Abstract
Spin scavenging combined with chromatographic and mass spectrometric procedures can, in principle, be employed to detect and identify protein-based radicals within complex biological matrices. This approach is based on the well-known ability of stable synthetic nitroxide radicals to scavenge carbon-centered radicals, forming stable diamagnetic addition products. Hence, characterization of these addition products would allow for the identification of specific free radicals. In the present work, we have explored the use of the stable nitroxide radical 4-hydroxy-2,2,6,6-tetramethylpiperidine 1-oxyl (TEMPOL) in scavenging protein-based radicals generated in a horse heart metmyoglobin/hydrogen peroxide (metMb/H2O2) system. Inclusion of a substoichiometric amount of TEMPOL in the metMb/H2O2 system resulted in a complete loss of peroxyl and tyrosyl radical signals and effectively inhibited the formation of oxidatively-damaged heme species, as monitored by electron paramagnetic resonance and reverse-phase liquid chromatography. Scavenging of globin radicals by TEMPOL did not lead to the formation of stable diamagnetic addition adducts; in fact, reverse-phase liquid chromatographic studies and oxygen electrode measurements indicated that TEMPOL acts as a catalyst and is recycled in this system. The oxoammonium cation generated in the course of this reaction initiated secondary reactions resulting in the formation of a free carbonyl on the N-terminal Gly-residue of the protein. This oxidative deamination was confirmed through the combined use of reverse-phase liquid chromatographic purification, tandem MS experiments and chemical analysis (e.g. by use of 2,4-dinitrophenyl hydrazine). The results reveal the pitfalls inherent in using stable nitroxide radicals such as TEMPOL to identify sites of radical formation on hemoproteins.
INTRODUCTION
Oxygen- and nitrogen-centered radicals and associated reactive species, such as hydrogen peroxide and peroxynitrite, are continuously produced in living organisms and are potentially damaging to cellular lipids, proteins, and nucleic acids (1). Due to their high abundance in cells and biological fluids, proteins are the most frequent targets for oxygen and nitrogen reactive species (2). Reactions of free radicals and associated reactive agents with proteins can lead to the formation of further radical species centered on the protein structure (2). Understanding the biochemistry of these protein-centered radicals has become of increasing interest as their roles in a wide range of diseases and disorders have become apparent (3). In addition, protein radicals have been implicated in the normal catalytic mechanisms of a growing number of enzymes (4).
Protein radicals have traditionally been studied either through direct electron paramagnetic resonance (EPR) or by spin trapping with EPR detection (5, 6). These methods suffer from a number of limitations including the rapid formation of diamagnetic addition products, which are not detectable by EPR. To overcome these limitations, analytical strategies based on mass spectrometric (MS) techniques have been developed. One highly sensitive technique that has been used by a number of groups (e.g., see ref (7) for a comprehensive list) combines spin trapping and coupled chromatographic/MS procedures to localize the actual protein residues labeled by the spin trap.
A variation of the above technique, the spin scavenging approach, is based on the well-known property of stable synthetic nitroxide radicals to react rapidly with carbon-centered radicals (108–109 M−1s−1) to yield diamagnetic addition products via radical-radical recombination (8, 9). Hence, characterization of these addition products using chromatographic and mass spectrometric analyses can, in principle, allow for the identification of specific free radicals. This approach has been used by a group of investigators (10) to examine the sites of protein radical formation in myoglobin and cytochrome c peroxidase exposed to H2O2. These authors reported the observation of ten nitroxide-labeled peptides following the treatment of the proteins with H2O2 in the presence of the nitroxide reagent 2,2,6,6-tetramethylpiperidine 1-oxyl (TEMPO), enzymatic digestion, and mass spectrometric analysis. Although the nature of the trapped amino acid residues was not unambiguously established by tandem mass spectrometry, the authors concluded that many of the TEMPO-peptide adduct species had survived the digestion and processing period and were stable under the enzymatic and mass spectrometric conditions used (10).
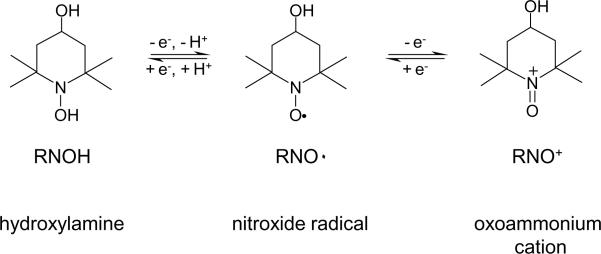
A potential complication of the spin scavenging approach is that nitroxides can `shuttle' among three oxidation states, being readily reduced to hydroxylamines or oxidized to oxoammonium species, as shown in Scheme 1 for 4-hydroxy-2,2,6,6-tetramethylpiperidine 1-oxyl (TEMPOL). The spin scavenging approach should be applicable to identification of peptide and protein-based radicals provided that these hydroxylamine or oxoammonium species do not initiate secondary reactions producing even more addition products. In particular, such secondary reactions may potentially generate the same diamagnetic addition products as the direct reaction of the nitroxide radical with a protein radical, which would lead to incorrect assignments of the protein-based radicals. That oxoammonium species can indeed generate stable addition products has been shown for electron-rich olefin systems such as vinyl ethers and enamines where an adduct was detected by mass spectrometry (11).
Scheme 1.
Nitroxides can `shuttle' between the nitroxide radical form, the reduced form known as the hydroxylamine, and the oxidized form known as the oxoammonium cation.
The in vitro reactions of the ferric (FeIII, met) form of myoglobins (metMb) with H2O2 have served as a useful model for both investigating the general properties of protein radicals and developing protein-based radical detection methods (7, 10, 12–16). Myoglobin protein-centered radicals (globin radicals) can be observed directly by EPR (17), can be spin trapped (18), and in the case of sperm whale myoglobin, can give rise to dityrosine cross-linked oligomers (19, 20). Globin radicals have been detected primarily on tyrosine (21, 22), tryptophan (22), and cysteine (23, 24) residues through the use of spin trapping in conjunction with site-directed mutagenesis and detailed EPR characterization of the trapped radicals. In addition, at least one lysine and two histidine residues have been reported as minor contributors to the radical pool formed in the protein upon oxidation as determined by spin trapping and mass spectrometry (7, 12).
All of these globin radicals are associated with the formation of ferryl (FeIV=O) heme species in the reaction of the ferric heme with H2O2 (25) and are thought to arise via the initial generation of a transient Compound I-like species in which the ferryl is coupled to a porphyrin radical rather than a protein radical (26). Electron transfer from the protein to the porphyrin radical cation combined with deprotonation then produces the observed protein radicals.
At neutral and basic pH the protein radical is quenched rapidly by unknown mechanisms, leaving a small residual radical signal and a relatively long-lived ferryl species that is slowly autoreduced back to the ferric state (27, 28). Under acidic conditions some globin radicals can lead to cross-linking reactions with the heme porphyrin with a concomitant rapid reduction of the heme iron from the ferryl to the ferric state (29). Formation of the cross-linked heme-to-protein species requires the participation of both the ferryl heme and the protein radical (29). The pH dependence of the yield of the cross-linked form indicates that the protonated ferryl form (FeIVOH), which is formally equivalent to a hydroxyl radical coordinated to the ferric heme, is the active species (30). The precise site of attachment of the protein to the heme group could involve Tyr103 (29), but has not been unambiguously defined. However, site-directed mutagenesis experiments indicate that the distal histidine is vital for the formation of the cross-link (31). In addition to heme-protein cross-linking, peroxide interactions with Mb produce oxidatively modified hemes that are not covalently linked to the protein. The structures of some of these oxidatively damaged heme species have been previously been reported (32).
In the present work, we have explored the use of the stable nitroxide radical TEMPOL in scavenging protein-based radicals generated in the horse heart metmyoglobin/H2O2 system. To date, there has been no investigation of the effect of nitroxide radical reagents on the formation of damaged heme species produced in the reaction of metMb and H2O2. Therefore, a preliminary step in this investigation consisted of assessing, through reverse-phase liquid chromatographic studies, the effect of a nitroxide reagent on the formation of these H2O2-damaged heme species. Following this assessment, we used EPR, reverse-phase liquid chromatographic purifications, and tandem MS experiments to identify specific residues of the polypeptide chain that are modified by TEMPOL in scavenging globin-based radicals.
EXPERIMENTAL PROCEDURES
Materials
Horse heart metmyoglobin (metMb) (USB, Cleveland, Ohio) was purified before use by passage through a Sephadex G-25 (PD-10) gel filtration column (GE Healthcare Bio-Sciences, Piscataway, NJ) and elution with 50 mM potassium phosphate buffer, pH 6.8, containing 50 μM diethylenetriaminepentaacetic acid (DTPA). The concentration of metHb was determined from the Soret maximum at 408 nm (εmetMb,408 = 188 mM−1cm−1) (33). The concentration of apoMb solutions was determined at 280 nm (εapoMb,280 = 13.5 mM−1cm−1) (34). Catalase from beef liver (suspension, 64000 units per mg) and chymotrypsin (from bovine pancreas, modified, sequencing grade) were obtained from Roche Molecular Biochemicals (Indianapolis, IN). All aqueous solutions were prepared using Picopure 2UV Plus system (Hydro Services and Supplies, Inc, RTP, NC) equipped with a 0.2 μm pore size filter. Diluted H2O2 solutions, prepared from a 30% solution (Fisher Scientific Co., Fairlawn, NJ), were used within 1 h of preparation. The H2O2 concentration was confirmed by absorbance measurements at 240 nm (εH2O2, 240 = 39.4 M−1cm−1) (35). NaOCl solutions were prepared immediately before use by dilution of 10% reagent grade sodium hypochlorite (Aldrich, Milwaukee, WI). The concentrations of NaOCl solutions were determined at 292 nm and pH 11, assuming a molar extinction coefficient of 350 M−1cm−1 (36). TEMPOL concentration was calculated using a molar extinction coefficient of 13.4 M−1cm−1 (37). The spin trap 3,5-dibromo-4-nitrosobenzene sulfonate (DBNBS) was synthesized according to the published procedure (38). All other chemicals were of analytical grade and were purchased from Sigma (St Louis, MO) or Roche Molecular Biochemicals (Indianapolis, IN). Absorption spectra were recorded on a Cary 100 UV-visible spectrometer (Varian Associates, Palo Alto, CA).
Standard Reaction Conditions
All incubations were 500 μM metMb in 50 mM potassium phosphate buffer, pH 6.8, and contained 50 μM diethylenetriaminepentaacetic acid (DTPA) to prevent trace-metal redox chemistry. Typically, the reactions were initiated by the addition of 1 eq of H2O2 and carried out at 25 °C in the presence or absence of an appropriate concentration of the nitroxide reagent or of 1 eq of the spin trap DBNBS, as indicated in the figure legends. Unless otherwise noted, the reactions were allowed to proceed for 30 s, after which the excess H2O2 was removed by adding catalase (250 units/ml). The resulting solutions were then quickly loaded onto a PD-10 gel filtration column and eluted with 50 mM potassium phosphate buffer, pH 6.8. This was done primarily to remove unreacted nitroxide reagent or DBNBS from the samples and to minimize the contribution of secondary reactions (i.e. reactions with hydroxylamine and oxoammonium species or the ene reaction that DBNBS is known to undergo with tryptophan (39)). In some experiments, the product mixtures were directly injected onto the HPLC column or into the ESI source of the mass spectrometer, without a preliminary purification through the PD10 gel filtration column.
Purification of nitroxide + H2O2-and DBNBS + H2O2-modified Mb
After passage through the PD-10 column, the samples were further purified by reverse phase chromatography on a C4 HPLC column, as described in the HPLC analyses section. HPLC fractions were collected and the fractions containing nitroxide + H2O2- or DBNBS + H2O2-modified Mb (as judged by the appearance of a new chromatographic peak at 214 nm eluting at 10.3 min) were pooled, lyophilized, and stored at −70 °C for subsequent analysis.
