SUMMARY
Ubiquitination is one of the most prevalent protein posttranslational modifications in eukaryotes, and its malfunction is associated with a variety of human diseases. Despite the significance of this process, the molecular mechanisms that govern the regulation of ubiquitination remain largely unknown. Here, we have used a combination of yeast proteome chip assays, genetic screening, and in vitro/in vivo biochemical analyses to identify and characterize eight novel in vivo substrates of the ubiquitinating enzyme Rsp5, a homolog of the human ubiquitin-ligating enzyme Nedd4 in yeast. Our analysis of the effects of a deubiquitinating enzyme, Ubp2, has demonstrated that an accumulation of K63-linked poly-ubiquitin chains results in processed forms of two substrates, Sla1 and Ygr068c. Finally, we have shown that the localization of another newly identified substrate, Rnr2, is Rsp5-dependent. We believe that our approach constitutes a paradigm for the functional dissection of an enzyme with pleiotropic effects.
INTRODUCTION
Post-translational modification (PTM), the covalent crosslinking of a modifying group to one or more amino acids of a protein, is of great interest because of its capacity to modulate the function, location, and stability of proteins as well as their interactions with other proteins [1]. Ubiquitination, one of the most prevalent PTMs in eukaryotes, has emerged as an important mechanism for intracellular signaling. Ubiquitin (Ub) is a highly conserved protein of ~8 kDa that is covalently attached to lysine (K) residues of target proteins, thereby drastically changing the fate of its substrates [2]. Ubiquitination occurs through a three-step process involving Ub-activating (E1), Ub-conjugating (E2) and Ub-ligating (E3) enzymes [3]. E3s determine substrate specificity, and mutations of these enzymes and/or their substrates can lead to a variety of human disorders, including neurodegenerative diseases and cancer [4].
The two major classes of E3 enzymes are the RING and HECT domain-containing E3s. HECT E3s differ from RING E3s in that they participate directly in the ubiquitination reaction by forming a ubiquitin-thioester intermediate and subsequently catalyzing the ubiquitination of the substrate [5]. Many of the E3s are indispensable for life because they serve as “hubs” to convey upstream signals and/or direct the fate of their downstream targets. Since the substrates of most E3 enzymes are unknown, we chose to take a proteome-wide approach to identifying these molecules, using yeast protein arrays as a platform [6]. We chose Rsp5 as our candidate E3 because it is essential for yeast viability, and it has pleiotropic effects on various intracellular pathways, including endocytosis [7], mitochondrial inheritance [8], maintenance of the actin cytoskeleton [9], drug resistance [10], biosynthesis of fatty acids [11], and protein sorting at the trans-Golgi network [12]. Rsp5 is also the closest yeast ortholog to Nedd4, a human HECT E3 that is involved in a congenital human hypertensive disorder known as Liddle's syndrome. Identification of the downstream targets of the yeast E3 enzyme should help identify the mechanisms by which Rsp5 signaling operates to regulate various crucial biologic functions in yeast.
EXPERIMENTAL PROCEDURES
Protein chip
The yeast protein chips were fabricated in-house as described previously [13].
Strains and constructs
The genotypes of the strains used in this study are listed in Table 1. Yeast strain FW1808 contains a temperature-sensitive (Ts) allele of Rsp5 (rsp5–1), derived from the wild-type (WT) strain FY56 [14]. To overproduce fusion proteins, constructs were transformed in strains FW1808 and FY56 using standard protocols [13]. UBP2 was deleted in strains FW1808 and FY56 using a standard yeast homologous recombination protocol [15, 16]. Deletion of UBP2 was confirmed by PCR analysis. Empty pEGH and pEGH-Rsp5 were used to transform the rsp5–1 and WT strains for the drug sensitivity experiments. Genes of interest were also chromosomally tagged with the 13×Myc epitope or C-terminal GFP (S65T) [17], on both the rsp5–1 and WT backgrounds.
TABLE 1.
Strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| FY56 | MATa ura3-52 his4-912ÆR5 lys2Æ128 | Huibregtse et al., 1997 |
| FW1808 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 | Huibregtse et al., 1997 |
| JYL01 | MATa ura3-52 his4-912ÆR5 lys2Æ128 RPN10∷URA3 | This study |
| JYL02 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 RPN10∷URA3 | This study |
| JYL03 | MATa ura3-52 his4-912ÆR5 lys2Æ128 RNR2∷URA3 | This study |
| JYL04 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 RNR2∷URA3 | This study |
| JYL05 | MATa ura3-52 his4-912ÆR5 lys2Æ128 NSL1∷URA3 | This study |
| JYL06 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 NSL1∷URA3 | This study |
| JYL07 | MATa ura3-52 his4-912ÆR5 lys2Æ128 NKP2∷URA3 | This study |
| JYL08 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 NKP2∷URA3 | This study |
| JYL09 | MATa ura3-52 his4-912ÆR5 lys2Æ128 SLA1∷URA3 | This study |
| JYL10 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 SLA1∷URA3 | This study |
| JYL11 | MATa ura3-52 his4-912ÆR5 lys2Æ128 SLA2∷URA3 | This study |
