This study sought to identify genes involved in the biosynthesis of the triterpene skeleton in Medicago truncatula using a comprehensive gene expression clustering analysis. The UGT73F3 gene was shown to function as a uridine diphosphate glycosyltransferase in the biosynthesis of triterpene saponins.
Abstract
Saponins, an important group of bioactive plant natural products, are glycosides of triterpenoid or steroidal aglycones (sapogenins). Saponins possess many biological activities, including conferring potential health benefits for humans. However, most of the steps specific for the biosynthesis of triterpene saponins remain uncharacterized at the molecular level. Here, we use comprehensive gene expression clustering analysis to identify candidate genes involved in the elaboration, hydroxylation, and glycosylation of the triterpene skeleton in the model legume Medicago truncatula. Four candidate uridine diphosphate glycosyltransferases were expressed in Escherichia coli, one of which (UGT73F3) showed specificity for multiple sapogenins and was confirmed to glucosylate hederagenin at the C28 position. Genetic loss-of-function studies in M. truncatula confirmed the in vivo function of UGT73F3 in saponin biosynthesis. This report provides a basis for future studies to define genetically the roles of multiple cytochromes P450 and glycosyltransferases in triterpene saponin biosynthesis in Medicago.
INTRODUCTION
Interest in plant natural products has recently increased due to the realization of their importance for animal and human health. Saponins, a group of natural products widespread throughout the plant kingdom, are glycosides of triterpenoid or steroidal aglycones (Abe et al., 1993; Osbourn, 2003; Vincken et al., 2007), and the corresponding aglycones are termed sapogenins. The name saponin is derived from the Latin word “sapo,” indicating that the plant contains a frothing agent when its extract is mixed with water. The foaming ability of saponins is caused by the combination of the hydrophobic sapogenin with the hydrophilic sugar substituent(s).
Triterpenoid saponins are widely found in the Leguminosae (Huhman and Sumner, 2002; Dixon and Sumner, 2003; Suzuki et al., 2005). Their biological activities can positively or negatively impact plant traits (Dixon and Sumner, 2003). For example, saponins confer protective functions to the plant due to their antimicrobial, antifungal, anti-insect, and antipalatability activities (Kendall and Leath, 1976; Tava and Odoardi, 1996; Osbourn, 2003) but can be toxic to monogastric animals and reduce forage digestibility in ruminants (Oleszek, 1996; Small, 1996; Oleszek et al., 1999). They also have potential health benefits for humans, and plants containing saponins have long been used in traditional medicine. Recent studies have illustrated useful pharmacological properties of saponins, including anticholesterolemic and anticancer activities (Waller and Yamasaki, 1996; Behboudi et al., 1999; Haridas et al., 2001; Chen et al., 2005). Saponin-based adjuvants have the unique ability to enhance immunity (Rajput et al., 2007). In addition to cosmetic and pharmaceutical products, saponins have also found wide applications in beverages and confectionery (Price et al., 1987; Petit et al., 1995; Uematsu et al., 2000; Sparg et al., 2004).
Most of the steps in the biosynthesis of triterpene saponins remain uncharacterized at the molecular level (Haralampidis et al., 2002; Dixon and Sumner, 2003). Triterpenoid saponins are synthesized via the isoprenoid pathway by cyclization of 2,3-oxidosqualene to yield the triterpenoid skeleton of β-amyrin (Haralampidis et al., 2002). This step is catalyzed by a specific cyclase, β-amyrin synthase (β-AS), which has been functionally characterized from several plants, including Arabidopsis thaliana (Shibuya et al., 2008), licorice (Glycyrrhiza echinata; Hayashi et al., 2001), oat (Avena sativa; Qi et al., 2004), Saponaria vaccaria (Caryophyllaceae; Meesapyodsuk et al., 2007), garden pea (Pisum sativum; Morita et al., 2000), and the model legume barrel medic (Medicago truncatula; Suzuki et al., 2002). However, little progress has been made in characterization of the enzymes involved in modification of the triterpenoid backbone; these include cytochrome P450-dependent monooxygenases, uridine diphosphate glycosyltransferases (UGTs), and occasionally other enzymes (Haralampidis et al., 2002).
Functional genomics approaches are powerful tools to facilitate the understanding of secondary metabolism in plants. For example, amplified fragment length polymorphism-based transcript profiling in combination with targeted metabolite analysis has been applied for the discovery of genes involved in secondary metabolism in tobacco (Nicotiana tabacum) cells (Goossens et al., 2003), and an integrated approach coupling transcriptome coexpression analysis with reverse genetics has been used for functional identification of members of a multigene family of flavonoid glycosyltransferases in Arabidopsis (Yonekura-Sakakibara et al., 2007). Genes encoding enzymes functioning in secondary metabolism are generally more divergent than those involved in primary metabolism. Although genes for most metabolic pathways in plants are not organized in gene clusters, a small but increasing number of operon-like gene clusters have been identified for synthesis of plant defense compounds (Frey et al., 1997; Qi et al., 2004; Wilderman et al., 2004; Shimura et al., 2007; Field and Osbourn, 2008; Osbourn and Field, 2009). Investigation of the genomic organization of metabolic pathways may therefore be a useful additional approach to ascribing potential function to candidate pathway genes as well as shedding further light on the evolution of chemical diversification in plants.
Root-derived cell suspension cultures of M. truncatula accumulate triterpene saponins after exposure to the wound signal methyl jasmonate (MJ) (Suzuki et al., 2005). Here, we use comprehensive clustering of MJ-induced transcript expression patterns, along with chromosomal location analysis, as tools to understand triterpene saponin biosynthesis, in particular, the potential involvement of specific P450 and UGT genes. As proof of concept, we expressed four candidate M. truncatula UGTs in Escherichia coli, one of which showed specificity for multiple sapogenins in vitro and was confirmed to be involved in saponin biosynthesis in vivo through genetic loss-of-function analysis. Such genes may have potential for improving the quality of forage legumes through metabolic engineering.
RESULTS
Ontology of Differentially Regulated Genes in M. truncatula Cell Cultures
Exposure of M. truncatula cell suspension cultures to MJ results in a strong increase in triterpene saponin levels (Suzuki et al., 2005). Transcriptional changes in response to the pathogen mimic yeast elicitor (YE) occur early (maximal at 2 h after elicitation), whereas the majority of gene expression changes in response to MJ occur later (maximal at 24 h after elicitation) (Suzuki et al., 2005; Naoumkina et al., 2007). Affymetrix microarray analysis was therefore performed to identify transcript changes in elicited cells at these two critical time points, with corresponding controls.
From 50,900 M. truncatula probe sets represented on the Affymetrix array, 7836 passed a statistical significance test (adjusted for false discovery rate) for being differentially expressed in response to YE or MJ. We applied the M. truncatula MedicCyc database (Urbanczyk-Wochniak and Sumner, 2007) to place the distribution of differentially expressed probe sets into functional categories (excluding those with unidentified function). In this database, nonplant pathways are excluded, and plant/legume-specific pathways such as isoflavonoid, triterpene saponin, and lignin are featured in detail. The frequencies of the groups of genes that are over represented in the upregulated and downregulated probe set categories were compared relative to the frequencies at which they occur on the microarray. More probe sets of all functional classes were upregulated at 2 h after YE elicitation, and more probe sets were downregulated at 24 h after MJ elicitation (see Supplemental Figure 1 online). Transcripts encoding genes classified in the secondary metabolism, signaling, miscellaneous enzyme, hormone metabolism, stress, and lipid metabolism categories were overrepresented among the upregulated probe sets, whereas genes classified in cell wall metabolism, signaling, and polyamine metabolism were overrepresented among those downregulated. Notably, the relative abundance of genes classified in secondary metabolism was 5 times higher among the upregulated probe sets than among the downregulated probe sets.
Transcriptional Reprogramming and Genomic Organization of Core Terpene Pathway Genes
Of the more than 30 different triterpene saponins detected in M. truncatula cell suspension cultures exposed to YE or MJ, many are strongly induced only by MJ (Suzuki et al., 2005). Such differential accumulation of saponins was reflected by differences in gene expression levels in the YE and MJ Affymetrix data sets (Figure 1; see Supplemental Data Set 1 online). These changes start with the primary metabolic pathways that feed into triterpene biosynthesis.
Figure 1.
Changes in Transcript Levels of Genes Potentially Involved in Terpenoid Biosynthesis in M. truncatula Cell Cultures.
Green/red color-coded heat maps represent relative transcript levels of different gene family members determined with Affymetrix arrays; red, upregulated; green, downregulated. After exposure to YE for 2 h (A); after exposure to MJ for 24 h (B). Data represent log2 scale ratios of transcript levels in elicited compared with control cells. MapMan (Thimm et al., 2004) visualization software was used to depict transcript levels. HMG-CoA, 3-hydroxy-3-methylglutaryl CoA; MVA, mevalonic acid; MVAP, mevalonic acid 5-phosphate; PMVK, phosphomevalonate kinase; MVD, MVA diphosphate decarboxylase; DMAPP, dimethylallyl diphosphate; GPP, geranyl diphosphate; FPP, farnesyl diphosphate; GGPP, geranylgeranyl diphosphate; SS, squalene synthase; 2,3-OSCs, 2,3-oxidosqualene cyclases; P450, Cytochrome P-450; GTs, glycosyltransferases.
The first steps of the acetate-mevalonate pathway to terpenes lead to the formation of isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (Lange and Ghassemian, 2003) (Figure 1). Thiolase and HMG-CoA synthase probe sets were upregulated 2 to 4 times by MJ, but not by YE. Of the two thiolase genes in the M. truncatula genome, Medtr5g106000 is located on chromosome 5 (see Supplemental Data Set 1 online; Figure 2A). 3-Hydroxy-3-methylglutaryl CoA reductases (HMGRs) are encoded by a multigene family in plants, and five isoforms are found in M. truncatula (Kevei et al., 2007). HMGR isoform 2 is represented by five copies located back-to-back on chromosome 5 (see Supplemental Data Set 1 online; Figure 2A). Four HMGR2b copies are identical, including the intron sequences, whereas HMGR2a has 98% nucleotide identity to these and a different intron sequence.
Figure 2.
Genetic Linkage and Coexpression of Potential Genes of Triterpene Metabolism in M. truncatula.
(A) Map positions of genes potentially involved in isoprenoid-triterpenoid pathways on chromosomes of M. truncatula.
(B) Microarray transcript level profiles of cluster of UGTs on chromosome 2.
(C) Microarray transcript level profiles of genes potentially involved in triterpene metabolism on chromosome 4.
