Abstract
The Paf1 complex (Paf1C), composed of the proteins Paf1, Ctr9, Cdc73, Rtf1, and Leo1, accompanies RNA polymerase II (pol II) from the promoter to the 3' end formation site of mRNA and snoRNA encoding genes; it is also found associated with RNA polymerase I (pol I) on rDNA. The Paf1C is found in simple and complex eukaryotes; in human cells hSki8 is also part of the complex. The Paf1C has been linked to a large and growing list of transcription related processes including: communication with transcriptional activators; recruitment and activation of histone modification factors; facilitation of elongation on chromatin templates; and the recruitment of 3' end processing factors necessary for accurate termination of transcription. Absence of, or mutations in, Paf1C factors result in alterations in gene expression that can result in misregulation of developmental programs and loss of control of cell division leading to cancer in humans. This review considers recent information that may help to resolve whether the Paf1C is primarily a “platform” on pol II that coordinates the association of many critical transcription factors, or if the complex itself plays a more direct role in one or more steps in transcription.
Keywords: RNA polymerase II, Paf1 complex, Transcription elongation, Transcription termination, Histone modifications, RNA 3' end formation
Introduction and Background
A major focus of this review is to present recent information about the RNA polymerase II (pol II)-associated Paf1 complex (Paf1C) in a context that will help to establish a functional role for the complex in transcription. The mechanistic questions to be addressed: how the complex is recruited to pol II; what processes require the Paf1C; what factors make direct contacts with the Paf1C; and how the contacts with pol II are reversed at the end of the transcription cycle, will be presented after a brief summary of how the Paf1C was identified and characterized in yeast, flies and humans.
Identification of the Paf1 Complex
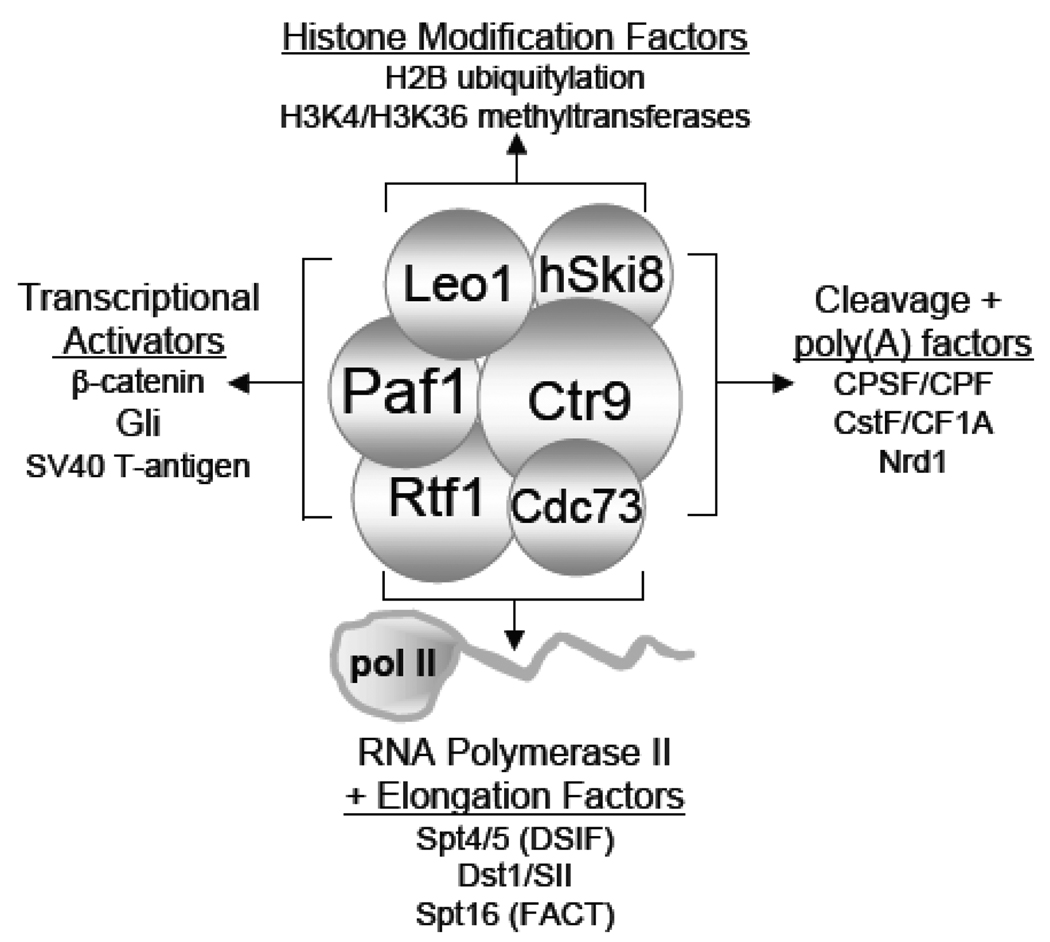
The proteins of the Paf1 complex were found nearly 15 years ago in a search for general transcription factors associated with yeast pol II [1]. The search was based on knowledge that mammalian general initiation factor TFIIF is a pol II binding protein [2]. By immobilizing an antibody directed against the C-terminal domain (CTD) of pol II to isolate Polymerase Associated Factors (PAF), we hoped to characterize the yeast equivalent of TFIIF as well as other critical initiation and elongation factors. The technique did in fact result in the isolation of yeast initiation factors TFIIF, TFIIB and the elongation factor TFIIS (Dst1/SII) [1]. Surprisingly, the proteins of the “mediator” complex, which had recently been described as associated with pol II in an activator responsive holoenzyme [3, 4], were not present in the PAF fraction. Instead, we found and began to characterize a new collection of pol II associated proteins, the first of which was previously uncharacterized by yeast genetics and so was named Paf1 [1]. In this review I will summarize what has been learned about Paf1 and the other factors in the eponymous Paf1C. Although much of the review will describe experiments done in yeast, the discovery of Paf1C homologues in more complex eukaryotes including plants, flies and humans has been a critical addition to our knowledge of the Paf1C. Throughout the review proteins or complexes from organisms other than yeast will be noted by an appropriate prefix (human Paf1 = hPaf1, Drosophila (fly) Cdc73 = dCdc73). As outlined in Fig. 1, the Paf1Cs from these different organisms have been shown to play an important role in communication with transcriptional activators, recruitment and activation of histone modification factors, and the association of transcriptional elongation, cleavage and polyadenylation (poly(A)) factors with pol II.
Figure 1. Overview of Paf1C Interactions with Transcriptional Activation, Histone Modification, Elongation and 3' End Formation Factors.
Details of the various interactions depicted are described in the text.
Paf1 and Cdc73 were the first proteins characterized in the yeast Paf1C, and we found that the genes encoding them are not essential, and that their loss results in changes of transcript abundance for only a small subset of yeast genes [5–8]. This led to the speculation that the Paf1C might be a form of the pol II holoenzyme, distinct from the Srb-containing mediator, involved in expression of a small number of yeast genes. However, compelling evidence has emerged over the last several years linking the Paf1C to transcriptional elongation of all genes. First, when the Paf1C was purified and the remaining subunits characterized we, and the Arndt and Greenblatt groups, found Ctr9, Leo1 and Rtf1 [9–11]. Although Rtf1 was originally isolated as a factor that affects transcriptional start site selection [12], it has extensive genetic and physical links to elongation factors including Spt4/5, Spt16/Pob3 and Dst1 (the yeast equivalents of elongation factors DSIF, FACT and TFIIS/SII, respectively [13]) [10, 11, 14]. Second, using the technique of chromatin immunoprecipitation (ChIP), the Young and Greenblatt labs localized the Paf1C subunits to the entire open reading frame of transcribed genes, a distribution consistent with their association with pol II, but distinctly different from that of the mediator and general initiation factors [11, 15].
The Paf1C in Complex Eukaryotes
The first report of the human form of the Paf1C came from the characterization of the parafibromin gene, found to be mutated in hyperparathyroidism jaw tumor syndrome (HPT-JT). Parafibromin is 27% identical to Cdc73 in its C-terminal region, and it co-purifies with the human homologues of Paf1, Ctr9 and Leo1 [16–19]. The hPaf1C also contains hSki8, a component of the human SKI complex important for 3'–5' mRNA degradation [19]. However, until recently, all reports of hPaf1C composition lacked the hRtf1 subunit. Because Rtf1 is so central to the function of the Paf1C this difference was puzzling. The puzzle has been resolved by the recent description of purified hPaf1C containing hRtf1 [20]. Roeder and coworkers created a cell line stably expressing tagged hPaf1, which was used to purify hPaf1C containing hPaf1, hCtr9, hLeo1, hCdc73, hSki8 and hRtf1. They found that Rtf1 exists in several forms, and the form associated with the hPaf1C encompasses residues 157–585. These investigators also created and characterized a fully recombinant hPaf1 containing all six proteins [20]. The association of hRtf1 with the hPaf1C appears to be less stable than that of the other factors, as observed for the yeast and fly Paf1Cs [21, 22], but hRtf1, and presumably its homologues from other organisms, are an integral part of the hPaf1C. This physical characterization of hPaf1C composition is consistent with the recent report that inactivation of any of the hPaf1C components, including hRtf1, results in similar effects on stem cell gene expression [23].
The hPaf1C is also associated with pol II, and human and fly Paf1Cs co-localize with pol II on the chromatin of actively transcribed genes [17, 20, 21, 24, 25]. Many aspects of Paf1C function in simple and complex eukaryotes are therefore likely to be similar. However, there are some intriguing differences. In flies, the dPaf1C makes direct contact with the transcriptional activator β-catenin through interactions with dCdc73 (Hyrax) [26]; dCdc73 also makes direct contacts with the Gli family of activators [27]. Whether these interactions result in changes to transcriptional initiation or elongation is not yet clear, but there are hints from other systems that the Paf1C may be important for direct effects on transcriptional activators, promoters and the initiation of transcription [28–30].