Preparation of oxoammonium cations
The oxoammonium derivative of TEMPOL (TEMPOL+) was prepared by hypochlorous treatment of TEMPOL in acidic phosphate buffer essentially as described previously by Dragutan and Mehlhorn (40) for acetylamino-TEMPO species. Briefly, TEMPOL (5 mM) was incubated with a subequivalent amount of NaOCl (1.5 mM) in 50 mM phosphate buffer, pH 2.0. This resulted in a TEMPOL+/TEMPOL molar ratio of around 1.5 assuming 1:2 (HOCl:TEMPOL) stoichiometry. After a 10 s incubation, this solution was diluted 10-fold with 50 mM phosphate buffer, pH 6.8, after which methionine (4 mM) and catalase (250 units/ml) were added successively to ensure complete removal of unreacted HOCl and H2O2. Methionine has both a free amino group and a thio ether, both of which have shown to be targets for oxidation by oxoammonium cations (40). A rate constant of 17.9 M−1s−1 was estimated by UV visible spectrometry for the reaction of TEMPOL+ with methionine at pH 6.8 (not shown). The half-life of the oxoammonium cation should therefore be around 9.7 s under our experimental conditions. Consequently, diluted TEMPOL+/TEMPOL solutions were used immediately after preparation.
Preparation and Purification of oxoammonium-modified Mb
Reactions were conducted at 25° C in 50 mM potassium phosphate buffer, pH 6.8, containing 500 μM Mb and 150 μM TEMPOL+/TEMPOL solution prepared as above. After a 5 min incubation, unreacted TEMPOL+/TEMPOL was removed from the sample by passage through a PD-10 column and oxoammonium-modified Mb was purified by HPLC, as described in the HPLC analyses section. Fractions containing oxoammonium-modified Mb (corresponding to a new chromatographic peak at 214 nm eluting at 10.3 min) were pooled, lyophilized, and stored at −70 °C for subsequent analysis.
Derivatization with 2,4-dinitrophenyl hydrazine and isolation of 2,4-dinitrophenyl hydrazine-labeled Mb
Carbonyl groups formed on Mb were detected using a modified procedure of Levine et al (41). Briefly, solutions of intact/unreacted apoMb (control 1), H2O2-modified Mb (control 2), and/or nitroxide + H2O2-modified Mb, prepared as above, were added to an equal volume of 2,4-dinitrophenyl hydrazine (2,4-DNPH; 10 mM in 2.5 M HCl); the resulting mixture was incubated in the dark with stirring for 15 min before centrifugation to remove insoluble materials. The pellet was discarded and the supernatant was passed over a PD-10 column to remove unreacted 2,4-DNPH from the sample. The DNPH-labeled Mb was then purified by HPLC, as described in the HPLC analyses section. The HPLC fractions containing the 2,4-DNPH-labeled Mb (corresponding to a new chromatographic peak at 400 nm eluting at 15.9 min) were pooled, lyophilized and stored at −70 °C for subsequent analysis.
Protein Digestion
Lyophilized samples of intact/unreacted apoMb (control), oxoammonium-modified Mb, 2,4-DNPH-labeled Mb, nitroxide + H2O2- and/or DBNBS + H2O2-modified Mb, prepared as above, were resuspended in 500 μl of the digestion buffer (100 mM Tris-HCl, 10 mM CaCl2, pH 7.8) to a final concentration of ~50 μM and digested using a 20:1 substrate-to-chymotrypsin ratio for 16 hours at 25 °C. Reactions were stopped by injecting the final mixtures directly onto the C18 reverse phase HPLC column, which was operated as described in the HPLC analyses section. HPLC fractions were collected with a fraction collector, lyophilized and stored at −70 °C for subsequent MS analysis.
Reversed-phase HPLC analyses and purification
All (analytical and semi-preparative) reversed phase (RP)-HPLC experiments were carried out on a Chemstation 1100 liquid chromatography system (Agilent Technologies, Palo Alto, CA), equipped with a control module, vacuum degasser, binary pump, manual injector, and diode-array UV-visible and fluorescence detectors. HPLC fractions were collected using a fraction collector (Model 2110, Bio-Rad, Richmond, CA). Separations of intact and/or oxidatively modified protein and heme species were conducted using a Vydac 214TP5410, 4.6 × 100-mm, C4 RP-HPLC column (Vydac, Hesperia, CA). Fractionations of protein digests were achieved using a Vydac 218TP54, 4.6 mm × 250 mm, C18 RP-HPLC column. The mobile phase used for gradient elution consisted of a mixture of solvent A (0.1% trifluoroacetic acid in water) and solvent B (0.85% trifluoroacetic acid in acetonitrile), the injection volume was 200 μl, and the separations were performed at a flow rate of 0.8 ml/min. The eluents were monitored with the UV-visible detector set at 214, 280 and 400 nm and with the fluorescence detector set at λex = 285 nm and λem = 360 nm. The gradient conditions were as follows:
i) For the analysis of intact and oxidatively-modified heme species: 0–1 min, 37–40% B; 1–11 min, 40–43% B; 11–15 min, 43–50% B; then 95% B for 5 min and 37% B for 5 min to regenerate the column.
ii) For the purification of the nitroxide + H2O2- and oxoammonium-modified Mb, purification of the 2,4-dinitrophenyl hydrazine-labeled Mb and/or DBNBS + H2O2-modified Mb: 0–1 min, 37–40% B; 1–11 min, 40–43% B; then 95% B for 5 min and 37% B for 5 min to regenerate the column.
iii) For the fractionation of protein digests: 0–80 min; 0–50% B; then 95% B for 5 min and 0% B for 5 min to regenerate the column.
Capillary RP-HPLC was carried out on a 75 μm C18 column, at a flow rate of 250 nl/min using an Eksigent nanopump (Eksigent, Dublin, CA).
EPR Spectroscopy
EPR spectra were obtained with a Bruker (Billerica, MA, USA) Elexsys 500 X-band EPR spectrometer equipped with an ER4122SHQ cavity. The incubations were conducted essentially as described above for the preparation of nitroxide + H2O2-modified Mb. The reactions were initiated by the addition of 1 eq of H2O2. The incubation mixtures were then rapidly vortex mixed, placed into a standard tuberculin syringe and snap-frozen in liquid nitrogen 10 s or 30 s after the initiation of the reaction, as indicated in the figure legends. The tip of the plastic syringe containing the sample was cut off and the contents were pushed out while still frozen. The icicle was then transferred to a 500 ml-finger dewar (Wilmad, NJ, USA) filled with liquid nitrogen and the EPR spectra were recorded. To reduce the EPR noise caused by nitrogen bubbling, an EPR-silent material such as filter paper was inserted into the gap between the icicle and the inner wall of the finger dewar. Samples were also analyzed by EPR spectrometry at room temperature (see the Supporting Information). The incubations were performed as described above for 3 min. The samples were then transferred into a standard aqueous flat cell and analyzed by EPR. All samples were measured under the following instrumental parameters: modulation frequency, 100 kHz; modulation amplitude, 2.0 G; time constant, 1.3 s; receiver gain, 2 × 104, microwave power, 0.2 mW, scan time, 670 s, number of scans, 1. Peak area was estimated by double integration using standard WINEPR software.
Electrospray Mass Spectrometry
Electrospray ionization (ESI) and tandem mass spectra (MS/MS) were acquired with a Micromass Q-TOF Micro (Waters Micromass Corporation, Manchester, UK) hybrid tandem mass spectrometer. All experiments were performed in the positive ion mode. Lyophilized HPLC fractions of nitroxide- and oxoammonium-modified samples were resuspended in a minimal volume (~50 μL) of 50:50 water:acetonitrile containing 0.1% formic acid and infused into the mass spectrometer at ~300 nL/min using a pressure injection vessel (42). The instrumental parameters were as follows: capillary voltage, ~3300 V; sample cone voltage, 40 V; collision energy, between 10 and 50 eV; source temperature, 80 °C. The DBNBS + H2O2-modified samples were subjected to on-line data dependent LC/MS/MS analysis on an Applied Biosystems QSTAR XL (Foster City, CA) hybrid tandem mass spectrometer: 1 s MS acquisitions were followed by 3 s collision induced dissociation experiments on computer-selected precursor ions; the collisions were set automatically according to the mass and the charge state of the precursor ion subjected to the analysis. Data analysis of the nitroxide + H2O2- and oxoammonium-modified samples was accomplished with a MassLynx data system and MaxEnt deconvolution software supplied by the manufacturer. ProteinProspector's MS isotope program was used to predict isotope patterns of DBNBS + H2O2-modified samples, while the MS Product feature of the same source was used for peptide fragment calculations (both at http://prospector.ucsf.edu).
RESULTS
TEMPOL Inhibits Formation of Oxidatively-Damaged Heme Species
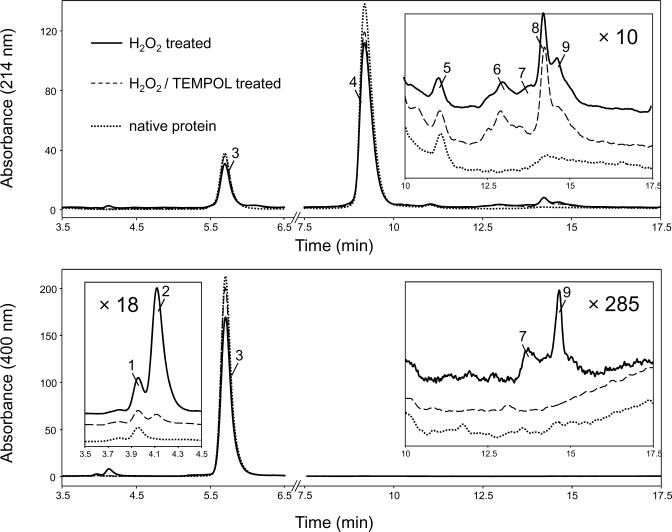
As previously reported (29, 30, 32), treatment of metMb with H2O2 produced a range of modifications in the heme group that was readily observed through HPLC analysis (Fig. 1). Detection at 400 nm predominantly measures the free heme whereas detection at 214 nm allows both heme and protein to be observed. The major component absorbing at 400 nm (peak 3) corresponds to the unmodified prosthetic heme moiety, which dissociates from the protein under the acidic conditions of the chromatography. The major component with absorption at 214 nm (peak 4) corresponds to the globin moiety. Minor protein impurities in the commercial preparation (peak 5) were difficult to remove but did not interfere with the subsequent studies.
Figure 1.
Detection of oxidatively-damaged heme species by reverse-phase HPLC. MetMb (500 μM) treated 30 s with one equivalent of H2O2 in the presence of 5 μM TEMPOL (dashed line). The controls are for metMb (500 μM) treated with one equivalent H2O2 for 30 s (solid line) and untreated sample (dotted line). Incubations were at pH 6.8 in 50 mM potassium phosphate and 25 °C. After 30 s, the reaction mixtures were diluted to a final concentration of 1 μM in heme and 200 μl of the diluted solutions were injected onto the reverse-phase C4 HPLC column. Insets: chromatograms off-set for clarity.
The HPLC profile of metMb after reaction with a stoichiometric amount of H2O2 (500 μM Mb, 500 μM H2O2) is shown in Fig. 1, solid line. A solution of native/untreated metMb was also analyzed as a control (dotted line). Upon treatment of metMb with H2O2, the peak corresponding to unmodified heme absorbing at 400 nm (peak 3) was decreased by approximately 18% relative to the control (i.e. unmodified heme content decreased by ~90 μM) and two altered `free' heme products with absorption at 400 nm (peaks 1 and 2) were observed eluting before the unmodified peak. Hemes that were covalently bound to the globin, as detected by their absorbance at both 400 and 214 nm, eluted between 13.7 and 14.7 min (peaks 7 and 9). These products were not present in the hydrolysates of samples of untreated myoglobin. In addition, the apoprotein peak (peak 4) was decreased by approximately 20% and several new peaks with absorption at 214 nm were observed (peaks 6 and 8), suggesting that peroxide treatment also induces significant alteration of the globin moiety; these results are in accord with previous observations (29, 30, 32).
