Abstract
The monopolar spindle-one-binder (Mob) family of kinase-interacting proteins regulate cell cycle and cell morphology, and their dysfunction has been linked to cancer. Models for Mob function are primarily based on studies of Mob1 and Mob2 family members in yeast. In contrast, the function of the highly conserved metazoan Phocein/Mob3 subfamily is unknown. We identified the Drosophila Phocein homolog (DMob4) as a regulator of neurite branching in a genome-wide RNA interference screen for neuronal morphology mutants. To further characterize DMob4, we generated null and hypomorphic alleles and performed in vivo cell biological and physiological analysis. We find that DMob4 plays a prominent role in neural function, regulating axonal transport, membrane excitability, and organization of microtubule networks. DMob4 mutant neuromuscular synapses also show a profound overgrowth of synaptic boutons, similar to known Drosophila endocytotic mutants. DMob4 and human Phocein are >80% identical, and the lethality of DMob4 mutants can be rescued by a human phocein transgene, indicating a conservation of function across evolution. These findings suggest a novel role for Phocein proteins in the regulation of axonal transport, neurite elongation, synapse formation, and microtubule organization.
Introduction
The monopolar spindle-one-binder (Mob) family of zinc binding proteins is conserved from yeast to humans (Stavridi et al., 2003; Ponchon et al., 2004; Li et al., 2006; Mrkobrada et al., 2006). Mobs are found in complexes with kinases and may function as activating subunits, similar to cyclins (Weiss et al., 2002; Devroe et al., 2004; Hergovich et al., 2005; Wei et al., 2007). The founding Mob members, Saccharomyces cerevisiae Mob1p and Mob2p, activate Dbf-2 kinases and facilitate exit from mitosis (Luca et al., 2001; Weiss et al., 2002; Stoepel et al., 2005; Mrkobrada et al., 2006). Mob isoforms have also been localized to neuronal dendrites in mammals, in which NDR (nuclear Dbf-2) kinases regulate dendritic branching (Zallen et al., 2000; Baillat et al., 2001; He et al., 2005; Emoto et al., 2006). These findings suggest that Mob proteins may have important functions within postmitotic neurons, in addition to regulating cell proliferation.
Whereas S. cerevisiae has two Mobs, Drosophila melanogaster, Caenorhabditis elegans, and Danio rerio have four, and Homo sapiens has seven (Li et al., 2006; Mrkobrada et al., 2006). Although roles in regulation of cell proliferation by Mob1 and cell polarity by Mob2 have been characterized, little is known about the third subfamily, Phocein/Mob3 (Colman-Lerner et al., 2001; Hou et al., 2003; Lai et al., 2005; Wei et al., 2007; Praskova et al., 2008). Phocein is highly expressed in Purkinje cell dendrites and spines (Baillat et al., 2001; Haeberlé et al., 2006). Biochemical studies indicate that Phocein interacts with various vesicle trafficking proteins, including Striatin, Eps-15, Dynamin-1, and Nucleoside-Diphosphate Kinase (NDPK) (Baillat et al., 2001, 2002). However, no loss-of-function analysis of Phocein has been conducted. In vivo knockdown of Striatin blocks dendrite formation, suggesting that the Phocein/Striatin complex may regulate neuronal morphology (Bartoli et al., 1999).
Phocein is also a component of the STRIPAK complex (for Striatin interacting phosphatase and kinase), which contains protein phosphatase 2A (PP2A), Ste-kinases, Dynein, and several other interactors (Goudreault et al., 2008). The presence of serine–threonine kinases and phosphatases in the STRIPAK complex suggests that it may act as an on/off switch that governs target phosphorylation states (Pi and Lisman, 2008). In neurons, PP2A associates with microtubules and regulates their dynamics by altering microtubule-associated protein (MAP) phosphorylation (Sontag et al., 1996; Tournebize et al., 1997; Aranda-Orgillés et al., 2008). These interactions suggest that the STRIPAK/PP2A complex may regulate neuronal microtubules through interactions with a variety of effector proteins, including Phocein. As such, Phocein may regulate a host of neuronal functions in vivo.
Here we describe the first in vivo knock-out analysis of phocein/DMob4 (hereafter referred to as DMob4) in metazoans. Within neurons, we find that DMob4 is required for axonal transport, synaptic development, and normal organization of microtubule networks. Genetic rescue of DMob4 mutant phenotypes with human phocein suggests a conserved role of this protein family in neurons. These findings highlight a new function for the metazoan-specific Phocein/Mob3 subfamily in the regulation of neuronal structure and function.
Materials and Methods
DMob4 mutant strains.
To isolate DMob4 (CG3403) mutant alleles, a P-element excision screen was conducted using y1,w67c23;P[EPgy2] EY23407 and the transposase source y1,w1118; amosTft/CyO, PBac[Δ2-3.Exel]2. EY23407 is a P-element insertion in the 5′ untranslated region (UTR) of DMob4 (in exon 1), and it is 33 bp upstream of the DMob4 start codon, as confirmed by inverse PCR and sequencing of the P-element ends. Putative w;EYΔ imprecise excision alleles arising from the screen were balanced over CyO and scored for homozygous lethality. A subsequent round of PCR screening of the homozygous lethal lines with oligonucleotides spanning the EY23407 insertion site (Pr3cdna forward, 5′ TCAGTCTAGAGCATACTACGGTGCAGGGAT; Pr3cdna reverse, 5′ TCGAGAATTCACGACTCCTTGATGGACACC) identified strains containing small deletions of the DMob4 locus. Using these oligonucleotide pairs, a 729 bp band is obtained in wild types. Lethal stocks that yielded PCR products of <729 bp were identified as being excision mutants. These bands were analyzed by DNA sequencing to map the excision breakpoints. DMob4EYΔL307 contains a 116 bp deletion starting at −33 and is missing part of exon 1 and the downstream intron; at the former EY23407 insertion site, a small 17 bp footprint of EY23407 remains (CATGATGAAATAA). The DMob4EYΔL3 allele contains a 357 bp deletion with the 5′ breakpoint also at −33. This deletion removes all of exon 1, the intervening intron, and part of exon 2. The EYΔHV1223 strain is a homozygous viable precise excision allele of y1,w67c23;P[EPgy2]EY23407.
DMob4 rescue strains and analysis.
The Drosophila DMob4 cDNA LD21194 (Drosophila Genomics Resource Center, Indiana University, Bloomington, IN) was PCR amplified with BglII/SpeI-tailed oligonucleotides (5′GAGATCTCATCGCGTCTTCGAGCGG/ 5′CACTAGTCATGCGAGTCTGACTAATCT) flanking the open reading frame of DMob4 and then directionally subcloned as a BglII/SpeI fragment into the pValum vector system (for vermillion attB LoxP UAS MCS), for the generation of transgenic lines (Markstein et al., 2008; Ni et al., 2008). For the human phocein rescue construct, we repeated the same strategy as for DMob4. The human phocein variant-1 (GenBank accession number NM_015387.2) cDNA clone (OriGene Technologies) was used to make the rescue construct, and the human phocein-specific BglII/SpeI-tailed oligonucleotides (5′GAGATCTACATCCGGGTACCGACTCCAG and 5′CACTAGTTGTTAATTATATGATCAGTAC) were used for PCR. DNA for the rescue constructs was sequenced to identify clones without polymerase errors; constructs were injected into an attP2 strain containing a source of phiC31-integrase (Genetic Services). To conduct the rescue experiments, yw;DMob4EYΔL3,actin-GAL4/CyO males were crossed to either yw;DMob4EYΔL3/CyO;UAS–DMob4,v+,attP2,y+ or yw;DMob4EYΔL3/CyO;UAS–human phocein variant-1,v+,attP2,y+ females at 21°C, and the progeny were examined for non-CyO escaper flies. The presence of the rescue transgene in the escapers was also confirmed with PCR using the BglII/SpeI primer pairs used in the generation of the rescue constructs.
Other Drosophila strains.
The P[EPgy2]EY23407 strain was obtained from Hugo Bellen (Baylor College of Medicine, Houston, TX). The Rab2C02699 stock was obtained from the Exelixis collection at Harvard Medical School (Boston, MA). The galactosidase-4 (GAL4) drivers used for rescue experiments [actin–GAL4, myosin heavy chain (MHC)–Gal4, and elavc155] were obtained from the Bloomington Drosophila Stock Center (Indiana University, Bloomington, IN), as was the Df(2R)42 strain.
Drosophila cell culture and morphology analysis.
The fly stock elavc155–GAL4, UAS–mCD8GFP/FM6;gcm–GAL4/CyO was used for the preparation of primary cell cultures for the DMob4 RNA interference (RNAi) experiments. RNAi knockdown of DMob4 was achieved using amplicons DRSC04993 and DRSC29567 (Drosophila RNAi Screening Center database at Harvard Medical School). DRSC4993 is a 369 bp amplicon directed against exon 2 of DMob4, whereas DRSC29567 is a 364 bp amplicon directed against exon 3 (see Fig. 2B). There are no predicted off-target fragments within the DRSC29567 amplicon and a single predicted off-target fragment for the DRSC4993 amplicon (matching a portion of Drosophila gene CG31107). The slight difference in the morphology phenotypes of the two Mob4-directed amplicons may be related to off-target effects of DRSC4993 (DRSC4993 vs DRSC29567, p = 0.002) (see Fig. 1). For double-stranded RNA (dsRNA) production, T7-tailed oligonucleotides (DRSC04993: 5′TAATACGACTCACTATAGGGAGATCTGTCGCTGGCCCG/5′TAATACGACTCACTATAGGGAGATGCTAGGGAAGTACTTGTTG; DRSC29567 5′TAATACGACTCACTATAGGGAGATGTGGAAGTACGAGCACCTG/ 5′TAATACGACTCACTATAGGGAGATGCGAGAAGATGCGATACAC) were used to PCR amplify the DRSC amplicons from w1118 genomic DNA. dsRNA was then synthesized from the T7-tailed amplicons using the MEGAshortscript T7 In Vitro Transcription kit according to the protocol of the manufacturer (Ambion). Synthesized dsRNAs were purified with RNeasy kits before use in cell culture experiments (Qiagen). The primary cultures, RNAi experiments, and morphometric analysis were all conducted as described previously (Sepp et al., 2008). For image analysis, we used custom algorithms that are described previously (Sepp et al., 2008). For each amplicon, we treated 72 wells and imaged four sites per well with an ImagExpress Micro automated microscope (Molecular Devices). To verify that dsRNA amplicons knock down DMob4 expression, 1 × 106 S2 cells were plated in serum-free media and treated with 15 μg of dsRNA for 30 min. Cells were then diluted 1:3 with serum-containing media and incubated for 5 d before Western blot analysis.
Figure 2.
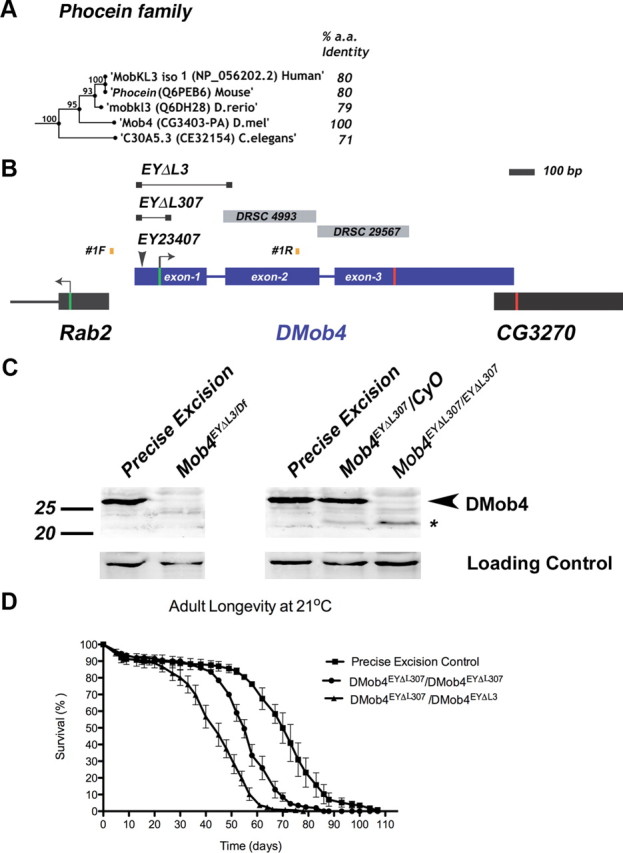
Molecular analysis of DMob4. A, Bootstrapping phylogenetic tree of the Phocein subfamily. The percentage amino acid conservation between DMob4 and Phocein proteins is noted to the right. Bootstrap scores are given at internodes. B, The DMob4 locus at band 42C5 on chromosome 2R is shown to scale. DMob4 (blue) has three exons (thick blue lines) that span ∼1.5 kb, including introns (thin blue lines). The ATG start codon is illustrated with a green bar in exon 1, whereas the stop codon is indicated with a red bar in exon 3. The breakpoints of the P-element excision mutants DMob4EYΔL3 and DMob4EYΔL307 are indicated (thin black lines with boxed ends). The insertion site of the progenitor P-element EY23407 is noted, as are the position of the oligonucleotides #1F and #1R used to identify excision mutants. Gray boxes DRSC4993 and DRSC29567 are dsRNA amplicons used for RNAi cell culture experiments. Rab2 and CG3270 flank the DMob4 locus. C, Western blot analysis of DMob4EYΔL3 and DMob4EYΔL307 excision mutants. The strain EYΔHV1223 was used as the precise excision control. Df refers to Df(2R)42, which is a small chromosomal deletion that spans the DMob4 locus. The expected molecular weight for DMob4 is 25.7 kDa. Dlg was used as a loading control. D, Adult longevity of the DMob4EYΔL307 hypomorphic excision mutant, DMob4EYΔL307/DMob4EYΔL3 transheterozygous deletions mutant, and precise excision control.