Since its discovery in HPT-JT, mutation of hCdc73 has also been connected with breast, renal and gastric cancer, and found to interact with SV40 T-antigen (reviewed in [31]). Consistent with its role as a tumor suppressor, over-expression of hCdc73 inhibits proliferation of NIH 3T3 and HEK293 cells [32]. hCdc73 is not the only hPaf1C component to be linked to cancer. As recently reviewed by Chaudhary et al., loss of hCtr9 and over-expression of hPaf1 are associated with pancreatic cancer [33]. In addition, over-expression of hPaf1 in NIH 3T3 cells results in an enhanced growth rate and tumor formation [34]. Components of the Paf1C are essential in complex eukaryotes, and conditional inactivation of hCdc73 results in changes in expression of a subset of human genes including several critical growth factors [24, 27, 35]. A further link to growth regulation is found in the recent report that expression of hPaf1 is cell cycle regulated and it is required for regulation of a subclass of genes important for cell cycle progression [36].
The Paf1C's Position in the Transcription Cycle
Pol II undergoes a cyclic association with the factors required for it to recognize, and initiate transcription from, promoters; factors that allow it to escape from its promoter-specific contacts; factors required to move the transcription complex through nucleosomal barriers; and factors necessary to cleave the nascent transcript and allow the enzyme to be released for another cycle (reviewed in [13, 37–42]). Fig. 2A shows a Paf1C centric and oversimplified view of some aspects of this cycle, a key feature of which is the modifications that occur on the pol II CTD (reviewed in [43–45]). The initiating form of pol II lacks phosphorylation of the serines (Ser) in the Y1S2P3T4S5P6S7 repeat of the CTD; initiation is accompanied by phosphorylation of Ser at repeat positions 5 and 7 (Ser5 and Ser7) by Kin28, a component of the TFIIH general initiation factor [46]. Although Ser5 phosphorylation (Ser5-P) is tightly linked to initiation, recent evidence suggests that this modification is not actually required for transcription, but instead, is necessary for mRNA capping [47, 48]. As pol II enters into elongation mode, Ser at position 2 (Ser2) is phosphorylated by the Ctk1 and Bur1 kinases (pTEFb in more complex eukaryotes) [49, 50]. Phosphorylation of Ser2 (Ser2-P) is required for the association of many factors with the elongating pol II; several of these will be described in more detail below. Ctk1 also has a kinase-independent function at an early point after pol II enters into elongation that results in the displacement of several general initiation factors [51].
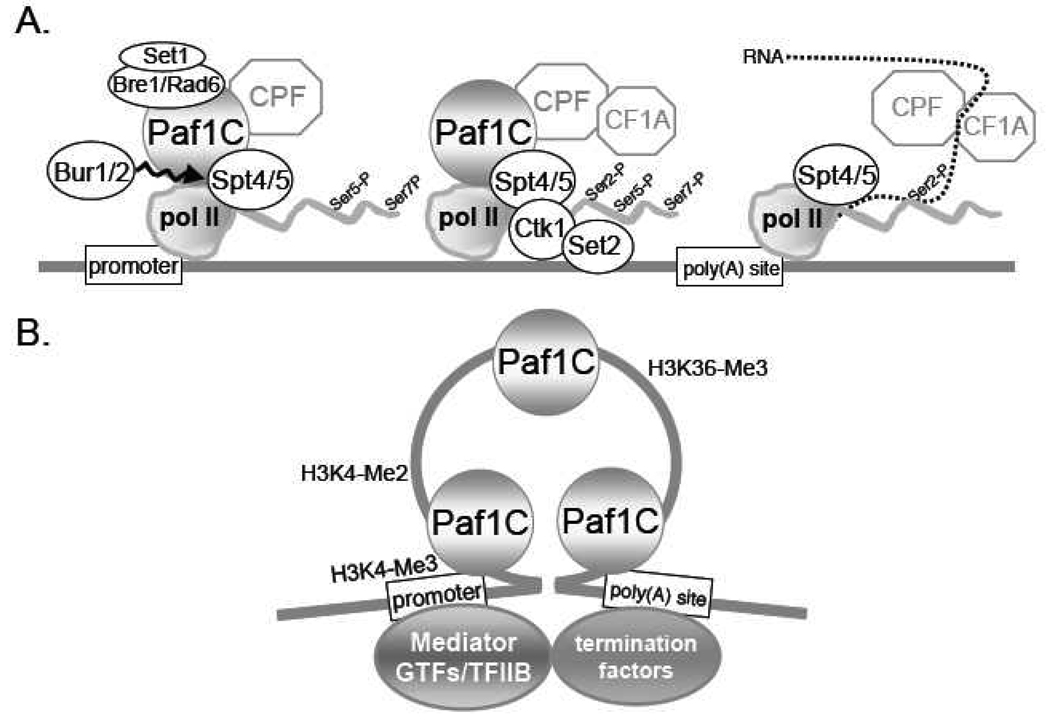
Figure 2. Dynamic Association of the Paf1C with Pol II During Transcription.
A. As described in the text the recruitment of the Paf1C minimally requires pol II and phosphorylation of Spt5 by the Bur1/2 cyclin/kinase. Many other factors are present on pol II during elongation including Bre1/Rad6 and the cleavage and poly(A) factor complexes CPF and CF1A. Increasing pol II CTD Ser2-P catalyzed by Ctk1 and Bur1 also helps to recruit histone modification and 3' end formation factors including Set2. Another transition occurs at the poly(A) site leading to release of the Paf1C and the kinases Bur1 and Ctk1 from pol II but retention of Spt4/5.
B. As described in the text a loop including a subset of initiation and 3' end formation factors forms between the promoter and poly(A) sites in yeast. The differential localization of histone H3 modifications is described in the text. The Paf1C is present throughout the region defined as the elongation loop but is absent up- and downstream where the general initiation factors, mediator complex and termination factors respectively are present.
Unlike the many factors that recognize the phosphorylation state of the CTD, the Paf1C associates with unphosphorylated, Ser5-P and Ser2-P forms of pol II [17], and it is found with pol II at or very near promoters and throughout the coding regions of genes [15, 20, 24, 52–55]. Intriguingly, as depicted in Fig. 2A, it detaches from pol II near the poly(A) site; the Ser2-P form of pol II continues on with elongation factors Spt4/5 until termination [21, 53, 54]. Another interesting way of thinking about the position of the Paf1C during the transcription cycle is diagrammed in Fig. 2B. The Hampsey and Proudfoot labs have presented evidence for a loop, created by interactions between some initiation (notably TFIIB) and termination factors, that connects the promoter and terminator regions of actively transcribed yeast genes [56–58]. Loop formation has been shown to be critical for maintenance of transcriptional memory, which allows the rapid reactivation of a regulated gene [59, 60] Using a model based on connection of the promoter and terminator region, the localization of the Paf1C correlates with the region of the loop. This may be the result of a series of mutually exclusive associations with a position on pol II occupied during the transcription cycle by either: the general initiation factors and/or the mediator; or the Paf1C; or termination factors. Hampsey has speculated that looping might be involved in moving active genes to the nuclear periphery thereby enhancing mRNA export [58], and the Proudfoot lab has recently shown that the loops interact with the nuclear pore complex [60]. These interactions are of particular interest in light of the known genetic and biochemical connections between the Paf1C and the TREX mRNA export complex (discussed in [52]).
The factors that recruit the Paf1C to pol II and the process of eviction at the poly(A) site will be discussed in more detail below. An important function of the Paf1C during the transcription cycle is to recruit histone modification factors to the transcribing pol II [61, 62]. This role has led to the use of static terms like “platform” [19, 61, 63] or “tether” [49]) to describe its properties. However, in this review I consider recent evidence that establishes a more active role for the Paf1C in the transcription cycle.
WHAT IS REQUIRED TO RECRUIT THE PAF1C?
The Paf1C is defined by its association with pol II, but we still do not know which pol II subunit(s) is involved, or how the association is restricted to only part of the transcription cycle (Fig. 2A). The original isolation of the Paf1C utilized an antibody specific for the unphosphorylated pol II CTD [1, 64]. Because the CTD was engaged with the antibody in this isolation, and because the Paf1C does not appear to be sensitive to CTD modification it is very probable that the Paf1C makes contacts with another part of pol II, although Cdc73 and Ctr9, presumably as part of the Paf1C, have been detected binding to a Ser2-P, Ser5-P doubly phosphorylated CTD peptide [65]. Because loss of either Cdc73 or Rtf1 results in the detachment of the remaining complex components from pol II, we have proposed that there are at least two contact points between the Paf1C and pol II [52, 55]. Consistent with one of these contact points, recombinant Cdc73 interacts directly with purified pol II, to form a stoichiometric complex [6]. Loss of Ctk1, and thereby the bulk of CTD Ser2-P, does not affect the recruitment of the Paf1C [55, 66]. Other factors that are recruited to elongating pol II, but appear to associate after the Paf1C recruitment step include Rad6, Bre1, Set1 and Dst1/SII [20, 55].
Bur1 Phosphorylation of Spt5 is Important for Paf1C Recruitment
Although the Ctk1 and Bur1 kinases are both related to mammalian pTEFb, they have distinctly different functions in yeast (reviewed in [49]). In particular, Bur1 and its cyclin partner Bur2 are required for Paf1C recruitment to chromatin and presumably to pol II [67], but the Paf1C itself does not seem to be the target of Bur1 phosphorylation [68]. Instead, two recent studies have identified Spt5 as a Bur1 kinase substrate. Phosphorylation of Spt5 in its C-terminal repeat by Bur1 stimulates recruitment of the Paf1C to chromatin as depicted in Fig. 2A [63, 69]. Phosphorylation of Spt5 by Bur1 is consistent with the phosphorylation of hSpt5 (DSIF) by pTEFb [49], which supports the prediction that the hPaf1C may also be recruited by phosphorylation of hSpt5. However, hSpt5 has recently been reported to interact with hPaf1 independent of phosphorylation [25]. It may be that phosphorylation is critical in the context of the ternary complex with pol II. Spt5 joins pol II after it has converted from the pre-initiation complex to an elongating form [63], so the issue of Paf1C recruitment moves upstream to discovering how Spt4/5 joins the elongation complex. The contact point between Spt5 and the Paf1C is not known, but Rtf1 is a likely candidate as described further below. Bur1 phosphorylation of Spt5 does not promote interaction with the Paf1C that can be detected in vitro in the absence of pol II and other factors, further supporting the idea that there are multiple connections with pol II [63].
