Abstract
Whole genome tiling arrays at a user specified resolution are becoming a versatile tool in genomics. Chromatin immunoprecipitation on microarrays (ChIP-chip) is a powerful application of these arrays. Although there is an increasing number of methods for analyzing ChIP-chip data, perhaps the most simple and commonly used one, due to its computational efficiency, is testing with a moving average statistic. Current moving average methods assume exchangeability of the measurements within an array. They are not tailored to deal with the issues due to array designs such as overlapping probes that result in correlated measurements. We investigate the correlation structure of data from such arrays and propose an extension of the moving average testing via a robust and rapid method called CMARRT. We illustrate the pitfalls of ignoring the correlation structure in simulations and a case study. Our approach is implemented as an R package called CMARRT and can be used with any tiling array platform.
Keywords: ChIP-chip, moving average, autocorrelation, false discovery rate
1. Background
Whole genome tiling arrays utilize array-based hybridization to scan the entire genome of an organism at a user specified resolution. Among their applications are ChIP-chip experiments for studying protein-DNA interactions. These experiments produce massive amounts of data and require rapid and robust analysis methods. Some of the commonly used methods are ChIPOTle,1 Mpeak,2 TileMap,3 HMMTiling,4 MAT5 and TileHGMM.6 Although these algorithms have been shown to be useful, they don't address the issues due to array designs. The most obvious issue is the correlation of the measurements from probes mapping to consecutive genomic locations.15 The basis for such a correlation structure is due to both overlapping probe design and fragmentation of the DNA sample to be hybridized on the array. There are several hidden Markov model (HMM) approaches to address the dependence among probes but the current implementations are limited to first order Markov dependence.4 Generalizations to higher orders increase the computational complexity immensely. We investigate the correlation structure of data from complex tiling array designs and propose an extension of the moving average approaches1,7 that carefully addresses the correlation structure. Our approach is based on estimating the variance of the moving average statistic by a detailed examination of the correlation structure and is applicable with any array platform. We illustrate the pitfalls of ignoring the correlation structure and provide several simulations and a case study illustrating the power of our approach CMARRT (Correlation, Moving Average, Robust and Rapid method on Tiling array).
2. Methods
Let Y1,…,YN denote measurements on the N probes of a tiling path. Yi could be an average log base 2 ratio of the two channels or (regularized) paired t-statistic for arrays with two channels (e.g., Nimblegen) and a (regularized) two sample t-statistic for single channel arrays (Affymetrix) at the i-th probe. These wide range of definitions of Y make our approach suitable for experiments with both single and multiple replicates per probe.
A common test statistic for analyzing ChIP-chip data is a moving average of Yi's over a fixed number of probes or fixed genomic distance.1,3,7 The parameter wi will be used to define a window size of 2wi+1, i.e., wi probes to the right and left of the i-th probe. In the case of moving average across a fixed number of probes for tiling arrays with constant probe length and resolution, the window size wi is calculated by L×(2wi+1)−2wi×O = FL, where L is the probe length, O is the overlap between two probes and FL is the average fragment size. Our framework also covers tiling arrays with non-constant resolution. In this case, wi will be different for each genomic interval and corresponds to the number of probes within a fixed genomic distance. For simplicity in presentation, we will utilize window size of fixed number of probes. We assume that the data has been properly normalized by potentially taking into account the sequence features, 8 and that E[Y] = μ and var(Y) = σ2. Consider the following moving average statistic
| (1) |
Then, standard variance calculation leads to
| (2) |
The standardized moving average statistic is given by
| (3) |
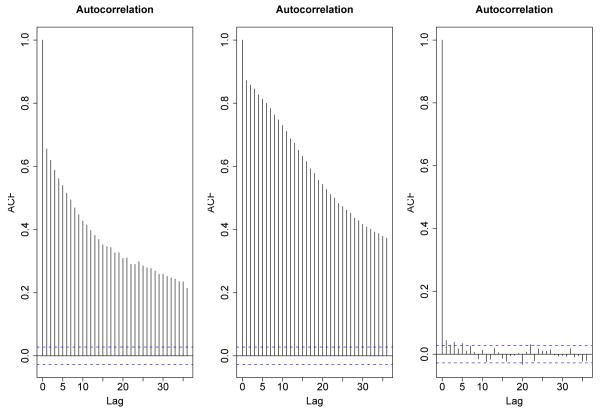
Standard practice of using moving average statistics relies on (1) estimating σ2 based on the observations that represent lower half of the unbound distribution; (2) ignoring the covariance term in equation (2); (3) and obtaining a null distribution under the hypothesis of no binding at probe i. In particular, ChIPOTle considers a permutation scheme where the probes are shuffled and the empirical distribution of the test statistic over several shufflings is used as an estimate of the null distribution. As an alternative, a Gaussian approximation is utilized assuming that Yi's are independent and identically distributed as normal random variables under the null distribution. As discussed by the authors of ChIPOTle, both approaches assume the exchangeability of the probes under the null hypothesis. Exchangeability implies that the correlation within any subset of the probes is the same. However, empirical autocorrelation plots from tiling arrays often exhibit evidence against this (Fig. 1). In particular, in the case of overlapping designs, a correlation structure is expected by design. When the spacing among the probes is large, correlation diminishes as expected (the right panel of Fig. 1), and this was the case for the dataset on which ChIPOTle was developed.