Figure 1.
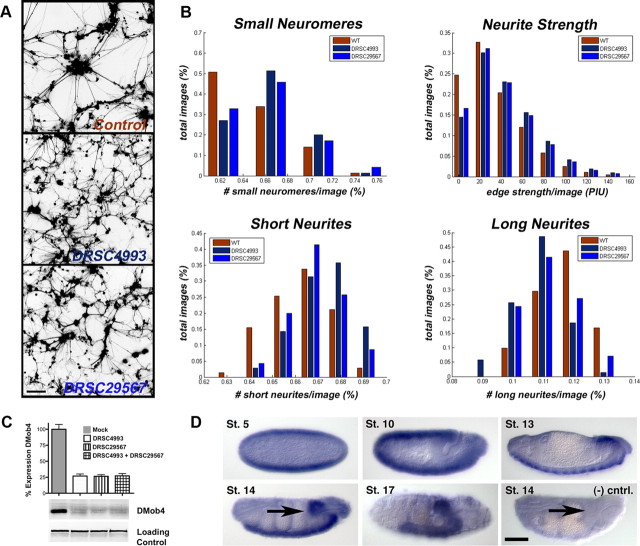
RNAi knockdown of DMob4 in primary cultures alters neurite morphology. A, Fluorescent microscopy images of Drosophila primary neural cultures derived from embryos expressing GFP. Cultures were either untreated (control) or subjected to DMob4 RNAi using DMob4-specific dsRNA amplicons (DRSC4993 and DRSC29567). B, Quantification of DMob4 RNAi phenotypes in primary neural cultures. Custom algorithms were used to extract and quantify image features from automated microscopy digital files (n = 192 per treatment group). For small neuromeres, a threshold of Loge area ≤5.6 μm2 was set. The number of small neuromeres per image was tabulated as a percentage, and images were binned into groups as displayed in the histogram (bins: 58–67, 67–70, 71–75, or 75–79%). For short neurites, a threshold was set at ≥6.5 and ≤16.1 μm (bins: 63–65, 65–67, 67–68, 68–69, and 69–70%). For long neurites, a threshold was set at ≥25.8 μm (bins: 8.9–9.9, 9.9–10.1, 10.1–12, 12–13, and 13–15%). For neurite strength, representing neurite thickness, bins were set at increments of 31.4 pixel intensity units (PIU). Analysis using all image features indicated that the control is statistically different from treatment groups (control vs DRSC4993, p = 2.22e−16; control vs DRSC29567, p = 1.41e−6), and the two treatment groups are also statistically different from each other (p = 0.002). WT, Wild type. C, Western blot analysis of cell cultures treated with DMob4 dsRNA amplicons. DMob4 protein levels are decreased by ∼75% with DRSC4993 or DRSC29567. No apparent increase in DMob4 knockdown is achieved using a combination of the two amplicons (n = 8). Dlg was used as a loading control. D, DMob4 mRNA in situ hybridization to embryos with DMob4 antisense transcript (blue). DMob4 expression is abundant in early embryos at stage 5 and widely expressed until stage 13, when it becomes enriched in the ventral nerve cord (arrow, stage 14). DMob4 negative control (sense transcript) fails to label the ventral nerve cord [arrow, stage 14, (−) cntrl]. Anterior is to the right, and ventral is down. Scale bars, 100 μm.
Generation of DMob4 antibody.
Full-length DMob4 was PCR amplified from the expressed sequence tag (EST) LD21194 (Drosophila Genomics Resource Center) using XhoI and KpnI tailed oligonucleotides (5′ primer, CCTCGAGATGAAGATGGCTGACGGCTCG; 3′ primer, GGTACCCTAAGCCTCGCTTTCGCCAGGG) and subcloned into the pGEM-T Easy vector system (Promega). DMob4 clones were sequence verified and subcloned into pTrcHis A (Invitrogen) as an XhoI/KpnI fragment for the production of N-terminal 6× His-tagged protein. His-tagged DMob4 was purified from BL-21 (DE3) Escherichia coli with a 1.0 ml HisTrapHP nickel column (GE Healthcare) and was injected into guinea pigs (Invitrogen) for antibody production. For downstream applications, the DMob4 antibody was affinity purified on HiTrap NHS-activated HP columns (1.0 ml) according to the specifications of the manufacturer.
RNA in situ hybridizations.
In situ hybridization studies of DMob4 in embryos were performed as described previously (Kearney et al., 2004). To produce a linearized template for digoxigenin (DIG)-labeled RNA probe synthesis, the full-length DMob4 EST LD21194 (Drosophila Genomics Resource Center) was PCR amplified, with oligonucleotides specific to the pOT2a backbone vector (OT2A forward, 5′GAACGCGGCTACAATTAATACA; OT2A reverse, 5′GCCGATTCATTAATGCAGGT). The resultant PCR product contains pOT2A-derived T7 and SP6 RNA polymerase initiation sites to enable in vitro antisense and sense (respectively) probe transcription. For DIG-labeled RNA probe synthesis and in situ analysis, the following reagents were used: proteinase K (Roche Molecular Biochemicals), DIG RNA Labeling kit (Sp6/T7) (Roche Molecular Biochemicals), T7 and SP6 RNA polymerase (New England Biolabs), anti-digoxigenin–alkaline phosphatase Fab fragments used at 1:2000 (Roche Molecular Biochemicals), and 5-bromo-4-chlor-indolyl-phosphate/nitroblue-tetrazolium-chloride color development substrate (Promega).
Immunohistochemistry.
Staining of embryos and larvae was performed according to standard protocols (Rothwell and Sullivan, 1999). In brief, embryos were fixed in PEM (0.1 m PIPES, 2 mm EGTA, 1 mm MgS04, pH 6.95) buffer and 4% formaldehyde for 20 min and washed/permeabilized in PBS plus 0.05% Tween 20. Larvae were reared at 21°C, and wandering third instars were dissected in calcium-free HL3.1 saline (Feng et al., 2004), and the larvae were fixed for 30 min in HL3.1 and 4% formaldehyde and washed/permeabilized in PBS and 0.1% Triton X-100. To obtain third-instar DMob4EYΔL3/Df(2R)42 mutants, homozygous mutants were collected as first instars from standard media vials and then nursed to the third-instar stage on molasses–agar plates containing semi-liquid yeast paste (Red Star inactive) supplemented with cold temperature-treated mashed banana to 50% and penicillin–streptomycin to a final concentration of 50 IU/ml and 50 μg/ml, respectively (Mediatech). Mutant larvae were transferred to fresh yeast/banana/penicillin–streptomycin media daily until they attained the third-instar stage. For neuromuscular junction (NMJ) morphology analysis, supernumerary boutons were defined as strings of five or fewer boutons that extend from the central NMJ axis. The following antibodies and concentrations were used: 22C10 mouse monoclonal antibody (mAb) at 1:5 (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA), mouse anti-acetylated α-Tubulin clone 6-11B-1 at 1:250 (T7451; Sigma-Aldrich), mouse anti-tyrosinated α-Tubulin TUB-1A2 at 1:1000 (T9028; Sigma-Aldrich), guinea pig anti-DMob4 at 1:600, rabbit anti-HRP rhodamine at 1:150 (Jackson ImmunoResearch), nc82 at 1:200 (Developmental Studies Hybridoma Bank), and rabbit anti-synaptotagmin-1 (Syt-1) at 1:500 (Littleton et al., 1993). Alexa Fluor 488- and Alexa Fluor 594-conjugated secondary antibodies were used at 1:400 (Invitrogen). Tissue preparations were imaged on a Leica TCS-SP2 confocal LSM microscope.
Western blot analysis and biochemical experiments.
For standard SDS-PAGE/Western blot analysis, 1.5 larvae per lane were loaded onto gels. For quantitative Western blot analysis, total protein levels were determined in lysates (10 larvae/100 μl lysis buffer: 20 mm HEPES, pH 7.5, 50 mm KCl, 2 mm EGTA, 1 mm MgCl2, and 0.2% NP-40 plus protease inhibitors) using a BCA protein assay kit (Pierce), and then 6.5 μg/lane was loaded onto gels. To reduce background bands on Western blots, the viscera of the third-instar larvae were removed before homogenization in sample buffer (standard SDS-PAGE) or lysis buffer (quantitative SDS-PAGE). For immunoblotting, antibodies were used at the following concentrations: mouse anti-Discs large (Dlg) (4F3; Developmental Studies Hybridoma Bank) at 1:1000, guinea pig anti-DMob4 at 1:2000, rabbit anti-Synaptogyrin at 1:10,000, mouse anti-α-Tubulin clone B-5-1-2 at 1:5000 (T5168; Sigma-Aldrich), and IR700- and IR800-conjugated secondary antibodies at 1:3000 (Rockland Immunochemicals). Western blots were imaged and quantified using an Odyssey infrared scanner (Li-Cor).
Adult behavioral analysis.
Viability assays were performed at 21°C. Virgin male and female Drosophila were collected in separate vials (15 flies per vial) and flipped onto fresh media three times per week. The number of deceased animals was noted at the time of each flip. Longevity plots were analyzed separately for the males and females of a given strain and then data were pooled, because there was no statistically significant difference between sexes. The numbers of flies used to obtain the longevity scores are as follows: DMob4EYΔL307/DMob4EYΔL307 (male, n = 520; female, n = 582), DMob4EYΔL307/DMob4EYΔL3 (male, n = 211; female, n = 286), and EYΔHV1223 (male, n = 507; female, n = 589).
Electron microscopy.
Wandering third-instar wild-type and DMob4 mutant larvae were dissected in calcium-free HL3.1 saline (Feng et al., 2004) and fixed in 1% glutaraldehyde and 0.2 m sodium phosphate buffer, pH 7.2, for 30 min. The larvae were washed in 0.2 m sodium phosphate buffer and then incubated for 1 h in 1% OsO4 in ddH2O. Osmium was removed, and larvae were washed in ddH20. The preparations were stained en bloc for 0.5 h in freshly prepared 2% uranyl acetate, washed in ddH20, and dehydrated in an increasing ethanol series and finally in propylene oxide; they were infiltrated with resin by incubation for 30 min in 1:3 ratio of propylene oxide/Araldite and overnight in pure Araldite. The larvae were embedded in fresh Araldite, and the resin was cured at 60°C for 24 h. All fixation and dehydration steps were conducted at room temperature. Ultrathin sections were imaged with a FEI Tecnai Spirit BioTwin transmission electron microscope (TEM) fitted with an Advanced Microscopy Techniques digital camera (Whitehead Institute, Massachusetts Institute of Technology, Cambridge, MA).
Electrophysiology.
Evoked postsynaptic currents were recorded from ventral longitudinal muscle 6 at abdominal segment A3 in third-instar larvae using two-microelectrode voltage clamp (OC725; Warner Instruments) at −80 mV holding potential (Acharya et al., 2006). All experiments were performed in modified HL3 solution composed of the following (in mm): 70 NaCl, 5 KCl, 4 MgCl2, 2 CaCl2, 10 NaHCO3, 115 sucrose, and 5 HEPES-Na, pH 7.2. Data acquisition and analysis were performed using pClamp software (Molecular Devices). For stimulation, nerves were cut close to the ventral ganglion and sucked into a pipette filled with working solution. The nerve was stimulated at frequencies indicated in each experiment using a programmable pulse stimulator (Master-8; A.M.P.I.) with a 0.1 ms duration. Nerve threshold for evoked release was estimated in each experiment (minimal stimulation), and the intensity of stimulation was increased twofold (maximal stimulation).
