Abstract
Phytochemicals are dietary phytoestrogens that may play a role in prostate cancer prevention. Forty percent of Americans use complementary and alternative medicines (CAM) for disease prevention and therapy. Ashwagandha (Withania somnifera) contains flavonoids and active ingredients like alkaloids and steroidal lactones which are called ‘Withanolides’. We hypothesize that the immunomodulatory and anti-inflammatory properties of Ashwagandha might contribute to its overall effectiveness as an anti-carcinogenic agent. The goal of our study was gain insight into the general biological and molecular functions and immunomodulatory processes that are altered as a result of Ashwagandha treatment in prostate cancer cells, and to identify the key signaling mechanisms that are involved in the regulation of these physiological effects using genomic microarray analysis in conjunction with quantitative real-time PCR and western blot analysis. Ashwagandha treatment significantly downregulated the gene and protein expression of proinflammatory cytokines IL-6, IL-1β, chemokine IL-8, Hsp70 and STAT-2, while a reciprocal upregulation was observed in gene and protein expression of p38 MAPK, PI3K, caspase 6, Cyclin D and c-myc. Furthermore, Ashwagandha treatment significantly modulated the JAK-STAT pathway which regulates both the apoptosis process as well as the MAP kinase signaling. These studies outline several functionally important classes of genes, which are associated with immune response, signal transduction, cell signaling, transcriptional regulation, apoptosis and cell cycle regulation and provide insight into the molecular signaling mechanisms that are modulated by Ashwagandha, thereby highlighting the use of this bioflavanoid as effective chemopreventive agent relevant to prostate cancer progression.
Keywords: Ashwagandha, bioflavonoids, genomics, immunomodulation, microarrays, prostate cancer, signal transduction
Introduction
The latest statistics from the American Cancer Society estimates that a total of 218 890 new cases of prostate cancer and 27 050 deaths from the disease are anticipated in the United States in 2007, making it the most frequent nondermatologic cancer among US males (1,2). Prostate cancer is about twice as common among African-American men as it is among white American men and exhibits tremendous differences in incidence among populations worldwide (1–3). This profound geographic and racial variation in prostate cancer risk has stimulated investigations into how diet affects the development and progression of prostate cancer (4,5).
Phytochemicals are dietary phytoestrogens that may play a role in prostate cancer prevention (6–10). Plant compounds inhibit carcinogenesis via several mechanisms including antioxidant activity, changes in carcinogen metabolism, modulation of the cell cycle, alterations in intracellular signaling, and inhibition of angiogenesis (10–15). Approximately 40% of Americans use complementary and alternative medicines (CAM) for disease prevention and therapy. Over 5000 flavonoids, the largest class of phenolics, have been described (16–19). Polyphenols that are effective when given during the post-initiation period, inhibiting tumor promotion and progression, appear to be more useful in preventing cancer in humans than polyphenols that are effective only when given before and during carcinogen exposure (20).
Ashwagandha (Withania somnifera)—known as Indian ginseng is cultivated in India and North America, and whose roots have been used for thousands of years by Ayurvedic practitioners (21,22). Ashwagandha's root contains flavonoids and many active ingredients like alkaloids and steroidal lactones which are commonly called Withanolides (16). The chemicals constituents of Ashwagandha include three natural powerful antioxidants-superoxide dismutase, catalase and glutathione peroxidase (17–19). Ashwagandha may have chemopreventive properties that make it a potentially useful adjunct for patients undergoing radiation and chemotherapy (15, 20).
Our previous studies have evaluated the antiproliferative properties of various flavonoids (23,24), and have indicated that flavonoids can modulate cell cycle regulation making them potential anti-cancer agents.
To obtain a comprehensive view of the possible molecular and biological processes that are modulated by Ashwagandha treatment to prostate cells, we performed genomic analysis using a cDNA microarray. The use of the microarray technology allowed us to rapidly screen large numbers of potentially relevant genes that are involved in the controlling of certain key biological processes, this gives us an insight into the general physiological and immunomodulatory processes that are altered as a result of Ashwagandha treatment of prostate cancer cells. In the present study, we have attempted to outline specific groups of genes whose expression is significantly regulated by Ashwagandha in a highly invasive prostate cancer cell line PC-3. These studies show that several functionally important classes of genes, which are associated with immune response, signal transduction, cell signaling and transcriptional regulation, apoptosis and cell cycle regulation, can be important targets in bioflavonoid therapy.
Material and Methods
Cell Culture
The human prostate cancer cell line, PC-3 is obtained from the American Type Culture Collection (Manassas, VA). Approximately 3 × 106 cells per 60 mm dish are cultured at 37°C in a humidified atmosphere of 5% CO2 and 95% air for 48 h. Only cell suspensions >90% viable are used for experiments.
Dietary Bioflavonoid
The alcoholic extract of Ashwagandha was a gift from Sami Labs Limited (Bangalore, India). In brief, dried root powder was extracted with ethyl alcohol at 60–70°C three times and all the three extracts combined. The combined extract was concentrated by distillation of the solvent in Rota vapor under reduced pressure. This extract was stored at –20°C and was stable for several months. For each assay, the extract was freshly solubilized in DMSO and utilized at a 0.5–1 μg/ml concentration.
RNA Extraction
Cytoplasmic RNA is extracted by an acid guanidinium-thiocyanate-phenol-chloroform method as described (25) using Trizol reagent and stored at –80°C until used.
cDNA Microarrays
The gene microarrays (Roswell Park Cancer Institute; Microarray and Genomics Core Facility) consisted of ∼5043 cDNA clones (Invitrogen, Carlsbad, CA) which were selected based on their association with cancer progression. A total of six RNA samples that included the three sets of Ashwagandha-treated (1 μg/ml) PC-3 cells and three sets of untreated controls. cDNA was synthesized from each of those RNA samples and the Ashwagandha-treated samples were labeled with Cy5, while the corresponding untreated controls were labeled with the Cy3 dye. To minimize intra-array variations, two hybridizations for each sample were performed, and additionally the labeling with the Cy dyes was inter-changed, so as to have technical replicates of the matched pairs of Ashwagandha-treated and untreated samples.
