Abstract
Probe-capture systems based on proteins and synthetic ligands have become important for new analytical and imaging applications. We have used kinetic measurements of luminescence and measurements of binding by isothermal calorimetry to determine essential rate and equilibrium constants for a system that permanently captures modified DOTA chelates for positron imaging. We used that information along with previous results to quantitatively characterize the behavior of this system in vitro and in vivo. Under physiological conditions at 37 °C, the equilibrium dissociation constant for yttrium S-2-(4-aminobenzyl)-1, 4, 7, 10-tetraazacyclododecanetetraacetate from antibody 2D12.5 is 2.0(±0.4) ×10−9 M and the dissociation rate constant is 7.0(±0.7) ×10−3 s−1, leading to an inferred association rate constant of 3.5 × 106 M−1s−1. Using these values to interpret data from earlier experiments leads to the rate constant 2.5 × 10−2 s−1 for covalent attachment of bound yttrium S-2-(4-acrylamidobenzyl)-1, 4, 7, 10-tetraazacyclododecanetetraacetate to the G54C mutant of antibody 2D12.5. These values lead to a model for the detailed behavior of the latter system for tumor imaging in vivo that is consistent with experimental observations.
Introduction
The capture of probe molecules by receptors has an established and increasing role in the biomedical applications of bioconjugate chemistry. Biotinylated probes bind to avidin or streptavidin (1); hexahistidine tags bind to NTA1 chelates (2); antibodies bind to haptens (3); and a number of engineered proteins bind to synthetic tags (4). These systems display a variety of properties, which dictate their applicability to specific problems. Their quantitative rates and equilibria of binding – and, if applicable, attachment – are fundamentally important but not always known. Here we describe measurements and interpretation of a set of values for the engineered antibody binding site 2D12.5 G54C, which captures labeled acrylamidobenzyl- DOTA (AABD) metal chelates with infinite affinity. This system is highly promising for pretargeted imaging and therapy applications in vivo, and has recently been demonstrated to have potential as a reporter gene (5).
Experimental Procedures
Reagents
DOTA and aminobenzyl-DOTA (ABD) were purchased from Macrocyclics (Dallas, TX). TEA and plastic backed thin layer silica (TLC) plates were purchased from EMD Chemicals. Rare earth salts were purchased from Sigma Aldrich (St Louis, MO). Fetal bovine serum and G418 selection antibiotic were also purchased from Sigma Aldrich. Biotechnical grade glycine, DEAE-cellulose, lithium dodecyl sulfate, and glycerol were purchased from Fisher Scientific (Waltham, MA). DTPA and EDTA were purchased from Acros Organics (Geel, Belgium). Microtiter plates were purchased from Greiner Bio-One (Monroe, NC). DMEM, PBS Based enzyme-free cell dissociation buffer, and 4–20% Tris-glycine Novex gels were purchased from Invitrogen (Carlsbad, CA). PVDF membrane was purchased from Millipore (Billerica, MA) and SuperSignal® West Dura Extended Duration substrate was purchased from Thermo Scientific (Waltham, MA). All buffers were made using 18 mΩ-cm water and thoroughly degassed for all experiments. Rabbit anti-human Fc fragment antibody was purchased from Jackson Immuno Research (West Grove, PA) while the horseradish peroxidase conjugated goat anti-rabbit antibody was purchased from Pierce (Rockford, IL). Tris-HCl was purchased from USB (Cleveland, OH). Bromophenol Blue was purchased from Bio-Rad (Hercules, CA).
Metallation of Chelates
The concentration of stock ABD and DOTA was measured using a Co-57 metal binding assay as previously described (6). Briefly, 200 μM stock YCl3 and Co(NO3)2 were prepared in metal-free 0.05N HCl. Next, 80 mM ABD (0.048 mmol, 600 μL) was chelated with 1.5 equivalent of YCl3 or Co(NO3)2 at 37 °C for 2 hr in triethylamine acetate, pH 6.0. Also, 80 mM DOTA (0.048 mmol, 600 μL) was chelated with 1.5 equivalent TbCl3 using same conditions. The percent chelation was measured using competitive metal-binding assay with Co-57. The unreacted excess metal was scavenged with DTPA (0.2 M, 300 μL). In order to remove excess salts, the metal chelates were passed through an anionic DEAE-cellulose column, eluted in ammonium acetate (gradient: 0.1 M to 1 M) and then lyophilized four times to remove ammonium acetate, being redissolved in water and relyophilized each day. The dry, desalted metal chelate was dissolved in 0.1 M sodium phosphate buffer, pH 7.0. The concentration of the metal chelate was confirmed by measuring its absorbance at 286 nm using ε286 = 1430 M−1cm−1, as determined from titrations of ABD with standard metal solutions.
Protein Preparation
Wild-Type 2D12.5 monoclonal antibody (whole IgG, 50 μM) was dialyzed into the same buffer as the chelates and filtered through a 0.22 micron. The concentration was measured by A280 as 48.3 μM (ε280 = 194,900) and diluted to the appropriate concentration in the sodium phosphate buffer.
Isothermal Titration Calorimetry
This was carried out according to the manufacturer’s instructions, using a VP-ITC titration calorimeter (Microcal Inc., Northampton, MA). All thermodynamic data were analyzed with MicroCal software, Origin 7.0, which performs non- linear least squares curve fitting.
Blank Experiment
ABD(Co), (150 μM) or ABD(Y) (128 μM) in the syringe was titrated in 46 injections into the sample buffer, 0.1M sodium phosphate (pH 7.0). The small heat change produced was used for blank subtraction from the analyte-ligand experiments.
Competitive Binding Experiments
The high affinity between ABD(Y) and 2D12.5 mAb led to results from direct binding experiments that were not sufficiently accurate to yield a reliable value for the equilibrium constant. The method of Velazquez et al. (7) employs a competitive or displacement method using a competing weaker ligand to lower the apparent affinity of the stronger ligand. This approach involves 2 sets of experiments, as described below.
Direct titration of weak competitor with 2D12.5 mAb
ABD(Co) (150 μM, 12 times molar concentration of the antibody) placed in the syringe was titrated in 45 injections into 12.5 μM 2D12.5 mAb (analyte in the cell, 1.45 mL). The volume for each injection was 6 μL. This experiment was carried out at 30 ºC and 37 ºC.
