Abstract
Glutathionylation of compounds is an important reaction in the detoxification of electrophilic xenobiotics and in the biosynthesis of endogenous molecules. The glutathione conjugates (GS conjugates) are further processed by peptidic cleavage reactions. In animals and plants, γ-glutamyl transpeptidases initiate the turnover by removal of the glutamate residue from the conjugate. Plants have a second route leading to the formation of γ-glutamylcysteinyl (γ-GluCys) conjugates. Phytochelatin synthase (PCS) is well known to mediate the synthesis of heavy metal-binding phytochelatins. In addition, the enzyme is also able to catabolize GS conjugates to the γ-GluCys derivative. In this study, we addressed the cellular compartmentalization of PCS and its role in the plant-specific γ-GluCys conjugate pathway in Arabidopsis (Arabidopsis thaliana). Localization studies of both Arabidopsis PCS revealed a ubiquitous presence of AtPCS1 in Arabidopsis seedlings, while AtPCS2 was only detected in the root tip. A functional AtPCS1:eGFP (enhanced green fluorescent protein) fusion protein was localized to the cytosolic compartment. Inhibition of the vacuolar import of GS-bimane conjugate via azide treatment resulted in both a strong accumulation of γ-GluCys-bimane and a massive increase of the cellular cysteine to GS-bimane ratio, which was not observed in PCS-deficient lines. These findings support a cytosolic action of PCS. Analysis of a triple mutant deficient in both Arabidopsis PCS and vacuolar γ-glutamyl transpeptidase GGT4 is consistent with earlier observations of an efficient sequestration of GS conjugates into the vacuole and the requirement of GGT4 for their turnover. Hence, PCS contributes specifically to the cytosolic turnover of GS conjugates, and AtPCS1 plays the prominent role. We discuss a potential function of PCS in the cytosolic turnover of GS conjugates.
Plants are sessile organisms that cannot evade unfavorable habitats. They have therefore developed effective means to overcome stress conditions such as herbivore and pathogen attack or abiotic challenges encountered by drought, light stress, high levels of metal ions, and exposure to herbicides. Under such conditions, the tripeptide glutathione (GSH) plays a number of pivotal roles in plants (Noctor et al., 2002; Schützendübel and Polle, 2002). GSH is present in the millimolar range and constitutes up to 90% of total nonprotein sulfur (Leustek et al., 2000). The tripeptide is an important component of the cellular redox system (Foyer et al., 2009) and is connected to the regulation of stress-responsive gene expression (Ball et al., 2004; Mullineaux and Rausch, 2005). The redox-coupled formation of mixed disulfides among plant proteins is considered to be an important mechanism for posttranslational regulation (Dixon et al., 2005). The conjugation of electrophilic molecules to GSH is catalyzed by the action of glutathione S-transferases (GSTs). In plants, GSTs are encoded by a large gene family with approximately 50 members in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa; Soranzo et al., 2004; Edwards and Dixon, 2005), highlighting the importance of GS conjugate formation for the metabolism of endogenous compounds and the detoxification of noxious compounds such as herbicides. GS conjugates are predominantly generated in the cytosol, with minor GST activities in the nucleus, chloroplast, and mitochondrion (Dixon et al., 2002; Dixon and Edwards, 2009).
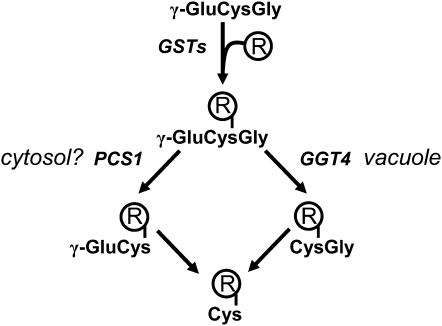
The rapid vacuolar sequestration of GS conjugates is mediated by ATP-binding cassette (ABC) transporters (Martinoia et al., 1993; Edwards et al., 2000). The ABC multidrug resistance-associated proteins (MRPs) AtMRP1, AtMRP2, and presumably AtMRP3 (Rea, 2007) have been characterized as GS conjugate transporters. In the vacuole, degradation of GS conjugates is initiated by the γ-glutamyl transpeptidase AtGGT4 (Fig. 1), yielding CysGly derivatives (Grzam et al., 2007; Ohkama-Ohtsu et al., 2007b). The cleavage of the unique isopeptidic bond of GS conjugates by GGTs, which corresponds to the first step in the catabolism of GS conjugates, and the subsequent enzymatic reactions are well characterized in animal cells (Martin et al., 2007). However, the enzymatic activity for the degradation of the resulting CysGly dipeptide conjugates to the Cys conjugates (Fig. 1) has not been unequivocally identified in plants. In yeast, the first degradation step is mediated by a single GGT, which is also required for extracellular secretion of Cys conjugates (Wünschmann et al., 2010). Plants possibly recruit the GGTs in a similar manner (Martin et al., 2007) or, alternatively, γ-glutamyl cyclotransferase and 5-oxoprolinase, which can convert the glutamyl moiety of γ-glutamylcysteinyl (γ-GluCys) into Glu (Ohkama-Ohtsu et al., 2008). Apart from AtGGT4, two other functional GGTs, AtGGT1 and AtGGT2, are expressed in Arabidopsis. While AtGGT4 plays a role in GS conjugate catabolism in the vacuole, AtGGT1 and AtGGT2 have an active site in the apoplast (Ohkama-Ohtsu et al., 2007a) and could be involved in the secretion of conjugates coupled with the cleavage of the glutamyl moiety from γ-GluCys and GS conjugates.
Figure 1.
Pathways of GS conjugate catabolism. Electrophilic compounds (R) are conjugated to glutathione (γ-GluCysGly) by GSTs. GS conjugates are sequestered in the vacuole and degraded to CysGly conjugates by the action of GGT4 in Arabidopsis. PCS has been shown to turn over GS conjugates to γ-GluCys conjugates. The cellular localization of PCS is not clear. The peptidic conjugates are processed to the Cys derivative.
A second mode of GS conjugate catabolism is initiated by the removal of the C-terminal Gly to the corresponding γ-GluCys product (Fig. 1). This catabolic step is common for GS conjugate degradation in plants (Lamoureux and Rusness, 1986, 1993). A vacuolar carboxypeptidase was postulated to catalyze the deglycination of GS-conjugated molecules of the herbicide alachlor in barley (Hordeum vulgare; Wolf et al., 1996). The removal of the C-terminal Gly from GS conjugates is efficiently catalyzed by phytochelatin synthase (PCS; EC 2.3.2.15; Beck et al., 2003; Blum et al., 2007). PCS is a specific γ-GluCys dipeptidyl transpeptidase (Grill et al., 1989; Vatamaniuk et al., 2004) known to generate the heavy metal-chelating phytochelatins with the general structure (γ-GluCys)n-Gly (n = 2–11) from GSH (Grill et al., 1985). Arabidopsis expresses two PCS proteins, AtPCS1 and AtPCS2 (Cazale and Clemens, 2001). Studies with the model xenobiotic monochlorobimane (MCB) suggest that AtPCS1 rather than AtPCS2 is the dominant player, as it has been shown that functional disruption of AtPCS1 but not of AtPCS2 leads to a decrease in the turnover of the fluorescent GS-bimane conjugate to γ-GluCys-bimane in Arabidopsis (Blum et al., 2007). Assessment of the PCS contribution to GS conjugate catabolism has been hampered by the existence of a second GGT-initiated pathway.
Glutathionylation of the herbicides alachlor, dimethenamid (Riechers et al., 1996), and fenoxaprop (Cummins et al., 2009) is followed by the deglycination of GSH to form γ-GluCys adducts. Similarly, the herbicide safener fenclorim is catabolized to γ-GluCys-fenclorim in the cytosol (Brazier-Hicks et al., 2008). Sulfur incorporation during the biosynthesis of glucosinolates and the detoxification of brassiglucosinolate-derived indolisothiocyanates possibly involve GS conjugate formation in the cytosol (Bednarek et al., 2009; Böttcher et al., 2009). Glucosinolate catabolism during pathogen attack generates an unknown signal required for callose deposition as part of the innate immune response (Clay et al., 2009). Both the glucosinolate catabolism and the immune response were impaired in the AtPCS1-deficient mutant. Collectively, these data support the existence of a cytosolic activity responsible for γ-GluCys conjugate formation in plants, which might be provided by PCS.