| JYL12 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 SLA2∷URA3 | This study |
| JYL13 | MATa ura3-52 his4-912ÆR5 lys2Æ128 SAR1∷URA3 | This study |
| JYL14 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 SAR1∷URA3 | This study |
| JYL15 | MATa ura3-52 his4-912ÆR5 lys2Æ128 TAF3∷URA3 | This study |
| JYL16 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 TAF3∷URA3 | This study |
| JYL17 | MATa ura3-52 his4-912ÆR5 lys2Æ128 YGR068C∷URA3 | This study |
| JYL18 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 YGR068C∷URA3 | This study |
| JYL19 | MATa ura3-52 his4-912ÆR5 lys2Æ128 RPN10∷URA3 ubp2Δ∷ KanMX | This study |
| JYL20 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 RPN10∷URA3 ubp2Δ∷ KanMX | This study |
| JYL21 | MATa ura3-52 his4-912ÆR5 lys2Æ128 RNR2∷URA3 ubp2Δ∷ KanMX | This study |
| JYL22 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 RNR2∷URA3 ubp2Δ∷ KanMX | This study |
| JYL23 | MATa ura3-52 his4-912ÆR5 lys2Æ128 NSL1∷URA3 ubp2Δ∷ KanMX | This study |
| JYL24 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 NSL1∷URA3 ubp2Δ∷ KanMX | This study |
| JYL25 | MATa ura3-52 his4-912ÆR5 lys2Æ128 NKP2∷URA3 ubp2Δ∷ KanMX | This study |
| JYL26 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 NKP2∷URA3 ubp2Δ∷ KanMX | This study |
| JYL27 | MATa ura3-52 his4-912ÆR5 lys2Æ128 SLA1∷URA3 ubp2Δ∷ KanMX | This study |
| JYL28 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 SLA1∷URA3 ubp2Δ∷ KanMX | This study |
| JYL29 | MATa ura3-52 his4-912ÆR5 lys2Æ128 SLA2∷URA3 ubp2Δ∷ KanMX | This study |
| JYL30 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 SLA2∷URA3 ubp2Δ∷ KanMX | This study |
| JYL31 | MATa ura3-52 his4-912ÆR5 lys2Æ128 [pEGH] | This study |
| JYL32 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 [pEGH] | This study |
| JYL33 | MATa ura3-52 his4-912ÆR5 lys2Æ128 [pJYL01] | This study |
| JYL34 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 [pJYL01] | This study |
| JYL35 | MATa ura3-52 his4-912ÆR5 lys2Æ128 ubp2Δ∷ KanMX | This study |
| JYL36 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 ubp2Δ∷ KanMX | This study |
| JYL37 | MATa ura3-52 his4-912ÆR5 lys2Æ128 RPN10-Myc∷KanMX6 | This study |
| JYL38 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 RPN10-MYC∷KanMX6 | This study |
| JYL39 | MATa ura3-52 his4-912ÆR5 lys2Æ128 RNR2-MYC∷KanMX6 | This study |
| JYL40 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 RNR2-MYC∷KanMX6 | This study |
| JYL41 | MATa ura3-52 his4-912ÆR5 lys2Æ128 NSL1-MYC∷KanMX6 | This study |
| JYL42 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 NSL1-MYC∷KanMX6 | This study |
| JYL43 | MATa ura3-52 his4-912ÆR5 lys2Æ128 SLA1-MYC∷KanMX6 | This study |
| JYL44 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 SLA1-MYC∷KanMX6 | This study |
| JYL45 | MATa ura3-52 his4-912ÆR5 lys2Æ128 TAF3-MYC∷KanMX6 | This study |
| JYL46 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 TAF3-MYC∷KanMX6 | This study |
| JYL47 | MATa ura3-52 his4-912ÆR5 lys2Æ128 YGR068C-MYC∷KanMX6 | This study |
| JYL48 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 YGR068C-MYC∷KanMX6 | This study |
| JYL49 | MATa ura3-52 his4-912ÆR5 lys2Æ128 RPN10-MYC∷KanMX6 ubp2Δ∷URA3 | This study |
| JYL50 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 RPN10-MYC∷KanMX6 ubp2Δ∷URA3 | This study |
| JYL51 | MATa ura3-52 his4-912ÆR5 lys2Æ128 RNR2-MYC∷KanMX6 ubp2Δ∷URA3 | This study |
| JYL52 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 RNR2-MYC∷KanMX6 ubp2Δ∷URA3 | This study |
| JYL53 | MATa ura3-52 his4-912ÆR5 lys2Æ128 NSL1-MYC∷KanMX6 ubp2Δ∷URA3 | This study |
| JYL54 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 NSL1-MYC∷KanMX6 ubp2Δ∷URA3 | This study |
| JYL55 | MATa ura3-52 his4-912ÆR5 lys2Æ128 SLA1-MYC∷KanMX6 ubp2Δ∷URA3 | This study |
| JYL56 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 SLA1-MYC∷KanMX6 ubp2Δ∷URA3 | This study |
| JYL57 | MATa ura3-52 his4-912ÆR5 lys2Æ128 TAF3-MYC∷KanMX6 ubp2Δ∷URA3 | This study |
| JYL58 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 TAF3-MYC∷KanMX6 ubp2Δ∷URA3 | This study |
| JYL59 | MATa ura3-52 his4-912ÆR5 lys2Æ128 YGR068C-Myc∷KanMX6 ubp2Δ∷URA3 | This study |
| JYL60 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 YGR068C-Myc∷KanMX6 ubp2Δ∷URA3 | This study |
| JYL61 | MATa ura3-52 his4-912ÆR5 lys2Æ128 RNR2-GFP∷KanMX6 | This study |
| JYL62 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 RNR2-GFP∷KanMX6 | This study |
| JYL63 | MATa ura3-52 his4-912ÆR5 lys2Æ128 RNR2-GFP∷KanMX6 ubp2Δ∷URA3 | This study |
| JYL64 | MATa rsp5-1 ura3-52 his4-912ÆR5 lys2Æ128 RNR2-GFP∷KanMX6 ubp2Δ∷URA3 | This study |
| Y258 | Mata, pep4-3, his4-580, ura3-52, leu2-3, 112 | Zhu et al., 2001 |
Protein purification
GST proteins were purified from 50 ml of culture at the desired temperature, as described previously [13]. The concentration of each purified protein was either estimated on gels stained with Coomassie blue (using bovine serum albumin [BSA] as a standard) or was determined using the BCA™ protein assay kit (Pierce). To remove the GST tag, GST fusion proteins were digested with thrombin (Sigma) at 22° C for 2.5 h according to the manufacturer's instruction.
