In this manuscript we describe the characterisation of human snoRNAs that co-purify with nucleoli and develop a new vector based system for targeted gene knock down. We demonstrate that this novel vector system (snoMEN) can deliver effective, sequence-specific knock down of endogenous cellular genes as well as GFP and GFP-fusion proteins.
Abstract
Human small nucleolar RNAs (snoRNAs) that copurify with nucleoli isolated from HeLa cells have been characterized. Novel fibrillarin-associated snoRNAs were detected that allowed the creation of a new vector system for the targeted knockdown of one or more genes in mammalian cells. The snoMEN (snoRNA modulator of gene expressioN) vector technology is based on snoRNA HBII-180C, which contains an internal sequence that can be manipulated to make it complementary to RNA targets. Gene-specific knockdowns are demonstrated for endogenous cellular proteins and for G/YFP-fusion proteins. Multiplex snoMEN vectors coexpress multiple snoRNAs in one transcript, targeted either to different genes or to different sites in the same gene. Protein replacement snoMEN vectors can express a single transcript combining cDNA for a tagged protein with introns containing cognate snoRNAs targeted to knockdown the endogenous cellular protein. We foresee applications for snoMEN vectors in basic gene expression research, target validation, and gene therapy.
INTRODUCTION
Small nucleolar RNAs (snoRNAs) comprise a family of nuclear RNAs that are present in all eukaryotic cells. snoRNAs are concentrated in nucleoli where they either function in the modification of rRNA or else participate in the processing of rRNA during ribosome subunit synthesis (Weinstein and Steitz, 1999; Kiss, 2001; Boisvert et al., 2007; Matera et al., 2007). There are two main classes of snoRNAs: the box C/D snoRNAs and the box H/ACA snoRNAs, which are involved in 2′-O-ribose methylation and pseudouridine modifications, respectively. snoRNAs function in vivo as RNA–protein complexes (snoRNPs), where the RNA moiety acts as a guide RNA that base-pairs to rRNA precursors, either to direct sites of modification to specific sequences on rRNA (in the case of box C/D and box H/ACA snoRNAs) or as chaperones to assist the maturation of nascent rRNA precursors (e.g., U3 snoRNA).
Box C/D snoRNAs are named after a common RNA motif in this subfamily that serves as a binding site for a group of box C/D proteins, including NOP56, NOP58, 15.5K, and the highly conserved protein fibrillarin, which has the specific 2′-O-methylase activity. Many of the Box C/D snoRNAs are encoded within the introns of mRNAs, particularly in genes encoding ribosomal protein genes or other efficiently expressed housekeeping genes (Kiss et al., 2006). A region of the box C/D snoRNA immediately 5′ to the stem II and box D and D′ region contains the “guide” sequence complementary to a specific site on rRNA that directs the fibrillarin 2′-O-methylase to add a methyl group at the desired ribose residue within the rRNA sequence that is complementary to the guide RNA region. A similar function has been shown to operate for Box C/D snoRNAs from yeast through to mammalian cells. Furthermore, it has been shown that transient expression in mammalian cells of recombinant snoRNAs, in which the guide sequence complementary to rRNA is altered to target mRNA, will cause 2′-O-methylation of the mRNA target (Cavaille et al., 1996).
Because of the conserved sequence elements it is possible to predict members of the human box C/D snoRNA family and predict their putative 2′-O-methylation target sites on rRNA through computational analysis. However, so far not all of these species have been identified or verified by direct experimentation. In addition to the modification of rRNA, two other functions have been proposed for certain box C/D snoRNAs. First, several family members are thought to specify methylation of 2′-O-ribose groups on other nonribosomal RNAs, including small nuclear RNAs, which are subunits of the nuclear pre-mRNA splicing machinery (Tycowski et al., 1998). This is basically the same modification reaction as seen for rRNA, but directed to different RNA substrates due to the specific guide RNA sequences they contain. Second, snoRNA HBII-52, a member of the box C/D snoRNA family encoded within a large transcript encoding multiple isoforms of HBII-52 RNA, has been reported to act as an alternative splicing factor that can cause a change in the choice of splice site, affecting exon 5 in the neuronal-specific transcript encoding serotonin receptor 2C (Kishore and Stamm, 2006).
Here we characterize human nucleolar snoRNAs that copurify with nucleoli isolated from HeLa cells. A novel group of related fibrillarin-associated box C/D snoRNAs were identified that allowed the development of a new vector-based technology for the targeted knockdown of expression of one or more proteins in mammalian cells. We demonstrate these snoMEN (snoRNA modulator of gene expressioN) vectors can be used for the convenient functional replacement of endogenous cell proteins with tagged and or mutated recombinant proteins in cultured cells.
MATERIALS AND METHODS
Construction of Nucleolar cDNA Library
HeLa cell nucleoli were purified using sucrose gradients, as previously described (Andersen et al., 2002, 2005; Lam et al., 2007). Nucleolar RNA was isolated from purified nucleoli by the TRIzol method with DNase I treatment according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). Nucleolar RNA was modified by addition of a 20–30 mer polyadenine sequence at the 3′-end using polyA polymerase (Invitrogen). In the case of the unamplified library, cDNA was synthesized from 10 μg of polyA-tailed nucleolar RNA by reverse transcription using an oligo dT primer. Second-strand cDNA was synthesized by the nick translation replacement technique using the Super Script Plasmid Synthesis Kit according to the manufacturer's instructions (Invitrogen). Double-stranded cDNA was cloned into pBluescript vector after creating blunt ends using T4 DNA polymerase. In the case of the amplified library, a nucleolar cDNA library was established using the GeneRacer Kit without the CIP step (Invitrogen). Small, double-stranded cDNA was amplified by short-time amplification PCR (e.g., 5-s polymerase reaction) using adaptor-specific primers. Amplified cDNA was cloned into the TOPO TA cloning vector according to the manufacturer's instruction.
Sequences and Visualization
All sequences and alignments were retrieved from the UCSC Genome Browser (Kent et al., 2002) using the March 2006 assembly of the human genome. The mammalian conservation of the HBII-180 snoRNA sequences was calculated and visualized using the Vertebrate Multiz Alignment and PhastCons Conservation utilities (Kent et al., 2003; Siepel et al., 2005). HBII-180D was identified as a molecule having high sequence identity to the known HBII-180 (A, B, and C) snoRNAs using BLAT (Kent et al., 2002) available from the University of California, Santa Cruz, (UCSC) Genome Browser Web site (http://genome.ucsc.edu/).