Results
DMob4 regulates neurite outgrowth in Drosophila primary cultures
We identified DMob4 (CG3403) in a genome-wide RNAi screen for genes required for neurite outgrowth or regulation of neuronal morphology (Sepp et al., 2008). We selected DMob4 for additional characterization because of its homology with other Mob proteins and its potential role in the regulation of endocytosis and vesicular trafficking with neurons (supplemental Fig. S1b, available at www.jneurosci.org as supplemental material). To confirm that DMob4 regulates neuronal morphology, we performed RNAi knockdown experiments in primary cultures prepared from Drosophila embryos in which green fluorescent protein (GFP) is expressed under the control of the pan-neuronal driver elav–Gal4. RNAi knockdown of DMob4 with two independent, nonoverlapping dsRNA amplicons, DRSC04993 and DRSC29567, altered neurite morphology relative to controls (control vs DRSC4993, p = 2.22e−16; control vs DRSC29567, p = 1.41e−6) (Fig. 1A,B). In control cultures, neuroblasts proliferate into multicell aggregates called neuromeres. Long and straight neurites typically extend from neuromere clusters as the cultures mature. DMob4 knockdown resulted in a disruption of neuronal morphology that was quantified with custom algorithms. Loss of DMob4 decreased the size of neuromere clusters but increased the complexity of neurite branching with shorter neurites and an increased thickness of neurite bundles (Fig. 1B). The two DMob4 dsRNA amplicons target distinct regions of DMob4 and are equally effective at knocking down DMob4 protein levels (Figs. 1C, 2B). The RNAi results indicate that reduced DMob4 levels within developing neurons causes defects in neurite branching and morphology in Drosophila primary neurons.
To analyze DMob4 expression within the nervous system, we performed in situ hybridization on Drosophila embryos using digoxigenin-labeled RNA probes (Fig. 1D). DMob4 transcripts were dynamically distributed in the embryo, with abundant maternal DMob4 expression in eggs and enrichment of zygotic transcripts in the CNS during development. Beginning at stage 10, DMob4 is present in the ventral neurogenic region and is abundant in the condensed ventral nerve cord by stage 17 (Fig. 1D, arrow). This staining pattern was not observed in embryos probed with negative control sense probes. Therefore, DMob4 is expressed widely in developing embryos and enriched in the nervous system.
DMob4 is a Phocein homolog
Crystal structures of yeast, Xenopus, and human Mob1 family members indicate that Mob proteins are composed of several α helical domains coordinated around a central zinc ion (supplemental Fig. S1A, available at www.jneurosci.org as supplemental material). In Drosophila, there are four Mob proteins: Mob1/mats (CG13852), Mob2 (CG11711), Mob3 (CG4946), and Mob4 (CG3403). Sequence alignment of the Drosophila Mobs with human Mob1 (Q9H8S9.4) demonstrates that the two histidine and two cysteine residues necessary for zinc coordination are conserved, suggesting that DMobs have a similar three-dimensional structure (supplemental Fig. S1B, available at www.jneurosci.org as supplemental material). Of the Drosophila Mobs, Mob1, Mob3, and Mob4 are each predicted to produce a single transcript encoding an ∼25 kDa polypeptide. Mob2 produces four transcripts encoding proteins of larger size (i.e., 78.3, 57.2, 45.8, and 58.9 kDa). DMob4 has a unique N-terminal domain and extensive sequence divergence across its entire length compared with other DMobs (supplemental Fig. S1B, available at www.jneurosci.org as supplemental material). Reciprocal-best basic local alignment search tool analysis indicates that DMob4 is the sole Drosophila homolog of vertebrate Phoceins, which constitute a subfamily of the Mob proteins. Rat Phocein, the first characterized member of this subfamily, is highly expressed in Purkinje cells of the cerebellum, which have elaborate dendritic trees. DMob4 shares ∼80% identity with mouse, rat, and human Phoceins (Fig. 2A).
Generation of DMob4 mutant alleles
The Drosophila Mob4 locus resides at cytological band 42C5 and spans 1.5 kb. DMob4 is closely flanked by Rab2 and CG3270 (Fig. 2B). To further characterize the role of DMob4, we generated null mutations in the locus using a P-element excision screen with the EY23407 strain, which contains a transposable element inserted into the 5′ UTR of DMob4. We mapped the EY23407 insertion site to 33 bp upstream of the DMob4 translation start codon by plasmid rescue. This line was designated DMob4EY23407. The DMob4EY23407 strain is homozygous lethal. Because the insertion does not disrupt the DMob4 open reading frame, we conducted an excision screen with DMob4EY23407 to generate definitive null alleles. Because the locus is compact, with Rab2 and CG3270 within a few hundred base pairs flanking DMob4 (Fig. 2B), we generated small deletions that had breakpoints entirely within the DMob4 gene. We screened 1329 white-eyed DMob4EY23407Δ/CyO excision strains for lethality (lines were designated EYΔL#) and used PCR to assay for desired deletions. We isolated two novel DMob4 excision mutants: DMob4EYΔL3 and DMob4EYΔL307 (Fig. 2B). Both excision lines have 5′ breakpoints that begin 33 bp upstream of the DMob4 initiator methionine, at the original insertion site of the EY23407 element. DMob4EYΔL3 contains a 357 bp deletion, whereas DMob4EYΔL307 contains a 116 bp deletion. Both excision lines disrupt the initiator methionine of DMob4. We isolated a precise excision allele EYΔHV1223, which is homozygous viable, and served as a control for future experiments.
We next performed Western blot analysis to determine whether DMob4EYΔL3 and DMob4EYΔL307 deletions alter levels of DMob4 protein expression. For these studies, we generated a guinea pig polyclonal antibody against the full-length DMob4 protein, which is 223 aa (25.7 kDa). The antibody recognizes a ∼25 kDa band on Western blots of larval extracts prepared from control animals (precise excision allele DMob4EYΔHV1223), consistent with the predicted molecular weight of DMob4. No DMob4 protein was observed on Western blots of larval extracts prepared from homozygous DMob4EYΔL3 third-instar larvae (Fig. 2C) or from DMob4EY23407 (data not shown), indicating that these alleles are protein nulls. DMob4-specific bands were observed on Westerns blots of extracts prepared from embryo lysates of the same mutants, likely representing DMob4 protein translated from maternally deposited transcripts observed by mRNA in situ hybridization (Fig. 1D). Western blot analysis of third-instar larvae prepared from the DMob4EYΔL307 strain revealed that this mutant produces an N-terminal truncated DMob4 protein. Heterozygous DMob4EYΔL307/CyO animals produce two bands on Western blots: one at 25 kDa representing full-length DMob4 protein, and a smaller fragment at 22 kDa representing the truncated protein (Fig. 2C). Homozygous DMob4EYΔL307 animals only produce the smaller 22 kDa DMob4 protein product (Fig. 2C). Analysis of the DMob4EYΔL307 DNA sequence revealed an alternative in-frame initiator methionine in exon 2, downstream of the 3′ breakpoint of the EYΔL307 deletion. The predicted translational product of this truncated transcript is 32 aa shorter than the full-length 223 aa protein, with a molecular weight of 22 kDa, in agreement with the observed molecular weight of DMob4 in homozygous DMob4EYΔL307. Expression levels of the homozygous DMob4EYΔL307 mutants appear reduced relative to controls, suggesting that the deletion has less efficient transcription. Thus, DMob4EYΔL307 is a hypomorph and produces N-terminal truncated DMob4, whereas DMob4EYΔL3 and DMob4EY23407 are protein nulls.
Lethal phase analysis and rescue of DMob4 mutants
To determine the lethal phase of DMob4 mutant alleles, we monitored the survival rates of allelic combinations (Table 1). We used the deletion strain Df(2R)42 in cis- to the mutants to control for the possibility of second site mutations on the DMob4EYΔL chromosomes. Df(2R)42 has breakpoints at 42C2–42D3 and spans the DMob4 locus. Lethal phase analysis revealed that DMob4EYΔL3 homozygous mutants die mostly as first- and second-instar larvae, although ∼10% can survive to the third-instar stage. Approximately 35% of homozygous DMob4EY23407 animals survive to the third-instar stage. The least severe allele, the N-terminal truncation mutant DMob4EYΔL307, survives to the adult stage and can be maintained as a homozygous stock.
Table 1.
Lethal phase analysis of DMob4 allelic combinations at 21°C
| EYΔHV1223 (precise excision) | EY23407 (5′ UTR insertion) | DMob4EYΔL3 (null) | DMob4EYΔL307 (hypomorph) | Df(2R)42 | |
|---|---|---|---|---|---|
| EYΔHV1223 (precise excision) | Adult viable | Adult viable | Adult viable | Adult viable | Adult viable |
| EY23407 (5′ UTR insertion) | ∼25% develop to third-instar larvae | 15% develop to third-instar larvae; 3% pupate but do not eclose | Adult viable | 35% develop to third-instar larve; ∼10% pupate but do not eclose | |
| DMob4EYΔL3 (null) | <10% survive to 3rd I.S. Majority are 1st and 2nd I.S. lethal | Adult viable | <10% survive to third-instar larvae; the majority are first- and second-instar lethal | ||
| DMob4EYΔL307 (hypomorph) | Adult viable | Adult viable | |||
| Df(2R)42 | Embryonic lethal |
Because Rab2 and DMob4 lie head-to-head on chromosome 2R and their start codons are separated by only a few hundred base pairs, we wanted to ascertain that the lethality observed for DMob4 mutants was not attributable to a loss of Rab2 activity. We therefore crossed DMob4EY23407 and DMob4EYΔL3 to the Rab2c02699 null allele. Although Rab2c02699 is homozygous lethal, the DMob4EY23407/Rab2c02699 and DMob4EYΔL3/Rab2c02699 transheterozygotes are viable to adult stages and have no obvious morphological or behavioral defects, indicating that DMob4EY23407 and DMob4EYΔL3 lesions are specific to DMob4.
The N terminus of Mob proteins is solvent exposed and flexible and does not appear to be an integral part of the conserved globular core (Stavridi et al., 2003; Ponchon et al., 2004; Mrkobrada et al., 2006). The N terminus is conserved in the Phocein family and therefore may have critical functions. In yeast, x-ray crystal structural analysis of Mob1p suggests that the N terminus can interact with the C-terminal core domain and is necessary for homodimer formation. Because the DMob4EYΔL307 is adult viable, we extended our longevity analysis into adulthood to more closely examine the physiological consequences of loss of the DMob4 N terminus. We found that DMob4EYΔL307 mutants have significantly decreased adult longevity (Fig. 2D). The precise excision allele was used as a control for longevity studies. DMob4EYΔL307 homozygous animals had an ∼20% decrease in adult longevity, whereas DMob4EYΔL307/DMob4EYΔL3 mutants had an ∼38% decrease in adult longevity. Collectively, these studies suggest that the N-terminal 32 aa are necessary for complete functionality of DMob4.
To confirm that the DMob4 phenotypes we observed were attributable to loss of DMob4, we conducted rescue experiments using a UAS–DMob4 (wild-type) transgene (Table 2; plus sign (+) refers to wild-type chromosome). We also conducted rescue experiments using the human homolog of DMob4: Phocein/Mobkl3 variant-1, to test for evolutionary conservation of function. The human Phocein protein is 80% identical to DMob4 at the amino acid level (Fig. 2A). For rescue experiments, we used the GAL4/UAS system to drive expression of the wild type constructs in the DMob4EYΔL3 null mutant background. To control for differences in expression levels of the upstream activating sequence (UAS) transgenes, we used the attP–attB integration system and targeted the UAS–DMob4 and human UAS–phocein attB rescue constructs to the attP2 integration site on chromosome 3 (Markstein et al., 2008; Ni et al., 2008). Using the actin–Gal4 ubiquitous driver, we were able to rescue DMob4EYΔL3 larval lethality with both DMob4 and human phocein transgenes to a comparative level (Table 2). We conducted PCR analysis on rescued animals to ensure that they were homozygous for the DMob4EYΔL3 mutant chromosome (supplemental Fig. S2, available at www.jneurosci.org as supplemental material). Rescued animals were able to pupate and eclose as adults (Table 2), suggesting that human phocein and Drosophila Mob4 are functionally equivalent orthologs.
Table 2.