In this contribution, we localized AtPCS1 to the cytosolic compartment and addressed the extent of PCS contribution to GS conjugate catabolism. Inhibition of GS conjugate transport into the vacuole resulted in a strong accumulation of γ-GluCys-bimane. The accumulation of the γ-GluCys conjugate required AtPCS1 activity, supporting a prominent role of PCS during cytosolic GS conjugate turnover. The analysis of GS-bimane degradation in PCS mutants and in a triple mutant deficient in PCS and GGT4 corroborated the major role of GGT4 in the vacuolar GS conjugate catabolism.
RESULTS
Blocking Vacuolar Transport by Azide
GS conjugates are transported to the vacuole, where they are degraded. The catabolism of GS conjugates may also occur in the cytoplasm. As PCS is known to catabolize GS conjugates to the γ-GluCys derivative, it is a prime candidate for mediating an alternative metabolic pathway for GS conjugates in the cytosol.
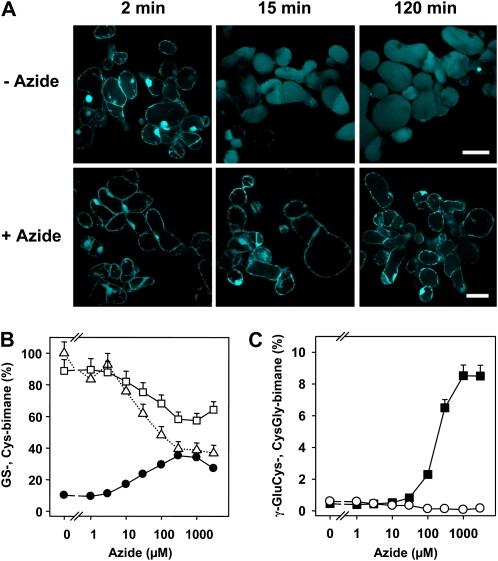
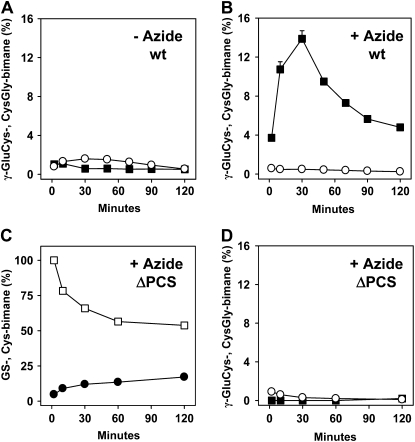
MCB is a model xenobiotic that has successfully been used to track the catabolism of GS-bimane conjugates in planta (Grzam et al., 2006, 2007; Blum et al., 2007). MCB is typically conjugated to GSH in the cytosol and subsequently sequestered in the vacuole. In order to evaluate the consequence of inhibiting the vacuolar sequestration of MCB, we treated cells with azide, a potent inhibitor of F-ATPases (Bowler et al., 2006) and of ABC transporters (Dallas et al., 2003; Jha et al., 2003). Confocal images of MCB-challenged cell suspension cultures of Arabidopsis show that the rapid accumulation of the fluorescent signal in the vacuole was prevented in the presence of azide (Fig. 2A), as has been reported in earlier studies (Meyer and Fricker, 2002; Grzam et al., 2006). We used feeding experiments with MCB in the presence of azide to assess whether blocking the uptake of bimane conjugates into vacuoles results in a conversion of the GS conjugate in the cytoplasm. To analyze the effect of azide on GS-bimane turnover, the optimal azide concentration was established. In the concentration range tested from 1 μm to 1 mm azide, an enhanced turnover of GS-bimane to γ-GluCys-bimane could be observed, with a maximum at 1 mm azide yielding 8.5% γ-GluCys-bimane of total bimane-labeled compounds compared with 0.5% γ-GluCys-bimane without azide 30 min after MCB challenge (Fig. 2, B and C). Interestingly, the conversion level of the GS conjugate to the Cys adduct was higher in the presence of 0.3 mm azide compared with noninhibited cells (Fig. 2B). In the presence of 0.3 mm azide, Cys-bimane accounted for 36% of bimane-labeled compounds, while total recovery dropped to approximately 40% in comparison with untreated cells. The lower recovery of inhibited cells may reflect stimulated secretion of bimane derivatives. Vital staining of cells with fluorescein diacetate provided no evidence for the loss of cellular integrity by azide treatment (Supplemental Fig. S1A). The determination of cellular GSH contents in the presence of various azide concentrations did not show significantly altered GSH/oxidized glutathione levels (Supplemental Fig. S1B). Subsequent time-course experiments revealed maximum γ-GluCys-bimane levels 30 min after a pulse of MCB challenge (Fig. 3, A and B). Arabidopsis cells exposed to azide yielded a 23-fold elevated γ-GluCys-bimane level compared with the control (13.9% and 0.6% of total bimane derivatives, respectively). Azide treatment (1 mm) did not prevent the degradation of GS-bimane to Cys-bimane in wild-type (Fig. 2B) and PCS-deficient cells (Fig. 3C). Suspension cells of the PCS double knockout ΔPCS, however, showed no increase in γ-GluCys-bimane levels (Fig. 3D).
Figure 2.
Effect of azide on GS-bimane catabolism. A, Confocal images of MCB-exposed suspension cells as in B and C in the absence (−azide) and in the presence (+azide) of 1 mm azide. The analysis reveals rapid accumulation of fluorescent bimane derivatives in the vacuole, while azide administration resulted in no significant fluorescence in the vacuole. The time points given indicate the incubation time after exposure to 5 μm MCB for 1 min and subsequent transfer of the cells to MCB-free medium. Bars = 50 μm. All images were taken from one experimental series with identical settings. B and C, Cells of an Arabidopsis suspension culture were exposed to different azide concentrations (5 min) prior to a 1-min exposure to MCB (5 μm). Subsequently, cells were incubated in MCB-free medium with the previous azide level. GS-bimane (white squares) and Cys-bimane (black circles) levels in B and CysGly-bimane (white circles) and γ-GluCys-bimane (black squares) amounts in C were determined 30 min after MCB challenge. For each inhibitor concentration, the sum of bimane derivatives including GS-bimane was set to 100%. The dotted line in B represents the values of the sum of bimane derivatives relative to the level of untreated cells (292 nmol g−1 dry weight). Data points represent means of duplicates ± sd. An independent experiment yielded comparable results.
Figure 3.
Time course of turnover of GS-bimane. A and B, Levels of CysGly-bimane (white circles) and γ-GluCys-bimane (black squares) in Arabidopsis wild-type (wt) cells in the absence (A) and presence (B) of 1 mm azide. C and D, Analysis of ΔPCS cell suspension culture for GS-bimane (white squares) and Cys-bimane (black circles) levels (C) and CysGly-bimane (white circles) and γ-GluCys-bimane (black squares) amounts (D). Cells were exposed to 1 mm azide. The initial amounts of GS-bimane and bimane derivatives were set to 100% at the onset of the experiment. The total levels were 448, 307, and 207 nmol g−1 dry weight bimane derivatives for the wild type (−azide and +azide) and ΔPCS (+azide), respectively. Data points represent means of duplicates (deviation < 10% of value), and two independent repetitions yielded similar results. Incubation conditions are as mentioned in Figure 2.