It has been reported that loss of Kin28, and hence loss of pol II CTD Ser5-P, results in a partial diminution of Paf1C association with chromatin [55]. This observation is consistent with a recent report that Kin28 enhances Bur1/2 recruitment, therefore stimulating Paf1C recruitment [50]. It is somewhat difficult to reconcile these observations with the reports that Kin28 is not necessary for transcription [47, 48], but apparently the fairly weak association of Bur1 with Ser5-P [63] helps to direct the kinase toward the 5' end of the transcription unit [50]. Loss of Bur1 also results in loss of CTD Ser2-P [50], but this is due in part to the coincident loss of the downstream Paf1C, which is known to be required for full levels of Ser2-P as described further below [22, 52]. Although the Paf1C does not depend on Ctk1 phosphorylation of Ser2 [55, 66], the observation that Bur1 phosphorylates Ser2 residues near the promoter leaves open the possibility that these Bur1 products may play a role in Paf1C recruitment [50]. Other factors that are earlier than the Paf1C during pol II's transition to elongation and may be important for recruitment include Spt6 [54], Spt16 as part of FACT [70], and the Ccr4-Not complex, which may act in a pathway parallel to Bur1 recruitment [71].
WHAT PROCESSES ARE DEPENDENT ON THE PAF1C?
Histone modifications
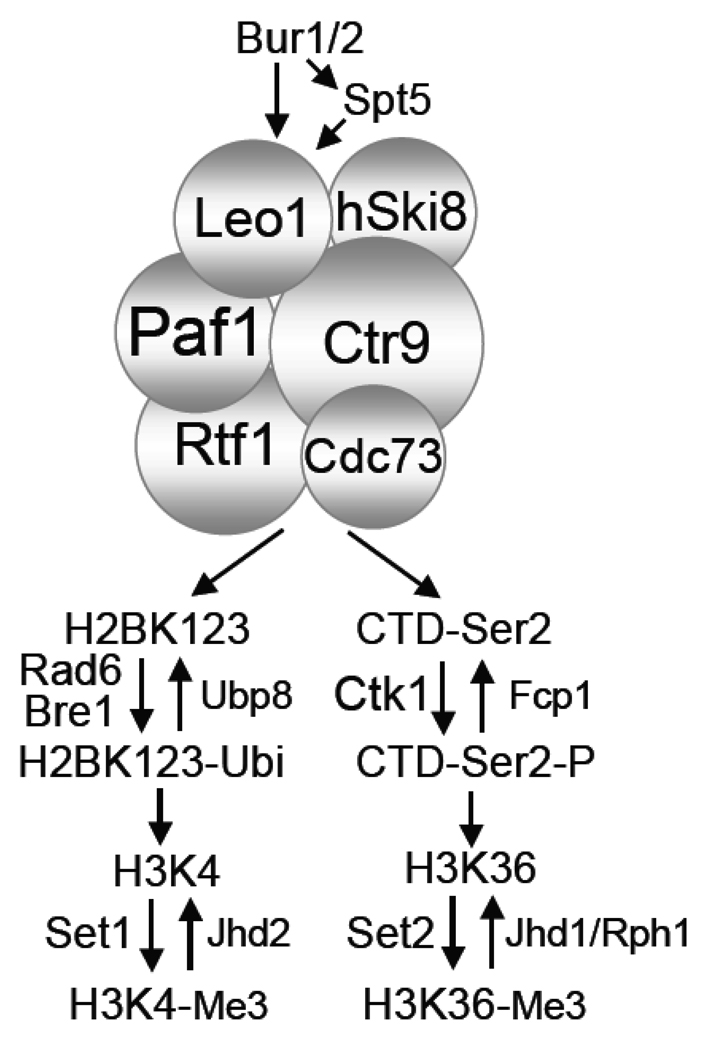
There has been a torrent of recent information about the pattern of post-translational modifications to nucleosomal histones delineating transcriptionally active and inactive regions of the genome, as well as many other structural and functional features of the chromosome. As recently extensively reviewed, some of these modifications depend on the Paf1C [62, 72–74]. I will focus in this section on a small subset of histone modifications linked to the Paf1C and to active transcription. In Fig. 2A+B, the promoter region is marked by the Set1-mediated trimethylation of lysine 4 of histone H3 (H3K4-Me3) (reviewed in [73]). The 5' region of the elongation region is defined by the appearance of H3K4-Me2, also created by Set1 [75]. Set2 trimethylates lysine 36 of H3 (H3K36-Me3), predominately in the 3' half of the transcription unit (reviewed in [72]). The connection of the Paf1C to two of these modifications is outlined in Fig. 3. Paf1C components are required for formation of both H3K4-Me3 and H3K36-Me3 [68, 76, 77]. However, these requirements lie downstream of two other Paf1C-dependent activities. First, the Paf1C has been shown to be necessary in both yeast and humans for the Rad6/Bre1-dependent ubiquitylation of histone H2B [67, 78–81]. The presence of ubiquitylated H2B is critical for the formation H3K4-Me3 (reviewed in [73]). Second, the Paf1C is required for full levels of pol II CTD Ser2-P [22, 52]. This modification is necessary for the recruitment of Set2 and the formation of H3K36-Me3 [82–84]. Because the presence of Set2 does not correlate completely with the formation of H3K36-Me3 [68], loss of the Paf1C may also affect demethylation of H3K36 or the activity of Set2. So the Paf1C and the Bur1 kinase, as outlined in Fig. 3, lie upstream of these two histone modifications that are unique to transcriptionally active regions of the chromosome.
Figure 3. Histone H3 Trimethylation is Dependent on the Paf1C.
Recruitment of the Paf1C to pol II requires the Bur1 kinase and the presence of Spt4/5 [63, 67, 69]. The Paf1C is not required for the initial recruitment of Rad6/Bre1[79] or known to be required for recruitment of Ctk1 [22], but it is required for the presence of the product of the two activities, H2BK123 ubiquitylation and CTD-Ser2 phosphorylation respectively. This could be due to a reduction in activity of the modifying enzymes in the absence of the Paf1C (without the Paf1C, Rad6/Bre1 does not spread into the coding region [79]), or to a stimulation of the modification removal activities (Ubp8 and Fcp1 as shown). Without modification of H2BK123 and the Ser2 of the pol II CTD there is no trimethylation of H3K4 by Set1 [78, 85] or H3K36 by Set2 [82, 83].
A requirement for the Paf1C in Set1-dependent formation of the recently identified H3K4-Me2 mark at the 5' end of genes [75] is not yet clear, although it seems probable based on the requirement for the Paf1C in Set1 recruitment and H3K4 methylation [85]. This modification results in recruitment of a histone deacetylase complex, which may play a role in the transition to elongation or in suppression of cryptic promoters [68, 75]. Loss of the Paf1C results in increased acetylation of H3 and H4 near the 5' ends of yeast genes [68], which may be the consequence of loss of the H3K4-Me2 mark [75]. Paf1C function in H3K4-Me3 formation is conferred by the Rtf1 subunit. Arndt and coworkers have mapped a region of Rtf1 critical for this modification distinct from the region involved in interaction with the rest of the Paf1C [86]. With the knowledge that Rtf1 from flies is also important for histone modifications [21, 87], it is indeed satisfying to know that Rtf1 appears to be an integral part of all eukaryotic Paf1Cs as described above. In contrast, H3K36-Me3 formation is less dependent on Rtf1, but is sensitive to loss of Paf1, Ctr9 and Cdc73, corresponding to the effects that loss of each these factors has on Ser2-P formation [22, 68].
In human cells hCdc73 appears to be required for full levels of H3K36-Me3, but not H3K4-Me3 [24], whereas in yeast Cdc73 is required for both modifications [68, 76, 77]. However, hCtr9 is important for H3K4-Me3 levels in human cells as it is in yeast [23] Future studies will reveal if Paf1C components in different organisms play different roles in directing histone modifications. Although it is still not clear how these histone modifications, which correlate with active transcription, might be involved with the regulation of that transcription, it is notable that several groups have identified plant Paf1C factors as genes that play important roles in the developmental program of flowering. These regulatory effects seem to be due to effects on histone lysine trimethylation of K4 and K36 of H3 [88–90]. In addition, the hPaf1C is important for maintaining embryonic stem cell identity and for HOX gene regulation; these roles are also correlated with effects on histone modifications [23, 80]
Telomere Length and Telomeric Silencing
Strains lacking the Paf1C have defects in telomeric silencing, which is a direct consequence of the alterations in chromatin modifications described above [76, 77]. Rtf1 is the central player in these processes with the same regions of the protein being responsible for both histone modifications and telomeric silencing [86]. The Paf1C is also important for normal expression of the telomerase RNA encoded by the TLC1 gene [30]. In this case, the role of the Paf1C appears to be separate from its effects on chromatin or 3' end formation. Loss of Cdc73 causes the greatest decrease in expression of TLC1, and Cech and co-workers speculate that the requirement for Paf1C components in TLC1 expression may be at the level of initiation [30]. It is therefore possible that in yeast as in flies that the Paf1C may be acting at promoters as well as in the body of genes to control expression.