Rescue of DMob4EYΔL3 lethality using tissue-specific drivers and Drosophila or human transgenes
| Strain | Genotype | Second-instar larvae collected (n) | Stage achieved |
|
|---|---|---|---|---|
| Pupae | Adults | |||
| Precise excision control | 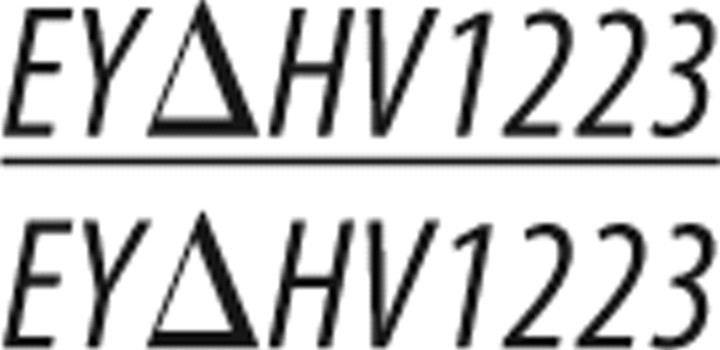 |
50 | 100% | 96% |
| DMob4 null | 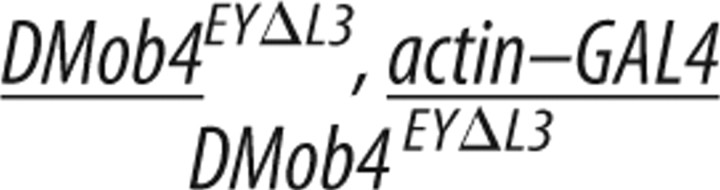 |
30 | 0% | 0% |
| Ubiquitous rescue (DMob4) | 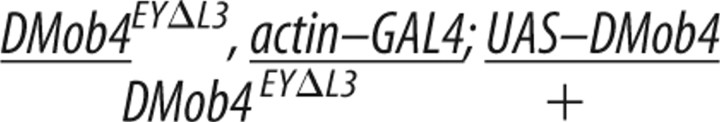 |
115 | 97% | 66% |
| Ubiquitous rescue (hPhocein) | 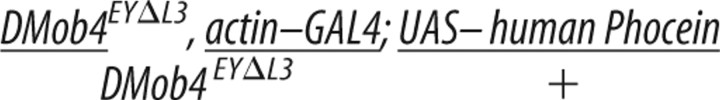 |
100 | 95% | 47% |
| Neuronal rescue (DMob4) | 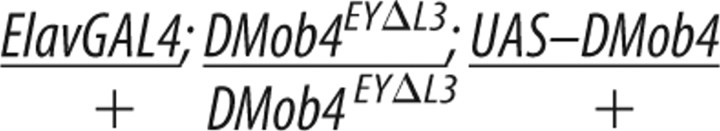 |
50 | 24% | 0% |
| Muscle rescue (DMob4) | 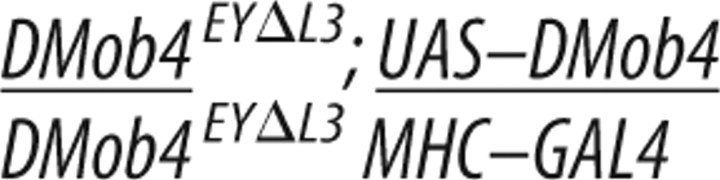 |
50 | 0% | 0% |
RNA in situ analysis revealed that DMob4 is expressed in the nervous system (Fig. 1D). We tested whether the observed DMob4EYΔL3 larval lethality could be solely attributed to loss of DMob4 function in the nervous system using rescue experiments with the nervous system-specific driver ElavC155. Driving expression of DMob4 in the nervous system of DMob4EYΔL3 null mutants partially rescued lethality. Twenty-four percent of ElavC155;UAS–DMob4 rescued null animals are able to pupate compared with 0% for mutants (Table 2). In contrast, expressing DMob4 in muscles using the MHC–GAL4 driver did not rescue the lethality. ElavC155;UAS–DMob4 rescued pupae failed to eclose, indicating that DMob4 also has essential functions outside the nervous system.
Tissue distribution and subcellular localization of DMob4
Anti-DMob4 antiserum was used for immunohistochemical analysis of embryos and larvae to examine the developmental expression of the protein. DMob4 is widely expressed in embryos and larvae. At the syncytial blastoderm stage, DMob4 staining is intense in the cell cortex, below the nuclei (supplemental Fig. S3, available at www.jneurosci.org as supplemental material). Zygotic genes are not yet active at this stage of development, suggesting that DMob4 protein, like the transcript, is maternally loaded into the egg. As cellularization advances, DMob4 localizes to the poles and mitotic spindles of dividing cells, together with a more widespread cytosolic distribution. In late-stage embryos, DMob4 is expressed in all tissues examined, including muscles, trachea, gut, peripheral glia, and neurons, with strong expression in the embryonic ventral nerve cord (supplemental Fig. S3, available at www.jneurosci.org as supplemental material). During larval development, the ubiquitous expression of DMob4 persists. At the NMJ, DMob4 fills synaptic boutons and surrounds nc82-positive active zones (Fig. 3A–C, concave arrowhead). This peri-active zone distribution is common for proteins implicated in endocytosis (Marie et al., 2004). In peripheral nerves, DMob4 is expressed at high levels in axons, but it is also present in the ensheathing glia (Fig. 3H). A fraction of DMob immunostaining in axons is punctate, suggesting that some DMob4 may be associated with vesicles undergoing transport. These staining patterns are abolished in null mutants (Fig. 3D–F), confirming specificity of the antisera.
Figure 3.
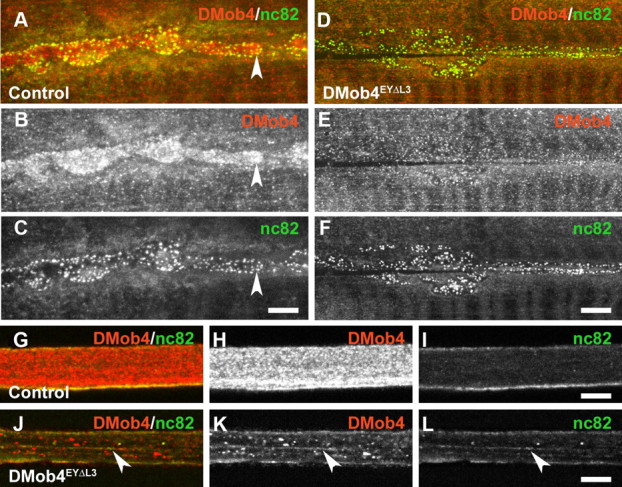
DMob4 expression in the peripheral nervous system of third-instar larvae. A–L, Confocal microscopy images of control (A–C, G–I) and DMob4EYΔL3 mutant (D–F, J–L) animals labeled with DMob4 antiserum (red) and counter-labeled with anti-bruchpilot (nc82) marker, which labels active zones at the NMJ and vesicular cargoes in peripheral nerves (green). Individual and merged confocal channels are presented as indicated. A–F, NMJs. DMob4 is present throughout synaptic boutons (B) and is enriched near active zones (concave arrowhead in A–C). DMob4 staining at the NMJ is absent in DMob4 mutants (E), indicating that NMJ immunoreactivity is specific to DMob4. Active zone staining persists in DMob4 mutants (F). G–L, Peripheral nerves. DMob4 is expressed at high levels in peripheral nerves and has both diffuse and punctate distribution characteristics (H). DMob4 staining of peripheral nerves is absent in DMob4 mutants (K), indicating that the DMob4 antiserum is specific. The distribution of nc82-positive vesicular cargo is altered in DMob4 mutants and not diffuse as observed in controls (arrowheads in J–L). Scale bars, 5 μm.
DMob4 mutants display synaptic overgrowth at the NMJ
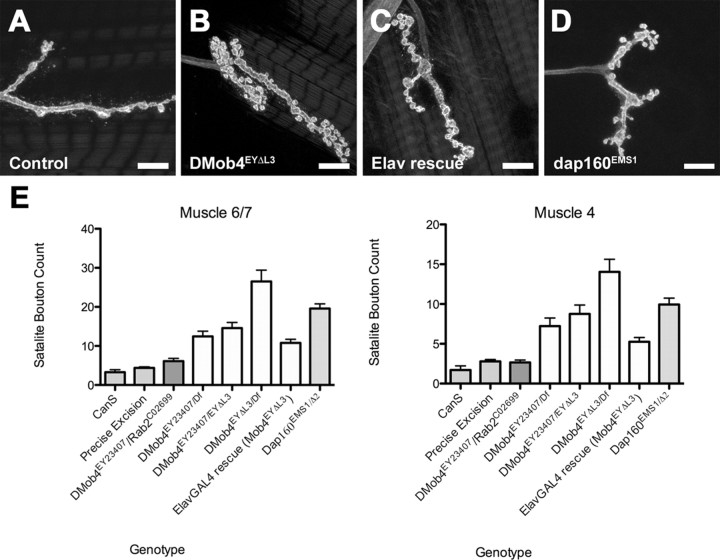
Given that RNAi knockdown of DMob4 in primary neural cultures results in excessive branching of neurite processes and that DMob4 is expressed at NMJs, we examined whether loss of the protein caused defects in synapse formation in vivo. We stained homozygous DMob4EYΔL3 and DMob4EY23407 larvae with the neuronal membrane marker anti-HRP to label NMJs. We observed a supernumerary bouton phenotype in the DMob4 mutants, similar to what has been reported previously for several endocytosis mutants, including Dap160 (Fig. 4A–D) (Koh et al., 2004; Dickman et al., 2006). We quantified the supernumerary bouton phenotype at muscle 6/7 and muscle 4 for various DMob4 allelic combinations and compared the phenotype with Dap160 endocytic mutants (Fig. 4E). At muscle 6/7 and muscle 4, DMob4EYΔL3 mutants have eightfold more supernumerary boutons than Canton S or the precise excision EYΔHV1223 animals, whereas Dap160 mutants have sixfold more. We also quantified the frequency of supernumerary boutons in Rab2c02699/DMob4EY23407 transheterozygotes and found no difference from controls, confirming that the observed supernumerary bouton phenotypes are specific to a loss of DMob4. To determine whether DMob4 acts cell autonomously in neurons to regulate synapse formation, we examined DMob4EYΔL3 homozygous mutants carrying the elavGAL4;UAS–DMob4 rescue transgene. Presynaptic expression of DMob4 partially suppresses the supernumerary bouton phenotype observed in nulls. The number of supernumerary boutons in the nulls is reduced by 2.6-fold when DMob4 is driven in the nervous system (Fig. 4B,C,E). This data indicates that DMob4 functions presynaptically to regulate normal synapse formation.
Figure 4.
DMob4 mutant third-instar larvae have supernumerary boutons at NMJs. NMJ morphology revealed by anti-HRP staining is shown for control (A), DMob4EYΔL3/Df(2R)42 (B), ElavGal4;DMob4EYΔL3;UAS–DMob4 rescue (C), and Dap160EMS1/Δ2 mutants (D). E, Mean satellite bouton number at muscle 6/7 and muscle 4 for various DMob4 allelic combinations and controls. Error bars are SEM. Scale bars, 10 μm.
Electron microscopy of DMob4 mutants
Because supernumerary boutons are a phenotype observed in several endocytic mutants and rat Phocein interacts with endocytosis/vesicle trafficking proteins (Eps-15, NDPK, and Dynamin-I), we performed electron microscopy on DMob4 nulls to investigate whether there are any anomalies in endocytosis and/or vesicle trafficking in the mutants. We focused on the NMJ because it is a region of active endocytosis of synaptic vesicles and DMob4 is localized in synaptic boutons. Similarly, we examined axons of peripheral nerves because vesicular cargoes are transported long distances along elaborate microtubule networks and Mob4 is highly expressed in axons. At the NMJ of DMob4EYΔL3 mutants, we observed the presence of multiple abnormal cisternae-like endocytic structures budding from the plasma membrane of presynaptic terminals (Fig. 5B–D). Such cisternae were absent from control animals (Fig. 5A), consistent with a requirement for DMob4 function in synaptic endocytosis. In peripheral nerves of DMob4 mutants, we observed accumulations of microtubule-associated vesicles with larger diameters than in controls (Fig. 5E–H). This may reflect the transport of large abnormal endocytic cisternae or perturbed axonal transport.
Figure 5.