Immunoprecipitation and Quantitative RT-PCR
Immunoprecipitations were prepared as previously described (Trinkle-Mulcahy et al., 2006). Nuclear lysates were prepared from HeLaYFP-fibrillarin and HeLaGFP stable cell lines. Purified nuclei were resuspended in RIPA buffer to solubilize proteins. Fluorescent proteins were immunoprecipitated using an anti-green fluorescent protein (GFP) mAb (Roche, Indianapolis, IN) covalently coupled to protein G-Sepharose, as previously described. Samples were divided in two, and for Input samples RNA was isolated from one-half of each nuclear lysate. RNA was isolated by the TRIzol method with DNase I treatment, according to manufacturer's instruction (Invitrogen). RT-PCR was performed to detect immunoprecipitated RNAs. RT and PCR were performed with the following gene-specific primers: HBII-180A: 5′-CCTCCATGATGTCCAGCACTG-3′ and 5′-CTCAGACCCCCGGGTGTCAA-3′; HBII-180B: 5′-GCACTGGGCTCTGACTGCCC-3′ and 5′-GAACCCCGGATGTCAAAGGT-3′; HBII-180C: 5′-CTCCCATGATGTCCAGCACT-3′ and 5′-CTCAGACCCCCAGGTGTCAA-3′; HBII-180D: 5′-GCACTGGGCTCTGATCACTC-3′ and 5′-GATCCCCAGGTAGCCATGGT-3′; C19orf48: 5′-CCGCGTTTCCTACTCTTTAAGC-3′ and 5′-TGGGTGGTCCTAAGCTTGAAGC-3′; U3: 5′-AGAGGTAGCGTTTTCTCCTGAGCG-3′ and 5′-ACCACTCAGACCGCGTTCTC-3′; E2: 5′-GGAGTTGAGGCTACTGACTGGC-3′ and 5′-CCACTCATTGGGCCAGAGACCC-3′; GAPDH: 5′-TTGCGTCGCCAGCCGAGCCACATC-3′ and 5′-CAATACGACCAAATCCGTTGACTCCGA-3′; pre-GAPDH: 5′-CGCATCTTCTTTTGCGTCGCCAG-3′ and 5′-GGTCAATGAAGGGGTCATTGATGGC-3′; U1: 5′-TACCTGGCAGGGGAGATACCATGATC-3′ and 5′-GCAGTCGAGTTTCCCACATTTGGGG-3′; 5S: 5′-ACGCGCCCGATCTCGTCTGAT-3′ and 5′-GCCTACAGCACCCGGTATTCCC-3′; β-actin: 5′-AGGCACCAGGGCGTGATGGTGG-3′ and 5′-GGTACTTCAGGGTGAGGATGCC-3′; B23: 5′-CTTTTCGGTTGTGAACTAAAGGCCG-3′ and 5′-CAGCCCCTAAACTGACCGTTCTTA-3′; HDAC1: 5′-TGCAGAGATTCAACGTTGGTGAGG-3′ and 5′-GCACTTGCCACAGAACCACCAG-3′; tubulin: 5′-CCTACGGTCATTGATGAGATCCGA-3′ and 5′-GGTCAATGATCTCCTTGCCAATGG-3′; hsp70: 5′-GAACCGGCATGGCCAAAGCCGC-3′ and 5′-TGGCGATGATCTCCACCTTGCC-3′; CREBBP: 5′-CCCCTGGGAAATAATCCAATGAAC-3′ and 5′-CGTTCATCAGTGGGTTTGTGGC-3′; snRNP70: 5′-GAGCTTAAAATGTGGGACCCTCAC-3′ and 5′-GTTGTGTCATAATTCACTCTCGCC-3′; PSP-1: 5′-TTGCTAACAACGACCCCTCGTCC-3′ and 5′-CTGCATCAGCTTCTCTGGCAAGC-3′; and HPRT1: 5′-CTGAAGAGCTATTGTAATGACCAGTC-3′ and 5′-GCCAGTGTCAATTATATCTTCCACAA-3, using the SuperScript one-step RT-PCR kit (Invitrogen). To decide linearity of cycles, we used real-time PCR using the Superscript III Platinum SYBR Green one-step quantitative RT-PCR Kit (Invitrogen) and Rotor-Gene RG-3000 system (Corbett Research, Cambridge, United Kingdom). The same amount of RNA for input and immunoprecipitated (IP) sRNA was used as templates for RT-PCR reactions. Each experiment was repeated three times independently.
Plasmid Construction and Transfections
The sequence spanning exon 2 to exon 3 of the C19orf48 gene was inserted 3′ of the cytomegalovirus (CMV) promoter in the pcDNA3.1 mammalian expression plasmid (Invitrogen), creating the HBII-180C expression vector. HBII-180A and B snoRNAs and mutant derivatives of HBII-180C snoRNA were established from the wild-type HBII-180C expression minigene construct by site-directed mutagenesis. The plasmids were transfected into either HeLa cells or U2OS cells using Effectine transfection regent (QIAGEN, Chatsworth, CA).
Microscopy
All cell images were recorded using the DeltaVision Spectris fluorescence microscope (Applied Precision, Issaquah, WA). Live cell images for HeLaYFP-fibrillarin cells (Leung et al., 2004) and HeLaGFP cells (Trinkle-Mulcahy et al., 2006) were prepared as previously described (http://www.lamondlab.com/f7protocols.htm). Cells were imaged using a 60× 1.4 NA Plan Apochromat objective. Twelve optical sections separated by 0.5 μm were recorded for each field and each exposure (SoftWoRx image processing software; Applied Precision).
Northern and High-Sensitivity RNA Blot Analysis
HeLa cell extracts were fractionated using sucrose gradients, as previously described (Andersen et al., 2002, 2005; Lam et al., 2007). Total HeLa cell RNA and RNA from separate cytoplasmic, nucleoplasmic, and nucleolar fractions was isolated using the TRIzol method, with DNase I treatment, according to the manufacturer's instructions (Invitrogen). Equal amounts of RNA from each sample were separated by 8 M urea polyacrylamide denaturing gel electrophoresis in 1× TBE buffer, and the RNA was transferred onto nylon membrane (Hybond-N; GE Healthcare, Little Chalfont, United Kingdom) by electroblotting. After UV cross-linking or chemical cross-linking, the membrane was hybridized with 32P 5′ end-labeled oligoribonucleotide probes specific for the following RNA species: HBII-180C: 5′-GUGCACUGUGUCCUCAGGGGUG-3′; tRNA-Ile: 5′-UGGUGGCCCGUACGGGGAUCGA-3′; chimera 1: 5′-GCAGCACGACUUCUUCAAGUC-3′; and chimera 2: 5′-GCAGAAGAACGGCAUCAAGGU-3′). High-sensitivity RNA blots were prepared as previously described (Pall et al., 2007).
Fluorescent In Situ Hybridization
The fluorescent in situ hybridization (FISH) procedure was performed as previously described (http://www.singerlab.org/protocols). HeLa cells were transfected with a plasmid vector containing the HBII-180C minigene expressed from the CMV promoter. The cells were fixed with 4% paraformaldehyde after prepermeabilization with 1% Triton X-100. After 70% ethanol treatment, a Cy-3–labeled HBII-180C–specific oligonucleotide probe (5′-AAAGGTCCTGGGGTGCACTGTGTCCTCAGGGGTGATCAGAGCCCAGTGCT-3′) was hybridized using standard procedures. The fluorescence signal was imaged using a DeltaVision Spectris fluorescence microscope (Applied Precision). The specific probes for chimera snoRNAs (chimera 1: 5′-GCAGCACGACUUCUUCAAGUC-3′; chimera 2: 5′-GCAGAAGAACGGCAUCAAGGU-3′; and chimera-3: 5′-CAGCCACAACGUCUAUAUCAU-3′) were also labeled with Cy-3.
siRNA Experiments
siRNA was transfectioned by Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instruction. Scrambled siRNA, which does not have an RNA target (Dharmacon, Boulder, CO), and Lamin siRNA (On-Targetplus SMART pool product, Dharmacon) were transfected as negative and positive controls, respectively. The Lamin M box siRNAs were targeted to the same intronic sequences as used for the Lamin snoMEN were synthesized and transfected (Lamin M box siRNA-1: 5′-CACCUGGGCGAACUCACCGCG-3′, Lamin M box siRNA-2: 5′-GGUAUUGCUAAAGAAGAGAGG-3′ (siMAX siRNA, MWG Operon, Ebersberg, Germany).
M Box Sequences
M box sequence of HBII-180C cDNA (5′-CACCCCTGAGGACACAGTGCA-3′) was modified to create complementary sequences to target genes as follows: chimera-1, -2, and -3: 5′-ATGATATAGACGTTGTGGCTG-3′; SMN1 (survival of motor neuron 1): 5′-ATTAGAACCAGAGGCTTGACG-3′ and 5′-GCACTGGCTGCGACCTCACCT-3′ and 5′-ATGTCAGAATCATCGCTCTGG-3′; Lamin: 5′-CACCTGGGCGAACTCACCGCG-3′ and 5′-GGTATTGCTAAAGAAGAGAGG-3′; and Coilin 5′-TCCCTGCGCCGCGCACCTGAG-3′ and 5′-TGAAATATACCTTAAATGCAA-3′.
RESULTS
Identification for HBII-180 box C/D snoRNAs
We have identified 118 human snoRNAs that copurify with nucleoli isolated from HeLa cells (Figure 1, a and b, and Supplementary Table). Two of these were closely related box C/D snoRNAs termed HBII-180B and C (Figure 2a), which are encoded within separate introns of the pre-mRNA transcript C19orf48 (Zhou et al., 2002). The same transcript also encodes a third related snoRNA (HBII-180A), which is difficult to detect in HeLa cells (Figure 2a). Phylogenetic comparison (Kent, 2002) of the homologous gene from different mammalian species revealed that the major regions of sequence conservation corresponded to the intron-encoded snoRNAs and not to the flanking exons (Figure 2a). All three show typical Box C/D snoRNA features, i.e., they are enriched within nucleoli, bind to fibrillarin, and contain conserved C and D boxes (Figure 2, b and c, and Supplementary Figure 1, a and b). Each has an identical guide sequence complementary to residues 3677-3686 of human 28S rRNA (Figure 2b), which contains a known 2′-O-ribose methylation site at residue 3680 (Maden, 1990).
