Abstract
Background/Aims:
Lp(a) levels have long been recognized as a potential risk factor for coronary heart disease that is almost completely under genetic control. Much of the genetics impacting Lp(a) levels has been attributed to the highly polymorphic LPA kringle IV-2 copy number variant, and most of the variance in Lp(a) levels in populations of European-descent is inversely correlated with kringle IV copy number. However, less of the variance is explained in African-descent populations for the same structural variation. African-descent populations have, on average, higher levels of Lp(a), suggesting other genetic factors contribute to Lp(a) level variability across populations.
Methods
To identify potential cis-acting factors, we re-sequenced the gene LPA for single nucleotide polymorphism (SNP) discovery in 23 European-Americans and 24 African-Americans. We also re-sequenced the neighboring gene plasminogen (PLG) and genotyped the kringle IV copy number variant in the same reference samples.
Results
These data are the most comprehensive description of sequence variation in LPA and its relationship with the kringle IV copy number variant. With these data, we demonstrate that only a fraction of LPA sequence diversity has been previously documented. Also, we identify several high frequency SNPs present in the African-American sample but absent in the European-American sample. Finally, we show that SNPs within PLG are not in linkage disequilibrium with SNPs in LPA, and we show that kringle IV copy number variation is not in linkage disequilibrium with either LPA or PLG SNPs.
Conclusions
Together, these data suggest that LPA SNPs could independently contribute to Lp(a) levels in the general population.
Key Words: African-American, Apo(a), Kringle, Lp(a), Plasminogen, Sequencing, Linkage disequilibrium
Introduction
Lipoprotein (a) [Lp(a)] particles, similar to low density lipoprotein (LDL) particles, contain a lipid core surrounded by unesterified cholesterol, phospholipids, and apolipoprotein B-100 [reviewed in [1]]. The factor distinguishing Lp(a) from LDL is the addition of the glycoprotein apolipoprotein(a), also known as LPA encoded by the gene LPA, linked to apolipoprotein B-100 via a disulfide bond [2]. LPA is expressed in the liver [3], and there is evidence that the assembly of apo(a) and apo B-100 to produce the final Lp(a) particle occurs on the surface of the hepatocytes [4]. Lp(a) levels are thought to be relatively constant throughout a person's lifetime; however, there are reports that Lp(a) levels are correlated with age [5], and intra-individual levels can vary significantly with repeated measurements [6, 7].
A remarkable feature of Lp(a) levels is its inter-individual variability, ranging from barely detectable to >250 nmol/l [8]. LPA is the major determinant of this plasma Lp(a) variability. LPA is structurally homologous to plasminogen [9], and both of their respective genes (LPA and PLG) lie on chromosome 6q26 within approximately 40 kilobases of one another, strongly suggesting that LPA arose from a duplication event. Furthermore, LPA is found only in Old World primates and hedgehog, and genomic evidence suggests this has occurred through convergent evolution [10].
LPA, compared with plasminogen, contains an inactive protease domain and a highly variable number of copies of the kringle IV-2 domain. It is the kringle IV-2 repeat (two exons flanking one intron totaling 5.5 kb per repeat unit) in the LPA gene that gives rise to the polymorphic DNA structure and extreme variability in Lp(a) plasma levels. Estimates suggest that the number of kringle IV-2 repeats alone contained within the Lp(a) complex explains 61–69% of the variability observed in Lp(a) levels for European-descent populations [11, 12]. Studies in non-European-descent populations estimate that the kringle IV-2 repeat explains less of the variance in Lp(a) levels (19–44%) compared with European-descent populations [13,14,15,16,17].
High Lp(a) levels are an independent risk factor for coronary heart disease [18] and future cardiovascular events [19], but not for coronary artery calcification [20]. Of the few studies that included African-descent populations, high Lp(a) levels are inconsistently associated with coronary artery disease risk [21, 22], yet African-descent populations have on average 2–3 times higher levels of Lp(a) compared with European-descent populations [16, 23]. The difference in Lp(a) levels between populations has yet to be explained. For European-descent populations, Lp(a) levels are inversely correlated with LPA kringle IV-2 repeat copy number [16, 24]. This inverse correlation is less striking in African-descent populations [13, 16, 17].
Genetic factors may play a role in the difference observed for Lp(a) levels in European- and African-descent populations. For all populations studied to date, Lp(a) levels are highly heritable [11, 14, 25, 26], and most [11, 14, 26] but not all studies [25] suggest that the LPA locus itself is the major determinant of Lp(a) levels. While the kringle IV-2 repeat polymorphism accounts for over half of the variance in Lp(a) levels, the remaining variance remains unexplained. Current evidence suggests that genetic variations (other than the kringle IV-2 repeat polymorphism) within or closely linked to LPA are responsible for Lp(a) level variation rather than trans-acting factors [27]. Thus, it may be these yet-undescribed cis-acting factors play a substantial role in shaping the Lp(a) trait distribution among human populations.
To begin the process of identifying these cis-acting genetic factors, we genotyped the kringle IV-2 repeat and re-sequenced the LPA and PLG genes in 23 European- and 24 African-American samples of presumably healthy men and women for single nucleotide polymorphism (SNP) discovery. With these data, we are able to describe here for the first time the natural variation contained within LPA as well as linkage disequilibrium across this locus containing the polymorphic kringle IV-2 repeat and across the flanking gene PLG. Collectively, these data and observations lay the foundation for the design and interpretation of the next generation Lp(a) genetic association studies.
Materials and Methods
Sequencing
LPA and PLG were re-sequenced by SeattleSNPs, a member of NHLBI's Program for Genomic Applications. A total of 47 samples from Coriell Cell Repositories were re-sequenced: 23 European-Americans (NA12560, NA12547, NA10845, NA10853, NA10860, NA10830, NA10842, NA10851, NA07349, NA10857, NA10858, NA10848, NA12548, NA10844, NA10854, NA10861, NA10831, NA10843, NA10850, NA07348, NA10852, NA06990, NA07019) and 24 African-Americans (members of the African-American panel of 50: NA17101-NA17116; NA17133-NA17140). The unrelated European-American samples represent 23 DNA samples from the original Centre d’Etude du Polymorphisme Humain (CEPH) reference panel, which consists of 61 large, presumably healthy families from the United States or France ascertained for the purpose of gene mapping [28]. Recent studies have suggested that genetic variation observed in both the CEPH and African-American panel DNA samples is representative of other U.S. populations of European- or African-descent [29,30,31,32,33].
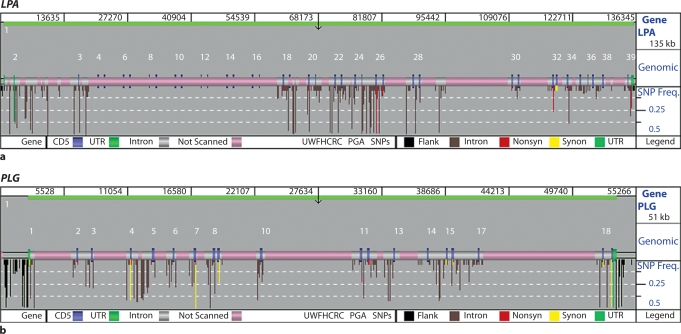
Sequencing was performed on an ABI3730 using standard Big Dye terminator chemistry. We targeted the genomic sequence of LPA for re-sequencing, but excluded the variable kringle regions and other regions because these genomic sequences mapped to several regions of LPA of similar sequence (as was the case for the kringle region) or did not uniquely map to LPA. From a target of 136,345 bp, we were able to re-sequence 56,071 bp (41%) for variation discovery. For PLG, approximately 24.5 kb of sequence was targeted from a total of 55.3 kb of sequence containing the gene. We also targeted the ∼40 kb of sequence between LPA and PLG for variation discovery. All DNA variation and genotype data for LPA and PLG were deposited in GenBank (accession numbers DQ452068 and AY192161, respectively), dbSNP, and the SeattleSNPs website (http://pga.gs.washington.edu). Location, rs numbers (where available), and sequence context of the DNA variations annotated in this discovery effort for LPA and PLG are also given in Supplementary tables 1 and 2 (www.karger.com/doi/10.1159/000143403), respectively. Figure 1 describes the sequence coverage for both LPA (a) and PLG (b) compared with the genomic sequence targeted for variation discovery.