Expression data were obtained from the M. truncatula Gene Expression Atlas database version 2 (MtGEAv2), which combines a large number of publicly available Medicago GeneChip microarrays (156 chips from 64 experiments). References with detailed descriptions of experiments presented in MtGEAv2 are available on the website http://bioinfo.noble.org/gene-atlas/v2/. HMGcAS, 3-hydroxy-3-methylglutaryl CoA synthase; HMGcAR, 3-hydroxy-3-methylglutaryl CoA reductase; MVK, mevalonate kinase; PMVK, phosphomevalonate kinase; IPPI, isopentenyl pyrophosphate isomerase; GGPS, geranylgeranyl pyrophosphate synthase; SSU, small subunit of GGPS; FPS1, farnesyl pyrophosphate synthase 1; SS, squalene synthase; UGT, uridine diphosphate glycosyltransferase. Up- and down-pointing arrows represent forward or reverse orientations on a chromosome, respectively.
HMGR isoforms 1 and 2 have 94% nucleotide sequence identity and are represented by the same probe sets, Mtr.10397.1.S1_at, on the Affymetrix chip. All five isoforms were strongly upregulated by MJ (5- to 30-fold at 24 h), whereas none of them were induced by YE (see Supplemental Data Set 1 online). HMGR isoforms 1 to 4, along with HMG-CoA synthase, are all located on chromosome 5, whereas HMGR isoform 5 is located on chromosome 8 (Figure 2A). HMGR isoforms 1 and 2 show the most similar expression pattern to that of β-AS, the entry point enzyme into the triterpene pathway (see Supplemental Figure 2 online).
A gene encoding mevalonate kinase (found on chromosome 7) was upregulated by MJ but not YE (see Supplemental Data Set 1 online; Figures 1 and 2A). However, phosphomevalonate kinase (on chromosome 3) and mevalonate diphosphate decarboxylase, which together convert phosphomevalonate to IPP, were not significantly upregulated. Microarray analysis also failed to detect significant changes in transcripts encoding IPP isomerase (chromosome 7), which converts IPP into dimethylallyl diphosphate.
The second phase of the terpenoid pathway involves condensation of allylic pyrophosphates with IPP to produce the higher prenyl pyrophosphates (Alonso and Croteau, 1993), which are converted to squalene for biosynythesis of triterpenes and sterols (Bramley, 1997; Figure 1). Two prenyl transferases (FPS1, Medtr2g032930, on chromosome 2; and SSU, Medtr5g100210, on chromosome 5) were upregulated by MJ (see Supplemental Data Set 1 online; Figure 2A). Only one squalene synthase gene, Medtr4g097000 (chromosome 4), was found in the Medicago genome, and this was induced 4.5-fold by MJ at 24 h after elicitation (Figures 1 and 2A; see Supplemental Data Set 1 online). Three squalene epoxidase (SE) genes most likely exist in Medicago; however, only one gene, Medtr4g122000, has been sequenced so far. Two SE genes, including the previously reported SE2 (Suzuki et al., 2002), were induced almost 2- to 3-fold at 24 h after MJ treatment (see Supplemental Data Set 1 online).
Identification and Induction of Medicago 2,3-Oxidosqualene Cyclases
The first committed step in the biosynthesis of triterpene saponins and steroids involves the initial cyclization of 2,3-oxidosqualene by 2,3-oxidosqualene cyclases into one of a number of different potential products (Haralampidis et al., 2002). There are two routes of cyclization of 2,3-oxidosqualene to sapogenins, either via the chair-chair-chair or the chair-boat-chair conformations (Vincken et al., 2007). Sterol cyclization proceeds via the chair-boat-chair conformation (Haralampidis et al., 2002; Vincken et al., 2007; Wang et al., 2008), catalyzed by cycloartenol synthase (CAS) or lanosterol synthase (LS) (see Supplemental Figure 3 online). Initial triterpene cyclization products with the chair-chair-chair conformation may be converted to dammarene-like or pentacyclic triterpenoids, including lupeol, α-amyrin, and β-amyrin (Haralampidis et al., 2002; Vincken et al., 2007). These cyclization events are catalyzed by dammarenediol synthase, lupeol synthase (LuS), α-amyrin synthase, or β-AS, respectively (see Supplemental Figure 3 online).
Eight genes, showing unique expression patterns and similarity to known 2,3-oxidosqualene cyclases, are present in the Medicago genome (see Supplemental Data Set 2 online). The expression of Medtr5g00886, represented by probe set Mtr.38244.1.S1_at, is quite evenly distributed among the tissues, likely reflecting a role in primary metabolism (see Supplemental Figure 4 online). Phylogenetic analysis places this gene into the CAS clade, with the closest homolog (92% amino acid identity) being the CAS from P. sativum (Figure 3).
Figure 3.
Phylogenetic Analysis of OSCs from M. truncatula and Other Plant Species.
GenBank accession numbers are provided on the tree leaves. Panax ginseng dammarenediol synthase represents an outgroup taxon. M. truncatula OSCs are indicated by blue font. Branches of different enzyme classes are colored: green, α/β amyrin synthases; maroon, cycloartenol synthases; sienna, lanosterol synthases; and black, lupeol synthases. A multiple alignment of the deduced amino acid sequences of OSCs and a phylogenetic tree were constructed using the A la Carte mode (Muscle 3.7 for multiple alignment; Gblocks 0.91b for alignment refinement; MrBayes 3.1.2 for phylogeny using maximum likelihood 6 number of substitution types, default substitution model, invariable + γ rates variations, MCMC 10,000 generations; TreeDyn 198.3 for Tree rending) of the Phylogeny.fr program (Dereeper et al., 2008). Posterior probabilities (marked by red font) are indicated near nodes (MrBayes, 10,000 generations). The bar indicates the branch length that corresponds to 0.1 substitutions per position.
TC132384 (genomic sequence not available) was represented by two probe sets, Mtr.38219.1.S1_at and Mtr.4710.1.S1_s_at, showing similar expression patterns with the highest expression level in nodules (shown for Mtr.38219.1.S1_ in Supplemental Figure 4 online). TC132384 corresponds to the gene GB# Y15366 identified in an M. truncatula root nodule cDNA library (Gamas et al., 1996) and annotated as encoding a CAS. However, phylogenetic analysis revealed that this gene belongs to the LuS clade, with highest homology (92%) to the LuS from Lotus japonicus (Figure 3; see Supplemental Data Set 3 online). The function of this gene, which was not induced by either YE or MJ treatments, may be questionable in Medicago species where lupeol derivatives have yet to be detected.
The probe set Mtr.9365.1.S1_at representing Medtr6g099000 showed expression specifically in roots and nodules (see Supplemental Figure 4 online). Phylogenetic analysis placed this gene in the LS clade (Figure 3). LS is conserved among the eudicots; however, its function in plants is still unknown. This gene was not induced in response to YE or MJ.
Oxidosqualene cyclase (Mtr.36453.1.S1_at) showed expression in leaves and petioles (see Supplemental Figure 4 online). This gene is represented by a single EST, and its genomic sequence is not yet available. It was not induced by YE or MJ.
Three genes, Medtr8g018540_50, Medtr8g018580_620, and Medtr8g018630_60, are situated back-to-back on chromosome 8. These genes are specifically expressed in flower tissue and showed similar expression patterns (shown only for Medtr8g018580_620 in Supplemental Figure 4 online). The amino acid sequence of each of them showed from 81 to 85% identity to that of a mixed AS from P. sativum (Morita et al., 2000). Phylogenetic analysis revealed that these genes belong to the α/β-amyrin cluster (Figure 3). None of these genes are expressed in response to either YE or MJ.
Only one β-AS gene, represented by probe set Mtr.18630.1.S1_at, was highly induced by MJ but not by YE (see Supplemental Data Set 1 online). This gene (GenBank accession number CAD23247) was functionally characterized previously by expression in yeast and the recombinant enzyme shown to convert 2,3-oxidosqualene to β-amyrin (Suzuki et al., 2002). It exists as two copies in the M. truncatula genome (Suzuki et al., 2002), although genomic sequence is currently available for only one copy on chromosome 4 (Medtr4g163370).
The M. truncatula β-AS gene clustered in the β-AS clade, with the closest putative ortholog from P. sativum (Figure 3). Mining the Medicago Gene Expression Atlas database (Benedito et al., 2008) showed that β-AS was most highly expressed in seeds at a late developmental stage and in roots (see Supplemental Figure 4 online).
The above studies define the biosynthetic potential of M. truncatula regarding cyclization of oxidosqualene. We next focused our attention on the identification of enzymes potentially involved in the hydroxylations of the carbon skeletons and transfer of sugar moieties to generate the structurally diverse complement of Medicago saponins.
Selection of Candidate Cytochrome P450s
Cytochrome P-450 enzymes are membrane-associated hemoprotein monooxygenases that are involved in a number of biosynthetic and detoxification pathways in plants (Werck-Reichhart et al., 2002). Two main classes of P450s, containing 10 clans and 62 families, have been identified in plants (Li et al., 2007). Recently, 151 putative P450 genes from M. truncatula were classified into nine clans and 44 families (Li et al., 2007). To find potential P450 candidates involved in saponin biosynthesis, we first searched the ongoing M. truncatula genome sequence (http://gbrowse.jcvi.org/cgi-bin/gbrowse/medicago_imgag/) and the probe sets of the Affymetrix Medicago chip (http://www.affymetrix.com). We found 184 P450 genes with probe sets present on the Affymetrix array and 37 probe sets for which genomic sequences are not yet available (some of which may be redundant; see Supplemental Data Set 4 online). Annotation of P450s was based on a domain search of the InterPro and Uniprot databases. The functional prediction of these genes was based on sequence similarity to previously characterized enzymes by BLASTX search of the plant nonredundant database.
Since MJ triggers saponin accumulation in Medicago, we first searched for MJ-inducible P450s; ∼10% of the P450 probe sets were upregulated by MJ (see Supplemental Data Set 4 online). Mtr.8618.1.S1_at (genomic sequence not available) corresponds to TC100810, which shares 90% homology at the amino acid level with β-amyrin and sophoradiol 24-hydroxylase (CYP93E1) from Glycine max (GenBank accession number BAE94181) (Shibuya et al., 2006), and was very strongly upregulated by MJ (532-fold at 24 h). Several highly MJ-induced P450s, such as Medtr4g032760, Medtr4g032910, Mtr.37299.1.S1_at, and Mtr.37298.1.S1_at, showed similarity with the CYP72A family. CYP72A1 from Catharanthus roseus was characterized as a secologanin synthase involved in the biosynthesis of terpene indole alkaloids (Irmler et al., 2000). Mtr.43018.1.S1_at, previously classified as CYP716A12 (GenBank accession number ABC59076; Li et al., 2007), shares 46% amino acid homology with CYP720B1 (GenBank accession number Q50EK6) from loblolly pine (Pinus taeda) an enzyme that catalyzes oxidations of multiple diterpene alcohol and aldehyde intermediates (Ro et al., 2005). Overall, the above similarities to the few functionally characterized P450s of terpenoid metabolism suggest that several of the genes in Supplemental Data Set 4 online are good candidates for involvement in triterpene hydroxylation.