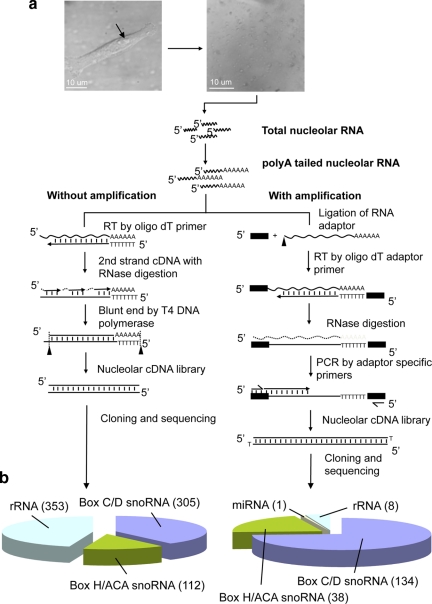
Figure 1.
Human gene C19 orf48 identified in nucleolar cDNA library. (a) Isolation and cloning of Nucleolar RNA from purified HeLa cell nucleolar fraction. Micrographs showing intact HeLa cells before fractionation (left) and isolated nucleoli (right). HeLa cell nucleoli were fractionated by the sucrose gradient method as previously described (Andersen et al., 2002, 2005; Lam et al., 2007), and then total nucleolar RNA was isolated from the nucleolar pellet. Total nucleolar RNA was modified by addition of poly adenosine at the 3′-end using poly A polymerase. Schematic diagram showing cDNA synthesis strategy, performed both with amplification (right) and without amplification (left). Nucleolar cDNAs were synthesized by reverse transcription using PolyA-tailed nucleolar RNA as the template. The library made without amplification (left) was mainly used to detect abundant nucleolar RNA species, whereas we used the amplification technique (right) to make a library including lower abundance nucleolar RNA species. Both of these libraries were independently cloned into general plasmid vectors and analyzed by DNA sequencing. (b) Summary of RNA species detected in the nucleolar cDNA libraries without amplification (left pie chart) and with amplification (right pie chart).
Figure 2.
Characterization for HBII-180 boxC/D snoRNAs. (a) Structure of the human C19orf48 gene. Exons are indicated as blue boxes with roman numerals. Predicted protein coding region is indicated in red. The intron-encoded snoRNAs HBII-180A, HBII-180B, and HBII-180C are indicated as green boxes. Data are derived from the C19orf48 chromosome position of UCSC Human Genome Browser (Kent, 2002). Vertebrate Multiz Alignment and conservation results (Kent et al., 2003; Siepel et al., 2005; Lestrade and Weber, 2006) are indicated as boxes and bar, respectively. Conserved regions between human and 11 other mammalian and marsupial species are indicated as bars (Conservation). The three most conserved regions correspond to the three detected HBII-180 snoRNAs. (b) Sequences of HBII-180 snoRNAs. HBII-180A, HBII-180B, HBII-180C, and HBII-180D sequences were aligned showing Box C and C′ (A/G)UGAUGA) and boxes D and D′ (CUGA) indicated as orange and green boxes, respectively. The predicted rRNA complementary guide sequence for ribose 2′-O-methylation is indicated as a pink box, as shown in snoRNABase (Lestrade and Weber, 2006). Identical sequences shared by the three snoRNAs are indicated with asterisks. (c) HBII-180 snoRNAs bind fibrillarin. Extracts were prepared from HeLa cell lines stably expressing either YFP-fibrillarin or free GFP and immunoprecipitated using a monoclonal anti-GFP antibody, with the specificity confirmed by Western blotting. Quantitative RT-PCR was used to detect coprecipitated HBII-180A, B, C, and D snoRNAs, using U3 snoRNA as a positive control and U1 snRNA, 5S rRNA, E2 H/ACA snoRNA, and GAPDH pre-mRNA as negative controls for fibrillarin-associated RNAs. For RNA blots equal amounts of material were loaded on the IP and Input lanes.
We observed that HBII-180C snoRNA contained a region 3′ to the methylation guide sequence that was complementary to fibroblast growth factor receptor 3 (FGFR3) pre-mRNA. The short length (∼21 nucleotides) and sequence complementarity of this region is similar to features seen in small regulatory RNAs, such as siRNAs and microRNAs (Bushati and Cohen, 2007; Chapman and Carrington, 2007). Given that human snoRNA HBII-52 acts as an alternative splicing factor to modulate the neuronal-specific splicing of serotonin receptor 2C (Kishore and Stamm, 2006), we decided to scan all human coding sequences, UTRs and introns to search for other potential mRNA targets that could be recognized by short complementary sequences within the HBII-180 snoRNAs.
A short internal region was detected at a similar position within each of the three HBII-180 snoRNAs, which we term the M box (Supplementary Figure 2a). The M box region had potential mRNA intron and exon targets in both cell-type specific and ubiquitously expressed genes. For example, the M box region in snoRNA HBII-180C was complementary to RNA sequences in the HIPPI (Huntington-interacting protein protein interactor) and FGFR3 (fibroblast growth factor receptor 3) genes (Supplementary Figure 2a). Analysis of the M box region and the effects of the HBII-180 snoRNAs on expression of endogenous RNAs will be reported elsewhere. Here we describe the adoption of the HBII-180C snoRNA backbone for development of a general gene knockdown vector system.
GFP and YFP Knockdown Mediated by the snoRNA M Box
To test whether snoRNAs can be used to modify the expression of targeted proteins, the M box sequence in HBII-180C was replaced with two alternative sequences to make it complementary to different regions in the coding sequence of GFP and yellow fluorescent protein (YFP; Figure 3a; for locations of targeted sequences, see also Figure 14). Plasmid vectors encoding either mCherry alone, mCherry linked to wild-type HBII-180C snoRNA, or mCherry linked to either the chimera 1 or chimera 2 HBII-180C snoRNAs, which are targeted to sites on G/YFP RNA, were transiently expressed in HeLa cell lines that stably express either free GFP (Trinkle-Mulcahy et al., 2006) or the FP-fusion proteins YFP-fibrillarin (Leung et al., 2004) or GFP-SMN (Trinkle-Mulcahy et al., 2008; Figure 3b and Supplementary Figure 2b; arrows indicate transfected cells). In each case mCherry was used a marker for detecting transfected cells. A reduced level of G/YFP fluorescence for free GFP and G/YFP-fusion proteins was observed specifically in cells expressing mCherry when a chimeric snoRNA was coexpressed, but not upon transient expression of controls corresponding to either mCherry alone, or mCherry plus WT HBII-180C snoRNA and a similar reduction was observed by Western blotting (Figure 3b and Supplementary Figures 2b and 3).
Figure 3.