Microarray Data Processing and Analysis
Expression data extracted from image files were first checked by an M (log2(Cy3/Cy5)) versus A((log2 (Cy3) + log2 (Cy5))/2) plot to see whether intensity-dependent expression bias existed between spots (genes) labeled with Cy3 and Cy5 on each individual slide. Data normalization was done to correct the intensity-dependent expression bias. After data normalization, the average intensity of individual gene from multiple spots on each slide was computed using an in-house developed PERL script. A total of 5043 average expression values were obtained. Paired t-test on normalized intensity with low P-values was used to generate a list of genes with significant change in expression between normal and Ashwagandha-treated samples. The false positive rate (FDR) of the significant genes was estimated by using the SAM algorithm. Detailed normalization approaches have been described in our previous publication (26–28).
Real-time, Quantitative RT-PCR
Gene expression was quantitated using real-time PCR. Relative expression of mRNA species was calculated using the comparative CT method (29). All data were controlled for quantity of RNA input by performing measurements on an endogenous reference gene, β-actin.
Western Blot Analysis
PC-3 cells were cultured with Ashwagandha (1 μg/ml) for 48 h, cells lysed and 30 µg of total extracted protein was separated on 4–20% gradient polyacrylamide gel (30) and electroblotted to PVDF membranes which were further probed with specific primary antibodies against the proteins of interest.
Cell Proliferation Assays
PC-3 cells are plated at a density of ∼1000 cells/well in 96-well plates and incubated with varying concentrations of Ashwagandha (0.5–1 μg/ml) in complete medium for 24 h. Cell viability is quantified using the MTS assay [Promega CellTiter 96® AQueous Non-Radioactive Cell Proliferation Assay (Cat.# G5421); Promega, Madison, WI] as described previously (31,32).
Results
Ashwagandha Inhibits Cell Proliferation
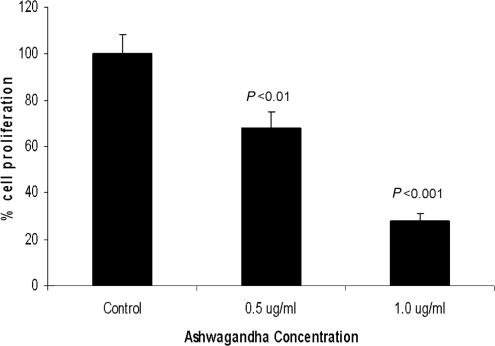
PC3 cells (1000 cells/well) were plated in a 96-well plate and treated with Ashwagandha extract at concentrations ranging from 0.5 to 1 μg/ml for 24 h. The results of our cell proliferation assay show that Ashwagandha treatment significantly inhibited cell proliferation in a dose-dependent manner (Fig. 1). The percentage inhibition at 0.5 and 1 μg/ml Ashwagandha concentrations was 39% and 68%, respectively. Supplementation of medium with 0.2% DMSO, which was used as the solvent had no detectable effect on cell proliferation or on the cell morphology.
Figure 1.
Ashwagandha treatment inhibits cell proliferation in PC-3 cells. Results show that treatment with Ashwagandha significantly decreased cell proliferation. Data are the mean ± SD of three separate experiments done in triplicate. Statistical significance was determined by ANOVA.
Modulation of Gene Expression in PC-3 Cells Treated with Ashwagandha
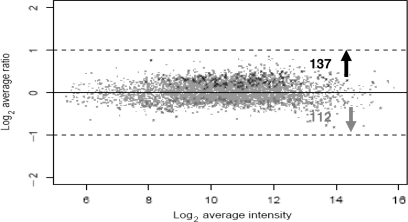
To compare gene expression levels between Ashwagandha-treated PC-3 cells and their respective untreated controls. Treatment of PC-3 cells with 0.2% DMSO had no detectable effect on PC-3 cells and consequently this group was not used in subsequent studies. We used the regularized paired t-test (28) to detect differentially expressed genes, which resulted in 266 significant genes with low P-values. For multiple test correction to control the FDR, these genes were further subjected to SAM analysis, which resulted in a shorter list of 249 genes with an estimated <1% FDR. Out of these 249 differentially expressed genes, 137 (55.02%) were up-regulated and 112 (44.57%) were down-regulated with Ashwagandha treatment, as shown in Fig. 2 in black and gray color, respectively. These results indicate that based on statistical analysis a large number of genes were differentially expressed between Ashwagandha-treated and untreated cells. The differentially expressed genes have been subdivided into various functional categories. Table 1 outlines the fold change in the gene expression of significantly modulated genes that are involved in cell cycle regulation, regulation of apoptosis, modulation of stress proteins, cytokine and chemokine regulation, regulation of signal transduction, regulation of structural proteins, and oncogene regulation.
Figure 2.
Distribution of differentially expressed genes shown as a scatter plot representation of the average log2 ratio between normal and Ashwagandha-treated PC-3 cells for the 249 genes. Statistically significant genes (P-value < 0.01) identified by two-step analyses of t-test and SAM are shown in color, including down-regulated genes (in grey) and up-regulated genes (in black). Dashed lines indicate two-fold changes.
Table 1.