Fig. 1.
A view of sequence coverage of LPA (a) and PLG (b) for genetic variation discovery in 47 DNA samples. Genome sequence targeted for re-sequencing is color-coded so that grey represents introns, blue represents coding sequence, and green represents untranslated regions. Targeted sequence not scanned for variation discovery is represented in pink. SNPs are marked by bars color-coded to represent the location and type of SNP (black for flanking, brown for intronic, red for nonsynymous, yellow for synonymous, and green for untranslated region), and the length of the bar represents the frequency of each SNP in the total population of 47 samples.
Preparation of Genomic DNA for Kringle Genotyping
We genotyped the kringle repeat in the 47 individuals that were re-sequenced for LPA and PLG variation discovery. GM cells were obtained from the Coriell Cell Repository and have the same identification number as the DNA samples listed for re-sequencing with the exception that the initials ‘GM’ denote cell lines while initials ‘NA’ denote DNA samples.
GM cells were propagated in RPMI-1640 with 10% FCS, harvested and washed with Mg2+-free PBS, and mixed with an equal volume of pre-warmed 1% InCert agarose (FMC) at the final cell concentration of 107/ml. The agarose was added to pre-cooled disposable plug molds (BioRad, Hercules, CA) at 100 μl/mold. After gel plugs forming, 1–6 plugs were treated with 1 ml ESP (0.5 M EDTA pH 9.5, 2% SLS, 2 mg/ml Proteinase K) for 48 h at 50°C and with fresh ESP for another 24 h. Plugs were stored in fresh ESP at 4°C.
Southern Blot Hybridization for Kringle Genotyping
DNA plugs were washed with TE buffer (1.8 ml/plug) and 1 mM PMSF for 30 min twice, followed by TE (5 ml/plug) wash for 30 min twice and TE (1 ml/plug) wash once for 30 min. After the TE washes, only 1/2 of a plug from each cell line was used in restriction enzyme digest. DNA plugs were first washed in 150 μl of restriction enzyme reaction buffer (1× NEBuffer 1, New England Biolabs) for 30 min. All washes were carried out at 4°C with slow shaking. DNA plugs were then incubated with 100 μl Kpn I digest solution (1× NEBuffer 1, 1 mg/ml BSA, 40 U Kpn I) at 37°C for 2 h. After digest, DNA plugs were run with size standards (NEB lambda DNA and MidRange I) on 1% agarose gel in the TAE buffer (40 mM Tris-acetate, and 1 mM EDTA, pH 8.0) by pulsed-field gel electrophoresis (PFGE) and the running condition of 6 V/cm, 120°C, 2.9–17.3 s ramping pulse time for 27 h at 14°C. After PFGE, the gels were stained with 0.5 μg/ml of EtBr for 30 min, treated with UV light on the UV transilluminator 2000 (BioRad) for 90 s, followed by denaturing buffer (3 M NaCl and 0.5 N NaOH) for 30 min, neutralizing buffer (0.5 M Tris-HCl pH 7.5, 1.5 M NaCl) for 30 min and 10× SSC (1.5 M NaCl and 0.15 M sodium citrate) for 30 min with shaking. Fractionated DNA was then blotted to the Hybond-N membrane (GE Healthcare, RPN1520N) for 4 h and immobilized by baking at 65°C for 2 h and cross-linking with the UV Stratalinker 2400 (Stratagene) for 30 s. The probe was generated by PCR (primers: 5′-TCCAGCAATTGGCAAATGTA-3′ and 5′-CTGCCCTGAAAAACTTGCTC-3′) that amplifies an 874-bp fragment of the human kringle IV-2 repeats. PCR product was purified from an agarose gel using the QIAquick gel extraction kit (QIAGEN). 200 ng of the probe DNA and 10 ng of lambda DNA were labeled with 32P-dCTP using the Megaprime DNA labeling system (GE Healthcare, RPN1606). The membranes were pre-hybridized with 15 ml hybridization buffer (0.5 M NaHPO4, pH 7.5, 1 mM EDTA, and 7% SDS) for 2 h at 50°C and then incubated with denatured 32P-labeled probe DNA in 25 ml hybridization buffer at 50°C overnight, followed by a series of washes with 100 ml 2× SSC and 1% SDS at room temperature for 15 min, 500 ml 1× SSC and 1% SDS at 58°C for 30 min, and 500 ml 0.5× SSC and 1% SDS at 58°C for 30 min. The membranes were exposed to Kodak X-ray films overnight at −80°C. The X-ray films were developed and the hybridized bands were sized using the Multi-Imager software (BioRad).
Statistical Analysis
Tajima's D was calculated according to Tajima [34]. Pair-wise linkage disequilibrium was calculated for diallelic sites >10% MAF using r2 available on the Genome Variation Server (http://gvs.gs.washington.edu/GVS/). Linkage disequilibrium between diallelic sites and kringle copy number was also calculated for repeat sizes with >10% frequency in the African-American (repeats 10, 11, 14, and 15) and European-Americans (repeats 10, 15, and 16) samples by dichotomizing the repeat allele distribution as either having the repeat or not for each allele. Sequence conservation across species was explored using the Evolutionary Conserved Region (ECR) Browser (http://ecrbrowser.dcode.org/) at default settings and the VISTA Browser (http://genome.lbl.gov/vista/index.shtml).
Results
Kringle Copy Number
The LPA kringle repeat was genotyped in 24 African-American and 23 European-American samples. Among the African-American samples, the mean and median kringle copy number was 13.5 and 20, and the copy number ranged from 6 to 25 repeats. The European-American samples were similar with respect to mean and median copy number (13.6 and 23) and range (5 to 22). The most frequent copy number was 16 for the European-Americans representing 20% of the chromosomes for that sample. For African-Americans, the copy numbers 10 and 14 were equally frequent in that sample each representing 13% of the chromosomes assayed. As expected based on the existing literature [35,36,37], both populations had high heterozygosities for the kringle copy number: 0.90 for the European-American sample and 0.91 for the African-American sample.
Sequence Diversity of LPA
Among the 47 samples re-sequenced, a total of 275 diallelic markers were annotated in 56,071 bp of sequence resulting in an average density of one diallelic marker every 204 bp (table 1a). Only 87 (31.64%) of the markers annotated here have previously been reported in dbSNP (Build 126). This collection of diallelic markers consists of both single nucleotide polymorphisms (95.6%) and insertion-deletion polymorphisms (4.4%). The 12 insertion-deletions annotated with LPA range in size from 1 to 4 bp with a mean (median) size of 2 bp (3 bp). The density of diallelic markers as well as the size distribution of insertion-deletions is consistent with previously described candidate genes [38, 39].
Table 1.