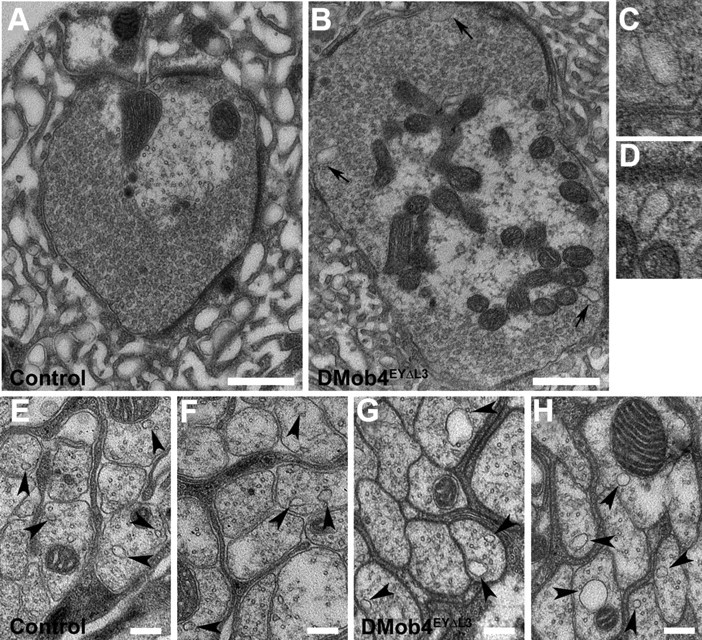
Transmission electron microscopy of DMob4 mutant NMJs and peripheral nerves. Control (A, E, F) and DMob4EYΔL3/Df(2R)42 mutant (B–D, G, H) NMJs. Large membrane-associated endocytic cisternae bud from DMob4 mutant NMJs (black arrows in B) but are absent in controls. C, D, High-magnification images of cisternae indicated in B. E–H, Cross-sectional profiles of axons from peripheral nerves of control (E, F) and DMob4EYΔL3/Df(2R)42 mutants (G, H). Microtubule-associated vesicles (black arrowheads) in control animals are smaller and less numerous than in DMob4EYΔL3/Df(2R)42 mutants. Scale bars: A, B, 500 nm; E–H, 200 nm.
DMob4 mutants manifest membrane excitability defects
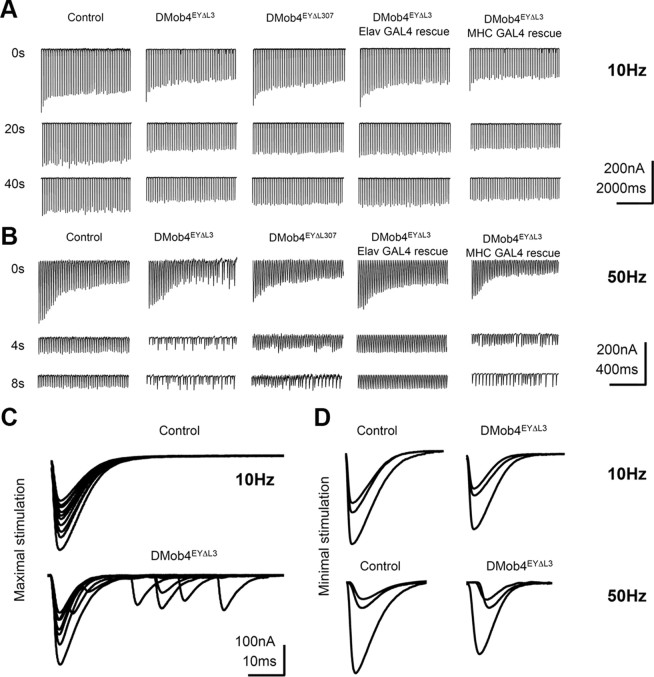
To quantitatively investigate endocytosis defects, we conducted electrophysiology experiments at third-instar larval NMJs of control and DMob4 mutants using two-electrode voltage clamp (Fig. 6). We first measured EPSCs at low frequency in 2 mm extracellular Ca2+. DMob4 mutant larvae displayed a small ∼15% reduction in evoked EPSC amplitude (control, 278 ± 52 nA, n = 4; DMob4, 237 ± 36 nA, n = 5), indicating that robust synaptic transmission persisted in the mutants. We next assayed for use-dependent alterations in synaptic function by recording EPSCs during stimulation trains of 10 or 50 Hz in 2 mm extracellular Ca2+ (Fig. 6A,B). Mutants with defective endocytosis typically display a gradual rundown of EPSCs with tetanizing stimuli as the synaptic vesicle pool depletes (Delgado et al., 2000). DMob4 mutants showed relatively normal synaptic depression during 10 Hz stimulation, but complete failures in EPSCs were observed after ∼4 s with a faster 50 Hz stimulation train (Fig. 6B). This phenotype was observed in both DMob4EYΔL3 nulls and DMob4EYΔL307 hypomorphs and was 100% penetrant in mutants and absent from controls. Mutant larvae that were allowed to recover from the high-frequency stimulation trains displayed normal EPSCs with subsequent 10 Hz stimulations.
Figure 6.
Electrophysiological analysis of DMob4 mutants. A–D, Voltage-clamp recordings at the NMJ of third-instar larvae. EPSCs were elicited at 10 Hz (A) or 50 Hz (B) stimulation from control, DMob4 (null and hypomorphic), and rescued (neural- and muscle-specific expression) animals. The EPSC traces represent responses to continuous tetanizing stimulus trains. To compress traces, short blocks of stimuli at different time points (as indicated) from a continuous stimulus train are presented. DMob4EYΔL3 and DMob4EYΔL307 mutants exhibit failures in EPSCs at 50 Hz stimulation after 4 s of sustained excitation of peripheral nerves. C, Overlaid EPSC responses from single stimuli at progressive time points in a 10 Hz stimulus train using a maximal (2× threshold) stimulation intensity. DMob4 mutants exhibit supernumerary responses late in a stimulus train. D, Overlaid EPSCs evoked at 10 and 50 Hz. DMob4 mutants exhibit a slight reduction in EPSC amplitude at 10 and 50 Hz stimulation and a delay (rightward shift) in EPSC response at 50 Hz.
Neuronal expression of DMob4 with Elavc155GAL4 completely suppressed the EPSC phenotype of DMob4EYΔL3 nulls (Fig. 6A,B), whereas muscle-specific expression did not, indicating that DMob4 is required presynaptically for normal membrane excitability. To further analyze synaptic transmission failures in DMob4 mutants, we tested potential contributing factors by altering stimulation intensities used to trigger action potentials. We observed supernumerary EPSC responses to single nerve stimuli in DMob4 mutants using stronger stimulation (2× nerve threshold) (Fig. 6C), suggesting abnormal membrane excitability in the absence of DMob4. Excitability defects were also found at minimal stimulation intensities required to trigger a response (Fig. 6D). Delays between the onset of EPSC responses were observed at 50 Hz, even early in stimulation trains, consistent with a slower propagation of action potentials in mutant animals. Similar perturbations in membrane excitability properties have been observed in a variety of ion channel mutants, including the Na+/K+ ATPase, and the Shaker and Shab potassium channels mutants (Jan et al., 1977; Jan and Jan, 1978; Ueda and Wu, 2006), suggesting that DMob4 is likely to modulate membrane excitability of neurons, in addition to its role in regulating morphology. The all-or-none failures in EPSCs observed in the DMob mutants likely mask any vesicle depletion phenotype characteristic of classical endocytotic mutants.
DMob4 mutants have defective axonal transport
While conducting lethal phase analysis on DMob4 mutants, we observed that mutant third-instar larvae have a tail-flip phenotype (Fig. 7A,B), with the posterior half of the mutant larva paralyzed. This phenotype has been described previously for microtubule motor mutants such as kinesin and dynein that disrupt axonal transport (Martin et al., 1999). We observed the tail-flip phenotype in all DMob4 strains, including DMob4EYΔL3, DMob4EY23407, and DMob4EYΔL307. The tail-flip phenotype was most severe in the DMob4EYΔL3 and DMob4EY23407 null strains.
Figure 7.
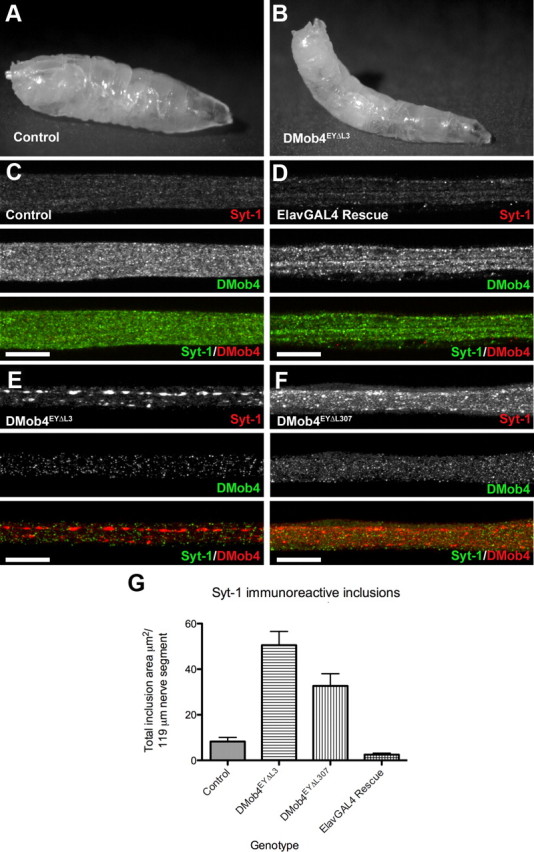
Axonal transport defects in DMob4 mutants. A, B, DMob4 mutant third-instar larvae have a tail-flip phenotype (B) compared with controls (A). C–F, Peripheral nerves of third-instar larvae coimmunolabeled for Syt-1 (red) and DMob4 (green). Blockade of axonal transport in DMob4EYΔL3/Df(2R)42 (null) and DMob4EYΔL307/Df(2R)42 (hypomorph) mutants results in accumulation of Syt-1 in large aggregates along the length of peripheral nerves (E, F). Axonal transport defects in DMob4EYΔL3 mutants can be rescued with the ectopic expression of UAS–DMob4 in neurons using the ElavGal4 driver (D). G, Quantification of Syt-1 immunoreactive inclusions present in peripheral nerves of DMob4 mutants, controls, and rescued animals. Scale bar, 20 μm.
Because microtubule motor mutants have defects in axonal transport and display a posterior tail-flip phenotype in third-instar larvae, we investigated whether DMob4 mutants also had defects in axonal transport. We immunolabeled homozygous DMob4EYΔL3 and DMob4EYΔL307 mutant third-instar larvae for the synaptic vesicle protein Syt-1 and counter-labeled for DMob4 (Fig. 7C–F). Syt-1 is transported in vesicles along microtubules and normally enriches at synapses. Syt-1 has a punctate distribution in the peripheral nerves of control animals (Fig. 7C). In contrast, DMob4EYΔL3 and DMob4EYΔL307 mutants accumulate large aggregates of Syt-1 along axons (Fig. 7E,F). We quantified the area of Syt-1 aggregate accumulation in control and DMob4 mutants and found that DMob4 nulls had sixfold more aggregates than controls, whereas the hypomorph had fourfold more aggregates (Fig. 7G). Neural-specific rescue of the DMob4 nulls with elavc155GAL4;UAS–DMob4 transgenes resulted in a complete elimination of the Syt-1 aggregates in the peripheral nerves (Fig. 7D,G). We also observed abnormal accumulation of the active zone marker bruchpilot (nc82) in peripheral nerves of DMob4 mutants (Fig. 3I). However, this marker is expressed at lower levels, and therefore axonal transport phenotypes were less apparent than with Syt-1 immunolabelings. Together, these results suggest that DMob4 is necessary for vesicle transport in peripheral nerve axons.
DMob4 mutants have abnormal microtubule organization at synapses, axons, and muscles
Microtubules serve as the main scaffold along which motors such as kinesin and dynein transport vesicular cargo. Microtubules are also necessary to stabilize presynaptic terminals during synaptic development. In mature presynaptic terminals, microtubules adopt a looped structure. Because DMob4 mutants have abnormal synapse development and defective axonal transport, we investigated whether microtubule networks at synapses and in axons were disrupted. To examine microtubules in control and DMob4 mutant third-instar larvae, we immunolabeled the animals for the neuronal microtubule-associated protein Futsch (mAb 22C10). We observed abnormal microtubule organization at the synapse and along peripheral nerves of DMob4 mutants (Fig. 8). In control animals, typical 22C10-positive microtubule loops at terminal boutons were present (Fig. 8A). In DMob4 homozygous mutant synapses, microtubule loops were either absent or had multiple breaks (Fig. 8B,C). In synapses in which microtubule loops were absent, the 22C10 staining either filled the boutons in a speckled pattern or was weak and diffuse. The microtubule organization along peripheral nerves was also altered in DMob4 mutant animals. In control animals, microtubule networks ran parallel along the length of the nerves (Fig. 8D). In DMob4 mutants, microtubule networks had a distorted appearance (Fig. 8E), suggesting that microtubule networks are disorganized at peripheral nerves, and synapses.
Figure 8.