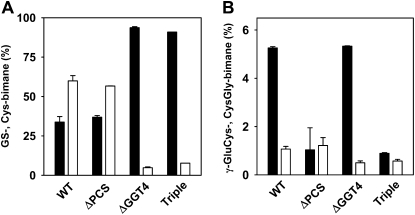
These data are consistent with an efficient PCS-dependent generation of the cleavage product in the cytosol. The GS to Cys conjugate conversion is slowed down in ΔPCS compared with the wild type in the presence of azide (Fig. 3C); however, the conversion in ΔPCS is little affected in the absence of the inhibitor (Blum et al., 2007), indicating a compensatory turnover of the xenobiotic in the vacuolar compartment. GGT4 has been identified as the critical player in the turnover of GS conjugates in the vacuole (Ohkama-Ohtsu et al., 2007b). To determine the contribution of vacuolar and cytosolic pathways to GS-bimane catabolism, we compared the GS-bimane conversion in the ΔGGT4 knockout mutant, the ΔPCS double mutant, and the triple mutant ΔPCSΔGGT4. Homozygous mutant and wild-type seedlings were infiltrated with MCB, and GS-bimane degradation was subsequently analyzed (Fig. 4). The GGT4 deficiency strongly inhibited GS conjugate catabolism and resulted in an approximately 10-fold decrease in Cys-bimane levels, while PCS deficiency in the ΔPCS line marginally affected Cys-bimane levels (Fig. 4A). The triple mutant and the ΔGGT4 line yielded comparable GS and Cys conjugate levels, but the γ-GluCys-bimane level was approximately 5-fold lower in the PCS-deficient triple mutant. CysGly-bimane levels were approximately 2-fold lower in GGT4-deficient lines (Fig. 4B). The comparative analysis corroborates the importance of the vacuolar GGT4 for efficient GS- to Cys-bimane catabolism. We conclude that PCS plays a major role in the metabolism of GS conjugates in the cytosol but only a minor role in the catabolism of GS conjugates that are efficiently sequestered in the vacuole.
Figure 4.
Catabolism of GS-bimane in PCS- and GGT4-deficient Arabidopsis. Five-day-old seedlings of the wild type (WT), PCS double mutant (ΔPCS), GGT4 knockout line (ΔGGT4), and the triple mutant ΔPCSΔGGT4 (Triple) were infiltrated and exposed to MCB (5 μm) for 4 h with a subsequent incubation period of 2 d prior to analysis of bimane derivatives. Values for GS-bimane (black bars) and Cys-bimane (white bars) in A and amounts of γ-GluCys-bimane (black bars) and CysGly-bimane (white bars) in B are presented as percentages of the sum of bimane derivatives (triplicate, ±sd). At the onset of the experiment, the bimane labeling of the seedlings was comparable in the different lines, yielding a total of approximately 90 nmol g−1 fresh weight bimane derivatives (±13%).
Complementation of PCS1 Deficiency by AtPCS1:eGFP
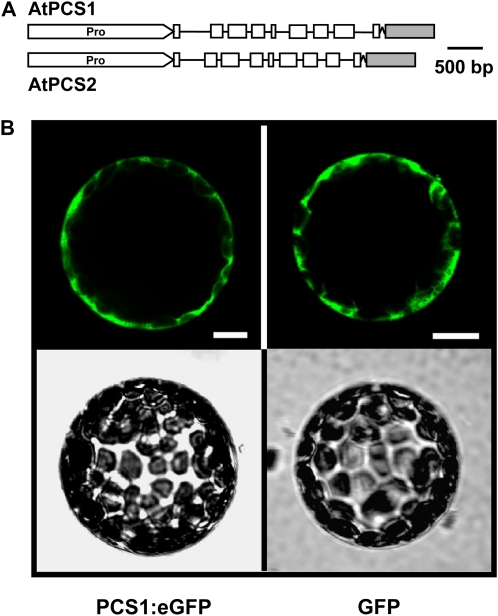
AtPCS1 initiates the GS conjugate catabolic pathway that presumably occurs in the cytosol. The enzyme, however, has not explicitly been shown to reside in the cytosol. To address this open question, Arabidopsis plants expressing PCS:eGFP (enhanced GFP) chimeric proteins were generated by in-frame fusion of the reporter gene to the last exon of the AtPCS1 and AtPCS2 genes (Fig. 5A). The AtPCS1:eGFP and AtPCS2:eGFP constructs encompass endogenous promoter sequences and the coding regions of the PCS genes. Transient expression analysis of AtPCS1:eGFP revealed a GFP signal in Arabidopsis protoplasts similar to cytosolic GFP expression under the control of a constitutive promoter (Fig. 5B). Similar experiments with AtPCS2:eGFP did not yield detectable GFP signals, consistent with no or a very low AtPCS2 expression in leaf cells. Subsequently, the fusion constructs were introduced into wild-type Arabidopsis plants (ecotype Columbia) as well as in a mutant deficient in AtPCS1 (ΔPCS1) and in the double PCS knockout line, ΔPCS. For AtPCS1:eGFP, 10 independent transgenic Arabidopsis lines were identified in the wild-type background (WT-P1 L1–L10) and seven each in ΔPCS1 (ΔPCS1-P1 L1–L7) and in the double knockout (ΔPCS-P1 L1–L7). Similarly, independent PCS2:eGFP transformants were recovered in the wild type (WT-P2 L1–L10) and in PCS-deficient lines (ΔPCS-P2 L1–L13). Lines were selected for a 3:1 segregation of kanamycin resistance in the F2 progeny, indicative of a single T-DNA insertion site, and used in further experiments.
Figure 5.
Arabidopsis PCS fusions with eGFP. A, Schematic presentation of genomic fragments of AtPCS1 and AtPCS2 encompassing the promoter region (Pro) linked to the eGFP cDNA. Exon and intron regions are indicated by white boxes and black lines, respectively. The eGFP cDNA (gray box) was joined in-frame to the 3′ terminus of the PCS gene via an oligonucleotide encoding a linker of 10 Ala residues. B, Expression of AtPCS1:eGFP and GFP in protoplasts of Arabidopsis. The GFP signals of single representative protoplasts are shown in the top row, while the bottom row depicts the corresponding bright-field images. Bars = 10 μm.
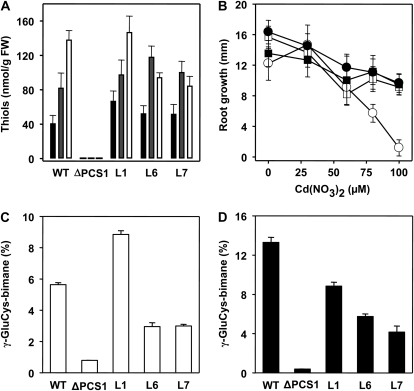
To assess the functionality of the AtPCS1:eGFP fusion, we tested its ability to rescue the mutant phenotype of the ΔPCS1 line, deficient in phytochelatin (PC) biosynthesis and impaired in GS conjugate catabolism. As AtPCS2 mutants have no reported phenotype, our complementation analysis focused on AtPCS1-deficient plants. Three ΔPCS1 lines homozygous for the AtPCS1:eGFP integration (L1, L6, and L7) were analyzed with respect to PC synthesis and cadmium tolerance (Fig. 6, A and B). Seedlings of these lines challenged to Cd2+ showed full restoration of PC synthesis, and generation of PC2, PC3, and PC4 occurred as in the wild type (Fig. 6A). The cadmium concentration chosen for induction of PC synthesis (50 μm) is just sufficient to impose a mild growth inhibition of wild-type roots in the range of 20% (Fig. 6B). At 100 μm Cd2+, residual growth was approximately 60% for the wild type and for both AtPCS1:eGFP lines, while ΔPCS1 showed only 9% of the control without cadmium. Expression of the PCS fusion protein restored cadmium tolerance in ΔPCS1 and also recovered catabolism of GS conjugate comparable to wild-type plants (Fig. 6, C and D). Turnover of the fluorescent GS-bimane conjugate in Arabidopsis generates γ-GluCys-bimane as a transient catabolite, primarily by the action of AtPCS1 (Blum et al., 2007). Protoplasts of the wild type, ΔPCS1, and three complemented ΔPCS1 plants were challenged with MCB and had γ-GluCys-bimane levels accounting for approximately 5.6%, 0.8%, 8.8%, 3.0%, and 3.0% of all recovered GS-bimane and bimane derivatives, respectively (Fig. 6C). The level of γ-GluCys catabolites is increased in the presence of PCS-activating heavy metal ions (Blum et al., 2007). Under such conditions, γ-GluCys-bimane constituted 13.3% of all recovered bimane conjugates in wild-type seedlings 4 h after exposure to MCB (Fig. 6D). In contrast, the γ-GluCys-bimane level in ΔPCS1 was below 0.5%, whereas in the PCS-expressing ΔPCS1 lines, the concentration increased to 4.2%, 5.8%, and 8.8%, respectively. Taken together, these findings document a successful complementation of ΔPCS1 by AtPCS1:eGFP.