Sequence variation
| Population | S | Common SNPs (MAF >10%) | π (Tajima's D) |
|---|---|---|---|
| aLPA | |||
| African-American (n = 24) | 232 | 78 | 40.74 (−0.8505) |
| European-American (n = 23) | 124 | 71 | 31.07 (0.3466) |
| Total | 275 | 101 | |
| bPLG | |||
| African-American (n = 24) | 173 | 83 | 17.18 (0.26) |
| European-American (n = 23) | 111 | 77 | 14.48 (1.43) |
| Total | 193 | 99 | |
S = Number of diallelic sites; MAF = minor allele frequency.
As expected, the African-American sample had a greater number of diallelic markers (232) compared with the European-American sample (124), and approximately 30% of the markers shared between the two samples (table 1a). Nucleotide diversity (as measured by π) was also higher in the African-American sample compared with the European-American sample (table 1a). Both the African-American and the European-American sample had Tajima's D values consistent with neutrality [40].
In comparing the allele frequencies of the LPA markers between European-American and African-American samples, we find that there are 26 high frequency (MAF >10%) markers in the African-American sample that are monomorphic in the European-American sample (supplementary fig. 1 (www.karger.com/doi/10.1159/000143403). At the extreme, intronic LPA SNPs 76022 (rs7755463) and 79217 (rs10601217) both have a minor allele frequency of 0.38 in the African-American sample and are monomorphic in the European-American sample. Conversely, three LPA markers are common among the European-American sample but monomorphic in the African-American sample. In comparing allele frequencies between the two population samples for 275 LPA markers, 30 SNPs had a MAF difference of ≥17%, and this MAF difference is statistically significant for each SNP comparison at p < 0.01. Two SNPs had a MAF difference of >35% between the two population samples, and this difference is statistically significant at p < 0.0001 for these two SNP comparisons. Overall, the correlation (R2) between European-American and African-American sample allele frequencies at LPA is 0.58, which is higher compared with an earlier estimate of 0.37 in an analysis of 50 candidate genes in the same two population samples [41].
Nineteen coding SNPs were annotated in LPA, 14 of which were nonsynonymous (table 2). Typically, the average candidate gene has an equal [39] or fewer [42] number of nonsynonymous SNPs compared with synonymous SNPs. The overall observed ratio of nonsynonymous to synonymous substitutions in this study was 2.8:1. For African-Americans, the observed ratio was 2.6:1 while the observed ratio in European-Americans was 7:0. If the expectation is that half of the coding SNPs are nonsynonmyous (that is, a ratio of 1:1 for each population sample), the observed ratio of nonsynonymous and synonymous SNPs in the African-American sample is not significantly different from expected (Fisher's exact; p = 0.305). However, for the European-American sample, there is evidence that an excess of nonsynonymous SNPs may exist compared with that expected (Fisher's exact p = 0.059).
Table 2.
Coding variation in LPA
| Site1 (residue) | Nucleotide change (rs number) | Amino acid change | Allele freq. (AA) | Allele freq. (EA) | Predicted effect (PolyPhen) | Predicted effect (SIFT) |
|---|---|---|---|---|---|---|
| 60672 (892) | A > G | Ser to Gly | 0.02 | 0.00 | benign | tolerated |
| 66185 (990) | G > A | Arg to Gln | 0.00 | 0.04 | possibly damaging | intolerant |
| 77653 (1298) | G > C | Trp to Ser | 0.02 | 0.00 | probably damaging | intolerant |
| 80733 (1349) | G > A | Thr to Thr | 0.07 | 0.00 | n/a | n/a |
| 80758 (1358) | C > G2 (rs7765803) | Leu to Val | 0.58 | 0.36 | benign | tolerated |
| 80800 (1372) | C > G3 (rs7765781) | Leu to Val | 0.57 | 0.36 | benign | tolerated |
| 82124 (1399) | A > C4 | Thr to Pro | 0.04 | 0.15 | possibly damaging | intolerant |
| 82191 (1421) | G > A4 | Arg to Gln | 0.02 | 0.02 | benign | tolerated |
| 90066 (1523) | T > C5 | Cys to Cys | 0.02 | 0.00 | n/a | n/a |
| 109817 (1586) | G > A | Val to Ile | 0.03 | 0.00 | benign | tolerated |
| 109854 (1598) | T > C5 | Met to Thr | 0.05 | 0.00 | benign | tolerated |
| 118667 (1679) | C > T6 (rs1801693) | Thr to Met | 0.14 | 0.39 | benign | tolerated |
| 118757 (1709) | T > A | Val to Asp | 0.02 | 0.00 | benign | tolerated |
| 119328 (1719) | C > T | Asp to Asp | 0.09 | 0.00 | n/a | n/a |
| 119421 (1750) | T > C | His to His | 0.02 | 0.00 | n/a | n/a |
| 121754 (1776) | C > T | Asp to Asp | 0.03 | 0.00 | n/a | n/a |
| 121755 (1777) | A > G | Ile to Val | 0.03 | 0.00 | benign | tolerated |
| 124522 (1822) | G > C | Gly to Ala | 0.08 | 0.00 | benign | tolerated |
| 135459 (2016) | C > T | Arg to Cys | 0.18 | 0.25 | probably damaging | intolerant |
AA =African-American; EA = European-American; freq = frequency.
SNPs are numbered based on the GenBank accession number DQ452068.
Kringle IV type 7 polymorphisms originally described by Prins et al. [63].
Kringle IV type 8 polymorphism originally described by Prins et al. [67].
Kringle IV type 8 polymorphism originally described by Ogorelkova et al. [53].
Kringle IV type 9 polymorphisms originally described by Ogorelkova et al. [53].
Linkage Disequilibrium within LPA
To describe linkage disequilibrium (LD) across LPA, we first calculated pair-wise LD (r2) for all SNPs with a MAF >10% for each population sample (table 3). As expected based on population history and demography [43], the European-American sample in general had a greater proportion of SNP comparisons with perfect (approximately 7%) or strong (12%) LD compared with the African-American sample. Likewise, the proportion of SNP comparisons with low LD (24%) was lower than that observed for the African-descent population.
Table 3.
Strength of linkage disequilibrium (r2) for common SNPs (minor allele frequency >10%) in LPA and across LPA/PLG in African-Americans and European-Americans
| Gene(s) | African-Americans (n = 24) |
European-Americans (n = 23) |
||||||
|---|---|---|---|---|---|---|---|---|
| # pair-wise comparisons | perfect LD1 | strong LD2 | weak LD3 | # pair-wise comparisons | perfect LD1 | strong LD2 | weak LD3 | |
| LPA | 2,926 | 6% | 9% | 40% | 2,485 | 7% | 12% | 24% |
| LPA/PLG | 15,225 | 2% | 3% | 7% | 10,878 | 4% | 9% | 35% |
LD = Linkage disequilibrium.
Perfect LD: r2 = 1.0;
strong LD: r2 = 0.80;
weak LD: r2 < 0.10.
Linkage Disequilibrium between LPA and PLG
Plasminogen (PLG) immediately flanks LPA with only ∼40 kb of sequence between the two genes. Because LPA is transcribed in the opposite direction compared with PLG, it is feasible that the two genes share a common regulatory element such as the transcription control region located between PLG and LPA[44,45,46]. Also, given the proximity of the two genes, LPA SNPs may be in strong LD with PLG SNPs, a factor that will affect the interpretation of genetic association studies correlating specific genetic variants to human disease or phenotypes. Because of the potential influence of PLG on LPA association studies, it is useful to characterize the LD structure of the genomic sequence that contains both LPA and PLG. To do this, we first re-sequenced and characterized genetic variation in PLG and the sequence between LPA and PLG among the same samples characterized for LPA (23 European-Americans and 24 African-Americans). We then calculated pair-wise LD (r2) across the region containing both LPA and PLG.