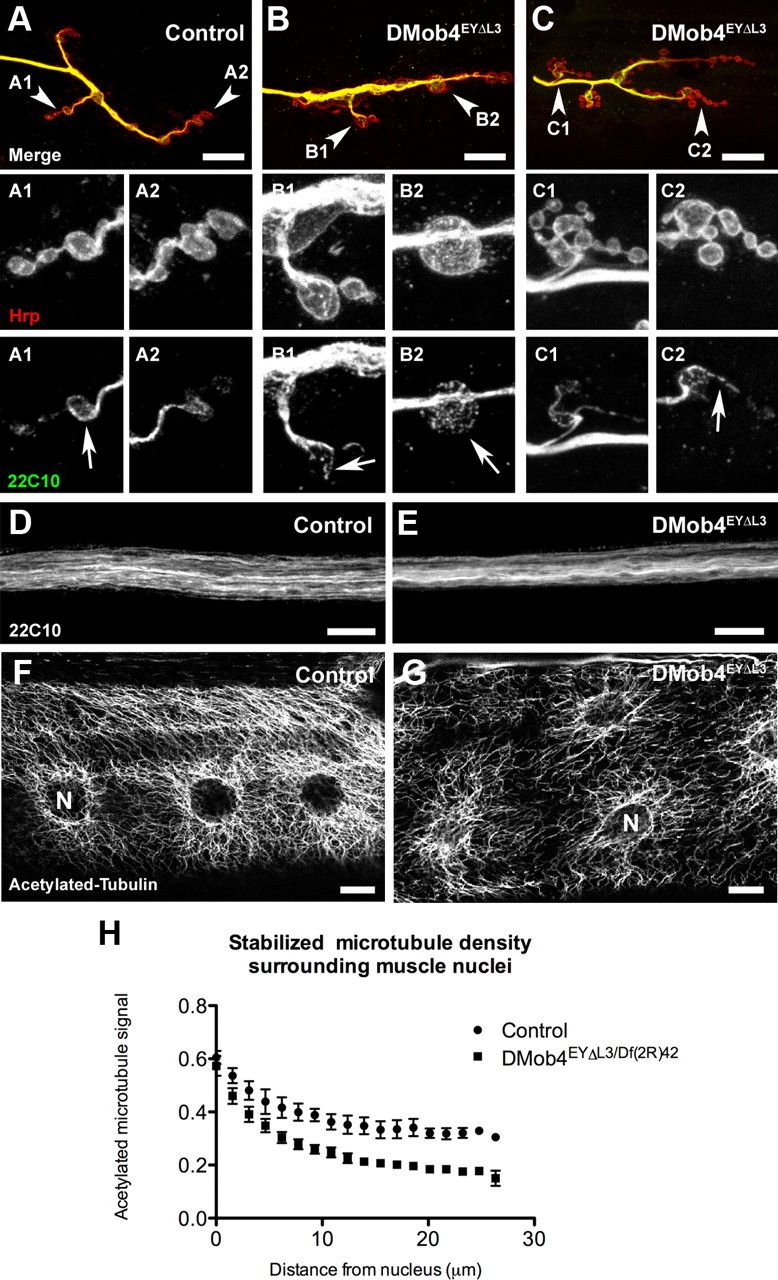
Abnormal microtubule morphology at NMJs, peripheral nerves, and muscles of DMob4 mutants. A–C, NMJs of third-instar larvae costained for HRP (red) to reveal neural membranes and anti-Futsch mAb 22C10 (green) to reveal microtubules. Arrowheads in merged images point to NMJs that are magnified and split into individual channels in subsequent panels. Looped microtubule structures are evident in the boutons of control (22C10; A1, A2). These structures are either discontinuous (22C10; B1, C1, C2) or absent (22C10; B2) in DMobEYΔL3 mutants. D, E, Peripheral nerves of control and DMob4 mutants stained for the microtubule-associated protein Futsch (22C10). D, Control animals have well defined microtubule bundles. E, DMob4 mutants have more poorly defined collapsed microtubule bundles compared with controls. F, G, Acetylated tubulin staining in larval muscle fibers. F, Control animals have extensive stabilized microtubule networks emanating from the nuclear membrane. G, DMob4 mutants have a decrease in stabilized microtubule networks. H, Quantification of acetylated tubulin signal around nuclei in control and DMob4 mutant muscle fibers. N, Nucleus. Scale bar, 10 μm.
To investigate whether the abnormal distribution of the microtubule-associated protein Futsch (mAb 22C10) in DMob4 mutants was a reflection of overall microtubule disorganization, we examined microtubules directly by immunolabeling for α-Tubulin. Posttranslational modifications of α-Tubulin can be used to monitor different populations of microtubules. Stabilized microtubules are enriched for acetylated α-Tubulin, whereas nascent microtubules are enriched for tyrosinated α-Tubulin (Palazzo et al., 2003; Fukushima et al., 2009). We examined both populations of α-Tubulin in DMob4 mutants.
In control animals, acetylated α-Tubulin immunolabeling gives a very stereotypic pattern in muscle fibers, with a high concentration around muscle nuclei and fibers radiating away from the nuclei in an elaborate meshwork (Fig. 8F). The muscle nuclei are also aligned along the longitudinal axis of the muscle fibers. In DMob4 mutants, there is a striking decrease in the extent of acetylated microtubule networks, and the muscle nuclei are more randomly located throughout muscle fibers (Fig. 8G). We quantified the acetylated microtubule signal in muscles as a function of distance from the nuclear membrane (Fig. 8H). In controls, the microtubule signal decreases by 38% at 20 μm from the nucleus, whereas in DMob4 mutants it is reduced by 70%. These data strongly suggest that stabilized microtubule networks are disorganized in multiple subcellular compartments in DMob4 mutants. To extend these findings, we stained control and DMob4 mutants for tyrosinated α-Tubulin, which labels newly formed microtubules (Fig. 9). We colabeled the larvae for Syt-1 to monitor vesicular cargoes in the peripheral nerves and the morphology of synapses. We observed that tyrosinated microtubules are decreased in peripheral nerves, NMJs, and muscles of DMob4 mutants (Fig. 9A–F). Because both acetylated and tyrosinated α-Tubulin networks are diminished in DMob4 mutants, we conclude that DMob4 plays a critical role in the overall organization of microtubule networks in multiple cellular compartments.
Figure 9.
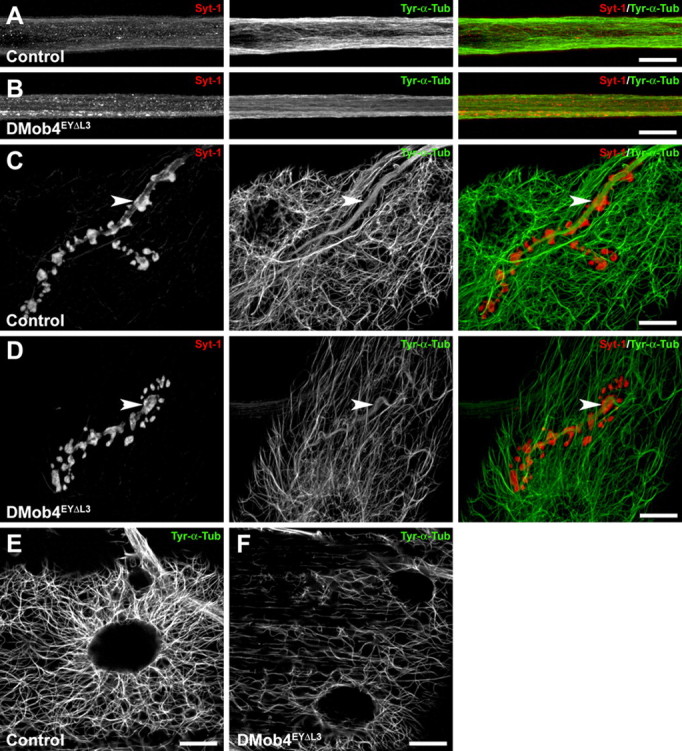
Diminished levels of tyrosinated microtubules in DMob4 mutants. A–E, Control and DMob4 mutants double stained for tyrosinated α-Tubulin (Tyr-α-Tub) using monoclonal antibody Tub-1A2 (green) and costained for Syt-1 (red). Individual and merged channels from confocal stacks are presented. A, B, Peripheral nerves. DMob4EYΔL3 homozygous mutants have decreased levels of Tyr-α-Tub, and individual microtubule bundles are less evident. In mutants, Tyr-α-Tub appears to have a smooth distribution. Syt-1 aggregates are evident in the DMob4 mutants (B). Stacks spanning half the diameter of a peripheral nerve are presented for increased resolution of microtubule networks. C, D, NMJs. Diminished levels of Tyr-α-Tub is observed in boutons of DMob4 mutant NMJs (compare arrowheads in green channel in C, D). Stacks of equivalent thickness spanning the NMJ are shown. E, F, Abdominal muscle. Microtubule network complexity, as well as Tyr-α-Tub levels are also reduced in the muscles of DMob4 mutants. Single confocal slices are presented. Scale bars, 20 μm.
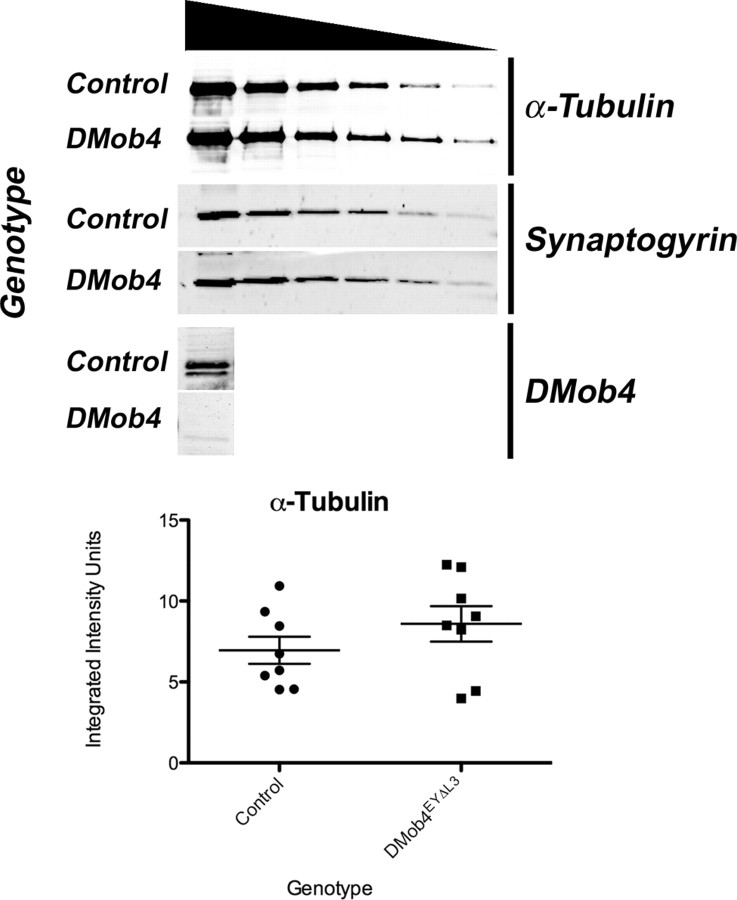
To examine whether the altered microtubule organization observed in DMob4 mutants is a consequence of decreased levels of total α-Tubulin, we conducted quantitative Western blot analysis. We found that total α-Tubulin levels are not significantly altered in DMob4 mutants compared with controls (Fig. 10). These data indicate that total levels of α-Tubulin are not limiting in DMob4 mutants and that DMob4 exerts its effect on microtubule organization through posttranslational mechanisms.
Figure 10.
Western blot analysis of total α-Tubulin levels in third-instar larvae. Serial dilutions of total protein isolated from control and DMob4 mutant larvae (indicated by gradient symbol) were immunoblotted for α-Tubulin and Synaptogyrin (loading control). To verify the genotypes of the samples analyzed, a DMob4 immunoblot is shown for the undiluted extract. Lanes 1–6, 6.5 μg protein/lane × 1, (3/7)1, (3/7)2, (3/7)3, (3/7)4, and (3/7)5. For quantitative analysis of α-Tubulin levels, protein samples (n = 8) at 6.5 μg × (3/7)3/lane were analyzed using an Odyssey infrared scanner with the data normalized to Synaptogyrin levels. Mean integrated intensity units: DMob4, 6.96 ± 0.84; control, 8.59 ± 1.09; p = 0.26. Error bars are SEM.