Ubiquitination reactions on the yeast proteome chips
A reaction mixture consisting of 5 μM E1 (Uba1), 25 μM E2 (UbcH5), and 0.04 μg/μL Ub, with or without the addition of 0.075 μg/μL E3 (GST-Rsp5 or Ubr1), was prepared in reaction buffer (25 mM Tris, pH 7.6, with 50 mM NaCl, 10 mM MgCl2, 4 mM ATP, and 0.5 mM DTT). A yeast proteome chip was incubated with 100 μL of the reaction mixture for 90 min at 37° C, and then subjected to three15-min washes with 0.5 M NaCl, followed by three 15-min washes with 0.5% SDS at room temperature. The chip was then probed with anti-Ub (3,000-fold dilution) (Covance) and anti-GST (5,000-fold dilution) (Chemicon) antibodies, and detected with Cy3- (1:200) and Cy5- (1:200) labeled secondary antibodies, respectively (Jackson ImmunoResearch). The signals were acquired and analyzed by using GenePix software to determine the relative ubiquitination levels of each of the proteins on the chip.
Dosage lethality/suppression interaction
The plasmid constructs of 86 candidate substrates and 64 random proteins were transformed into strains FW1808 and FY56. The growth of each transformant was monitored by plating five-fold serial dilutions of the cells in quadruplicate on SC-Ura agar containing either 2% glucose or 2% galactose at 30° C (permissive temperature) or 34° C (semi-permissive temperature) for 3–4 days.
In-liquid ubiquitination
In-liquid ubiquitination reactions were carried out in test tubes using the buffer system mentioned above at 37° C for 90 min. The outcome of the ubiquitination reactions was determined by immunoblot analysis using an anti-GST antibody.
In vivo ubiquitination
After the yeast culture had been shifted from 30° C to 37° C for 2 h, the GST-tagged proteins of interest were purified from rsp5–1, ubp2Δ, rsp5–1, ubp2Δ, and WT cells as described previously, followed by immunoblot analysis using the anti-Ub antibody. The same blot was later stripped and re-probed with the anti-GST antibody as a quantity control.
Protein turnover analysis
Cells expressing Myc-tagged Rnr2 were grown to log phase and treated with 100 mg/mL cycloheximide. The relative Rnr2-Myc amounts at the indicated time points (0', 5', 10', 30', 60', 90', 120', and 150') were determined by immunoblot analysis using an anti-c-Myc (9E10) antibody (Santa Cruz).
Drug screen
The drug sensitivity of the rsp5–1 and WT strains was assessed by plating five-fold serial dilutions of the cells in quadruplicate on agar with or without the drugs (Table 4) [18–20] for 3–4 days at 30° C and 34° C. Strains containing the empty vector were used as controls. Meanwhile, the rsp5–1 cells were transformed with a low-copy plasmid carrying RSP5 to determine whether the hypersensitivity of rsp5–1 to HU could be reversed at the semi-permissive temperature.
TABLE 4.
Drugs used in this study
| Drug | Conc. | Mechanism | Reference | Rsp5-dependent sensitivities |
|---|---|---|---|---|
| HU | 0.2M | ribonucleotide reductase dependent inhibition of DNA synthesis | Mulder K W, et al., 2005 | Yes |
| MMS | 0.03% | DNA-damaging alkylating agents | Lundin C, et al., 2005 | No |
| CPT | 40μg/ml | DNA-damaging topoisomerase-I inhibition | Wang LF, et al., 1997 | No |
FACS analysis
Approximately 1× 107 yeast cells were harvested at mid-log phase and fixed in 70% (v/v) ethanol overnight at 4° C. Fixed cells were sequentially incubated with 2 mg/ml RNase A solution for 2 h at 37° C, then 5 mg/mL pepsin solution (in 4.5 μl/ml HCl) for 1 h at 37° C and 50 μg/mL (1×) propidium iodide (PI; in 0.1 M Tris, pH 7.5, with180 mM NaCl and 70 mM MgCl2) overnight at 4° C. The samples were then resuspended in 0.1× PI, sonicated twice on low power for 5 sec, and analyzed using a Becton Dickinson FACSCalibur. Data were collected on 20,000 cells per sample.
RESULTS
Identification of in vitro substrates of Rsp5
We first took advantage of a combination of protein chip technology, genetic screening, and biochemical assays to identify and characterize in vivo substrates of Rsp5 (Fig. 1A). After optimizing surface chemistries and detection methods, we chose a FullMoon surface for the reactions and anti-Ub antibodies for detection. Each ubiquitination reaction was set up by incubating a proteome chip with a mixture of Ub monomer, ATP, and the E1 (Uba1), E2 (UbcH5), and E3 (Rsp5) enzymes (Fig. S1A) [13]. To ensure that only covalently bound ubiquitins were detected, the chips were washed under highly stringent and denaturing conditions after the reactions took place. To measure the Ub signals and the relative amounts of the spotted proteins, the chips were incubated with anti-Ub and -GST antibodies, followed by incubation with Cy3- and Cy5-labeled secondary antibodies to detect the anti-Ub and -GST antibodies, respectively (Fig. 1B, Fig. S1A). As a negative control, a separate proteome chip was incubated with the same reaction mixture lacking Rsp5. We also performed the ubiquitination reaction using Ubr1, a RING domain-containing E3 ligase, as an additional control. Each assay was performed in duplicate to ensure reproducibility.
Fig. 1. Identification of Rsp5 substrates using yeast proteome chips.
(A) Scheme of the study. Eighty-six candidate substrates identified by the chip assays and 64 randomly chosen proteins were subjected to the genetic interaction screening in parallel. Twenty-eight of the 86 showed dosage lethality/suppression, namely genetic interaction with Rsp5, while only two of the 64 showed a genetic interaction with Rsp5. Eight of the 28 that showed positive genetic interaction, and three of the 28 that did not show genetic interaction were confirmed as in vitro substrates of Rsp5 using traditional in-liquid assays. Further in vivo analysis confirmed a total of eight in vivo substrates of Rsp5. (B) An example of a specific substrate identified in the proteome chip analysis. Ygr068c was strongly ubiquitinated by Rsp5 (upper left panel), whereas it remained unmodified when Rsp5 was either not included (upper right panel) or was replaced by Ubr1 (lower left panel). In addition, its ubiquitination signals were independent of the amount of protein on the chip (lower right panel). (C) Gene ontology (GO) analysis of the top 100 candidate substrates of Rsp5. N and P indicate the number of substrates in each category and P-values of enrichment, respectively.