Functional classification of significantly modulated genes
| Gene Acession no | Gene nomenclature | Physiological function | Fold change in gene expression | |
|---|---|---|---|---|
| Cell cycle regulation | ||||
| AI340905 | Cyclin D3 | D-type cyclins are essential for progression from G1 to S-phase. They bind to and activate both CDK4 and CDK6. | 1.32 | ↑ |
| AA489042 | Cell division cycle 2-like 5 | Cell division controller | 1.09 | ↑ |
| AA704459 | Chromatin assembly factor 1, subunit A (p150) | One of the replication-coupled assembly factors involved in the reconstitution of S-phase chromatin | 0.85 | ↓ |
| AA450265 | Proliferating cell nuclear antigen | PCNA gene is induced by p53 and plays a key role in the control of eukaryotic DNA replication | 0.86 | ↓ |
| Regulation of apoptosis | ||||
| AA626710 | Caspase 6 | Apoptosis-related cysteine protease | 1.21 | ↑ |
| AA453766 | CASP8 and FADD-like apoptosis regulator | A caspase-like molecule known to bind to Fas-associated protein with death domain | 0.87 | ↓ |
| Modulation of stress proteins | ||||
| AA456298 | H2B histone family, member Q | Basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes | 1.41 | ↑ |
| AI276745 | Prostaglandin E receptor 2 | PGE(2) regulates cellular immune responses through Distinct EP receptors | 1.25 | ↑ |
| AA398410 | Heat shock transcription factor 2 binding protein | Heat-shock transcription factors (HSFs) bind to heat-shock elements (HSE) that are promoter sites for heat-shock proteins; important in differentiation and development | 1.24 | ↑ |
| H50114 | Glutamate receptor, ionotropic, N-methyl D-aspartate 2C | Regulates NMDA receptor and cell trafficking and ion channel properties | 1.23 | ↑ |
| AA620511 | Heat shock 70kD protein 8 | Molecular chaperone. | 0.91 | ↓ |
| R33642 | Glutathione S-transferase pi | GSTs bind flavonoid natural products in the cytosol prior to their deposition in the vacuole; are components of ultraviolet-inducible cell signalling pathways and are potential regulators of apoptosis; cell survival by GST-P is due to alleviation of oxidative stress and/or JNK activition | 0.87 | ↓ |
| Cytokines and chemokine regulation | ||||
| AA128153 | Interleukin 1 receptor-like 1 | Receptor for interleukin-33 (IL-33), its stimulation recruits MYD88, IRAK1, IRAK4, and TRAF6, followed by phosphorylation of MAPK3/ERK1 and/or MAPK1/ERK2, MAPK14, and MAPK8 | 1.30 | ↑ |
| AA015914 | Fibroblast growth factor 1 (acidic) | Potent mitogen and is involved in the regulation of key cellular process such as angiogenesis, differentiation, and morphogenesis | 1.25 | ↑ |
| AA630120 | Vascular endothelial growth factor B | Up-regulation of urokinase Plasminogen Activator and its inhibitor plasminogen activator inhibitor-1 (PAI-1), which suggests a role in endothelial cell migration and matrix remodeling | 1.18 | ↑ |
| AA102526 | Interleukin 8 | Pro-inflammatory cytokine involved in the cellular response to inflammation, being a powerful chemoattractant for neutrophils | 0.86 | ↓ |
| AI668847 | Small inducible cytokine subfamily B (Cys-X-Cys) | RNA binding, RNA-dependent DNA replication, cell-cell signaling, chemokine activity, chemotaxis, extracellular, immune response, inflammatory response, signal transduction, transferase activity | 0.59 | ↓ |
| T72877 | Interleukin 1 receptor antagonist | Neutralizes biological activity of IL1 α & β in physiologic and pathophysiologic immune and inflammatory responses | 0.80 | ↓ |
| AI074784 | Colony stimulating factor 3 (granulocyte) | Cytokines and Inflammatory Response, Regulation of hematopoiesis by cytokines | 0.75 | ↓ |
| AA150507 | Interleukin 1, beta | Pro-inflammatory cytokine that mediates acute phase response | 0.75 | ↓ |
| Signal transduction modulation | ||||
| H80263 | Phosphatidylinositol-4-phosphate 5-kinase, type II, beta | Cell surface receptor linked signal transduction | 1.34 | ↑ |
| AA913804 | Receptor-interacting serine-threonine kinase 2 | Regulation of apoptosis and protein amino acid phosphorylation | 1.34 | ↑ |
| H18633 | Protein tyrosine phosphatase, receptor type, B | Regulation of tyrosine kinase signals | 1.28 | ↑ |
| AA134814 | TRAF family member-associated NFKB activator | NF-kappaB activation through IKK-i-dependent I-TRAF/TANK phosphorylation | 1.26 | ↑ |
| AI381043 | Inositol 1,4,5-trisphosphate 3-kinase A | Serves both as a second messenger and as a substrate for inositol polyphosphate kinases | 1.24 | ↑ |
| AA489245 | Mitogen-activated protein kinase 8 interacting protein 3 | Activation of JNK via IL-1β; JNK-mediated proapoptotic stimuli. | 1.22 | ↑ |
| AA709036 | Regulator of G-protein Signalling 10 | Regulator of G protein Signaling, RGS4 type | 1.21 | ↑ |
| AA434420 | Protein tyrosine phosphatase, non-receptor type 9 | Regulates responses to extracellular signals by regulating the phosphotyrosine content of specific intracellular proteins | 1.14 | ↑ |
| AA927490 | Signal transducer and activator of transcription 2, 113kD | JAK-STAT signaling pathway | 0.89 | ↓ |
| AA182847 | Mitogen-activated protein kinase kinase kinase kinase 5 | MAPKS are extracellular signal-regulated kinases (ERKs) that act as an integration point for multiple biochemical signals because they are activated by a wide variety of extracellular signals, are rapidly phosphorylated on threonine and tyrosine residues | 0.84 | ↓ |
| AA039851 | Protein tyrosine phosphatase type IVA, member 3 | Prenylated protein tyrosine phosphatases (PTPs); regulatory role in a variety of cellular processes; modulates angiotensin-II induced cell calcium mobilization and cell growth | 0.83 | ↓ |
| AA022666 | IKK-related kinase epsilon; inducible IkappaB kinase | Transcription factor that plays an important role in the immune, anti-apoptotic and inflammatory responses. | 0.74 | ↓ |
| AA419268 | Phosphoinositide-3-kinase, | Control the activity and subcellular localisation of a diverse array of signal transduction molecules; implicated in a wide range of cellular processes, including cell cycle progression, cell growth, cell motility, cell adhesion and cell survival | 0.73 | ↓ |
| AA487426 | Rho GDP dissociation inhibitor | Caspase Cascade in Apoptosis, FAS and TNFR1 Signaling Pathway | 0.68 | ↓ |
| Oncogene regulation | ||||
| AA020001 | C-myc binding protein | C-myc is a cellular proto-oncogene strongly implicated in the control of cellular proliferation, programmed cell death, and differentiation | 1.23 | ↑ |
| W46900 | GRO1 oncogene | Regulates growth factor activity | 0.56 | ↓ |
| Regulation of structural proteins | ||||
| AA598978 | Filamin A, alpha | Actin-binding protein-280 | 0.89 | ↓ |
| W31391 | Zona occludens 2 | Regulation of tight junctions | 0.86 | ↓ |
| AA001432 | Laminin, alpha 3 | Basement membrane protein | 0.74 | ↓ |
Categorization of Differentially Expressed Genes Based on the Biological and Molecular Function
To enhance biological interpretation of the differentially expressed genes from our cDNA microsarray studies, we sought to see if any of the biological processes or molecular function were over-represented by the differentially expressed genes. We therefore performed a function enrichment analyses for these genes using a functional classification tool. DAVID 2.0 (33). Our results indicated that, although there were many over-represented biological function categories as can been seen from Table 2, majority of them are related to a few major function categories. The most relevant function categories include immune response, protein kinase cascade, regulation of apoptosis, and regulation of cell cycle.