Discussion
Here we describe the first in vivo functional characterization of a Phocein protein in the nervous system using Drosophila. Phocein is a member of the Mob family of zinc-binding proteins that are enriched in Purkinje cell dendrites. A function for DMob4 in regulation of neurite branching was suggested from a genome-wide RNAi screen designed to identify genes necessary for neurite outgrowth and morphology (Sepp et al., 2008). Our mutant analysis of Dmob4 loss-of-function alleles revealed a host of nervous system defects, including abnormal synaptic development with extensive satellite bouton formation, disrupted axonal transport, disorganized microtubules, and action potential failure during high-frequency stimulation. DMob4 is essential for viability, because null alleles are larval lethal and hypomorphic alleles have decreased adult longevity. The larval lethality of nulls can be rescued by expression of DMob4 or human phocein, indicating functional conservation and orthology of DMob4 across species. Although other Mob family members have been found to function in mitotic cells to enable cell cycle progression, facilitate apoptosis, or regulate cell morphogenesis (Luca et al., 2001; Weiss et al., 2002; Hou et al., 2003; He et al., 2005; Lai et al., 2005; Praskova et al., 2008), we find that Phocein/DMob4 has a unique role in postmitotic neurons to regulate synapse formation, axonal transport, and microtubule organization in the nervous system.
In yeast, Mobs function as activating subunits of the Dbf-2 family of protein kinases (Komarnitsky et al., 1998; Ho et al., 2002). There are two Drosophila Dbf-2 homologs, Tricornered and Warts, which interact with Mob1 (mats) and Mob2 (Justice et al., 1995; He et al., 2005; Lai et al., 2005). The kinase binding partners for Drosophila Mob3 and Mob4 are not known. Mutations in warts and mats cause overproliferation phenotypes, whereas mutations in tricornered results in a split denticle belt phenotype (Justice et al., 1995; He et al., 2005). We did not observe these phenotypes in DMob4 mutants, suggesting that it is unlikely to function as an activating subunit for Tricornered or Warts. What might be the target kinase(s) for DMob4? NDPK associates with rat Phocein (Baillat et al., 2002) and is an interesting candidate kinase, given its established roles in endocytosis and microtubule dynamics (Biggs et al., 1990; Krishnan et al., 2001). NDPK is the main enzyme that synthesizes GTP from GDP in many species, and a large fraction of cellular NDPK is associated with microtubules (Postel, 1998). During microtubule polymerization, GTP is bound to tubulin dimers and is necessary for tubulation. During endocytosis, GTP binding by dynamin triggers oligomerization at the necks of clathrin-coated vesicles to drive fission. Based on studies in mammals, Phocein is also part of the STRIPAK protein complex, which contains multiple STE-kinases (Goudreault et al., 2008). As such, several Drosophila STRIPAK STE-kinase homologs might also be regulated by DMob4. The mammalian/Drosophila STRIPAK complex is likely to be well conserved, because many of the non-kinase components, including Striatin (Drosophila Cka), FGF receptor (e.g., Drosophila Heartless), Dynein, and PP2A, are present in Drosophila.
DMob4 function in endocytosis and vesicular traffic
Phocein was found previously to localize to the Golgi apparatus and dendritic spines of Purkinje cells (Baillat et al., 2001; Haeberlé et al., 2006). The association of Phocein with proteins that have well established roles in vesicular traffic and endocytosis (Eps-15, NDPK, and Dynamin-1), in addition to its subcellular localization to sites of active endocytosis, lead to a hypothesis that Phocein may function in membrane budding and vesicle trafficking (Baillat et al., 2001, 2002; Haeberlé et al., 2006). Our in vivo characterization of DMob4 mutants supports a role for the protein in endocytosis and vesicular traffic. In Drosophila, many endocytosis mutants have been identified that show a supernumerary bouton phenotype, including endophilin, synaptojanin, dynamin, AP180, and Dap-160 (Dickman et al., 2006). DMob4 mutants have a supernumerary bouton phenotype comparable with Dap-160 mutants. The excess synaptic growth in these mutants is predicted to arise from defective endocytotic processing of retrogradely released synaptic growth factors such as the TGFβ homolog, Glass Bottom Boat, resulting in excessive signaling and enhanced synapse formation. TEM analysis of DMob4 mutant NMJs also reveal endocytic cisternae that are characteristic of defective endocytosis (Kosaka and Ikeda, 1983). Similar cisternae have been reported for eps-15 and dap-160 mutants (Koh et al., 2007). In mutants with severely impaired endocytosis, such as shibire, cisternae occur in large numbers and elongated tubules are evident (Kosaka and Ikeda, 1983). We did not observe such tubules in DMob4, implying that the protein is not absolutely required for endocytosis but likely plays a regulatory role. Our observation that N-terminal truncation mutants (DMob4EYΔL307) undergo temperature-sensitive paralysis (data not shown) further supports a role in endocytosis, because shi, dap-160, and eps-15 mutants show similar phenotypes (Koh et al., 2004). TEM analysis revealed an increase in the number and size of vesicles associated with axonal microtubules. This phenotype may reflect endocytosis defects or abnormal membrane budding events from other cellular compartments, in addition to defects in axonal transport. Many proteins that are integral to endocytosis at the synapse also function in budding of vesicles from the Golgi apparatus, including Clathrin, Dynamin, and Eps-15 (Baillat et al., 2002; McNiven and Thompson, 2006; Soldati and Schliwa, 2006).
DMob4 function in axonal transport and microtubule dynamics
In addition to its role in endocytosis, phocein has been suggested to function during mitosis. GFP–DMob4 transgenes associate with centrioles and kinetochores in dividing Drosophila S2 cells (Trammell et al., 2008). Microtubules attach to these structures to generate force to push/pull chromosomes apart and enable their segregation to daughter cells. RNAi knockdown of DMob4 in S2 cells results in a mono-aster spindle phenotype that arises from a failure of microtubule minus-ends to focus at centrioles. RNAi knockdown of other Drosophila Mobs (DMob1–DMob3) does not affect spindle focusing, suggesting that DMob4/Phocein has a unique role in regulating microtubule dynamics. Although we did not examine mitotic defects in our mutants, we found that microtubule networks are disorganized in multiple cellular compartments, including NMJs, axons, and muscle fibers. How DMob4 functions to stabilize microtubule networks is currently unclear, but could be mediated through several pathways. The decrease in complexity of acetylated and tyrosinated tubulin networks we observe in DMob4 muscle fibers may reflect improper minus-end anchoring on muscle nuclei during development, consistent with the observations in S2 cells. The microtubule phenotypes could also result from abnormal microtubule severing, transport, or sorting (Baas et al., 2005; Roll-Mecak and Vale, 2006). Microtubule severing by AAA–ATPases and active sorting of fragments by microtubule motors can rapidly change microtubule architecture. The supernumerary bouton phenotypes observed in DMob4 may also reflect contributions from altered microtubule dynamics, in addition to defective endocytosis (Dickman et al., 2006). Spastin mutants, which lack an AAA–ATPase with microtubule-severing activity, have elaborate supernumerary NMJ boutons, similar to DMob4 (Sherwood et al., 2004; Trotta et al., 2004). Microtubule phenotypes in DMob4 could also result from altered association of microtubule associated proteins (MAPs). Microtubule cross-linkers such as Tau modulate microtubule stability, and DMob4 may alter their in vivo activity through regulation of PP2A activity. Phocein has been reported to associate with PP2A in many proteomic studies, and our preliminary studies also indicate that DMob4 associates with PP2A (data not shown). Additional studies in DMob4 mutants will be required to assess these alternative causes for disorganized microtubule networks.
Regardless of the mechanism of microtubule disorganization in DMob4, kinase/phosphatase imbalances likely contribute. Because certain Mobs have been shown to function as Dbf-2 kinase activators and their overall three-dimensional structure is likely to be conserved, including key residues for kinase binding (He et al., 2005), it is likely that DMob4 will also function as a kinase activator. As a component of the STRIPAK complex, which contains several serine/threonine kinases and a serine/threonine phosphatase (PP2A), loss of DMob4 could alter the balance of STRIPAK complex activity and substrate specificity (Goudreault et al., 2008). Indeed, PP2A is known to regulate the phosphorylation state of Tau and other MAPs (Sontag et al., 1996; Schild et al., 2006). Linkages between microtubule motors and their cargoes are also regulated by kinase/phosphatase switches. Jun kinases, for example, control the linkage between Kinesin and vesicular cargoes (Horiuchi et al., 2007). The STRIPAK complex may similarly regulate the association of Dynein with its cargoes, and Dynein is known to be differentially phosphorylated (Mische et al., 2008). Thus, removal of DMob4 from the STRIPAK complex is likely to produce pleiotropic phenotypes as observed in DMob4 mutants.
Our in vivo analysis of DMob4 has identified a unique role for Phoceins in the regulation of microtubule organization and axonal transport. Defects in axonal transport or microtubule organization have been linked to many neurodegenerative diseases, including Huntington's disease, hereditary spastic paraplegias, amyotrophic lateral sclerosis, and Alzheimer's disease (Duncan and Goldstein, 2006). It will be interesting to determine whether disruption of phocein in mammals leads to neurodegenerative disease. Additional studies of Drosophila Mob4 mutants should provide critical insights into how this important protein regulates axonal transport, endocytosis, and microtubule organization in vivo.
Footnotes
This work was supported by National Institutes of Health Grants R01 EB007042 (P.H.) and RO1 NS40296 and R01 NS052203 (J.T.L.). We are grateful to Hugo Bellen and the Bloomington Stock Center for Drosophila stocks, the Drosophila Genomics Resource Center for ESTs, the Developmental Studies Hybridoma Bank for antibodies, the Drosophila RNAi Screening Center and staff for assistance with RNAi, Norbert Perrimon for use of microscopes, the National Center for Biotechnology Information for Cn3D crystal structure image of Human Mob1 protein, and Sonal Jhaveri for proofreading and editing of this manuscript.