The HPLC profile of myoglobin obtained after inclusion of 5 μM TEMPOL directly before the addition of peroxide in the above reaction system is shown in Fig. 1, dashed line. Comparison of this HPLC profile to that of native metMb showed that the unmodified heme content was similar to that of the untreated metMb (compare dashed and dotted lines, peak 3) and the two altered heme products eluting before the unmodified peak were just barely detectable above the baseline in the HPLC trace at 400 nm (peaks 1 and 2). Relative to the chromatogram for the H2O2-treated Mb, only minor changes were observed in the HPLC trace at 214 nm as a result of TEMPOL pretreatment (compare solid and dashed lines, peaks 6 and 8). These observations demonstrate that TEMPOL effectively inhibits the formation of oxidatively-damaged heme species but does not protect the globin moiety against peroxide-mediated damage. A relatively low TEMPOL concentration (5 μM) was sufficient to almost completely block the formation of substantially higher concentrations (~90 μM) of oxidatively-damaged heme species. This supports the view that TEMPOL acts as a catalyst and is recycled in this system.
Detection of Nitroxide + H2O2 -modified Mb
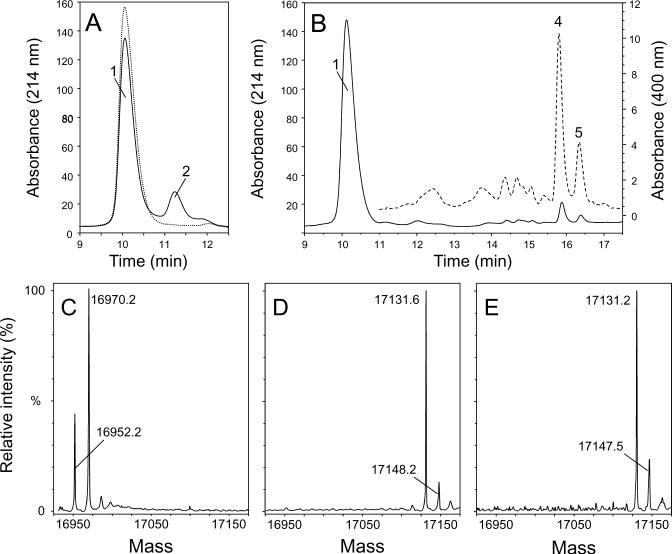
Additional experiments were performed using large excess of TEMPOL to increase our chance to scavenge radicals generated on the globin moiety. When a 10-fold excess of TEMPOL relative to Mb was added to the reaction mixture directly before the addition of peroxide (e.g. the same nitroxide:protein ratio than used by Wright and English in ref. (10)), a new peak with absorption at 214 nm was observed (Fig. 2A solid line, peak 2) along with a decrease in the intensity of the apoprotein peak (peak 1), suggesting that this type of treatment induces significant alteration of the globin moiety. Area integration of the chromatogram indicates that approximately 30% of the globin was converted to this form. In the absence of either TEMPOL or H2O2 (data not shown) or in presence of substoichiometric amounts of TEMPOL relative to Mb (Fig. 1, dashed line), this new peak was not observed. A time course study showed that this peak reached a maximum yield after 60 sec of incubation (data not shown).
Figure 2.
Detection of nitroxide + H2O2-modified Mb and deconvoluted electrospray mass spectra of HPLC purified Mb. (A) C4 RP-HPLC separation of nitroxide + H2O2-modified Mb. MetMb (500 μM) was treated 30 s with one equivalent of H2O2 in the presence of 5 mM TEMPOL. The negative control (dashed line) is for an untreated metMb sample. Incubation conditions were the same as in figure 1. The reaction mixture was injected onto the HPLC column 30 s after the addition of H2O2. (B) C4 RP-HPLC separation of 2,4-DNPH-treated nitroxide + H2O2-modified Mb. (C) Deconvoluted mass spectrum of nitroxide modified Mb (corresponding to peak 2 in Fig. 2A); (D) Deconvoluted mass spectrum of 2,4-DNPH-derivatized Mb eluting at 15.8 min (corresponding to peak 4 in Fig. 2B); (E) Deconvoluted mass spectrum of 2,4-DNPH-derivatized Mb eluting at 16.3 min (corresponding to peak 5 in Fig. 2B). Chromatography was performed at 214 nm for peptides bonds (Panel A and B) and 400 nm for 2,4-DNPH derivatives (Panel B, dashed line). The trace at 400 nm was obtained after injecting 5 times more protein onto the C4-HPLC column.
Previous investigations have established that the reaction of many proteins with oxygen and nitrogen radical species often lead to the formation of amino acid carbonyls (43). This type of modification can easily be assessed using 2,4-dinitrophenyl hydrazine (2,4-DNPH), a reagent which reacts with carbonyl groups to produce hydrazones (41). To evaluate the presence of carbonyl groups in the nitroxide + H2O2-modified globin sample, the above Mb/TEMPOL/H2O2 reaction mixture was passed over a PD-10 gel filtration column to remove unreacted TEMPOL from the sample. PD-10 fractions containing Mb were then pooled and reacted with 2,4-DNPH; the reaction was allowed to proceed for 15 min, after which the mixture was centrifugated to remove insoluble materials, passed on a PD-10 column to remove unreacted 2,4-DNPH, and analyzed by reversed-phase C4 HPLC.
The HPLC profile of nitroxide + H2O2-modified globin samples after incubation with 2,4-DNPH is shown in Fig. 2B. Upon 2,4-DNPH treatment, the nitroxide + H2O2-modified globin peak (peak 2 in Fig. 2A at 11.2 min) was not detected, whereas two new peaks with characteristic absorbance at 400 nm (peaks 4 and 5 in Fig. 2B) were observed. The UV spectra of the two peaks, as monitored on the HPLC diode-array detector, were typical of a 2,4-DNPH product with λmax at 365 nm (data not shown). No reactivity of 2,4-DNPH was observed with the native Mb (data not shown).
The reversed-phase C4 HPLC fractions containing the unreacted Mb (Fig. 2A and 2B, peak 1), the nitroxide + H2O2-modified globin (Fig 2A, peak 2) and the 2,4-DNPH-labeled Mb (Fig. 2B, peaks 4 and 5) were collected and further analyzed by electrospray ionization mass spectrometry. The deconvoluted ESI mass spectrum of unreacted Mb (data not shown) exhibits a molecular mass of 16952.4 (Mr calculated = 16952.5). The deconvoluted mass spectrum of nitroxide + H2O2-modified Mb (Fig. 2C) shows an abundant molecular mass of Mr = 16970.2 and a less abundant mass of Mr = 16952.2, similar to that of the intact protein. The spectra of 2,4-DNPH-labeled Mb (Fig. 2D and 2E) show a molecular mass of Mr = 17131.6 and of 17131.2, respectively. This represents a mass increase of 179 Da over the native protein, which is consistent with 2,4-DNPH-reactive proteins that have undergone an oxidative deamination before derivatization with the 2,4-DNPH reagent (44, 45). Thus, the observation of these peaks provides the first solid evidence that the nitroxide + H2O2-modified Mb has undergone an oxidative deamination in which some amino groups were converted to free carbonyls. In addition, low abundance peaks corresponding in mass to the oxidized forms of the 2,4-DNPH-reacted proteins were observed (Mr = 17148.2 and 17147.5). Interestingly, no ions corresponding in mass to the Mb-TEMPOL adduct were detected under the experimental conditions used in this study.
Detection of Chemically Modified Peptide Fragments: Spin-scavenging versus Spin-trapping
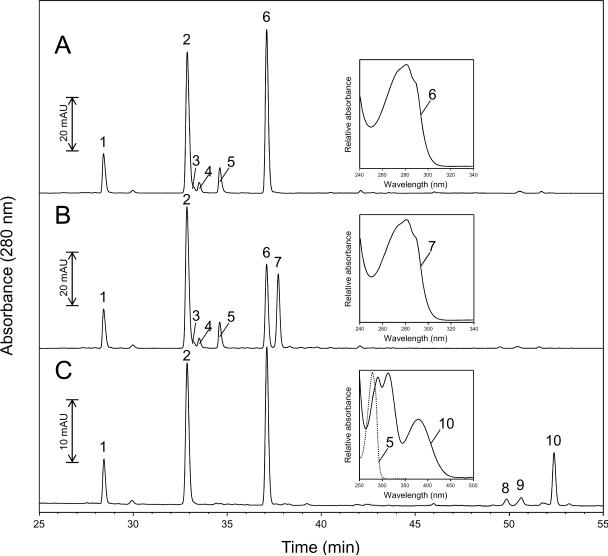
To characterize amino acid residues that had undergone an alteration in the Mb/nitroxide/H2O2 reaction mixtures, the HPLC-purified globins were subjected to chymotryptic digestion. The resulting peptide mixtures were then separated by C18 RP-HPLC with simultaneous detection at 214 and 280 nm or fluorescence detection at 360 nm (λex = 285 nm). Representative peptide maps at 280 nm of native Mb (the negative control) and nitroxide + H2O2-modified Mb are shown in Fig. 3A and B, respectively. It has been established by several groups that Tyr-103 is the primary site of radical formation in horse heart myoglobin(12, 21, 46). It has also been shown that the spin trap DBNBS traps almost exclusively at Tyr-103 (13), although there is some controversy about this point (22). Therefore, as an additional control, a peptide map of Mb modified by the spin trap DBNBS is also presented (Fig. 3C). This control helps to establish that Tyr-103-containing peptides can be identified by peptide map comparisons.
Figure 3.
C18 RP-HPLC separation of peptides produced by chymotryptic mapping of purified Mb. (A), Untreated native metMb; (B), nitroxide + H2O2-modified Mb (corresponding to peak 2 in Fig. 2A); (C), DBNBS + H2O2-modified Mb. Chromatograms show only the time window in which peptides exhibiting a strong absorbance at 280 nm eluted. Insets: absorbance spectra recorded by the diode-array detector in the effluent of the HPLC system. The spectra are scaled arbitrarily.
It can be seen that the chromatograms from both the nitroxide + H2O2- and the DBNBS + H2O2- modified samples show distinct differences compared with the chromatogram from the globins from untreated native metMb: the peak eluting at 37.3 min in the control (Fig. 3A, peak 6) was decreased by approximately 50% in the nitroxide + H2O2-modified globin sample (Fig. 3B), and a new peak eluting at 37.9 min was formed (Fig. 3B, peak 7). Both peaks give nearly identical absorbance (Fig. 3A and 3B, insets) and fluorescence emission spectra (data not shown) typical of tryptophan-containing peptides. Three peaks eluting between 33.5 and 35 min (Fig 3A and 3B, peaks 3, 4 and 5) were dramatically attenuated and three additional peaks are observed (Fig. 3C, peaks 8, 9 and 10) for the DBNBS + H2O2-modified globin sample. The absorbance spectra of peaks 3, 4 and 5 are typical of tyrosine-containing peptides (absorbance of peak 5, Fig. 3C, inset). The peaks eluting between 49 and 53 min (peaks 8, 9 and 10) show a lack of Tyr absorbance maximum at 276 nm and appearance of new absorption bands at 288, 312 and 380 nm (absorbance spectrum of peak 10, Fig. 3C, inset). An increased conjugation of the chromophores associated with the binding of the nitroso group of DBNBS to the aromatic ring of a tyrosine residue could conceivably cause such an effect. No additional differences were detected by comparing the peptide maps at 214 nm or by comparing the peptide maps with fluorescence detection at 360 nm (data not shown).
All spin trapping experiments described above were allowed to proceed for 30 s, after which the reaction mixtures were quickly loaded onto PD-10 gel filtration column and eluted with phosphate buffer, pH 6.8. This was done primarily to remove unreacted DBNBS from the sample and to minimize artifacts due to the non radical addition reaction that DBNBS is known to undergo with tryptophan (39). DBNBS adducted peptides were detected exclusively in the samples containing metMb, H2O2 and DBNBS. No DBNBS adducted peptides were detected by comparing the peptide maps of any of the control samples lacking DBNBS or H2O2 (see the Supporting Information, Fig. S1). Therefore, these data suggest little, if any, formation nonradical addition products under the conditions used in this study.