In re-sequencing 24,465 bp of the sequence for PLG, a total of 193 diallelic sites were annotated in the European- and African-American samples combined (table 1b). Like LPA, the African-American sample had more sites (173) compared with the European-American sample (111), and the resulting estimates of nucleotide diversity for both population samples was consistent with neutrality (table 1b). PLG had nearly equal numbers of synonymous (10) and nonsynonymous (7) SNPs in the combined samples, and neither the African-American nor European-American sample had a significant excess of nonsynonymous SNPs (5 and 3, respectively) compared with synonymous SNPs (3 and 8, respectively) in PLG.
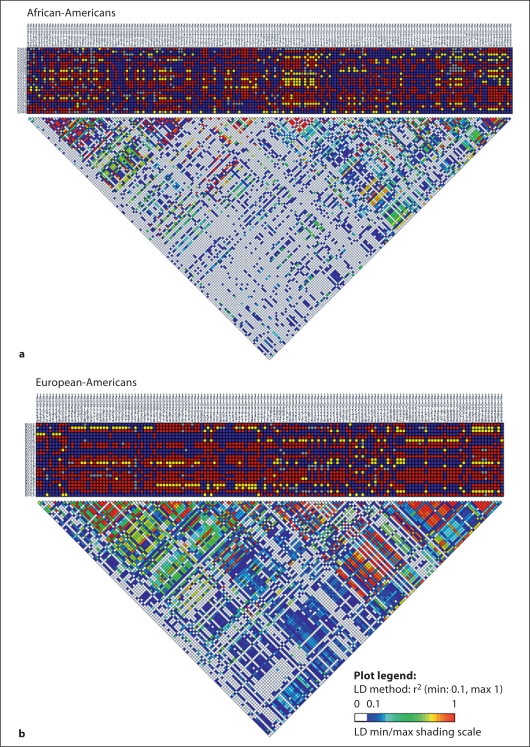
We calculated pair-wise LD (r2) for the combined LPA/PLG datasets for each population sample separately for all common SNPs (MAF >10%; fig. 2 and table 3). For the African-American sample, none of the LPA SNPs were in strong LD with PLG SNPs. Unlike the African-American sample, the European-American sample did have a few LPA SNPs in strong LD with PLG SNPs. More specifically PLG SNPs 1412 (rs4252051) and 1417 (rs4252052) were correlated with or ‘tagged’ PLG SNP 1470 (rs4252053) and LPA SNPs 3264, 17836, 73803, and 82124. Interestingly, PLG SNP 1470 and LPA SNP 82124 were 116 kb apart, and the latter SNP was a nonsynonymous SNP in LPA exon 6 while the former SNP (as well as the other two PLG SNPs) was in the PLG 5′ flanking region that is conserved with rhesus macaque.
Fig. 2.
Pair-wise linkage disequilibrium (r2) calculated across LPA and PLG in two populations. Common SNPs (minor allele frequency > 10%) are numbered across the top of the figure, and samples are numbered to the left side of the figure. SNPs are numbered according to their chromosomal position based on NCBI Build 36. Each square represents the individual's genotype for a specific SNP, and each square is color-coded so that blue represents being homozygous for the common allele, red represents being heterozygous, and yellow represents being homozygous for the rare allele. Gray represents missing data. a African-Americans. b European-Americans
Association between LPA and PLG SNPs and Kringle Copy Number
There is a well-established inverse relationship between the highly variable kringle size polymorphism and Lp(a) levels in humans [47,48,49]. Not so well-established are associations between specific LPA SNPs and Lp(a) levels in humans. For the few reported studies with significant associations between LPA SNPs and Lp(a) levels [50,51,52,53,54] or clinical phenotypes [55,56,57], none has had the complete or near complete LPA sequence data to assess whether or not the genotyped SNP is in LD with kringle IV-2 repeat copy number in their population sample. Thus, it is unclear whether an LPA SNP associated with Lp(a) levels is due the well-established association between the kringle IV-2 repeat polymorphism and Lp(a) levels or a novel SNP association. Because we have both re-sequencing data for SNP discovery and kringle copy number in the same samples, we are able to establish whether or not kringle copy number is associated with specific LPA or PLG SNPs in European- or African-Americans.
Since the kringle polymorphism is multi-allelic, we dichotomized the repeat distribution as having the repeat or not for common repeats (frequency >10%) in either the African-American or European-American samples. In the African-American sample, no kringle repeat was in strong LD (r2 ≥ 0.80) with any LPA or PLG SNPs in this dataset. The highest r2 in African-Americans was 0.57 for kringle repeat 14 and LPA SNP 72373. Similar to the African-American population, no kringle repeat was in strong LD with either LPA or PLG SNPs in the European-American samples. The highest r2 in European-Americans was 0.42 for kringle repeat 10 and LPA SNP 74970. Thus, for either population sample, no single LPA or PLG SNP was in strong LD with specific kringle copy number polymorphisms.
Discussion
We describe here the first in-depth characterization of the natural sequence variation present in the candidate gene LPA and its relationship with the well-known, highly polymorphic kringle IV-2 repeat. Previous reports of SNP discovery in LPA have been published; however, many of these reports describe only a fraction of natural variation in the LPA reference sequence. The majority of reports, in fact, concentrated polymorphism discovery efforts to the promoter/5′ upstream region [58,59,60,61] or to coding regions of LPA[52, 53,62,63,64,65,66,67,68,69,70]. A few SNPs in the intronic or flanking region of LPA have also been described [71].
In re-sequencing 47 presumably healthy individuals (23 European-Americans and 24 African-Americans), we discovered and characterized vastly more LPA variation compared with that published in the current literature or reported in dbSNP (Build 126). Based on previous SNP discovery efforts using the same DNA samples and subsequent large-scale genotyping efforts in various U.S. populations [30,31,32,33], we expect that these results are representative for Americans of European- and African-descent. These data, although unlinked to either Lp(a) levels or cardiovascular phenotypes, are important and necessary for the study of Lp(a)'s role in human disease for several reasons. First, as described above, all published variation discovery efforts for LPA save for two [50, 53] included only European-descent populations. Given the well-known difference in Lp(a) levels between these two populations [16, 23], our data can serve as the foundation in understanding these differences at the DNA sequence level. Indeed, we demonstrate that only 30% of LPA SNPs are shared between the two samples, and we have identified several high-frequency SNPs in the African-American sample that are monomorphic in the European-American sample warranting further study in samples linked to phenotypes. Also, within populations, these data can help determine why LPA kringle IV-2 alleles with the same size are associated with very different Lp(a) levels among unrelated individuals [71, 72].
Second, these data provide the genotype data for LPA and PLG variation necessary for calculating linkage disequilibrium within these candidate genes as well as across the genomic region containing both these genes. Similar to previous reports [37], we do find strong pair-wise linkage disequilibrium for SNPs found on relatively opposite ends of LPA. In general, though, we find that most LPA pair-wise comparisons for either European-American or African-American sample resulted in moderate to weak LD (fig. 1). Also, we found that LPA is not in strong linkage disequilibrium with its neighboring relative, PLG. Finally, contrary to previous reports [64], specific LPA SNPs are not correlated with specific LPA kringle IV-2 copy numbers. Kraft and colleagues [64] presented evidence for linkage disequilibrium between LPA SNP 118667 (Met to Thr) and kringle repeat 18 in a sample of Austrians. In our European-American dataset, kringle repeat 18 occurred at a frequency of only 4% and was excluded from the calculations. LPA SNP 118667 is not in strong linkage disequilibrium with the kringle repeats 10, 15, and 16 in the European-American sample tested here (r2 = 0.10, 0.04, and 0.01, respectively).