Full levels of CTD Ser2-P
Loss of the Paf1C is associated with a significant reduction in phosphorylation of pol II CTD Ser2 on chromatin [52]. Not only are chromatin-associated Ser2-P levels reduced by loss of Paf1, but loss of Rtf1, which serves to detach the Paf1C from chromatin, also lowers both chromatin-associated and bulk levels of Ser2-P [22, 52]. The mechanism for the Ser2-P reduction is not yet known, but because Ctk1 is the major Ser2 kinase, it is possible that loss of the Paf1C either affects recruitment or activity of Ctk1. This speculation is consistent with the fact that the Paf1C and Ctk1 have very similar distributions on the chromatin of transcribed genes, and that Ctk1 lies downstream of the Paf1C in terms of recruitment to pol II and chromatin [53, 66]. It is interesting in this regard that Berger and co-workers have shown that Ctk1 interacts directly with H2A/H2B in the non-ubiquitylated form [91], leading these investigators to speculate that Ctk1 localization to chromatin of coding regions is due to interactions with pol II and with the de-ubiquitylated histones. But without the Paf1C there is no H2B-ub as described above (Fig. 3), so Ctk1 recruitment might actually be enhanced by loss of the Paf1C, potentially resulting in higher Ser2-P levels rather than the 10-fold lower levels observed [22]. Perhaps instead the absence of the Paf1C from pol II creates a more easily accessed substrate for the Ser2-P phosphatase Fcp1 (Fig. 3). This speculation is of particular interest because loss of Rtf1 is lethal in combination with an allele of Fcp1 [14].
Bur1 also phosphorylates Ser2 near promoters [50], so it is possible that loss of the Paf1C reduces Bur1 derived Ser-2P. Hinnebusch and coworkers reported that loss of Bur1 affects the activity of Ctk1 [50], but, as described above, Bur1 is upstream of the Paf1C, which is required for full levels of Ser2-P, so this could be a secondary effect. To resolve this issue it will be necessary to map the location of the residual Ser2-P and measure the association of Ctk1 and Fcp1 with chromatin and pol II in the absence of the Paf1C. If the Paf1C is required for Ctk1 association with chromatin, it will also be of interest to ask if loss of the Paf1C results in the spreading of initiation factors into the coding region as observed for loss of Ctk1 [51].
Elongation through chromatin
Distribution of the Paf1C on chromatin of actively transcribed genes is consistent with a role in transcriptional elongation, and in fact the complex has been referred to as an elongation complex for some time [11]. However, evidence that the Paf1C is actually important for elongation has been inconsistent. For example, loss of Paf1C factors leads to phenotypes associated with elongation defects, but only in some yeast strains and only in some assays (discussed in [92]). The Paf1C was originally found with pol II and elongation factor Dst1/SII [1], but more extensive purification disrupted the Paf1C/Dst1 connection [6]. The Paf1C components have been identified in complexes with other elongation factors [10, 11], but loss of the Paf1C factors does not seem to affect the density or distribution of pol II on DNA in yeast or in flies as might be expected for a critical elongation factor [21, 52, 68]. In addition, in flies, under conditions where loss of dSpt6 reduces the rate of elongation, loss of dPaf1C factors has little effect [93].
Aguilera and coworkers reported that the Paf1C was required for elongation in vitro, supported by apparent elongation defects from extracts isolated from paf1 and cdc73 mutant strains in transcription assays using a non-chromatin form of G-less cassettes [94]. But loss of Rtf1, which removes the Paf1C from pol II [52], did not cause an elongation defect in these assays. In addition, the in vivo assay used in this work, expression from a bacterial lacZ reporter, is known in many cases to give artificially low responses due to the presence of cryptic poly(A) sites which can result in premature termination [95–97]. As described in more detail below, loss of Paf1 has been shown to alter poly(A) site utilization [24, 98, 99], and we have found that many promoters that appear to be down-regulated by loss of Paf1 in a lacZ construct, have normal expression levels when the reporter is changed to luciferase ([98] and unpublished observations). Recently the hPaf1C has been reported to act in combination with hSpt5 to stimulate elongation in an in vitro transcription system [25].
A recent report has extended the role of the Paf1C in transcription to pol I and rRNA expression [100]. In this study, the role of the Paf1C has been linked to elongation of pol I; however, loss of Paf1 results in reduced expression of several factors critical for rRNA synthesis and maturation [98]. Therefore it is difficult to evaluate whether the effects on pol I elongation are primary or secondary. Again as for pol II, loss of Paf1 does not result in changes to density or distribution of pol I on rDNA [100]. Reinberg and co-workers reported that purified hPaf1C lacking hRtf1 stimulates transcriptional elongation in vitro on a chromatin template, in the presence of the FACT complex [70]. This stimulation was linked to ubiquitylation of H2B, which facilitates FACT function. So again the function of the Paf1C was linked to its effects on histone modifications.
In light of all of these tantalizing partial links to transcriptional elongation, it is very satisfying that Roeder and co-workers have recently isolated highly purified forms of native and recombinant human Paf1C and demonstrated that they act on their own and in concert with known elongation factor hDst1/SII to stimulate elongation in vitro on chromatin templates independent of any effects on histone modification [20]. The synergistic effect of the hPaf1C is greatest at limiting hDst1/SII concentrations. Using an elongation-specific assay involving extension of a short pre-made transcript, the hPaf1C stimulates elongation on its own and synergizes with hDst1/SII. The hPaf1C is also required for full levels of hDst1/SII association with chromatin. The authors interpret these data as proof that the hPaf1C has intrinsic transcription elongation activity and that it cooperates with hDst1/SII.
mRNA 3' End Formation and Recruitment of 3' End Processing Factors
After identifying yeast genes dependent on the Paf1C for full expression we determined that Paf1C sensitivity was not conferred by promoters, but instead by changes in utilization of 3' end formation sites [98]. These changes resulted in the formation of unstable transcripts leading to reduced transcript abundance for the Paf1-dependent genes. Because expression profiles of strains lacking Paf1, Bur2 and COMPASS component Sdc1 are similar [67], it is likely that all of these yeast genes may be affecting 3' end formation rather than transcriptional initiation. This is of particular interest because another COMPASS component, Swd2 is also part of the cleavage and poly(A) complex [101]. In addition to changes in the efficiency of 3' end formation, loss of the Paf1C results in a reduction in the length of poly(A) tails on yeast mRNAs [52]. Winston and coworkers have also reported links between the Paf1C and transcriptional termination. They found that recruitment of Ctr9, and presumably the Paf1C, is dependent on Spt6 [54]. Loss of Spt6, and therefore also Ctr9, results in read through of the GAL10 3' end/termination site into the downstream GAL7 gene. In addition, the recent description of Paf1C effects on phenotypic expression of the yeast prion Psi were shown to be due to altered poly(A) site usage resulting in changes in translation efficiency [99].
A molecular explanation for these changes in mRNA 3' end formation can be found in our observation that loss of the Paf1C reduces recruitment of cleavage and poly(A) factor Pcf11 to chromatin [52]. However, Pcf11 is known to interact with the Ser2-P form of pol II [102]. Therefore, some of the reduction in cleavage and poly(A) factors could be a secondary effect of reduced Ser2-P in the absence of the Paf1C. However, we have also described direct non-pol II mediated interactions between the Paf1C and cleavage and poly(A) factor Cft1 that are important for recruitment of Cft1 to the Ser5-P form of pol II [22]. In addition, Rozenblatt-Rosen and coworkers recently reported that the human cleavage and poly(A) factors, including CPSF-30, 73, 100, 160 (hCft1) and CstF-64, 77 as well as Symplekin and Fip1, associate with the hPaf1C [24]. This association is resistant to RNAse indicating that it is based on protein/protein contacts. Immunodepletion of hCdc73 from an RNA processing extract also depletes CPSF and CstF, which reduces the extract's ability to cleave pre-mRNA with no effect on splicing. Reduction of hCdc73 by siRNA results in reduced expression of a small number of human mRNAs, including the INTS6 transcript, without altering the amount of pol II associated with the affected genes [24]. Instead, loss of hCdc73 results in a 2-to 3-fold decreased association of cleavage and poly(A) factors with chromatin of the INTS6 gene and increased read-through of proximal 3' processing sites. These results argue that in human cells, as in yeast, the connection between Paf1C and cleavage and poly(A) factors is important for proper 3' end formation.
snoRNA 3' end formation
In addition to its role in mRNA 3' end formation, the Paf1C is also required for correct formation of the non-polyadenylated 3' ends of snoRNAs [103]. This role does not appear to be a consequence of changes in histone modifications, but instead is the result of reduced recruitment of snoRNA 3' end formation factor Nrd1. Although 3' end formation of both mRNAs and snoRNAs is dependent on the Paf1C, the function of Ctk1 may be different for the two processes. Loss of Ctk1 results in increased snoRNA read-through [104], but it has little effect on 3' end formation of an mRNA that requires the Paf1C [22]. In contrast, loss of Cdc73 has little effect on snoRNA read-through [103], but is as important as Rtf1 for mRNA 3' end formation [22]. Clearly, each Paf1C component has a unique spectrum of functions.
WHAT MAKES DIRECT CONTACTS WITH THE PAF1C?
Intracomplex Interactions and Connections to Pol II
Although as described above, Paf1Cs with very similar protein compositions have been isolated from both yeast and human cells, until very recently there has been no real information about the internal interactions within the complex. The Paf1C components do show interesting interdependencies. For example, loss of yeast Ctr9 results in decreased abundance of all the other subunits [52]. Ctr9, Cd73 and Rtf1, but not Leo1, require Paf1 for normal levels, and loss of Cdc73 results in lower Rtf1 abundance [10, 52]. Arndt and co-workers identified a region in Rtf1's C-terminus critical for interactions with the Paf1C [86]. It has also been reported that loss of hCtr9 or hSki8 causes a reduction of hPaf1 and hLeo1 [19], and hCdc73 is required for full levels of hPaf1 and hLeo1 [29].