Detection at 280 nm predominantly measures Tyr and Trp residues. The latter have been reported as major contributors to the radical pool formed in the protein upon oxidation. Therefore, fractions exhibiting strong absorbance at 280 nm were collected, lyophilized and characterized by mass spectrometry. The results of these analyses are summarized in Table 1. Horse heart Mb has two tryptophan residues (Trp-7 and Trp-14) and two tyrosine residues (Tyr-103 and Tyr-146). As shown in the table, peptides containing each of these four aromatic residues could clearly be identified in the HPLC chromatogram of the hydrolyzed samples.
Table 1.
Summary of Tyr- and Trp-containing-peptides identified in the chymotryptic maps of native and nitroxide-modified Mb.
| Peaka | Elution Time (min) | Peptideb | Amino acid sequence | Trp and Tyrc | Observedd (m/z) | Calculatedd (m/z) | Change in concentratione (%) |
|---|---|---|---|---|---|---|---|
| 1 | 28.61 | Y10 | Arg139-Tyr146 | Tyr146 | 950.44 (+1) | 950.51 (+1) | (+)1.62 ± 0.05 |
| 2 | 32.94 | Y2* | Asn12-Trp14 | Trp14 | 418.18 (+1) | 418.20 (+1) | (+)2.8 ± 1.4 |
| 3 | 33.48 | Y6* | Lys96-Tyr103 | Tyr103 | 513.70 (+2) | 513.83 (+2) | (−) 3.2 ± 5.4f |
| 4 | 33.79 | Y6* | His97-Tyr103 | Tyr103 | 449.75 (+2) | 449.78 (+2) | (−) 3.2 ± 5.4f |
| 5 | 34.84 | Y6* | Lys98-Tyr103 | Tyr103 | 381.17 (+2) | 381.25 (+2) | (−) 3.2 ± 5.4f |
| 6 | 37.09 | Y1 | Gly1-Trp7 | Trp7 | 763.30 (+1) | 763.32 (+1) | (−) 48.8 ± 0.1 |
| 7 | 37.76 | Y1** | Gly1-Trp7 | Trp7 | 762.27 (+1) | 763.32 (+1) | (+) 46.8 ± 0.1g |
Peak numbers are assigned in the order of retention time as shown in Fig. 3.
Peptides are labeled Yx according to the expected chymotrypsin cleavage sites (C-terminal to F, Y and W residues); *, non-specific site of cleavage at the N-terminus; **, peptide Y1 modified at the N-terminus.
Trp and Tyr residues present in the chymotryptic peptides.
Observed and calculated m/z values: (+1), +1 ions; (+2), +2 ions.
Changes in concentrations are for nitroxide-modified Mb relative to untreated samples as estimated from the HPLC peak areas at 280 nm. Seven peaks eluting between 40 and 60 min and absorbing at 214 nm were chosen as the internal standards. Values are mean ± S.D. (n = 3).
Changes in concentrations are for all Tyr-103-containing peptides (e.g. sum of peaks 3, 4 and 5) assuming similar extinction coefficients.
The % gain of peak 7 was calculated relative to that of peak 6 in untreated samples assuming similar extinction coefficients.
The data in Table 1 and in Fig. 3 indicate that the bulk of Tyr-103-containing peptides in the control sample elute from the column between 33.5 and 35 min (Fig. 3A, peaks 3, 4 and 5). Interestingly, apart from the absence of Tyr-containing peptides between 33.5 and 35 min and three additional peaks at 49.8, 50.5 and 52.3 min (Fig. 3C, peaks 8, 9 and 10), no significant difference was observed between peptide maps of the DBNBS + H2O2-modified globin sample and the control. MS/MS analyses of the three peaks eluting at 50–53 min revealed a series of peptides 96KHKIPIK(Y*)L104 (peak 8), 97HKIPIK(Y*)L104 (peak 9) and 98KIPIK(Y*)L104 (peak 10) each modified at Tyr-103 (data not shown). Therefore, our data further suggests that DBNBS traps almost exclusively at Tyr-103 (13).
MS/MS Fragmentation of Chemically Modified Peptides
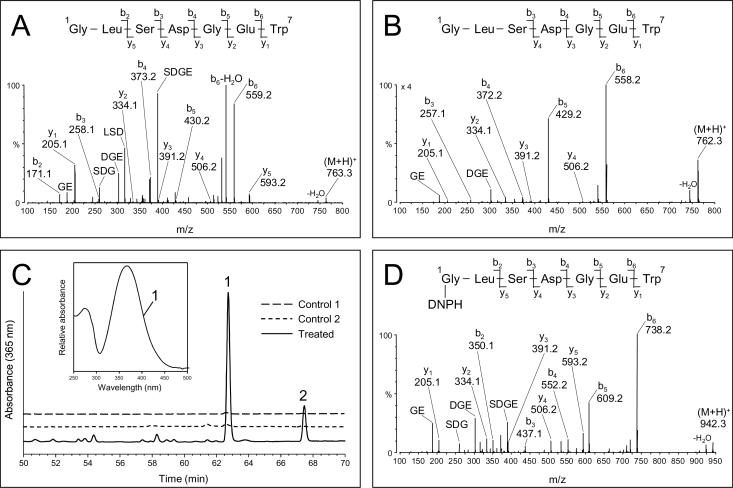
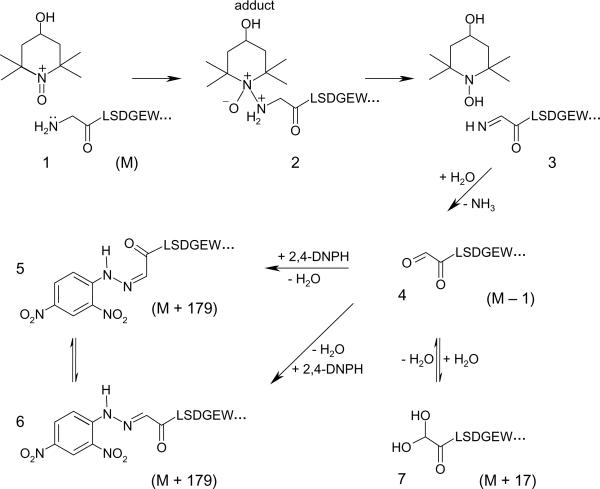
The data in Table 1 and in Fig. 3 indicate that TEMPOL/H2O2 treatment of Mb caused a substantial and exclusive modification of a Trp-containing peptide (e.g. a decrease in intensity of peak 6 and its quantitative conversion into peak 7). This outcome was unexpected because in their original report, Wright and English (10) tentatively assigned Tyr-103 as the site of radical scavenging by TEMPO in equine Mb. One peptide isolated from the HPLC fraction corresponding to peak 6 shows an abundant (M + H)+ ion of m/z 763.3 (data not shown), which corresponds in mass to the N-terminal chymotryptic peptide Y1. The MS/MS fragmentation spectrum of this ion (Fig. 4A) shows a nearly complete series of both carboxy-terminal y ions (y1 to y5) and amino terminal b ions (b2 to b6), thus confirming the amino acid sequence in the Gly1-Trp7 peptide. The mass spectrum of peak 7 exhibits two abundant (M + H)+ ions of m/z 762.2 and of m/z 780.2 (data not shown). These ions correspond in mass to a 1 Da decrease and a 17 Da increase, respectively, compared to the (M + H)+ ion of unmodified Y1 peptide. Interestingly, the MS/MS data of these two ions displayed very similar fragmentation patterns (MS/MS spectrum acquired from the molecular ion of m/z 762.2 shown in Fig. 4B): the observed series of b ions, starting from b3, shows 1 Da differences relative to the control Y1 peptide (Fig. 4A), whereas the masses of the y ions are consistent with those observed in the MS/MS spectrum of Y1. These data suggest that the 1 Da mass loss is located within the first three residues (e.g. Gly, Leu, Ser). The one Dalton mass difference may possibly be accounted for by the net addition of O (16) and loss of NH3 (17) from the unmodified peptide. The localization of the 1 Da loss within the first three residues (Gly, Leu, Ser) of the N-terminal peptide, the absence of a residue susceptible to oxidative deamination at positions 2 and 3 in the peptide (Leu and Ser), and the presence of a free carbonyl on the nitroxide + H2O2-modified Mb all suggest that the modified peptide was oxidatively deaminated at the N-terminus. The observed (M + H)+ ion of m/z 780.2 (data not shown) corresponds in mass to the addition of one molecule of water to the oxidatively deaminated peptide, and is possibly due to hydration of the N-terminal aldehyde function. Equilibrium between aldehydes and their hydrated forms occurs in solution and can be detected by ESI mass spectrometry (47–49).
Figure 4.
MS/MS fragmentation spectra of HPLC purified peptides and detection of 2,4-DNPH-derivatized peptides. (A), MS/MS spectrum of the N-terminal chymotryptic peptide Y1 eluting at approximately 37.3 min (corresponding to peak 6 in Fig. 3). The MS/MS spectrum was acquired from a parent ion of m/z 763.3 (+1); (B), MS/MS spectrum of a nitroxide + H2O2-modified peptide eluting at approximately 37.9 min (corresponding to peak 7 in Fig. 3). The MS/MS spectrum was acquired from a parent ion of m/z 762.2 (+1); (C), C18 RP-HPLC separation of peptides produced by chymotryptic mapping of nitroxide + H2O2-modified globin after reaction with 2,4-DNPH (solid line). The negative controls are for native metMb after reaction with 2,4-DNPH (control 1) and for H2O2-reacted Mb after reaction with 2,4-DNPH (control 2). Insets: absorbance spectrum recorded by the diode-array detector in the effluent of the HPLC system. The spectrum is scaled arbitrarily. (D), MS/MS spectrum of a 2,4-DNPH-derivatized peptide eluting at approximately 63.1 min (corresponding to peak 1 in Fig. 4C). The MS/MS spectrum was acquired from a parent ion at m/z 942.3 (+1).
To determine the nature of the products formed from the nitroxide + H2O2-modified globin sample after reaction with 2,4-DNPH (Fig. 2B), the modified proteins were subjected to chymotryptic digestion and HPLC purification, as above. Two chromatographic peaks showing absorption at 365 nm were observed, which were absent in any of the control samples lacking TEMPOL and/or H2O2 (Fig. 4C, peak 1 and 2). The UV spectra of these two peaks are typical of a 2,4-DNPH product with λmax at 365 nm (absorbance of peak 1, Fig. 4C, inset). The mass spectra of these fractions show an abundant singly charged ion of m/z 942.3 which corresponds in mass to the addition of 179 Da to the unmodified N-terminal peptide [(M + H)+ = 763.3)] (data not shown). The addition of 179 Da is consistent with the addition of one 2,4-DNPH group to the peptide.
The MS/MS spectra acquired from the ion of m/z 942.3 in the two chromatographic peaks displayed very similar fragmentation patterns (MS/MS data for peak 1 shown in Fig. 4D). The masses of the b2–b6 series of ions correspond to an increase of 179 Da when compared to the same series of ions in the MS/MS spectrum of the unmodified peptide (Fig. 4A). In addition, a series of y ions (y1–y5) are also observed and the mass of these ions are similar to the masses of these ions in the MS/MS data of the unmodified peptide. These data indicate that either Gly-1 or Leu-2 is the site of formation of the 2,4-DNPH hydrazone adduct. Lacking a free amino group, Leu-2 is not susceptible to oxidative deamination. Therefore, the data are most consistent with the formation of an 2,4-DNPH adduct at the N-terminal residue (Gly-1). Because the two chromatographic peaks show similar mass spectral data, two stereoisomers of the 2,4-DNPH adduct are likely formed. These data are in agreement with the MS/MS fragmentation data of the peptide minus 1 Da (Fig. 4B) and support the view that the Mb N-terminus has undergone an oxidative deamination in which the Gly N-terminal amino group was replaced with a free carbonyl group.
