Abstract
Bioassay-guided fractionation of the extract of a consortium of a marine cyanobacterium and a red alga (Rhodophyta) led to the discovery of a novel compound, palmyramide A, along with the known compounds curacin D and malyngamide C. The planar structure of palmyramide A was determined by one- and two-dimensional NMR studies and mass spectrometry. Palmyramide A is a cyclic depsipeptide which features an unusual arrangement of three amino acids and three hydroxy acids; one of the hydroxy acids is the rare 2,2-dimethyl-3-hydroxyhexanoic acid unit (Dmhha). The absolute configurations of the six residues were determined by Marfey’s analysis, chiral HPLC analysis and GC/MS analysis of the hydrolysate. Morphological and phylogenetic studies revealed the sample to be composed of a Lyngbya majuscula-Centroceras sp. association. MALDI-imaging analysis of the cultured L. majuscula indicated that it was the true producer of this new depsipeptide. Pure palmyramide A showed sodium channel blocking activity in neuro-2a cells and cytotoxic activity in H-460 human lung carcinoma cells.
Cyanobacteria are well known to produce a wide variety of secondary metabolites displaying significant structural diversity and biological activity.1,2 Because the production of secondary metabolites depends both on the collection location as well as specific species, our program has focused on collections from a wide variety of tropical and subtropical locations including Panama, Papua New Guinea, Curaçao, Jamaica and Puerto Rico, and this has generally been a productive approach.2 In this vein, we recently had the opportunity to examine the unique marine flora and cyanobacteria of Palmyra Atoll in the Northern Pacific Ocean, a remote Pacific island with a unique biogeography.3 Bioassay-guided fractionation of the extract of a cyanobacterial/red algal consortium led to the discovery of a novel cyclic depsipeptide, palmyramide A (1), along with the known compounds, curacin D and malyngamide C.4,5 This paper describes the isolation, structural determination including stereostructure, biological activity, and identification of the producing organism of 1.
Results and Discussion
The crude extract of the consortium of cyanobacterium and red alga collected at Palmyra Atoll was subjected to silica gel vacuum liquid chromatography and assayed for sodium channel blocking activity and cytotoxic activity. The bioactive fraction eluted with 100% EtOAc and was further purified by C18 SPE cartridge and repetitive reversed-phase HPLC to yield palmyramide A (1) (Figure 1) as a colorless glassy solid.
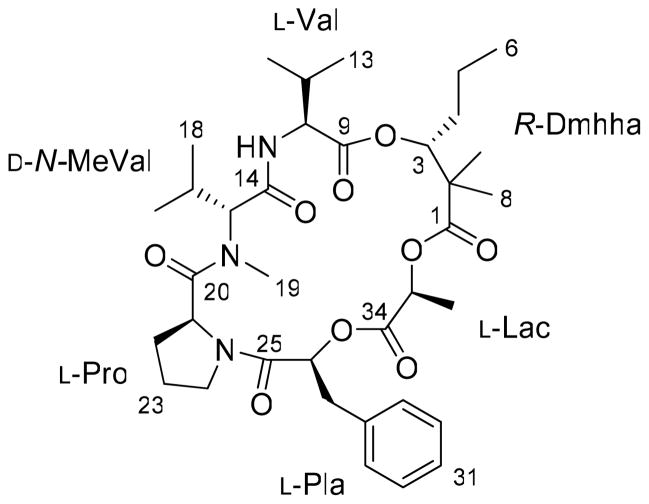
Figure 1.
Structure of palmyramide A (1).
High-resolution ESIMS of 1 gave [M+H]+ and [M+Na]+ peaks at m/z 672.3852 and 694.3669, respectively, indicating a molecular formula of C36H53N3O9 and requiring 12 degrees of unsaturation. The 1H NMR spectrum possessed five aromatic protons at δH 7.23–7.28, an amide NH proton at δH 6.55, an N-methyl group at δH 2.85, and two singlet methyl groups at δH 1.16 and 1.21. Correspondingly, the 13C NMR spectrum revealed the presence of six ester/amide carbonyls and a monosubstituted phenyl ring at δC 127.4, 128.6 (2C), 129.7 (2C) and 134.9, accounting for 10 of the 12 degrees of unsaturation.
Analysis of 1- and 2D-NMR spectra including COSY, TOCSY, HSQC, and HMBC led to the assignments of three amino acids [valine (Val), N-methylvaline (N-MeVal), and proline (Pro)] and three hydroxy acids (3-phenyllactic acid (Pla), lactic acid (Lac), and 2,2-dimethyl-3-hydroxyhexanoic acid (Dmhha)) (Table 1). The N-methyl group of N-MeVal was revealed by HMBC correlations (H-19/C-15, H-15/C-19) and the chemical shifts of the NMe (δH 2.85, δC 30.5). In the Pla residue, the proton chemical shifts at δH 7.23–7.28 (monosubstituted benzene ring), δH 3.13 and 2.95 (methylene), and δH 5.02 (methine) were very similar to those reported for phenylalanine. However, the carbon chemical shift of the methine (δC 70.9) was consistent with that of an oxymethine, indicating that this residue was not phenylalanine but rather 3-phenyllactic acid. The proton chemical shifts of the Lac residue at δH 1.47 (methyl) and 4.81 (methine) are also very similar to those reported for alanine, but once again the carbon chemical shift of the methine (δC 69.0) indicated that this residue was an α-hydroxy acid, namely lactic acid. The identity of the Dmhha residue was deduced through analysis of TOCSY, COSY and HMBC correlations as follows: the proton spin system from H-3 to H-6 was disclosed by TOCSY, and two singlet methyl proton resonances H-7 (δH 1.21) and H-8 (δH 1.15) gave HMBC cross peaks to C-1, C-2, and C-3. Oxygenation at C-3 was once again inferred from its 13C NMR chemical shift at δC 77.2. The remaining required degree of unsaturation suggested the overall monocyclic nature of 1.