Table 2.
Functional annotation classification based on physiological function of significantly regulated genes.
| Classifications | gene count (%) | P-value |
|---|---|---|
| Biological Processes | ||
| Response to stress | 29.0 | 1.4E−5 |
| Immune response | 26.1 | 5.7 E−5 |
| Regulation of biological process | 47.8 | 3.0E−5 |
| Protein kinase cascade | 11.6 | 2.3E−4 |
| Intracellular signaling cascade | 21.7 | 3.0E−4 |
| Inflammatory response | 10.1 | 3.3E−4 |
| Regulation of signal transduction | 10.1 | 4.5E−4 |
| Cell surface receptor linked signal transduction | 26.1 | 5.2E−4 |
| Regulation of I-kappaB kinase/NF-kappaB cascade | 7.2 | 5.3E−4 |
| Regulation of cell proliferation | 10.1 | 2.0E−3 |
| Regulation of cellular physiological process | 37.7 | 3.0E−3 |
| Regulation of cell cycle | 11.6 | 4.3E−3 |
| Regulation of apoptosis | 10.1 | 4.5E−3 |
| Regulation of cell adhesion | 4.3 | 1.2E−2 |
| Molecular functions | ||
| Signal transducer activity | 47.8 | 3.0E−8 |
| Protein binding | 52.2 | 4.1E−6 |
| Cytokine activity | 11.6 | 3.8E−5 |
| Receptor binding | 17.4 | 5.0E−5 |
| Growth factor activity | 10.1 | 7.9E−5 |
| Protein kinase activity | 17.4 | 1.4E−3 |
| Protein tyrosine phosphatase activity | 5.8 | 6.7E−3 |
| Inositol or phosphatidylinositol kinase activity | 4.3 | 9.5E−3 |
| Chemokine receptor binding | 4.3 | 1.4E−2 |
| Chemokine activity | 4.3 | 1.4E−2 |
| G-protein-coupled receptor binding | 4.3 | 2.0E−2 |
| Protein serine/threonine kinase activity | 8.7 | 2.7E−2 |
Analysis of Microarray Data Using Functional Annotational Clustering
We further analyzed our microarray data using functional annotation clustering for our differentially expressed genes to see if any of these genes could be clustered together, thus establishing biological links between genes. The resulting gene clusters are shown in Table 3, in which the group enrichment scores which rank the genes based on their biological significance are also listed. The results of this analysis outline, two groups of genes that are tightly coupled. The first group of genes included caspase recruitment domain family, member 4; receptor-interacting serine-threonine kinase 2; casp8 and fadd-like apoptosis regulator; mitogen-activated protein kinase kinase 5; cell division cycle 2-like 5, while the second group of genes displaying tight coupling included signal transducer and activator of transcription 2 and c-myc binding protein.
Table 3.
Functional annotation clustering
| Gene Accesssion no | P-value | |
|---|---|---|
| Enrichment Score: 1.95 | ||
| AA699441 | Caspase recruitment domain family, member 4 | 1.5E−3 |
| AA913804 | Receptor-interacting serine-threonine kinase 2 | 1.9E−3 |
| AA453766 | casp8 and fadd-like apoptosis regulator | 2.6E−3 |
| AA182847 | Mitogen-activated protein kinase kinase kinase kinase 5 | 1.3E−3 |
| AA489042 | Cell division cycle 2-like 5 | 1.5E−2 |
| Enrichment Score: 1.55 | ||
| AA927490 | Signal transducer and activator of transcription 2 | 1.3E−3 |
| AA020001 | c-myc binding protein | 4.5E−2 |
Using the DAVID 2.0 annotation system, functional annotational clustering of the significantly regulated genes. The group enrichment score represents the geometric mean (in log scale) of member's P-values in a corresponding annotation cluster. This is used to rank their biological significance. The Functional Annotation Clustering integrates the techniques of Kappa statistics to measure the degree of the common genes between two annotations, and fuzzy heuristic clustering to classify the groups of similar annotations according to kappa values. Thus, the more common genes annotations share, the higher chance they will be grouped together. The P-values associated with each annotation terms inside each clusters are obtained using the Fisher's Exact test or/EASE Score. In DAVID annotation system, the Fisher's Exact test is adopted to measure the gene-enrichment in annotation terms.
Mapping Signaling Pathways Using Functional Genomics Data
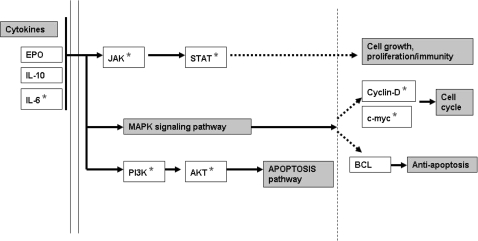
Biological implications can also be deduced from biochemical pathways triggered by the list of genes modulated by Ashwagandha treatment. By looking at the over-representation of genes in pathways, we could obtain direct or indirect insight into the physiological functions of these significantly modulated genes. Using yet another feature of the DAVID functional annotation tool called DAVID Pathway Viewer, a short list of significant pathways (Table 4) was obtained which displays key functional genes from the entire list of significantly modulated genes (249 in this case) on pathway maps thus facilitating biological interpretation in a network context. Our results show that our genes were significantly enriched along three distinct pathways. They are JAK-STAT pathway, the apoptosis pathway and the MAPK signaling pathway. Among these the JAK-STAT pathway appears to be the key because it not only modulates both the apoptosis process and the MAPK signaling but also is the signaling pathway that is most significantly modulated as indicated by the P-value <0.0037 (Table 4 and Fig. 3).