Targeted suppression of free GFP and YFP fusion proteins using a modified snoRNA expression vector. (a) chimera expression plasmid targeting sequences shared by GFP and YFP. The sequence in WT HBII-180C snoRNA that is complementary to FGFR3 pre-mRNA was changed from 5′-CACCCCTGAGGACACAGTGCA-3′ to either 5′-GACTTGAAGAAGTCGTGCTGC-3′ (chimera 1) or to 5′-ACCTTGATGCCGTTCTTCTGC-3′ (chimera 2). The resulting chimera 1 and 2 snoRNAs were subcloned into the 3′ region of the vector with mCherry fluorescent protein cDNA. Diagram shows complementary regions of chimera 1 and chimera 2 targeting two separate regions of GFP, both of which are also present in the related YFP gene. (b) The effect of chimera constructs on GFP/YFP-fibrillarin expression in the HeLaGFP/HeLaYFP-fibrillarin stable cell line, which expresses unfused GFP alone (left panel) or YFP fused at the amino terminus of fibrillarin (right panel). Images show the effect of transfecting either empty mCherry expression plasmid mCherry-C1 (mCherry-C1), expression plasmid HBII-180C chimera 1, or expression plasmid HBII-180C chimera 2 in the HeLaGFP/HeLaYFP-fibrillarin stable cell lines. Top panels, GFP fluorescence signals of images recorded from fixed cells (green). Bottom panels, merged images combining the GFP (green) and mCherry (red) signals. Scale bar, 10 μm. The arrows indicate transfected cells, and arrowheads show untransfected cells. Note clear reduction in GFP expression specifically in the cells transfected with the expression plasmids encoding either HBII-180C chimera 1 or 2. (c) Detection of RNA levels for YFP-fibrillarin after transfection of HeLaYFP-fibrillarin stable cell lines using either the wild-type HBII-180C with mCherry expression plasmid pmCherry-HBII-180C (control: lane 1), expression plasmid pHBII-180C chimera 1 (lane 2), or expression plasmid pHBII-180C chimera 2 (lane 3). An equivalent amount of HeLa total RNA was loaded for each lane, and the RNA was separated by denaturing agarose gel electrophoresis, electroblotted onto membrane, and probed for G/YFP, GAPDH, β-actin mRNAs, and 18S rRNA as a loading control. The graphs show average signal intensity for three independent experiments; error bars, SD. YFP-fibrillarin signal ratio was normalized to the 18S rRNA signal. Note reduction in YFP-fibrillarin RNA levels, but not GAPDH and β-actin, specifically with the expression plasmids encoding either HBII-180C chimera 1 or 2.
Figure 14.
The RNA target sites of snoMEN vectors. The targeted regions on the RNAs for each of the snoMEN vectors used in this study are shown in a schematic diagram.
Analysis of RNA by Northern blot showed that both chimera 1 and chimera 2 snoRNAs reduced YFP-fibrillarin mRNA levels with no reduction in the levels of either GAPDH or β-actin RNAs (Figure 3c). The mCherry plus WT HBII-180C snoRNA control vector did not reduce either YFP fluorescence or YFP-fibrillarin RNA levels. Western blot analysis confirmed that there was also a reduction in the corresponding level of YFP-fibrillarin protein, consistent with the reduced level of YFP fluorescence, specifically after expression of either chimera 1 or chimera 2 snoRNAs (Supplementary Figure 3). Similar results were obtained for knockdown of free GFP (Supplementary Figure 3). To investigate further the specificity of the knockdown, the levels of 10 nontargeted RNAs were compared with YFP-fibrillarin RNA levels by quantitative RT-PCR in HeLa cells expressing either control WT HBII-180C snoRNA, or the chimera 1 or chimera 2 snoRNAs (Supplementary Figure 4). Expression of either chimera 1 or chimera 2 snoRNAs caused a specific reduction in YFP-fibrillarin RNA levels with little or no off target effects on the levels of the other 10 RNAs.
The data indicate that the snoRNA vector based on HBII-180C can modulate both RNA and protein expression levels of targeted genes mediated via short internal regions of complementary sequence in the M box region. Furthermore, the chimeric snoRNAs exhibit the same localization and protein-binding properties as bona fide box C/D snoRNAs. Thus, a combination of cell fractionation, RNA blotting, FISH, and coimmunoprecipitation experiments showed that the endogenous HBII-180C snoRNA and the chimera 1 and chimera 2 snoRNAs that knockdown GFP and YFP-fibrillarin levels are nuclear, accumulate in the nucleolus in a pattern similar to other Box C/D snoRNAs, and bind to the Box C/D-specific protein fibrillarin (Supplementary Figure 1, a and b).
Analysis of snoRNA Sequence Determinants
The knockdown ability of vectors expressing HBII-180C snoRNAs with altered M box regions could either arise from expression of processed snoRNA or, alternatively, could be formed independently of snoRNA processing directly from the C19orf48 pre-mRNA (Figure 2a). The latter possibility is suggested by the reports of intron-encoded microRNAs processed independently of Drosha (Ruby et al., 2007). To distinguish between these possibilities, a series of three base mutations were introduced at eight separate regions of an HBII-180C minigene and the expression of full-length snoRNA compared between the WT and mutant forms of HBII-180C (Figure 4a). Expression of the WT HBII-180C minigene vector elevates the level of full-length HBII-180C snoRNA over endogenous levels, but the empty vector does not (Figure 4a, cf. lane 1 with lanes 2 and 3). Multiple mutations, including sequence changes within either of the conserved C or D boxes, reduce or eliminate expression of exogenous full-length HBII-180C snoRNA, leaving only the endogenous level of snoRNA (Figure 4a, lanes 4–11).
Figure 4.
Analysis of HBII-180C and chimera snoRNA mutations. (a) The sequence of snoRNA HBII-180C is shown with the positions of the eight separate three nucleotide AAA mutations indicated as m1–m8 (shown on the sequence). Boxes C, D, and D′ are indicated with boxes. The predicted guide sequence complementary to 28S rRNA is indicated by an underline, as shown in snoRNABase (Lestrade and Weber, 2006). The M box region is indicated by a bold underline. HeLa cells were transiently transfected with either an empty plasmid vector (lane 2), with plasmid vectors expressing wild-type HBII-180C (lane 3), or with HBII-180C snoRNA mutants m1–m8 (lanes 4–11). The same amount of total HeLa cell RNA was loaded for each lane (Ctrl: without transfection). The bands show the results of high-sensitivity RNA blotting using 32P-labeled oligonucleotide probes to detect HBII-180C snoRNA, M box fragment, and tRNA, respectively. Transfection efficiency was confirmed by semiquantitative RT-PCR using C19 orf48 and GAPDH specific primer sets (C19orf48 and GAPDH). * The C19orf48 primers detect a region shared between the endogenous C19orf48 transcript and the HBII-180C minigene. (b) Wild-type plasmid (CM2WT) and mutant plasmids (CM2m1 and CM2m7) were transiently transfected into HeLa cells either stably expressing GFP alone (HeLaGFP) or expressing a GFP-SMN fusion protein (HeLaGFP-SMN). Scale bar, 15 μm. Arrow, transfected; arrowhead, untransfected. Note the mutations for core regions box C (CM2m1) and box D (CM2m7) of HBII-180C snoRNA on chimera 2 plasmid showed no knockdown effect on GFP or GFP-SMN expression levels.
We next tested whether mutations in regions known to be required for snoRNA processing and expression also affect knockdown efficiency, using HeLa cells expressing either GFP alone, or a GFP-SMN fusion protein (Figure 4b). Thus, mutants within either the conserved box C or box D regions, which both disrupt snoRNA production (Figure 4a, cf. lane 3 with lanes 4 and 10), also fail to knockdown fluorescence levels of either GFP or GFP-SMN (Figure 4b). Further systematic comparison of multiple mutations in snoRNA chimera 2 showed a clear correlation between mutations affecting snoRNA processing and expression and the ability to knockdown a target GFP gene. For example, the m3 mutation in the conserved D′ box decreased snoRNA formation and reduced knockdown efficiency (Figures 4a and 5, a and b; see also Supplementary Figure 5). Whenever a mutation reduced the level of exogenous chimera 2 snoRNA expression, there was a corresponding decrease in the level of knockdown (Figure 5, a and b), indicating that knockdown is dependent on correct snoRNA processing from the vector transcript. This argues against a snoRNA-independent knockdown mechanism, arising directly from parallel processing of the C19orf48 pre-mRNA transcript, as would be the case if a separate small regulatory RNA was encoded within the snoRNA. Furthermore, none of the mutations that destroy base pairing between 28S rRNA and the snoRNA guide sequence, either prevent snoRNA formation, or reduce knockdown efficiency (Figures 4a, lane 5, and 5, a and b; see also Supplementary Figure 5, mutants m2-1 and m2-2). Therefore, we infer that the ability of M box–modified snoRNAs to knockdown expression of target genes does not depend on the snoRNA binding and methylating rRNA.
Figure 5.
Summary of chimera 2 mutagenesis constructs. (a) Left, motifs of HBII-180C snoRNA and mutated regions for each plasmid; right, plasmid name and mutated position (b) Quantification of knockdown level for different snoRNA constructs in transfected cells. The ratio of GFP-SMN fluorescence signals between transfected cells/nontransfected cells was calculated using SoftWoRx image processing software (Applied Precision). Graph shows average ratio between each mutant construct; error bars, SD; n > 10.