Table 1.
NMR Spectroscopic Data (600 MHz, CDCl3) for Palmyramide A.
| Residue | Position | δC | mult | δH (mult, J in Hz) | HMBC |
|---|---|---|---|---|---|
| Dmhha | 1 | 174.9 | qC | ||
| 2 | 46.4 | qC | |||
| 3 | 77.2 | CH | 5.57 (dd, 9.6, 1.8) | C-1, C-2, C-4, C-5, C-7, C-8 | |
| 4a | 31.6 | CH2 | 1.43 (m) | C-3, C-5, C-6 | |
| 4b | 1.52 (m) | C-5, C-6 | |||
| 5 | 19.6 | CH2 | 1.36 (m) | C-3, C-4, C-6 | |
| 6 | 13.9 | CH3 | 0.92 (t, 7.2) | C-4, C-5 | |
| 7 | 16.9 | CH3 | 1.21 (s) | C-1, C-2, C-3, C-8 | |
| 8 | 24.3 | CH3 | 1.16 (s) | C-1, C-2, C-3, C-7 | |
| Val | 9 | 170.1 | qC | ||
| 10 | 58.3 | CH | 4.46 (dd, 5.4, 3.0) | C-9, C-12, C-13 | |
| 11 | 30.5 | CH | 2.38 (dqq, 3.0, 7.2, 7.2) | C-9, C-10, C-12, C-13 | |
| 12 | 17.8 | CH3 | 0.89 (d, 7.2) | C-10, C-11, C-13 | |
| 13 | 18.3 | CH3 | 0.98 (d, 7.2) | C-10, C-11, C-12 | |
| NH | 6.55 (d, 5.4) | C-10, C-14 | |||
| N -MeVal | 14 | 169.7 | qC | ||
| 15 | 61.7 | CH | 4.42 (d, 10.8) | C-14, C-16, C-17, C-18, C-19, C-20 | |
| 16 | 27.3 | CH | 2.13 (m) | C-14, C-15, C-17, C-18 | |
| 17 | 18.8 | CH3 | 0.70 (d, 6.6) | C-15, C-16, C-18 | |
| 18 | 19.3 | CH3 | 0.94 (d, 6.6) | C-15, C-16, C-17 | |
| 19 (N -CH3) | 30.5 | CH3 | 2.85 (s) | C-15, C-20 | |
| Pro | 20 | 171.2 | qC | ||
| 21 | 56.7 | CH | 3.22 (dd, 7.8, 3.0) | C-22, C-23 | |
| 22 | 30.7 | CH2 | 1.50 (m) | C-20, C-23, C-24 | |
| 23a | 22.0 | CH2 | 1.56 (m) | C-22, C-24 | |
| 23b | 1.68 (m) | C-22, C-24 | |||
| 24a | 46.7 | CH2 | 3.42 (ddd, 12.0, 6.6, 6.6) | C-22, C-23, C-25 | |
| 24b | 3.57 (ddd, 12.0, 8.4, 4.8) | C-22, C-23 | |||
| Pla | 25 | 167.2 | qC | ||
| 26 | 70.9 | CH | 5.02 (dd, 12.0, 4.8) | C-25, C-27, C-28, C-34 | |
| 27a | 39.2 | CH2 | 2.95 (dd, 12.0, 4.8) | C-25, C-26, C-28, C-29/33 | |
| 27b | 3.13 (dd, 12.0, 4.8) | C-25, C-26, C-28, C-29/33 | |||
| 28 | 134.9 | qC | |||
| 29/33 | 129.7 | CH | 7.23 (m) | C-27, C-31 | |
| 30/32 | 128.6 | CH | 7.28 (m) | C-28, C-30/32 | |
| 31 | 127.4 | CH | 7.25 (m) | C-29/33 | |
| Lac | 34 | 168.3 | qC | ||
| 35 | 69.0 | CH | 4.81 (q, 6.6) | C-34, C-36, C-1 | |
| 36 | 16.7 | CH3 | 1.47 (d, 6.6) | C-34, C-35 |
The sequencing of these residues in 1 was accomplished by interpretation of HMBC correlations (Table 1). Correlations from the NH proton or N-methyl protons to the neighboring carbonyl carbons were observed between Val/N-MeVal and N-MeVal/Pro. Correlations from methine or methylene protons to carbonyl carbons of the neighboring amino or hydroxy acids were also observed between Pro/Pla, Pla/Lac and Lac/Dmhha. Thus, the planar structure of 1 was determined as shown in Figure 1.
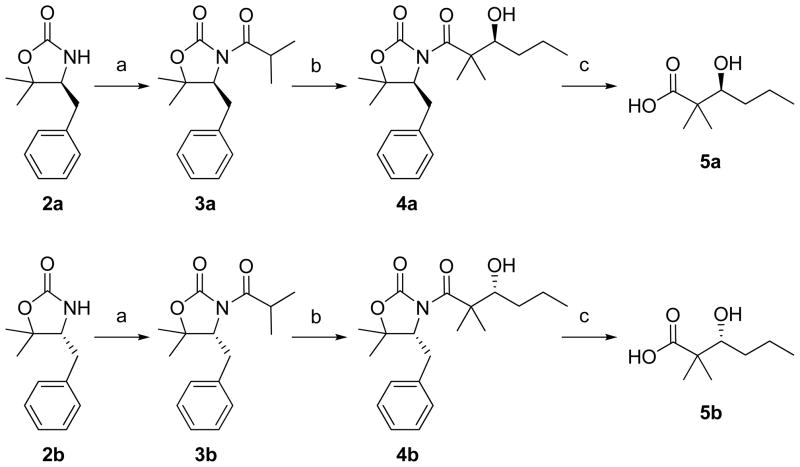
The absolute configurations of the three amino acids in palymramide A (1) were determined by HPLC analysis of the acid hydrolysate derivatized with Marfey’s reagent [Nα-(2,4-dinitro-5-fluorophenyl)-L-valinamide: DFVA].6 LC/MS analysis of the derivatives eluting by a linear gradient MeCN/H2O (0.1% HCOOH) revealed the presence of D-N-MeVal and L-Pro, whereas the configuration of the Val residue could not be determined because of the overlap between the peaks of L-Val and Marfey’s reagent. After exploring a variety of HPLC conditions, LC/MS analysis using a linear gradient of MeOH/H2O (0.1% HCOOH) and a Merck LiChrospher 100 RP-18 column, enabled separation of the peaks of L-Val and the Marfey’s reagent, and thus allowed determination of the Val residue in 1 to be L. The absolute configurations of Pla and Lac were determined as L by chiral HPLC analysis. The absolute configuration of the β-hydroxy group of Dmhha was deduced by chromatographic comparisons with authentic S- and R-Dmhha which were stereoselectively synthesized via a tertiary aldol reaction (Figure 2).7 The absolute configuration of the intermediate 4a and 4b formed in these syntheses were confirmed by the Mosher ester method8 and optical rotations. Chiral GC/MS analysis of the Dmhha methyl ester liberated from 1 by acid hydrolysis revealed only the presence of R-Dmhha. Thus, the absolute configuration of palmyramide A (1) was determined as shown in Figure 1.