Figure 6.
Functional complementation of the ΔPCS1 mutant by AtPCS1:eGFP. A, Cadmium-induced PC formation. Arabidopsis seedlings were exposed to 50 μm Cd2+ for 24 h prior to PC analysis. The level of thiol is given for PC2 (black columns), PC3 (gray columns), and PC4 (white columns). The error bars indicate sd (n = 3). B, Root growth assay for Cd2+ sensitivity. Root growth of 4-d-old seedlings of the wild type (WT; white squares), ΔPCS1 (white circles), and two independent, nonsegregating transgenic ΔPCS1 lines transformed with AtPCS1:eGFP, L1 (black squares) and L7 (black circles), was determined after transfer of the seedlings onto solidified nutritional medium containing Cd2+ as indicated. The values of root extension within 3 d are presented (n = 13; sd values are shown). Analysis of the complemented line L6 yielded comparable results as shown for L1 and L7. C and D, Analysis of GS-bimane turnover in the wild type, ΔPCS1, and three independent transgenic ΔPCS1 lines (L1, L6, and L7). Leaf protoplasts (C) and seedlings (D) of these lines were exposed to 5 μm MCB for 4 h prior to analysis of fluorescent bimane conjugates. The seedlings were also challenged with 10 μm Cd2+. Values of triplicates with sd are given as percentages of the sum of bimane derivatives analyzed (84.2 nmol g−1 fresh weight [FW] bimane derivatives ± 12%).
Localization of PCS Proteins in Arabidopsis
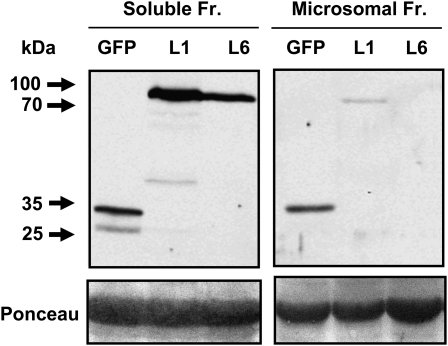
PCS might be a soluble or a membrane-associated protein. To address the issue of intracellular localization of AtPCS1, cell-free extracts of AtPCS1:eGFP-expressing seedlings were separated into a soluble protein fraction and a microsome-enriched fraction by differential centrifugation. Comparable protein levels from both fractions were analyzed for the presence of the tagged AtPCS1 protein by immunodetection (Fig. 7). The AtPCS1 fusion protein was primarily detected in the soluble protein fraction at a molecular mass of approximately 80 kD, which corresponds to the sum of 55 kD for AtPCS1 and 26 kD for eGFP. The microsomal fraction yielded low levels of AtPCS1:eGFP, possibly due to a weak interaction of the fusion protein with membranes or to a contamination of the fraction with soluble proteins, as observed for the GFP control and indicated by the presence of Rubisco (Fig. 7).
Figure 7.
Immunodetection of the AtPCS1 fusion protein. Abundance of AtPCS1:eGFP in the soluble versus microsome-enriched protein fraction (Fr.). Cell-free extracts of ΔPCS1 plants transformed with AtPCS1:eGFP, lines L1 and L6, and of a cytosolic GFP-expressing control line (GFP) were separated into the two protein fractions by differential centrifugation. Top, The protein samples (20 μg per lane) were analyzed by gel electrophoresis and by western-blot analysis using GFP-specific antibodies. Extracts of the GFP line served as a control. The arrows indicate the positions of protein standards, and their molecular masses are given in kD. Bottom, The Ponceau-stained membrane prior to immunodetection shows the large subunit of Rubisco as a prominent band and reveals contamination of the microsome-enriched fraction with soluble proteins.
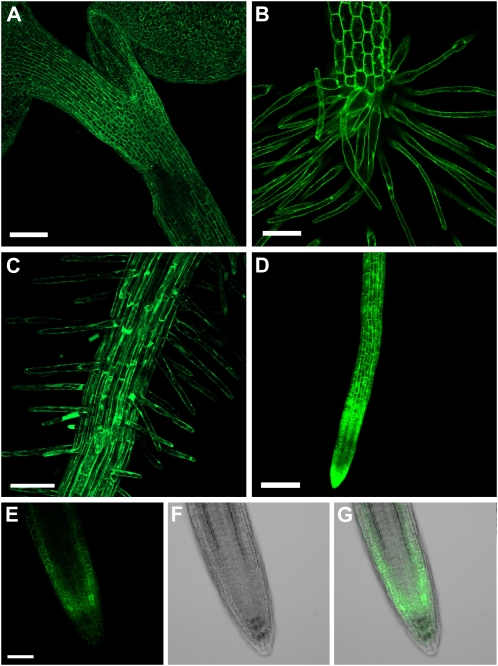
Analyses of PCS:eGFP expression by confocal imaging revealed a major difference between AtPCS1- and AtPCS2-expressing lines. AtPCS1:eGFP was readily detectable throughout the seedling (Fig. 8, A–D). In contrast, the AtPCS2:eGFP signal was consistently low in more than 20 independent transgenic lines analyzed, and the fusion protein was only detected in the root tip, preferentially in the tunica (Fig. 8, E–G). The low expression levels of AtPCS2, however, might have prevented the detection of the fusion protein in other parts of the seedling and also made it difficult to assess the intracellular localization of AtPCS2, other than to say that it resembled that of AtPCS1 described below (Fig. 9).
Figure 8.
Expression of both PCS proteins in Arabidopsis. The presence of AtPCS1 (A–D) and AtPCS2 (E–G) in seedlings of ΔPCS1 transformed with AtPCS1:eGFP and seedlings of ΔPCS transformed with AtPCS2:eGFP is indicated by the fluorescence signal. AtPCS1 expression was observed in the shoot, including cotyledons and hypocotyl (A and B), and throughout the root, with a prominent signal in the root hairs (C and D). AtPCS2 expression was detected only in the root tip, primarily in the cortex. The GFP signal is presented in E, the corresponding bright-field image in F, and the overlay of both photographs in G. The images in A to D are composite presentations of confocal Z scans. Bars = 100 μm (A and D) and 50 μm (B, C, and E).
Figure 9.
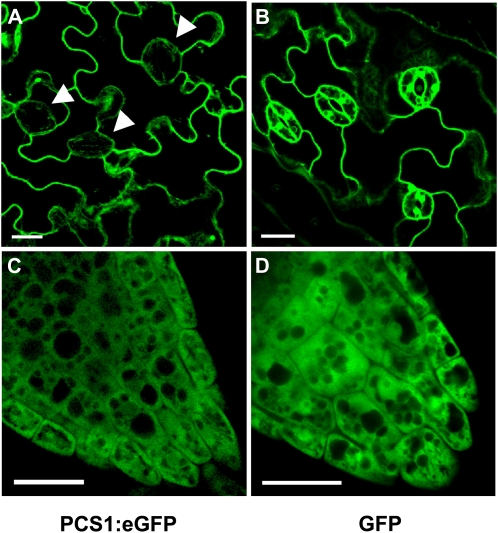
Intracellular localization of AtPCS1. Analysis of the GFP signal in the epidermis (A and B) and root tip (C and D) of stably transformed Arabidopsis seedlings is shown. The expression of AtPCS1:eGFP and GFP is presented. Arrowheads mark the positions of stomates. Bars = 20 μm.
AtPCS1 was predominantly expressed in the epidermal layers of the shoot and root, including root hairs, but weakly expressed in guard cells (Figs. 8 and 9A). In epidermal cells, AtPCS1:eGFP was found in the cytosol, where it could be seen in the cytoplasmic strands of the highly vacuolated cells (Fig. 9A). Comparison of the AtPCS1:eGFP lines with marker lines in which GFP is targeted to specific organelles (Cutler et al., 2000) pointed to a similarity to soluble GFP, which is found in the cytosol (Fig. 9B). The only difference observed was that whereas soluble GFP can be clearly detected in the nucleus of the control line, the AtPCS1:eGFP fusion was not found in the nucleus. In some of the primary transformants, AtPCS1:eGFP labeled diffuse reticulate structures in addition to the cytosol. The analysis of meristematic, largely avacuolate cells of the root tip supported a cytosolic localization of AtPCS1 (Fig. 9, C and D).