To extend the results obtained with TEMPOL, experiments were also performed with the nitroxides 2,2,6,6-tetramethylpiperidine 1-oxyl (TEMPO) and 4-carboxy-2,2,6,6-tetramethylpiperidine 1-oxyl (COOH-TEMPO). The modified globins formed in Mb/nitroxide/H2O2 reaction mixtures were identically purified and subjected to chymotryptic digestion, as described above. Consistent with the HPLC results observed with TEMPOL, the peptides maps showed a substantial decrease in the amount of N-terminal chymotryptic peptide Y1 (amino acid residue 1–7) (peptide map for the COOH-TEMPO + H2O2-treated sample: not shown; peptide map for the TEMPO + H2O2-treated sample: see the Supporting Information, Fig. S3). In both cases a new HPLC peak was observed. Reaction of this new peak with 2,4-DNPH yielded evidence of formation of hydrazone adducts with a characteristic absorbance at 365 nm. MS/MS analyses of the hydrazone adducts revealed either Gly-1 or Leu-2 as the site of formation of the 2,4-DNPH adducts in the TEMPO/COOH-TEMPO systems (data not shown).
Suppression of the EPR signals of globin-centered radicals by TEMPOL
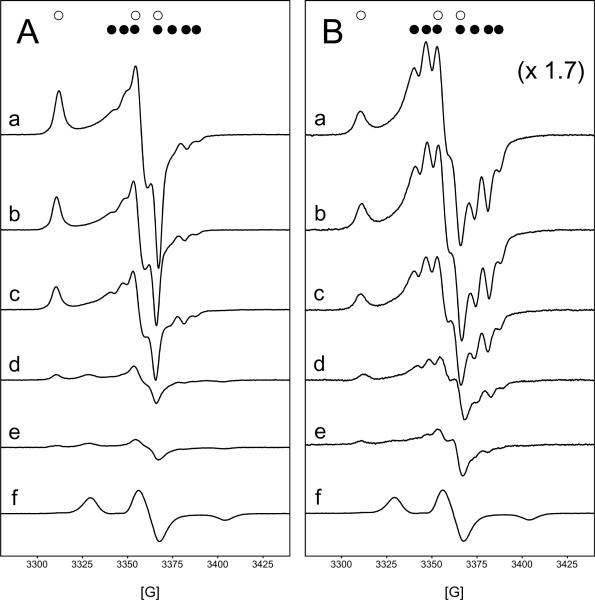
Reaction of horse heart metmyoglobin with H2O2 produces protein radicals (globin radicals). As already described (25, 46), the globin radicals can be detected by freezing the reaction mixture at specific times after the addition of H2O2 and recording their low-temperature EPR spectra. Fig 5 shows the freeze-quenched EPR spectra recorded 10 s (panel A, trace a) and 30 s (panel B, trace a) after initiation of the reaction with a stoichiometric amount of H2O2 (500 μM Mb, 500 μM H2O2) at 25 °C. The anisotropic signals recorded at 77 K arise from the combination of two species similar to those previously assigned to the Trp-14 peroxyl radical (50, 51) and Tyr-103 phenoxyl radical (46, 52). With a 10 s time period between initiation of the reaction and freezing, the major EPR-detectable species was the Trp-14 peroxyl radical. With a 30 s interval, the characteristic septet signal from the Tyr-103 phenoxyl radical became more prominent. Both radical species have short lifetimes and were not detected after more than 5 minutes of incubation at room temperature prior to freezing (data not shown); this result is in accord with previous observations (46, 50–52).
Figure 5.
Low temperature EPR spectra obtained from the reaction of metMb with H2O2 in the absence or presence of increasing concentrations of TEMPOL. MetMb (500 μM) was reacted with H2O2 (500 μM) in 50 mM phosphate buffer, pH 6.8 containing 50 μM DTPA at 25 °C. Times between initiation of the reaction with H2O2 and freeze quenching to 77 K to halt the reaction were 10 s (Panel A) or 30 s (Panel B). Different concentrations of TEMPOL were added to metMb directly before the addition of peroxide. (a), 0 μM; (b), 1.25 μM; (c), 2.5 μM; (d), 3.75 μM; (e), 5 μM; (f), 10 μM. Signals are assigned to the formation of the peroxyl radical on the indole ring of the Trp-14, marked (○), and the phenoxyl radical formed at Tyr-103, marked (●).
The reactivity of globin-centered radicals (or their precursor) with varying amount of TEMPOL was assessed using 10 s (Fig. 5A) and 30 s (Fig. 5B) time periods between initiation of the reaction and freezing. TEMPOL was added to the Mb directly before the addition of peroxide. Inclusion of TEMPOL in the metMb/H2O2 reaction system at concentrations lower than 5 μM resulted in a loss of both the peroxyl and the tyrosyl radical signals in a concentration dependent manner (trace b–d). When the concentration of TEMPOL was 5 μM or higher (trace e–f), the EPR signals from the globin-centered radicals were virtually absent and a new anisotropic triplet spectrum characteristic of a highly immobilized nitroxide radical arising from the presence of unconsumed TEMPOL was observed; these results are also in accord with previous observations (53).
After more than 3 min of incubation, and depending on its initial concentration, TEMPOL was totally or partially converted into EPR silent species, as monitored by the EPR spectra of the final incubation mixtures recorded at room temperature (see the Supporting Information, Fig. S2).
Effect of TEMPOL on Ferrylmyoglobin Decay
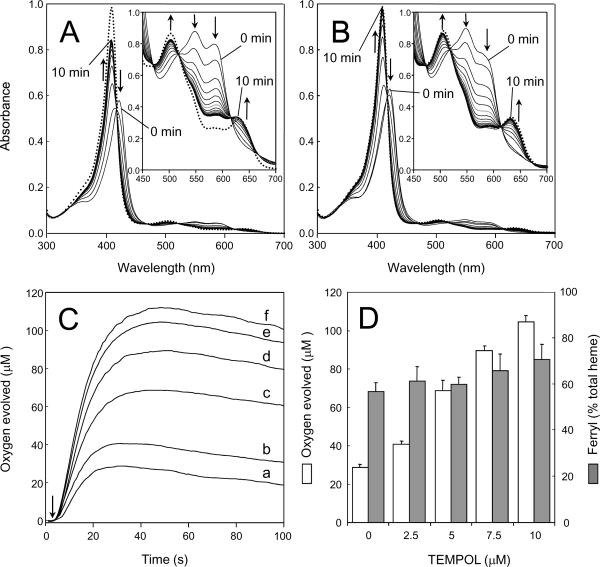
Incubation of metMb with H2O2 led to the formation of a ferryl (FeIV=O) species that was spectrophotometrically identifiable (Fig. 6A). The spectrum obtained for native metMb before the addition of H2O2 shows the typical features of the ferric form (λmax at 409, 503, and 632 nm). The addition of H2O2 resulted in a shift of the Soret peak from 409 to 422 nm concomitant with the appearance of peaks at 547 and 579 nm. These changes in spectral properties are consistent with formation of a ferryl Mb derivative (25). After 10 s, the excess H2O2 was removed by adding catalase. Once H2O2 was consumed, e.g. ~ 10 s after the addition of catalase, 10 μM TEMPOL was added to the reaction mixture. This caused the rapid conversion of part of the ferryl Mb back to the metMb form (t1/2 = 71 s). During the first 2 minutes following the addition of TEMPOL, the Soret intensity increased quickly. Subsequent spectra showed that the Soret intensity rose for over 5 minutes, after which it remained at a constant level. Importantly, the final product of the decay has a spectrum which differs slightly from that of the native protein due to heme degradation. In particular, the Soret band height is decreased by approximately 14% relative to that of metMb before the addition of H2O2.
Figure 6.
Effect of TEMPOL on ferryl Mb decay and on time course of O2 evolution. (A), ferrylMb decay measured with a UV-visible spectrophotometer. The reaction was initiated by the addition of H2O2. After 10 s incubation, excess H2O2 was removed by addition of catalase. TEMPOL was added 10 s after the addition of catalase. Absorption spectra were recorded every 45 s after the addition of H2O2. Arrows indicate the directions of the absorption changes. (B), same as (A) but TEMPOL added directly before the addition of peroxide. (C), time course of O2 formation and decay measured by a Clark-type oxygen electrode. Different concentrations of TEMPOL were added to metMb directly before the addition of peroxide. (a), 0 μM; (b), 2.5 μM; (c), 5 μM; (d), 7.5 μM; (e), 10 μM; (f), 25 μM. The arrow indicates when H2O2 was added. Time course traces are representative recordings obtained from more than three experiments. (D), Amount of O2 produced (white bars) and extent of ferryl formation (gray bars) 30 s after addition of H2O2 into metMb/TEMPOL reaction mixtures. Error bars represent SD obtained from at least three experiments. Experimental conditions: (A, B) 5.3 μM metMb (main panels) or 85 μM metMb (inserts), 600 μM H2O2, 10 μM TEMPOL, 250 units/ml catalase in 50 mM phosphate buffer, pH 6.8 at 25 °C.; (C, D) incubations and concentrations of reagents were the same as in figure 5.
Similar changes in the spectra were observed when TEMPOL was added to the Mb directly before the addition of peroxide (Fig. 6B). However, the spectrum recorded 10 min after the start of the reaction was almost identical to that obtained for metMb before the addition of H2O2. This observation confirms that inclusion of TEMPOL directly before the addition of peroxide effectively inhibits heme degradation.
Stimulation of oxygen evolution by TEMPOL
Myoglobin shows a limited catalatic activity (i.e. catalase-like oxygen production) in the presence of H2O2, which can be stimulated by nitroxide free radicals (54, 55). The effect of TEMPOL on the time course of H2O2-induced O2 evolution is shown in Fig. 6C. These experiments were performed under conditions identical to those used for the EPR measurements. In the absence of TEMPOL, incubation of metMb and H2O2 resulted in the rapid evolution of oxygen. The evolution of oxygen reached a maximum in 30 s. No further O2 was released when catalase was added after this interval, indicating that all H2O2 had been consumed (data not shown). After O2 production had plateaued, a second phase was observed in which oxygen was consumed, consistent with incorporation of oxygen into the protein and/or heme moiety. The addition of TEMPOL changed the time course of O2 formation, enhancing the amount of oxygen formed. Oxygen consumption during the second phase of the reaction, however, was not significantly modified by TEMPOL.
The relationship between the amount of O2 produced and the extent of ferryl formation 30 s after addition of H2O2 into a metMb/TEMPOL reaction mixture is shown in Fig. 6D. These experiments were performed under conditions identical to those used for the EPR measurements. The results indicate that TEMPOL increased the amount of oxygen formed in a dose-dependent manner (white bars). However, the addition of TEMPOL did not significantly affect the amount of ferryl myoglobin (gray bars). Addition of relatively low concentrations of TEMPOL (2.5–10 μM) resulted in the production of substantially higher amounts of oxygen (12–76 μM). This latter result is in agreement with previous reports, (54, 55) demonstrating that TEMPOL acts as a catalyst and is recycled in this system.