Next, insertion and deletion mutants of the M box sequence in modified snoRNA chimera 2 were constructed to test whether knockdown ability correlated with M box sequence length (Figure 5, a and b, and Supplementary Figure 6). All three insertion mutants that increased sequence complementary to G/YFP mRNA retained knockdown activity (Figure 5, a and b, In1-3). Interestingly, insertion mutant CM2In-3, which has an additional eight bases of complementary sequence to G/YFP mRNA, enhanced knockdown efficiency (Figure 5a and b). In contrast, 2 and 3 base deletion mutants in the M box (CM2Del-1 and -2) and the seven-base deletion mutant CM2Del-3, which all reduced the length of sequence complementary to G/YFP, significantly reduced or eliminated knockdown activity, compared with wild-type chimera 2 (Figure 5, a and b).
To investigate further the relationship between the sequence in the M box region of the snoRNA and the RNA target sequence, a comparison was made of six mutations that systematically increased the number of mismatches in the G/YFP complementary sequence and each assayed for knockdown ability (Figure 5, a and b and Supplementary Figure 7). This showed that up to 1-3 base mismatch mutations in the M box (CM2X-1 and -3) still retained some knockdown ability, albeit less efficient compared with wild-type chimera 2 snoRNA (Figure 5, a and b). However, increasing the number of mismatches up to four to six bases (CM2X-4 to -6), either reduced, or eliminated, knockdown activity, compared with wild-type chimera 2 snoRNA (Figure 5, a and b). A dose-response analysis was also conducted to compare the GFP and mCherry fluorescence levels in HeLa cells stably expressing GFP-SMN (Supplementary Figure 8). The analysis compared transient expression of two negative control snoRNA vectors that do not cause knockdown of GFP (HBII-180C WT and box D mutant CM2m7) and two positive control snoRNA vectors that do knockdown GFP (chimera 2 and CM2-In3). This shows that low GFP fluorescence levels are clearly associated with high mCherry fluorescence levels specifically for the two vectors that knockdown GFP.
The data above all support a knockdown mechanism that depends on base pairing between the snoRNA M box and the target RNA. To test rigorously that the knockdown mechanism requires base pairing between the snoRNA M box and target RNA, a compensatory mutation analysis was also performed (Figure 6; see also Figure 14). This showed that the ability of chimera 2 snoRNA to knockdown GFP-SMN in HeLa cells could be prevented by introducing either three or five mismatch mutations into the target site for chimera 2 snoRNA on GFP in the GFP-SMN fusion transcript (GFPX3-SMN and GFPX5-SMN). However, expressing variants of snoRNA chimera 2 containing the corresponding compensatory three- or five-base M box changes, which restored base pairing with the mutated GFP-SMN target genes (CM2-X3 and CM2-X5), restored knockdown, as judged both by decreased GFP fluorescence levels and by decreased GFP-SMN protein levels (Figure 6). We conclude that the snoRNA gene knockdown mechanism is mediated by hybridization affinity between the M box region and the complementary sequence on the target RNA.
Figure 6.
Compensatory mutation analysis. Top, the structure of each plasmid. Two compensatory GFP-SMN constructs (reporter plasmids: GFPX3-SMN1 and GFPX5-SMN1), which are complementary to chimera 2 mutant plasmids that have either three or five point mutations in the M box (snoMEN: CM2X-3 and CM2X-5, Figure 4a) were established and transiently cotransfected into HeLaGFP-SMN cells. The graphs show average signal intensity for three independent experiments; error bars, SD. GFP signal ratio was normalized to the tubulin signal. An equivalent amount of HeLa extract was loaded for each lane, and the proteins were separated by SDS PAGE, electroblotted, and probed both with a monoclonal anti-GFP antibody and with anti-tubulin as a loading control.
Multiple snoRNAs Can Be Expressed in One Vector Transcript
As many endogenous RNA transcripts encode multiple snoRNAs in different introns, we next investigated the design of multiplex vectors that can deliver more than one M box–modified snoRNA from a single transcript. Thus, a triplet chimera snoRNA vector was constructed that encodes mCherry as a transfection marker and three M box–modified snoRNAs, each targeted to different positions in the G/YFP mRNA sequence (Figure 7a). Transient transfection of the triple chimera vector showed knockdown of fluorescence in cells expressing either free GFP or G/YFP-fusion proteins (Figure 7b and Supplementary Figure 9). Quantification of relative fluorescence levels indicated that the knockdown efficiency using the triple chimera vector was higher than with the vectors expressing a single chimera snoRNA (Figure 9b and Supplementary Figure 10a). FISH analysis, using specific probes for each chimera snoRNA, showed that all three M box–modified chimera 1-3 snoRNAs were expressed after transfection and localized in the nucleus in a similar pattern to wild-type HBII-180C (Figure 7a).
Figure 7.
Gene knockdown using triplet chimera snoRNA vector. (a) Structure of triplet chimera snoRNA construct and schematic diagram of targeted GFP/YFP knock down. FISH showing nucleolar localization of wild-type HBII-180C and chimera snoRNAs (Cy3). DNA is stained by DAPI. Scale bar,15 μm. Top, the structure of the triplet chimera snoRNA construct. Arrows, nucleolus. (b) The same experiment as for Figure 3b except the chimera triplet plasmid was transfected. pmCherry-triple-HBII-180C is a negative control that has three repeats of wild-type HBII-180C snoRNA cloned 3′ of mCherry cDNA. Scale bar, 10 μm. Arrow, transfected; arrowhead, untransfected.
Figure 9.
Gene knockdown using psnoMENv1. (a) This shows the same experiment as Figure 7b, except the triple chimera vector psnoMENv1 was transfected. Scale bar, 15 μm. Note that the psnoMENv1 vector showed the same knockdown level as the wild-type chimera triplet plasmid. (b) This is the same quantification analysis as shown in Figure 5B, except single and triple snoMEN vectors were transfected. The ratio of GFP-SMN fluorescence signal between transfected cells and nontransfected cells was calculated by SoftWoRx image-processing software (Applied Precision). Graph shows average ratio between each mutant construct; error bars, SD; n > 10.
These results suggested that the efficiency of targeted knockdown can be increased, at least for some target RNAs, by multiplexing, i.e., by expressing multiple snoRNAs in one transcript. Moreover, we noted that transient expression of vectors encoding both mCherry and one or more snoRNAs able to knockdown G/YFP, resulted in transfected cells now expressing mCherry red fluorescent protein instead of the G/YFP and G/YFP fusion proteins (Figure 7b and Supplementary Figure 9). These results demonstrated effective replacement in these cell lines of the stably expressed GFP and G/YFP-fusion proteins with mCherry, i.e., a simultaneous knockdown and “protein replacement” or “protein knockin” effect delivered via a single transcript using the snoRNA vectors.
Next, we investigated the structure of the snoRNA backbone. A derivative was constructed from the HBII-180C snoRNA that combined multiple mutations that neither prevented snoRNA production, nor reduced knockdown efficiency (see Figure 5, a and b), with the aim of improving the vector design. A vector encoding three snoRNAs, each incorporating the mutations m2 (28S rRNA complementary region), m4 and m5, showed either a similar or slightly improved knockdown efficiency, as judged by fluorescence microscopy, to a triple chimera plasmid vector encoding three snoRNAs without these mutations, i.e., with wild-type snoRNA sequence outside of the M box, (Figures 8 and 9, a and b, and Supplementary Figure 9). FISH analysis using chimera 1–, -2–, and -3–specific probes showed that this combination mutant plasmid (triple chimera psnoMENv1), expressed all three chimera snoRNAs, as also seen with the wild-type triple chimera plasmid (Figure 8). These mutant analyses indicate that the knockdown ability of snoRNAs can be affected both by mutations within the target RNA complementary M box sequence as well as by mutations in other regions of the snoRNA backbone. For example, mutating the snoRNA guide region that binds rRNA may potentially increase the pool of snoRNA available for knockdown interactions.
Figure 8.
Gene knockdown using mutant triplet chimera snoRNA vector. Top, mutant m2, m4, and m5 sequences were combined into chimera triple snoRNA vector (psnoMENv1). Bottom, FISH using Cy3-labeled chimera snoRNA specific probes (Cy3) showing nucleolar localization. DNA is stained by DAPI. Scale bar, 15 μm. Arrow, nucleolus.