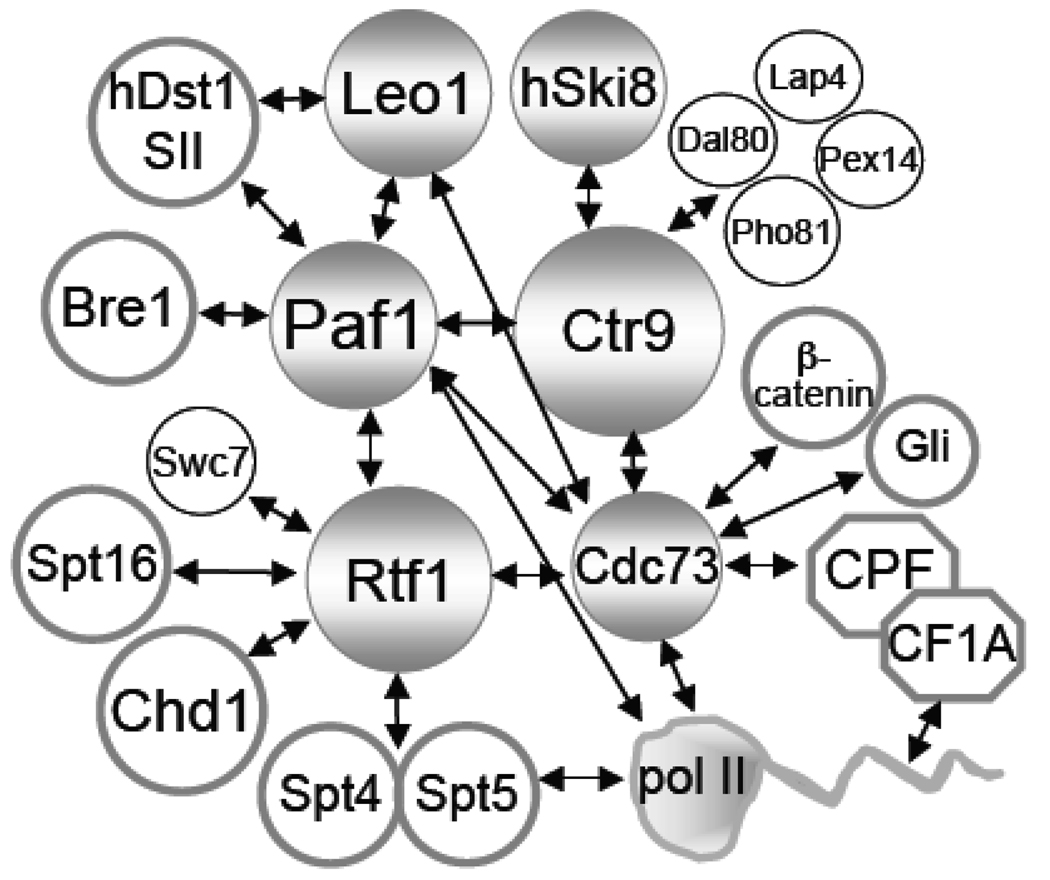
As summarized in Fig. 4, Roeder and co-workers have recently created an interaction map of the hPaf1C factors by expressing pairwise combinations of hPaf1C factors in insect cells and assaying for co-immunoprecipitation [20]. They found that hCtr9 is essential for hSki8 association, and that hPaf1 interacts directly with each of the other components. In addition, hCdc73 was found to interact with hCtr9, hRtf1 and hLeo1. It will be interesting to determine if similar interactions hold the yeast Paf1C together. Although the genes encoding the Paf1C are recognizably similar between yeast and humans, there are also many differences. This may explain why Kim et al. observe direct interactions between hPaf1 and hLeo1 and pol II [20], but no direct interactions between hCdc73 and pol II, in contrast to previous observations in yeast [6]. Both pol II connections are indicated in Fig. 4 and await further molecular characterization.
Figure 4. A Summary of Known Physical Interactions Within the Paf1C and Between the Paf1C and Other Factors.
Interactions from a global two-hybrid screen shown as circles with thin black lines: Paf1/Leo1, Rtf1/Swc7, Ctr9/ Pex14-Pho81-Lap4-Dal80 [109]. All other interactions shown with thick gray borders: yeast factor interactions- Cdc73/pol II [6]; Rtf1/Chd1-Paf1-Ctr9 [86, 107]; Rtf1/Spt4/5-Spt16 [10, 11]; Cdc73/CPF-CF1A [22]; human factor interactions-hPaf1/hBre1-hDst1-hLeo1-hCtr9-hRtf1-hCdc73, hCdc73/hRtf1-hCtr9, and hCtr9/hSki8 [20, 106]; hCdc73/CPSF-CstF [24]; fly factor interactions- dCdc73/β-catenin [26], dCdc73/Gli [27]. A comprehensive listing of genetic and physical interactions can be found in the Saccharomyces Genome Database (http://www.yeastgenome.org).
In yeast a second point of contact with pol II is probably through Rtf1 via contacts with Spt5 [10]. When either Cdc73 or Rtf1 is lost, the rest of the Paf1C components still associate with each other, but no longer interact with pol II or chromatin and relocalize as a unit from the nucleus to the nucleolus [22, 52, 55, 105]. Although the direct Spt5/Rtf1 interaction has not been completely established, the extensive genetic and physical interactions between these two factors lead to this speculation [10]. It is probable that the well-conserved central region of Rtf1, shown to be required both for association with ORFs and for the Spt5-related Spt- phenotype, is important for this contact with pol II [86]. As described above in the section about how the Paf1C is loaded onto pol II, there are probably multiple points of contact controlled by different molecular transitions that act synergistically to effect the transition from the initiating to the elongating form of pol II.
Connections to Other Factors
Interactions between Paf1C factors and other proteins have been reported, but in some cases are difficult to interpret. Because the Paf1C is defined by its association with pol II, it will necessarily be found together with other pol II -associated factors. Components of the Paf1C have been reported to co-purify/co-precipitate with: elongation factors Dst1/SII, Spt4/5 and the Spt16/Pob3 FACT complex [1, 10, 11, 25]; components of the THO and Ccr4/NOT complex [7]; and the Set1-containing COMPASS complex [76, 85]. All of these putative interactions are consistent with extensive genetic interactions between these factors, but not all are necessarily direct physical interactions. For example, as described above (Fig. 3), Set1-directed methylation of H3K4 does not depend directly on the Paf1C but instead requires the Paf1C-dependent ubiquitylation of H2B. Recently Kim and Roeder have demonstrated direct interactions between the yeast Paf1C and the Bre1 component of the Rad6/Bre1 H2B ubiquitylation complex [106], as well as a direct interaction between hPaf1 and hBre1 as marked in Fig. 4 [81]. Therefore, although both the Paf1C and COMPASS are found on pol II during elongation, they may well not be in direct contact. There is also no confirmation of direct interactions between the Paf1C and the THO and Ccr4-NOT complexes. However a recent report provides intriguing evidence that the Ccr4-NOT complex is required for Paf1C recruitment parallel to or downstream of the action of Bur1 [71].
In contrast, extensive allele-specific interactions between Spt5 and components of the Paf1C [10], plus the recent elucidation of the importance of Spt5 as a Bur1 kinase substrate for Paf1C recruitment [63, 69], make it very likely that Spt5 is a direct contact as indicated in Fig. 4. The evidence for direct contacts between the Paf1C and the Spt16 component of elongation factor FACT are somewhere in between these two examples. As described in the section on recruitment of the Paf1C, genetic and physical links have been established between the two complexes [10, 11, 107]. The connection in Fig. 4 is identified as being through Rtf1, but the evidence is not as clear as for Spt5.
Recently, direct contacts between the hPaf1C and elongation factor hDst1/SII have been clearly established using recombinant proteins [20]. Purified hPaf1C interacts selectively with purified hDst1/SII; the interaction appears to be through the hPaf1 and hLeo1 subunits as shown in Fig. 4. The interaction between these factors leads to cooperative binding to pol II and synergistic effects on transcription elongation of chromatin templates in vitro as described above [20]. The connection between hDst1/SII and the hPaf1C therefore may define another point of contact with pol II. It will be interesting to determine if a similar contact point also exists for the yeast Paf1C, although loss of Dst1/SII does not affect Paf1C recruitment to chromatin, and presumably pol II, in yeast [55], and the distribution of the two factors on both yeast and human chromatin appears to be different [15, 20]. The fact that a double dst1/paf1 mutant is lethal [10] supports the idea that these two factors function in parallel pathways in transcription elongation.
Chd1 definitely interacts physically with Rtf1 as noted in Fig. 4, because it was identified in a two-hybrid screen using Rtf1 as bait [107]. Chd1 has nucleosome remodelling activity and colocalizes with the Paf1C on transcribed chromatin [107]. Chd1 is also a transcription termination factor [42, 108]; therefore, the direct interaction between Chd1 and Rtf1 is another argument for the Paf1C functioning in both nucleosome remodeling and transcription termination. Despite several extensive two-hybrid screens of yeast protein interactions, only one intra-complex interaction has been reported; yeast Paf1 interacts with Leo1 as found for the human complex (Fig. 4, [109]). This same screen, of the several that have been published, also identified the only interactions with non-Paf1C components, represented by the smaller, thin lined circles in Fig. 4. The interaction identified between Swc7 and Rtf1 is intriguing, because this protein is linked to deposition of the histone variant H2A.Z into nucleosomes [72], establishing another link between Rtf1 and chromatin remodeling factors. The significance of the four proteins interacting with Ctr9 is difficult to determine at this time. Pho81 and Dal80 are nuclear proteins involved in transcriptional regulation, so these may link the Paf1C to regulation in yeast, as has been demonstrated for flies [26]. Pex14 and Lap4 are peroxisomal and vacuolar, respectively; their possible connection with nuclear Ctr9 is not clear. One additional two-hybrid interaction has been reported between the fly Cdc73 homologue Hyrax and β-catenin [26]; dCdc73 also interacts with the Gli transcriptional activators [27].