DISCUSSION
GSH is of pivotal importance for metal ion homeostasis, detoxification of xenobiotics, and the metabolism of sulfur compounds. PCS is involved in these processes by generating the metal-binding PCs from GSH and catabolizing GS conjugates. In this paper, we localize AtPCS1 to the cytosol and provide evidence for a cytosolic action of the enzyme. While PCS is responsible for cytosolic γ-GluCys conjugate formation, the γ-glutamyl transpeptidase GGT4 plays a prominent role in GS-bimane catabolism in the vacuole. In fact, comparative analysis revealed an almost complete block of GS-bimane turnover in both the GGT4 knockout line and the PCS/GGT4 triple mutant, consistent with an efficient sequestration and catabolism of GS-bimane conjugates in the vacuole (Grzam et al., 2006, 2007; Ohkama-Ohtsu et al., 2007b).
For both xenobiotics and endogenous substrates, there is evidence for a cytosolic GS conjugate metabolism leading to the formation of the γ-GluCys intermediate. The herbicide safener fenclorim, for instance, is subjected to glutathionylation in the cytosol (Brazier-Hicks et al., 2008). The first conjugation to GSH is followed by the turnover to γ-GluCys-fenclorim to Cys-fenclorim, the former a presumed product of PCS activity (Brazier-Hicks et al., 2008). The metabolic reactions are reminiscent of cytosolic steps of the phytoalexin and glucosinolate biosynthetic pathways postulated to involve GSH conjugation and deglycination of the GS conjugate (Bednarek et al., 2009; Böttcher et al., 2009; Clay et al., 2009). This reaction pathway is supported by several observations. GSH deficiency of the Arabidopsis pad2/cad2 mutants leads to a deficiency in the biosynthesis of glucosinolates (Schlaeppi et al., 2008) and to reduced levels of camalexin, the most abundant phytoalexin of Arabidopsis (Parisy et al., 2007). The precursor of camalexin, indoleacetonitrile, is conjugated to GSH, and both γ-GluCys- and Cys-indoleacetonitrile accumulated in the pad3 mutant deficient in the formation of the sulfur-heterocyclic thiazole (Böttcher et al., 2009). A similar process may be responsible for sulfur incorporation into phytoalexin precursors via conjugation to GSH (Bednarek et al., 2009). PCS-deficient plants, however, revealed higher glucosinolate levels and lower levels of catabolites thereof such as methylindole (Clay et al., 2009). This finding does not support a PCS function in the biosynthesis but argues for a critical role of PCS in glucosinolate catabolism involving GS conjugate formation (Bednarek et al., 2009). The functional redundancy of the PCS- and γ-glutamyl transpeptidase-mediated GS conjugate turnover, however, may explain the presence of glucosinolates in the ΔPCS knockout despite a role of PCS in glucosinolate biosynthesis. Analysis of the PCS/GGT4 triple mutant for glucosinolate levels could clarify this issue.
In our analysis with the xenobiotic bimane, we forced the GS-bimane conjugates to remain in the cytosol by azide-mediated inhibition of vacuolar import. The treatment resulted in the strong PCS-dependent accumulation of γ-GluCys conjugate, a more than 2-fold loss of detectable bimane derivatives, and a shift from GS-bimane toward the Cys adduct in plant cells. The absolute amount of Cys-bimane was only slightly increased by a factor of 1.4 in azide-treated compared with control cells, pointing to a specific drop in GS-bimane levels. This observation may either reflect a stimulated conversion of the GS conjugate to not detected derivatives or a more efficient export of GS conjugates out of the cells, since cell integrity was not detectably affected by the inhibitor treatment. A stimulated Cys-bimane formation in Arabidopsis cells by azide has previously been reported (Grzam et al., 2006); however, the authors failed to detect elevated γ-GluCys-bimane levels, which may reflect metabolic differences between leaf pieces and a cell suspension culture. In our hands, the azide-mediated and PCS-dependent γ-GluCys-bimane increase was consistent and prominent. Conversion of γ-GluCys-bimane to the cysteinyl derivative is mediated either by a γ-glutamyl transpeptidase or γ-glutamyl cyclotransferase activity present in the cytoplasm (Ohkama-Ohtsu et al., 2008).
In yeast, the γ-glutamyl transpeptidase activity is required for extracellular secretion of GS conjugates, releasing CysGly- and Cys-bimane into the culture medium (Wünschmann et al., 2010). The secreted Cys conjugates might reenter the cell. The plasma membrane-localized γ-glutamyl transpeptidases of Arabidopsis, GGT1 and GGT2, might fulfill a similar function in the extracellular transport of GS conjugates and their metabolites.
The assays using azide showed that AtPCS1 is active in the cytosol, and our localization studies revealed that it resides in the cytosol throughout the plant. The abundance of AtPCS2 is much lower compared with AtPCS1, and AtPCS2:eGFP was only detected in the root tip. These findings are in agreement with earlier studies that have characterized AtPCS1 as the predominant PCS activity and reported low levels of AtPCS2 expression in Arabidopsis, which was not sufficient to contribute significantly to PC biosynthesis or GS conjugate turnover (Cazale and Clemens, 2001; Lee and Kang, 2005; Blum et al., 2007). The function of AtPCS2 is still a conundrum. The low expression of AtPCS2 might be caused by a missing consensus TATA box in the promoter (Lee and Kang, 2005), nonoptimal codons at the translation start site (Joshi, 1987; Jaiswal and Rangan, 2007), and/or the long 3′ untranslated region (UTR) of AtPCS2 transcript comprising 1,204 nucleotides versus 167 bases for the 3′ UTR of AtPCS1. Long stretches of 3′ UTR are reported to destabilize transcripts (Schwartz et al., 2006). While the low expression level of AtPCS2 prevented the clear assignment of the intracellular compartmentalization in this study, presumably the cytosol, AtPCS1 has been localized to the cytosol based on confocal analysis, subcellular fractionation experiments, and inhibitor studies.
Taken together, PCS is the prominent activity for initiating the cytosolic catabolism of GS conjugates (Fig. 1). Simultaneously, PCS functions in the generation of the metal-binding PCs. Both enzymatic activities are stimulated by heavy metal ions (Beck et al., 2003; Blum et al., 2007), and these findings raise the question of how the two different reactions are regulated. PCs are generated by the transpeptidation reaction of PCS, in which the products, PCs, are scavenging the PCS-activating heavy metal ions. PCS is able to degrade metal-free PCs into γ-GluCys dipeptides by its hydrolase activity; however, PC-metal complexes are no or poor substrates for PCS and therefore stable (Loeffler et al., 1989). GS conjugates with bulky residues do not serve as substrates for PCS-catalyzed transpeptidation; hence, the hydrolase activity (i.e. transfer of GluCys onto a molecule water) predominates. PCS under resting conditions has a low enzymatic activity, which is limited by the availability of heavy metal ions. The high efficiency of copper ions to activate PCS (Beck et al., 2003) implies that PCS is a copper-containing enzyme. Copper mobilization might occur under biotic interactions in plants similar to bacteria and macrophage interaction, where it plays a prominent role in bacterial killing (White et al., 2009). Copper mobilization during biotic interaction could activate PCS. Release of glucosinolates and the generation of catabolites such as isothiocyanates and GS conjugates are part of the biotic defense response in Arabidopsis, and the defense reaction requires PCS function (Bednarek et al., 2009; Clay et al., 2009).
GS conjugation is necessary not only for the detoxification of xenobiotics but also for the transport, biosynthesis, and catabolism of a number of endogenous compounds (Dixon et al., 2002). Glutathionylation has been reported for phenolic plant secondary metabolites such as anthocyanins (Alfenito et al., 1998; Mueller et al., 2000), cinnamic acid, p-coumaric acid (Dean et al., 1995), and caftaric acid in addition to fatty acids (Dixon and Edwards, 2009), the terpenoid gibberthione, a sulfur-containing metabolite of gibberellic acid (Edwards et al., 2000), and possibly metabolites of porphyrin (Dixon et al., 2008). Whether PCS has a critical function in the metabolism of these endogenous compounds remains to be determined.