After normalizing the Cy3 (ubiquitin) signals against the Cy5 (GST) signals and removing regional artifacts in the data using Lowess normalization, we determined the degree of Rsp5-dependent ubiquitination by comparing the normalized signals between the Rsp5 and the negative control experiments (without Rsp5) (Table S1). The example in Fig. 1B indicates that Ygr068c was clearly ubiquitinated by Rsp5, whereas it remained unmodified when Rsp5 was not included or was replaced by Ubr1. We decided to focus on the top 100 in vitro substrates of Rsp5 for further analysis and characterization. By comparing these hits to the top 40 substrates of Ubr1, as determined in the same fashion, we found that only Vma6 and Nkp2 were shared by both enzymes; this result indicates that specific substrates could be identified using in vitro ubiquitination reactions on a proteome chip.
Gene ontology and statistical analyses revealed no significant protein motifs (e.g., PXY motifs) shared by the substrate candidates. Forty-two proteins shared the same subcellular localization with Rsp5; however, Rsp5 has been localized to multiple subcellular compartments, including the Golgi, cytoplasm, endosomal membrane, plasma membrane, and mitochondria. Furthermore, none of them shared the same biological process with Rsp5 (Fig. 1C). Among the 145 proteins that had previously been shown to bind to either the full-length or the WW domains of Rsp5 [21, 22], only five (Sla2, Met12, Bna5, Ygr068c, and Yjl084c) were found on the hit list, and four (excluding Sla2) of them contain a PXY motif [23]. Therefore, these data were unlikely to help us generate a robust hit list for further validation. These results therefore prompted us to conduct genetic and alternative in vitro assays before carrying out the more rigorous in vivo investigations.
Many in vitro substrates interact genetically with RSP5
We picked 86 top candidates from the hit list and 64 other proteins at random to evaluate in terms of their potential synthetic dosage lethality or suppression interaction with RSP5 (Fig. 1A). The 150 genes we chose for this analysis were then overexpressed on both RSP5 temperature-sensitive (rsp5–1) and wild-type (WT) strain backgrounds (Table 1, Table 2) [14], in order to monitor potential differences in colony growth at both a semi-permissive temperature (34° C) and permissive temperature (30° C). Of the 86 candidates, 28 (32.6%) showed an obvious synthetic growth defect or suppression (Figs. 2 and S1C). Among these, Sla2 and Ygr068c have known physical interactions with Rsp5 [9, 24], while Sla1 and Taf3 could be co-purified with Rsp5 [25, 26]. In contrast, only two (Rim11 and Slt2) of the 64 (3.1%) random genes showed dosage lethality/suppression interaction with Rsp5 (data not shown). This dramatic difference in the likelihood of observing dosage lethality interactions suggests that combining the results for protein chip assays and genetic screening may significantly improve the probability of identifying in vivo substrates.
TABLE 2.
Plasmids used in this study.
| Plasmid | Details | Reference |
|---|---|---|
| pEGH | RGS-HisX6 (pEG(KG[2 micron/URA3]) | Zhu et al., 2001 |
| pJYL01 | RSP5 (pEGH[2 micron/URA3]) | This study |
| pFA6a-13Myc-kanM X6 | See reference | Longtine et al., 1998 |
| pFA6a-GFP(S65T)-kanMX6 | See reference | Longtine et al.,1998 |
| pRS406 | See reference | Wach et al. 1994 |
| pRS400 | [I/KanMX4] | Brachmann et al., 1998 |
Fig. 2. Dosage lethality/suppression interaction between the candidate substrates and Rsp5.
WT and rsp5–1 mutant were transformed with plasmids containing the GST-tagged candidate substrates of Rsp5 identified using the protein chip assays. The transformed strains were grown at 30° C or 34° C on 2% glucose (to inhibit overexpression of candidate substrates) or galactose (to induce over-expression of candidate substrates). The figure illustrates some examples of strains with (RPN10, RNR2, NSL1, SLA1, SLA2 and YGR068C), or without (NKP2) a synthetic dosage lethality/suppression interaction with RSP5.
Eight proteins are confirmed as in vivo substrates of Rsp5
The authenticity of the 28 identified proteins with positive dosage lethality/suppression interactions, as well as 28 other proteins from the 86 top candidates, was examined by in vitro ubiquitination assays. The extent of ubiquitination of each protein was determined by immunoblot analysis with anti-GST antibodies (Fig. 3, Fig. S1C). Eight proteins (Bro1, Nsl1, Rnr2, Rpn10, Sla1, Sla2, Taf3, and Ygr068c) of the positive group and three (Nkp2, Ygr206c, and Bna5) of the negative group were readily ubiquitinated by Rsp5 in solution.
Fig. 3. In vitro validation of the chip results.
Candidate substrates that showed positive genetic interactions with RSP5 were purified and subjected to traditional in-liquid ubiquination reactions. The outcomes of these reactions were determined by immunoblot analysis with anti-GST antibodies. Several candidate substrates, including Rpn10, Rnr2, Nsl1, Nkp2, Sla1, Sla2, Taf3, and Ygr068c, showed significant GST ladders after the addition of Rsp5.