Figure 2.
Stereoselective synthesis of S- and R-Dmhha. a) 1. n-BuLi, THF, −78 °C. 2. isobutyryl chloride. b) 1. LDA, THF, −78 °C. 2. Ti(O-i-Pr)3Cl. 3. Butanal. c) 1. 30% aq. H2O2. 2. LiOH/H2O, THF/H2O (4:1).
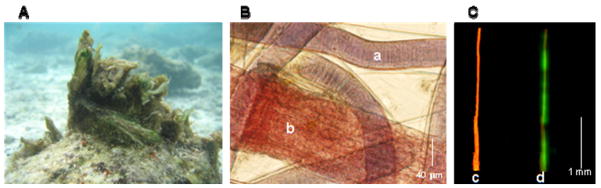
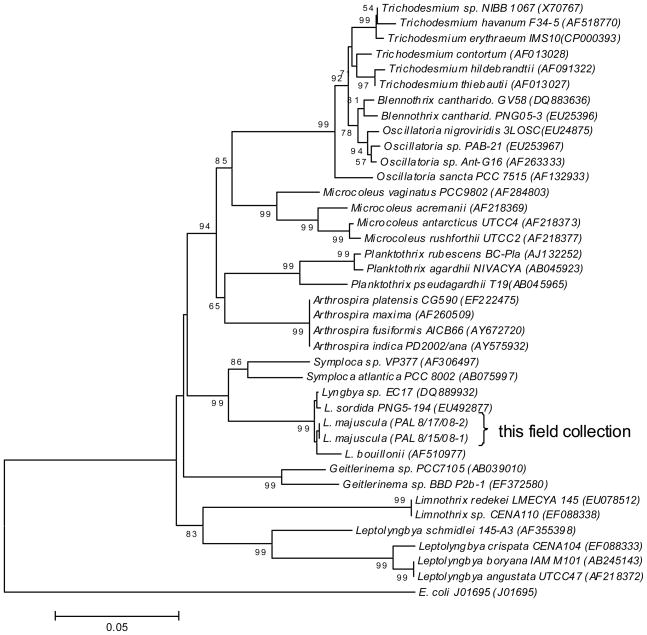
As noted, palmyramide A (1) was isolated from an environmental sample composed of a consortium of a cyanobacterium and a red alga (Figure 3A and 3B), and obtained from two different populations collected from different sampling sites at Palmyra Atoll. Morphological and phylogenetic studies revealed that both samples were of similar composition and composed of L. majuscula Harvey ex Gomont (Cyanophyta) and Centroceras sp. (Rhodophyta). The L. majuscula appeared as blackish tufts approximately 10 cm in height (Figure 3A). Microscopically, the Lyngbya filaments were long (>1 cm), straight or slightly waved, cylindrical, and 40.2 ± 3.4 μm (n = 3) wide. The trichomes were enclosed with distinct visible, colorless sheaths. The cells were disk-shaped (39.1 ± 2.8 μm wide and 3.5 ± 0.9 μm long; n = 30) with no or only very small constrictions at the cross-walls. The terminal cells were rounded, non-capitated, and lacked calyptras. Taxonomic identifications and evolutionary histories were inferred for both the L. majuscula and Centroceras sp. components of this consortium using the conserved 16S/18S (SSU) rRNA genes and the more variable RNA polymerase γ-subunit (rpoC1) gene. Furthermore, both genetic markers were obtained from the consortium obtained at both sampling sites, and confirmed that these were the same two organisms in each case. An identical 16S rRNA gene (1,373 base pairs or ~95% of the gene coverage; GenBank Acc. No. GQ231522) and rpoC1 gene (866 base pairs; GenBank Acc. No. GQ231521) were obtained from the L. majuscula of both populations. The evolutionary history was inferred from the 16S rRNA gene using the Minimum Evolution and MrBayes algorithms which resulted in conserved tree topologies and similar bootstrap-support. These collections of L. majuscula claded tightly (<1% sequence divergence) with other Pacific strains of Lyngbya within the “marine Lyngbya lineage” (Figure 4). The 18S rRNA genes (206 bp; GenBank Acc. No. GQ246180) obtained from the two collections of Centroceras sp. were also identical in sequence.
Figure 3.
MALDI-TOF-imaging of the palmyramide A (1) producing cyanobacterium Lyngbya majuscula. (A) Photograph of the consortium of Lyngbya majuscula and Centroceras sp. at the time of collection; (B) Photomicrograph of the consortium of Lyngbya majuscula (a) and Centroceras sp.; (b) (C) Epifluorescent image of the cultured Lyngbya majuscula at 590 nm (c, orange) and MALDI image of m/z 672 (= palmyramide A, 1, d, green).