Interactions with Cleavage and Poly(A) Factors
As stated above, co-immunoprecipitation of the many factors associated with pol II does not necessarily establish direct physical interactions. However, by detaching the Paf1C from pol II using mutation of either Cdc73 or Rtf1 we were able to detect a direct interaction between the Paf1C and cleavage and poly(A) factor Cft1, equivalent to human CPSF-160 [22]. As described above, Rozenblatt-Rosen et al. found the components of both the human CPSF and CstF cleavage and poly(A) complexes co-purifying with the hPaf1C [24]. Like the interactions between Cft1 and the yeast Paf1C, the hPaf1-CPSF-CstF interactions are not mediated by RNA [22, 24]. In addition, hCdc73 and FACT component hSpt16 were identified in a global analysis of factors found in the purified human pre-mRNA 3' end-processing complex [110]. In Fig. 4 the connection between CPF and CF1A (the yeast homologues of CPSF and CstF [40]) is depicted as through Cdc73. Despite finding only hCdc73 in the human 3' end formation complex, this is undoubtedly an oversimplification because in the absence of Cdc73, Cft1 still associates with the remaining Paf1C [22]. Determining the actual points of contact between the Paf1C and the cleavage and poly(A) factors, as well as the other proteins described above, may help to further elucidate the function of the Paf1C in the transcription cycle.
WHAT CAUSES THE PAF1C TO EXIT POL II AT THE POLY(A) SITE?
As schematized in Fig. 2, the Paf1C is no longer associated with pol II downstream of the poly(A) site, coinciding with increased association between pol II and the cleavage and poly(A) factors [53]. Because there are probably multiple points of contact between the Paf1C and pol II it may be that breaking any of these contacts triggers dissociation. Although Spt4/5 remain associated with pol II beyond the poly(A) site, Bur1 levels drop, providing an explanation for the departure of the Paf1C, because the phosphorylated form of Spt5 is very labile [53, 63, 111]. What triggers this transition is not yet clear. Cleavage of the transcript at this point may play a role, and although Paf1C association with pol II is not RNA dependent [22], it may be that Bur1 is sensitive to the state of the nascent transcript. The poly(A) site itself is clearly important based on work from the Winston lab. When the poly(A) site is mutated, the Paf1C component Ctr9 persists with pol II into the intergenic region [54]. It could be that factors known to recognize the poly(A) site [40] are required for disrupting connections to Bur1, resulting in the loss of the Paf1C. Or perhaps the contacts between the cleavage and poly(A) factors and the Paf1C described above result in a handoff of the Paf1C at the poly(A) site.
Ctk1, which behaves much like the Paf1C in this transition [53], is also found in the cytoplasm playing a role in translation [112]. It is therefore interesting to ask if the Paf1C also has a cytoplasmic role. As described above (Fig. 4), Ctr9 interacts in two-hybrid constructs with cytoplasmic proteins [109]. In addition, detaching the Paf1C from pol II allows it to appear in the nucleolus, which may be part of the mRNP export pathway [105]. Another possible link to cytoplasmic mRNP transactions is the presence of hSki8, found in both the nucleus and the cytoplasm, in the hPaf1C [19]. It will be important to determine both the triggers for release of the Paf1C from pol II and its ultimate fate after it has left the transcription complex.
CONCLUSIONS AND UNANSWERED QUESTIONS
It is my hope that this review has further established the Paf1C as a major player in pol II transcription. In addition to its function in recruiting histone modification and mRNA 3' end formation factors, the Paf1C is also important for facilitating transcription elongation through nucleosomal templates. Although the Paf1Cs from different organisms have intriguing differences in composition and possibly function, the overall conservation of the complex from yeast to humans is striking. Of course, many questions about the structure and function of the Paf1C remain. Some that I find of particular interest are:
Are the hPaf1C-linked effects on gene expression leading to cancer due to alterations in histone modifications, effects on elongation, changes in utilization of 3' end formation sites, or to as yet uncharacterized defects in another stage of transcription?
How many contact points are involved in the recruitment of the Paf1C to pol II and are they the same or different in different organisms?
What are the transitions at the poly(A) site that lead to dissociation of Bur1 and the Paf1C?
Leo1 and Ctr9 have not been as extensively studied as Paf1, Cdc73 and Rtf1, but the human homologues are linked to cancerous transformations [33]. What are these conserved factors doing, and how does loss of Leo1 suppress the loss of Rtf1 and Paf1 [9]?
What is the internal structure of the complex? Is it similar between yeast and humans?
Yeast Ski8 has not been found in the Paf1C, but it does have genetic interactions with Cdc73 [113] and a physical interaction with a pol II subunit [114]. Is it, like its human counterpart connected with the Paf1C?
How does loss of the Paf1C reduce Ser2-P levels? Does loss of the hPaf1C also reduce Ser2-P levels?
Why are most of the transcripts found to be sensitive to loss of Paf1 from essential genes [105]? Does this define a class of 3 'ends, a new regulatory pathway?
Is the major role of the Paf1C in elongation or 3' end formation? Are these roles different in different organisms?
Answering these questions will surely raise others, but their resolution will help to clarify the role or roles the Paf1C plays in eukaryotic transcription and to further establish its importance in the expression of genes.
Acknowledgements
I would like to thank P. Megee, J. Betz, K. Arndt and T. Blumenthal for discussions that helped to shape the points made in this review and for their comments on the manuscript. I also thank R. Roeder, J. Kim, M. Meyerson and O. Rozenblatt-Rosen for communicating results prior to publication. I am grateful to P. Megee and R. Garcea for providing quiet places to work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wade PA, Werel W, Fentzke RC, Thompson NE, Leykam JF, Burgess RR, Jaehning JA, Burton ZF. A novel collection of accessory factors associated with yeast RNA polymerase II. Protein Expr. Purif. 1996;8:85–90. doi: 10.1006/prep.1996.0077. [DOI] [PubMed] [Google Scholar]
- 2.Burton ZF, Killeen M, Sopta M, Ortolan LG, Greenblatt J. RAP30/74: a general initiation factor that binds to RNA polymerase II. Mol. Cell. Biol. 1988;8:1602–1613. doi: 10.1128/mcb.8.4.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 4.Koleske AJ, Young RA. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 5.Shi X, Finkelstein A, Wolf AJ, Wade PA, Burton ZF, Jaehning JA. Paf1p, an RNA polymerase II-associated factor in Saccharomyces cerevisiae, may have both positive and negative roles in transcription. Mol. Cell. Biol. 1996;16:669–676. doi: 10.1128/mcb.16.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi X, Chang M, Wolf AJ, Chang CH, Frazer-Abel AA, Wade PA, Burton ZF, Jaehning JA. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol. Cell. Biol. 1997;17:1160–1169. doi: 10.1128/mcb.17.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang M, French-Cornay D, Fan HY, Klein H, Denis CL, Jaehning JA. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol. Cell. Biol. 1999;19:1056–1067. doi: 10.1128/mcb.19.2.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porter SE, Washburn TM, Chang M, Jaehning JA. Theyeast pafl-RNA polymerase II complex is required for full expression of a subset of cell cycle-regulated genes Eukaryot. Cell. 2002;1:830–884. doi: 10.1128/EC.1.5.830-842.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mueller CL, Jaehning JA. Ctr9, Rtf1, and Leo1 Are Components of the Paf1/RNA Polymerase II Complex. Mol.Cell. Biol. 2002;22:1971–1980. doi: 10.1128/MCB.22.7.1971-1980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Squazzo SL, Costa PJ, Lindstrom DL, Kumer KE, Simic R, Jennings JL, Link AJ, Arndt KM, Hartzog GA. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 2002;21:1764–1774. doi: 10.1093/emboj/21.7.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stolinski LA, Eisenmann DM, Arndt KM. Identification of RTF1, a novel gene important for TATA site selection by TATA box-binding protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:4490–4500. doi: 10.1128/mcb.17.8.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arndt KM, Kane CM. Running with RNA polymerase: eukaryotic transcript elongation. Trends Genet. 2003;19:543–550. doi: 10.1016/j.tig.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 14.Costa PJ, Arndt KM. Synthetic lethal interactions suggest a role for the Saccharomyces cerevisiae Rtf1 protein in transcription elongation. Genetics. 2000;156:535–547. doi: 10.1093/genetics/156.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pokholok DK, Hannett NM, Young RA. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell. 2002;9:799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- 16.Carpten JD, Robbins CM, Villablanca A, Forsberg L, Presciuttini S, Bailey-Wilson J, Simonds WF, Gillanders EM, Kennedy AM, Chen JD, Agarwal SK, Sood R, Jones MP, Moses TY, Haven C, Petillo D, Leotlela PD, Harding B, Cameron D, Pannett AA, Hoog A, Heath H, 3rd, James-Newton LA, Robinson B, Zarbo RJ, Cavaco BM, Wassif W, Perrier ND, Rosen IB, Kristoffersson U, Turnpenny PD, Farnebo LO, Besser GM, Jackson CE, Morreau H, Trent JM, Thakker RV, Marx SJ, Teh BT, Larsson C, Hobbs MR. HRPT2, encoding parafibromin, is mutated in hyperparathyroidism-jaw tumor syndrome. Nat. Genet. 2002;32:676–680. doi: 10.1038/ng1048. [DOI] [PubMed] [Google Scholar]