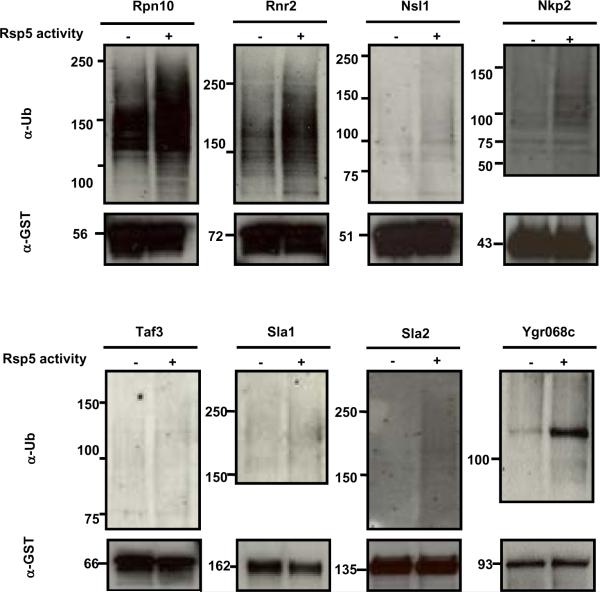
To better evaluate each of the validation steps performed thus far and to identify bona fide Rsp5 substrates, we decided to include all the 28 proteins that were positive in terms of dosage lethality/suppression interactions, the three proteins that were positive in liquid assays but negative for dosage lethality, and Rim11 and Slt2, which were positive in the genetic screens but negative in protein chip assays, in our further experiments to determine whether their ubiquitination is Rsp5-dependent in vivo. These 33 proteins were purified from both WT and rsp5–1 strains grown at the non-permissive temperature, and equal amounts of the purified proteins were then subjected to immunoblot analysis using anti-ubiquitin antibodies to detect their ubiquitinated forms (Fig. 4 and Fig. S1D). The same blot was then stripped and re-probed with anti-GST to visualize all their forms (Fig. 4 and Fig. S1D). The ubiquitinated substrates migrate slower than the unmodified forms, and this shift always correlates with the molecular weight of the unmodified form, excluding the possibility of contamination by other ubiquitinated proteins during the purification. Eight proteins (Rpn10, Rnr2, Nsl1, Nkp2, Sla1, Sla2, Taf3, and Ygr068c) showed Rsp5-dependent ubiquitination (Figs. 4 and S1D, Table 3). Therefore, these proteins are confirmed to be novel, in vivo substrates of Rsp5. In contrast, neither Rim11 nor Slt2 showed Rsp5-dependent ubiquitination, suggesting that positive results in dosage lethality/suppression screening can reflect indirect effects.
Fig. 4. In vivo validation of Rsp5 substrates.
GST-tagged candidate proteins were purified from WT and rsp5–1 strains grown at 37° C (non-permissive temperature) for 2 h. Ubiquitination was detected with anti-Ub antibodies (top panel); the same blot was stripped and re-probed with anti-GST antibody as a loading control (bottom panel). Eight proteins were confirmed to be in vivo substrates of Rsp5 by comparison of the ubiquitin signals in the presence and absence of the Rsp5 activity in vivo.
TABLE 3.
Eight Rsp5 substrates
| Substrate | Knockout phenotype* | Ranking on Chip | PPI* | GI** | Ubr1*** | Function |
|---|---|---|---|---|---|---|
| Rpn10 | Reduced fitness | 9 | − | S | − | Ubiquitin-dependent protein catabolism |
| Rnr2 | Inviable | 10 | − | S | − | DNA replication/ribonucleoside-diphosphate reductase activity |
| Nsl1 | Inviable | 31 | − | S | − | Chromosome segregation/Essential component of the MIND kinetochore complex |
| Nkp2 | Reduced fitness | 12 | − | N | + | Non-essential kinetochore protein |
| Sla1 | ts | 94 | + | W | + | Cytoskeletal protein binding protein |
| Sla2 | ts | 5 | + | S | − | required for assembly of the cortical actin cytoskeleton |
| Taf3 | Inviable | 66 | + | W | − | Transcription initiation from RNA polymerase II promoter |
| Ygr068c | Viable | 73 | + | S | − | Unknown |
Knockout phenotype and PPI (protein protein interaction) data base: YPD and SGD
GI (genetic interaction): dosage lethality/suppression defect induced by over-expression of potential substrates in this study; S indicates strong, W indicates weak, N indicates no genetic interaction.
Ubr1 data is based on protein chip results in this study.
Rsp5 regulates the processing of Sla1 and Ygr068c via K63-linked poly-Ub chains
The deubiquitinating enzyme Ubp2 has been reported to form a complex with Rsp5 and to antagonize Rsp5-dependent poly-ubiquitination by removing the K63-linked poly-Ub chains [5]. We predicted that the deletion of UBP2 would cause an accumulation of ubiquitinated substrates of Rsp5. To test this hypothesis, we determined the proportion of the total protein that was ubiquitinated for each of the eight validated substrates expressed on the rsp5–1, rsp5–1ubp2Δ, WT, and ubp2Δ backgrounds. Deletion of UBP2 in cells with intact Rsp5 activity resulted in a significant and specific increase in the ubiquitination signals in all eight of the substrates (Table 1, Figs. 5A and S2A).
Fig. 5. Effects of an Rsp5 antagonizing enzyme, Ubp2.
(A) GST-tagged substrates were over-expressed and purified from WT and ubp2Δ cells grown at 37° C (non-permissive temperature) for 2 h. The eight Rsp5 substrates all showed a significant increase in ubiquitin signals in ubp2Δ cells. Ubiquitination was detected with the anti-Ub antibodies (top panel), and the total amount of each protein loaded was determined with anti-GST antibody (bottom panel). Sla1 and Ygr068c were processed to specific smaller products when UBP2 was deleted (arrows). (B) When overexpressed, two substrates (Sla1 and Ygr068c), were processed to very specific smaller products in ubp2Δ but not in rsp5–1 ubp2Δ, indicating that the processing was Rsp5-dependent.
Intriguingly, two of the substrates (Sla1 and Ygr068c), when overexpressed, were processed in ubp2Δ strains to specific smaller products; however, they were not processed when Rsp5 function was dampened, indicating that this processing is Rsp5-dependent (Fig. 5B). These results suggest that Ubp2 specifically protects these two nonessential proteins from a previously unidentified type of processing, by removing poly-ubiquitin chains added by Rsp5.