Selection of Candidate UGTs
To date, only two Medicago UGTs with specificity for the triterpene aglycones hederagenin, soyasapogenols B and E, and medicagenic acid have been biochemically characterized (Achnine et al., 2005), and the triterpene-specific UGT74M1 catalyzes C-28 glycosylation for formation of monodesmosides in S. vaccaria (Meesapyodsuk et al., 2007). There are no reliable methods to identify the substrate specificity of plant glycosyltransferases based on sequence similarity alone (Modolo et al., 2007). One hundred sixty-three probe sets (based on domain searching of genome and Affymetrix probe sets) were therefore selected for correlation analysis with candidate P450s and β-AS; ∼15% of the UGTs were highly MJ inducible (see Supplemental Data Set 5 online). Most were annotated as (iso)flavonoid glycosyltransferases. The gene Medtr4g032770, corresponding to the previously reported UGT73K1 with specificity for hederagenin and soyasapogenols B and E (Achnine et al., 2005), was upregulated 359-fold by MJ at 24 h after elicitation.
Cluster Analysis for Gene Function Prediction
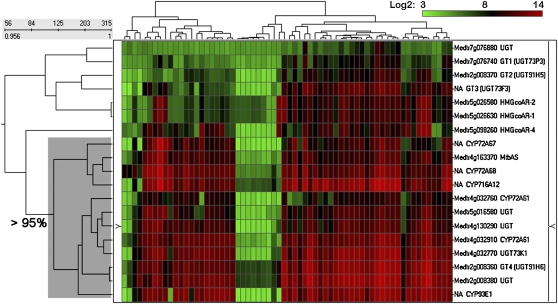
Enzymes involved in the same function will be temporally and spatially coexpressed. In some cases, such coexpression is associated with the operation of metabolic channels (Jorgensen et al., 2005). Based on this principle, we applied clustering analysis to find P450s and UGTs with the most similar expression pattern to β-AS, the entry point enzyme into the triterpene saponin pathway. For clustering analysis, we used expression data obtained from the Medicago truncatula Gene Expression Atlas database version 2 (MtGEAv2), which combines a large number of publicly available Medicago GeneChip microarrays (156 chips from 64 experiments; http://bioinfo.noble.org/gene-atlas/v2/). Signal intensities were converted into log2 scale; data for P450s and UGTs are provided in Supplemental Data Sets 4 and 5 online. Genes that did not show a detectable level of expression were excluded from analysis to reduce noise. Three hundred and fifteen probe sets, including β-AS, the P450s, UGTs, and early pathway enzymes (HMGS, HMGRs, squalene synthases, and SEs), were interrogated by hierarchical cluster analysis based on Pearson's correlation (with ranges from +1 to −1 where +1 is the highest correlation). Profiles with identical shapes have maximum positive correlation. Perfectly mirrored profiles have the maximum negative or inverse correlation.
Since, here, we are interested only in triterpene metabolism, the cluster with similar profiles to β-AS is provided in this study (Figure 4). This cluster includes nine UGTs, six P450s, β-AS, and three HMGR probe sets, which exhibited correlation values of 0.956 or greater.
Figure 4.
The β-AS Cluster: Hierarchical Clustering Analysis of Gene Expression Patterns.
Transcript levels were measured in the different tissues (microarray data were obtained from Atlas database version 2, MtGEAv2, http://bioinfo.noble.org/gene-atlas/v2/). The dendrogram represents hierarchical similarity in microarray-determined transcript profiles of genes potentially involved in triterpene metabolism. The vertical axis of the dendrogram consists of the individual records, and the horizontal axis represents the clustering level. The scale above the row dendrogram is the cluster slider. The numbers above the scale refer to the number of clusters at different positions in the dendrogram. The numbers below the scale refer to the calculated similarity measures. The color scale above the cluster reflects the signal intensity converted to log2. The part of the dendrogram shadowed in gray passed a cluster significance test, using Pvclust analysis (Suzuki and Shimodaira, 2006), with approximately unbiased and bootstrap probability values of >95% (i.e., the hypothesis that “the cluster does not exist” is rejected; P value <0.05).
It has recently been reported that operon-like gene clusters in Arabidopsis and oat are required for triterpene biosynthesis and that such clusters assemble de novo under evolutionary pressure (Field and Osbourn, 2008). To test this concept for triterpene metabolism in M. truncatula, we interrogated the ongoing M. truncatula genome sequence (http://www.tigr.org). Five genes likely associated with triterpene production were located on chromosome 4, including β-AS (Medtr4g163370), two CYP72A61s (Medtr4g032760 and Medtr4g032910), UGT73K1 (Medtr4g032770), and UGT (Medtr4g130290; Figure 2A). Although all these five genes are not tightly linked on chromosome 4, their expression profiles are very similar (Figure 2C) and the two CYP72A61s, Medtr4g032760 and Medtr4g032910, and UGT73K1 (Medtr4g032770) were closely linked. UGT73K1 is known to glycosylate multiple sapogenins (Achnine et al., 2005), and it is tempting to speculate that the linked P450 introduces the hydroxyl group that is subsequently glycosylated. We also found that three UGTs, Medtr2g008360 (GT4), Medtr2g008370 (GT2), and Medtr2g008380, are linked on chromosome 2 (Figure 2A). The expression profiles of these three UGTs are very similar (Figure 2B), and GT4 and Medtr2g008380 share 93% nucleotide sequence identity. Two UGTs found on chromosome 7 (Figure 2A) also showed a similar expression pattern.
Functional Characterization of a Predicted Triterpene UGT in Vitro
To confirm that coexpression analysis can correctly predict genes involved in triterpene saponin biosynthesis, we selected four currently uncharacterized UGT genes from the β-AS expression cluster described above. Although the significance of the clustering had a P value of >0.05 for some of these genes (Figure 4), they were included because none was expressed in a tissue or treatment in which β-AS was not expressed (the significance level is reduced because some of the UGTs are root-specific, whereas saponins, and therefore β-AS expression, are also found in the aerial parts in M. truncatula). We selected UGT candidates over P450s because UGTs are easy to express in E. coli, and it is not possible to predict UGT function based on sequence similarity alone. The selection was mainly based on availability of full-length sequences. The genomic sequence of GT1 (GenBank accession number FJ477889) is available, locus Medtr7g076740 on chromosome 7; GT2 (FJ477890) and GT4 (FJ477892) are from the cluster of three UGTs on chromosome 2 (Figure 2B); the genomic sequence of GT3 (FJ477891) is not yet available, but the corresponding TC94916 represents a full-length sequence. BLAST analysis revealed that all four UGTs contain the conserved PSPG domain characteristic of the nucleotide sugar binding site of small molecule UGTs. The four selected UGTs have low amino acid sequence identity to each other, except for GT2 and GT4, which shared 56% identity. GT1 shared 53% amino acid identity to an (iso)flavonoid UGT from M. truncatula (ABI94026); GT2 and GT4 shared 47 and 43% sequence identity, respectively, to a putative UDP-rhamnose, rhamnosyltransferase from Fragaria x ananassa (AAU09445); and GT3 shared 65% sequence identity to an isoflavonoid UGT from G. echinata (BAC78438). These UGTs have subsequently been assigned as UGT73P3 (GT1), UGT91H5 (GT2), UGT73F3 (GT3), and UGT91H6 (GT4) by the UGT Nomenclature Committee (Mackenzie et al., 1997).
To determine whether the selected UGTs encode functional enzymes, their His-tagged fusion proteins were expressed in E. coli Rosetta 2 (DE3) pLysS cells and affinity purified. An SDS-PAGE gel of purified His-tagged GT3 and GT4 fusion proteins is shown in Supplemental Figure 5 online. The glycosyltransferase activities of the recombinant proteins were first tested using UDP-glucose as the sugar donor and a range of potential acceptor substrates, including hormones, steroids, triterpenoids, and flavonoids (see Supplemental Figure 6 online). This selection was based on the fact that cytokinins (some), gibberellins, abscisic acid, cucurbitanes, brassinosteroids, and triterpenes are products of the terpenoid pathway and that at least one UGT active with triterpenes can also glycosylate flavonoids (Achnine et al., 2005).
GT3 (UGT73F3) showed activity with UDP-glucose as donor and triterpene aglycones or the flavonol kaempferol as sugar acceptors. The other three UGTs did not show activity with any of the tested acceptors using either UDP-glucose, UDP-galactose, or UDP-glucuronic acid as donors. This could be because expression in a prokaryotic system resulted in inactive enzymes, the range of tested sugar donors and acceptors was not extensive enough, or the UGTs function to add additional sugars to a mono- or diglycosidic saponin.
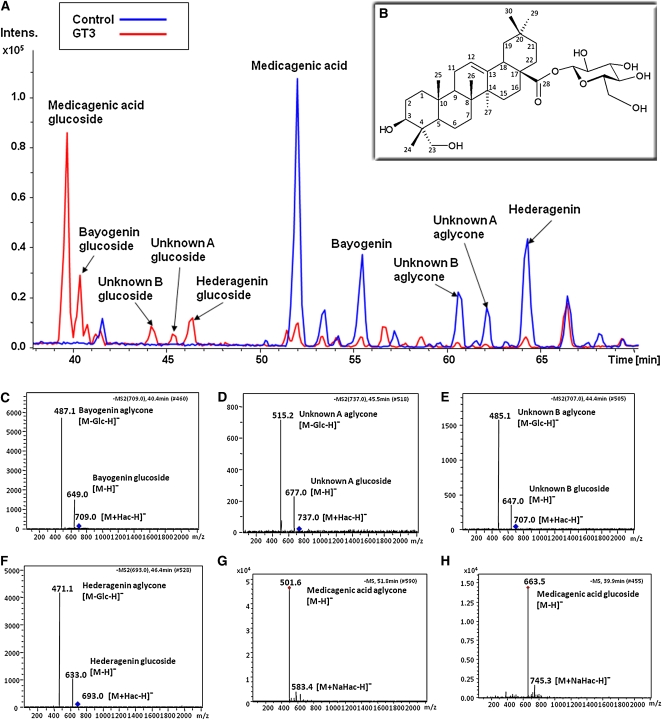
Products from the reaction of UGT73F3 with a crude mixture of sapogenins were analyzed by HPLC–electrospray ionization–mass spectrometry (ESI-MS). Figure 5A shows a selected base peak chromatogram in which the sapogenin substrates are color coded blue and their glucosylated derivatives are color coded red. Seven major peaks were detected in the crude sapogenin extract, three of which were identified as medicagenic acid (MS 502-H), bayogenin (MS 488-H), and hederagenin (MS 472-H). Five compounds were glucosylated by UGT73F3, including the above three (Figure 5). No products were observed with UDP-galactose as sugar donor.