Competitive binding with excess of stronger competitor in the syringe
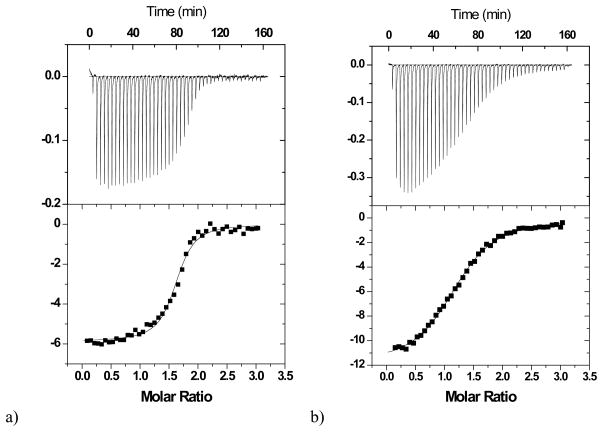
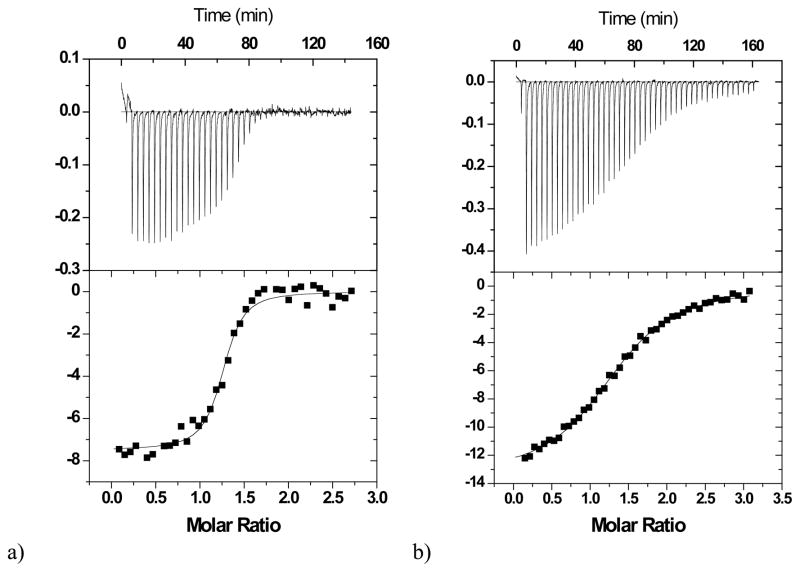
After the first direct titration of the monoclonal antibody 2D12.5 with ABD(Co), the cell was cleaned out and a second titration involving the strong ligand was carried out. 150 μM ABD(Y) in the syringe was titrated in 46 injections against a mixture of 100 μM ABD(Co)-16.1 μM 2D12.5 (analyte in the cell, 1.45 mL). The volume for each injection was 6 μL. Experiments were carried out at 30 ºC and 37 ºC. Titrations are shown in Figure 1 and Figure 2.
Figure 1.
Calorimetric titrations of wild-type 2D12.5 monoclonal antibody (12.5 μM for ABD(Co) or 16.1 μM for competitive titration) with a) the chelate ABD(Y) (150 μM) in the presence of ABD(Co) (100 μM) (competitive titration), b) chelate ABD(Co) (150 μM). The upper vertical axis represents the raw differential power in μcal/sec between the reference and sample cells, while the lower vertical axis represents the normalized kcal/mole of injected ABD chelate. The experiments were performed at 30 °C in 0.1 M sodium phosphate, pH 7.0.
Figure 2.
Calorimetric titrations of wild-type 2D12.5 monoclonal antibody (15.6 μM for ABD(Co) or 12.4 μM for competitive titration) with a) the chelate ABD(Y) (150 μM) in the presence of ABD(Co) (100 μM) (competitive titration), b) chelate ABD(Co) (150 μM). The upper vertical axis represents the raw differential power in μcal/sec between the reference and sample cells, while the lower vertical axis represents the normalized kcal/mole of injected ABD chelate. The experiments were performed at 37 °C in 0.1 M sodium phosphate, pH 7.0.
Time-Resolved Fluorescence
Time-resolved fluorescence measurements were made on a Fluostar® Fluorescence microplate reader (BMG Lab Technologies) using the 280 nm excitation and 550 nm emission filters, each with ±10nm bandpass.
Off-rate determination using 2D12.5 enhanced luminescence of Tb3+
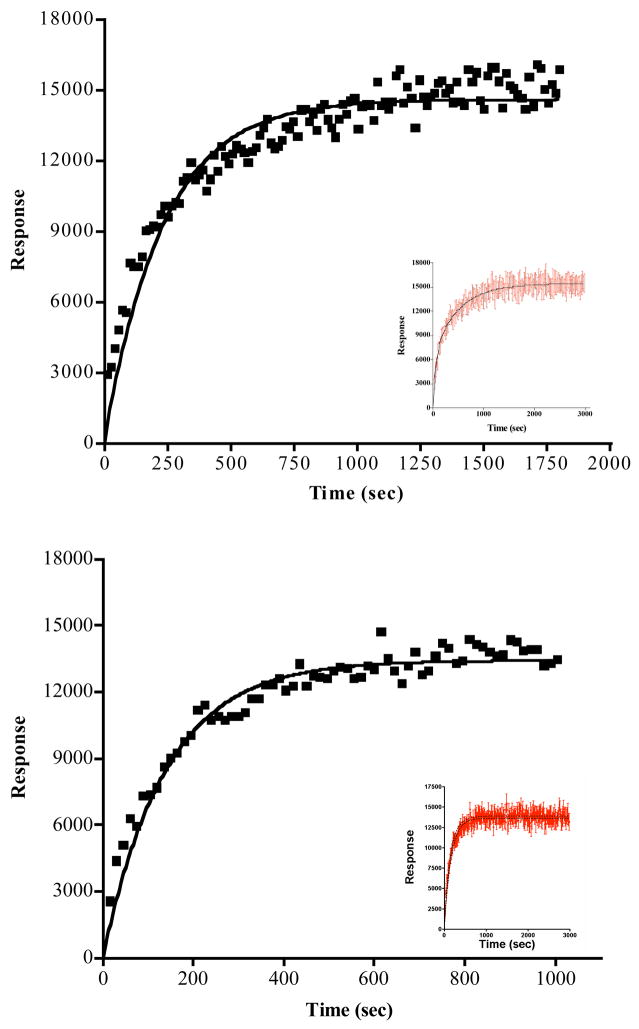
The UV excitation of the 2D12.5-DOTA(Tb) complex at 280 nm leads to energy transfer from aromatic amino-acid side chains in the antibody to the bound DOTA(Tb), producing an enhanced green Tb luminescence at 545 nm (8). We used this phenomenon to measure the dissociation rate of ABD(Y) from the antibody. Monoclonal antibody 2D12.5 (10 μM) was incubated with 100 μM ABD(Y) overnight to ensure that equilibrium association was attained. At the beginning of the luminescence experiment, the DOTA(Tb) complex was quickly added in large excess, so that each ABD(Y) dissociating from the 2D12.5 antibody binding pocket would be replaced immediately by DOTA(Tb). In the experiment, 50 μL of 2 mM DOTA(Tb) was autoinjected into 50 μL of 10 μM 2D12.5+ABD(Y) in TBS (Tris buffered saline, 150mM NaCl), pH 7.4. This was done at 30 °C and also at 37 °C in 96-well black round bottom microplates with 5 sets of controls: 2D12.5, 2D12.5+ABD(Y), 2D12.5+DOTA(Tb), DOTA(Tb) only, and TBS buffer only. In all the wells, total volume was kept at 100 μL and the final concentration of 2D12.5 mAb kept at 5 μM in the appropriate wells. The time-resolved fluorescence was read at 550±10 nm, 40 μs delay, with 200 scans every 15 s. Each reading was blanked with the buffer and DOTA(Tb) readings. Also, increasing concentrations of DOTA(Tb) (2 mM and 4 mM final concentrations) were evaluated in order to assure that sufficient excess was being used. The experiments were carried out in triplicate. As a control, non-specific mouse IgG was incubated with ABD(Y) and analyzed the same way as 2D12.5 mAb. Data are presented in Figure 3.