Intermediacy of the oxoammonium cations
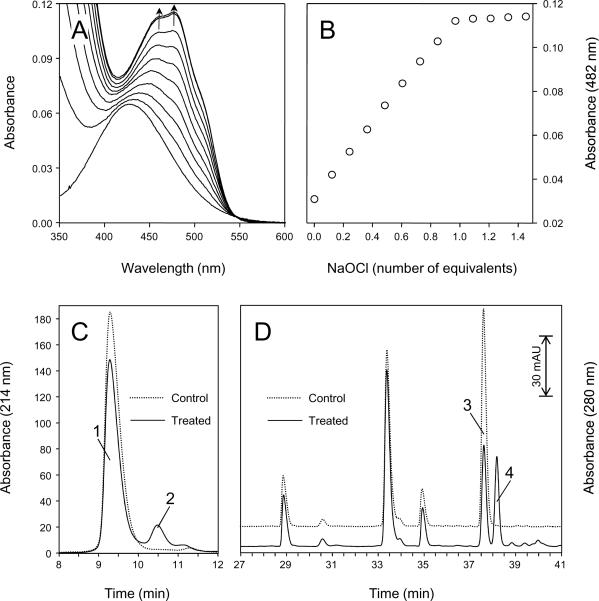
The nitroxide-facilitated catalase-like activity of Mb has been studied in some detail (54, 55) and appears to involve a one-electron oxidation of the nitroxide to the oxoammonium cation, which then reacts further to release oxygen and the nitroxide. Oxoammonium compounds are strong oxidants that can react with a large variety of organic and inorganic molecules. Formation of nitroxide-modified globins from a direct reaction between metMb and the oxoammonium derivative of TEMPOL (TEMPOL+) was, therefore, investigated. TEMPOL+ was prepared by hypochlorous acid treatment of TEMPOL in acidic phosphate buffer. The procedure is illustrated in Fig. 7A, which shows the conversion of TEMPOL (λmax 429 nm) to TEMPOL+ (λmax 458 and 482 nm) as a function of the amount of NaOCl used in the assay. At the equivalence point of the titration, the spectrum is essentially identical to that of freshly dissolved oxoammonium salts (40). A titration curve at 482 nm (Fig. 7B) indicates a photometric end point close to the exact 1:1 equivalence point of titration assuming 1:2 (HOCl:TEMPOL) stoichiometry. These data indicate that, at the equivalence point, essentially all of the HOCl has been converted to chloride and water. Beyond the equivalence point, unreacted HOCl accumulates in the system. Therefore, TEMPOL+/TEMPOL stock solutions were prepared by mixing a subequivalent amount of NaOCl with TEMPOL. As an additional precaution, the resulting mixtures were then further reacted with methionine and catalase to ensure complete removal of unreacted HOCl or H2O2.
Figure 7.
Conversion of TEMPOL to TEMPOL+ as a function of NaOCl concentration and detection of oxoammonium-modified peptide fragments. (A), spectral changes in the visible region upon treatment with increments of 0.12 equivalents of NaOCl. Experimental conditions were as follows: TEMPOL (4.8 mM) in 50 mM phosphate buffer, pH 2.0 at 25 °C. NaOCl was added stepwise as indicated, and each spectrum was recorded < 10 s after the addition of NaOCl. The directions of spectral changes are indicated by the arrows. (B), titration curve at 482 nm (absorption maximum of TEMPOL+). The curve indicates a photometric end point close to the exact 1:1 equivalence point of titration assuming 1:2 (HOC1:TEMPOL) stoichiometry. (C), C4 RP-HPLC separation monitored at 214 nm of metMb (500 μM) treated with TEMPOL+/TEMPOL solution (150 μM) for 5 minutes (TEMPOL+/TEMPOL ratio around 1.5). The negative control (dashed line) is for an untreated metMb sample. (D), C18 RP-HPLC separation monitored at 280 nm of peptides produced by chymotryptic mapping of oxoammonium-modified Mb (corresponding to peak 2, in Fig. 7C). The negative control (dashed line) is for an untreated metMb sample.
Fig. 7C shows the C4 RP-HPLC profile of myoglobin before (dotted line) and after the reaction with TEMPOL+/TEMPOL solution (solid line). When metMb was reacted for 5 minutes with a freshly prepared TEMPOL+/TEMPOL solution, the apoprotein peak (peak 1) decreased by approximately 20% and a new peak with absorption at 214 nm was observed (peak 2). Only negligible changes in the HPLC traces at 214 nm were detected after the reaction of metMb for 5 min with a reagent solution including all the reactants except TEMPOL (data not shown). The stock reagent solution used in this control experiment was prepared by mixing NaOCl with methionine and catalase using similar experimental conditions (i.e. same concentrations of reactants and same incubation times) as were used when preparing the TEMPOL+/TEMPOL solution. Therefore, this control experiment clearly indicates that the new peak with absorbance at 214 nm (Fig. 7C, peak 2) is not due to reactions between metMb and HOCl or H2O2 possibly present in the TEMPOL+/TEMPOL solution. Instead, the data are consistent with a direct reaction between the oxoammonium cation and the globin moiety.
The oxoammonium modified globin HPLC peak formed in the reaction of Mb with TEMPOL+/TEMPOL solution (Fig. 7C, peak 2) was collected and subjected to chymotryptic digestion. Consistent with the results seen with TEMPOL, the C18 RP-HPLC chromatogram of the oxoammonium-treated sample (Fig. 7D) showed a decrease in the amount of N-terminal chymotryptic peptide Y1 eluting at 37.6 min (peak 3) and an additional peak eluting at approximately 38.2 min (peak 4). Reaction of this new peak with 2,4-DNPH yielded evidence of formation of hydrazone adducts that displayed a mass gain of 179 Da compared with the nominal N-terminal chymotryptic peptide Y1 upon ESI/MS analyses (data not shown). These data are consistent with the interpretation that, upon incubation with the oxoammonium cation, the Mb N-terminus had undergone an oxidative deamination.
DISCUSSION
TEMPOL Inhibits Formation of Oxidatively-Damaged Heme Species in a Catalytic Fashion
The ability of stable cyclic nitroxide radicals to reduce ferryl heme species produced in the reaction of metMb and H2O2 has been known for some time (54–56). However, there is a lack of information regarding the effect of nitroxide radical reagents on the formation of H2O2-damaged heme species. Here we have demonstrated that a short incubation (30 s) with substoichiometric amounts of TEMPOL can effectively inhibit the formation of damaged heme species (Fig. 1).
In the absence of a clear and detailed mechanism for the generation of H2O2-damaged heme species, one can only speculate about the mechanism of protection by TEMPOL. Our finding that TEMPOL, at 5 μM, was sufficient to almost completely block the formation of substantially higher concentrations (~90 μM) of oxidatively damaged heme species (Fig. 1) leads to the conclusion that TEMPOL acts as a catalyst and is recycled in this system. This is confirmed by parallel experiments carried out with an oxygen electrode (Fig. 6D, white bars), which revealed that low concentrations of TEMPOL (2.5–10 μM) resulted in the production of substantially higher amounts of oxygen (12–76 μM).
Previous studies (54, 55) have shown that Mb shows a limited catalase-like activity in the presence of H2O2, which can be stimulated by nitroxide free radical. This reaction has been studied in some detail (54, 55) and appears to involve a one-electron oxidation of the nitroxide (RNO·) to the oxoammonium cation (RNO+), which then reacts further to release oxygen and the nitroxide (reactions 1–8):
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
| (8) |
where Por•+ refers to a porphyrin radical cation and globin represents a globin radical. Values of some rate constants for the above sequence of reactions have been determined experimentally; these are as follows: k1 = 1.4 × 102 M−1s−1 (57), k2 = 3.8 × 103 s−1 (58) and k4 = 3.4 × 10 M−1s−1 (54) for horse myoglobin , k5 = 1.4 × 108 M−1s−1 for TEMPO+ (59), k-5 = 1.2 × 108 M−1s−1 for TEMPO (59), k6 = 3.4 × 109 M−1s−1 for TEMPO+ (59) and 1.5 × 1010 M−1s−1 for TEMPOL+ (60).
TEMPOL could function to inhibit the formation of the damaged heme species due to its capacity, as a one-electron reductant, to scavenge protein radicals (reaction 3). The rate constant for this reaction has never been determined but is likely to be very large, as this is a radical-radical recombination reaction rather than a reaction between a radical and a diamagnetic molecule. An alternative pathway might involve a one-electron reduction of the Compound I transient porphyrin cation radical to yield a ferryl species that no longer has a radical state:
| (9) |
This later reaction could also contribute to the protective properties of TEMPOL because, if reaction 9 is faster than reaction 2, no protein radical is expected to accumulate. The finding that inclusion of TEMPOL in the metMb/H2O2 reaction system even at low doses (5 μM) resulted in a complete loss of both the Trp-14 peroxyl and Tyr-103 tyrosyl EPR radicals signals (Fig. 5) may indicate that the nitroxide reagent reacts directly with the globin radicals (reaction 3) or, alternatively, that it reacts with its precursor (reaction 9). Thus, no definitive conclusion can be drawn from EPR data presented in this paper about the pathways of porphyrin and/or globin radical decay.
Nitroxides have been shown to react with the ferryl state of myoglobin (reaction 4) (54–56). However, the rate constant reported for this reaction (3.4 × 10 M−1s−1) (54) is order of magnitude lower than reported for the reaction of nitroxides with carbon centered radicals (~ 108–109 M−1s−1) (8, 9). We showed that under the employed conditions (e.g. substochiometric amounts of TEMPOL relative to Mb), TEMPOL did not significantly affect the level of ferryl heme species (Fig. 6D, gray bars). It is thus reasonable to conclude that the one-electron reduction of the ferryl heme species by TEMPOL is not responsible for the protective properties of the nitroxide reagent.
Oxoammonium Cation-induced Modifications
Whatever the pathways of ferryl, porphyrin, and/or globin radical decay in the above reaction system, there is solid evidence (54, 55) that nitroxides are able to enhance the catalase-like activity of Mb by shuttling between two oxidation states, the nitroxide radical and the oxoammonium cation. Oxoammonium compounds are strong oxidants that can react with a large variety of organic and inorganic molecules (40). The present study was initially undertaken with the aim of identifying specific residues that are modified by nitroxide radical reagents in protein radical scavenging. As mentioned before, the applicability of the spin scavenging approach combined with chromatographic/mass spectrometric procedures to identify protein-based radicals requires that i) the desired diamagnetic addition products be sufficiently stable to survive under the normal workup conditions used for protein denaturation, proteolysis and LC/MS analysis and ii) the diamagnetic addition products be exclusively formed via radical combinations between the nitroxide and the protein-centered radical. In particular, it is critical that the other probable products, which can arise during the reaction, not initiate complicating secondary reactions producing additional products.
Here, we have demonstrated that reaction of the oxoammonium derivative of TEMPOL (TEMPOL+) with the globin moiety resulted in the formation of a free carbonyl on the N-terminal Gly-residue without any other significant modification of the protein. This oxidative deamination was confirmed through the combined use of reverse-phase liquid chromatographic purifications and tandem MS experiments (Figs. 3 and 4). In the case of the free amino acid, deamination of glycine by oxoammonium cations has been described previously (40). However, to our knowledge the present study constitutes the first documented case for a protein.
The results presented in this study would be consistent with a pathway involving imine formation, followed by spontaneous hydrolysis to yield an aldehyde specifically at the N-terminus (scheme 2). Additional support for such a pathway is provided by the trapping of the aldehyde with concomitant formation of a hydrazone adduct following treatment of the mixture with 2,4-DNPH (Fig. 4D), the detection of hydrated aldehydes (data not shown), and the detection of ammonia release, in the case of the free amino acid (40).
Scheme 2.
Proposed mechanism for oxidative deamination by oxoammonium cations occurring at the N-terminus of horse heart myoglobin. The actual oxidant is the oxoammonium ion, easily generated from TEMPOL on oxidation by myoglobin. The oxoammonium ion reacts with the lone pair of the primary amine at the N-terminus (1) to form an adduct (2). Deprotonation and fragmentation of the adduct give rise to the reduced form of TEMPOL and an imine intermediate at the N-terminus (3). The imine is unstable and readily undergoes spontaneous hydrolysis to the corresponding aldehyde (4), thereby decreasing the peptide molecular mass by 1 Da. 2,4-DNPH treatment of the aldehyde (4) yields isomeric hydrazone adducts (5 and 6), resulting in mass increase of 179 Da to the peptide. In aqueous medium, the aldehyde (4) is in equilibrium with its hydrated form (7), a diol (i.e., a mass increase of 17 Da to the peptide).
Two chromatographic peaks showing absorbance at 365 nm and corresponding to each 2,4-dinitrophenyl hydrazone isomeric derivative were detected following treatment of the Mb/nitroxide/H2O2 mixture with 2,4-DNPH (Fig. 4C). The formation of isomeric 2,4-dinitrophenyl hydrazone by-products from unsymmetrical carbonyl compounds is well documented (61, 62). Presumably, the isomerization occurs during sample preparation and involves addition of a nucleophile (water or phosphate ion) to the protonated (E)- or (Z)-isomers (61, 62).