The characterization of linkage disequilibrium and the inclusion of the kringle IV-2 copy number variation are necessary for the interpretation of previous LPA genetic association studies. For example, Kraft et al. [54] reported a significant association with the +93 C/T polymorphism (LPA SNP 2995; rs1853021) and Lp(a) concentrations in Africans but not Europeans. The lack of association in the European population has often been interpreted as a consequence of the linkage disequilibrium between +93 C/T polymorphism and intermediate size kringle IV-2 repeats in that population [54]. We offer an alternative explanation: the +93 C/T polymorphism in African-Americans is in linkage disequilibrium (r2 = 1) with the nonsynonymous 77653 (Trp to Ser), among other LPA SNPs, described here (table 2). LPA SNP 77653 was predicted by SIFT and PolyPhen to be intolerant/probably damaging and was not found among European-Americans. In contrast, no nonsynonymous or potentially functional SNPs were in linkage disequilibrium with +93 C/T polymorphism in the European-American sample presented here. Based on these new data, we propose that the association with the +93 C/T polymorphism and Lp(a) levels in Africans may be due to the nonsynonymous LPA SNP 77653. Further LPA SNP discovery efforts in diverse populations will be needed to interpret and design studies that include individuals of other racial/ethnic backgrounds [37, 73].
Despite the amount of sequence represented in this study, a weakness of this approach lies in the fact that we did not attempt to re-sequence the LPA kringle IV-2 repeat region for variation discovery as it was difficult to locate enough unique sequence for successful primer design. Based on previous reports of sequence conservation in European-descent populations [52, 62, 70], we do not expect the lack of coverage in this area to significantly impact our overall analyses and conclusions. Because there are no data available on kringle IV-2 repeat variation in African-descent populations, we cannot predict whether or not the addition of this information would alter our conclusions drawn for the African-American samples here.
Other limitations include the fact that only a small proportion of the sequence scanned for variation discovery includes flanking sequence for either LPA or PLG. Thus, there could be SNPs distal to these re-sequenced flanking regions that are in strong LD with LPA or PLG SNPs or kringle copy numbers. Finally, this study is limited to a small sample of European-Americans and African-Americans. While the study is powered to detect common genetic variation, the study is underpowered to detect SNPs with minor allele frequencies between 1 and 5% [74]. Further re-sequencing in greater numbers of individuals is required to complete the catalogue of rare genetic variations [75, 76].
Despite these limitations, we were able to catalogue common variation in LPA and PLG in the same DNA samples and describe the level of linkage disequilibrium between both of these candidate genes important in cardiovascular research. We also demonstrate that kringle copy number variation in LPA is not in strong linkage disequilibrium with LPA or PLG SNPs, which provides invaluable data for the interpretation of association studies for these candidate genes. Collectively, these data demonstrate that similar to the genome-wide effort of cataloguing and integrating copy-number variation into SNP datasets [77], effort must be made to catalogue and integrate kringle IV-2 repeat polymorphism and sequence variation data for LPA in all populations for future genetic association studies relevant to cardiovascular disease.
Supplementary Material
Comparison of minor allele frequencies for LPA markers between European-American and African-American samples. The numbers on the X-axis represent the minor allele frequency observed in the African-American sample (n=24). The numbers on the Y-axis represent the minor allele frequency observed in the European-American sample (n=23). A total of 275 SNPs, represented by circles, were compared between the two samples. The number of circles on the graph does not equal the number of total LPA SNPs because many SNPs have the same estimated minor allele frequency.
Location, rs number, and sequence context of SNPs in LPA. SNPs are numbered based on the GenBank accession number DQ452068.
Location, rs number, and sequence context of SNPs in PLG. SNPs are numbered based on the GenBank accession number AY192161.
Acknowledgements
We thank Dr. Mark Wurfel and Jeanna Strout (University of Washington) for maintaining the cell lines used in the kringle genotyping. This work was funded by grants from the National Heart, Lung, and Blood Institute's Program for Genomic Applications (U01 HL66682 and U01 HL66728) and the National Institute of Environmental Health Science's Environmental Genome Project (N01 ES15478).
References
- 1.Scanu AM. Lp(a) lipoprotein – coping with heterogeneity. N Engl J Med. 2003;349:2089–2090. doi: 10.1056/NEJMp038128. [DOI] [PubMed] [Google Scholar]
- 2.Koschinsky ML, Cote GP, Gabel B, van der Hoek YY. Identification of the cysteine residue in apolipoprotein(a) that mediates extracellular coupling with apolipoprotein B-100. J Biol Chem. 1993;268:19819–19825. [PubMed] [Google Scholar]
- 3.Kraft HG, Menzel HJ, Hoppichler F, Vogel W, Utermann G. Changes of genetic apolipoprotein phenotypes caused by liver transplantation. Implications for apolipoprotein synthesis. J Clin Invest. 1989;83:137–142. doi: 10.1172/JCI113849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White AL, Lanford RE. Cell surface assembly of lipoprotein(a) in primary cultures of baboon hepatocytes. J Biol Chem. 1994;269:28716–28723. [PubMed] [Google Scholar]
- 5.Rotimi CN, Cooper RS, Marcovina SM, McGee D, Owoaje E, Ladipo M. Serum distribution of lipoprotein(a) in African Americans and Nigerians: Potential evidence for a genotype-environmental effect. Genet Epidemiol. 1997;14:157–168. doi: 10.1002/(SICI)1098-2272(1997)14:2<157::AID-GEPI5>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 6.Marcovina SM, Gaur VP, Albers JJ. Biological variability of cholesterol, triglyceride, low- and high-density lipoprotein cholesterol, lipoprotein(a), and apolipoproteins A- I and B. Clin Chem. 1994;40:574–578. [PubMed] [Google Scholar]
- 7.Cobbaert C, Arentsen JC, Mulder P, Hoogerrugge N, Lindemans J. Significance of various parameters derived from biological variability of lipoprotein(a), homocysteine, cysteine, and total antioxidant status. Clin Chem. 1997;43:1958–1964. [PubMed] [Google Scholar]