Figure 3.
Curve fits from the determination of the dissociation rate constant between ABD(Y) and parental 2D12.5 antibody. Upper panel, 30 °C; lower panel 37 °C (note: time scales are different). One, 2 and 4 mM DOTA(Tb) was used to competitively replace the ABD(Y) when it dissociated from the 2D12.5 binding site. Triplicate experiments were done at each DOTA(Tb) concentration, and all the data were combined for the fits and plots. Insets show longer total time courses, which indicate that equilibrium has been reached.
Western blots to evaluate the number of binding sites on DAbR1-transfected U-87 glioma cells
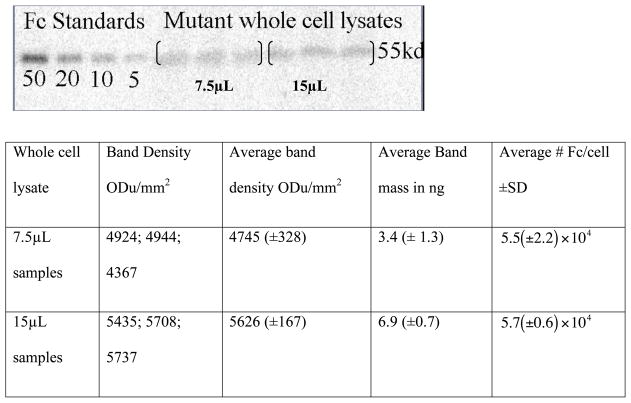
Glioblastoma (U87) cells stably transfected with the DAbR1 gene and the scFv-human Fc minibody standard protein were prepared as described (5). Cells were maintained at 37 °C in a humidified CO2 incubator (5% CO2) in complete Dulbecco’s modified Eagle’s medium containing 10% (v/v) heat inactivated fetal bovine serum and G418 selection antibiotic (250ug/mL). Cells were dissociated for western blots using PBS based enzyme-free cell dissociation buffer and pelleted by centrifugation (1,000 × g for 5 minutes). Cell pellets were re- suspended in phosphate-buffered saline and diluted 1:1 with 5× concentrated SDS PAGE sample application buffer consisting of 125mM Tris-HCl pH 8, 0.5M EDTA, 10% (w/v) lithium dodecyl sulfate, 50% (v/v) glycerol, 0.05% (w/v) bromophenol blue (0.05%w/v) and 0.5% (v/v) β-mercaptoethanol. Re-suspended cells were boiled for 10 min and sonicated in an ice bath in 5-sec bursts until samples were completely soluble (5 to 6 min). Reduced whole cell lysates were loaded into 4–20% Tris-glycine Novex gel and electrophoresed at 150V for 1.5 h. Protein was transferred to a polyvinylidene difluoride membrane and blocked overnight at 4 ºC in Tris- buffered saline, pH 7.4, with 5% nonfat milk. The membranes were probed for 2 h with rabbit anti-human Fc antibody (1:5,000) in TBST. The membranes were washed 6× with 50 mL of TBST and probed for 2 h with a horseradish peroxidase conjugated goat anti-rabbit antibody (1:5,000). Blots were washed 6× with 50 mL of TBST and were treated with SuperSignal® West Dura Extended Duration substrate according to manufacturer instructions and imaged with a Typhoon™ Trio variable mode imaging system (GE Healthcare Life Sciences) using the chemiluminescent setting. The band density of triplicate whole cell lysate samples were measured and compared with a human IgG Fc standard curve using Bio-Rad Quantity One® software. Results are shown in Figure 4.
Figure 4.
Western blot of reduced whole cell lysate vs. human Fc standard. Calculations were performed for 4.4 × 104cell/μL and a formula weight of 110 kDa for the protein, using the equation y= 0.004x − 15.63 obtained by fitting the standards. There are 2 binding sites per 110 kDa protein, implying ≈105 binding sites per cell.
Mathematical modeling of kinetic behavior
The systems of differential equations 1–4 and 5–9 (see below) were solved numerically, and Figure 5 – Figure 8 were prepared, using Mathematica® 7 software.
Figure 5.