CONCLUSION
A cytosolic GS conjugate turnover occurs during the biosynthesis of secondary metabolites and the detoxification of reactive compounds. The pathway leads to the formation of γ-GluCys conjugates, and AtPCS1 represents the major enzymatic activity in Arabidopsis responsible for the generation of γ-GluCys conjugates in the cytosol. Future work is required to address the role of AtPCS1 in biotic and abiotic stress responses in addition to its well-established function in heavy metal detoxification.
MATERIALS AND METHODS
Plant Material and Chemicals
For Agrobacterium tumefaciens-mediated transformation and protoplast preparation, Arabidopsis (Arabidopsis thaliana ecotype Columbia) plants were grown in pots on a perlite-soil mixture at 23°C under long-day conditions with 16 h of light (250 μE m−2 s−1). Transient transfection of protoplasts was carried out as described (Blum et al., 2007). The transformation of wild-type and mutant plants (Blum et al., 2007) was carried out using the Agrobacterium strain GV3101pMP90 as described (Clough, 2005). For the construct AtPCS1:eGFP, 10 independent transgenic Arabidopsis lines were identified in the wild-type background (WT-P1 L1–L10) and seven each in ΔPCS1 (ΔPCS1-P1 L1–L7) and in the double knockout (ΔPCS-P1 L1–L7). Similarly, independent AtPCS2:eGFP transformants were recovered in the wild type (WT-P2 L1–L10) and in PCS-deficient lines (ΔPCS-P2 L1–L13). F2 seeds, obtained from selfed transformants, were examined for segregation of the kanamycin resistance marker in a ratio of 3:1, indicative of a single insertion site. Plants of such lines were selected, and nonsegregating, homozygous lines were used in the experiments. The transgenic Arabidopsis line expressing cytoplasmic GFP (Cutler et al., 2000) was a gift from Farhah Assaad. The ΔGGT4 knockout line (Ohkama-Ohtsu et al., 2007b) was obtained from the Ds transposon insertion lines (Parinov et al., 1999) at the Nottingham Arabidopsis Stock Centre (http://nasc.nott.ac.uk; stock no. N161036). Crossing ΔPCS with ΔGGT4 was performed to generate the triple mutant ΔPCSΔGGT4. Plants homozygous for all three homozygous mutations were screened among the F2 generation by PCR using AtPCS- and AtGGT4-specific primers as described (Blum et al., 2007; Ohkama-Ohtsu et al., 2007b).
Seedlings were raised under sterile conditions on agar-containing Murashige and Skoog (MS) solution (Murashige and Skoog, 1962) and used for induction of PC synthesis and confocal fluorescence microscopy analysis. Cell cultures of Arabidopsis were maintained as reported (Blum et al., 2007). All chemicals and standards used were of analytical grade. MCB (Calbiochem; http://www.emdbiosciences.com) was dissolved in acetonitrile to yield a 100 mm stock solution. For the MCB levels used, no inhibitory effect of acetonitrile was observed on the growth of cell suspension cultures. All experiments were carried out in duplicate or triplicate and reproduced in at least two independent analyses.
Plasmid Constructs
The genes encompassing the coding regions of AtPCS1 and AtPCS2 were amplified from genomic DNA and fused upstream of the eGFP cDNA present in a modified version of the plasmid pEZT-NL (http://deepgreen.stanford.edu/), containing a 10-Ala residue-encoding linker (a gift from Ram Yadav). The primers 5′-TATATATAGGCGCGCCATGGCTATGGCGAGTTTA-3′ and 5′-TATATATAGACGTCGCATAGGCAGGAGCAGCG-3′ were used for AtPCS1 amplification and 5′-TATATATAGGCGCGCCATGTCTATGGCGAGTTTG-3′ and 5′-TATATATAGACGTCGCAGGAGCAGAGAGTTCT-3′ for AtPCS2. The promoter sequences (2 kb) of AtPCS1 and AtPCS2 were amplified from genomic DNA by PCR and using the primers 5′-TATATAGGATCCATGTAGGGAATCTGAGAACC-3′/5′-TATATAACGCGTAGAAAGAGCATTCAACACC-3′ and 5′-TATATAGGATCCTGGCCCAATATGTTGTTTCC-3′/5′-TATATAAGATCTCTCACCAACAACTAAACTTG-3′, respectively. The endogenous promoter sequences were cloned upstream of the PCS:eGFP construct using the unique endogenous restriction sites MluI in AtPCS1 and BglII in AtPCS2. The expression cassettes AtPCS1:eGFP and AtPCS2:eGFP were excised via BamHI and SacI and integrated into a modified pSK (pSK-AscI) and thereafter to a modified pBI121 (pBI121-AscI) via AscI restriction (Moes et al., 2008).
Stress Exposure and Labeling with MCB
Arabidopsis seedlings were raised on agar-solidified MS plates and used for stress exposure and MCB labeling experiments 5 d after germination. For the analysis of PCs, seedlings were transferred to 20 mL of liquid MS medium containing 50 μm Cd(NO3)2 and incubated for 48 h prior to extraction and HPLC analysis. Cadmium sensitivity was examined by determination of root growth within 3 d after transfer of the seedlings on MS plates containing up to 100 μm Cd(NO3)2. For the analysis of bimane conjugates, seedlings were vacuum infiltrated for 10 min in MS medium containing 5 μm MCB.
Arabidopsis protoplasts were prepared from 3-week-old rosette leaves as described (Himmelbach et al., 2002), and approximately 5 × 104 protoplasts (0.5 mL) were incubated with 5 μm MCB for 4 h prior to extraction of bimane derivatives (Blum et al., 2007). For experiments with azide (NaN3), 9.6 g (fresh weight) of Arabidopsis cell culture material was transferred to 100 mL of fresh Linsmaier and Skoog medium (Linsmaier and Skoog, 1965) and allowed to recover for 24 h. The cells were preincubated with 1 mm azide for 5 min, subsequently pulsed with 5 μm MCB for 1 min, washed (25 mL), and then incubated in MCB-free medium in the presence or absence of azide as indicated. Aliquots of 10 mL of cell suspension were removed at different time points. Harvested plant material (0.3 g fresh weight) was used for extraction of bimane derivatives, and the remaining material was used for the determination of dry weight. For analysis of the GSH and oxidized glutathione content, 30 mg of cell material was transferred to 0.2 mL of 20 mm Tris-Cl buffer, pH 8, and homogenized by sonification for 15 min. After centrifugation, the cell-free supernatant was incubated (1 h, 37°C) with or without 1 mm dithiothreitol (DTT). The samples were diluted 1:10 with the Tris-Cl buffer (total volume of 50 μL) and incubated with 1 mm MCB (30 min, 37°C). The pH value was adjusted to approximately 3 by administration of 3 μL of 0.36 m HCl prior to HPLC analysis (see below).
GS Conjugate Catabolism and PC Biosynthesis
GS-bimane and its catabolites were analyzed by HPLC (Summit ASI-100; Dionex; http://www.dionex.com) as reported (Newton and Fahey, 1995; Beck et al., 2003). Briefly, seedlings (0.15 g fresh weight) were frozen in liquid nitrogen and ground to powder, and the material was incubated with extraction buffer (80 mm Gly, 20 mm NaCl, and 10 mm EDTA, pH 3). Protoplasts (5 × 104) were directly lysed in 0.4 mL of extraction buffer. Precipitated proteins were removed by centrifugation (10 min, 16,000g), and the deproteinated extract was subjected to HPLC analysis (Blum et al., 2007). PCs were analyzed by online derivatization of sulfhydryl groups with Ellman reagent (Grill et al., 1987).