Fig. 1. Example autocorrelation plots from ChIP-chip data.
The left, middle and right panels are from the data in Krig et al.,14 Landick Lab and Kim et al.2 respectively. The autocorrelation plots for Krig et al.14 and Landick Lab clearly show the presence of correlations among probes. The autocorrelation plot for Kim et al.2 shows that the correlation structure diminish with increasing spacing between probes. The data from Krig et al.14 and Landick Lab are from tiling arrays with overlappping probes, whereas the design in Kim et al.2 have subtantial spacing between probes (i.e., probe length = 50 bp and resolution = 100 bp).
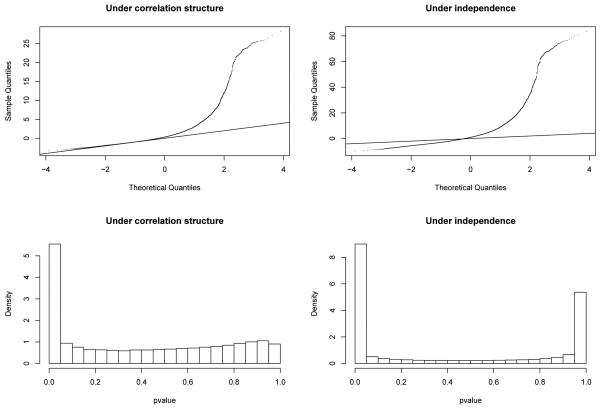
We illustrate the problem with ignoring the correlation structure on a ChIP-chip dataset from an E-coli RNA Polymerase II experiment utilizing a Nimblegen isothermal array (Landick Lab, Department of Bacteriology, UW-Madison). The probe lengths vary between 45 and 71 bp, tiled at a 22 bp resolution. Approximately half of the probes are of length 45 bp. We compute the standardized moving average statistic Si (assuming cov(Yj; Yk) ≠ 0) and (assuming independence of Yi's). A method of estimating cov(Yj, Yk) is described in the next section. The p-values for each Si and are obtained from the standard Gaussian distribution under the null hypothesis. We expect the quantiles of Si and for unbound probes to fall along a 45° reference line against the quantiles from the standard Gaussian distribution, whereas the quantiles for bound probes to deviate from this reference line. As evident in Fig. 2, if the correlation structure is ignored, the distribution of 's for unbound probes deviates from the standard Gaussian distribution. Since the data is obtained from a RNA Polymerase II experiment, we expect a larger number of points, corresponding to promoters, to deviate from the reference line. An additional diagnostic tool is the histogram of the p-values. If the underlying distributions for Si and are correctly specified, the p-values obtained should be a mixture of uniform distribution between 0 and 1 and a non-uniform distribution concentrated near 0. The histograms of the p-values (Fig. 2) again illustrate that the distribution for Ti is misspecified.
Fig. 2. Normal quantile-quantile plots (qqplot) and histograms of p-values.
The left panels show the qqplot of Si and distribution of p-values under correlation structure. The top right panel shows that if the correlation structure is ignored, the distribution of 's for unbound probes deviates from the standard Gaussian distribution. The bottom right panel shows that if the correlation structure is ignored, the distribution of p-values for unbound probes deviates from the uniform distribution for larger p-values.