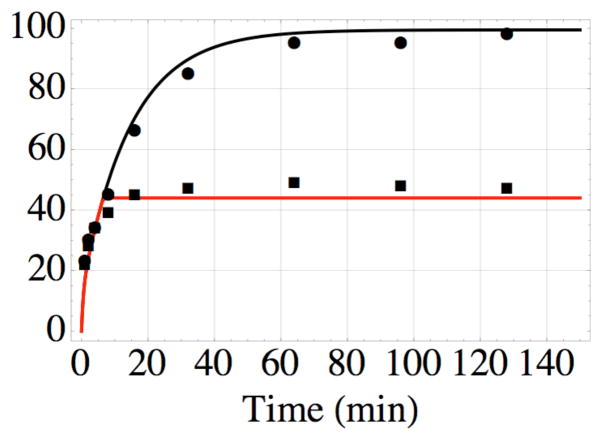
Formation of the covalent bond between AABD(Y) and the G54C mutant of DOTA- binding Fab 2D12.5 in vitro at 37 °C, pH 7.5, as a function of time; nonlinear least-squares fit of data from reference 11 using equations 1–4. The top data (●) are for bond formation in the absence of competitor, while the bottom data (■) are the result of adding excess unlabeled competitor after 6 min. The values kon = 3.5× 106 M−1s−1 and koff = 7.0× 10−3 s−1 determined in this work lead to the nonlinear least-squares value kirr = 2.5 × 10−2 s−1 for formation of the covalent bond, and probability x = 0.2 of binding in a reactive conformation, all of which were used to construct the black curve. The red curve was drawn using the same values, with the added constraint of no association or reassociation after 6 min.
Figure 8.
Predicted speciation of the probe in the tumor at two positions, as a function of time. Top: near the capillary surface (12μ from the center of the coordinate system, for which the capillary radius is 10μ). Bottom: near the outer periphery of the tumor cord (distance 110μ from the center of the coordinate system, 100μ from the edge of the capillary). The curves represent product P (black), non-reactive complex CNR (red), and, practically superimposed, the reactive complex CR (blue) and free probe AABD(Y) (green). The product P dominates the concentrations of the other species after about 20 min.
Results and Discussion
In what follows, we build up a simple mathematical description of the behavior of this probe- capture system that is consistent with all the data available. This begins with calorimetric measurements of the equilibrium thermodynamics of reversible binding of aminobenzyl- DOTA(Y) to mouse 2D12.5 IgG. We turned to calorimetry after surface plasmon resonance experiments failed to give reliable results; this appeared to result primarily from the small size of the DOTA chelate, which led to a lack of sensitivity to binding. Alternative surface plasmon resonance strategies were not successful in our hands. On the other hand, calorimetry generally is suitable for small molecules as well as macromolecules.
Thermodynamic Parameters obtained from ITC
Using isothermal titration calorimetry, we measured the binding affinity and other thermodynamic parameters involved in the reversible interaction of ABD(Y) and parental 2D12.5 monoclonal antibody. From previous reports (9), we expected that the KD should be on the order of 10−8 M, indicating that the binding interaction is strong. In the direct titration of ABD(Y) with 2D12.5, the binding curve was difficult to fit because of high binding affinity. However, as shown by Velazquez et al. (7), the apparent affinity of a strong ligand can be lowered by the presence of a weaker ligand so that a more accurate binding affinity can be measured for the strong ligand. Sigurskjold (10) provides a complete mathematical analysis of the displacement or competitive isothermal titration calorimetric technique. These experiments were carried out at 30 °C (Figure 1) and 37 ºC (Figure 2); results are summarized in Table 1.
Table 1.
Thermodynamic parameters for the reversible association of aminobenzyl-DOTA chelates to Antibody 2D12.5 at 30 °C and 37 °C. (Standard deviations in parentheses.)a
| Chelate | T, °C | KA, M−1 | ΔG, kcal/mol | ΔH, kcal/mol | ΔS, cal/mol-K | KD, M |
|---|---|---|---|---|---|---|
| ABD(Co) | 30 | 8.2(±0.9) ×105 | −8.2(±0.1) | −12.2(±0.4) | −13.2(±1.3) | 1.2(±0.1) ×10−6 |
| ABD(Co) | 37 | 7.1(±0.4) ×105 | −8.3(±0.1) | −12.4(±4.8) | −13.3(±4.8) | 1.4(±0.1) ×10−6 |
| ABD(Y) | 30 | 3.3(±2.1) ×108 | −11.7(±0.4) | −18.2(±0.5) | −21.5(±2.9) | 3.1(±2.0) ×10−9 |
| ABD(Y) | 37 | 5.2(±0.9) ×108 | −12.4(±0.2) | −20.1(±1.2) | −24.9(±4.1) | 2.0(±0.4) ×10−9 |
Having the equilibrium constant in hand, we measured the dissociation rate constant koff at 30 °C and 37 °C; we did this indirectly, by measuring the luminescence of DOTA(Tb) as it bound to the antibody when ABD(Y) dissociated, as described below.
Statistical analysis of fluorescence time-resolved data
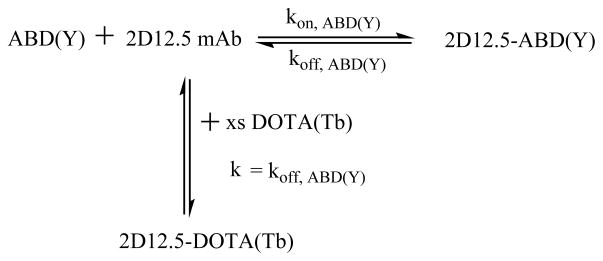
As shown in Scheme 1, the rate of association of the excess DOTA(Tb) with 2D12.5 was taken to equal the rate of dissociation of ABD(Y) from 2D12.5 mAb. The terbium luminescence response at 545 nm for DOTA(Tb) bound to 2D12.5 mAb (8) was monitored as a function of time. To assure that excess DOTA(Tb) was present, three different concentrations were used. Similar results were obtained in all cases, and all three sets of triplicate results were fitted to one curve. The koff was obtained from a fit of the following equation that describes the pseudo-first order association kinetics: Y = Ymax(1−e−kt), where Y is the terbium luminescence intensity and the pseudo-first-order rate constant for binding DOTA(Tb) is k = koff s−1 (with t in sec).
Scheme 1.