References
- Acharya U, Edwards MB, Jorquera RA, Silva H, Nagashima K, Labarca P, Acharya JK. Drosophila melanogaster Scramblases modulate synaptic transmission. J Cell Biol. 2006;173:69–82. doi: 10.1083/jcb.200506159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda-Orgillés B, Aigner J, Kunath M, Lurz R, Schneider R, Schweiger S. Active transport of the ubiquitin ligase MID1 along the microtubules is regulated by protein phosphatase 2A. PLoS ONE. 2008;3:e3507. doi: 10.1371/journal.pone.0003507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas PW, Karabay A, Qiang L. Microtubules cut and run. Trends Cell Biol. 2005;15:518–524. doi: 10.1016/j.tcb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Baillat G, Moqrich A, Castets F, Baude A, Bailly Y, Benmerah A, Monneron A. Molecular cloning and characterization of phocein, a protein found from the Golgi complex to dendritic spines. Mol Biol Cell. 2001;12:663–673. doi: 10.1091/mbc.12.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillat G, Gaillard S, Castets F, Monneron A. Interactions of phocein with nucleoside-diphosphate kinase, Eps15, and Dynamin I. J Biol Chem. 2002;277:18961–18966. doi: 10.1074/jbc.M108818200. [DOI] [PubMed] [Google Scholar]
- Bartoli M, Ternaux JP, Forni C, Portalier P, Salin P, Amalric M, Monneron A. Down-regulation of striatin, a neuronal calmodulin-binding protein, impairs rat locomotor activity. J Neurobiol. 1999;40:234–243. [PubMed] [Google Scholar]
- Biggs J, Hersperger E, Steeg PS, Liotta LA, Shearn A. A Drosophila gene that is homologous to a mammalian gene associated with tumor metastasis codes for a nucleoside diphosphate kinase. Cell. 1990;63:933–940. doi: 10.1016/0092-8674(90)90496-2. [DOI] [PubMed] [Google Scholar]
- Colman-Lerner A, Chin TE, Brent R. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell. 2001;107:739–750. doi: 10.1016/s0092-8674(01)00596-7. [DOI] [PubMed] [Google Scholar]
- Delgado R, Maureira C, Oliva C, Kidokoro Y, Labarca P. Size of vesicle pools, rates of mobilization, and recycling at neuromuscular synapses of a Drosophila mutant, shibire. Neuron. 2000;28:941–953. doi: 10.1016/s0896-6273(00)00165-3. [DOI] [PubMed] [Google Scholar]
- Devroe E, Erdjument-Bromage H, Tempst P, Silver PA. Human Mob proteins regulate the NDR1 and NDR2 serine-threonine kinases. J Biol Chem. 2004;279:24444–24451. doi: 10.1074/jbc.M401999200. [DOI] [PubMed] [Google Scholar]
- Dickman DK, Lu Z, Meinertzhagen IA, Schwarz TL. Altered synaptic development and active zone spacing in endocytosis mutants. Curr Biol. 2006;16:591–598. doi: 10.1016/j.cub.2006.02.058. [DOI] [PubMed] [Google Scholar]
- Duncan JE, Goldstein LS. The genetics of axonal transport and axonal transport disorders. PLoS Genet. 2006;2:e124. doi: 10.1371/journal.pgen.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto K, Parrish JZ, Jan LY, Jan YN. The tumour suppressor Hippo acts with the NDR kinases in dendritic tiling and maintenance. Nature. 2006;443:210–213. doi: 10.1038/nature05090. [DOI] [PubMed] [Google Scholar]
- Feng Y, Ueda A, Wu CF. A modified minimal hemolymph-like solution, HL3.1, for physiological recordings at the neuromuscular junctions of normal and mutant Drosophila larvae. J Neurogenet. 2004;18:377–402. doi: 10.1080/01677060490894522. [DOI] [PubMed] [Google Scholar]
- Fukushima N, Furuta D, Hidaka Y, Moriyama R, Tsujiuchi T. Post-translational modifications of tubulin in the nervous system. J Neurochem. 2009;109:683–693. doi: 10.1111/j.1471-4159.2009.06013.x. [DOI] [PubMed] [Google Scholar]
- Goudreault M, D'Ambrosio LM, Kean MJ, Mullin M, Larsen BG, Sanchez A, Chaudhry S, Chen GI, Sicheri F, Nesvizhskii AI, Aebersold R, Raught B, Gingras AC. A PP2A phosphatase high-density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol Cell Proteomics. 2009;8:157–171. doi: 10.1074/mcp.M800266-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeberlé AM, Castets F, Bombarde G, Baillat G, Bailly Y. Immunogold localization of phocein in dendritic spines. J Comp Neurol. 2006;495:336–350. doi: 10.1002/cne.20895. [DOI] [PubMed] [Google Scholar]
- Hergovich A, Bichsel SJ, Hemmings BA. Human NDR kinases are rapidly activated by MOB proteins through recruitment to the plasma membrane and phosphorylation. Mol Cell Biol. 2005;25:8259–8272. doi: 10.1128/MCB.25.18.8259-8272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Emoto K, Fang X, Ren N, Tian X, Jan YN, Adler PN. Drosophila Mob family proteins interact with the related tricornered (Trc) and warts (Wts) kinases. Mol Biol Cell. 2005;16:4139–4152. doi: 10.1091/mbc.E05-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Horiuchi D, Collins CA, Bhat P, Barkus RV, Diantonio A, Saxton WM. Control of a kinesin-cargo linkage mechanism by JNK pathway kinases. Curr Biol. 2007;17:1313–1317. doi: 10.1016/j.cub.2007.06.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou MC, Wiley DJ, Verde F, McCollum D. Mob2p interacts with the protein kinase Orb6p to promote coordination of cell polarity with cell cycle progression. J Cell Sci. 2003;116:125–135. doi: 10.1242/jcs.00206. [DOI] [PubMed] [Google Scholar]
- Jan YN, Jan LY. Genetic dissection of short-term and long-term facilitation at the Drosophila neuromuscular junction. Proc Natl Acad Sci U S A. 1978;75:515–519. doi: 10.1073/pnas.75.1.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan YN, Jan LY, Dennis MJ. Two mutations of synaptic transmission in Drosophila. Proc R Soc Lond B Biol Sci. 1977;198:87–108. doi: 10.1098/rspb.1977.0087. [DOI] [PubMed] [Google Scholar]
- Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- Kearney JB, Wheeler SR, Estes P, Parente B, Crews ST. Gene expression profiling of the developing Drosophila CNS midline cells. Dev Biol. 2004;275:473–492. doi: 10.1016/j.ydbio.2004.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh TW, Verstreken P, Bellen HJ. Dap160/intersectin acts as a stabilizing scaffold required for synaptic development and vesicle endocytosis. Neuron. 2004;43:193–205. doi: 10.1016/j.neuron.2004.06.029. [DOI] [PubMed] [Google Scholar]
- Koh TW, Korolchuk VI, Wairkar YP, Jiao W, Evergren E, Pan H, Zhou Y, Venken KJ, Shupliakov O, Robinson IM, O'Kane CJ, Bellen HJ. Eps15 and Dap160 control synaptic vesicle membrane retrieval and synapse development. J Cell Biol. 2007;178:309–322. doi: 10.1083/jcb.200701030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky SI, Chiang YC, Luca FC, Chen J, Toyn JH, Winey M, Johnston LH, Denis CL. DBF2 protein kinase binds to and acts through the cell cycle-regulated MOB1 protein. Mol Cell Biol. 1998;18:2100–2107. doi: 10.1128/mcb.18.4.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka T, Ikeda K. Reversible blockage of membrane retrieval and endocytosis in the garland cell of the temperature-sensitive mutant of Drosophila melanogaster, shibirets1. J Cell Biol. 1983;97:499–507. doi: 10.1083/jcb.97.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan KS, Rikhy R, Rao S, Shivalkar M, Mosko M, Narayanan R, Etter P, Estes PS, Ramaswami M. Nucleoside diphosphate kinase, a source of GTP, is required for dynamin-dependent synaptic vesicle recycling. Neuron. 2001;30:197–210. doi: 10.1016/s0896-6273(01)00273-2. [DOI] [PubMed] [Google Scholar]
- Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, Ho LL, Li Y. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Li H, Coghlan A, Ruan J, Coin LJ, Hériché JK, Osmotherly L, Li R, Liu T, Zhang Z, Bolund L, Wong GK, Zheng W, Dehal P, Wang J, Durbin R. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 2006;34:D572–D580. doi: 10.1093/nar/gkj118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littleton JT, Bellen HJ, Perin MS. Expression of synaptotagmin in Drosophila reveals transport and localization of synaptic vesicles to the synapse. Development. 1993;118:1077–1088. doi: 10.1242/dev.118.4.1077. [DOI] [PubMed] [Google Scholar]
- Luca FC, Mody M, Kurischko C, Roof DM, Giddings TH, Winey M. Saccharomyces cerevisiae Mob1p is required for cytokinesis and mitotic exit. Mol Cell Biol. 2001;21:6972–6983. doi: 10.1128/MCB.21.20.6972-6983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie B, Sweeney ST, Poskanzer KE, Roos J, Kelly RB, Davis GW. Dap160/intersectin scaffolds the periactive zone to achieve high-fidelity endocytosis and normal synaptic growth. Neuron. 2004;43:207–219. doi: 10.1016/j.neuron.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40:476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Iyadurai SJ, Gassman A, Gindhart JG, Jr, Hays TS, Saxton WM. Cytoplasmic dynein, the dynactin complex, and kinesin are interdependent and essential for fast axonal transport. Mol Biol Cell. 1999;10:3717–3728. doi: 10.1091/mbc.10.11.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNiven MA, Thompson HM. Vesicle formation at the plasma membrane and trans-Golgi network: the same but different. Science. 2006;313:1591–1594. doi: 10.1126/science.1118133. [DOI] [PubMed] [Google Scholar]
- Mische S, He Y, Ma L, Li M, Serr M, Hays TS. Dynein light intermediate chain: an essential subunit that contributes to spindle checkpoint inactivation. Mol Biol Cell. 2008;19:4918–4929. doi: 10.1091/mbc.E08-05-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrkobrada S, Boucher L, Ceccarelli DF, Tyers M, Sicheri F. Structural and functional analysis of Saccharomyces cerevisiae Mob1. J Mol Biol. 2006;362:430–440. doi: 10.1016/j.jmb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Ni JQ, Markstein M, Binari R, Pfeiffer B, Liu LP, Villalta C, Booker M, Perkins L, Perrimon N. Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster. Nat Methods. 2008;5:49–51. doi: 10.1038/nmeth1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo A, Ackerman B, Gundersen GG. Cell biology: tubulin acetylation and cell motility. Nature. 2003;421:230. doi: 10.1038/421230a. [DOI] [PubMed] [Google Scholar]
- Pi HJ, Lisman JE. Coupled phosphatase and kinase switches produce the tristability required for long-term potentiation and long-term depression. J Neurosci. 2008;28:13132–13138. doi: 10.1523/JNEUROSCI.2348-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponchon L, Dumas C, Kajava AV, Fesquet D, Padilla A. NMR solution structure of Mob1, a mitotic exit network protein and its interaction with an NDR kinase peptide. J Mol Biol. 2004;337:167–182. doi: 10.1016/j.jmb.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Postel EH. NM23-NDP kinase. Int J Biochem Cell Biol. 1998;30:1291–1295. doi: 10.1016/s1357-2725(98)00087-9. [DOI] [PubMed] [Google Scholar]
- Praskova M, Xia F, Avruch J. MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr Biol. 2008;18:311–321. doi: 10.1016/j.cub.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A, Vale RD. Making more microtubules by severing: a common theme of noncentrosomal microtubule arrays? J Cell Biol. 2006;175:849–851. doi: 10.1083/jcb.200611149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell W, Sullivan W. Fluorescent analysis of Drosophila embryos. In: Sullivan W, Ashburner M, Hawley R, editors. Drosophila protocols. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1999. [Google Scholar]
- Schild A, Ittner LM, Götz J. Altered phosphorylation of cytoskeletal proteins in mutant protein phosphatase 2A transgenic mice. Biochem Biophys Res Commun. 2006;343:1171–1178. doi: 10.1016/j.bbrc.2006.03.066. [DOI] [PubMed] [Google Scholar]
- Sepp KJ, Hong P, Lizarraga SB, Liu JS, Mejia LA, Walsh CA, Perrimon N. Identification of neural outgrowth genes using genome-wide RNAi. PLoS Genet. 2008;4:e1000111. doi: 10.1371/journal.pgen.1000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood NT, Sun Q, Xue M, Zhang B, Zinn K. Drosophila spastin regulates synaptic microtubule networks and is required for normal motor function. PLoS Biol. 2004;2:e429. doi: 10.1371/journal.pbio.0020429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati T, Schliwa M. Powering membrane traffic in endocytosis and recycling. Nat Rev Mol Cell Biol. 2006;7:897–908. doi: 10.1038/nrm2060. [DOI] [PubMed] [Google Scholar]
- Sontag E, Nunbhakdi-Craig V, Lee G, Bloom GS, Mumby MC. Regulation of the phosphorylation state and microtubule-binding activity of Tau by protein phosphatase 2A. Neuron. 1996;17:1201–1207. doi: 10.1016/s0896-6273(00)80250-0. [DOI] [PubMed] [Google Scholar]
- Stavridi ES, Harris KG, Huyen Y, Bothos J, Verwoerd PM, Stayrook SE, Pavletich NP, Jeffrey PD, Luca FC. Crystal structure of a human Mob1 protein: toward understanding Mob-regulated cell cycle pathways. Structure. 2003;11:1163–1170. doi: 10.1016/s0969-2126(03)00182-5. [DOI] [PubMed] [Google Scholar]
- Stoepel J, Ottey MA, Kurischko C, Hieter P, Luca FC. The mitotic exit network Mob1p-Dbf2p kinase complex localizes to the nucleus and regulates passenger protein localization. Mol Biol Cell. 2005;16:5465–5479. doi: 10.1091/mbc.E05-04-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournebize R, Andersen SS, Verde F, Dorée M, Karsenti E, Hyman AA. Distinct roles of PP1 and PP2A-like phosphatases in control of microtubule dynamics during mitosis. EMBO J. 1997;16:5537–5549. doi: 10.1093/emboj/16.18.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trammell MA, Mahoney NM, Agard DA, Vale RD. Mob4 plays a role in spindle focusing in Drosophila S2 cells. J Cell Sci. 2008;121:1284–1292. doi: 10.1242/jcs.017210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta N, Orso G, Rossetto MG, Daga A, Broadie K. The hereditary spastic paraplegia gene, spastin, regulates microtubule stability to modulate synaptic structure and function. Curr Biol. 2004;14:1135–1147. doi: 10.1016/j.cub.2004.06.058. [DOI] [PubMed] [Google Scholar]
- Ueda A, Wu CF. Distinct frequency-dependent regulation of nerve terminal excitability and synaptic transmission by IA and IK potassium channels revealed by Drosophila Shaker and Shab mutations. J Neurosci. 2006;26:6238–6248. doi: 10.1523/JNEUROSCI.0862-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Shimizu T, Lai ZC. Mob as tumor suppressor is activated by Hippo kinase for growth inhibition in Drosophila. EMBO J. 2007;26:1772–1781. doi: 10.1038/sj.emboj.7601630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss EL, Kurischko C, Zhang C, Shokat K, Drubin DG, Luca FC. The Saccharomyces cerevisiae Mob2p-Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell-specific localization of Ace2p transcription factor. J Cell Biol. 2002;158:885–900. doi: 10.1083/jcb.200203094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen JA, Peckol EL, Tobin DM, Bargmann CI. Neuronal cell shape and neurite initiation are regulated by the Ndr kinase SAX-1, a member of the Orb6/COT-1/warts serine/threonine kinase family. Mol Biol Cell. 2000;11:3177–3190. doi: 10.1091/mbc.11.9.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]