Figure 5.
HPLC-MS Analysis of the Products of GT3 Activity with a Crude Sapogenin Extract from Medicago Roots.
(A) Selected base peak chromatogram of the crude sapogenin extract (blue) and glucosylated products (red).
(B) Structure of hederagenin 28-O-β-d-glucopyranoside.
(C) to (H) Negative-ion HPLC-ESI-MS/MS of sapogenins and their glucosides produced by the action of UGT73F3 on a crude sapogenin extract from M. truncatula: bayogenin glucoside (C), unidentified A glucoside (D), unidentified B glucoside (E), hederagenin glucoside (F), medicagenic acid aglycone (G), and medicagenic acid glucoside (H). Explanation of masses for molecules indicated in square brackets: M, molecule; Glc, β-d-glucopyranosyl; Hac, acetic acid; Na, sodium; and H, hydrogen.
A radioactive assay using UDP-[U-14C] glucose was used to determine the kinetic properties of purified recombinant UGT73F3 with commercially available triterpene sapogenins and kaempferol (Table 1). UGT73F3 was highly efficient with hederagenin as sugar acceptor, exhibiting the highest Kcat/Km ratio and turnover rate (Kcat value). The lowest Km was observed for soyasapogenol A as substrate. However, the turnover of this substrate was quite slow (Table 1). The substrate specificity of UGT73F3 was approximately the same toward soyasapogenols A and B (Table 1). We could not determine the Km value for kaempferol since the saturation curve displayed a sigmoidal rate substrate concentration relationship.
Table 1.
Kinetic Parameters for UGT73F3
| Substrate | Vmax (μmol/min) | Km (μM) | Kcat (s−1) | Kcat/Km (s−1 M−1) |
| Hederagenin | 20.4 ± 0.98 | 220.3 | 0.190 | 862.5 |
| Soyasapogenol B | 1.8 ± 0.23 | 92.7 | 0.017 | 183.4 |
| Soyasapogenol A | 0.8 ± 0.03 | 46.7 | 0.008 | 171.3 |
Errors represent sd from triplicate measurements.
To determine the regiospecificity of UGT73F3, we performed NMR analysis of the glucosylated product of hederagenin. The combination of TOCSY, gCOSY, gHSQC, and gHMBC spectra enabled us to determine all of the proton and carbon chemical shifts of the hederagenin glucoside. The assignment of the peaks is given in Supplemental Tables 1 and 2 online. The proton and carbon chemical shifts of the anomeric position of the glucose residue indicate that it is O-linked to C-28 of hederagenin in an ester linkage (Figure 5B).
Genetic Loss-of-Function Analysis of UGT73F3
To determine whether UGT73F3 functions in triterpene saponin biosynthesis in vivo, we performed loss-of-function genetic analyses by screening pooled DNA from the M. truncatula Tnt1 retrotransposon insertion population (Tadege et al., 2008) with primers specific for UGT73F3. Two mutant lines, NF8981 and NF5746, were isolated. NF8981 was found to have the Tnt1 insertion at position 185 relative to the translation start site of UGT73F3, whereas line NF5746 contained an insertion at position 650 (see Supplemental Figure 7A online). Only two plants germinated from seven seeds of line NF5746, and one of them was confirmed to be heterozygous (see Supplemental Figure 7B online). PCR analysis of genomic DNA of seven plants (R1 progeny) of line NF8981, using combinations of gene-specific primers for UGT73F3 and the Tnt1 retrotransposon, revealed that lines 6 and 7 were homozygous, whereas lines 1 and 3 were heterozygous (see Supplemental Figure 7B online). RT-PCR confirmed no expression of UGT73F3 in homozygous NF8981 lines, while expression levels in heterozygous lines were not significantly different from the wild type (t test, P value >0.14) (see Supplemental Figure 7C online). Flanking sequence tag analysis of two homozygous plants of line NF8981 detected seven Tnt1 retrotransposon insertions in the genome of line 6 and 15 insertions in the genome of line 7 (see Supplemental Data Set 6 online). Five insertions were the same in both lines but did not interrupt any known protein except UGT73F3. BLASTX analysis against the nonredundant protein sequence database revealed that in most cases the Tnt1 retrotransposon incorporated into hypothetical or putative proteins of unknown function.
Unexpectedly, homozygous plants were retarded in growth and never reached the size of normal plants, whereas heterozygous plants did not show any morphological differences compared with the wild type (see Supplemental Figure 7D online). However, the homozygous plants could flower and produce a few pods. The seeds did not show any visible morphological changes but took an unusually long time to germinate (at least 3 weeks). Root growth was severely affected in homozygous lines; roots of these plants were very short and less branched compared with the wild type.
Four plants of one homozygous line, NF8981-6, have survived and show the same dwarf phenotype (Figure 6). Fifty seeds of heterozygous line NF5746 have been screened and two homozygous plants were detected; only one of them survived, and this R2 generation plant showed the same dwarf phenotype (Figure 6).
Figure 6.
Growth Phenotype of UGT73F3 Mutants.
Eight-week-old UGT73F3 tnt1 homozygous mutant lines NF5746 and NF8981 (R2 generation plants) are stunted in growth compared with wild-type M. truncatula (R108 var) plants.
[See online article for color version of this figure.]
Saponin content was evaluated in root and leaf tissue of 8-week-old plants by HPLC-ESI-MS analysis. There were no significant changes in leaf saponin levels between controls and mutants. This was not surprising since the main site of UGT73F3 expression is roots, and the saponins in the aerial parts of M. truncatula are primarily glucuronic acid conjugates (Huhman et al., 2005; Kapusta et al., 2005b).
Roots from each of two individual plants (from four plants in total) of line NF8981 were pooled into one sample to obtain sufficient tissue for saponin analysis. Six different saponins have been identified in M. truncatula root extracts according to previously published work (Huhman and Sumner, 2002; Huhman et al., 2005; Kapusta et al., 2005a, 2005b), and mass data for some are provided in Supplemental Figure 8 online. Levels of Rha-Hex-Hex-Hex-hederagenin, Hex-Hex-Hex-bayogenin, 3-Glc-28-Glc-medicagenic acid, 3-Glc-Ara-28-Glc hederagenin, and Hex-Hex-Hex-soyasapogenol E were significantly (P value <0.05) reduced in mutant lines compared with controls, by around threefold overall (Figures 7A and 7C). Only one saponin, 3-Glc-28-Ara-Rha-Xyl-medicagenic acid, was significantly increased in the mutant lines. Reduction of 3-Glc-28-Glc-medicagenic acid and 3-Glc-Ara-28-Glc hederagenin levels in knockout lines is consistent with the in vitro regiospecificity of UGT73F3 for C-28 and confirms in vivo the involvement of UGT73F3 in glucosylation of multiple sapogenins in Medicago. Although UGT73F3 was active with kaempferol in vitro, neither kaempferol nor its conjugates were detected in M. truncatula roots.
Figure 7.
Targeted Metabolic Profiles of Roots of Wild-Type and Mutant M. truncatula.
(A) and (B) Full scan (A) and selected negative-ion HPLC-ESI-MS chromatograms (B) at m/z 269 of M. truncatula root extract of control line R108-1 (black line) and UGT73F3 tnt1 knockdown line NF8981-1 (gray dashed line). F, formononetin; FG, formononetin 7-O-β-d-glucoside (ononin); FGM, formononetin 7-O-β-d-glucoside-6”-O-malonate; MG, medicarpin 3-O-β-d-glucoside; MGM, medicarpin 3-O-β-d-glucoside-malonate; RHHHHed, Rha-Hex-Hex-Hex-hederagenin; HHHBay, Hex-Hex-Hex-Bayogenin; 3G28GMed, 3-Glc-28-Glc-medicagenic acid; 3G28ARXMed, 3-Glc-28-Ara-Rha-Xyl-medicagenic acid; 3GA28GHed, 3-Glc-Ara-28-Glc hederagenin; and HHHSoyE, Hex-Hex-Hex-Soyasapogenol E. Names of saponins (in [A]) are represented by black font and isoflavonoids by gray font. The inset in (B) shows the positive ion mass spectrum of the MGM peak.
(C) and (D) Relative content of saponins and isoflavonoids, respectively, based on peak areas in roots of control and UGT73F3 tnt1 mutant lines of M. truncatula. The controls represent three independent plants of M. truncatula R108. Root tissues from each of two individual plants of the R2 generation UGT73F3 tnt1 mutant line NF8981 were pooled into two samples, NF8981-1 and NF8981-2. The pooling was necessary due to the limited amount of tissue from the dwarf mutant lines. Only one homozygous plant (R2 generation) was available for line NF5746 (no pooling for this line). Error bars indicate se from three biological replicates. All identified metabolites accumulated significantly differently in controls compared with mutant plants with a P value of <0.05. Identification of saponins and isoflavonoids was based on previously published work (Huhman and Sumner, 2002; Huhman et al., 2005; Kapusta et al., 2005a, 2005b; Farag et al., 2007). Explanations of masses for saponins are provided in Supplemental Figure 8 online.
In 3-Glc-28-Ara-Rha-Xyl-medicagenic acid, the 28-position is substituted with arabinose, and glucosylation occurs only at the C-3 hydroxyl position. The large (10-fold) increase of this compound in UGT73F3 knockout lines suggests that the UDP-glucose pool is being diverted toward increased formation of non-C-28-glucosylated sapogenins. Levels of the isoflavone formononetin and its conjugates were also significantly (P value <0.05) increased in UGT73F3 knockout lines (Figures 7A and 7D).
DISCUSSION
Chromosomal Location and Coexpression of Saponin Biosynthetic Genes in Medicago
The early enzymes common to multiple branches of terpenoid metabolism are often encoded by multigene families in plants. To assess those family members most likely associated with triterpene saponin biosynthesis in Medicago, we compared both chromosomal localization and gene expression pattern for all possible candidates.
HMGR is often regarded as the key regulatory gene in terpenoid metabolism. Five HMGR genes are present in the M. truncatula genome, and the appearance of four identical HMGR2b genes could be due to recent duplication events. The Arabidopsis genome contains only two HMGR genes. The expansion of HMGR genes in the M. truncatula genome might be related to the expansion of triterpene metabolism in this species. Interestingly, several of the genes encoding the early steps in terpene biosynthesis (thiolase, HMG-CoA synthase, most of the HMGRs, and some IPP synthases) are found on chromosome 5. Medicago HMGR1 has been shown to be critical for nodulation (Kevei et al., 2007); since both isoforms 1 and 2 are tightly coexpressed with β-AS, either might be involved in triterpene biosynthesis, although functions for triterpenes in nodulation have not been demonstrated.