Table 4.
DAVID Pathway Viewer lists key signaling pathways that are modulated by ashwangandha treatment
| Signaling pathways | Genes (%) | P-value |
|---|---|---|
| JAK-STAT Signaling pathway | 11.6 | 3.7E−3 |
| Apoptosis | 7.2 | 1.3E−2 |
| MAPK Signaling pathway | 10.1 | 1.8E−2 |
Figure 3.
Schematic representation of the genes regulated by Ashwagandha extract in JAK-STAT signaling pathway. The genes highlighted with an asterisk (*) are significantly regulated by Ashwagandha and are important in the regulation of cell proliferation, regulation of apoptosis and immunomodulation.
Ashwagandha Modulates Gene and Protein Expression of a Group of Cytokines, Chemokines, Apoptotic Markers, Cell Cycle Proteins and Signal Transduction Molecules
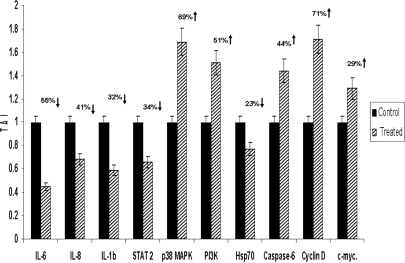
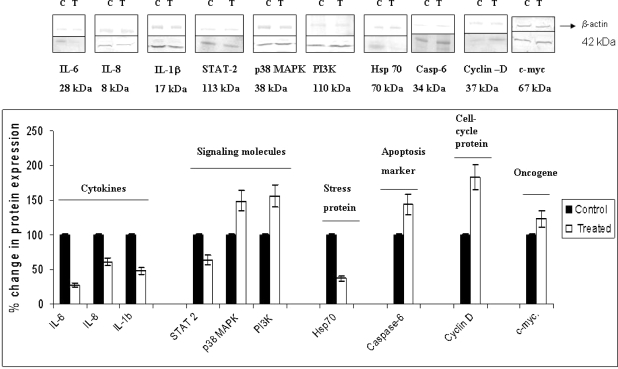
Based on the functional annotation analysis using DAVID as described above, a group of genes that modulate the JAK-STAT signaling pathway and regulate key biological and molecular functions were selected. They included IL-6, IL-8, IL-1β, STAT 2, p38 MAPK, PI3K, Caspase 6, Hsp70, Cyclin D, and c-myc. These genes are significantly modulated by Ashwagandha treatment as shown by gene array analysis. To further validate our findings, we performed quantitative real-time PCR using specific PCR primers and western blot analysis for protein expression for these key regulatory genes. Our results presented in Fig. 4 show that PC-3 cells treated with 1 μg/ml Ashwagandha displayed down-regulation of gene expression of the proinflammatory cytokines IL-6 (55% down-regulation, P < 0.002), IL-1β (41% down-regulation, P < 0.05), chemokine IL-8 (32% down-regulation, P < 0.04) and stress heat shock protein Hsp70 (23% down-regulation, P < 0.02). Ashwagandha treatment also down-regulated Signal Transducer and Activator of Transcription 2 (STAT-2) (34% down-regulation, P < 0.01), a dual function protein that regulates signal transduction and activates transcription as part of a phosphorylation cascade. The genes that were significantly upregulated with Ashwagandha treatment were p38 MAPK (69% upregulation, P < 0.013); PI3K (51% upregulation, P < 0.04) caspase 6 (44% upregulation, P < 0.03); Cyclin D (71% upregulation, P < 0.006) and c-myc (29% upregulation, P < 0.043). The respective percent increases in protein expression for each these genes are shown in Fig. 5. Our results show that PC-3 cells treated with 1 μg/ml Ashwagandha downregulated the protein expression of the proinflammatory cytokines IL-6 (72% downregulation, P < 0.01), IL-1β (39% downregulation, P < 0.05), chemokine IL-8 (52% downregulation, P < 0.03), Hsp70 (63% downregulation, P < 0.01) and STAT-2 (38% downregulation, P < 0.05). The proteins that were significantly upregulated with Ashwagandha treatment were p38 MAPK (49% upregulation, P < 0.03); PI3K (56% upregulation, P < 0.02) caspase 6 (44% upregulation, P < 0.05); Cyclin D (81% upregulation, P < 0.01) and c-myc (23% upregulation, P = 0.06). We observed concordance between gene expression and protein expression levels in the Ashwagandha-treated PC-3 cells. Furthermore, these results showed similar trends in gene expression as those obtained by our microarray analysis thus validating our gene microarray results presented in Table 1.
Figure 4.
Ashwagandha treatment significantly modulates gene expression in PC-3 cells. Graphical representation of quantitative changes in gene expression by real-time PCR. Relative mRNA species expression was quantitated and expressed as transcript accumulation index (TAI= 2–ΔΔCT), calculated using the comparative CT method. All data were controlled for quantity of RNA input by performing measurements on an endogenous reference gene, β-actin. In addition, results on RNA from Ashwagandha-treated samples were normalized to results obtained on RNA from the control, untreated sample. Data are the mean ± SD of three separate experiments done in triplicate. Statistical significance was determined by ANOVA.
Figure 5.
Ashwagandha treatment significantly modulates protein expression in PC-3 cells. Graphical representation of quantitative changes in protein expression by Western blot analysis. Data are the mean ± SD of three separate experiments done in triplicate. Statistical significance was determined by ANOVA.
Statistical Analysis
All statistical analysis for the above data was performed using analysis of variance (ANOVA), following the Mann–Whitney U-test to determine the statistical difference between control and treated groups. P < 0.05 was considered significant.