Immunoblot Analysis
For immunodetection of eGFP-tagged AtPCS1, protein extracts from Arabidopsis seedlings were analyzed essentially as reported for fibrillin (Yang et al., 2006). Briefly, transgenic seedlings (0.1 g fresh weight) were frozen in liquid nitrogen and ground to powder, and the material was incubated for 10 min in 0.4 mL of homogenization buffer (50 mm Tris-Cl, pH 8, 10 mm DTT, 1 mm 4-[2-aminoethyl]-benzolsulfonylfluoride, and 2% SDS). Cell debris was separated by centrifugation (10 min, 16,000g), and the supernatant was subjected to acetone precipitation (80% final acetone level). After centrifugation (10 min, 7,500g), the protein pellets were washed with 80% acetone, resuspended in 100 μL of sample buffer (60 mm Tris-Cl, pH 6.8, 2% SDS, 10% glycerol, 100 mm DTT, and 0.05% bromphenol blue), and heated for 5 min at 95°C. Separation of protein samples was carried out by SDS-PAGE using a 4% stacking gel and a 12% separation gel, followed by western-blot analysis with mouse anti-eGFP antibodies (dilution 1:1,000; Santa Cruz Biotechnology; http://www.scbt.com/) and goat anti-mouse secondary antibodies (dilution 1:10,000; Thermo Scientific Pierce; http://www.piercenet.com/). Signals were detected using the Pierce Super Signal West Femto Trial Kit (Thermo Scientific Pierce) and a CCD camera (Hamamatsu Photonics; http://www.hamamatsu.com).
Confocal Analysis
For eGFP analysis in transgenic plants, 5-d-old seedlings were examined using a confocal laser scanning microscope (Fluoview FV1000; Olympus; http://www.olympus-global.com). A 40× water-immersion 0.9 numerical aperture objective (Olympus) was used, and scanning was carried out with 2-fold or without electronic magnification. Images were acquired and processed with the Fluoview FV1000 acquisition software. Three-dimensional projections of Z stacks of GFP expression in seedlings were generated with the Imaris 6.3 software (Bitplane Scientific Software; http://www.bitplane.com).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Effect of azide on Arabidopsis suspension cells.
Supplementary Material
Acknowledgments
We thank Farhah Assaad for critical reading of the manuscript. The technical assistance of Johanna Berger is gratefully acknowledged. We thank Caroline Klaus and Lisa Held for tending to our plants and Ram Yadav for providing the pEZT plasmid.
References
- Alfenito MR, Souer E, Goodman CD, Buell R, Mol J, Koes R, Walbot V. (1998) Functional complementation of anthocyanin sequestration in the vacuole by widely divergent glutathione S-transferases. Plant Cell 10: 1135–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball L, Accotto GP, Bechtold U, Creissen G, Funck D, Jimenez A, Kular B, Leyland N, Mejia-Carranza J, Reynolds H, et al. (2004) Evidence for a direct link between glutathione biosynthesis and stress defense gene expression in Arabidopsis. Plant Cell 16: 2448–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck A, Lendzian K, Oven M, Christmann A, Grill E. (2003) Phytochelatin synthase catalyzes key step in turnover of glutathione conjugates. Phytochemistry 62: 423–431 [DOI] [PubMed] [Google Scholar]
- Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, et al. (2009) A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323: 101–106 [DOI] [PubMed] [Google Scholar]
- Blum R, Beck A, Korte A, Stengel A, Letzel T, Lendzian K, Grill E. (2007) Function of phytochelatin synthase in catabolism of glutathione-conjugates. Plant J 49: 740–749 [DOI] [PubMed] [Google Scholar]
- Böttcher C, Westphal L, Schmotz C, Prade E, Scheel D, Glawischnig E. (2009) The multifunctional enzyme CYP71B15 (PHYTOALEXIN DEFICIENT3) converts cysteine-indole-3-acetonitrile to camalexin in the indole-3-acetonitrile metabolic network of Arabidopsis thaliana. Plant Cell 21: 1830–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler MW, Montgomery MG, Leslie AG, Walker JE. (2006) How azide inhibits ATP hydrolysis by the F-ATPases. Proc Natl Acad Sci USA 103: 8646–8649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazier-Hicks M, Evans KM, Cunningham OD, Hodgson DR, Steel PG, Edwards R. (2008) Catabolism of glutathione conjugates in Arabidopsis thaliana: role in metabolic reactivation of the herbicide safener fenclorim. J Biol Chem 283: 21102–21112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazale AC, Clemens S. (2001) Arabidopsis thaliana expresses a second functional phytochelatin synthase. FEBS Lett 507: 215–219 [DOI] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ. (2005) Floral dip: Agrobacterium-mediated germ line transformation. Methods Mol Biol 286: 91–102 [DOI] [PubMed] [Google Scholar]
- Cummins I, Bryant DN, Edwards R. (2009) Safener responsiveness and multiple herbicide resistance in the weed black-grass (Alopecurus myosuroides). Plant Biotechnol J 7: 807–820 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. (2000) Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97: 3718–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas S, Zhu X, Baruchel S, Schlichter L, Bendayan R. (2003) Functional expression of the multidrug resistance protein 1 in microglia. J Pharmacol Exp Ther 307: 282–290 [DOI] [PubMed] [Google Scholar]
- Dean JV, Devarenne TP, Lee IS, Orlofsky LE. (1995) Properties of a maize glutathione S-transferase that conjugates coumaric acid and other phenylpropanoids. Plant Physiol 108: 985–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Edwards R. (2009) Selective binding of glutathione conjugates of fatty acid derivatives by plant glutathione transferases. J Biol Chem 284: 21249–21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Lapthorn A, Edwards R. (2002) Plant glutathione transferases. Genome Biol 3: Reviews3004.1–Reviews3004.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Lapthorn A, Madesis P, Mudd EA, Day A, Edwards R. (2008) Binding and glutathione conjugation of porphyrinogens by plant glutathione transferases. J Biol Chem 283: 20268–20276 [DOI] [PubMed] [Google Scholar]
- Dixon DP, Skipsey M, Grundy NM, Edwards R. (2005) Stress-induced protein S-glutathionylation in Arabidopsis. Plant Physiol 138: 2233–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R, Dixon DP. (2005) Plant glutathione transferases. Methods Enzymol 401: 169–186 [DOI] [PubMed] [Google Scholar]
- Edwards R, Dixon DP, Walbot V. (2000) Plant glutathione S-transferases: enzymes with multiple functions in sickness and in health. Trends Plant Sci 5: 193–198 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G, Buchanan B, Dietz KJ, Pfannschmidt T. (2009) Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antioxid Redox Signal 11: 861–905 [DOI] [PubMed] [Google Scholar]
- Grill E, Löffler S, Winnacker EL, Zenk MH. (1989) Phytochelatins, the heavy-metal-binding peptides of plants, are synthesized from glutathione by a specific gamma-glutamylcysteine dipeptidyl transpeptidase (phytochelatin synthase). Proc Natl Acad Sci USA 86: 6838–6842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill E, Winnacker EL, Zenk MH. (1985) Phytochelatins: the principal heavy-metal complexing peptides of higher plants. Science 230: 674–676 [DOI] [PubMed] [Google Scholar]
- Grill E, Winnacker EL, Zenk MH. (1987) Phytochelatins, a class of heavy-metal-binding peptides from plants, are functionally analogous to metallothioneins. Proc Natl Acad Sci USA 84: 439–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzam A, Martin MN, Hell R, Meyer AJ. (2007) γ-Glutamyl transpeptidase GGT4 initiates vacuolar degradation of glutathione S-conjugates in Arabidopsis. FEBS Lett 581: 3131–3138 [DOI] [PubMed] [Google Scholar]
- Grzam A, Tennstedt P, Clemens S, Hell R, Meyer AJ. (2006) Vacuolar sequestration of glutathione S-conjugates outcompetes a possible degradation of the glutathione moiety by phytochelatin synthase. FEBS Lett 580: 6384–6390 [DOI] [PubMed] [Google Scholar]