The identical oxidative deamination of the globin moiety can be produced by inclusion of TEMPO or COOH-TEMPO, instead of TEMPOL, in the metMb/H2O2 reaction system. It can thus reasonably be assumed that several oxoammonium species, having different substituent groups on their rings, are capable of mediating the oxidative deamination observed.
Absence of Stable Addition Products
Nitroxides were earlier reported to be capable of scavenging carbon-centered and peroxyl radicals to give relatively stable diamagnetic addition products (9, 63–65). However, these stable addition adducts have not been detected in our experiments. If significant adduct formation were to occur in the presence of TEMPOL, the nitroxide reagent would have been consumed irreversibly, which would have preclude its continuous recycling in presence of H2O2. It is, thus, concluded that scavenging of the globin radicals via radical-radical recombination, leading to the formation of stable addition adducts, does not occur to any appreciable extent under the experimental conditions used in this study.
Eventually, once H2O2 is consumed and depending on its initial concentration, TEMPOL was totally or partially converted into EPR silent species, as monitored by the EPR spectra of the final incubation mixtures recorded at room temperature (see the Supporting Information, Fig. S2). We interpret these observations to indicate that in absence of H2O2, the EPR-silent oxoammonium cations are not recycled back to TEMPOL, which reduces dramatically the EPR signal intensity. Other investigators using a similar reaction system have shown that the magnitude of the initial EPR signal can completely by restored by treatment of Mb/TEMPOL/H2O2 mixture with large excess of sodium borohydride (NaBH4) and potassium ferricyanide (K3Fe(CN)6) (54). NaBH4 is a two electrons reductant that converts the oxoammonium cation to the hydroxylamine whereas K3Fe(CN)6 acts as a one-electron oxidant that reacts with the hydroxylamine to regenerate the nitroxide. Therefore the complete restoration of EPR signal of TEMPOL by the two reactants implies that TEMPOL+ was originally present in the reaction mixtures and suggests little, if any, formation of stable TEMPOL-Mb adducts in this reaction system.
In their original report, Wright and English (10) tentatively assigned Tyr-103 as the preferred site of radical coupling by TEMPO in equine Mb. A control experiment performed here with the spin trap DBNBS (Fig. 3C) clearly demonstrates that Tyr-103-containing peptides can be identified by peptide map comparisons. If TEMPO formed an adduct with Tyr-103 then a significant decrease in intensity of the HPLC peaks corresponding to Tyr-103-containing peptides would be expected for the TEMPO + H2O2-modified globin samples (e.g. a decrease of intensity of peaks 3, 4 and 5 as observed for DBNBS + H2O2-modified globin samples). However, such a decrease was not observed; i.e. none of the chromatographic peaks corresponding to the Tyr-103-containing peptides showed significant decreases in intensity relative to controls upon TEMPO/H2O2 treatment (see the Supporting Information, Fig. S3). These results strongly suggest that either Tyr-103 is not the preferred site of radical coupling by TEMPO in equine Mb or, alternatively and most likely, that the Tyr-103-TEMPO adduct is short-lived and therefore remained undetected under our experimental conditions.
Supplementary Material
ACKNOWLEDGMENT
This work has been supported by the Intramural Research Program of the National Institutes of Health and the National Institute of Environmental Health Sciences, the National Center for Research Resources Grants RR001614, and RR012961 (to D.A.M., K.F.M., and director A.L.Burlingame), and GM32488 (P.R.O.M.). The authors thank Dr. Arno Siraki and Dr. Marcelo Bonini for their careful review of the manuscript and Mrs. Anne Froment, Ms. Jean Corbett, Mrs. Mary J. Mason and Dr. Ann Motten for their valuable assistance in the preparation of this manuscript. We also thank Dr. Jinjie Jiang for advice and for kindly providing us training and access to the EPR spectrometer (to O.M.L.) and Mr. Robert Sik for synthesis of the spin trap DBNBS.
Abbreviations
- EPR
electron spin resonance spectrometry
- TEMPO
2,2,6,6-tetramethylpiperidine 1-oxyl
- TEMPOL
4-Hydroxy-2,2,6,6-tetramethylpiperidine 1-oxyl
- COOH-TEMPO
4-carboxy-2,2,6,6-tetramethylpiperidine 1-oxyl
- DBNBS
3,5-dibromo-4-nitrosobenzene sulfonate
- DNPH
2,4-dinitrophenyl hydrazine
- ESI
electrospray ionization
- Mb
myoglobin
- metMb
metmyoglobin
- heme
iron protoporphyrin IX regardless of the oxidation and ligation states
Footnotes
SUPPORTING INFORMATION AVAILABLE EPR spectra obtained at room temperature from the reaction of metMb with H2O2 in the absence or presence of increasing concentrations of TEMPOL. Peptide maps at 280 nm of native Mb, for TEMPO + H2O2- modified and for H2O2-modified globin samples, and for Mb after incubation with 1 eq DBNBS in absence of H2O2. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- (2).Davies MJ, Fu S, Wang H, Dean RT. Stable markers of oxidant damage to proteins and their application in the study of human disease. Free Radic Biol Med. 1999;27:1151–1163. doi: 10.1016/s0891-5849(99)00206-3. [DOI] [PubMed] [Google Scholar]
- (3).Dalle-Donne I, Scaloni A, Giustarini D, Cavarra E, Tell G, Lungarella G, Colombo R, Rossi R, Milzani A. Proteins as biomarkers of oxidative/nitrosative stress in diseases: the contribution of redox proteomics. Mass Spectrom Rev. 2005;24:55–99. doi: 10.1002/mas.20006. [DOI] [PubMed] [Google Scholar]
- (4).Stubbe J, van Der Donk WA. Protein Radicals in Enzyme Catalysis. Chem Rev. 1998;98:2661–2662. doi: 10.1021/cr980059c. [Chem. Rev. 1998, 98, 705minus sign762. [DOI] [PubMed] [Google Scholar]
- (5).Davies MJ, Hawkins CL. EPR spin trapping of protein radicals. Free Radic Biol Med. 2004;36:1072–1086. doi: 10.1016/j.freeradbiomed.2003.12.013. [DOI] [PubMed] [Google Scholar]
- (6).Augusto O, Muntz Vaz S. EPR spin-trapping of protein radicals to investigate biological oxidative mechanisms. Amino Acids. 2007;32:535–542. doi: 10.1007/s00726-006-0429-4. [DOI] [PubMed] [Google Scholar]
- (7).Lardinois OM, Detweiler CD, Tomer KB, Mason RP, Deterding LJ. Identifying the site of spin trapping in proteins by a combination of liquid chromatography, ELISA, and off-line tandem mass spectrometry. Free Radic Biol Med. 2007;44:893–906. doi: 10.1016/j.freeradbiomed.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Beckwith AL, Bowry VW, Ingold KU. Kinetics of nitroxide radical trapping. 1. Solvent effects. J Am. Chem. Soc. 1992;114:4983–4991. [Google Scholar]
- (9).Bowry VW, Ingold KU. Kinetics of nitroxide radical trapping. 2. Structural effects. J Am. Chem. Soc. 1992;114:4992–4996. [Google Scholar]
- (10).Wright PJ, English AM. Scavenging with TEMPO* to identify peptide-and protein-based radicals by mass spectrometry: advantages of spin scavenging over spin trapping. J Am Chem Soc. 2003;125:8655–8665. doi: 10.1021/ja0291888. [DOI] [PubMed] [Google Scholar]
- (11).Takata T, Tsujino Y, Nakanishi S, Nakamura K, Yoshida E, Endo T. Electrophilic 1,2-addition of oxoammonium salts to olefins. Chem. Lett. 1999:937–938. [Google Scholar]
- (12).Fenwick CW, English AM. Trapping and LC-MS identification of protein radicals formed in the horse heart metmyoglobin-H2O2 reaction. J Am. Chem. Soc. 1996;118:12236–12237. [Google Scholar]
- (13).Harris MN, Burchiel SW, Winyard PG, Engen JR, Mobarak CD, Timmins GS. Determining the site of spin trapping of the equine myoglobin radical by combined use of EPR, electrophoretic purification, and mass spectrometry. Chem Res Toxicol. 2002;15:1589–1594. doi: 10.1021/tx025594t. [DOI] [PubMed] [Google Scholar]
- (14).Detweiler CD, Lardinois OM, Deterding LJ, de Montellano PR, Tomer KB, Mason RP. Identification of the myoglobin tyrosyl radical by immuno-spin trapping and its dimerization. Free Radic Biol Med. 2005;38:969–976. doi: 10.1016/j.freeradbiomed.2004.12.031. [DOI] [PubMed] [Google Scholar]
- (15).Deterding LJ, Bhattacharjee S, Ramirez DC, Mason RP, Tomer KB. Top-down and bottom-up mass spectrometric characterization of human myoglobin-centered free radicals induced by oxidative damage. Anal Chem. 2007;79:6236–6248. doi: 10.1021/ac070935z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Detweiler CD, Deterding LJ, Tomer KB, Chignell CF, Germolec D, Mason RP. Immunological identification of the heart myoglobin radical formed by hydrogen peroxide. Free Radic Biol Med. 2002;33:364–369. doi: 10.1016/s0891-5849(02)00895-x. [DOI] [PubMed] [Google Scholar]
- (17).Gibson JF, Ingram DJ, Nicholls P. Free radical produced in the reaction of metmyoglobin with hydrogen peroxide. Nature. 1958;181:1398–1399. doi: 10.1038/1811398a0. [DOI] [PubMed] [Google Scholar]
- (18).Davies MJ. Detection of myoglobin-derived radicals on reaction of metmyoglobin with hydrogen peroxide and other peroxidic compounds. Free Radic Res Commun. 1990;10:361–370. doi: 10.3109/10715769009149905. [DOI] [PubMed] [Google Scholar]
- (19).Tew D, Ortiz de Montellano PR. The myoglobin protein radical. Coupling of Tyr-103 to Tyr-151 in the H2O2-mediated cross-linking of sperm whale myoglobin. J Biol Chem. 1988;263:17880–17886. [PubMed] [Google Scholar]
- (20).Lardinois OM, Ortiz de Montellano PR. Intra- and intermolecular transfers of protein radicals in the reactions of sperm whale myoglobin with hydrogen peroxide. J Biol Chem. 2003;278:36214–36226. doi: 10.1074/jbc.M304726200. [DOI] [PubMed] [Google Scholar]
- (21).Gunther MR, Tschirret-Guth RA, Witkowska HE, Fann YC, Barr DP, Ortiz De Montellano PR, Mason RP. Site-specific spin trapping of tyrosine radicals in the oxidation of metmyoglobin by hydrogen peroxide. Biochem J. 1998;330(Pt 3):1293–1299. doi: 10.1042/bj3301293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Gunther MR, Tschirret-Guth RA, Lardinois OM, Ortiz de Montellano PR. Tryptophan-14 is the preferred site of DBNBS spin trapping in the self-peroxidation reaction of sperm whale metmyoglobin with a single equivalent of hydrogen peroxide. Chem Res Toxicol. 2003;16:652–660. doi: 10.1021/tx0256580. [DOI] [PubMed] [Google Scholar]