A preliminary conclusion from our analyses of both the early and (predicted) late genes of triterpene saponin biosynthesis is that some of the genes might be clustered as a result of gene duplication in M. truncatula but that, overall, these genes are not assembled into operon-like clusters, as has been reported for genes involved in some branches of triterpene biosynthesis in oat and Arabidopsis (Qi et al., 2004; Field and Osbourn, 2008). Chromosome 4 is the most likely site for the genes involved specifically in triterpene biosynthesis in Medicago since, apart from the presence of several of the early enzyme genes on chromosome 5, most of the critical enzymes are found there. These genes are not tightly linked; however, the maintenance of their location on the same chromosome during evolution suggests that there may be some benefit from having these genes physically associated. It is possible that regulatory effects can operate over relatively long distances on this region of chromosome 4.
Medicagenic acid conjugates are major constituents in the aerial parts of M. truncatula, whereas soyasapogenol conjugates are major constituents in roots (Huhman et al., 2005; Kapusta et al., 2005a). However, it is likely that a single β-AS is responsible for production of most triterpene saponins in this species. A truncated copy of a gene with high similarity to β-AS was found on chromosome 8, next to three mixed ASs (see Supplemental Data Set 2 online; Figure 2A); this may explain the two β-AS copies observed on DNA gel blot analysis (Suzuki et al., 2002).
Our analysis of potential OSC genes predicts genes involved in formation of triterpene classes yet to be discovered in Medicago. Compounds with the α-amyrin skeleton have not yet been reported in Medicago species. Further studies specifically targeting α-amyrin–derived saponins in Medicago are clearly warranted, since >90 different triterpene skeletal types can theoretically be generated by cyclization of 2,3-oxidosqualene (Morita et al., 2000).
Determination of Candidate P450s and UGTs for Triterpene Saponin Biosynthesis
Clustering analysis of transcript and metabolite profiles is becoming a powerful technique for identifying candidate genes in complex plant secondary metabolic pathways (Yonekura-Sakakibara et al., 2007, 2008; Saito et al., 2008; Shulaev et al., 2008). In this study, the transcript profiles of a large number of P450 and UGT genes clustered tightly with β-AS in regards to both tissue-specific and elicitor-inducible expression. One of these genes, encoding the glucosyltransfrease UGT73K1 with specificity for hederagenin and soyasapogenols B and E (Achnine et al., 2005), had already been identified as being involved in triterpene saponin biosynthesis, and TC100810 shares 90% amino acid identity with CYP93E1, a β-amyrin and sophoradiol 24-hydroxylase from soybean (Shibuya et al., 2006). These observations, along with the subsequent identification of UGT73F3 as a triterpene UGT, validate the clustering approach in Medicago and suggest that more of the UGT and P450 genes listed in Figures 2 and 4 and Supplemental Table 1 online are strong candidates for involvement in the saponin pathway. Our work therefore provides the basis for future studies to define genetically the roles of P450s and UGTs in triterpene saponin biosynthesis in Medicago.
UGT73F3 Is a Saponin Glycosyltransferase
Five different triterpene aglycones, medicagenic acid, bayogenin, hederagenin, and soyasapogenols B and E, have been determined as base skeletons for >30 M. truncatula saponins described previously (Huhman and Sumner, 2002; Suzuki et al., 2002; Huhman et al., 2005; Kapusta et al., 2005a, 2005b). UGT73F3 showed activity with at least four of these compounds in vitro and also glucosylated the flavonol kaempferol. We would not expect kampferol to be a natural substrate for UGT73F3 in vivo since flavonoid glucoside levels were not increased in response to MJ treatment of Medicago cell cultures (Farag et al., 2008), and kampferol was not detected in roots, the main site of UGT73F3 expression. Similarly, kaempferol is a good substrate for Medicago UGT71G1 in vitro (Shao et al., 2005), although this enzyme is also active with triterpene sapogenins (Achnine et al., 2005).
Typically, glucosyltransferases exhibit substrate regiospecificity rather than absolute specificity for a particular compound in vitro. For example, UGT85B1 from Sorghum bicolor showed a broad activity spectrum in vitro with different families of acceptors (including cyanohydrins, terpenoids, phenolics, and hexanol derivatives) that was influenced by the stereochemistry and/or interactive chemistry of the substituents on the hydroxyl-bearing carbon atom (Hansen et al., 2003). However, this may not reflect the in vivo situation. For example, the glucosyltransferase UGT78G1, which showed a strong preference for isoflavonoid substrates in vitro, appears to function as an anthocyanin glycosyltransferase in Medicago in vivo (Modolo et al., 2007; Peel et al., 2009). Another example is TOGT1 from N. tabacum, which showed activity with salicylic acid in vitro but not in vivo (Chong et al., 2002).
M. truncatula and alfalfa sapogenins are glycosylated at the C-3 and C-28 positions (Huhman and Sumner, 2002; Huhman et al., 2005; Kapusta et al., 2005a, 2005b). Medicagenic acid and bayogenin, like hederagenin, have carboxyl groups at the C-28 position (see Supplemental Figure 5 online), which may therefore be glucose esterified through the action of UGT73F3. NMR analysis indicated that the glucoside produced by the action of UGT73F3 with hederagenin as sugar acceptor was the C-28 ester. It is therefore likely that medicagenic acid and bayogenin are also glycosylated at the 28 position by UGT73F3. Soyasapogenols A, B, and E have no hydroxyl group at C-28; therefore, only the hydroxyl at C-3 is available for glycosylation of these compounds. Since UGT73F3 can glycosylate soyasapogenols A and B, albeit relatively weakly, the enzyme is not strictly regiospecific for triterpene glycosylation.
Phenotypic and Biochemical Effects of Loss of UGT73F3 Function
The reduction in levels of C-28 glycosylated triterpenes in M. trunctula lines harboring a retrotransposon insertion in UGT73F3 is strong evidence in support of a function for this enzyme in saponin glycosylation in vivo. However, other characteristics of these mutant lines require explanation. For example, isoflavone glucosides are among the major secondary metabolites in Medicago roots (Farag et al., 2007), and total isoflavone content was ∼2 times higher in UGT73F3 knockout lines than in corresponding controls. There are two possible interpretations for this observation. First, blocking saponin glycosylation might increase the endogenous pool of the sugar donor UDP-glucose, thus leading to preferential synthesis of other glycosides. Alternatively, accumulation of nonglycosylated sapogenins might be toxic, and the increased isoflavone accumulation might be a nonspecific response to toxic stress (von Rad et al., 2001; Bowles et al., 2005, 2006; Mylona et al., 2008).
The most striking phenotype of the UGT73F3 knockout lines is the strong decrease in plant growth. Although our data do not conclusively prove that this is a direct result of loss of function of UGT73F3, the importance of glycosylation in cell division, growth, and development in animals and plants has been supported by many research reports (Bowles et al., 2005, 2006), some of which emphasize protective functions of glycosylation against toxic compounds, including saponins. Thus, the sad3 and sad4 mutants of oat, deficient in a UGT, accumulate the saponin monodeglucosyl avenacin A-1, which disrupts membrane trafficking and causes degeneration of the epidermis, with consequential effects on root hair formation (Mylona et al., 2008). Loss-of-function mutations in an Arabidopsis UGT required for glucosinolate biosynthesis gave a leaf chlorosis phenotype that has been ascribed to the accumulation of toxic levels of thiohydroximate, the substrate for this enzyme (Grubb et al., 2004). A root-expressed pea UDP-glycosyltransferase, UGT1, that glycosylates flavonoids has been shown to be essential for plant development, possibly via regulation of the cell cycle (Woo et al., 1999).
In this work, we predicted at least nine UGTs, including the previously characterized UGT73K1 (Achnine et al., 2005), that might be involved in triterpene saponin biosynthesis. Assuming that the growth phenotype is indeed the result of loss of function of UGT73F3, it is hard to understand how dysfunction of only one UGT will cause such severe growth phenotypes. Do the various triterpene UGTs perform specific functions in glycosylation of only one or at most a few compounds in vivo, or do they have broader specificity? If the latter, why does redundancy not protect the plant from adverse effects of downregulation of a single enzyme? Either the 28-glycosylation of saponins is especially critical for biological activity, or perhaps UGT73F3 has yet to be discovered functions beyond the triterpene pathway. These questions can only be answered when the remaining enzymes of triterpene substitution have been characterized. This work sets the stage for this endeavor.
METHODS
Plant Material
Details of the initiation and elicitation of Medicago truncatula Gaerth ‘Jemalong’ (line A17) cell suspension cultures have been given previously (Broeckling et al., 2005; Suzuki et al., 2005; Naoumkina et al., 2007; Farag et al., 2008).
M. truncatula R108 tnt1 mutant lines were grown in 6.5-inch-diameter pots containing Professional blend soil (Sun Gro Horticulture) at a temperature of 20°C/19°C (day/night), 16 h/8 h light/dark regime, and 40% relative humidity. Plants were fertilized at the time of watering using a commercial fertilizer mix [Peters Professional 20-10-20 (N-P-K) General Purpose; The Scotts Company].
Chemicals and Biological Materials
UDP-[U-14C] glucose (300 mCi/mmol) was purchased from American Radiolabeled Chemicals. Hederagenin and soyasapogenols A and B were from Chromadex. Auxins (4-chlorophenoxyacetic acid, 2,4-D, and indole-3-acetic acid), cytokinins (kinetin, 2-isopentenyl adenine, and zeatin), gibberellic acid, abscisic acid, campesterol, and kaempferol were from Sigma-Aldrich. Cucurbitacin D was from Extrasynthese (Z.I. Lyon Nord). Medicarpin was extracted and purified from alfalfa roots as described previously (Modolo et al., 2007). Sapogenin extracts for profiling were obtained from M. truncatula (Jemalong, cv A17) and Medicago sativa (cv Radius, Kleszczewska, and Apollo) roots using a solid phase extraction technique described previously (Huhman and Sumner, 2002).
Saponins for profiling were obtained from M. truncatula R108 tnt1 mutant lines by extraction of freeze-dried ground shoots tissue with 80% methanol.