- Himmelbach A, Hoffmann T, Leube M, Hohener B, Grill E. (2002) Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J 21: 3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal M, Rangan L. (2007) Context sequence for transcription factors surrounding start codon in model crops. Curr Sci 93: 215–218 [Google Scholar]
- Jha S, Karnani N, Dhar SK, Mukhopadhayay K, Shukla S, Saini P, Mukhopadhayay G, Prasad R. (2003) Purification and characterization of the N-terminal nucleotide binding domain of an ABC drug transporter of Candida albicans: uncommon cysteine 193 of Walker A is critical for ATP hydrolysis. Biochemistry 42: 10822–10832 [DOI] [PubMed] [Google Scholar]
- Joshi CP. (1987) An inspection of the domain between putative TATA box and translation start site in 79 plant genes. Nucleic Acids Res 15: 6643–6653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamoureux GL, Rusness DG. (1986) Xenobiotic conjugation in higher plants. Paulson GH, Caldwell J, Hutson DH, Menn JJ, , Xenobiotic Conjugation Chemistry, Vol 299 American Chemical Society, Washington, DC, 62–105 [Google Scholar]
- Lamoureux GL, Rusness DG. (1993) Glutathione in the metabolism and detoxification of xenobiotics in plants. De Kok LJ, Stulen I, Rennenberg H, Brunold C, Rauser WE, , Sulfur Nutrition and Assimilation in Higher Plants: Regulatory, Agricultural and Environmental Aspects. SPB Academic Publishing, The Hague, The Netherlands, 221–237 [Google Scholar]
- Lee S, Kang BS. (2005) Expression of Arabidopsis phytochelatin synthase 2 is too low to complement an AtPCS1-defective Cad1-3 mutant. Mol Cells 19: 81–87 [PubMed] [Google Scholar]
- Leustek T, Martin MN, Bick JA, Davies JP. (2000) Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu Rev Plant Physiol Plant Mol Biol 51: 141–165 [DOI] [PubMed] [Google Scholar]
- Linsmaier EM, Skoog F. (1965) Organic growth factor requirements of tobacco tissue cultures. Physiol Plant 18: 100–127 [Google Scholar]
- Loeffler S, Hochberger A, Grill E, Winnacker EL, Zenk MH. (1989) Termination of the phytochelatin synthase reaction through sequestration of heavy metals by the reaction product. FEBS Lett 258: 42–46 [Google Scholar]
- Martin MN, Saladores PH, Lambert E, Hudson AO, Leustek T. (2007) Localization of members of the gamma-glutamyl transpeptidase family identifies sites of glutathione and glutathione S-conjugate hydrolysis. Plant Physiol 144: 1715–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinoia E, Grill E, Tommasini R, Kreuz K, Amrhein N. (1993) An ATP-dependent glutathione S-conjugate “export” pump in the vacuolar membrane of plants. Nature 364: 247–249 [Google Scholar]
- Meyer AJ, Fricker MD. (2002) Control of demand-driven biosynthesis of glutathione in green Arabidopsis suspension culture cells. Plant Physiol 130: 1927–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moes D, Himmelbach A, Korte A, Haberer G, Grill E. (2008) Nuclear localization of the mutant protein phosphatase abi1 is required for insensitivity towards ABA responses in Arabidopsis. Plant J 54: 806–819 [DOI] [PubMed] [Google Scholar]
- Mueller LA, Goodman CD, Silady RA, Walbot V. (2000) AN9, a petunia glutathione S-transferase required for anthocyanin sequestration, is a flavonoid-binding protein. Plant Physiol 123: 1561–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullineaux PM, Rausch T. (2005) Glutathione, photosynthesis and the redox regulation of stress-responsive gene expression. Photosynth Res 86: 459–474 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassay with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Newton GL, Fahey RC. (1995) Determination of biothiols by bromobimane labeling and high performance liquid chromatography. Methods Enzymol 251: 148–166 [DOI] [PubMed] [Google Scholar]
- Noctor G, Gomez L, Vanacker H, Foyer CH. (2002) Interactions between biosynthesis, compartmentation and transport in the control of glutathione homeostasis and signalling. J Exp Bot 53: 1283–1304 [DOI] [PubMed] [Google Scholar]
- Ohkama-Ohtsu N, Oikawa A, Zhao P, Xiang C, Saito K, Oliver DJ. (2008) A γ-glutamyl transpeptidase-independent pathway of glutathione catabolism to glutamate via 5-oxoproline in Arabidopsis. Plant Physiol 148: 1603–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkama-Ohtsu N, Radwan S, Peterson A, Zhao P, Badr AF, Xiang C, Oliver DJ. (2007a) Characterization of the extracellular gamma-glutamyl transpeptidases, GGT1 and GGT2, in Arabidopsis. Plant J 49: 865–877 [DOI] [PubMed] [Google Scholar]
- Ohkama-Ohtsu N, Zhao P, Xiang C, Oliver DJ. (2007b) Glutathione conjugates in the vacuole are degraded by gamma-glutamyl transpeptidase GGT3 in Arabidopsis. Plant J 49: 878–888 [DOI] [PubMed] [Google Scholar]
- Parinov S, Sevugan M, Ye D, Yang WC, Kumaran M, Sundaresan V. (1999) Analysis of flanking sequences from dissociation insertion lines: a database for reverse genetics in Arabidopsis. Plant Cell 11: 2263–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisy V, Poinssot B, Owsianowski L, Buchala A, Glazebrook J, Mauch F. (2007) Identification of PAD2 as a gamma-glutamylcysteine synthetase highlights the importance of glutathione in disease resistance of Arabidopsis. Plant J 49: 159–172 [DOI] [PubMed] [Google Scholar]
- Rea PA. (2007) Plant ATP-binding cassette transporters. Annu Rev Plant Biol 58: 347–375 [DOI] [PubMed] [Google Scholar]
- Riechers DE, Fuerst EP, Miller KD. (1996) Initial metabolism of dimethenamid in safened and unsafened wheat shoots. J Agric Food Chem 44: 1558–1564 [Google Scholar]
- Schlaeppi K, Bodenhausen N, Buchala A, Mauch F, Reymond P. (2008) The glutathione-deficient mutant pad2-1 accumulates lower amounts of glucosinolates and is more susceptible to the insect herbivore Spodoptera littoralis. Plant J 55: 774–786 [DOI] [PubMed] [Google Scholar]
- Schützendübel A, Polle A. (2002) Plant responses to abiotic stresses: heavy metal-induced oxidative stress and protection by mycorrhization. J Exp Bot 53: 1351–1365 [PubMed] [Google Scholar]
- Schwartz AM, Komarova TV, Skulachev MV, Zvereva AS, Dorokhov YL, Atabekov JG. (2006) Stability of plant mRNAs depends on the length of the 3′ untranslated region. Biochemistry (Mosc) 71: 1377–1384 [DOI] [PubMed] [Google Scholar]
- Soranzo N, Sari Gorla M, Mizzi L, De Toma G, Frova C. (2004) Organisation and structural evolution of the rice glutathione S-transferase gene family. Mol Genet Genomics 271: 511–521 [DOI] [PubMed] [Google Scholar]
- Vatamaniuk OK, Mari S, Lang A, Chalasani S, Demkiv LO, Rea PA. (2004) Phytochelatin synthase, a dipeptidyltransferase that undergoes multisite acylation with gamma-glutamylcysteine during catalysis: stoichiometric and site-directed mutagenic analysis of Arabidopsis thaliana PCS1-catalyzed phytochelatin synthesis. J Biol Chem 279: 22449–22460 [DOI] [PubMed] [Google Scholar]
- White C, Lee J, Kambe T, Fritsche K, Petris MJ. (2009) A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J Biol Chem 284: 33949–33956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf AE, Dietz KJ, Schroder P. (1996) Degradation of glutathione S-conjugates by a carboxypeptidase in the plant vacuole. FEBS Lett 384: 31–34 [DOI] [PubMed] [Google Scholar]
- Wünschmann J, Krajewski M, Letzel T, Huber EM, Ehrmann A, Grill E, Lendzian KJ. (2010) Dissection of glutathione conjugate turnover in yeast. Phytochemistry 71: 54–61 [DOI] [PubMed] [Google Scholar]
- Yang Y, Sulpice R, Himmelbach A, Meinhard M, Christmann A, Grill E. (2006) Fibrillin expression is regulated by abscisic acid response regulators and is involved in abscisic acid-mediated photoprotection. Proc Natl Acad Sci USA 103: 6061–6066 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.