- (23).Witting PK, Douglas DJ, Mauk AG. Reaction of human myoglobin and H2O2. Involvement of a thiyl radical produced at cysteine 110. J Biol Chem. 2000;275:20391–20398. doi: 10.1074/jbc.M000373200. [DOI] [PubMed] [Google Scholar]
- (24).Witting PK, Mauk AG. Reaction of human myoglobin and H2O2. Electron transfer between tyrosine 103 phenoxyl radical and cysteine 110 yields a protein-thiyl radical. J Biol Chem. 2001;276:16540–16547. doi: 10.1074/jbc.M011707200. [DOI] [PubMed] [Google Scholar]
- (25).King NK, Winfield ME. The mechanism of metmyoglobin oxidation. J Biol Chem. 1963;238:1520–1528. [PubMed] [Google Scholar]
- (26).Egawa T, Shimada H, Ishimura Y. Formation of compound I in the reaction of native myoglobins with hydrogen peroxide. J Biol Chem. 2000;275:34858–34866. doi: 10.1074/jbc.M004026200. [DOI] [PubMed] [Google Scholar]
- (27).Kroger-Ohlsen MV, Andersen ML, Skibsted LH. Acid-catalysed autoreduction of ferrylmyoblobin in aqueous solution studied by freeze quenching and ESR spectroscopy. Free Radic Res. 1999;30:305–314. doi: 10.1080/10715769900300331. [DOI] [PubMed] [Google Scholar]
- (28).Lardinois OM, Ortiz de Montellano PR. Autoreduction of ferryl myoglobin: discrimination among the three tyrosine and two tryptophan residues as electron donors. Biochemistry. 2004;43:4601–4610. doi: 10.1021/bi036241b. [DOI] [PubMed] [Google Scholar]
- (29).Catalano CE, Choe YS, Ortiz de Montellano PR. Reactions of the protein radical in peroxide-treated myoglobin. Formation of a heme-protein cross-link. J Biol Chem. 1989;264:10534–10541. [PubMed] [Google Scholar]
- (30).Reeder BJ, Svistunenko DA, Sharpe MA, Wilson MT. Characteristics and mechanism of formation of peroxide-induced heme to protein cross-linking in myoglobin. Biochemistry. 2002;41:367–375. doi: 10.1021/bi011335b. [DOI] [PubMed] [Google Scholar]
- (31).Reeder BJ, Cutruzzola F, Bigotti MG, Watmough NJ, Wilson MT. Histidine and not tyrosine is required for the peroxide-induced formation of haem to protein cross-linked myoglobin. IUBMB Life. 2007;59:477–489. doi: 10.1080/15216540601178083. [DOI] [PubMed] [Google Scholar]
- (32).Sugiyama K, Highet RJ, Woods A, Cotter RJ, Osawa Y. Hydrogen peroxide-mediated alteration of the heme prosthetic group of metmyoglobin to an iron chlorin product: evidence for a novel oxidative pathway. Proc Natl Acad Sci U S A. 1997;94:796–801. doi: 10.1073/pnas.94.3.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Antonini E, Brunori M. Hemoglobin and Myoglobin in the Reactions with Ligands. Elsevier; Amsterdam, The Netherlands, North-Holland: 1971. [Google Scholar]
- (34).Bismuto E, Sirangelo I, Irace G, Gratton E. Pressure-induced perturbation of apomyoglobin structure: fluorescence studies on native and acidic compact forms. Biochemistry. 1996;35:1173–1178. doi: 10.1021/bi951163g. [DOI] [PubMed] [Google Scholar]
- (35).Nelson DP, Kiesow LA. Enthalpy of decomposition of hydrogen peroxide by catalase at 25 degrees C (with molar extinction coefficients of H2O2 solutions in the UV) Anal Biochem. 1972;49:474–478. doi: 10.1016/0003-2697(72)90451-4. [DOI] [PubMed] [Google Scholar]
- (36).Morris JC. The acid ionization constant of HOCl from 5 to 35°. J Phys Chem. 1966;70:3798–3805. [Google Scholar]
- (37).Kooser R, Kirchmann E, Matkov T. Measurements of spin concentration in electron paramagnetic resonance spectrometry. Preparation of standard solutions from optical absorption. Concepts Magn. Res. 1992:145–152. [Google Scholar]
- (38).Kaur H, Leung KHW, Perkins MJ. A water-soluble, nitroso-aromatic spin-trap. J Chem Soc Chem Commun. 1981;3:142–143. [Google Scholar]
- (39).Hiramoto K, Hasegawa Y, Kikugawa K. Appearance of ESR signals by the reaction of 3,5-dibromo-4-nitrosobenzenesulfonate (DBNBS) and non-radical biological components. Free Radic Res. 1994;21:341–349. doi: 10.3109/10715769409056586. [DOI] [PubMed] [Google Scholar]
- (40).Dragutan I, Mehlhorn RJ. Modulation of oxidative damage by nitroxide free radicals. Free Radic Res. 2007;41:303–315. doi: 10.1080/10715760601089356. [DOI] [PubMed] [Google Scholar]
- (41).Levine RL, Williams JA, Stadtman ER, Shacter E. Carbonyl assays for determination of oxidatively modified proteins. Methods Enzymol. 1994;233:346–357. doi: 10.1016/s0076-6879(94)33040-9. [DOI] [PubMed] [Google Scholar]
- (42).Deterding LJ, Moseley MA, Tomer KB, Jorgenson JW. Coaxial continuous flow fast atom bombardment in conjunction with tandem mass spectrometry for the analysis of biomolecules. Anal Chem. 1989;61:2504–2511. doi: 10.1021/ac00197a011. [DOI] [PubMed] [Google Scholar]
- (43).Levine RL. Carbonyl modified proteins in cellular regulation, aging, and disease. Free Radic Biol Med. 2002;32:790–796. doi: 10.1016/s0891-5849(02)00765-7. [DOI] [PubMed] [Google Scholar]
- (44).Levine J, Weickert M, Pagratis M, Etter J, Mathews A, Fattor T, Lippincott J, Apostol I. Identification of a nickel(II) binding site on hemoglobin which confers susceptibility to oxidative deamination and intramolecular cross-linking. J Biol Chem. 1998;273:13037–13046. doi: 10.1074/jbc.273.21.13037. [DOI] [PubMed] [Google Scholar]
- (45).Handelman GJ, Nightingale ZD, Dolnikowski GG, Blumberg JB. Formation of carbonyls during attack on insulin by submolar amounts of hypochlorite. Anal Biochem. 1998;258:339–348. doi: 10.1006/abio.1998.2592. [DOI] [PubMed] [Google Scholar]
- (46).Davies MJ. Identification of a globin free radical in equine myoglobin treated with peroxides. Biochim Biophys Acta. 1991;1077:86–90. doi: 10.1016/0167-4838(91)90529-9. [DOI] [PubMed] [Google Scholar]
- (47).Bure C, Gobert W, Lelievre D, Delmas A. In-source fragmentation of peptide aldehydes and acetals: influence of peptide length and charge state. J Mass Spectrom. 2001;36:1149–1155. doi: 10.1002/jms.220. [DOI] [PubMed] [Google Scholar]
- (48).Bure C, Le Falher G, Lange C, Delmas A. Fragmentation study of peptide acetals and aldehydes using in-source collision-induced dissociation. J Mass Spectrom. 2004;39:817–823. doi: 10.1002/jms.661. [DOI] [PubMed] [Google Scholar]
- (49).De Nino A, Mazzotti F, Perri E, Procopio A, Raffaelli A, Sindona G. Virtual freezing of the hemiacetal-aldehyde equilibrium of the aglycones of oleuropein and ligstroside present in olive oils from Carolea and Coratina cultivars by ionspray ionization tandem mass spectrometry. J Mass Spectrom. 2000;35:461–467. doi: 10.1002/(SICI)1096-9888(200003)35:3<461::AID-JMS961>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- (50).Kelman DJ, DeGray JA, Mason RP. Reaction of myoglobin with hydrogen peroxide forms a peroxyl radical which oxidizes substrates. J Biol Chem. 1994;269:7458–7463. [PubMed] [Google Scholar]
- (51).DeGray JA, Gunther MR, Tschirret-Guth R, Ortiz de Montellano PR, Mason RP. Peroxidation of a specific tryptophan of metmyoglobin by hydrogen peroxide. J Biol Chem. 1997;272:2359–2362. doi: 10.1074/jbc.272.4.2359. [DOI] [PubMed] [Google Scholar]
- (52).Gunther MR, Sturgeon BE, Mason RP. A long-lived tyrosyl radical from the reaction between horse metmyoglobin and hydrogen peroxide. Free Radic Biol Med. 2000;28:709–719. doi: 10.1016/s0891-5849(00)00164-7. [DOI] [PubMed] [Google Scholar]
- (53).Lam MA, Pattison DI, Bottle SE, Keddie DJ, Davies MJ. Nitric oxide and nitroxides can act as efficient scavengers of protein-derived free radicals. Chem Res Toxicol. 2008;21:2111–2119. doi: 10.1021/tx800183t. [DOI] [PubMed] [Google Scholar]
- (54).Mehlhorn RJ, Swanson CE. Nitroxide-stimulated H2O2 decomposition by peroxidases and pseudoperoxidases. Free Radic Res Commun. 1992;17:157–175. doi: 10.3109/10715769209068163. [DOI] [PubMed] [Google Scholar]
- (55).Krishna MC, Samuni A, Taira J, Goldstein S, Mitchell JB, Russo A. Stimulation by nitroxides of catalase-like activity of hemeproteins. Kinetics and mechanism. J Biol Chem. 1996;271:26018–26025. doi: 10.1074/jbc.271.42.26018. [DOI] [PubMed] [Google Scholar]
- (56).De Bono D, Yang WD, Symons MC. The effect of myoglobin on the stability of the hydroxyl-radical adducts of 5,5 dimethyl-1-pyrolline-N-oxide (DMPO), 3,3,5,5 tetramethyl-1-pyrolline-N-oxide (TMPO) and 1-alpha-phenyl-tert-butyl nitrone (PBN) in the presence of hydrogen peroxide. Free Radic Res. 1994;20:327–332. doi: 10.3109/10715769409145632. [DOI] [PubMed] [Google Scholar]
- (57).Yonetani T, Schleyer H. Studies on cytochrome c peroxidase. IX. The reaction of ferrimyoglobin with hydroperoxides and a comparison of peroxide-induced compounds of ferrimyoglobin and cytochrome c peroxidase. J Biol Chem. 1967;242:1974–1979. [PubMed] [Google Scholar]
- (58).Zhang R, Chandrasena RE, Martinez E, 2nd, Horner JH, Newcomb M. Formation of compound I by photo-oxidation of compound II. Org Lett. 2005;7:1193–1195. doi: 10.1021/ol050296j. [DOI] [PubMed] [Google Scholar]
- (59).Goldstein S, Merenyi G, Russo A, Samuni A. The role of oxoammonium cation in the SOD-mimic activity of cyclic nitroxides. J Am Chem Soc. 2003;125:789–795. doi: 10.1021/ja028190w. [DOI] [PubMed] [Google Scholar]
- (60).Zhang R, Goldstein S, Samuni A. Kinetics of superoxide-induced exchange among nitroxide antioxidants and their oxidized and reduced forms. Free Radic Biol Med. 1999;26:1245–1252. doi: 10.1016/s0891-5849(98)00328-1. [DOI] [PubMed] [Google Scholar]
- (61).Behforouz M, Bolan J, Flynt M. Erratum: 2,4-Dinitrophenylhydrazones: A modified method for the preparation of these derivatives and an explanation of previous conflicting results. J. Org. Chem. 1985;50:1186–1189. (Journal of Organic Chemistry (1985) 50, (1187-1189)) [Google Scholar]
- (62).Uchiyama S, Matsushima E, Aoyagi S, Ando M. Measurement of acid-catalyzed isomerization of unsaturated aldehyde-2,4-dinitrophenylhydrazone derivatives by high-performance liquid chromatography analysis. Anal. Chim. Acta. 2004;523:157–163. [Google Scholar]
- (63).Barton DHR, Le Gloahec VN, Smith J. Study of a new reaction: Trapping of peroxyl radicals by TEMPO. Tetrahedron Lett. 1998;39:7483–7486. [Google Scholar]
- (64).Kieber DJ, Blough NV. Fluorescence detection of carbon-centered radicals in aqueous solution. Free Radic Res Commun. 1990;10:109–117. doi: 10.3109/10715769009145940. [DOI] [PubMed] [Google Scholar]
- (65).Johnson CG, Caron S, Blough NV. Combined liquid chromatography/mass spectrometry of the radical adducts of a fluorescamine-derivatized nitroxide. Anal Chem. 1996;68:867–872. doi: 10.1021/ac951006n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.