HPLC-ESI-MS Analysis
An Agilent 1100 series II HPLC system (Hewlett-Packard) equipped with a photodiode array detector was coupled to a Bruker Esquire ion-trap mass spectrometer via an ESI source. UV spectra were obtained by scanning from 200 to 600 nm. HPLC separation used a reverse-phase, C18, 5-μm, 4.6 × 250-mm column (J.T. Baker) eluted with 0.1% aqueous acetic acid (eluent A) and acetonitrile (eluent B) with a linear gradient of 5 to 90% B (v/v) over 90 min. The flow rate was 0.8 mL min−1, and the temperature of the column was maintained at 28°C. Negative-ion ESI mass spectra were acquired. Nebulization was aided with a coaxial nitrogen sheath gas provided at a pressure of 60 p.s.i. Desolvation was assisted using a countercurrent nitrogen flow set at a pressure of 12 p.s.i. and a capillary temperature of 300°C. Mass spectra were recorded over the range 50 to 2200 m/z. The Bruker ion-trap mass spectrometer was operated using an ion current control of ∼10,000 with a maximum acquire time of 100 ms. Tandem mass spectra were obtained in manual mode for targeted masses using an isolation width of 2.0, fragmentation amplitude of 2.2, and threshold set at 6000. HPLC-MS data files were analyzed using Bruker Daltonics esquireLC.
DNA Microarray Analysis
RNA samples for analysis using the Affymetrix GeneChip Medicago Genome Array were prepared from cells exposed to YE or MJ for 2 or 24 h, along with the corresponding nonelicited controls. Two biological replicates, with analytical duplicates, were used for minimal statistical treatment, and mean values for each treatment were divided by the corresponding control baseline values. Full details of the experimental procedures have been presented elsewhere (Naoumkina et al., 2007). Differentially expressed genes in treatment/control experiments were selected using associative analysis as described (Dozmorov and Centola, 2003). Type I family-wise error rate was reduced using a Bonferroni-corrected P value threshold of 0.05/N, where N represents the number of probe sets present on the chip. The gene selections were further confirmed by significance analysis of microarrays (Tusher et al., 2001). The false discovery rate was monitored and controlled by calculating the Q value (false discovery rate) using extraction of differential gene expression (http://www.biostat.washington.edu/software/jstorey/edge/) (Storey and Tibshirani, 2003; Leek et al., 2006). The complete Affymetrix data set is publicly available at ArrayExpress (http://www.ebi.ac.uk/arrayexpress; ID = E-MEXP-1092).
Cluster Analysis
Expression data for selected genes were obtained from the Medicago truncatula Gene Expression Atlas database version 2 (MtGEAv2), which combine a large number of publicly available Medicago GeneChip microarrays (156 chips from 64 experiments). References with a detailed description of the experiments presented in MtGEAv2 are available on the website http://bioinfo.noble.org/gene-atlas/v2/. Hierarchical clustering analysis was performed with Spotfire DecisionSite 8.1. Data were transformed to log2 and clustered using Pearson correlation analysis (Zar, 1999). Statistical analysis of uncertainty in hierarchical clustering was performed with Pvclust software (Suzuki and Shimodaira, 2006).
RNA Isolation, Cloning, and Expression of UGTs
Total RNA was isolated from 0.5 g of frozen, ground M. truncatula suspension cells using 5 mL of Tri-Reagent (Molecular Research Center) following the manufacturer's protocol. One microgram of total RNA was used in a first-strand synthesis using SuperScript III reverse transcriptase (Invitrogen) in a 20-μL reaction with oligo(dT) primers according to the manufacturer's protocol. A 2-μL aliquot of the first-strand reaction was then PCR amplified for 30 cycles at 60°C annealing temperature using KOD Hot Start DNA polymerase (EMD Chemicals) according to the manufacturer's protocol.
Candidate UGTs were cloned into Gateway pENTR cassettes (Invitrogen). The inserts were transferred into the destination vector pDEST17 for expression in Escherichia coli using the LR recombination reaction. Primers for Gateway cloning were as follows: GT1, forward 5′-CACCATGGAGTCTCAACAATCCCATAAC-3′, reverse 5′-CTAATCTGCTTTCACACCAAGTGCCTTA-3′; GT2, forward 5′-CACCATGGATAACAAGAAAAACAAACCTCTTCA-3′, reverse 5′-CTATGAATTGTGATTTTGAAGTGAAGAAATGAAGT-3′; GT3, forward 5′-CACCATGGAAGGTGTTGAAGTTGAACAA-3′, reverse 5′-TTAATCATCCAGCTTGAGGTCTCTCAATCT-3′; GT4, forward 5′-CACCATGGGTTCTACTGTTAATGAAGAAGA-3′, reverse 5′-TTAGTTGTTGGAATTGGAAGGAACCCTATAC-3′. E. coli Rosetta 2 (DE3) pLysS cells (Novagen) harboring the expression construct were grown to an OD600 of 0.4 to 0.5, and expression was initiated by addition of isopropyl 1-thio-β-d-galactopyranoside to a final concentration of 0.2 mM, with further incubation with shaking overnight at 16°C. The recombinant proteins were purified using the MagneHis Protein Purification System according to the manufacturer's protocol (Promega).
Screening the M. truncatula Tnt1 Retrotransposon Insertion Population for Identification of UGT73F3 Loss-of-Function Mutants
The M. truncatula R108 Tnt1 population (Tadege et al., 2008) was screened for insertions in the UGT73F3 sequence using the following pairs of primers: GT3F1 forward, 5′-ATGGAAGGTGTTGAAGTTGAACAACC-3′; GT3R1 reverse, 5′-TTAATCATCCAGCTTGAGGTCTCTCA-3′; GT3F2 forward, 5′-TATGTTTGCATCCCGTGGCCAGCAAG-3′; and GT3R2 reverse, 5′-CTCTCGATCTTTTAAGTTCGTCAATC-3′. The line NF8981 was found to have the Tnt1 insertion at position 185 relative to the translation start site of UGT73F3.
Ten seeds of NF8981 were scarified with concentrated sulfuric acid and germinated for 5 d on moist sterile filter paper. Only seven seeds germinated, and they were then planted in soil and screened by PCR for identification of homozygous lines using the gene-specific primers GT3F1-GT3R1 (shown above). To confirm the Tnt1 insertion, plants were screened by PCR using one gene-specific primer, GT3R1, and one retrotransposon-specific primer, Tnt1R, 5′- CAGTGAACGAGCAGAACCTGTG-3′.
RT-PCR
RT-PCR was performed using a Quantum RNA 18S internal standard kit (Ambion) according to the manufacturer's protocol. RNA was isolated from leaf tissue as described above. GT3F1 and GT3R1 primers (sequences shown above) were used to determine UGT73F3 transcript levels in Tnt1 mutant lines. Actin (reference gene) was amplified by the following: actin forward, 5′-GGCTGGATTTGCTGGAGATGATGC-3′; and actin reverse, 5′-CAATTTCTCGCTCTGCTGAGGTGG-3′. Each RT-PCR reaction was repeated three times independently. PCR products were separated in a 1% agarose gel and stained with Syber Green (Invitrogen). The fluorescence signal was captured using a UVP Bioimaging system. Analysis of signal intensity of products was performed with Image Quant TL software (Amersham Biosciences). Transcript abundance was determined by ratio to actin.
Enzyme Assays
Enzyme reactions were performed with 5 μg of enzyme in a total volume of 50 μL containing 50 mM Tris-HCl, pH 9.5, for GT3 (optimum) and pH 7.0 for GT1, GT2, and GT4, 5.0 mM UDP-glucose, UDP-galactose, or UDP-glucuronic acid, and 250 μM acceptor substrate or 2 μg crude sapogenin extract at 30°C for 30 min. Samples were extracted with 250 μL of ethyl acetate, and 225-μL aliquots were taken to dryness using a rotary evaporator, diluted in 50 μL of methanol, and products analyzed by HPLC-ESI-MS as described above.
For kinetic analysis of UGT73F3 (using three analytical replicates), 5 μg of purified enzyme was added to reaction mixtures (50 μL final volume) containing 50 mM Tris-HCl, pH 9.5, 1.7 μM UDP-[U-14C]-glucose (0.3 Ci/mmol), 250 μM UDP-glucose (unlabeled), and 0 to 250 μM acceptor substrate. Reactions were incubated for 30 min at 30°C. Samples were extracted with 250 μL of ethyl acetate, and 200 μL was taken for liquid scintillation counting (Beckman LS6500). Data were analyzed using Hyper32 software (http://www.liv.ac.uk/∼jse/software.html).
NMR Spectroscopy of Hederagenin Glucoside
Hederagenin glucoside was generated by enzymatic reaction with 10 mg of UGT73F3 protein in a total volume 100 mL containing 50 mM Tris-HCl, pH 9.5, 5 mM UDP-glucose, and 220 μM hederagenin overnight at 30°C. The product was extracted with 3 volumes of ethyl acetate, dried under nitrogen, and diluted in 8% methanol. Hederagenin glucoside was purified using C18 SPE cartridges (Waters). The mobile phases consisted of eluent A (0.1% aqueous acetic acid) and eluent B (acetonitrile). The SPE cartridges were equilibrated with three column volumes of 5% B (v/v). The sample (half column volume) was loaded to the SPE cartridge, which was washed with one column volume of 35% B (v/v). Hederagenin glucoside was eluted with one column volume of 45% B (v/v) and dried under a stream of nitrogen. Product (1.3 mg) was collected, deuterium exchanged by lyophilization from D2O, and dissolved in 0.3 mL pyridine-d5. One- and two-dimensional NMR spectra were acquired on a Varian Inova-800 MHz spectrometer at 298K (25°C). Proton chemical shifts were measured relative to the most upfield pyridine-d5 singlet (δH = 7.22 and δC = 123.87 ppm).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers FJ477889 (UGT73P3), FJ477890 (UGT91H5), FJ477891 (UGT73F3), and FJ477892 (UGT91H6). Microarray data are available at ArrayExpresss (http://www.ebi.ac.uk/arrayexpress, ID = E-MEXP-1092).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Functional Categories of Genes That Are Up- or Downregulated in Response to YE or MJ in M. truncatula Suspension Cells.
Supplemental Figure 2. M. truncatula β-AS (Mtr.18630.1.S1_at) and HMGR1-2 (Mtr.1039.1.S1_at) Expression Profiles (Atlas/v2).
Supplemental Figure 3. Cyclization of 2,3-Oxidosqualene to Sterols and Triterpene Saponins.
Supplemental Figure 4. Microarray Analysis of the Tissue-Specific Expression of Medicago 2,3-Oxidosqualene Cyclases.
Supplemental Figure 5. SDS-PAGE Gel of Purified Histidine-Tagged Fusion Proteins Encoded by GT3 (UGT73F3) and GT4 (UGT91H6) Genes.
Supplemental Figure 6. Structures of Substrates Tested with Recombinant Medicago UGT73F3 Expressed in E. coli.