2.1. Estimating the correlation structure
Although it is desirable to develop a structured statistical model that captures the correlations, developing such a model is both theoretically and computationally challenging due to the complex, heterogeneous data generated by tiling array experiments. We propose a fast empirical method that estimates the correlation structure based on sample autocorrelation function. The covariance cov(Yj, Yj+k) can be estimated from the sample autocorrelation ρ̂(k) and sample variance σ̂2,10
| (4) |
The following strategy is used in CMARRT for estimating the correlation structure. The top M% of outlying probes which roughly correspond to bound probes are excluded in the estimation of ρ̂(k). For the remaining probes, the sample autocorrelation at lag k ρ̂j(k)) is computed for each segment j consisting of at least N consecutive probes. Genomic regions flanking a large gap or repeat masked regions will be considered as two separate segments. For any lag k, we let ρ̂(k) to be the average of ρ̂j(k) over j. Here, N can be considered as a tuning parameter and our initial experiments with ENCODE datasets suggest that N = 500 works well in practice based on the diagnostic plots discussed in Section 1. M is an anti-conservative preliminary estimate of the percentage of bound probes which can be obtained under the assumption of independence among probes (usually ∼ 1 – 5%, depending on the type of ChIP-chip experiment).
3. Simulation studies
In this section, we investigate the performance of CMARRT, the conventional normal approximation approach under the independence assumption (Indep) and the HMM option in TileMap under various scenarios where we know the true bound regions in terms of sensitivity and specificity while controlling FDR at various levels used in practice.
Simulation I: Autoregressive model
We consider the following model
| (5) |
where Ni is the autoregressive background component and Ri is the real signal. We generate 100,000 Ni from AR(p) to represent the background component under the assumption of cor(Ni, Ni+k) = ρ0.4(k−1)+1 and randomly choose 500 peak start sites. We let the size of a peak to be 10 probes, so that ∼ 5% of the probes belong to bound regions. To design scenarios similar to what we have observed in practice, we also allow for ∼ 3 outliers within a bound region. The data is simulated from various p (AR order), ρ (cor(Ni, Ni+k)) and σ (var(Ni)) for the background component, and strength c for the real signal.
Simulation II: Hidden Markov model
In this scenario, the data is simulated from hidden markov models (HMMs)12 with explicit state duration distribution to introduce direct dependencies at the probe level observations. Let the duration HMM densities be pSi (di) ∼Geometric(pSi). The transition probabilities (aij) and the parameters pSi in the duration HMM densities are chosen such that ∼ 5% of the probes belong to bound regions. We consider the joint observation density fNi (Y1, Y2, …Yd1) ∼ MV N(0;ΣN) for the unbound regions and fBi (Y1, Y2, …Yd1) ∼ MV N(μ, ΣB); μ > 0 for the bound regions, where MV N denotes the multivariate normal distribution. The parameters μ, ΣN and ΣB are chosen such that generated data resembles observed ChIP-chip data exhibiting correlations at the observation level.