Rescue of a Lethal Gene Knockdown by Protein Replacement
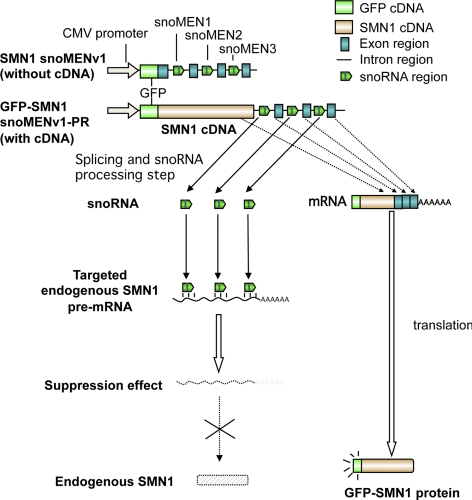
Two vectors expressing M box–modified snoRNAs targeted to endogenous SMN1 pre-mRNA were constructed, using the design from plasmid triple chimera psnoMENv1 (Figure 8). Plasmid pSMN1snoMENv1 encodes GFP and three M box–modified snoRNAs, each targeted to different exon–intron junction positions within endogenous SMN1 pre-mRNA (Figure 10and see Figure 14). The second plasmid, pGFP-SMN1snoMENv1-PR, encodes a GFP-SMN1 fusion protein and the same three M-box modified snoRNAs as pSMN1snoMENv1. Transient transfection analysis on HeLa cells showed that although control vectors encoding either GFP alone, or mCherry plus triple G/YFP-targeted snoRNAs, were not cytotoxic, the pSMN1snoMENv1 plasmid killed cells when transfected (Figure 11a, arrowhead). Western blot analysis showed that the endogenous SMN1 protein was knocked down after transfecting cells with the plasmid pSMN1snoMENv1 (Figure 11b). This cytotoxic effect was consistent with previous reports showing a lethal phenotype for SMN1 knockdown by siRNA, which induced apoptosis (Gonsalvez et al., 2007). Also, knockout of the SMN1 gene in mice is known to be embryonic lethal (Hsieh-Li et al., 2000). To test whether this lethal phenotype could be rescued by a protein replacement strategy, HeLa cells were transiently transfected with the pGFP-SMN1snoMENv1-PR plasmid (Figure 11a). In this case the HeLa cells were not killed, despite expressing the same three M box–modified snoRNAs targeted to SMN1 pre-mRNA as expressed from plasmid pSMN1snoMENv1. The cells transfected with pGFP-SMN1snoMENv1-PR expressed GFP-SMN in the same localization pattern as seen in the HeLaGFP-SMN1 stable cell line (Figure 11a). These results indicate that expression of endogenous SMN1 protein can be suppressed by M box–modified snoRNAs targeted to SMN1 pre-mRNA and that the resulting cytotoxic effect of SMN1 depletion can be rescued by protein replacement with the GFP-SMN1 fusion protein expressed from the same vector transcript that encodes the snoRNAs.
Figure 10.
Protein replacement of endogenous SMN1 protein. Structures for targeted endogenous SMN1 gene knockdown plasmid (pSMN1snoMENv1) and SMN1 protein replacement plasmid (pGFP-SMN1snoMENv1-PR). These constructs have three snoMEN sequences, as in the triple chimera psnoMENv1 plasmid (Figure 8), except that the M box sequences are complementary to endogenous SMN1 pre-mRNA sequences (see Materials and Methods).
Figure 11.
Protein replacement of endogenous SMN1 protein. (a) Targeted endogenous SMN1 plasmid (pSMN1snoMENv1) and protein replacement plasmid (pGFP-SMN1snoMENv1-PR) were transfected into HeLa cells. EGFP-C1, expressing GFP cDNA and mCherry-triple chimera plasmid (Figure 7a) were also transiently transfected into HeLa cells as controls (GFP and Triple chimera). Scale bar, 10 μm. Note that plasmid pSMN1snoMENv1 showed a cytotoxic phenotype, which was not seen with control plasmids. This cytotoxic effect was rescued by expression of the GFP-SMN1 fusion protein using pGFP-SMN1snoMENv1-PR plasmid. Arrow, transfected cells; arrowhead: cytotoxic phenotype cells. (b) Detection of protein levels for endogenous SMN1 after transfection of HeLa cells using either mCherry triple chimera (Control: lane1) and SMN1 snoMENv1 (lane 2). An equivalent amount of HeLa extract was loaded for each lane and the proteins separated by SDS PAGE, electroblotted, and probed both with a monoclonal anti-SMN1 antibody and with anti-B23 as a loading control. Graph shows SMN1 signal intensity normalized to the B23 signal.
Multiplexed snoRNAs Can Simultaneously Knock Down Multiple Target Genes
Having established that a single vector transcript can express multiple M box–modified snoRNAs, each targeted to different regions of the same protein, we next tested whether a multiplex approach can be used to simultaneously knockdown expression of more than one target gene using a single vector. Therefore, a vector was constructed from the triple chimera psnoMENv1 plasmid, encoding four M box–modified snoRNAs, targeted, respectively, to Coilin and LaminA/C pre-mRNAs (Figure 12a). Neither Coilin, nor LaminA/C, show a lethal phenotype upon siRNA knockdown (Elbashir et al., 2001; Lemm et al., 2006). Plasmid pCoilin/LaminsnoMENv1 encodes mCherry cDNA as a transfection marker and in total four M box–modified snoRNAs, two targeted to different positions within exon–intron junction sequences of endogenous Coilin pre-mRNA and two targeted to LaminA/C pre-mRNA (Figures 12a and 14). Transient transfection of HeLa cells with pCoilin/LaminsnoMENv1 resulted in simultaneous knockdown of both Coilin and LaminA/C proteins, as judged both by fluorescence microscopy (Figure 12b) and Western blotting (Supplementary Figure 10b).
Figure 12.
Multiplex knockdown for endogenous Coilin and Lamin proteins using a single snoMEN plasmid. (a) Structure of targeted endogenous Coilin and LaminA/C gene knockdown plasmid (pCoilin/LaminsnoMENv1). This construct has four tandem snoMEN sequences, with the respective M box sequences complementary to either endogenous Coilin, or LaminA/C pre-mRNA sequences (see Supplementary Materials and Supplementary Methods). (b) Targeted endogenous Coilin/Lamin plasmid (pCoilin/Lamin snoMENv1) was transfected into both HeLa and U2OS cells. mCherry-triple chimera plasmid (see Figure 7a) was also transiently transfected as a control (data not shown but the same information as in Figure 11a). Scale bar, 10 μm. Arrow, transfected cells.
The data above show that M box–modified snoRNAs can knockdown expression of endogenous cell proteins when targeted to sequences within introns of pre-mRNAs and intron–exon junction sequences that are not present in the mature mRNA. We compared this with the ability of siRNA oligoribonucleotides to knockdown expression of both the LaminA/C and Coilin proteins when targeted against the same intronic sequences. Therefore, for both LaminA/C and Coilin, two siRNA oligonucleotides per gene complementary to the same exon–intron sequences in either LaminA/C or Coilin pre-mRNAs as targeted by the M box–modified snoRNAs in pCoilin/LaminsnoMENv1, were transfected into HeLa cells. Both intron-targeted siRNAs failed to show knockdown of LaminA/C, as did a further negative control siRNA (Figure 13, a and b). As a positive control, another siRNA targeted to a LaminA/C exon sequence, resulted in knockdown. A similar result was obtained using two siRNAs targeted to the intron of Coilin, which also failed to knock down, as did a further negative control siRNA, whereas a positive control siRNA targeted to a Coilin exon sequence did reduce Coilin levels (Supplementary Figure 11, a and b). The siRNA results for both LaminA/C and Coilin were confirmed both by fluorescence microscopy and by protein blotting (Figure 13b and Supplementary Figure 11b). We conclude that the snoRNA vectors can knock down genes by targeting RNA sequences that are not amenable to siRNA knockdown. The combined results indicate that the mechanism of knockdown is dependent on snoRNA expression, that snoRNAs are specifically nuclear and that they can knock down target RNA sequences that are not expressed in cytoplasmic mRNA. This, supports the view that knockdown is distinct from the siRNA pathway.
Figure 13.