- 8.Marcovina SM, Koschinsky ML, Albers JJ, Skarlatos S. Report of the National Heart, Lung, and Blood Institute workshop on lipoprotein(a) and cardiovascular disease: recent advances and future directions. Clin Chem. 2003;49:1785–1796. doi: 10.1373/clinchem.2003.023689. [DOI] [PubMed] [Google Scholar]
- 9.McLean JW, Tomlinson JE, Kuang WJ, Eaton DL, Chen EY, Fless GM, Scanu AM, Lawn RM. cDNA sequence of human apolipoprotein(a) is homologous to plasminogen. Nature. 1987;330:132–137. doi: 10.1038/330132a0. [DOI] [PubMed] [Google Scholar]
- 10.Boffelli D, Cheng J-F, Rubin EM. Convergent evolution in primates and an insectivore. Genomics. 2004;83:19–23. doi: 10.1016/s0888-7543(03)00148-4. [DOI] [PubMed] [Google Scholar]
- 11.Boerwinkle E, Leffert CC, Lin J, Lackner C, Chiesa G, Hobbs HH. Apolipoprotein(a) gene accounts for greater than 90% of the variation in plasma lipoprotein(a) concentrations. J Clin Invest. 1992;90:52–60. doi: 10.1172/JCI115855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boomsma DI, de Knijff P, Labeur C, Martin NG, Havekes LM, Princen HMG. The effect of apolipoprotein(a)-, apolipoprotein E-, and apolipoprotein A4-polymorphisms on quantitative lipoprotein(a) concentrations. Twin Res. 2000;3:152–158. doi: 10.1375/136905200320565436. [DOI] [PubMed] [Google Scholar]
- 13.Kraft HG, Lingenhel A, Pang RW, Delport R, Trommsdorff M, Vermaak H, Janus ED, Utermann G. Frequency distributions of apolipoprotein(a) kringle IV repeat alleles and their effects on lipoprotein(a) levels in Caucasian, Asian, and African populations: The distribution of null alleles is non-random. Eur J Hum Genet. 1996;4:74–87. doi: 10.1159/000472175. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt K, Kraft HG, Parson W, Utermann G. Genetics of the Lp(a)/apo(a) system in an autochthonous Black African population from the Gabon. Eur J Hum Genet. 2006;14:190–201. doi: 10.1038/sj.ejhg.5201512. [DOI] [PubMed] [Google Scholar]
- 15.Ali S, Bunker CH, Aston CE, Ukoli FA, Kamboh MI. Apolipoprotein A kringle 4 polymorphism and serum lipoprotein(a) concentrations in African blacks. Hum Biol. 1998;70:477–490. [PubMed] [Google Scholar]
- 16.Marcovina SM, Albers JJ, Jacobs DR, Jr, Perkins LL, Lewis CE, Howard BV, Savage P. Lipoprotein[a] concentrations and apolipoprotein[a] phenotypes in Caucasians and African Americans. The CARDIA study. Arterioscler Thromb. 1993;13:1037–1045. doi: 10.1161/01.atv.13.7.1037. [DOI] [PubMed] [Google Scholar]
- 17.Sandholzer C, Hallman DM, Saha N, Sigurdsson G, Lackner C, Csaszar A, Boerwinkle E, Utermann G. Effects of the apolipoprotein(a) size polymorphism on the lipoprotein(a) concentration in 7 ethnic groups. Hum Genet. 1991;86:607–614. doi: 10.1007/BF00201550. [DOI] [PubMed] [Google Scholar]
- 18.Danesh J, Collins R, Peto R. Lipoprotein(a) and coronary heart disease. Meta-analysis of prospective studies. Circulation. 2000;102:1082–1085. doi: 10.1161/01.cir.102.10.1082. [DOI] [PubMed] [Google Scholar]
- 19.Danik JS, Rifai N, Buring JE, Ridker PM. Lipoprotein(a), measured with an assay independent of apolipoprotein(a) isoform size, and risk of future cardiovascular events among initially healthy women. JAMA. 2006;296:1363–1370. doi: 10.1001/jama.296.11.1363. [DOI] [PubMed] [Google Scholar]
- 20.Guerra R, Yu Z, Marcovina S, Peshock R, Cohen JC, Hobbs HH. Lipoprotein(a) and apolipoprotein(a) isoforms: no association with coronary artery calcification in the dallas heart study. Circulation. 2005;111:1471–1479. doi: 10.1161/01.CIR.0000159263.50305.BD. [DOI] [PubMed] [Google Scholar]
- 21.Moliterno DJ, Jokinen EV, Miserez AR, Lange RA, Willard JE, Boerwinkle E, Hillis LD, Hobbs HH. No association between plasma lipoprotein(a) concentrations and the presence or absence of coronary atherosclerosis in African-Americans. Arterioscler Thromb Vasc Biol. 1995;15:850–855. doi: 10.1161/01.atv.15.7.850. [DOI] [PubMed] [Google Scholar]
- 22.Paultre F, Pearson TA, Weil HFC, Tuck CH, Myerson M, Rubin J, Francis CK, Marx HF, Philbin EF, Reed RG, Berglund L. High levels of Lp(a) with a small apo(a) isoform are associated with coronary artery disease in African American and white Men. Arterioscler Thromb Vasc Biol. 2000;20:2619–2624. doi: 10.1161/01.atv.20.12.2619. [DOI] [PubMed] [Google Scholar]
- 23.Marcovina SM, Albers JJ, Wijsman E, Zhang ZH, Chapman NH, Kennedy H. Differences in Lp(a) concentrations and apo(a) polymorphs between black and white Americans. J Lipid Res. 1996;37:2569–2585. [PubMed] [Google Scholar]
- 24.Boerwinkle E, Menzel HJ, Kraft HG, Utermann G. Genetics of the quantitative Lp(a) lipoprotein trait. III. Contribution of Lp(a) glycoprotein phenotypes to normal lipid variation. Hum Genet. 1989;82:73–78. doi: 10.1007/BF00288277. [DOI] [PubMed] [Google Scholar]
- 25.Scholz M, Kraft HG, Lingenhel A, Delport R, Vorster EH, Bickeboller H, Utermann G. Genetic control of lipoprotein(a) concentrations is different in Africans and Caucasians. Eur J Hum Genet. 1999;7:169–178. doi: 10.1038/sj.ejhg.5200290. [DOI] [PubMed] [Google Scholar]
- 26.Mooser V, Scheer D, Marcovina SM, Wang J, Guerra R, Cohen J, Hobbs HH. The Apo(a) gene is the major determinant of variation in plasma Lp(a) levels in African Americans. Am J Hum Genet. 1997;61:402–417. doi: 10.1086/514851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barkley RA, Brown AC, Hanis CL, Kardia SL, Turner ST, Boerwinkle E. Lack of genetic linkage evidence for a trans-acting factor having a large effect on plasma lipoprotein[a] levels in African Americans. J Lipid Res. 2003;44:1301–1305. doi: 10.1194/jlr.M300163-JLR200. [DOI] [PubMed] [Google Scholar]
- 28.Dausset J, Cann H, Cohen D, Lathrop M, Lalouel JM, White R. Centre d'etude du polymorphisme humain (CEPH): collaborative genetic mapping of the human genome. Genomics. 1990;6:575–577. doi: 10.1016/0888-7543(90)90491-c. [DOI] [PubMed] [Google Scholar]
- 29.Meucci MA, Marsh S, Watters JW, McLeod HL. CEPH individuals are representative of the European American population: implications for pharmacogenetics. Pharmacogenomics. 2005;6:59–63. doi: 10.1517/14622416.6.1.59. [DOI] [PubMed] [Google Scholar]
- 30.Reiner AP, Carlson CS, Rieder MJ, Siscovick DS, Liu K, Chandler WL, Green D, Schwartz SM, Nickerson DA. Coagulation factor VII gene haplotypes, obesity-related traits, and cardiovascular risk in young women. J Thromb Haemost. 2007;5:42–49. doi: 10.1111/j.1538-7836.2006.02279.x. [DOI] [PubMed] [Google Scholar]