Each simulation scenario is repeated 50 times. A probe is declared as bound if its adjusted p-value11 is smaller than a pre-specified FDR level α when analyzing with CMARRT and Indep. For TileMap, we use the direct posterior probability approach13 to control the FDR.
3.1. Results of simulations I and II
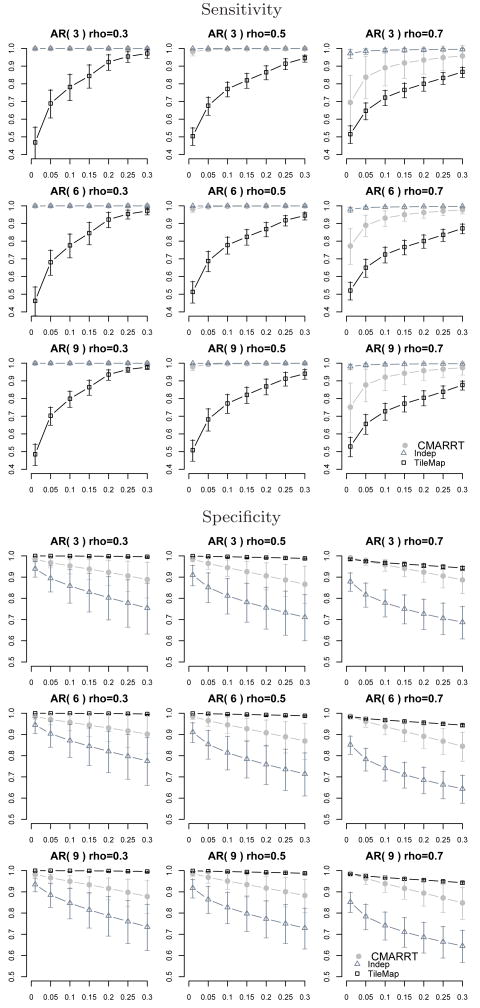
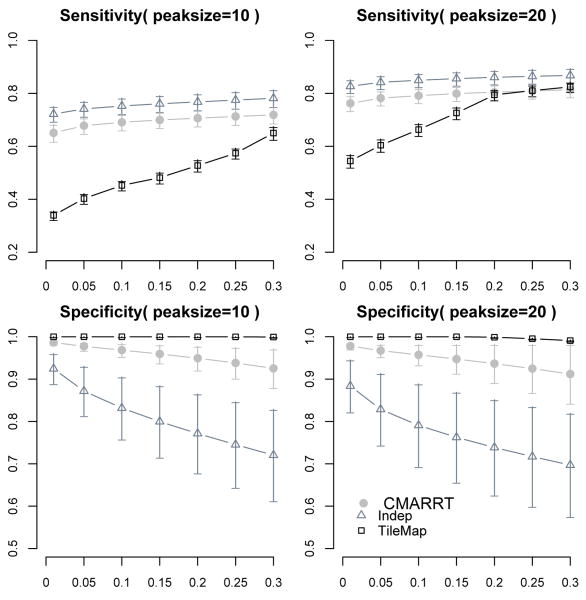
In Fig. 3, we summarize the sensitivity at the peak level and the specificity at various FDR thresholds from Simulation I for CMARRT, Indep, and TileMap. CMARRT is able to identify most of the bound regions at FDR of 0.05 and above while TileMap tends to be more conservative in declaring bound regions as shown in the sensitivity plots. Although Indep has the highest sensitivity, it also has a high proportion of false positives. The specificity of Indep is significantly lower compared to CMARRT, even under the case of low correlation among the probes. Similar results are obtained in Simulation II under the duration HMM (Fig. 4). The left panels show the sensitivity and specificity for the case of smaller peaks with an average peak size of 10 probes while the right panels are for the case of larger peaks of size 20 probes on average. These results illustrate the superior performance of CMARRT in terms of both sensitivity and specificity even when the data is generated from a complex model. The heuristic way of estimating the correlation structure in CMARRT is able to reduce the false positives (specificity) significantly, but not at the expense of increasing false negatives (sensitivity). On the other hand, ignoring the correlation structure results in a higher proportion of false positives. Additionally, the HMM option in TileMap is more conservative than the moving average approach when the FDR is controlled at the same level.
Fig. 3. Sensitivity at peak level (top figure) and specificity (bottom figure) at various FDR control (x-axis).
The background N is generated from various autoregressive models with sd(Ni)=0.3, Yi = Ni + 1.5, p = {3, 6, 9} and ρ = {0.3, 0.5, 0.7}. Vertical lines are error bars. CMARRT is able to identify most of the bound regions at FDR of 0.05 and above. TileMap tends to be more conservative in declaring bound regions. Although Indep gives the highest sensitivity, it also has the highest proportion of false positives. The specificity for CMARRT is significantly higher than the Indep approach.
Fig. 4. Sensitivity and specificity at various FDR control (x-axis).
The left panels are the results under duration HMM simulation with average peak size of 10 probes. The right panels correspond to using average peak size of 20 probes. TileMap tends to be more conservative and has the lowest sensitivity and highest specificity. CMARRT is able to achieve a balance between sensitivity and specificity at each FDR threshold. Indep tends to identify many false positives.