Supplemental Figure 7. Growth Phenotype and PCR/RT-PCR Analysis of Mutants Harboring a Transposon Insertion in UGT73F3.
Supplemental Figure 8. Negative-Ion HPLC/ESI/MS/MS of Saponins Extracted from Roots of M. truncatula Lines Harboring a Transposon Insertion in UGT73F3.
Supplemental Table 1. NMR Chemical Shift Data for the Carbohydrate Portion of the Hederagenin Glycoside.
Supplemental Table 2. NMR Chemical Shift Data for the Aglycone Portion of the Hederagenin Glycoside.
Supplemental Data Set 1. Chromosomal Positions of M. truncatula Genes Involved in Terpenoid Metabolism.
Supplemental Data Set 2. 2,3-Oxidosqualene Cyclase Gene Names and Probe Sets Available on the Medicago Affymetrix Chip.
Supplemental Data Set 3. Amino Acid Sequences of OSCs Used for Phylogenetic Analysis.
Supplemental Data Set 4. Microarray Analysis of M. truncatula P450s.
Supplemental Data Set 5. Microarray Analysis of M. truncatula UGTs.
Supplemental Data Set 6. Flanking Sequence Tags in the Genomes of Two Homozygous Plants (6 and 7) of Line NF8981.
Supplementary Material
Acknowledgments
We thank Lahoucine Achnine (BASF Plant Sciences, Research Triangle Park, NC) and Xiaoqiang Wang (Noble Foundation) for critical reading of the manuscript and Parastoo Azadi (University of Georgia at Athens) for NMR analysis. This work was supported by the National Science Foundation Plant Genome Program Research Award DBI-0109732 and by the Samuel Roberts Noble Foundation. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. NMR analysis of hederagenin glucoside was supported in part by the Department of Energy–funded (DE-FG09-93R-20097) Center for Plant and Microbial Complex Carbohydrates at the University of Georgia.
References
- Abe I., Rohmer M., Prestwich G.D. (1993). Enzymatic cyclization of squalene and oxidosqualene to sterols and triterpenes. Chem. Rev. 93: 2189–2206 [Google Scholar]
- Achnine L., Huhman D.V., Farag M.A., Sumner L.W., Blount J.W., Dixon R.A. (2005). Genomics-based selection and functional characterization of triterpene glycosyltransferases from the model legume Medicago truncatula. Plant J. 41: 875–887 [DOI] [PubMed] [Google Scholar]
- Alonso W.R., Croteau R. (1993). Prenyltransferases and cyclases. Methods in Plant Biochemistry. Enzymes of Secondary Metabolism, Dey P.M., Harborne J.B., (London: Academic Press; ), pp. 239–260 [Google Scholar]
- Behboudi S., Morein B., Villacres Eriksson M.C. (1999). Quillaja saponin formulations that stimulate proinflammatory cytokines elicit a potent acquired cell-mediated immunity. Scand. J. Immunol. 50: 371–377 [DOI] [PubMed] [Google Scholar]
- Benedito V.A., et al. (2008). A gene expression atlas of the model legume Medicago truncatula. Plant J. 55: 504–513 [DOI] [PubMed] [Google Scholar]
- Bowles D., Isayenkova J., Lim E.K., Poppenberger B. (2005). Glycosyltransferases: managers of small molecules. Curr. Opin. Plant Biol. 8: 254–263 [DOI] [PubMed] [Google Scholar]
- Bowles D., Lim E.K., Poppenberger B., Vaistij F.E. (2006). Glycosyltransferases of lipophilic small molecules. Annu. Rev. Plant Biol. 57: 567–597 [DOI] [PubMed] [Google Scholar]
- Bramley P.M. (1997). Isoprenoid metabolism. Plant Biochemistry, Dey P.M., Harborne J.B., (London: Academic Press; ), pp. 417–434 [Google Scholar]
- Broeckling C.D., Huhman D.V., Farag M., Smith J.T., May G.D., Mendes P., Dixon R.A., Sumner L.W. (2005). Metabolic profiling of Medicago truncatula cell cultures reveals effects of biotic and abiotic elicitors on primary metabolism. J. Exp. Bot. 56: 323–336 [DOI] [PubMed] [Google Scholar]
- Chen J.C., Chiu M.H., Nie R.L., Cordell G.A., Qiu S.X. (2005). Cucurbitacins and cucurbitane glycosides: structures and biological activities. Nat. Prod. Rep. 22: 386–399 [DOI] [PubMed] [Google Scholar]
- Chong J., Baltz R., Schmitt C., Beffa R., Fritig B., Saindrenan P. (2002). Downregulation of a pathogen-responsive tobacco UDP-Glc:phenylpropanoid glucosyltransferase reduces scopoletin glucoside accumulation, enhances oxidative stress, and weakens virus resistance. Plant Cell 14: 1093–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.F., Guindon S., Lefort V., Lescot M., Claverie J.M., Gascuel O. (2008). Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36: W465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R.A., Sumner L.W. (2003). Legume natural products. Understanding and manipulating complex pathways for human and animal health. Plant Physiol. 131: 878–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozmorov I., Centola M. (2003). An associative analysis of gene expression array data. Bioinformatics 19: 204–211 [DOI] [PubMed] [Google Scholar]
- Farag M.A., Huhman D.V., Dixon R.A., Sumner L.W. (2008). Metabolomics reveals novel pathways and differential mechanistic and elicitor-specific responses in phenylpropanoid and isoflavonoid biosynthesis in Medicago truncatula cell cultures. Plant Physiol. 146: 387–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag M.A., Huhman D.V., Lei Z., Sumner L.W. (2007). Metabolic profiling and systematic identification of flavonoids and isoflavonoids in roots and cell suspension cultures of Medicago truncatula using HPLC-UV-ESI-MS and GC-MS. Phytochemistry 68: 342–354 [DOI] [PubMed] [Google Scholar]
- Field B., Osbourn A.E. (2008). Metabolic diversification - Independent assembly of operon-like gene clusters in different plants. Science 320: 543–547 [DOI] [PubMed] [Google Scholar]
- Frey M., Chomet P., Glawischnig E., Stettner C., Grun S., Winklmair A., Eisenreich W., Bacher A., Meeley R.B., Briggs S.P., Simcox K., Gierl A. (1997). Analysis of a chemical plant defense mechanism in grasses. Science 277: 696–699 [DOI] [PubMed] [Google Scholar]
- Gamas P., Niebel Fde C., Lescure N., Cullimore J. (1996). Use of a subtractive hybridization approach to identify new Medicago truncatula genes induced during root nodule development. Mol. Plant Microbe Interact. 9: 233–242 [DOI] [PubMed] [Google Scholar]
- Goossens A., Hakkinen S.T., Laakso I., Seppanen-Laakso T., Biondi S., De Sutter V., Lammertyn F., Nuutila A.M., Soderlund H., Zabeau M., Inze D., Oksman-Caldentey K.M. (2003). A functional genomics approach toward the understanding of secondary metabolism in plant cells. Proc. Natl. Acad. Sci. USA 100: 8595–8600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb C.D., Zipp B.J., Ludwig-Muller J., Masuno M.N., Molinski T.F., Abel S. (2004). Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J. 40: 893–908 [DOI] [PubMed] [Google Scholar]
- Hansen K.S., Kristensen C., Tattersall D.B., Jones P.R., Olsen C.E., Bak S., Moller B.L. (2003). The in vitro substrate regiospecificity of recombinant UGT85B1, the cyanohydrin glucosyltransferase from Sorghum bicolor. Phytochemistry 64: 143–151 [DOI] [PubMed] [Google Scholar]
- Haralampidis K., Trojanowska M., Osbourn A.E. (2002). Biosynthesis of triterpenoid saponins in plants. Adv. Biochem. Eng. Biotechnol. 75: 31–49 [DOI] [PubMed] [Google Scholar]
- Haridas V., Higuchi M., Jayatilake G.S., Bailey D., Mujoo K., Blake M.E., Arntzen C.J., Gutterman J.U. (2001). Avicins: Triterpenoid saponins from Acacia victoriae (Bentham) induce apoptosis by mitochondrial perturbation. Proc. Natl. Acad. Sci. USA 98: 5821–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H., Huang P., Kirakosyan A., Inoue K., Hiraoka N., Ikeshiro Y., Kushiro T., Shibuya M., Ebizuka Y. (2001). Cloning and characterization of a cDNA encoding beta-amyrin synthase involved in glycyrrhizin and soyasaponin biosyntheses in licorice. Biol. Pharm. Bull. 24: 912–916 [DOI] [PubMed] [Google Scholar]
- Huhman D.V., Berhow M.A., Sumner L.W. (2005). Quantification of saponins in aerial and subterranean tissues of Medicago truncatula. J. Agric. Food Chem. 53: 1914–1920 [DOI] [PubMed] [Google Scholar]
- Huhman D.V., Sumner L.W. (2002). Metabolic profiling of saponins in Medicago sativa and Medicago truncatula using HPLC coupled to an electrospray ion-trap mass spectrometer. Phytochemistry 59: 347–360 [DOI] [PubMed] [Google Scholar]
- Irmler S., Schroder G., St-Pierre B., Crouch N.P., Hotze M., Schmidt J., Strack D., Matern U., Schroder J. (2000). Indole alkaloid biosynthesis in Catharanthus roseus: New enzyme activities and identification of cytochrome P450 CYP72A1 as secologanin synthase. Plant J. 24: 797–804 [DOI] [PubMed] [Google Scholar]
- Jorgensen K., Rasmussen A.V., Morant M., Nielsen A.H., Bjarnholt N., Zagrobelny M., Bak S., Moller B.L. (2005). Metabolon formation and metabolic channeling in the biosynthesis of plant natural products. Curr. Opin. Plant Biol. 8: 280–291 [DOI] [PubMed] [Google Scholar]
- Kapusta I., Janda B., Stochmal J., Oleszek W. (2005a). Determination of saponins in aerial parts of barrel medic (Medicago truncatula) by liquid chromatography-electrospray ionization/mass spectrometry. J. Agric. Food Chem. 53: 7654–7660 [DOI] [PubMed] [Google Scholar]
- Kapusta I., Stochmal A., Perrone A., Piacente S., Pizza C., Oleszek W. (2005b). Triterpene saponins from barrel medic (Medicago truncatula) aerial parts. J. Agric. Food Chem. 53: 2164–2170 [DOI] [PubMed] [Google Scholar]
- Kendall W.A., Leath K.T. (1976). Effect of saponins on palatability of alfalfa to meadow voles. Agron. J. 68: 473–476 [Google Scholar]
- Kevei Z., et al. (2007). 3-Hydroxy-3-methylglutaryl coenzyme a reductase 1 interacts with NORK and is crucial for nodulation in Medicago truncatula. Plant Cell 19: 3974–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange B.M., Ghassemian M. (2003). Genome organization in Arabidopsis thaliana: A survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Mol. Biol. 51: 925–948 [DOI] [PubMed] [Google Scholar]
