuolar biogenesis in Saccharomyces cerevisiae. Genetics 165: 467–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura S., Kanesaki Y., Ohnuma M., Inouye T., Sekine Y., Fujiwara F., Kuroiwa T., Tanaka K. (2009). R2R3-type MYB transcription factor, CmMYB1, is a central nitrogen assimilation regulator in Cyanidioschyzon merolae. Proc. Natl. Acad. Sci. USA (In press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz A., Kinner A., Kölling R. (2001). A family of small coiled-coil-forming proteins functioning at the late endosome in yeast. Mol. Biol. Cell 12: 711–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa T. (1998). The primitive red algae: Cyanidium Caldarium and Cyanidioschyzon merolae as model systems for investigation the dividing apparatus of mitochondria and plastids. Bioessays 20: 344–354 [Google Scholar]
- Kutsuna N., Kumagai F., Sato M.H., Hasezawa S. (2003). Three-dimensional reconstruction of tubular structure of vacuolar membrane throughout mitosis in living Tobacco cells. Plant Cell Physiol. 44: 1045–1054 [DOI] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., Stock J. (1991). Predicting coiled coils from protein sequences. Science 252: 1162–1164 [DOI] [PubMed] [Google Scholar]
- Marty F. (1999). Plant vacuoles. Plant Cell 11: 587–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M., et al. (2004). Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428: 653–665 [DOI] [PubMed] [Google Scholar]
- Misumi O., Matsuzaki M., Nozaki H., Miyagishima S.Y., Mori T., Nishida K., Yagisawa F., Yoshida Y., Kuroiwa H., Kuroiwa T. (2005). Cyanidioschyzon merolae genome. A tool for facilitating comparable studies on organelle biogenesis in photosynthetic eukaryotes. Plant Physiol. 137: 567–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima S., Itoh R., Toda K., Kuroiwa H., Nishimura M., Kuroiwa T. (1999a). Microbody proliferation and segregation cycle in the single-microbody alga Cyanidioschyzon merolae. Planta 208: 326–336 [Google Scholar]
- Miyagishima S., Takahara M., Mori T., Kuroiwa H., Higashiyama T., Kuroiwa T. (2001). Plastid division is driven by a complex mechanism that involves differential transition of the bacterial and eukaryotic division rings. Plant Cell 13: 2257–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima S.Y., Itoh R., Aita S., Kuroiwa H., Kuroiwa T. (1999b). Isolation of dividing chloroplasts with intact plastid-dividing rings from a synchronous culture of the unicellular red alga Cyanidioschyzon merolae. Planta 209: 371–375 [DOI] [PubMed] [Google Scholar]
- Miyagishima S.Y., Nishida K., Mori T., Matsuzaki M., Higashiyama T., Kuroiwa H., Kuroiwa T. (2003). A plant-specific dynamin-related protein forms a ring at the chloroplast division site. Plant Cell 15: 655–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K., Misumi O., Yagisawa F., Kuroiwa H., Nagata T., Kuroiwa T. (2004). Triple immunofluorescent labeling of FtsZ, dynamin, and EF-Tu reveals a loose association between the inner and outer membrane mitochondrial division machinery in the red alga Cyanidioschyzon merolae. J. Histochem. Cytochem. 52: 843–849 [DOI] [PubMed] [Google Scholar]
- Niwa Y., Hirano T., Yoshimoto K., Shimizu M., Kobayashi H. (1999). Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. Plant J. 18: 455–463 [DOI] [PubMed] [Google Scholar]
- Nozaki H., et al. (2007). A 100%-complete sequence reveals unusually simple genomic features in the hot-spring red alga Cyanidioschyzon merolae. BMC Biol. 5: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnuma M., Misumi O., Fujiwara T., Watanabe S., Tanaka K., Kuroiwa T. (2009). Transient gene suppression in a red alga, Cyanidioschyzon merolae 10D. Protoplasma 236: 107–112 [DOI] [PubMed] [Google Scholar]