4. Case study: ZNF217 ChIP-chip data
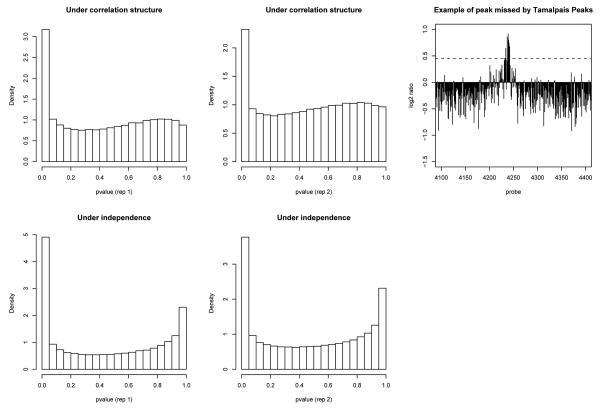
We provide an illustration of CMARRT with a ZNF217 ChIP-chip data tiling the ENCODE regions (available from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/)14 with accession number GSE6624). The ENCODE regions were tiled at a density of one 50-mer every 38 bp, leading to ∼ 380,000 50-mer probes on the array. We analyze two different replicates of this dataset separately and compare the analysis on these single replicates. In Krig et al.,14 the bound regions were identified with the Tamalpais Peaks program,9 which requires a bound region to have at least 6 consecutive probes in the top 2% of the log base 2 ratios. This criteria tends to be too stringent and fails to identify bound regions which contain a few outlier probes with log base 2 ratios below the top 2% threshold and may result in a higher level of false negatives. In the top right panel of Fig. 5, we show one potential peak missed by the Tamalpais Peaks program. In such cases, the sliding window approach is more powerful for finding peaks. Moreover, this method also assumes the observations are independent. As evident in the left panel of Fig. 1, observations from nearby probes in this tiling array are correlated. As shown in Fig 5, the histograms of p-values for the unbound probes under the independence assumption deviates from the expected distribution in both replicates. Similar problem is present in the normal quantile-quantile plots (online supp. mat.) when the correlation structure is ignored.
Fig. 5. Histograms of p-values for replicates 1 and 2 and an example of peak missed by the Tamalpais Peaks program.
The distributions of the probes for unbound regions deviates from uniform distribution when the correlation structure is not taken into account (bottom panels). The dotted line in top right panel is the 98-th percentile of the log base 2 ratios. Tamalpais Peaks requires a peak to have at least 6 probes in a row to be in the top 2 %.
As in Krig et al.,14 we require the number of consecutive probes in each bound region to be at least 6. A set of peaks is obtained for each replicate at a given FDR control. We assess the extent of overlaps between the set of peaks in these two replicates. The results are summarized in Table 1. All the methods identified more peaks in replicate 1 than replicate 2. Therefore, using the peaks from rep 1 as reference, the common peaks are defined as the percentage of overlapping peaks in replicate 2. For all FDR thresholds (except 0.01), CMARRT has the highest value of common peaks, followed by Indep and TileMap, which illustrates the consistency of the peaks identified by CMARRT.
Table 1.
Distance of ZNF217-binding sites relative to TSS.
| FDR=0.01 | CMARRT | Indep | TileMap |
| Common peaks | 0.803(791/935) | 0.819(1423/1736) | 0.718(799/1113) |
| % of peaks within ±2kb | 0.334 | 0.278 | 0.136 |
| % of peaks within ±10kb | 0.619 | 0.565 | 0.442 |
| % of peaks within ±100kb | 0.911 | 0.903 | 0.824 |
| FDR=0.05 | CMARRT | Indep | TileMap |
| Common peaks | 0.806(1023/1269) | 0.790(1796/2272) | 0.714(978/1370) |
| % of peaks within ±2kb | 0.321 | 0.267 | 0.134 |
| % of peaks within ±10kb | 0.589 | 0.565 | 0.431 |
| % of peaks within ±100kb | 0.903 | 0.900 | 0.826 |
| FDR=0.10 | CMARRT | Indep | TileMap |
| Common peaks | 0.805(1209/1491) | 0.779(2096/2689) | 0.703(1071/1524) |
| % of peaks within ±2kb | 0.300 | 0.265 | 0.135 |
| % of peaks within ±10kb | 0.579 | 0.561 | 0.428 |
| % of peaks within ±100kb | 0.904 | 0.894 | 0.821 |
| FDR=0.15 | CMARRT | Indep | TileMap |
| Common peaks | 0.794(1333/1678) | 0.763(2301/3051) | 0.701(1171/1671) |
| % of peaks within ±2kb | 0.284 | 0.259 | 0.136 |
| % of peaks within ±10kb | 0.564 | 0.552 | 0.434 |
| % of peaks within ±100kb | 0.899 | 0.890 | 0.827 |
As an independent validation, we determine the location of bound regions relative to the transcription start site (TSS) of the nearest gene using GENECODE genes from UCSC Genome Browser as in Krig et al.14 (Table 1). For a given FDR control, the percentage of peaks located within ±2kb, ±10kb and ±100kb of the TSS is the highest in CMARRT, followed by Indep and TileMap. As expected, these numbers decrease as we increase the FDR threshold for all the three methods. These results illustrate the power of CMARRT in detecting biologically more plausible bound regions of ZNF217.