Figure 4.
Phylogenetic tree of the cyanobacterial order Oscillatoriales based on the 16S rRNA gene and placement of this field collection of Lyngbya majuscula.
In order to determine which organism was responsible for production of palmyramide A (1), a natural product MALDI-TOF-imaging (npMALDI-I) approach was applied to the environmental samples stored in either EtOH/H2O (1:1) at −20 °C or in RNA stabilization solution (RNAlater®) at −20 °C.9,10 In principal, the npMALDI-I approach can visualize the spatial distribution of secondary metabolites in intact tissues of marine organisms. However, in this case the molecular weight of 1 was not detected in any of the Lyngbya majuscula or Centroceras sp. samples. We reasoned that this surprising result might be due to the instability of the palmyramide in the preserved biological samples. However, we were able to adapt into laboratory culture the cyanobacterial component of this consortium, and npMALDI-I of fresh Lyngbya filaments possessed a robust signal for a compound with the molecular weight of 1, indicating that L. majuscula is the producer of this new depsipeptide (Figure 3C).
Pure palmyramide A (1) was assayed for sodium channel blocking and cancer cell cytotoxic activities (presumably due the presence of one N-methyl amide in 1, it occurs as a 95:5 mixture of major and minor conformers by 1H NMR; see Supporting Information).11,12 In the sodium channel blocking assay, 1 inhibited the veratridine and ouabain induced sodium overload and resulting cytotoxicity in neuro2a cells, presumably by blocking the voltage gated sodium channel with an IC50 value of 17.2 μM. In H-460 human lung carcinoma cells, palmyramide A (1) showed modest cytotoxic effects with an IC50 value of 39.7 μM.
Palmyramide A (1) is a novel cyclic depsipeptide which features an unusual arrangement of three amino acids and three hydroxy acids. Among them, Dmhha is the most unusual hydroxy acid and has only previously been reported in guineamide E and F, two metabolites isolated from Papua New Guinea collections of Lyngbya majuscula.13 Since environmental samples of marine organisms are almost always collected as a consortium, mixtures or with symbionts, it is necessary to determine the true source of interesting secondary metabolites by other approaches. The npMALDI-I method is well suited to directly observing the physical location of metabolites in intact marine organisms of assemblages.10 In this case, palmyramide A (1) was isolated from a consortium of two organisms, a cyanobacterium and a red alga, however, analysis by npMALDI-I showed that L. majuscula is the true producer. Indeed, the cyanobacterium L. majuscula is one of the most prolific producers of natural products with more than 185 reported compounds to date.2 In this study, our investigation of L. majuscula from a new geographical region was rewarded by the discovery of a new sodium channel blocking and cytotoxic cyclic lipopeptide, palmyramide A (1), and suggests that a continued investigation of this organism from new locations will be productive in the discovery of novel natural compounds with pharmaceutical potential.
Experimental Section
General Experimental Procedures
Optical rotations were measured with a JASCO P-2000 polarimeter. IR and UV spectra were recorded with a Nicolet IR-100 FT-IR and Beckman-Coulter DU800 spectrophotometer, respectively. NMR spectra were recorded on Varian Inova 500 and Bruker Avance III DRX600 with the solvent CDCl3 (δH at 7.26, δC at 77.0) used as an internal standard. High-resolution mass spectra were obtained on a Thermo Scientific LTQ-XL Orbitrap mass spectrometer. LCMS analysis was carried out on a Finnigan LCQ advantage mass spectrometer with a Finnigan Surveyor HPLC system. HPLC was performed using a Waters 515 pumps and a Waters 996 photodiode array spectrometer.
Collection
The consortium of a cyanobacterium and a red alga was collected by hand from shallow water (<1 m) at two different sites in the Western lagoon South of Strawn Island, Palmyra Atoll, USA, in August, 2008 (GPS coordinates: 05′53.04N, 162′05.161W and 05′52.172N, 162′05.047W). The environmental samples were stored in EtOH/H2O (1:1) at −20°C while the genetic materials were preserved in RNA stabilization solution at −20°C (RNAlater®, Ambion Inc.). Voucher specimens are available from WHG as collection number PAL 8/17/08-2 and PAL 8/15/08-1.
Extraction and Isolation
Approximately 417.1 g (dry weight) of the consortium was extracted repeatedly with CH2Cl2-MeOH (2:1) to yield 7.1 g of crude extract. A portion of the extract (1.5 g) was subjected to silica gel vacuum liquid chromatography (VLC, hexanes/EtOAc/MeOH) to produce nine chemically distinct fractions. The fraction eluted with EtOAc was subjected to C18 SPE (MeOH-H2O, stepwise) and repetitive reversed-phase HPLC (Phenomenex Jupiter C18, 10 × 250 mm, 70% MeCN, 3 mL/min) to obtain 6.1 mg of palmyramide A (1, 20.1 min) and 2.0 mg of malyngamide C (14.1 min). The fraction eluting with 40% EtOAc in hexanes was purified by silica gel column chromatography [hexane-EtOAc (6:1)] to yield 28.6 mg of curacin D.
Palmyramide A (1)
colorless glass; [α]D +19.6 (c 0.25, CHCl3); UV (MeOH) λmax (log ε) 203 (4.27); IR (neat) νmax 3409, 2965, 2876, 1741, 1660, 1504, 1454, 1387, 1261, 1197, 1146, 1100 cm−1; 1H and 13C NMR, see Table 1; HRESIMS m/z 672.3852 [M+H]+ (calcd for C36H54N3O9, 672.3855).