Competition experiment to measure koff by forming the luminescent DOTA(Tb) complex with antibody 2D12.5. First the antibody was saturated with ABD(Y), then it was mixed with a large excess of DOTA(Tb).
The dissociation rate constant of ABD(Y) with 2D12.5 antibody was determined from the plot of the fluorescence signal growth of 2D12.5-DOTA(Tb) complex with time (Figure 3). Attempts to fit the data to more than one rate constant gave inferior results. Having measured koff and KD, we can calculate kon = (koff/KD) M−1s−1 (Table 2).
Table 2.
Kinetic parameters for the dissociation of ABD(Y) ligand from Antibody 2D12.5.b
| Experiment | koff, s−1 | 37°C Half-life, s | Calc kon, M−1s−1 | koff, s−1 | 30°C Half-life, s | Calc kon, M−1s−1 |
|---|---|---|---|---|---|---|
| Avg(SD) | 7.0(±0.7) ×10−3 | 97±9 | 3.5 × 106 | 4.0(±0.3) ×10−3 | 161±13 | 1.3 ×106 |
Data from luminescence experiments in Figure 3.
With these results we are now able to model the behavior of this infinite affinity system, explaining previous observations in vitro and in vivo. Corneillie et al. (11) studied the rate of formation of the covalent bond between the acryloyl group of AABD(Y) and the cysteine of the G54C mutant of antibody 2D12.5. As shown in Figure 5, we were able to fit Corneillie’s results with the following set of kinetic equations:
| (1) |
| (2) |
| (3) |
| (4) |
In order to fit the complex kinetic behavior observed in reference 11, it is necessary to assume that AABD(Y) can form either a reactive complex CR (which can form the product P) or a non- reactive complex CNR with the G54C antibody binding site. This is consistent with the observation of two modes of binding in crystal structures of the parental antibody complex (12). The fit in Figure 5 gives the value kirr =1.5 min−1 = 2.5 × 10−2 s−1 derived using the nonlinear least-squares method. The partitioning of complex formation between the non-reactive CNR, with probability 1 − x = 0.8, and the reactive CR, with probability x = 0.2, is also necessary to fit these data.
It is expected that the different binding modes should have different values for kon and koff. However, the values of kon and koff are not sufficiently different to be measured separately by the methods available to us. The crystal structures show two binding modes in which the antibody-DOTA interactions are almost identical but the DOTA side chain is oriented differently (12). Based on the proximity of the DOTA side chain to residue 54 in one structure but not the other, these could plausibly correspond to the reactive and unreactive binding observed kinetically in reference 11. The goodness of fit in Figure 5 justifies the use of this minimal set of rate constants to model the processes of tumor uptake discussed below.
We have found that the 2D12.5 G54C Fab expressed in D. melanogaster S2 cells (11) is largely cysteinylated (has a cysteine monomer attached to Cys-54 by a disulfide bond), which protects the Cys-54 side chain from reaction with electrophiles such as AABD(Y) (13). The experiments in reference 11 were carried out using an excess of radiolabeled AABD(Y) to detect the rate of covalent bond formation by Michael addition; thus the results were sensitive only to protein with a free thiol at G54C, and the value of kirr is not affected by the presence of cysteinylated protein.
Reviewing the values of these new rate constants shows that the second-order association reaction controlled by kon = 3.5 × 106 M−1s−1 is rather rapid compared to the more typical values of kon ≈105 M−1s−1 observed for antibody-protein association (14). However, it is not as fast as the biotin-streptavidin association, for which the rate constant is reported to be kon = 7.5 × 107 M−1s−1 at 25 °C (15). This comparison is important because the streptavidin-biotin system has been widely used for pretargeted delivery of small probes for imaging and therapy of cancer (16). However, streptavidin is immunogenic in human patients (17), so the irreversible antibody capture system may provide a desirable alternative: currently, making antibodies minimally immunogenic is less challenging than doing the same for streptavidin (5).
The rate constant for reversible dissociation of nonelectrophilic ABD(Y) from parental antibody 2D12.5, koff = (7.0 ± 0.7) × 10−3 s−1, is quite fast compared to that for dissociation of biotin from streptavidin, which is reported to be koff = (4.1± 0.3) × 10−5 s−1 at 37 °C under comparable conditions (18). This rapid dissociation, combined with the effects of irreversible binding and the branching between reactive and non-reactive complexes, makes the behavior of the reactive AABD(Y) with the G54C binding site quite unusual. The results above imply that in a homogeneous solution approximately of the AABD(Y) molecules will form permanent bonds with the receptor site the first time they bind, without ever dissociating, while the majority (≈ 80%) will dissociate, diffuse, and rebind. Because the results in vivo will depend on additional factors such as effective target site concentration on cell surfaces and transport behavior of the probe in a tumor, it was important to observe this system in an animal model to see if it would exhibit biological properties useful for imaging and perhaps therapy.
Wei et al. (5) prepared an engineered version of the 2D12.5 G54C antibody with infinite affinity, termed DAbR1, in which the binding site was expressed as part of an scFv-Fc fusion protein on the surface of U-87 human glioma cells implanted in the flanks of SCID mice. The behavior of AABD(86Y) after injection into the tail vein was monitored by microPET imaging, with highly successful results; some experimental data from reference 5 are used in Figure 6. Having determined values for kon, koff, kirr, and x under physiological conditions in vitro, we sought to use these numbers to fit the data for in vivo tumor uptake in order to understand how this system behaves relative to reversible ligand-receptor pairs.
Figure 6.