When we assessed the endogenous protein levels of six of the substrates (Rpn10, Rnr2, Nsl1, Sla1, Taf3, and Ygr068c), we found that only Nsl1 increased slightly when Rsp5 function was impaired (Fig. S2C). Protein turnover analysis revealed that the protein levels of Rnr2 remained stable, even after 150 min of treatment with cycloheximide, in the case of both the rsp5–1 mutant and WT, indicating a role for Rsp5 beyond targeting proteins for degradation (Fig. S2D).
A novel function can be assigned to Rsp5
Two of the in vivo substrates identified in this study, Rnr2 and Nsl1, reside in pathways not currently known to be regulated by Rsp5. We also found that the rsp5–1 mutant was hypersensitive to hydroxyurea (HU), a specific inhibitor of ribonucleotide reductase (RNR), but not to methyl methanesulfonate (MMS) and camptothecin (CPT) (Fig. 6A), all of which lead to DNA damage responses by different mechanisms (Table 4) [18–20]. Moreover, introduction of low-copy-number plasmids containing the wild-type Rsp5 completely suppressed the hypersensitivity to HU in rsp5–1 strains at 34° C (Fig. S3A). Hypersensitivity to HU on an rsp5–1 mutant background could not be explained by cell cycle arrest (Fig. S3B), nor could it be explained by the ubiquitination status of Rnr2 (Fig. S3C).
Fig. 6. New functions can be assigned to Rsp5.
(A) Yeast viability was examined on YPD and YPD agar containing HU, MMS or CPT. Cells were incubated at the indicated temperatures for 3–4 days. The rps5–1 mutant, when grown at the semi-permissive temperature, showed hypersensitivity to HU, but not to two other DNA damaging agents, MMS and CPT. HU, hydroxyurea; MMS, methyl methanesulfonate; CPT, camptothecin. (B) Subcellular localization of Rnr2 is dependent on Rsp5 activity. The WT and rsp5–1 strains were grown to early log phase and split in half: one half of the culture was treated with HU and the other was left untreated, while the temperature was shifted to 37° C for 2 h. Rnr2 was seen to be mostly confined to the nucleus in rsp5–1, but was redistributed to the cytoplasm after HU treatment. However, when Rsp5 activity was restored by introducing a low-copy-number plasmid carrying RSP5 into the rsp5–1 strain, the localization of Rnr2 showed the same pattern as in the WT strain: Rnr2 was localized to both the cytoplasm and the nucleus, and it did not show any obvious redistribution after HU treatment. DNA was visualized by DAPI staining. (C) Rsp5 serves as a hub for regulating various crucial biological functions by ubiquitinating its downstream substrates, which exist in protein complexes involved in DNA repair (RNR complex), chromosome segregation (MIND and Ctf19 complex), actin assembly (Lap17-associated complex), and transcription initiation (TAF complex and 19S proteasome lid).
To further elucidate how Rsp5 regulates Rnr2, we asked whether the Rnr2 localization depends on Rsp5 activity. Using chromosomally GFP-tagged RNR2 in WT and rsp5–1 strains, we assessed the subcellular localization of Rnr2 at permissive and non-permissive temperatures and examined the effect of treatment with HU. We found that the majority of the Rnr2 molecules were redistributed from the nuclei to the cytoplasm in rsp5–1 in the presence of HU at the non-permissive temperature (Fig. 6B, Fig. S4). However, when Rsp5 was active, the same dose of HU had no effect on the subcellular localization of Rnr2 (data not shown). In WT cells, the localization of Rnr2 was not affected by either the temperature shift or HU treatment (Fig. 6B, Fig. S4). In the presence of HU, the pattern of Rnr2 localization in rsp5–1 obviously differed from that for the WT at the non-permissive temperature. Furthermore, when Rsp5 activity was restored by introducing a low-copy-number plasmid carrying RSP5 in the rsp5–1 strain, the localization of Rnr2 showed the same pattern as in the WT strain (Fig. 6B). Taken together, these results demonstrate that the Rsp5-depnedent ubiquitination of Rnr2 contributes to the substrate's resistance to HU, perhaps by regulating the subcellular localization of Rnr2.
DISCUSSION
Using traditional techniques to elucidate the molecular function of an enzyme with multiple roles in many pathways has always been challenging; identifying all the downstream substrates of such enzymes usually requires a systematic approach. The emerging protein chip technology offers a new tool for globally identifying in vitro substrates of various enzymes. Like other types of large-scale, high-throughput screening (e.g., yeast two-hybrid screening and gene expression profiling), investigators using this approach now face two challenges: how to identify bona fide, direct in vivo targets and how to establish a biological connection between a new target and its upstream modulator. Data integration has been proposed as a means of improving the accuracy of the “hits” derived from large-scale screening [27], but this strategy does not always work when obvious enrichment is lacking. Therefore, careful examination and evaluation of the robustness, reliability, and inherited bias of the proteomic approach is important for identifying the true substrates of an enzyme.
In this study, the use of chip assay allowed us to quickly narrow down the potential substrates from 5,800 to about 100 proteins. By using genetic screening and a less sensitive, solution-based ubiquitination reaction, we were able to rapidly reduce the number of candidates to eight; seven (87.5%) of these were further validated as true substrates of Rsp5 by more rigorous in vivo analyses. This combination of the three methods dramatically improved the probability of identifying bona fide substrates of Rsp5.
Of the yeast strains harboring knockout mutations of the eight in vivo substrates identified in this study, three (rnr2Δ, nsl1Δ, and taf3Δ) are lethal, two (slaΔ1 and sla2Δ) are temperature sensitive, and two (rpn10Δ and nkp2Δ) show reduced fitness (Table 3) [28]. It is intriguing that many downstream targets of Rsp5 are also essential for viability. Although previous studies have suggested that the essential requirement for Rsp5 is related to the oleic acid pathway [29], our data seem to indicate that the vital importance of Rsp5 is correlated with its effects on several additional essential pathways. On the basis of the known functions of the substrates we have identified, it is likely that Rsp5 plays a pivotal role in a complicated network that regulates various crucial downstream events, including proteasome function, DNA synthesis, chromosome segregation, cytoskeleton assembly/ endocytosis, and transcription (Fig. 6C). These results should encourage in-depth studies related to the function of ubiquitin E3 ligase.