Discussion
Withania somnifera Dunal, which is commonly known as Ashwagandha is reported to have antitumor, anti-stress, immunomodulatory, anti-inflammatory and antibacterial effects (11–15). The roots of W. somnifera contain several alkaloids, withanolides, a few flavanoids and reducing sugars (14–19). Several studies support the role of ashwagandha as an effective cancer chemopreventive agent, however limited information is available regarding the mechanism of regulation of these chemopreventive effects (20). The goal of our study was gain insight into the general biological and molecular functions and immunomodulatory processes that are altered as a result of Ashwagandha treatment in prostate cancer cells, and to identify the key signaling mechanisms that are involved in the regulation of these physiological effects. Since the alcohol extract of Ashwagandha can contain numerous molecules that are antiproliferative the biological effect that we observed could be due to one or many compounds. The effect could also be additive in nature. However, on initial reverse phase HPLC analysis of the alcohol extract we observed that the anti-proliferative effect was confined to a compound that eluted at 10.7 min, giving a peak at m/z of 567 (data not given). Therefore, it is our conviction that the majority of the antiproliferative and the effects on gene expression that we observed in this study is due to this particular compound; further characterization of the compound is ongoing in our laboratory.
Our results showed that Ashwagandha treatment modulated several functionally important classes of genes, which are associated with cell cycle regulation, regulation of apoptosis, modulation of stress proteins, cytokine and chemokine regulation, regulation of signal transduction, and oncogene regulation in prostate cancer cells (PC-3). Additionally, a good concordance was observed between our microarray data, and the results obtained using both our validation methodologies, namely the real time PCR analysis and the Western blot analysis, however some of the protein expression levels did not show the extent of changes as seen in the gene expression level. This could possibly be due to the post-transcriptional or post-translational modification associated with protein expression. Analysis of our microarray data using functional annotation clustering showed significantly modulated gene clusters and provides group enrichment scores which rank the genes based on their biological significance. These analyses further confirm the important role played by genes that regulate apoptosis, cell signaling and signal transduction in the Ashwagandha-mediated immunomodulation.
Using the function annotation tool DAVID Pathway Viewer, we were able to highlight key signaling pathways via which the immunomodulatory effects of Ashwagandha are mediated. Our results show three distinct pathways that are significantly modulated; they are JAK-STAT pathway, the apoptosis pathway and the MAPK signaling pathway. Among these the JAK-STAT pathway appears to be key because it also modulates both the apoptosis process and the MAPK signaling.
Ashwagandha treatment significantly downregulated the gene and protein expression of proinflammatory cytokines IL-6, IL-1β, chemokine IL-8, Hsp70 and STAT-2, while a reciprocal upregulation was observed in gene and protein expression of p38 MAPK, PI3K, caspase 6, Cyclin D and c-myc. Several in vitro studies show that polyphenols can modulate oncogenes, tumor suppressor genes, cell cycling, apoptosis, angiogenesis and associated signal transduction pathways (34,35). Tyagi et al. (36) showed that the anticancer effects of grape seed extract (GSE) on moderately aggressive DU-145 prostate cancer cells were due to activation of the JNK pathway, resulting in growth inhibition and apoptosis. Recent studies demonstrated that flavonoids can inhibit proliferation of other cancers by interfering with tubulin binding and subsequent microtubule assembly (37), among the 249 genes that were significantly modulated by Ashwagandha, several were structural proteins, like filamin A, laminin and tight junction protein-zona occludens 2. The significant anti-tumor effects of flavonoids are attributed to the modulation of protein tyrosine phosphorylation (38). Suppression of carcinogenesis by tumor suppressor genes is mediated by constraining tumor cell proliferation and colony formation. The tumor suppressor genes, suppresses tumor growth by modulating the phosphatidylinositol 3-kinase (PI3K) and causes G1 cell cycle arrest and cell death (39–41). The cyclin D gene encodes a regulatory subunit of the CDK4 and CDK6 holoenzyme complex that phosphorylates and inactivates the tumor suppressor protein, pRB, thereby modulating cell cycle progression into late G1 and S phases (41–43).
Ashwangandha treatment down regulates the expression of the proinflammatory cytokines IL-6, IL-8 and IL-1β hereby acting as a immunomodulator, via the dynamic regulation of these cytokines. These cytokines are crucial to innate and adaptive inflammatory responses, cell growth and differentiation, cell death, angiogenesis and developmental as well as repair processes. The janus kinase/signal transducer and activator of transduction (JAK/STAT) pathway is a intracellular signal-transducing pathway that is activated by oxygen radicals, various cytokines, and growth factors in various disease states (44,45). Originally described as the regulator for cytokine signaling, JAK/STAT pathway is now recognized as an important membrane-to-nucleus signaling pathway for a variety of stress responses and oxidative stress. STAT proteins have the dual function of signal transduction and activation of transcription as part of a phosphorylation cascade (44). Ashwagandha downregulates the expression of STAT2 which is a potent transactivator and forms stable homodimers which complex with adaptor protein p48 and bind to the interferon-stimulated response element (ISRE) thereby modulating cell proliferation and immunity.
Stress-inducible heat-shock protein 70 (Hsp70) has gained plenty of attention as a putative target for tumor therapy since it is an important regulator of cancer cell growth. Expression of Hsp70 in tumor cells has been proposed to enhance their immunogenicity and prevent tumor cell death (46). Ashwagandha treatment significantly decreases the gene and protein expression of Hsp70, which may result in the generation of a specific immune response by promoting apoptosis. The oncogene c-myc gene participates in most aspects of cellular function, including replication, growth, metabolism, differentiation, and apoptosis (34,35). Our studies highlight that treatment of PC-3 cells with Ashwagandha results in the activation of c-Jun N-terminal kinase (JNK) signaling, indicating that it may be the key signaling pathway involved in the process leading to activation of apoptosis in these cancer cells. Thus, plant-derived antioxidants can modulate biochemical and physiological processes that may affect cancer progression. Based on its pro-apoptotic and its immunomodulatory effects, Ashwagandha can be considered to be useful as novel complementary therapy for integrative oncology care. Furthermore, based on this preliminary finding we would like to characterize the active ingredient using reverse phase HPLC separation. The eluted compound which shows the presence of active ingredients would be further characterized using the NMR and the mass spectroscopy to elucidate the structure of the molecule.