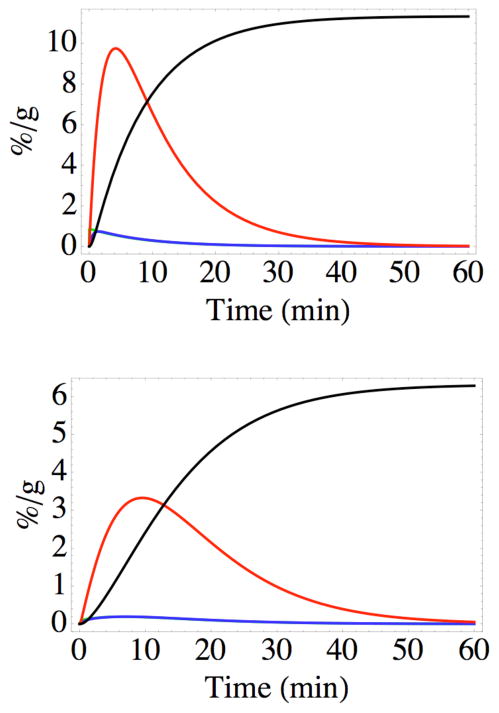
Experimental tumor uptake data (●±SD) from reference 5, showing the results of injection of AABD(86Y) into the tail veins of SCID mice bearing xenograft U-87 human glioma tumors expressing the DAbR1 reporter gene, plotted as average % dose per gram tissue vs time. The black curve shows how these data are fit by the solutions to the concentrations of all labeled species in equations 5–9. The red curve shows the behavior predicted if irreversible binding did not occur (kirr → 0), and the blue curve shows the behavior predicted if there were no binding of the probe at all (kon → 0), which is consistent with experimental observations of mice bearing xenograft U-87 human glioma tumors that do not carry the DAbR1 gene.
It is convenient to describe the basic unit of a glioma tumor as a “cord,” a cylindrical array of cancer cells surrounding a blood capillary (19), from which the tumor tissue receives nutrients and also small molecules such as the PET probe AABD(86Y). Characterizing the behavior of cells around one capillary can lead to a useful description of the main features of the tumor (20). Our goal here is to consider the major consequences of irreversible binding of the probe to its receptor, in order to gain insight into the essentials of this chemistry in a living organism.
The immediate requirement is the formulation and solution of a set of partial differential equations to describe not only the chemistry already considered in vitro but also the diffusion of the AABD(Y) probe in vivo to its receptors on the tumor cell surface. Assuming cylindrical symmetry and considering a small segment of a long cylinder reduces the geometry to one dimension: the radial distance r from the center of the capillary (20). The appropriate reaction-diffusion equations are given below as equations 5–9; only the probe diffuses, but the concentration of each species depends on both r and t:
| (5) |
| (6) |
| (7) |
| (8) |
| (9) |
We sought realistic solutions to these equations by fitting the in vivo tumor uptake data from reference 5, using nonlinear least-squares with the concentration of AABD(Y) at the periphery of the capillary as the only adjustable parameter, along with literature values for the typical radius of a capillary (10μ) and a tumor cord (≈100μ) (20, 21), the diffusion coefficient of a metal chelate in brain tissue (3.4 × 10−6 cm2/s) (22), and the observed ≈5 min circulatory half-life of the AABD(Y) probe (5). The calculated behavior is compared to experimental data in Figure 6; it is important to emphasize that the values for kon, koff, kirr, and x were those determined from in vitro experiments described above, with no adjustments. An important underlying assumption throughout is that, consistent with the crystal structures of parental Fab-DOTA complexes (12), the complex tolerates molecular changes at positions peripheral to the sites of van der Waals contact between protein and DOTA. Thus the most important interactions between receptor and ligand persist through genetic substitution of Cys for Gly at position 54, reformatting of the mouse IgG to chimeric G54C Fab or to G54C scFv, and chemical changes at the para position of the DOTA side chain.
Figure 6 also illustrates the predicted effects of eliminating irreversible binding of the AABD(Y) probe in the tumor while maintaining reversible binding (setting kirr → 0), or eliminating probe binding altogether (setting kon → 0). The fit to the in vivo experimental data in Figure 6 was made with the assumption that all the G54C binding sites expressed on cell surfaces have the side chain of Cys-54 free to react with the acryloyl group of AABD(Y), which would not be true for the highly cysteinylated antibody expressed in S2 cells (13). Changing the calculation to assume a significant degree of blocked Cys-54 leads to curves that resemble the red curve (kirr → 0) in Figure 6, diverging markedly from experimental observation. Thus the binding sites expressed on the surface of U-87 cells in vivo exhibit little or no blockage of the Cys-54 side chain. Because there is no established way to change the oxidation state of the expressed G54C side chain on cell surfaces in vivo, this information is crucial to the design of future experiments. It also illustrates the diversity of posttranslational modification in eukaryotic cells.
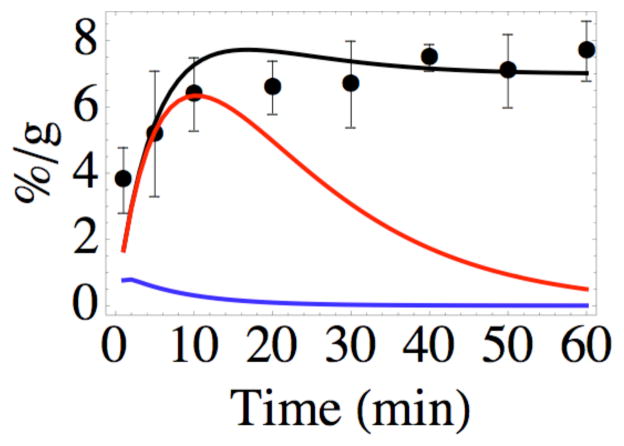
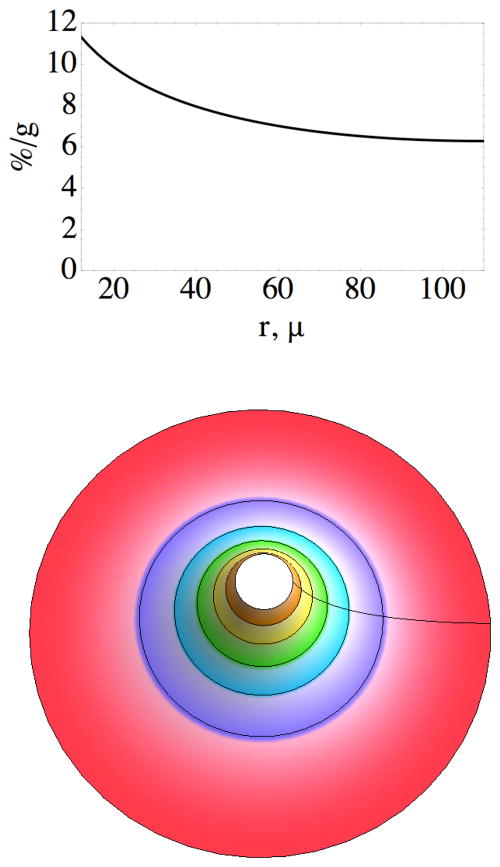
One of the central issues of molecular targeting is the transport of probe molecules, whether large or small, from the blood into the target tissue (21). A number of interesting experiments have been carried out to demonstrate the effects of various parameters on this process (e.g., 23, 24). The results in (5) demonstrate rapid, durable uptake in a xenograft tumor but lack the spatial resolution to provide information about whether the irreversible binding occurs at the edge of the capillary or throughout the tumor. The model developed here predicts that probe molecules present in the tumor 1 hr after administration will all be irreversibly bound to the engineered receptor, with a spatial distribution plotted in Figure 7. While the average concentration of probe in the tumor at 1 hr was observed to be approximately 7%/g, the predicted concentration near the capillary is almost 12%/g, declining to slightly more than 6%/g near the periphery of the tumor cord.