- 31.Reiner AP, Carty CL, Carlson CS, Wan JY, Rieder MJ, Smith JD, Rice K, Fornage M, Jaquish CE, Williams OD, Tracy RP, Lewis CE, Siscovick DS, Boerwinkle E, Nickerson DA. Association between patterns of nucleotide variation across the three fibrinogen genes and plasma fibrinogen levels: the Coronary Artery Risk Development in Young Adults (CARDIA) study. J Thromb Haemost. 2006;4:1279–1287. doi: 10.1111/j.1538-7836.2006.01907.x. [DOI] [PubMed] [Google Scholar]
- 32.Carlson CS, Aldred SF, Lee PK, Tracy RP, Schwartz SM, Rieder M, Liu K, Williams OD, Iribarren C, Lewis EC, Fornage M, Boerwinkle E, Gross M, Jaquish C, Nickerson DA, Myers RM, Siscovick DS, Reiner AP. Polymorphisms within the C-reactive protein (CRP) promoter region are associated with plasma CRP levels. Am J Hum Genet. 2005;77:64–77. doi: 10.1086/431366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crawford DC, Sanders CL, Qin X, Smith JD, Shephard C, Wong M, Witrak L, Rieder MJ, Nickerson DA. Genetic variation is associated with C-reactive protein levels in the Third National Health and Nutrition Examination Survey. Circulation 12–5–. 2006;114:2458–2465. doi: 10.1161/CIRCULATIONAHA.106.615740. [DOI] [PubMed] [Google Scholar]
- 34.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lackner C, Boerwinkle E, Leffert CC, Rahmig T, Hobbs HH. Molecular basis of apolipoprotein(a) isoform size heterogeneity as revealed by pulsed-field gel electrophoresis. J Clin Invest. 1991;87:2077–2086. doi: 10.1172/JCI115248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaw A, Boerwinkle E, Cohen JC, Hobbs HH. Comparative analysis of the Apo(a) gene, apo(a) glycoprotein, and plasma concentration of Lp(a) in three ethnic groups. J Clin Invest. 1994;93:2526–2534. doi: 10.1172/JCI117263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puckey LH, Lawn RM, Knight BL. Polymorphisms in the apolipoprotein(a) gene and their relationship to allele size and plasma lipoprotein(a) concentration. Hum Mol Genet. 1997;6:1099–1107. doi: 10.1093/hmg/6.7.1099. [DOI] [PubMed] [Google Scholar]
- 38.Bhangale TR, Rieder MJ, Livingston R, Nickerson DA. Comprehensive identification and characterization of diallelic insertion-deletion polymorphisms in 330 human candidate genes. Hum Mol Genet. 2005;14:59. doi: 10.1093/hmg/ddi006. [DOI] [PubMed] [Google Scholar]
- 39.Crawford DC, Akey DT, Nickerson DA. The patterns of natural variation in human genes. Annu Rev Genomics Hum Genet. 2005;6:287–312. doi: 10.1146/annurev.genom.6.080604.162309. [DOI] [PubMed] [Google Scholar]
- 40.Biswas S, Akey JM. Genomic insights into positive selection. Trends Genet. 2006;22:437–446. doi: 10.1016/j.tig.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 41.Carlson CS, Eberle MA, Rieder MJ, Smith JD, Kruglyak L. Additional SNPs and linkage-disequilibrium analyses are necessary for whole-genome association studies in humans. Nat Genet. 2003;33:518. doi: 10.1038/ng1128. [DOI] [PubMed] [Google Scholar]
- 42.Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- 43.Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC. Linkage disequilibrium in the human genome. Nature. 2001;411:199. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- 44.Yang Z, Boffelli D, Boonmark N, Schwartz K, Lawn R. Apolipoprotein(a) gene enhancer resides within a LINE element. J Biol Chem. 1998;273:891–897. doi: 10.1074/jbc.273.2.891. [DOI] [PubMed] [Google Scholar]
- 45.Huby T, Afzal V, Doucet C, Lawn RM, Gong EL, Chapman MJ, Thillet J, Rubin EM. Regulation of the expression of the apolipoprotein(a) gene: Evidence for a regulatory role of the 5′ distal apolipoprotein(a) transcription control region enhancer in yeast artificial chromosome transgenic mice. Arterioscler Thromb Vasc Biol. 2003;23:1633–1639. doi: 10.1161/01.ATV.0000084637.01883.CA. [DOI] [PubMed] [Google Scholar]
- 46.Wade DP, Puckey LH, Knight BL, Acquati F, Mihalich A, Taramelli R. Characterization of multiple enhancer regions upstream of the apolipoprotein(a) gene. J Biol Chem. 1997;272:30387–30399. doi: 10.1074/jbc.272.48.30387. [DOI] [PubMed] [Google Scholar]
- 47.Kraft HG, Kochl S, Menzel HJ, Sandholzer C, Utermann G. The apolipoprotein(a) gene: A transcribed hypervariable locus controlling plasma lipoprotein(a) concentration. Hum Genet. 1992;90:220–230. doi: 10.1007/BF00220066. [DOI] [PubMed] [Google Scholar]
- 48.Utermann G, Menzel HJ, Kraft HG, Duba HC, Kemmler HG, Seitz C. Lp(a) glycoprotein phenotypes. Inheritance and relation to Lp(a)-lipoprotein concentrations in plasma. J Clin Invest. 1987;80:458–465. doi: 10.1172/JCI113093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gavish D, Azrolan N, Breslow JL. Plasma Lp(a) concentration is inversely correlated with the ratio of kringle IV/kringle V encoding domains in the apo(a) gene. J Clin Invest. 1989;84:2021–2027. doi: 10.1172/JCI114395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chretien JP, Coresh J, Berthier-Schaad Y, Kao WH, Fink NE, Klag MJ, Marcovina SM, Giaculli F, Smith MW. Three single-nucleotide polymorphisms in LPA account for most of the increase in lipoprotein(a) level elevation in African Americans compared with European Americans. J Med Genet. 2006;43:917–923. doi: 10.1136/jmg.2006.042119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suzuki K, Kuriyama M, Saito T, Ichinose A. Plasma lipoprotein(a) levels and expression of the apolipoprotein(a) gene are dependent on the nucleotide polymorphisms in its 5′-flanking region. J Clin Invest. 1997;99:1361–1366. doi: 10.1172/JCI119295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mancini FP, Mooser V, Guerra R, Hobbs HH. Sequence microheterogeneity in apolipoprotein(a) gene repeats and the relationship to plasma Lp(a) levels. Hum Mol Genet. 1995;4:1535–1542. doi: 10.1093/hmg/4.9.1535. [DOI] [PubMed] [Google Scholar]
- 53.Ogorelkova M, Kraft HG, Ehnholm C, Utermann G. Single nucleotide polymorphisms in exons of the apo(a) kringles IV types 6 to 10 domain affect Lp(a) plasma concentrations and have different patterns in Africans and Caucasians. Hum Mol Genet. 2001;10:815–824. doi: 10.1093/hmg/10.8.815. [DOI] [PubMed] [Google Scholar]
- 54.Kraft HG, Windegger M, Menzel HJ, Utermann G. Significant impact of the +93 C/T polymorphism in the apolipoprotein(a) gene on Lp(a) concentrations in Africans but not in Caucasians: confounding effect of linkage disequilibrium. Hum Mol Genet. 1998;7:257–264. doi: 10.1093/hmg/7.2.257. [DOI] [PubMed] [Google Scholar]
- 55.Ichinose A, Kuriyama M. Detection of polymorphisms in the 5′-flanking region of the gene for apolipoprotein(a) Biochem Biophys Res Comm. 1995;209:372–378. doi: 10.1006/bbrc.1995.1513. [DOI] [PubMed] [Google Scholar]