Marfey’s Analysis
Palmyramide A (0.5 mg) in 0.5 mL of 6 N HCl was heated in a sealed tube at 110 °C for 17 h, and the reaction mixture extracted with EtOAc. The aqueous layer was dried under N2, and the residue dissolved in 400 μL of H2O. Subsequently, 200 μL of this solution was treated with 40 μL of 1% DFVA solution in acetone and 8.5 μL of 1 M NaHCO3, and heated at 90 °C for 5 min. After cooling to room temperature, 20 μL of 1 N HCl was added and the solution was analyzed by LC/MS on a Merck LiChrospher 100 RP-18 (4×125 mm) at a flow rate of 0.8 mL/min using a linear gradient from MeCN/H2O (30:70, 0.1% HCOOH) to MeCN/H2O (70:30, 0.1% HCOOH) over 60 min. The DFVA derivatives from the acid hydrolysate were identified by comparing the retention times with those of authentic standards, as follows: D-N-MeVal 20.9 min (L-N-MeVal 15.8 min) and L-Pro 8.0 min (D-Pro 10.6 min). The configuration of Val could not be determined in the above solvent system because L-Val and the Marfey’s reagent co-eluted. However, employing a linear gradient from MeOH/H2O (30:70, 0.1% HCOOH) to MeOH/H2O (70:30, 0.1% HCOOH) over 60 min cleanly resolved these peaks, and identified the configuration of Val in 1 to be L (L 34.1 min, D 51.5 min).
Chiral HPLC Analysis
Palmyramide A (0.5 mg) in 0.5 mL of 6 N HCl was heated in a sealed tube at 110 °C for 17 h, and the reaction mixture extracted with EtOAc. The organic layer was dried under N2, and the residue dissolved in 200 μL of MeOH. The hydrolysate was analyzed by chiral HPLC on a Phenomenex Chirex 3126 (4.6 × 250 mm) at a flow rate of 0.7 mL/min with UV detection at 254 nm. The isocratic solvent system of 2.0 mM CuSO4 in H2O was used to determine the configuration of L-Lac (L 16.4 min, D 20.3 min). The isocratic solvent system of 2.0 mM CuSO4 in H2O/2-propanol (85:15) was used to determine the configuration of L-Pla (L 53.2 min, D 74.1 min).
(S)-4-Benzyl-3-isobutyryl-5,5-dimethyloxazolidin-2-one (3a)
Oxazolidinone 2a (102 mg, 0.497 mmol) was dissolved in dry THF (1.3 mL) and treated with n-BuLi (1.42 M in hexanes, 350 μL, 0.497 mmol, 1 eq) dropwise at −78°C. After 1 h, isobutyryl chloride (130 μL, 1.242 mmol, 2.5 eq) in THF (0.8 mL) was added at −78°C. The reaction went to completion within 1 h, as monitored by TLC, and was quenched with H2O (2.1 mL). The mixture was extracted with Et2O (4 × 4 mL), dried over MgSO4, and concentrated in vacuo. The product was purified by flash chromatography on silica gel 60 (Et2O/hexanes, 1:19) yielding the title compound as a nearly colorless oil (132.5 mg, 97% yield):[α]D −34.0 (c 0.87, CHCl3); 1H NMR (CDCl3) δ 1.15 (3H, d), 1.17 (3H, d), 1.36 (3H, s), 1.38 (3H, s), 2.89 (1H, dd), 3.09 (1H, dd), 3.76 (1H, m), 4.51 (1H, dd), 7.21–7.32 (5H, m); HRMS m/z 275.1516 [M]+ (calcd for C16H21NO3, 275.1516).
(R)-4-Benzyl-3-isobutyryl-5,5-dimethyloxazolidin-2-one (3b)
Oxazolidinone 3b was prepared in the same way as oxazolidinone 3a from the D-phenylalanine derived oxazolidinone 2b (81% yield, due to a partial accidental loss of product):[α]D +33.6 (c 1.08, CHCl3); 1H NMR (CDCl3) δ 1.16 (3H, d), 1.17 (3H, d), 1.36 (3H, s), 1.38 (3H, s), 2.89 (1H, dd), 3.09 (1H, dd), 3.76 (1H, m), 4.51 (1H, dd), 7.21–7.32 (5H, m); HRMS m/z 275.1517 [M]+ (calcd for C16H21NO3, 275.1516).
(S)-4-Benzyl-3-(3-hydroxy-2,2-dimethylhexanoyl)-5,5-dimethyloxazolidin-2-one (4a)
Oxazolidinone 3a (47.6 mg, 0.173 mmol) was dissolved in dry THF (2 mL) and added dropwise to a solution of LDA (1.5 eq) in dry THF (1.76 mL) at −78°C [prepared by addition of 1.35 M n-BuLi (192 μL, 0.259 mmol, 1.5 eq) to a solution of diisopropylamine (39 μL, 0.277 mmol, 1.6 eq) in dry THF at −78°C, the solution was warmed to 0°C for 15 min, then cooled to −78°C]. After 30 min, a solution of chlorotriisopropoxy titanium IV (1 M in THF, 692 μL, 0.691 mmol, 4 eq) was added dropwise, and the solution was warmed to −40°C. After 1 h, the solution was cooled to −78°C and butyraldehyde (46.5 μL, 0.519 mmol, 3 eq) in THF (2 mL) was added dropwise, and the solution was warmed to −40°C. After 3 h, the reaction was quenched with satd. NH4Cl (4 mL) and stirred with Celite until warmed to room temperature. The filtrate was extracted with EtOAc (4 × 4 mL), washed with brine (4 mL), dried over Na2SO4, and concentrated in vacuo. The product was purified by flash chromatography on silica gel 60 (DCM/Hexanes/MeCN, 49.5:49.5:1) yielding the title compound as a nearly colorless oil (32.3 mg, 54% yield): [α]D −33.0 (c 0.48, CHCl3); 1H NMR (CDCl3) δ 0.94 (3H, t), 1.28 (2H, m), 1.33 (3H, s), 1.34 (3H, s), 1.35 (3H, s), 1.37 (3H, s), 1.60 (2H, m), 2.39 (1H, brd), 2.90 (1H, dd), 3.10 (1H, dd), 4.11 (1H, m), 4.56 (1H, dd), 7.20–7.35 (5H, m); HRMS m/z 370.1992 [M+Na]+ (calcd for C20H29NO4Na, 370.1989).