- 17.Rozenblatt-Rosen O, Hughes CM, Nannepaga SJ, Shanmugam KS, Copeland TD, Guszczynski T, Resau JH, Meyerson M. The parafibromin tumor suppressor protein is part of a human Paf1 complex. Mol. Cell. Biol. 2005;25:612–620. doi: 10.1128/MCB.25.2.612-620.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yart A, Gstaiger M, Wirbelauer C, Pecnik M, Anastasiou D, Hess D, Krek W. The HRPT2 tumor suppressor gene product parafibromin associates with human PAF1 and RNA polymerase II. Mol. Cell. Biol. 2005;25:5052–5060. doi: 10.1128/MCB.25.12.5052-5060.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu B, Mandal SS, Pham AD, Zheng Y, Erdjument-Bromage H, Batra SK, Tempst P, Reinberg D. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 2005;19:1668–1673. doi: 10.1101/gad.1292105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J, Guermah M, Roeder RG. The Human Paf Complex Acts in Chromatin Transcription Elongation Both Independently and Cooperatively with SII/TFIIS. Cell. 2010 doi: 10.1016/j.cell.2009.12.050. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adelman K, Wei W, Ardehali MB, Werner J, Zhu B, Reinberg D, Lis JT. Drosophila Paf1 modulates chromatin structure at actively transcribed genes. Mol.Cell. Biol. 2006;26:250–260. doi: 10.1128/MCB.26.1.250-260.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nordick K, Hoffman MG, Betz JL, Jaehning JA. Direct interactions between the Paf1 complex and a cleavage and polyadenylation factor are revealed by dissociation of Paf1 from RNA polymerase II. Eukaryot. Cell. 2008;7:1158–1167. doi: 10.1128/EC.00434-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, de Vries I, Kittler R, Junqueira M, Shevchenko A, Schulz H, Hubner N, Doss MX, Sachinidis A, Hescheler J, Iacone R, Anastassiadis K, Stewart AF, Pisabarro MT, Caldarelli A, Poser I, Theis M, Buchholz F. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell stem cell. 2009;4:403–415. doi: 10.1016/j.stem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 24.Rozenblatt-Rosen O, Nagaike T, Francis JM, Kaneko S, Glatt KA, Hughes CM, LaFramboise T, Manley JL, Meyerson M. The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3' mRNA processing factors. Proc. Natl. Acad. Sci. U.S.A. 2009;106:755–760. doi: 10.1073/pnas.0812023106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y, Yamaguchi Y, Tsugeno Y, Yamamoto J, Yamada T, Nakamura M, Hisatake K, Handa H. DSIF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation. Genes Dev. 2009;23:2765–2777. doi: 10.1101/gad.1834709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mosimann C, Hausmann G, Basler K. Parafibromin/Hyrax activates Wnt/Wg target gene transcription by direct association with beta-catenin/Armadillo. Cell. 2006;125:327–341. doi: 10.1016/j.cell.2006.01.053. [DOI] [PubMed] [Google Scholar]
- 27.Mosimann C, Hausmann G, Basler K. Therole of Parafibromin/Hyrax as a nuclear Gli/Ci-interacting protein in Hedgehog target gene control. Mech. Dev. 2009;126:394–405. doi: 10.1016/j.mod.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 28.Youn MY, Yoo HS, Kim MJ, Hwang SY, Choi YW, Desiderio SV, Yoo JY. hCTR9, a component of PAF1 complex, participates in the transcription of IL-6 responsive genes through regulation of STAT3-DNA interactions. J. Biol. Chem. 2007;282 doi: 10.1074/jbc.M705411200. 34727-34724. [DOI] [PubMed] [Google Scholar]
- 29.Lin L, Zhang JH, Panicker LM, Simonds WF. The parafibromin tumor suppressor protein inhibits cell proliferation by repression of the c-myc proto-oncogene. Proc. Natl. Acad. Sci. U.S.A. 2008;105:17420–17425. doi: 10.1073/pnas.0710725105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mozdy AD, Podell ER, Cech TR. Multiple yeast genes, including Paf1 complex genes, affect telomere length via telomerase RNA abundance. Mol. Cell. Biol. 2008;28:4152–4161. doi: 10.1128/MCB.00512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newey PJ, Bowl MR, Thakker RV. Parafibromin--functional insights. J. Intern. Med. 2009;266:84–98. doi: 10.1111/j.1365-2796.2009.02107.x. [DOI] [PubMed] [Google Scholar]
- 32.Woodard GE, Lin L, Zhang JH, Agarwal SK, Marx SJ, Simonds WF. Parafibromin, product of the hyperparathyroidism-jaw tumor syndrome gene HRPT2, regulates cyclin D1/PRAD1 expression. Oncogene. 2005;24:1272–1276. doi: 10.1038/sj.onc.1208274. [DOI] [PubMed] [Google Scholar]
- 33.Chaudhary K, Deb S, Moniaux N, Ponnusamy MP, Batra SK. Human RNA polymerase II-associated factor complex: dysregulation in cancer. Oncogene. 2007;26:7499–7507. doi: 10.1038/sj.onc.1210582. [DOI] [PubMed] [Google Scholar]
- 34.Moniaux N, Nemos C, Schmied BM, Chauhan SC, Deb S, Morikane K, Choudhury A, Vanlith M, Sutherlin M, Sikela JM, Hollingsworth MA, Batra SK. The human homologue of the RNA polymerase II-associated factor 1 (hPaf1), localized on the 19q13 amplicon, is associated with tumorigenesis. Oncogene. 2006;25:3247–3257. doi: 10.1038/sj.onc.1209353. [DOI] [PubMed] [Google Scholar]
- 35.Wang P, Bowl MR, Bender S, Peng J, Farber L, Chen J, Ali A, Zhang Z, Alberts AS, Thakker RV, Shilatifard A, Williams BO, Teh BT. Parafibromin a component of the human PAF complex, regulates growth factors and is required for embryonic and development survival in adult mice. Mol. Cell. Biol. 2008;28:2930–2940. doi: 10.1128/MCB.00654-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moniaux N, Nemos C, Deb S, Zhu B, Dornreiter I, Hollingsworth MA, Batra SK. The human RNA polymerase II-associated factor 1 (hPaf1): a new regulator of cell-cycle progression. PloS one. 2009;4:e7077. doi: 10.1371/journal.pone.0007077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krishnamurthy S, Hampsey M. Eukaryotic transcription initiation. Curr. Biol. 2009;19:R153–R156. doi: 10.1016/j.cub.2008.11.052. [DOI] [PubMed] [Google Scholar]
- 38.Sikorski TW, Buratowski S. The basal initiation machinery: beyond the general transcription factors. Curr. Opin. Cell Biol. 2009;21:344–351. doi: 10.1016/j.ceb.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shilatifard A. Transcriptional elongation control by RNA polymerase II: a new frontier. Biochim. Biophys. Acta. 2004;1677:79–86. doi: 10.1016/j.bbaexp.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 40.Proudfoot N. New perspectives on connecting messenger RNA 3' end formation to transcription. Curr. Opin. Cell. Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 41.Rosonina E, Kaneko S, Manley JL. Terminating the transcript: breaking up is hard to do. Genes Dev. 2006;20:1050–1056. doi: 10.1101/gad.1431606. [DOI] [PubMed] [Google Scholar]
- 42.Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes Dev. 2009;23:1247–1269. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 44.Hirose Y, Ohkuma Y. Phosphorylation of the C-terminal domain of RNA polymerase II plays central roles in the integrated events of eucaryotic gene expression. J. Biochem. 2007;141:601–608. doi: 10.1093/jb/mvm090. [DOI] [PubMed] [Google Scholar]
- 45.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol. Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akhtar MS, Heidemann M, Tietjen JR, Zhang DW, Chapman RD, Eick D, Ansari AZ. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol. Cell. 2009;34:387–393. doi: 10.1016/j.molcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanin EI, Kipp RT, Kung C, Slattery M, Viale A, Hahn S, Shokat KM, Ansari AZ. Chemical inhibition of the TFIIH-associated kinase Cdk7/Kin28 does not impair global mRNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 2007;104:5812–5817. doi: 10.1073/pnas.0611505104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hong SW, Hong SM, Yoo JW, Lee YC, Kim S, Lis JT, Lee DK. Phosphorylation of the RNA polymerase II C-terminal domain by TFIIH kinase is not essential for transcription of Saccharomyces cerevisiae genome. Proc. Natl. Acad. Sci. U.S.A. 2009;106:14276–14280. doi: 10.1073/pnas.0903642106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wood A, Shilatifard A. Bur1/Bur2 and the Ctk complex in yeast: the split personality of mammalian P-TEFb. Cell Cycle. 2006:1066–1068. doi: 10.4161/cc.5.10.2769. [DOI] [PubMed] [Google Scholar]
- 50.Qiu H, Hu C, Hinnebusch AG. Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol. Cell. 2009;33:752–762. doi: 10.1016/j.molcel.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ahn SH, Keogh MC, Buratowski S. Ctk1 promotes dissociation of basal transcription factors from elongating RNA polymerase II. Embo J. 2009;28:205–212. doi: 10.1038/emboj.2008.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mueller CL, Porter SE, Hoffman MG, Jaehning JA. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol. Cell. 2004;14:447–456. doi: 10.1016/s1097-2765(04)00257-6. [DOI] [PubMed] [Google Scholar]
- 53.Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S. Transitions in RNA polymerase II elongation complexes at the 3' ends of genes. EMBO J. 2004;23:354–364. doi: 10.1038/sj.emboj.7600053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaplan CD, Holland MJ, Winston F. Interaction between transcription elongation factors and mRNA 3'-end formation at the Saccharomyces cerevisiae GAL10-GAL7 locus. J. Biol. Chem. 2005;280:913–922. doi: 10.1074/jbc.M411108200. [DOI] [PubMed] [Google Scholar]
- 55.Qiu H, Hu C, Wong CM, Hinnebusch AG. The Spt4p subunit of yeast DSIF stimulates association of the Paf1 complex with elongating RNA polymerase II. Mol. Cell. Biol. 2006;26:3135–3148. doi: 10.1128/MCB.26.8.3135-3148.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O'Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ. Gene loops juxtapose promoters and terminators in yeast. Nat. Genet. 2004;36:1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- 57.Ansari A, Hampsey M. A role for the CPF 3'-end processing machinery in RNAP II-dependent gene looping. Genes Dev. 2005;19:2969–2978. doi: 10.1101/gad.1362305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh BN, Hampsey M. A Transcription-Independent Role for TFIIB in Gene Looping. Mol. Cell. Biol. 2007;27:806–816. doi: 10.1016/j.molcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 59.Laine JP, Singh BN, Krishnamurthy S, Hampsey M. A physiological role for gene loops in yeast. Genes Dev. 2009;23:2604–2609. doi: 10.1101/gad.1823609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 2009;23:2610–2624. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gerber M, Shilatifard A. Transcriptional elongation by RNA polymerase II and histone methylation. J. Biol. Chem. 2003;278:26303–26306. doi: 10.1074/jbc.R300014200. [DOI] [PubMed] [Google Scholar]