Figure 7.
Top: predicted concentration of the probe 60 min after injection, as a function of distance from just outside the boundary of the capillary (12μ) to the outer periphery of the tumor cord (110μ from the center of the coordinate system). Bottom: contour plot of probe concentration in a transaxial slice of the tumor from 12μ (≈12%/g) to 110μ (≈ 6%/g); color of contour changes at approximately 1%/g intervals. The capillary (radius 10μ) is within the empty space in the center, and the concentration profile along the radial line is given by the graph above.
It is informative to use the solutions of equations 5–9 to gain a sense of how the concentrations of the different probe-containing species vary with time and position in the tissue. The curves in Figure 8 imply that the free probe AABD(Y) is quickly incorporated into complexes CNR and CR, with the latter rapidly converted to P. Because of the short lifetime of either complex, the product P becomes the dominant species within a short period. Rapid clearance of the probe from the circulation leads to disappearance of the other species from the tumor before 60 minutes have elapsed. The behavior illustrated in Figure 8 for positions near the capillary and near the outer periphery of the tumor cord is predicted to be qualitatively similar at intermediate distances from the capillary, with no remarkable differences except that the final concentration of P varies with r according to Figure 7, and the concentrations of all the species are predicted to change more slowly farther from the capillary.
The interplay between molecular transport and ligand-receptor binding provides an almost inexhaustible range of possibilities for molecular imaging probes. This is above all true when irreversible binding may take place, as in the case of antibodies with infinite affinity (3, 25). In the most extreme manifestation, an irreversible probe molecule would encounter its receptor and become permanently attached immediately after entering tumor tissue; in that case the curve in Figure 7 would be a narrow peak at the left side, falling to zero with increasing distance. In the case examined here, the probe has both reactive and non-reactive modes of binding to the receptor, and the latter is more probable; non-reactive binding and dissociation can serve to further spread the probe through the tissue before it becomes permanently fixed on a receptor.
If we compare this to purely reversible binding, we see the possibility that it can offer a useful middle ground encompassing the most desirable aspects of opposing extremes. Very strong (though reversible) binding such as provided by the streptavidin-biotin pair would be expected to yield a zone of stably bound probe molecules near the capillary, as long as streptavidin is in excess over biotinylated probe molecules. This nonuniform distribution of probes in tissue, referred to as the binding-site barrier, could be ameliorated if the concentration of avidin molecules were too low to efficiently capture the probes, or if the concentration of probe molecules were large enough to saturate the streptavidin sites, allowing other probe molecules to diffuse past. On the other hand, weaker reversible binding naturally allows small probe molecules to penetrate into tissue by virtue of repeated dissociation, diffusion, and association. This might eventually lead to uniform distribution in tissue, but since the binding is not durable, the concentration of the probe falls to zero in an inconveniently short time. The contour plot in Figure 7 implies a combination of durable binding and broad distribution that would be difficult to produce by other means.
Supplementary Material
Acknowledgments
We thank Dr. Andrew Fisher for sharing his isothermal calorimeter and instructions for its use, Dr. Mark McCoy and Dr. Michael Toney for insightful discussion and help on the kinetics experiments, and Dr. Tove Olafsen for the human IgG standard. Supported by Research Grant CA016861 to C.F.M. from the National Cancer Institute, National Institutes of Health.
Footnotes
Abbreviations: NTA, Nitrilotriacetic acid; ABD, S-2-(4-Aminobenzyl)-1, 4, 7, 10-tetraazacyclododecanetetraacetic acid; AABD, S-2-(4-acrylamidobenzyl)-1, 4, 7, 10-tetraazacyclododecanetetraacetatic acid; DOTA, 1,4,7,10-tetraazacyclododecane N,N′,N″,N‴-tetraacetate; DTPA, Diethylenetriaminepentaacetic acid; DMEM, Dulbecco’s modified Eagle’s medium; TEA, Triethylamine; DTPA, Diethylenetriaminepentaacetic acid; DAbR1, DOTA antibody reporter 1; PVDF, Polyvinylidine difluoride; TBST, Tris-buffered saline with 0.5% (v/v) Tween 20.