(R)-4-Benzyl-3-(3-hydroxy-2,2-dimethylhexanoyl)-5,5-dimethyloxazolidin-2-one (4b)
Prepared in the same way as 4a from 3b (35% yield):[α]D +32.4 (c 0.69, CHCl3); 1H NMR (CDCl3) δ 0.94 (3H, t), 1.28 (2H, m), 1.33 (3H, s), 1.34 (3H, s), 1.35 (3H, s), 1.37 (3H, s), 1.60 (2H, m), 2.39 (1H, brd), 2.90 (1H, dd), 3.10 (1H, dd), 4.11 (1H, m), 4.56 (1H, dd), 7.20–7.35 (5H, m); HRMS m/z 370.1991 [M+Na]+ (calcd for C20H29NO4Na, 370.1989).
(S)-3-Hydroxy-2,2-dimethylhexanoic acid (5a)
Hydrogen peroxide solution (30%, 13 μL, 0.115 mmol, 3.6 eq) and lithium hydroxide monohydrate (2.1 mg, 0.051 mmol, 1.6 eq), dissolved in H2O (61 μL), were added successively to a solution of 4a (11 mg, 0.031 mmol) in 4:1 THF:H2O (275 μL) at 0°C. After stirring 1 h, 20 min at 0°C, sodium sulfite (16.6 mg, 0.132 mmol, 4.2 eq) was added. THF was removed in vacuo and the residual aqueous solution was partitioned between CH2Cl2 (3 × 1 mL) and H2O. The combined aqueous layers were acidified to pH 1 with 1 N HCl. The aqueous layer was extracted with Et2O (3 × 1 mL), dried over MgSO4, filtered, and concentrated in vacuo yielding the title compound as a colorless oil (3.8 mg, 75% yield):[α]D −35.5 (c 0.24, CHCl3); 1H NMR (CDCl3) δ 0.95 (3H, t), 1.20 (3H, s), 1.25 (3H, s), 1.25–1.70 (4H, m), 3.66 (1H, dd); HRMS m/z 183.0995 [M+Na]+ (calcd for C8H16O3Na, 183.0992).
(R)-3-Hydroxy-2,2-dimethylhexanoic acid (5b)
Prepared in the same as 5a from 4b (69% yield):[α]D +26.5 (c 0.28, CHCl3); 1H NMR (CDCl3) δ 0.95 (3H, t), 1.20 (3H, s), 1.25 (3H, s), 1.25–1.70 (4H, m), 3.66 (1H, dd); HRMS m/z 183.0994 [M+Na]+ (calcd for C8H16O3Na, 183.0992).
Chiral GC/MS Analysis
Palmyramide A (0.5 mg) in 0.5 mL of 6N HCl was heated in a sealed tube at 110 °C for 17 h, and the reaction mixture extracted with EtOAc. The organic layer was dried under N2, and half of the residue was dissolved in 600 μL 1:1 Et2O:MeOH and treated with diazomethane for 25 min. Solvent and excess diazomethane were removed with N2 gas and the residue was resuspended in CH2Cl2. Standards of the R-Dhhma and S-Dhhma methyl esters were prepared similarly. Capillary GC/MS analysis was conducted using a Cyclosil B column (Agilent Technologies J&W Scientific, 30 m × 0.25 mm) under the following conditions: the initial oven temperature was 40°C, followed by an immediate ramp from 40°C to 90°C at a rate of 5°C/min, and held at 90°C for 55 min. The Dhhma methyl ester derived from 1 eluted at 42.00 min, while the R-Dhhma methyl ester eluted at 41.90 min. The S-Dhhma methyl ester eluted at 39.82 min. Co-injection of the Dhhma methyl ester derived from compound 1 with the R-Dhhma methyl ester yielded a single peak at 41.95 min, while coinjection with the S-Dhhma methyl ester yielded a peak at 39.87 min and a peak at 42.00 min, confirming that the β-ester linkage of the Dhhma residue of 1 is R.
Mosher’s Method
To a solution of 4a (5.4 mg, 0.016 mmol) in CH2Cl2 (1 mL) were added Et3N (0.010 mL, 0.078 mmol), R-MTPA chloride (0.015 mL, 0.078 mmol) and DMAP (2.0 mg, 0.017 mmol). The mixture was stirred at room temperature overnight, followed by purification on a silica gel column to yield the S-MTPA ester 6a (5.7 mg, yield 62.5%). The same procedure for the synthesis of 6a was used for the synthesis of the R-MTPA ester 6b (2.2 mg. yield 29.6%). The absolute configuration of oxymethine carbon of 4a was determined to be S by Δδ values for the protons adjacent to the MTPA esters moieties.
6a
1H NMR (CDCl3) δ 0.83 (3H, t), 1.14 (2H, m), 1.30 (3H, s), 1.32 (3H, s), 1.34 (3H, s), 1.42 (3H, s), 1.50 (1H, m), 1.61 (1H, m), 2.85 (1H, dd), 3.12 (1H, dd), 3.54 (3H, s), 4.42 (1H, dd), 6.15 (1H, dd), 7.20–7.56 (10H, m); HRMS m/z 586.2389 [M+Na]+ (calcd for C30H36 F3NO6Na, 586.2387)
6b
1H NMR (CDCl3) δ 0.88 (3H, t), 1.26 (2H, m), 1.28 (3H, s), 1.30 (3H, s), 1.33 (3H, s), 1.38 (3H, s), 1.48 (1H, m), 1.56 (1H, m), 2.82 (1H, dd), 3.04 (1H, dd), 3.55 (3H, s), 4.44 (1H, dd), 6.36 (1H, dd), 7.20–7.56 (10H, m); HRMS m/z 586.2385 [M+Na]+ (calcd for C30H36 F3NO6Na, 586.2387).