References
- 1.American Cancer Society. Cancer Facts and Figures 2007. Atlanta, GA: American Cancer Society; 2007. [Google Scholar]
- 2.Stanford JL, Stephenson RA, Coyle LM, Cerhan J, Correa R, Eley JW, et al., editors. Bethesda, Md: National Cancer Institute; 1999. Prostate Cancer Trends 1973–1995. NIH Pub. No. 99-4543. [Google Scholar]
- 3.Miller BA, Kolonel LN, Bernstein L, Young JL Jr, Swanson GM, West D, et al., editors. Bethesda, MD: National Cancer Institute; 1996. Racial/Ethnic Patterns of Cancer in the United States 1988–1992. NIH Pub. No. 96-4104. [Google Scholar]
- 4.Kolonel LN. Fat, meat, and prostate cancer. Epidemiol Rev. 2001;23:72–81. doi: 10.1093/oxfordjournals.epirev.a000798. [DOI] [PubMed] [Google Scholar]
- 5.Heinonen OP, Albanes D, Virtamo J, Taylor PR, Huttunen JK, Hartman AM, et al. Prostate cancer and supplementation with alpha-tocopherol and beta-carotene: incidence and mortality in a controlled trial. J Natl Cancer Inst. 1998;90:440–6. doi: 10.1093/jnci/90.6.440. [DOI] [PubMed] [Google Scholar]
- 6.Chan JM, Giovannucci EL. Vegetables, fruits, associated micronutrients, and risk of prostate cancer. Epidemiol Rev. 2001;23:82–6. doi: 10.1093/oxfordjournals.epirev.a000799. [DOI] [PubMed] [Google Scholar]
- 7.Giovannucci E, Rimm EB, Liu Y, Stampfer MJ, Willett WC. A prospective study of tomato products, lycopene, and prostate cancer risk. J Natl Cancer Inst. 2002;94:391–8. doi: 10.1093/jnci/94.5.391. [DOI] [PubMed] [Google Scholar]
- 8.Chan JM, Giovannucci EL. Dairy products, calcium, and vitamin D and risk of prostate cancer. Epidemiol Rev. 2001;23:87–92. doi: 10.1093/oxfordjournals.epirev.a000800. [DOI] [PubMed] [Google Scholar]
- 9.Platz EA, Helzlsouer KJ. Selenium, zinc, and prostate cancer. Epidemiol Rev. 2001;23:93–101. doi: 10.1093/oxfordjournals.epirev.a000801. [DOI] [PubMed] [Google Scholar]
- 10.Barnes S. Role of phytochemicals in prevention and treatment of prostate cancer. Epidemiol Rev. 2001;23:102–5. doi: 10.1093/oxfordjournals.epirev.a000773. [DOI] [PubMed] [Google Scholar]
- 11.Chandrashekar A, Sharada P, Solomon E, Umadevi P, Nayanabhirama U, Srinivasan K. Antitumor and radiosensitizing effects of withaferin A on mouse Erhlich ascites carcinoma in vivo. Acta Oncol. 1996;35:95–100. doi: 10.3109/02841869609098486. [DOI] [PubMed] [Google Scholar]
- 12.Devi PU, Akagi K, Ostapenko V, Tanaka Y, Sugahara T. Withaferin A. a new radiosensitizer from the Indian medicinal plant Withania somnifera. Int J Radiat Biol. 1996;69:193–7. doi: 10.1080/095530096146020. [DOI] [PubMed] [Google Scholar]
- 13.Archana R, Namasivayam A. Antistressor effect of Withania somnifera. J Ethnopharmocol. 1999;64:91–3. doi: 10.1016/s0378-8741(98)00107-x. [DOI] [PubMed] [Google Scholar]
- 14.Ziauddin M, Phansalkar N, Patki P, Diwanay S, Patwardhan B. Studies on the immunomodulatory effects of Ashwagandha. J Ethnopharmacol. 1996;50:69–76. doi: 10.1016/0378-8741(95)01318-0. [DOI] [PubMed] [Google Scholar]
- 15.Umadevi P. Withania somnifera dunal (Ashwagandha): potential plant source of promising drug for cancer chemotherapy and radiosensitization. Indian J Exp Biol. 1996;34:927–32. [PubMed] [Google Scholar]
- 16.Ganzera M, Choudhary MI, Khan IA. Quantitative HPLC analysis of withanolides in Withania somnifera. Fitoterapia. 2003;74:68–76. doi: 10.1016/s0367-326x(02)00325-8. [DOI] [PubMed] [Google Scholar]
- 17.Jayaprakasam B, Zhang Y, Seeram NP, Nair MG. Growth inhibition of human tumor cell lines by withanolides from Withania somnifera leaves. Life Sci. 2003;74:125–32. doi: 10.1016/j.lfs.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 18.Kaur P, Sharma M, Mathur S, Tiwari M, Divekar HM, Kumar R, et al. Effect of 1-oxo-5beta, 6beta-epoxy-witha-2-ene-27-ethoxy-olide isolated from the roots of Withania somnifera on stress indices in Wistar rats. J Altern Complement Med. 2003;9:897–907. doi: 10.1089/107555303771952244. [DOI] [PubMed] [Google Scholar]
- 19.Matsuda H, Murakami T, Kishi A, Yoshikawa M. Structures of withanosides I, II, III, IV, V, VI, and VII, new withanolide glycosides, from the roots of Indian Withania somnifera DUNAL and inhibitory activity for tachyphylaxis to clonidine in isolated guinea-pig ileum. Bio Org Med Chem. 2001;9:1499–507. doi: 10.1016/s0968-0896(01)00024-4. [DOI] [PubMed] [Google Scholar]
- 20.Tenborg M. Modulation of cytokine expression by traditional medicines: a review of herbal immunomodulators.(Cytokines/Herbs)-Alternative Medicine Review 2006. http://www.encyclopedia.com/doc/1G1-148424513.html. [PubMed]
- 21.Sangwan RS, Chaurasiya ND, Lal P, Misra L, Uniyal GC, Tuli R, et al. Withanolide A Biogeneration in in Vitro shoot cultures of ashwagandha (Withania somnifera DUNAL), a main medicinal plant in ayurveda. Chem Pharm Bull. 2007;55:1371–5. doi: 10.1248/cpb.55.1371. [DOI] [PubMed] [Google Scholar]
- 22.Bhattacharya SK, Bhattacharya A, Sairam K, Ghosal S. Anxiolytic-antidepressant activity of Withania somnifera glycowithanolides: an experimental study. Phytomedicine. 2000;7:463–9. doi: 10.1016/S0944-7113(00)80030-6. [DOI] [PubMed] [Google Scholar]