- Ohnuma M., Yokoyama T., Inouye T., Sekine Y., Tanaka K. (2008). Polyethylene glycol (PEG)-mediated transient gene expression in a red alga, Cyanidioschyzon merolae 10D. Plant Cell Physiol. 49: 117–120 [DOI] [PubMed] [Google Scholar]
- Ohta N., Matsuzaki M., Misumi O., Miyagishima S.Y., Nozaki H., Tanaka K., Shin-I T., Kohara Y., Kuroiwa T. (2003). Complete sequence and analysis of the plastid genome of the unicellular red alga Cyanidioschyzon merolae. DNA Res. 10: 67–77 [DOI] [PubMed] [Google Scholar]
- Ohta N., Sato N., Kuroiwa T. (1998). Structure and organization of the mitochondrial genome of the unicellular red alga Cyanidioschyzon merolae deduced from the complete nucleotide sequence. Nucleic Acids Res. 2: 5190–5198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakajiri T., Asano K., Hirooka S., Tashiro K., Misumi O., Fujiwara T., Kuroiwa T. (2008). Microarray analysis reveals S-adenosylmethionine (SAM) synthetase involvement in salt tolerance of Cyanidioschyzon merolae. Cytologia (Tokyo) 73: 341–368 [Google Scholar]
- Shim J.H., et al. (2006). CHMP5 is essential for late endosome function and down-regulation of receptor signaling during mouse embryogenesis. J. Cell Biol. 172: 1045–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Ehara T., Osafune T., Kuroiwa H., Kawano S., Kuroiwa T. (1994). Behavior of mitochondria, chloroplasts and their nuclei during the mitotic cycle in the ultramicroalga Cyanidioschyzon merolae. Eur. J. Cell Biol. 63: 280–288 [PubMed] [Google Scholar]
- Takahara M., Takahashi H., Matsunaga S., Miyagishima S., Takano H., Sakai A., Kawano S., Kuroiwa T. (2000). A putative mitochondrial ftsZ gene is present in the unicellular primitive red alga Cyanidioschyzon merolae. Mol. Gen. Genet. 264: 452–460 [DOI] [PubMed] [Google Scholar]
- Takahashi H., Takano H., Yokoyama A., Hara Y., Kawano S., Toh-e A., Kuroiwa T. (1995). Isolation, characterization and chromosomal mapping of an actin gene from the primitive red alga Cyanidioschyzon merolae. Curr. Genet. 28: 484–490 [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. (1997). The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisman L.S. (2006). Organelles on the move: Insights from yeast vacuole inheritance. Nat. Rev. Mol. Cell Biol. 7: 243–252 [DOI] [PubMed] [Google Scholar]
- Yagisawa F., Nishida K., Kuroiwa H., Nagata T., Kuroiwa T. (2007). Identification and mitotic partitioning strategies of vacuoles in the unicellular red alga Cyanidioschyzon merolae. Planta 226: 1017–1029 [DOI] [PubMed] [Google Scholar]
- Yagisawa F., Nishida K., Yoshida M., Ohnuma M., Shimada T., Fujiwara T., Yoshida Y., Misumi O., Kuroiwa H., Kuroiwa T. (2009). Identification of novel proteins in isolated polyphosphate vacuoles in the primitive red alga Cyanidioschyzon merolae. Plant J. 60: 882–893 [DOI] [PubMed] [Google Scholar]
- Yoshida Y., Kuroiwa H., Hirooka S., Fujiwara T., Ohnuma M., Yoshida M., Kuroiwa T. (2009). The bacterial ZapA-like protein ZED is required for mitochondrial division. Curr. Biol. 19: 1491–1497 [DOI] [PubMed] [Google Scholar]
- Yoshida Y., Kuroiwa H., Misumi O., Nishida K., Yagisawa F., Fujiwara T., Nanamiya H., Kawamura F., Kuroiwa T. (2006). Isolated chloroplast division machinery can actively constrict after stretching. Science 313: 1435–1438 [DOI] [PubMed] [Google Scholar]
- Yin D., Ji Y. (2002). Genomic analysis using conditional phenotypes generated by antisense RNA. Curr. Opin. Microbiol. 5: 330–333 [DOI] [PubMed] [Google Scholar]