Morphological Characterization
Taxonomic identification of the cyanobacterium was performed in accordance with bacteriological systems14 and current phycological systems.15 Morphological characterizations were performed using an Olympus IX51 epifluorescent microscope (100× objective) equipped with an Olympus U-CMAD3 camera. Size measurements were calculated as an average with standard deviation of ten neighboring cells from three different filaments of a population.
DNA extraction, PCR-Amplification, and Cloning
DNA was extracted from approximately 40 mg of cleaned cyanobacterial filaments using the Wizard® Genomic DNA Purification Kit (Promega Inc.) following the manufacturer’s specifications. The isolated DNA was further purified using a Genomic-tip 20/G kit (Qiagen Inc). DNA concentration and purity was measured on a DU® 800 spectrophotometer (Beckman Coulter Inc.). The 16S rRNA genes were PCR-amplified using the cyanobacterial-specific primers 106F and 1509R,16 the rpoC1 genes using the degenerate primers LrpoC1-F (5′-CYTGYTTNCCYTCDATDATRTC-3′) and LrpoC1-R (5′-YTNAARCCNGARATGGAYGG-3′s), and the 18S rRNA gene using the universal primers U1F and U1R as previously described.17 The PCR reaction volumes were 25 μL containing 0.5 μL (50 ng) of DNA, 2.5 μL of 10 × PfuUltra IV reaction buffer, 0.5 μL of dNTP mix (25 mM each of dATP, dTTP, dGTP, and dCTP), 0.5 μL of each primer (10 μM), 0.5 μL of PfuUltra IV fusion HS DNA polymerase and 20.5 μL dH2O. The PCR reactions were performed in an Eppendorf® Mastercycler® gradient as follows: initial denaturation for 2 min at 95°C, 25 cycles of amplification followed by 20 sec at 95°C, 20 sec at 50°C and 1.5 min at 72°C, and final elongation for 3 min at 72°C. PCR-products were analyzed on a (1%) agarose-gel in SB-buffer and visualized by EtBr-staining. PCR products were subcloned using the Zero Blunt® TOPO® PCR Cloning Kit (Invitrogen) into the pCR®-Blunt IV TOPO® vector, and then transformed into TOPO® cells and cultured on LB-kanamycin plates. Plasmid DNA was isolated using the QIAprep® Spin Miniprep Kit (Qiagen) and sequenced with pCR®-Blunt IV TOPO® vector specific primers M13F and M13R. Sequencing of the 16S rRNA genes middle regions were improved using the internal primers 359F and 781R.16 Gene sequences were analyzed for anomalies using the Pintail software with the cut-off size set at >600 bp before submission to GenBank/EMBL/DDBJ.18 The gene sequences are available in the GenBank/EMBL/DDBJ databases under accession numbers: L. majuscula 16S rRNA gene (GQ231522), L. majuscula rpoC1 (GQ231521), and the Centroceras sp. 18S rRNA gene (GQ246180).
Phylogenetic Analyisis
The gene sequences were aligned in ClustalW XXL in MEGA 4.0 with standard gap opening and extension penalties without gaps.19 The evolutionary histories of the cyanobacterial 16S rRNA genes were inferred using the Minimum Evolution (ME) algorithm in MEGA 4.0 as well as the Bayesian (MrBayes algorithm) method using TOPALi v2.5.20 All algorithms were performed with 1000 bootstrap replicates. The evolutionary distances (pair-wise sequence divergence) were computed using the Maximum Composite Likelihood method. The ME tree was searched using the Close-Neighbor-Interchange (CNI) algorithm at a search level of 1.21 All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option) for a total of 855 bp.
Culture
Specimens for culturing were isolated under microscopy and cleaned by running individual filaments through 1% agar SWBG-11 plates. The isolated specimens were cultured in SWBG-11 medium at 28°C with 35 g/L Instant Ocean (Aquarium Systems Inc.). The cultures were kept in a 16 h light/8 h dark cycle with a light intensity of ~7 μmol photon/s/m2 provided by 40W cool white fluorescent lights.
Filament Sample Preparation
Filaments were removed from the parent culture using small sterile, blunt tip tweezers and transferred to a Petri dish containing a small amount of distilled H2O in order to remove excess salt and media; this process was repeated. Using the same tweezers, the filament was carefully removed and laid horizontally on a Bruker MSP 96 stainless steel microflex target plate. Excess liquid on the surface was carefully blotted using the corner of a Kimwipe®. The plate and filament were allowed to dry at room temperature until visibly dry.
Image Acquisition
After the filament had dried on the target plate, a photograph (Nikon Coolpix, 3 megapixel) of the plate was taken to use as spatial teach reference for the Bruker Microflex MALDI instrument. Next, epifluorescent images of the length of the filament were acquired at 4× magnification using an Olympus IX511 Microscope with an excitation filter of 590 nm. The resulting images were tiled together using Photoshop CS to create a single, epifluorescent image of the filament.
MALDI Matrix Deposition
After image acquistion, MALDI matrix composed of 35 mg/mL α-cyanohydroxycinnamic acid, 35 mg/mL DHB, 75% MeCN and 0.2% TFA was coated onto the MALDI MSP 96 plate using a Paasche airbrush (www.paascheairbrush.com) and repeated side-to-side strokes until an even, thin crystalline layer occluded the background of the plate. The target plate containing the sample and matrix was placed in an empty Petri dish until analysis.