- 23.Nair HK, Rao KV, Aalinkeel R, Mahajan S, Chawda R, Schwartz SA. Inhibition of prostate cancer cell colony formation by the flavonoid quercetin correlates with modulation of specific regulatory genes. Clin Diagn Lab Immunol. 2004;11:63–9. doi: 10.1128/CDLI.11.1.63-69.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nair MP, Mahajan S, Reynolds JL, Aalinkeel R, Nair H, Schwartz SA. The flavonoid quercetin inhibits proinflammatory cytokine (tumor necrosis factor alpha) gene expression in normal peripheral blood mononuclear cells via modulation of the NF-kappa beta system. Clin Vaccine Immunol. 2006;13:319–28. doi: 10.1128/CVI.13.3.319-328.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chomczynski P, Saachi N. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–9. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 26.Mahajan SD, Hu Z, Reynolds JL, Aalinkeel R, Schwartz SA, Nair MP. Methamphetamine modulates gene expression patterns in monocyte derived mature dendritic cells: implications for HIV-1 pathogenesis. Mol Diagn Ther. 2006;10:257–69. doi: 10.1007/BF03256465. [DOI] [PubMed] [Google Scholar]
- 27.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baldi P, Long AD. A Bayesian Framework for the Analysis of Microarray Expression Data: Regularized t-test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–19. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 29.Shively L, Chang L, LeBon JM, Liu Q, Riggs AD, Singer-Sam J. Real-time PCR assay for quantitative mismatch detection. Biotechniques. 2003;34:498–502. doi: 10.2144/03343st01. 504. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Malich G, Markovic B, Winder C. The sensitivity and specificity of the MTS tetrazolium assay for detecting the in vitro cytotoxicity of 20 chemicals using human cell lines. Toxicol. 1997;124:179. doi: 10.1016/s0300-483x(97)00151-0. [DOI] [PubMed] [Google Scholar]
- 32.Barile FA, DierickX PJ, Kristen U. In vitro cytotoxicity testing for prediction of acute human toxicity. Cell Biol Toxicol. 1994;10:155. doi: 10.1007/BF00757558. [DOI] [PubMed] [Google Scholar]
- 33.Dennis G Jr, Sherman BT, Hopsack DA, Yang J, GAO W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 34.Elend M, Eilers M. Cell growth: downstream of Myc – to grow or to cycle? Curr Biol. 1999;9:R936–8. doi: 10.1016/s0960-9822(00)80109-8. [DOI] [PubMed] [Google Scholar]
- 35.Hoffman B, Liebermann DA. The proto-oncogene c-myc and apoptosis. Oncogene. 1998;17:3351–7. doi: 10.1038/sj.onc.1202592. [DOI] [PubMed] [Google Scholar]
- 36.Tyagi A, Agarwal R, Agarwal C. Grape seed extract inhibits EGF-induced and constitutively active mitogenic signaling but activates JNK in human prostate carcinoma DU145 cells: possible role in antiproliferation and apoptosis. Oncogene. 2003;22:1302–16. doi: 10.1038/sj.onc.1206265. [DOI] [PubMed] [Google Scholar]
- 37.Gupta K, Panda D. Perturbation of microtubule polymerization by quercetin through tubulin binding: a novel mechanism of its antiproliferative activity. Biochemistry. 2002;41:13029–38. doi: 10.1021/bi025952r. [DOI] [PubMed] [Google Scholar]
- 38.Ferry DR, Smith A, Malkhandi J, Fyfe DW, deTakats PG, Anderson D, Baker J, et al. Phase I clinical trial of the flavonoid quercetin: pharmacokinetics and evidence for in vivo tyrosine kinase inhibition. Clin Cancer Res. 1996;2:659–68. [PubMed] [Google Scholar]
- 39.Shay JW, Pereira-Smith OM, Wright WE. A role for both RB and p53 in the regulation of human cellular senescence. Exp Cell Res. 1991;196:33–9. doi: 10.1016/0014-4827(91)90453-2. [DOI] [PubMed] [Google Scholar]
- 40.Sherr CJ. G1 phase progression: cycling on cue. Cell. 1994;79:551–5. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- 41.Wright WE, xPereira-Smith WE, Shay JW. Reversible cellular senescence: implications for immortalization of normal human diploid fibroblasts. Mol Cell Biol. 1989;9:3088–92. doi: 10.1128/mcb.9.7.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hunter T, Pines J. Cyclins and cancer. Cell. 1991;66:1071–4. doi: 10.1016/0092-8674(91)90028-w. [DOI] [PubMed] [Google Scholar]
- 43.Weng LP, Smith WM, Dahia PL, Ziebold U, Gil E, Lees JA, et al. PTEN suppresses breast cancer cell growth by phosphatase activity-dependent G1 arrest followed by cell death. Cancer Res. 1999;59:5808–14. [PubMed] [Google Scholar]
- 44.Hebenstreit D, Horejs-Hoeck J, Duschl A. JAK/STAT-dependent gene regulation by cytokines. Drug News Perspect. 2005;18:243–9. doi: 10.1358/dnp.2005.18.4.908658. [DOI] [PubMed] [Google Scholar]
- 45.O'Shea JJ, Park H, Pesu M, Borie D, Changelian P. New strategies for immunosuppression: interfering with cytokines by targeting the Jak/Stat pathway. Curr Opin Rheumatol. 2005;17:305–11. doi: 10.1097/01.bor.0000160781.07174.db. [DOI] [PubMed] [Google Scholar]
- 46.Drysdale MJ, Brough PA, Massey A, Jensen MR, Schoepfer J. Targeting Hsp90 for the treatment of cancer. Curr Opin Drug Discov Devel. 2006;9:483–95. [PubMed] [Google Scholar]