Among the 145 reported Rsp5-interacting proteins containing the PXY motif [30], only five were ubiquitinated in vitro by Rsp5 on the protein chip, and two of the five were confirmed as in vivo substrates of Rsp5. This situation may be explained by the notion that a significant portion of these proteins acts as adaptors for Rsp5. Emerging evidence suggests that many Rsp5-interacting proteins recruit Rsp5 to particular subcellular compartments to facilitate the ubiquitination of their substrates [31]. Moreover, the WW domains of Rsp5 may interact only with phosphorylated PXY motifs, and some adaptor proteins may mediate substrate interactions from which proline-rich PXY motifs are absent [32]. Therefore, it would be useful to carry out the Rsp5 ubiquitination on protein chips in the presence of an adaptor protein or after pre-treatment with specific kinases.
Previous studies have identified 11 proteins as bona fide substrates of Rsp5, as determined by Rsp5-dependent ubiquitination in vivo [12, 14, 24, 26, 29, 33]. Most of these substrates either have a low protein abundance on chips because they are membrane proteins and are therefore difficult to express and purify (Fur4, Gap1, Lsb1, Sna4, and Ydl203c), or because they have proved to be unstable in separate attempts at purification (Rpb1 and Ste2). For the rest (Rvs167, Mga2, Sna3, and Ydl203c), we observed only moderate ubiquitin signals for Rvs167 and Sna3, suggesting that the protein chip approach has its own bias against certain proteins. Moreover, Gupta et al. used a similar proteome-wide approach but found a different spectrum of substrates (Fig. S5) [24]. The discrepancy can conceivably be explained by the different strategies used to validate the candidates. In our study, we found that combining the result of on-chip biochemical experiments with genetic interaction profiling significantly increased the probability of identifying biologically relevant substrates (Fig. 1A). Our results further suggest that protein-protein interaction is not required for substrate identification, since only four of the eight validated substrates have been previously shown to interact with Rsp5.
Among the validated in vivo substrates of Rsp5, Rnr2 is of particular interest. Rnr2 is a highly conserved ribonucleotide reductase (RNR) that coverts nucleotides to deoxynucleotides in a reaction dependent on a diferric-tyrosyl cofactor [34]. A heterozygous null mutant of RNR2 is associated with hypersensitivity to DNA damage and to treatment with HU, a chemical inhibitor of the RNRs [19]. After DNA damage, Rnr4 is redistributed within cells, perhaps reflecting an as yet-unidentified posttranslational mechanism [35]. Our results suggest that Rnr2 localization is determined by Rsp5 activity as well as HU treatment. Rnr2 was found in both the nucleus and the cytoplasm in the WT strain, but the majority of the Rnr2 was localized to the nucleus in the rsp5–1 mutant. The fact that the RNR complex needs to be present in the cytoplasm in order to be functional [34] may help explain why the rsp5–1 mutant is hypersensitive to HU at the semi-permissive temperature.
We conclude that a combination of proteome microarray-based biochemical assays and genetic interaction screens offers a powerful platform for identifying bona fide substrates of enzymes involved in various cellular pathways and our approach constitutes a paradigm for the functional dissection of an enzyme with pleiotropic effects.
Supplementary Material
ACKNOWLEDGMENTS
This manuscript is dedicated to the memory of Dr. Cecile Pickart. We thank Drs. Jef Boeke and Eric Cooper for critical comments and helpful discussion, Dr. Andrew Emili for providing strains FW1808 and Fy56, and Dr. Deborah McClellan for editorial assistance. This work is supported in part by funding from the NIH (U54RR020839, EY015684, GM28470). JYL is supported in part by NTU and NTUH.
The abbreviations used are
- PTM
post-translational modification
- K
lysine
- Ub
ubiquitin
- RING
really interesting new gene-ankyrin
- HECT
homologous to the E6-AP carboxyl terminus
- GST
glutathione S-transferase
- GFP
green fluorescent protein
REFERENCES
- 1.Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat Biotechnol. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 2.Haglund K, Dikic I. Ubiquitylation and cell signaling. Embo J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phizicky E, Bastiaens PI, Zhu H, Snyder M, Fields S. Protein analysis on a proteomic scale. Nature. 2003;422:208–215. doi: 10.1038/nature01512. [DOI] [PubMed] [Google Scholar]
- 4.Robinson PA, Ardley HC. Ubiquitin-protein ligases--novel therapeutic targets? Curr Protein Pept Sci. 2004;5:163–176. doi: 10.2174/1389203043379800. [DOI] [PubMed] [Google Scholar]
- 5.Kee Y, Lyon N, Huibregtse JM. The Rsp5 ubiquitin ligase is coupled to and antagonized by the Ubp2 deubiquitinating enzyme. Embo J. 2005;24:2414–2424. doi: 10.1038/sj.emboj.7600710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ardley HC, Robinson PA. E3 ubiquitin ligases. Essays Biochem. 2005;41:15–30. doi: 10.1042/EB0410015. [DOI] [PubMed] [Google Scholar]
- 7.Gajewska B, Shcherbik N, Oficjalska D, Haines DS, Zoladek T. Functional analysis of the human orthologue of the RSP5-encoded ubiquitin protein ligase, hNedd4, in yeast. Curr Genet. 2003;43:1–10. doi: 10.1007/s00294-003-0371-x. [DOI] [PubMed] [Google Scholar]
- 8.Fisk HA, Yaffe MP. A role for ubiquitination in mitochondrial inheritance in Saccharomyces cerevisiae. J Cell Biol. 1999;145:1199–1208. doi: 10.1083/jcb.145.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaminska J, Gajewska B, Hopper AK, Zoladek T. Rsp5p, a new link between the actin cytoskeleton and endocytosis in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 2002;22:6946–6948. doi: 10.1128/MCB.22.20.6946-6958.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andoh T, Hirata Y, Kikuchi A. PY motifs of Rod1 are required for binding to Rsp5 and for drug resistance. FEBS Lett. 2002;525:131–134. doi: 10.1016/s0014-5793(02)03104-6. [DOI] [PubMed] [Google Scholar]