MALDI MS and Imaging
The target plate containing the sample was inserted into a Microflex Bruker Daltonics mass spectrometer outfitted with Compass 1.2 software suite (Consists of FlexImaging 2.0, FlexControl 3.0, and FlexAnalysis 3.0). The sample was run in positive mode, with 80 μm raster intervals in X and Y and 20–25 % absolute laser power. Briefly, a photomicrograph of the sample to be imaged by mass spectrometry was loaded onto the Fleximaging command window. Three teach points were selected in order to align the background image with the sample target plate. After the target plate calibration was complete, the AutoXecute command was used to run the sample. The settings under the FlexControl panel were used as previously described.10
Image Visualization and Analysis
Using the Bruker FlexImaging 2.0 software, a window containing the m/z values corresponding to the palmyramide A (1) isotope cluster was selected and assigned a bright green color. It was clear that the spatial distribution of the specified mass window co-localized precisely with the filament contained in the teach image. These visual results were exported and overlaid on the epifluorescent images of the filament acquired before matrix application using Photoshop CS.
Biological Activity
Sodium channel blocking activity was measured as previously described.11 Neuro2a mouse neuroblastoma cells were seeded in 96-well plates at 3.0 × 105 cells/mL in 200 μL of RPMI medium with FBS. After 16 h, the test compounds were dissolved in DMSO and diluted into medium without FBS and then added to wells. A solution of 10 mM of ouabain and 1 mM of veratridine was applied to wells to cause sodium overload cytotoxicity. After 24 h, cell viability was determined by the MTT assay and sodium channel blocking activity was calculated as percentage of recovery from the toxicity of ouabain and veratridine.
Cytotoxicity was measured in NCI H-460 human lung tumor cells using the MTT assay.12 Cells were seeded in 96-well plates at 3.3 × 104 cells/mL in 180 μL of RPMI medium with FBS. After 16 h, the test compounds were dissolved in DMSO and diluted into medium without FBS and then added to wells. After 48 h, cell viability was determined by MTT assay.
Supplementary Material
Acknowledgments
We thank R. C. Coates for help with the collection of the organisms, J. Smith for identification of the red alga, Y. Su for accurate mass measurement, and T. L. Suyama and A. Pereira for helpful discussions. Financial support for this work was provided by NS053398.
Footnotes
Supporting Information Available: 1H NMR, 13C NMR, COSY, TOCSY, HSQC and HMBC spectra in CDCl3 for palmyramide A (1). This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.Tan LT. Phytochemistry. 2007;68:954–979. doi: 10.1016/j.phytochem.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 2.Tidgewell K, Clark BR, Gerwick WH. Comprehensive Natural Products Chemistry. Vol. 8. Pergamon Press; 2009. in press. [Google Scholar]
- 3.Dinsdale EA, Pantos O, Smriga S, Edwards RA, Angly F, Wegley L, Hatay M, Hall D, Brown E, Haynes M, Krause L, Sala E, Sandin SA, Thurber RV, Willis BL, Azam F, Knowlton N, Rohwer F. PLoS ONE. 2008;3:e1584. doi: 10.1371/journal.pone.0001584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marquez B, Verdier-Pinard P, Hamel E, Gerwick WH. Phytochemistry. 1998;49:2387–2389. doi: 10.1016/s0031-9422(98)00365-3. [DOI] [PubMed] [Google Scholar]
- 5.Ainslie RD, Barchi JJ, Kuniyoshi M, Moore RE, Mynderse JS. J Org Chem. 1985;50:2859–2862. [Google Scholar]
- 6.Marfey P. Carlsberg Res Commun. 1984;49:591–596. [Google Scholar]
- 7.Nunnery JK, Suyama TL, Gerwick WH. manuscript in preparation. [Google Scholar]
- 8.Hoye TR, Jeffrey CS, Shao F. Nature Protocols. 2007;2:2451–2458. doi: 10.1038/nprot.2007.354. [DOI] [PubMed] [Google Scholar]
- 9.Simmons TL, Coates RC, Clark BR, Engene N, Gonzalez D, Esquenazi E, Dorrestein PC, Gerwick WH. Proc Natl Acad Sci U S A. 2008;105:4587–4594. doi: 10.1073/pnas.0709851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esquenazi E, Coates C, Simmons L, Gonzalez D, Gerwick WH, Dorrestein PC. Mol Biosyst. 2008;4:562–570. doi: 10.1039/b720018h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manger RL, Leja LS, Lee SY, Hungerford JM, Wekell MM. Anal Biochem. 1993;214:190–194. doi: 10.1006/abio.1993.1476. [DOI] [PubMed] [Google Scholar]
- 12.Alley MC, Scudiero DA, Monks A, Hursey ML, Czerwinski MJ, Fine DL, Abbott BL, Mayo JG, Shoemaker RH, Boyd MR. Cancer Res. 1988;38:589–601. [PubMed] [Google Scholar]
- 13.Tan LT, Sitachitta N, Gerwick WH. J Nat Prod. 2003;66:764–771. doi: 10.1021/np020492o. [DOI] [PubMed] [Google Scholar]
- 14.Castenholz RW. Bergey’s Manual of Systematic Bacteriology. Vol. 1. Springer; New York: 2001. pp. 473–553. [Google Scholar]
- 15.Komárek J. Süsswasserflora von Mitteleuropa. Elsevier/Spektrum; Heidelberg: 2005. K. Anagnostidis; pp. 1–759. [Google Scholar]
- 16.Nübel U, Garcia-Pichel F, Muyzer G. Appl Environ Microbiol. 1997;63:3327–3332. doi: 10.1128/aem.63.8.3327-3332.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rivas R, Velázquez E, Zurdo-Piñeiro JL, Mateos PF, Molina EM. J Microbiol Methods. 2003;56:413–426. doi: 10.1016/j.mimet.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 18.Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman A. Appl Env Microbiol. 2005;71:7724–7736. doi: 10.1128/AEM.71.12.7724-7736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamura K, Dudley J, Nei M, Kumar S. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 20.Milne I, Lindner D, Bayer M, Husmeier D, McGuire G, Marshall DF, Wright F. Bioinformatics. 2009;25:126–127. doi: 10.1093/bioinformatics/btn575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nei M, Kumar S. Molecular Evolution and Phylogenetics. Oxford University Press; New York: 2000. pp. 99–108. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.