SiRNA knockdown targeted to endogenous LaminA/C. (a) The same pre-mRNA sequence of LaminA/C as targeted by the snoMEN vector was targeted by siRNA oligoribonucleotides. Scrambled siRNA (Control siRNA) and Lamin siRNA (Dharmacon) were transfected as a negative and a positive control, respectively. Lamin M box siRNA-1 and Lamin M box siRNA-2 have the same target sequence as Lamin snoMEN set 1 and Lamin snoMEN set 2, respectively (Figure 14). Scale bar, 5 μm. Arrow, cells not showing knockdown; arrowhead, cells showing knockdown. (b) Western blot analysis for siRNA experiments. Detection of protein levels for endogenous Lamin after transfection of HeLa cells using either Scrambled siRNA (Control: lane 1), Lamin siRNA (lane 2), Lamin M box siRNA-1 (lane 3) and Lamin M box siRNA-2 (lane 4). An equivalent amount of HeLa extract was loaded for each lane, and the proteins were separated by SDS PAGE, electroblotted onto membrane, and probed both with a monoclonal anti-Lamin antibody and with anti-tubulin as a loading control (left). Right, graph shows Lamin signal intensity normalized to the tubulin signal.
DISCUSSION
In this study we characterize human snoRNAs that copurify with nucleoli isolated from HeLa cells and present a new gene knockdown vector system, designed for use in mammalian cells, derived from a novel human box C/D snoRNA (HBII-180C) that was identified. We demonstrate that both wild-type and mutant derivatives of the HBII-180C snoRNA backbone can modulate expression levels of targeted cellular genes and G/YFP-fusion proteins. Gene knockdown is mediated via a short internal snoRNA region, termed the M box, that can be manipulated to make it complementary to a target RNA sequence of choice. M box–modified snoRNAs, like endogenous box C/D snoRNAs, are encoded within introns of RNA polymerase II transcripts and expressed efficiently from plasmid vectors. Multiple M box–modified snoRNAs can be delivered simultaneously in a single multiplex vector transcript and targeted either to different regions of the same target RNA, to different RNA targets, or to both. We call this new vector system snoMEN.
The snoMEN vector system offers several potential advantages for targeted gene knockdown that make it a useful addition to the widely used siRNA-based approaches and that expand the repertoire of gene knockdown technologies available. The snoMEN system benefits from the fact that endogenous box C/D snoRNAs are highly abundant nuclear RNAs that are efficiently processed from within introns of many different protein coding cellular pre-mRNAs. M box–modified snoRNAs are thus processed efficiently from vector transcripts with a minimal chance of overloading the endogenous snoRNA processing machinery. It is also convenient for multiplex delivery of a pool of knockdown M box–modified snoRNAs from a single vector, where expression of all snoRNAs can be controlled from a single vector promoter, each encoded within introns of a single transcript. This is similar in principle to cotransfecting pools of mixed siRNA oligoribonucleotides, but here delivered via an expression vector. Although in this study the vectors used a constitutive promoter to drive expression of the modified snoRNAs, they can also be combined with a wide range of existing constitutive and regulated RNA pol II promoters. This is more flexible than using RNA pol III promoters to drive expression of artificial hairpin RNAs. Although it is possible to design vectors encoding siRNAs within introns (Greber and Fussenegger, 2007; Greber et al., 2008), it remains to be established whether these will be processed as efficiently as snoRNAs, especially when expressed at high levels as may be required for gene therapy or other applications.
Another potential advantage we foresee for the snoMEN system is that it may be possible to target a distinct, and potentially larger, “sequence space” than is available using siRNAs or microRNAs. In particular, snoMEN vectors can knock down genes by targeting pre-mRNA intron sequences, which are exclusively nuclear and do not appear in cytoplasmic, mature mRNAs. This is illustrated here for the Coilin, LaminA/C, and SMN1genes. In the case of both the LaminA/C and Coilin genes, we demonstrate no knockdown of LaminA/C or Coilin proteins was obtained using two separate siRNA oligonucleotides for each gene targeted to the same intron sequences as used in the snoMEN vector. However, siRNAs targeted to exon sequences of LaminA/C or Coilin did knockdown expression. If this proves to be a general feature, it indicates that the snoMEN vectors can open up the possibility to target a large range of intron sequences for knockdown that are currently not accessible using other approaches. Considering that pre-mRNAs can contain ∼10–30-fold more intron than exon sequences, this could prove to be very useful. For example, intron sequences vary more than exon sequences between closely related genes and in gene families with homologous protein isoforms. Therefore, targeting pre-mRNA intron sequences, rather than exon or other sequences expressed in mature cytoplasmic mRNA, may aid specific knockdown of closely related protein isoforms. The snoMEN system differs from the recently described synthetic U1 adaptor system for targeted gene knockdown, which uses a bifunctional oligonucleotide to recruit U1 snRNP to a target pre-mRNA, thereby inhibiting polyadenylation and resulting in destabilization of the mRNA (Goraczniak et al., 2009). Although the U1 adaptors can also target pre-mRNAs, they are not designed for efficient delivery in a plasmid or vector system or for protein replacement, as discussed below. Nonetheless, the U1 adaptor system also operates via complementary base pairing to the target RNA, similar to the snoMEN system, and a genome-wide microarray analysis showed that such directed base pairing with pre-mRNAs caused minimal off-target effects.
We note that the ability of snoMEN to target pre-mRNA sequences not expressed in mature mRNA also facilitates their use as convenient protein replacement vectors for transient protein knock-in. This was shown here for both replacement of endogenous SMN1 with GFP-SMN1 and replacement of stably expressed free GFP with free mCherry. Thus, the same snoMEN vector transcript can encode a tagged or otherwise modified protein coding cDNA, together with one or more M box–modified snoRNAs that knockdown expression of the cognate endogenous cellular protein. The ability to target intron sequences in the endogenous gene, that are not present in the corresponding cDNA, simplifies the design of protein replacement vectors and avoids the need to create cDNA with altered codons to vary the RNA sequence sufficiently to escape it being knocked down along with the targeted cellular RNA.
Another consequence of the convenient multiplex design of snoMEN vectors is that it makes it easier for a single vector to deliver knockdown of two or more different target genes simultaneously. This was demonstrated here by simultaneous knockdown of both Coilin and LaminA/C proteins, using a combination of four M box–modified snoRNAs delivered from the same transcript. This can be important for many functional studies in human cells, for example, as an approach to overcome genetic redundancy. Considering that endogenous transcripts have been characterized that encode as many as 48 tandem copies of box C/D snoRNAs (Smith and Steitz, 1998; Cavaille et al., 2000), we envisage that large arrays of introns encoding many different M box–modified snoRNAs can also be constructed in snoMEN vectors to expand such applications. For example, the combined ability of the snoMEN vectors to simultaneously knock down two or more gene targets and to express a tagged or mutated protein, all from a single vector–encoded transcript, may be particularly useful for creating stable cell lines where several enzymes or receptors with overlapping substrate specificity can be replaced in the cell by a single prototypic protein or a mutated version thereof. This can facilitate characterization of the properties of the protein and its response to inhibitors and cell perturbations.
From the analysis presented it is apparent that the snoMEN vectors can reduce expression levels of both protein and RNA from targeted genes via a mechanism that critically depends upon base pairing between the snoRNA M box and a complementary sequence in the RNA target. This model is supported by the analysis of many separate mutations in the snoRNA M box that demonstrate loss of knockdown when the ability to base pair with the target RNA is reduced or eliminated. It is confirmed also by the compensatory mutational analysis, where it was shown that loss of knockdown of a GFP-SMN1 fusion protein when the target GFP RNA sequence has either three or five mismatch mutations introduced to inhibit base pairing with the corresponding M box–modified snoRNAs, was overcome by expressing snoRNAs with compensatory mutations in the M box region that restored the ability to base pair with the mutated GFP-SMN target RNA. The M box differs from the region in snoRNA HBII-52 shown to modulate alternative splicing of serotonin receptor 2C (Kishore and Stamm, 2006) and also differs from the snoRNA guide sequence region, which was previously shown to have the ability to mediate targeted mRNA methylation when modified to make it complementary to an exogenously expressed globin mRNA target (Cavaille et al., 1996). The ability to base pair with the target, rather than overall length, of the M box sequence appears to the most important feature and the knockdown efficiency could be enhanced by increasing the length of the complementary region by eight bases. However, much larger M box sequences are not tolerated, likely because they disrupt the processing or maturation of a mature snoRNA (our unpublished observations). This is consistent with the effects shown for mutations in the conserved C and D boxes that disrupt snoRNA processing, all of which inhibit knockdown. Further studies should help to optimize vector design with respect to the length of the M box, the best choice of sequences in the target RNA and potentially also further changes in the backbone structure of the snoRNA to enhance knockdown efficiency.