- 11.Kaliszewski P, Ferreira T, Gajewska B, Szkopinska A, Berges T, Zoladek T. Enhanced levels of Pis1p (phosphatidylinositol synthase) improve the growth of Saccharomyces cerevisiae cells deficient in Rsp5 ubiquitin ligase. Biochem J. 2006;395:173–181. doi: 10.1042/BJ20051726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horak J. The role of ubiquitin in down-regulation and intracellular sorting of membrane proteins: insights from yeast. Biochim Biophys Acta. 2003;1614:139–155. doi: 10.1016/s0005-2736(03)00195-0. [DOI] [PubMed] [Google Scholar]
- 13.Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidlingmaier S, Houfek T, Mitchell T, Miller P, Dean RA, Gerstein M, Snyder M. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 14.Huibregtse JM, Yang JC, Beaudenon SL. The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin-protein ligase. Proc Natl Acad Sci U S A. 1997;94:3656–3661. doi: 10.1073/pnas.94.8.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol Cell Biol. 2005;25:637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 17.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 18.Lundin C, North M, Erixon K, Walters K, Jenssen D, Goldman AS, Helleday T. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 2005;33:3799–3811. doi: 10.1093/nar/gki681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mulder KW, Winkler GS, Timmers HT. DNA damage and replication stress induced transcription of RNR genes is dependent on the Ccr4-Not complex. Nucleic Acids Res. 2005;33:6384–6392. doi: 10.1093/nar/gki938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang LF, Ting CY, Lo CK, Su JS, Mickley LA, Fojo AT, Whang-Peng J, Hwang J. Identification of mutations at DNA topoisomerase I responsible for camptothecin resistance. Cancer Res. 1997;57:1516–1522. [PubMed] [Google Scholar]
- 21.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fleischer TC, Weaver CM, McAfee KJ, Jennings JL, Link AJ. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 2006;20:1294–1307. doi: 10.1101/gad.1422006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hesselberth JR, Miller JP, Golob A, Stajich JE, Michaud GA, Fields S. Comparative analysis of Saccharomyces cerevisiae WW domains and their interacting proteins. Genome Biol. 2006;7:R30. doi: 10.1186/gb-2006-7-4-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta R, Kus B, Fladd C, Wasmuth J, Tonikian R, Sidhu S, Krogan NJ, Parkinson J, Rotin D. Ubiquitination screen using protein microarrays for comprehensive identification of Rsp5 substrates in yeast. Mol Syst Biol. 2007;3:116. doi: 10.1038/msb4100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Auty R, Steen H, Myers LC, Persinger J, Bartholomew B, Gygi SP, Buratowski S. Purification of active TFIID from Saccharomyces cerevisiae. Extensive promoter contacts and co-activator function. J Biol Chem. 2004;279:49973–49981. doi: 10.1074/jbc.M409849200. [DOI] [PubMed] [Google Scholar]
- 26.Stamenova SD, Dunn R, Adler AS, Hicke L. The Rsp5 ubiquitin ligase binds to and ubiquitinates members of the yeast CIN85-endophilin complex, Sla1-Rvs167. J Biol Chem. 2004;279:16017–16025. doi: 10.1074/jbc.M313479200. [DOI] [PubMed] [Google Scholar]
- 27.Demichelis F, Sboner A, Barbareschi M, Dell'Anna R. TMABoost: an integrated system for comprehensive management of tissue microarray data. IEEE Trans Inf Technol Biomed. 2006;10:19–27. doi: 10.1109/titb.2005.855540. [DOI] [PubMed] [Google Scholar]
- 28.Deutschbauer AM, Jaramillo DF, Proctor M, Kumm J, Hillenmeyer ME, Davis RW, Nislow C, Giaever G. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics. 2005;169:1915–1925. doi: 10.1534/genetics.104.036871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shcherbik N, Zoladek T, Nickels JT, Haines DS. Rsp5p is required for ER bound Mga2p120 polyubiquitination and release of the processed/tethered transactivator Mga2p90. Curr Biol. 2003;13:1227–1233. doi: 10.1016/s0960-9822(03)00457-3. [DOI] [PubMed] [Google Scholar]
- 30.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 31.Sullivan JA, Lewis MJ, Nikko E, Pelham HR. Multiple interactions drive adaptor-mediated recruitment of the ubiquitin ligase rsp5 to membrane proteins in vivo and in vitro. Mol Biol Cell. 2007;18:2429–2440. doi: 10.1091/mbc.E07-01-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harty RN, Brown ME, Wang G, Huibregtse J, Hayes FP. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc Natl Acad Sci U S A. 2000;97:13871–13876. doi: 10.1073/pnas.250277297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kus B, Gajadhar A, Stanger K, Cho R, Sun W, Rouleau N, Lee T, Chan D, Wolting C, Edwards A, Bosse R, Rotin D. A high throughput screen to identify substrates for the ubiquitin ligase Rsp5. J Biol Chem. 2005;280:29470–29478. doi: 10.1074/jbc.M502197200. [DOI] [PubMed] [Google Scholar]
- 34.An X, Zhang Z, Yang K, Huang M. Cotransport of the heterodimeric small subunit of the Saccharomyces cerevisiae ribonucleotide reductase between the nucleus and the cytoplasm. Genetics. 2006;173:63–73. doi: 10.1534/genetics.105.055236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao R, Zhang Z, An X, Bucci B, Perlstein DL, Stubbe J, Huang M. Subcellular localization of yeast ribonucleotide reductase regulated by the DNA replication and damage checkpoint pathways. Proc Natl Acad Sci U S A. 2003;100:6628–6633. doi: 10.1073/pnas.1131932100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.