5. Discussion
We have investigated and illustrated the pitfalls of ignoring the correlation structure due to tiling array design in ChIP-chip data analysis. We proposed an extension of the moving average approaches in CMARRT to address this issue. CMARRT is a robust and fast algorithm that can be used with any tiling platform and any number of replicates. Both the simulation results and the case study illustrate that CMARRT is able to reduce false positives significantly but not at the expense of increasing false negatives, thereby giving a more confident set of peaks. We have recently became aware of the work of Bourgon15 who carefully studies the correlation structure in ChIP-chip arrays and proposes a fixed order autoregressive moving average model (ARMA(1, 1)) and we are in the process of comparing CMARRT with this approach.
CMARRT is developed using the Gaussian approximation approach and the diagnostic plots illustrated can be utilized to detect whether a given dataset violates this assumption. One possible relaxation of this assumption is a constrained permutation approach that aims to conserve the correlation structure among the probes under the null distribution. Implementation of such an approach efficiently is a challenging future research direction.
Supplementary Material
Acknowledgments
We thank Professor Robert Landick for providing the E-coli ChIP-chip data for our analysis. Supplementary materials are available at http://www.stat.wisc.edu/∼keles/CMARRT.sm.pdf. This research has been supported in part by a PhARMA Foundation Research Starter Grant (P.K. and S.K.) and NIH grants 1-R01-HG03747-01 (S.K.) and 4-R37-GM038660-20 (H.C.).
References
- 1.Buck MJ, Nobel AB, Lieb JD. ChIPOTle: a user-friendly tool for the analysis of ChIP-chip data. Genome Biol. 2005;6(11) doi: 10.1186/gb-2005-6-11-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmand TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ji H, Wong WH. TileMap: create chromosomal map of tiling array hybridizations. Bioinformatics. 2005;21(18):3629–3636. doi: 10.1093/bioinformatics/bti593. [DOI] [PubMed] [Google Scholar]
- 4.Li W, Meyer CA, Liu XS. A hidden Markov model for analyzing ChIP-chip experiments on genome tiling arrays and its application to p53 binding sequences. Bioinformatics. 2005;21(1):i274–i282. doi: 10.1093/bioinformatics/bti1046. [DOI] [PubMed] [Google Scholar]
- 5.Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS. MAT: Model-based Analysis of Tiling-arrays for ChIP-chip. Proc Natl Acad Sci USA. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keles S. Mixture modeling for genome-wide localization of transcription factors. Biometrics. 2006;63(1):10–21. doi: 10.1111/j.1541-0420.2005.00659.x. [DOI] [PubMed] [Google Scholar]
- 7.Keles S, van der Laan MJ, Dudoit S, Cawley SE. Multiple Testing Methods for ChIP-Chip High Density Oligonucleotide Array Data. J of Comp Bio. 2006;13(3):579–613. doi: 10.1089/cmb.2006.13.579. [DOI] [PubMed] [Google Scholar]
- 8.Royce TE, Rozowsky JS, Gerstein MB. Assessing the need for sequence-based normalization in tiling microarray experiments. Bioinformatics. 2007 doi: 10.1093/bioinformatics/btm052. [DOI] [PubMed] [Google Scholar]
- 9.Bieda M, Xu X, Singer MA, Green R, Farnham PJ. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome. 2007 doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Box GP, Jenkins GM. Time series analysis forecasting and control. Holden-Day; 1976. [Google Scholar]
- 11.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. JRSS-B. 1995;57:289–300. [Google Scholar]
- 12.Rabiner LR. A Tutorial on Hidden Markov Models and Selected Applications in Speech Recognition. Proceedings of the IEEE. 1989;77(2):257–286. [Google Scholar]
- 13.Newton MA, Noueiry A, Sarkar D, Ahlquist P. Detecting differential gene expression with a semiparametric hierarchical mixture method. Biostatistics. 2004;5:155–176. doi: 10.1093/biostatistics/5.2.155. [DOI] [PubMed] [Google Scholar]
- 14.Krig SR, Jin VX, Bieda MC, O'Geen H, Yaswen P, Green R, Farnham PJ. Identification of genes directly regulated by the oncogene ZNF217 using ChIP-chip assays. J Biol Chem. 2007;282(13):9703–9712. doi: 10.1074/jbc.M611752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bourgon RW. Ph D Thesis. UC Berkeley; 2006. Chromatin immunoprecipitation and high-density tiling microarrays: a generative model, methods for analysis and methodology assessment in the absence of a “gold standard”. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.