- Leek J.T., Monsen E., Dabney A.R., Storey J.D. (2006). EDGE: Extraction and analysis of differential gene expression. Bioinformatics 22: 507–508 [DOI] [PubMed] [Google Scholar]
- Li L., Cheng H., Gai J., Yu D. (2007). Genome-wide identification and characterization of putative cytochrome P450 genes in the model legume Medicago truncatula. Planta 226: 109–123 [DOI] [PubMed] [Google Scholar]
- Mackenzie P.I., et al. (1997). The UDP glycosyltransferase gene superfamily: Recommended nomenclature update based on evolutionary divergence. Pharmacogenetics 7: 255–269 [DOI] [PubMed] [Google Scholar]
- Meesapyodsuk D., Balsevich J., Reed D.W., Covello P.S. (2007). Saponin biosynthesis in Saponaria vaccaria. cDNAs encoding beta-amyrin synthase and a triterpene carboxylic acid glucosyltransferase. Plant Physiol. 143: 959–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modolo L.V., Blount J.W., Achnine L., Naoumkina M.A., Wang X., Dixon R.A. (2007). A functional genomics approach to (iso)flavonoid glycosylation in the model legume Medicago truncatula. Plant Mol. Biol. 64: 499–518 [DOI] [PubMed] [Google Scholar]
- Morita M., Shibuya M., Kushiro T., Masuda K., Ebizuka Y. (2000). Molecular cloning and functional expression of triterpene synthases from pea (Pisum sativum) new alpha-amyrin-producing enzyme is a multifunctional triterpene synthase. Eur. J. Biochem. 267: 3453–3460 [DOI] [PubMed] [Google Scholar]
- Mylona P., Owatworakit A., Papadopoulou K., Jenner H., Qin B., Findlay K., Hill L., Qi X., Bakht S., Melton R., Osbourn A. (2008). Sad3 and sad4 are required for saponin biosynthesis and root development in oat. Plant Cell 20: 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoumkina M., Farag M.A., Sumner L.W., Tang Y., Liu C.J., Dixon R.A. (2007). Different mechanisms for phytoalexin induction by pathogen and wound signals in Medicago truncatula. Proc. Natl. Acad. Sci. USA 104: 17909–17915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleszek W. (1996). Alfalfa Saponins: Structure, Biological Activity, and Chemotaxonomy. (New York: Plenum Press; ). [DOI] [PubMed] [Google Scholar]
- Oleszek W., Junkuszew M., Stochmal A. (1999). Determination and toxicity of saponins from Amaranthus cruentus seeds. J. Agric. Food Chem. 47: 3685–3687 [DOI] [PubMed] [Google Scholar]
- Osbourn A.E. (2003). Saponins in cereals. Phytochemistry 62: 1–4 [DOI] [PubMed] [Google Scholar]
- Osbourn A.E., Field B. (2009). Operons. Cell. Mol. Life Sci. 66: 3755–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel G.J., Pang Y., Modolo L.V., Dixon R.A. (2009). The LAP1 MYB transcription factor orchestrates anthocyanidin biosynthesis and glycosylation in Medicago. Plant J. 59: 136–149 [DOI] [PubMed] [Google Scholar]
- Petit P.R., Sauvaire Y.D., Hillaire-Buys D.M., Leconte O.M., Baissac Y.G., Ponsin G.R., Ribes G.R. (1995). Steroid saponins from fenugreek seeds: Extraction, purification, and pharmacological investigation on feeding behavior and plasma cholesterol. Steroids 60: 674–680 [DOI] [PubMed] [Google Scholar]
- Price K.R., Johnson I.T., Fenwick G.R. (1987). The chemistry and biological significance of saponins in foods and feedstuffs. Crit. Rev. Food Sci. Nutr. 26: 27–135 [DOI] [PubMed] [Google Scholar]
- Qi X., Bakht S., Leggett M., Maxwell C., Melton R., Osbourn A. (2004). A gene cluster for secondary metabolism in oat: implications for the evolution of metabolic diversity in plants. Proc. Natl. Acad. Sci. USA 101: 8233–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajput Z.I., Hu S.H., Xiao C.W., Arijo A.G. (2007). Adjuvant effects of saponins on animal immune responses. J. Zhejiang Univ. Sci. B 8: 153–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro D.K., Arimura G., Lau S.Y., Piers E., Bohlmann J. (2005). Loblolly pine abietadienol/abietadienal oxidase PtAO (CYP720B1) is a multifunctional, multisubstrate cytochrome P450 monooxygenase. Proc. Natl. Acad. Sci. USA 102: 8060–8065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Hirai M.Y., Yonekura-Sakakibara K. (2008). Decoding genes with coexpression networks and metabolomics - 'Majority report by precogs'. Trends Plant Sci. 13: 36–43 [DOI] [PubMed] [Google Scholar]
- Shao H., He X., Achnine L., Blount J.W., Dixon R.A., Wang X. (2005). Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell 17: 3141–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya M., Hoshino M., Katsube Y., Hayashi H., Kushiro T., Ebizuka Y. (2006). Identification of beta-amyrin and sophoradiol 24-hydroxylase by expressed sequence tag mining and functional expression assay. FEBS J. 273: 948–959 [DOI] [PubMed] [Google Scholar]
- Shibuya M., Katsube Y., Otsuka M., Zhang H., Tansakul P., Xiang T., Ebizuka Y. (2008). Identification of a product specific beta-amyrin synthase from Arabidopsis thaliana. Plant Physiol. Biochem. 47: 26–30 [DOI] [PubMed] [Google Scholar]
- Shimura K., et al. (2007). Identification of a biosynthetic gene cluster in rice for momilactones. J. Biol. Chem. 282: 34013–34018 [DOI] [PubMed] [Google Scholar]
- Shulaev V., Cortes D., Miller G., Mittler R. (2008). Metabolomics for plant stress response. Physiol. Plant. 132: 199–208 [DOI] [PubMed] [Google Scholar]
- Small E. (1996). Adaptations to herbivory in alfalfa (Medicago sativa). Can. J. Bot. 74: 807–822 [Google Scholar]
- Sparg S.G., Light M.E., van Staden J. (2004). Biological activities and distribution of plant saponins. J. Ethnopharmacol. 94: 219–243 [DOI] [PubMed] [Google Scholar]
- Storey J.D., Tibshirani R. (2003). Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Achnine L., Xu R., Matsuda S.P., Dixon R.A. (2002). A genomics approach to the early stages of triterpene saponin biosynthesis in Medicago truncatula. Plant J. 32: 1033–1048 [DOI] [PubMed] [Google Scholar]
- Suzuki H., Reddy M.S.S., Naoumkina M., Aziz N., May G.D., Huhman D.V., Sumner L.W., Blount J.W., Mendes P., Dixon R.A. (2005). Methyl jasmonate and yeast elicitor induce differential genetic and metabolic re-programming in cell suspension cultures of the model legume Medicago truncatula. Planta 220: 698–707 [DOI] [PubMed] [Google Scholar]
- Suzuki R., Shimodaira H. (2006). Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22: 1540–1542 [DOI] [PubMed] [Google Scholar]
- Tadege M., Wen J., He J., Tu H., Kwak Y., Eschstruth A., Cayrel A., Endre G., Zhao P.X., Chabaud M., Ratet P., Mysore K.S. (2008). Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 54: 335–347 [DOI] [PubMed] [Google Scholar]
- Tava A., Odoardi M. (1996). Saponins from Medicago Spp.: Chemical characterization and biological activity against insects. Adv. Exp. Med. Biol. 405: 97–109 [DOI] [PubMed] [Google Scholar]
- Thimm O., Blasing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Muller L.A., Rhee S.Y., Stitt M. (2004). MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Tusher V.G., Tibshirani R., Chu G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu Y., Hirata K., Saito K. (2000). Spectrophotometric determination of saponin in Yucca extract used as food additive. J. AOAC Int. 83: 1451–1454 [PubMed] [Google Scholar]
- Urbanczyk-Wochniak E., Sumner L.W. (2007). MedicCyc: A biochemical pathway database for Medicago truncatula. Bioinformatics 23: 1418–1423 [DOI] [PubMed] [Google Scholar]
- Vincken J.-P., Heng L., de Groot A., Gruppen H. (2007). Saponins, classification and occurrence in the plant kingdom. Phytochemistry 68: 275–297 [DOI] [PubMed] [Google Scholar]
- von Rad U., Huttl R., Lottspeich F., Gierl A., Frey M. (2001). Two glucosyltransferases are involved in detoxification of benzoxazinoids in maize. Plant J. 28: 633–642 [DOI] [PubMed] [Google Scholar]
- Waller G.R., Yamasaki K. (1996). Saponins Used in Traditional and Modern Medicine. Advances in Experimental Medicine and Biology. (New York: Plenum Press,). [Google Scholar]
- Wang N., Li Z., Song D., Li W., Fu H., Koike K., Pei Y., Jing Y., Hua H. (2008). Lanostane-type triterpenoids from the roots of Kadsura coccinea. J. Nat. Prod. 71: 990–994 [DOI] [PubMed] [Google Scholar]
- Werck-Reichhart D., Bak S., Paquette S. (2002). Cytochromes P450. The Arabidopsis Book, Somerville C.R., Meyerowitz E.M., (Rockville, MD: American Society of Plant Biologists; ), doi/http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilderman P.R., Xu M., Jin Y., Coates R.M., Peters R.J. (2004). Identification of syn-pimara-7,15-diene synthase reveals functional clustering of terpene synthases involved in rice phytoalexin/allelochemical biosynthesis. Plant Physiol. 135: 2098–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H.H., Orbach M.J., Hirsch A.M., Hawes M.C. (1999). Meristem-localized inducible expression of a UDP-glycosyltransferase gene is essential for growth and development in pea and alfalfa. Plant Cell 11: 2303–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K., Tohge T., Matsuda F., Nakabayashi R., Takayama H., Niida R., Watanabe-Takahashi A., Inoue E., Saito K. (2008). Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell 20: 2160–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K., Tohge T., Niida R., Saito K. (2007). Identification of a flavonol 7-O-rhamnosyltransferase gene determining flavonoid pattern in Arabidopsis by transcriptome coexpression analysis and reverse genetics. J. Biol. Chem. 282: 14932–14941 [DOI] [PubMed] [Google Scholar]
- Zar J.H. (1999). Biostatistical Analysis. (Upper Saddle River, NJ: Prentice Hall; ). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.