- 56.Luke MM, Kane JP, Liu DM, Rowland CM, Shiffman D, Cassano J, Catanese JJ, Pullinger CR, Leong DU, Arellano AR, Tong CH, Movsesyan I, Naya-Vigne J, Noordhof C, Feric NT, Malloy MJ, Topol EJ, Koschinsky ML, Devlin JJ, Ellis SG. A Polymorphism in the protease-like domain of apolipoprotein(a) is associated with severe coronary artery disease. Arterioscler Thromb Vasc Biol. 2007;27:2030–2036. doi: 10.1161/ATVBAHA.107.141291. [DOI] [PubMed] [Google Scholar]
- 57.Simo JM, Joven J, Vilella E, Ribas M, Figuera L, Virgos C, Sundaram IM, Hoover-Plow J. Polymorphisms in human apolipoprotein(a) kringle IV-10 and coronary artery disease: Relationship to allele size, plasma lipoprotein(a) concentration, and lysine binding activity. J Mol Med. 2001;79:294–299. doi: 10.1007/s001090100203. [DOI] [PubMed] [Google Scholar]
- 58.Wade DP, Clarke JG, Lindahl GE, Liu AC, Zysow BR, Meer K, Schwartz K, Lawn RM. 5′ control regions of the apolipoprotein(a) gene and members of the related plasminogen gene family. Proc Natl Acad Sci USA. 1993;90:1369–1373. doi: 10.1073/pnas.90.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu JH, Lee IN. Studies of apolipoprotein (a) promoter from subjects with different plasma lipoprotein (a) concentrations. Clin Biochem. 2003;36:241–246. doi: 10.1016/s0009-9120(03)00002-x. [DOI] [PubMed] [Google Scholar]
- 60.Zysow BR, Lindahl GE, Wade DP, Knight BL, Lawn RM. C/T polymorphism in the 5′ untranslated region of the apolipoprotein(a) gene introduces an upstream ATG and reduces in vitro translation. Arterioscler Thromb Vasc Biol. 1995;15:58–64. doi: 10.1161/01.atv.15.1.58. [DOI] [PubMed] [Google Scholar]
- 61.Puckey LH, Knight BL. Sequence and functional changes in a putative enhancer region upstream of the apolipoprotein(a) gene. Atherosclerosis. 2003;166:119–127. doi: 10.1016/s0021-9150(02)00315-5. [DOI] [PubMed] [Google Scholar]
- 62.Rosby O, Alestrom P, Berg K. High-degree sequence conservation in LPA kringle IV-type 2 exons and introns. Clin Genet. 1997;52:293–302. doi: 10.1111/j.1399-0004.1997.tb04346.x. [DOI] [PubMed] [Google Scholar]
- 63.Prins J, Leus FR, Bouma BN, van Rijn HJ. The identification of polymorphisms in the coding region of the apolipoprotein(a) gene. Thromb Haemost. 1999;82:1709–1717. [PubMed] [Google Scholar]
- 64.Kraft HG, Haibach C, Lingenhel A, Brunner C, Trommsdorff M, Kronenberg F, Muller HJ, Utermann G. Sequence polymorphism in kringle IV 37 in linkage disequilibrium with the apolipoprotein(a) size polymorphism. Hum Genet. 1995;95:275–282. doi: 10.1007/BF00225193. [DOI] [PubMed] [Google Scholar]
- 65.van der Hoek YY, Wittekoek ME, Beisiegel U, Kastelein JJ, Koschinsky ML. The apolipoprotein(a) kringle IV repeats which differ from the major repeat kringle are present in variably-sized isoforms. Hum Mol Genet. 1993;2:361–366. doi: 10.1093/hmg/2.4.361. [DOI] [PubMed] [Google Scholar]
- 66.Scanu AM, Pfaffinger D, Lee JC, Hinman J. A single point mutation (Trp72 → Arg) in human apo(a) kringle 4–37 associated with a lysine binding defect in Lp(a) Biochim Biophys Acta. 1994;1227:41–45. doi: 10.1016/0925-4439(94)90104-x. [DOI] [PubMed] [Google Scholar]
- 67.Prins J, Leus FR, van der Hoek YY, Kastelein JJ, Bouma BN, van Rijn HJ. The identification and significance of a Thr → Pro polymorphism in kringle IV type 8 of apolipoprotein(a) Thromb Haemost. 1997;77:949–954. [PubMed] [Google Scholar]
- 68.Ogorelkova M, Gruber A, Utermann G. Molecular basis of congenital lp(a) deficiency: A frequent apo(a) ‘null’ mutation in Caucasians. Hum Mol Genet. 1999;8:2087–2096. doi: 10.1093/hmg/8.11.2087. [DOI] [PubMed] [Google Scholar]
- 69.Parson W, Kraft HG, Niederstatter H, Lingenhel AW, Kochl S, Fresser F, Utermann G. A common nonsense mutation in the repetitive Kringle IV-2 domain of human apolipoprotein(a) results in a truncated protein and low plasma Lp(a) Hum Mutat. 2004;24:474–480. doi: 10.1002/humu.20101. [DOI] [PubMed] [Google Scholar]
- 70.Rosby O, Alestrom P, Berg K. Sequence conservation in kringle IV-type 2 repeats of the LPA gene. Atherosclerosis. 2000;148:353–364. doi: 10.1016/s0021-9150(99)00285-3. [DOI] [PubMed] [Google Scholar]
- 71.Cohen JC, Chiesa G, Hobbs HH. Sequence polymorphisms in the apolipoprotein(a) gene. Evidence for dissociation between apolipoprotein(a) size and plasma lipoprotein(a) levels. J Clin Invest. 1993;91:1630–1636. doi: 10.1172/JCI116370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Perombelon YFN, Soutar AK, Knight BL. Variation in lipoprotein(a) concentration associated with different apolipoprotein(a) alleles. J Clin Invest. 1994;93:1481–1492. doi: 10.1172/JCI117126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim JH, Roh KH, Nam SM, Park HY, Jang Y, Kim DK, Song KS. The apolipoprotein(a) size, pentanucleotide repeat, C/T(+93) polymorphisms of apolipoprotein(a) gene, serum lipoprotein(a) concentrations and their relationship in a Korean population. Clin Chim Acta. 2001;314:113–123. doi: 10.1016/s0009-8981(01)00683-0. [DOI] [PubMed] [Google Scholar]
- 74.Kruglyak L, Nickerson DA. Variation is the spice of life. Nat Genet. 2001;27:234. doi: 10.1038/85776. [DOI] [PubMed] [Google Scholar]
- 75.Crawford DC, Yi Q, Smith JD, Shephard C, Wong M, Witrak L, Livingston RJ, Rieder MJ, Nickerson DA. Allelic spectrum of the natural variation in CRP. Hum Genet. 2006;119:496–504. doi: 10.1007/s00439-006-0160-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Glatt CE, DeYoung JA, Delgado S. Screening a large reference sample to identify very low frequency sequence variants: comparisons between two genes. Nat Genet. 2001;27:435. doi: 10.1038/86948. [DOI] [PubMed] [Google Scholar]
- 77.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, Gonzalez JR, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Zhang J, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of minor allele frequencies for LPA markers between European-American and African-American samples. The numbers on the X-axis represent the minor allele frequency observed in the African-American sample (n=24). The numbers on the Y-axis represent the minor allele frequency observed in the European-American sample (n=23). A total of 275 SNPs, represented by circles, were compared between the two samples. The number of circles on the graph does not equal the number of total LPA SNPs because many SNPs have the same estimated minor allele frequency.
Location, rs number, and sequence context of SNPs in LPA. SNPs are numbered based on the GenBank accession number DQ452068.
Location, rs number, and sequence context of SNPs in PLG. SNPs are numbered based on the GenBank accession number AY192161.