The data are consistent with an antisense-type mechanism acting directly on pre-mRNA sequences in the nucleus, mediated by base pairing between the M box–modified snoRNA and its target. This could affect expression of target genes either by changing the stability of the pre-mRNA, by inducing degradation, by inhibiting either splicing or transport of the RNA, or by a combination of these effects. All of the localization and cell fractionation studies indicate that the M-box–modified snoRNAs are concentrated in the nucleus and do not reach the cytoplasm. It is unlikely, therefore, that M box–modified snoRNAs block cytoplasmic translation of mRNAs, as is the case for inhibitory mechanisms involving microRNAs. The M box–modified snoRNAs show a steady-state accumulation in nucleoli but still are able to affect expression of pre-mRNAs. A likely explanation for this effect is provided by the observations from photobleaching studies that many nucleolar factors, including fibrillarin and other snoRNP proteins, are not static within nucleoli (Phair and Misteli, 2000; Chen and Huang, 2001). Instead, there is a constant flux of snoRNPs between the nucleoplasm and nucleoli, and this may be sufficient to provide snoRNAs with access to pre-mRNA pools that can be recognized via base-pairing interactions. It is also possible that specific pre-mRNAs may be recruited into nucleoli through interactions with snoRNAs. There is no evidence that snoRNA-mediated gene knockdown either requires, or involves, formation of a small siRNA-type product from the M box–modified snoRNA. Although we cannot exclude that this can occur, the data show that the knockdown effect from the snoMEN vectors cannot occur independently of snoRNA processing and together with the ability of the snoRNAs to target pre-mRNA intron sequences, suggest that a mechanism distinct from siRNA processing is predominantly involved.
We foresee future applications for snoMEN vectors in basic gene expression research, in drug screening and target validation studies and possibly also for gene therapy. All of these applications can benefit from the ability to deliver knockdown and protein replacement RNAs from a single vector encoding a single transcript. The snoMEN vectors expand the repertoire of technologies available for manipulating gene expression in mammalian cells and can provide new opportunities for overcoming limitations in alternative technologies.
Supplementary Material
ACKNOWLEDGMENTS
We thank our colleagues in the Lamond group for helpful discussions and suggestions and Elizabeth Farrell for technical assistance. A.I.L. is a Wellcome Trust Principal Research Fellow. M.S. is supported by a postdoctoral fellowship from the Canadian Institutes of Health Research. Funding for this research was provided by a Wellcome Trust Programme grant to A.I.L. (Ref: 073980/Z/03/Z), by an MRC Milstein award to A.I.L. (Ref: G0801738) and by a Scottish Research Council grant to the Scottish Bioinformatics Research Network.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-01-0078) on March 10, 2010.
REFERENCES
- Andersen J. S., Lam Y. W., Leung A. K., Ong S. E., Lyon C. E., Lamond A. I., Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- Andersen J. S., Lyon C. E., Fox A. H., Leung A. K., Lam Y. W., Steen H., Mann M., Lamond A. I. Directed proteomic analysis of the human nucleolus. Curr. Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- Boisvert F. M., van Koningsbruggen S., Navascues J., Lamond A. I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Bushati N., Cohen S. M. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Cavaille J., Buiting K., Kiefmann M., Lalande M., Brannan C. I., Horsthemke B., Bachellerie J. P., Brosius J., Huttenhofer A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci. USA. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaille J., Nicoloso M., Bachellerie J. P. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996;383:732–735. doi: 10.1038/383732a0. [DOI] [PubMed] [Google Scholar]
- Chapman E. J., Carrington J. C. Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- Chen D., Huang S. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J. Cell Biol. 2001;153:169–176. doi: 10.1083/jcb.153.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S. M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Gonsalvez G. B., Tian L., Ospina J. K., Boisvert F. M., Lamond A. I., Matera A. G. Two distinct arginine methyltransferases are required for biogenesis of Sm-class ribonucleoproteins. J. Cell Biol. 2007;178:733–740. doi: 10.1083/jcb.200702147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goraczniak R., Behlke M. A., Gunderson S. I. Gene silencing by synthetic U1 adaptors. Nat. Biotechnol. 2009;27:257–263. doi: 10.1038/nbt.1525. [DOI] [PubMed] [Google Scholar]
- Greber D., El-Baba M. D., Fussenegger M. Intronically encoded siRNAs improve dynamic range of mammalian gene regulation systems and toggle switch. Nucleic Acids Res. 2008;36:e101. doi: 10.1093/nar/gkn443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber D., Fussenegger M. Multi-gene engineering: simultaneous expression and knockdown of six genes off a single platform. Biotechnol. Bioeng. 2007;96:821–834. doi: 10.1002/bit.21303. [DOI] [PubMed] [Google Scholar]
- Hsieh-Li H. M., Chang J. G., Jong Y. J., Wu M. H., Wang N. M., Tsai C. H., Li H. A mouse model for spinal muscular atrophy. Nat. Genet. 2000;24:66–70. doi: 10.1038/71709. [DOI] [PubMed] [Google Scholar]
- Kent W. J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W. J., Baertsch R., Hinrichs A., Miller W., Haussler D. Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl. Acad. Sci. USA. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore S., Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20:3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T., Fayet E., Jady B. E., Richard P., Weber M. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb. Symp. Quant. Biol. 2006;71:407–417. doi: 10.1101/sqb.2006.71.025. [DOI] [PubMed] [Google Scholar]
- Lam Y. W., Lamond A. I., Mann M., Andersen J. S. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr. Biol. 2007;17:749–760. doi: 10.1016/j.cub.2007.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemm I., Girard C., Kuhn A. N., Watkins N. J., Schneider M., Bordonne R., Luhrmann R. Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol. Biol. Cell. 2006;17:3221–3231. doi: 10.1091/mbc.E06-03-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestrade L., Weber M. J. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006;34:D158–D162. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A. K., Gerlich D., Miller G., Lyon C., Lam Y. W., Lleres D., Daigle N., Zomerdijk J., Ellenberg J., Lamond A. I. Quantitative kinetic analysis of nucleolar breakdown and reassembly during mitosis in live human cells. J. Cell Biol. 2004;166:787–800. doi: 10.1083/jcb.200405013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden B. E. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 1990;39:241–303. doi: 10.1016/s0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- Matera A. G., Terns R. M., Terns M. P. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- Pall G. S., Codony-Servat C., Byrne J., Ritchie L., Hamilton A. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 2007;35:e60.s. doi: 10.1093/nar/gkm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair R. D., Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- Ruby J. G., Jan C. H., Bartel D. P. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. M., Steitz J. A. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol. Cell. Biol. 1998;18:6897–6909. doi: 10.1128/mcb.18.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L., Andersen J., Lam Y. W., Moorhead G., Mann M., Lamond A. I. Repo-Man recruits PP1 gamma to chromatin and is essential for cell viability. J. Cell Biol. 2006;172:679–692. doi: 10.1083/jcb.200508154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L., et al. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol. 2008;183:223–239. doi: 10.1083/jcb.200805092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski K. T., You Z. H., Graham P. J., Steitz J. A. Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol. Cell. 1998;2:629–638. doi: 10.1016/s1097-2765(00)80161-6. [DOI] [PubMed] [Google Scholar]
- Weinstein L. B., Steitz J. A. Guided tours: from precursor snoRNA to functional snoRNP. Curr. Opin. Cell Biol. 1999;11:378–384. doi: 10.1016/S0955-0674(99)80053-2. [DOI] [PubMed] [Google Scholar]
- Zhou X. D., Liu L. Z., Qian G. S., Huang G. J., Chen J. [Cloning and sequence analysis of a new, full-length cDNA fragment of drug resistance-related gene in human lung adenocarcinoma] Ai Zheng. 2002;21:341–345. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.