Supporting Information Available: Key Mathematica instructions for solving equations 1–9. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Liu G, Hnatowich DJ. A semiempirical model of tumor pretargeting. Bioconjugate Chem. 2008;19:2095–2104. doi: 10.1021/bc8002748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang Z, Hwang P, Watson DS, Cao L, Szoka FC., Jr Tris-nitrilotriacetic acids of subnanomolar affinity toward hexahistidine tagged molecules. Bioconjugate Chem. 2009;20:1667–1672. doi: 10.1021/bc900309n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chmura AJ, Orton MS, Meares CF. Antibodies with infinite affinity. Proc Natl Acad Sci USA. 2001;98:8480–8484. doi: 10.1073/pnas.151260298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chidley C, Mosiewicz K, Johnsson K. A designed protein for the specific and covalent heteroconjugation of biomolecules. Bioconjugate Chem. 2008;19:1753–1756. doi: 10.1021/bc800268j. [DOI] [PubMed] [Google Scholar]
- 5.Wei LH, Olafsen T, Radu C, Hildebrandt IJ, McCoy MR, Phelps ME, Meares CF, Wu AM, Czernin J, Weber WA. Engineered antibody fragments with infinite affinity as reporter genes for PET imaging. J Nucl Med. 2008;49:1828–1835. doi: 10.2967/jnumed.108.054452. [DOI] [PubMed] [Google Scholar]
- 6.Meares CF, McCall MJ, Reardan DT, Goodwin DA, Diamanti CI, McTigue M. Conjugation of antibodies with bifunctional chelating agents: isothiocyanate and bromoacetamide reagents, methods of analysis, and subsequent addition of metal ions. Anal Biochem. 1984;142:68–78. doi: 10.1016/0003-2697(84)90517-7. [DOI] [PubMed] [Google Scholar]
- 7.Velazquez-Campoy A, Freire E. Isothermal titration calorimetry to determine association constants for high-affinity ligands. Nature Protocols. 2006;1:186–191. doi: 10.1038/nprot.2006.28. [DOI] [PubMed] [Google Scholar]
- 8.Corneillie TM, Whetstone PA, Fisher AJ, Meares CF. A rare earth- DOTA-binding antibody: probe properties and binding affinity across the lanthanide series. J Am Chem Soc. 2003;125:3436–3437. doi: 10.1021/ja029363k. [DOI] [PubMed] [Google Scholar]
- 9.Goodwin DA, Meares CF, Watanabe N, McTigue M, Chaovapong W, Ransone CM, Renn O, Greiner DP, Kukis DL, Kronenberger SI. Pharmacokinetics of pretargeted monoclonal antibody 2D12.5 and Y-88-Janus-2-(p-nitrobenzyl)-1,4,7,10- tetraazacyclododecanetetraacetic acid (DOTA) in balb/c mice with KHJJ mouse adenocarcinoma - a model for Y-90 radioimmunotherapy. Cancer Res. 1994;54:5937–5946. [PubMed] [Google Scholar]
- 10.Sigurskjold BW. Exact analysis of competition ligand binding by displacement isothermal titration calorimetry. Anal Biochem. 2000;277:260–266. doi: 10.1006/abio.1999.4402. [DOI] [PubMed] [Google Scholar]
- 11.Corneillie TM, Lee KC, Whetstone PA, Wong JP, Meares CF. Irreversible engineering of the multielement-binding antibody 2D12.5 and its complementary ligands. Bioconjugate Chem. 2004;15:1392–1402. doi: 10.1021/bc049824m. [DOI] [PubMed] [Google Scholar]
- 12.Corneillie TM, Fisher AJ, Meares CF. Crystal structures of two complexes of the rare-earth-DOTA-binding antibody 2D12.5: ligand generality from a chiral system. J Am Chem Soc. 2003;125:15039–15048. doi: 10.1021/ja037236y. [DOI] [PubMed] [Google Scholar]
- 13.Miao Z, McCoy MR, Singh DD, Barrios B, Hsu OL, Cheal SM, Meares CF. Cysteinylated protein as reactive disulfide: an alternative route to affinity labeling. Bioconjugate Chem. 2008;19:15–19. doi: 10.1021/bc700330j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramakrishnan M, Kandimalla KK, Wengenack TM, Howell KG, Poduslo JF. Surface plasmon resonance binding kinetics of Alzheimer’s disease amyloid β peptide- capturing and plaque-binding monoclonal antibodies. Biochemistry. 2009;48:10405–10415. doi: 10.1021/bi900523q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hyre DE, Le Trong I, Merritt EA, Eccleston JF, Green NM, Stenkamp RE, Stayton PS. Cooperative hydrogen bond interactions in the streptavidin-biotin system. Protein Sci. 2006;15:459–67. doi: 10.1110/ps.051970306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pagel JM, Orgun N, Hamlin DK, Wilbur DS, Gooley TA, Gopal AK, Park SI, Green DJ, Lin Y, Press OW. A comparative analysis of conventional and pretargeted radioimmunotherapy of B-cell lymphomas by targeting CD20, CD22, and HLA-DR singly and in combinations. Blood. 2009;113:4903–4913. doi: 10.1182/blood-2008-11-187401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pagel JM, Hedin N, Subbiah K, Meyer D, Mallet R, Axworthy D, Theodore LJ, Wilbur DS, Matthews DC, Press OW. Comparison of anti-CD20 and anti- CD45 antibodies for conventional and pretargeted radioimmunotherapy of B-cell lymphomas. Blood. 2003;101:2340–2348. doi: 10.1182/blood-2002-03-0874. [DOI] [PubMed] [Google Scholar]
- 18.Klumb LA, Chu V, Stayton PS. Energetic roles of hydrogen bonds at the ureido oxygen binding pocket in the streptavidin–biotin complex. Biochemistry. 1998;37:7657–7663. doi: 10.1021/bi9803123. [DOI] [PubMed] [Google Scholar]
- 19.Thalhauser CJ, Sankar T, Preul MC, Kuang Y. Explicit separation of growth and motility in a new tumor cord model. Bull Math Biol. 2009;71:585–601. doi: 10.1007/s11538-008-9372-8. [DOI] [PubMed] [Google Scholar]
- 20.Eikenberry S. A tumor cord model for Doxorubicin delivery and dose optimization in solid tumors. Theor Biol Med Model. 2009;6:16. doi: 10.1186/1742-4682-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujimori K, Covell DG, Fletcher JE, Weinstein JN. Modeling analysis of the global and microscopic distribution of immunoglobulin G, F(ab′)2, and Fab in tumors. Cancer Res. 1989;49:5656–5663. [PubMed] [Google Scholar]
- 22.Syková E, Nicholson C. Diffusion in brain extracellular space. Physiol Rev. 2008;88:1277–1340. doi: 10.1152/physrev.00027.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adams GP, Schier R, McCall AM, Simmons HH, Horak EM, Alpaugh RK, Marks JD, Weiner LM. High affinity restricts the localization and tumor penetration of single-chain Fv antibody molecules. Cancer Res. 2001;61:4750–4755. [PubMed] [Google Scholar]
- 24.Dennis MS, Jin H, Dugger D, Yang R, McFarland L, Ogasawara A, Williams S, Mary J, Cole Ross S, Schwall R. Imaging tumors with an albumin-binding Fab, a novel tumor-targeting agent. Cancer Res. 2007;67:254–261. doi: 10.1158/0008-5472.CAN-06-2531. [DOI] [PubMed] [Google Scholar]
- 25.Butlin NG, Meares CF. Antibodies with infinite affinity: origins and applications. Acc Chem Res. 2006;39:780–787. doi: 10.1021/ar020275e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.