- 62.Fuchs SM, Laribee RN, Strahl BD. Protein modifications in transcription elongation. Biochim. Biophys. Acta. 2009;1789:26–36. doi: 10.1016/j.bbagrm.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Y, Warfield L, Zhang C, Luo J, Allen J, Lang WH, Ranish J, Shokat KM, Hahn S. Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol. Cell. Biol. 2009;29:4852–4863. doi: 10.1128/MCB.00609-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jones JC, Phatnani HP, Haystead TA, MacDonald JA, Alam SM, Greenleaf AL. C-terminal repeat domain kinase I phosphorylates Ser2 and Ser5 of RNA polymerase II C-terminal domain repeats. J. Biol. Chem. 2004;279:24957–24964. doi: 10.1074/jbc.M402218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Phatnani HP, Jones JC, Greenleaf AL. Expanding the functional repertoire of CTD kinase I and RNA polymerase II: novel phosphoCTD-associating proteins in the yeast proteome. Biochemistry. 2004;43:15702–15719. doi: 10.1021/bi048364h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3' end processing. Mol. Cell. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- 67.Laribee RN, Krogan NJ, Xiao T, Shibata Y, Hughes TR, Greenblatt JF, Strahl BD. BUR kinase selectively regulates H3 K4 trimethylation and H2B ubiquitylation through recruitment of the PAF elongation complex. Curr. Biol. 2005;15:1487–1493. doi: 10.1016/j.cub.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 68.Chu Y, Simic R, Warner MH, Arndt KM, Prelich G. Regulation of histone modification and cryptic transcription by the Bur1 and Paf1 complexes. Embo J. 2007;26:4646–4656. doi: 10.1038/sj.emboj.7601887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou K, Kuo WH, Fillingham J, Greenblatt JF. Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc. Natl. Acad. Sci. U.S.A. 2009;106:6956–6961. doi: 10.1073/pnas.0806302106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 71.Mulder KW, Brenkman AB, Inagaki A, van den Broek NJ, Timmers HT. Regulation of histone H3K4 tri-methylation and PAF complex recruitment by the Ccr4-Not complex. Nucleic Acids Res. 2007;35:2428–2439. doi: 10.1093/nar/gkm175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 73.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr. Opin. Cell Biol. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Z, Schones DE, Zhao K. Characterization of human epigenomes. Curr. Opin. Genet. Dev. 2009;19:127–134. doi: 10.1016/j.gde.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim T, Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5' transcribed regions. Cell. 2009;137:259–272. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, Greenblatt JF, Shilatifard A. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 77.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 78.Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 2003;278:34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- 79.Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD. Histone H2B Ubiquitylation Is Associated with Elongating RNA Polymerase II. Mol. Cell. Biol. 2005;25:637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol. Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 81.Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, Strahl BD. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 2003;17:654–663. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, Shilatifard A, Buratowski S, Greenblatt J. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell. Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ng HH, Dole S, Struhl K. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 2003;278:33625–33628. doi: 10.1074/jbc.C300270200. [DOI] [PubMed] [Google Scholar]
- 86.Warner MH, Roinick KL, Arndt KM. Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol. Cell. Biol. 2007;27:6103–6115. doi: 10.1128/MCB.00772-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tenney K, Gerber M, Ilvarsonn A, Schneider J, Gause M, Dorsett D, Eissenberg JC, Shilatifard A. Drosophila Rtf1 functions in histone methylation, gene expression, and Notch signaling. Proc. Natl. Acad. Sci. U.S.A. 2006;103:11970–11974. doi: 10.1073/pnas.0603620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Oh S, Zhang H, Ludwig P, van Nocker S. A mechanism related to the yeast transcriptional regulator Paf1c is required for expression of the Arabidopsis FLC/MAF MADS box gene family. The Plant Cell. 2004;16:2940–2953. doi: 10.1105/tpc.104.026062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.He Y, Doyle MR, Amasino RM. PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 2004;18:2774–2784. doi: 10.1101/gad.1244504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu L, Zhao Z, Dong A, Soubigou-Taconnat L, Renou JP, Steinmetz A, Shen WH. Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol. Cell. Biol. 2008;28:1348–1360. doi: 10.1128/MCB.01607-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, Eick D, Hughes TR, Krogan NJ, Strahl BD, Berger SL. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex Mol. Cell. 2007;27:275–288. doi: 10.1016/j.molcel.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 92.Betz JL, Chang M, Washburn TM, Porter SE, Mueller CL, Jaehning JA. Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis, and lipid and nucleic acid metabolism. Mol. Genet. Genomics. 2002;268:272–285. doi: 10.1007/s00438-002-0752-8. [DOI] [PubMed] [Google Scholar]
- 93.Ardehali MB, Yao J, Adelman K, Fuda NJ, Petesch SJ, Webb WW, Lis JT. Spt6 enhances the elongation rate of RNA polymerase II in vivo. Embo J. 2009;28:1067–1077. doi: 10.1038/emboj.2009.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rondon AG, Gallardo M, Garcia-Rubio M, Aguilera A. Molecular evidence indicating that the yeast PAF complex is required for transcription elongation. EMBO Rep. 2004;5:47–53. doi: 10.1038/sj.embor.7400045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.West RWJ, Kruger B, Thomas S, Ma J, Milgrom E. RLR1 (THO2), required for expressing lacZ fusions in yeast, is conserved from yeast to humans and is a suppressor of SIN4. Gene. 2000 Feb 8;243:195–205. doi: 10.1016/s0378-1119(99)00510-7. [DOI] [PubMed] [Google Scholar]
- 96.West RW, Milgrom E. DEAD-box RNA helicase Sub2 is required for expression of lacZ fusions in Saccharomyces cerevisiae and is a dosage-dependent suppressor of RLR1 (THO2) Gene. 2002;288:19–27. doi: 10.1016/s0378-1119(02)00482-1. [DOI] [PubMed] [Google Scholar]
- 97.Russnak R, Nehrke KW, Platt T. REF2 encodes an RNA-binding protein directly involved in yeast mRNA 3'-end formation. Mol. Cell. Biol. 1995;15:1689–1697. doi: 10.1128/mcb.15.3.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Penheiter KL, Washburn TM, Porter SE, Hoffman MG, Jaehning JA. A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol. Cell. 2005;20:213–223. doi: 10.1016/j.molcel.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 99.Strawn LA, Lin CA, Tank EM, Osman MM, Simpson SA, True HL. Mutants of the Paf1 complex alter phenotypic expression of the yeast prion [PSI+] Mol. Biol. Cell. 2009;20:2229–2241. doi: 10.1091/mbc.E08-08-0813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang Y, Sikes ML, Beyer AL, Schneider DA. The Paf1 complex is required for efficient transcription elongation by RNA polymerase I. Proc. Natl. Acad. Sci. U.S.A. 2009;106:2153–2158. doi: 10.1073/pnas.0812939106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cheng H, He X, Moore C. The essential WD repeat protein Swd2 has dual functions in RNA polymerase II transcription termination and lysine 4 methylation of histone H3. Mol. Cell. Biol. 2004;24:2932–2943. doi: 10.1128/MCB.24.7.2932-2943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Meinhart A, Cramer P. Recognition of RNA polymerase II carboxy-terminal domain by 3'-RNA-processing factors. Nature. 2004;430:223–226. doi: 10.1038/nature02679. [DOI] [PubMed] [Google Scholar]
- 103.Sheldon KE, Mauger DM, Arndt KM. A Requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3' end formation. Mol. Cell. 2005;20:225–236. doi: 10.1016/j.molcel.2005.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Steinmetz EJ, Conrad NK, Brow DA, Corden JL. RNA-binding protein Nrd1 directs poly(A)-independent 3'-end formation of RNA polymerase II transcripts. Nature. 2001;413:327–331. doi: 10.1038/35095090. [DOI] [PubMed] [Google Scholar]
- 105.Porter SE, Penheiter KL, Jaehning JA. Separation of the Saccharomyces cerevisiae Paf1 Complex from RNA Polymerase II Results in Changes in Its Subnuclear Localization. Eukaryot. Cell. 2005;4:209–220. doi: 10.1128/EC.4.1.209-220.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim J, Roeder RG. Direct Bre1-Paf1 complex interactions and RING finger-independent Bre1-Rad6 interactions mediate H2B Ubiquitylation in yeast. J. Biol. Chem. 2009;284:20582–20592. doi: 10.1074/jbc.M109.017442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, Johnson AD, Hartzog GA, Arndt KM. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 2003;22:1846–1856. doi: 10.1093/emboj/cdg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Alen C, Kent NA, Jones HS, O'Sullivan J, Aranda A, Proudfoot NJ. A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol. Cell. 2002;10:1441–1452. doi: 10.1016/s1097-2765(02)00778-5. [DOI] [PubMed] [Google Scholar]
- 109.Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, Hao T, Rual JF, Dricot A, Vazquez A, Murray RR, Simon C, Tardivo L, Tam S, Svrzikapa N, Fan C, de Smet AS, Motyl A, Hudson ME, Park J, Xin X, Cusick ME, Moore T, Boone C, Snyder M, Roth FP, Barabasi AL, Tavernier J, Hill DE, Vidal M. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, 3rd, Frank J, Manley JL. Molecular architecture of the human pre-mRNA 3' processing complex. Mol. Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Keogh MC, Podolny V, Buratowski S. Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol. Cell. Biol. 2003;23:7005–7018. doi: 10.1128/MCB.23.19.7005-7018.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rother S, Strasser K. The RNA polymerase II CTD kinase Ctk1 functions in translation elongation. Genes Dev. 2007;21:1409–1421. doi: 10.1101/gad.428407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wilmes GM, Bergkessel M, Bandyopadhyay S, Shales M, Braberg H, Cagney G, Collins SR, Whitworth GB, Kress TL, Weissman JS, Ideker T, Guthrie C, Krogan NJ. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol. Cell. 2008;32:735–746. doi: 10.1016/j.molcel.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tarassov K, Messier V, Landry CR, Radinovic S, Serna Molina MM, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]