Abstract
Eukaryotic chromosomes replicate with defined timing patterns. However, the mechanism that regulates the timing of replication is unknown. In particular, there is an apparent conflict between population experiments, which show defined average replication times, and single-molecule experiments, which show that origins fire stochastically. Here, we provide a simple simulation that demonstrates that stochastic origin firing can produce defined average patterns of replication firing if two criteria are met. The first is that origins must have different relative firing probabilities, with origins that have relatively high firing probability being likely to fire in early S phase and origins with relatively low firing probability being unlikely to fire in early S phase. The second is that the firing probability of all origins must increase during S phase to ensure that origins with relatively low firing probability, which are unlikely to fire in early S phase, become likely to fire in late S phase. In addition, we propose biochemically plausible mechanisms for these criteria and point out how stochastic and defined origin firing can be experimentally distinguished in population experiments.
Keywords: replicationtiming, stochastic origin firing, origin regulation, origin efficiency
Introduction
Eukaryotic genomes replicate with characteristic timing patterns; some parts of a genome replicate early in S phase and other parts replicate late. These patterns correlate with patterns of transcriptional regulation and chromosome structure, and they change as cells differentiate, suggesting an intimate relation between replication timing and other important aspects of chromosome metabolism (Goren and Cedar 2003). Patterns of replication timing have long been recognized in mammalian cells, where the R bands (regions of active transcription and higher CG content) replicate early and the G bands (regions that tend to be heterochromatic) replicate late (Holmquist et al. 1982; Taylor 1989). It has been widely assumed that the reproducible patterns of replication timing reflect reproducible firing times of the replication origins in these regions (Goren and Cedar 2003). Consistent with this prediction, recent work has shown the individual human origins fire on average at defined times during S phase (Cadoret et al. 2008).
Similar patterns of replication timing are observed in budding yeast (Fangman et al. 1983; McCarroll and Fangman 1988; Reynolds et al. 1989; Raghuraman et al. 2001). The advantage of studying budding yeast is that the location of origins are known genome wide, and the average firing time of each origin has been mapped (Raghuraman et al. 2001; Yabuki et al. 2002). These studies show that origins in budding yeast fire at characteristic times, with some origins firing on average earlier and others firing on average later. As with the mammalian studies, the reproducible average replication times of budding yeast origins were interpreted to demonstrate that origins fire at predetermined times in S phase.
In mammalian cells, individual metaphase chromosomes can be observed. Therefore, it is known that the patterns of replication timing are similar in all cells (Taylor 1989). However, the spatial resolution of this data is low, averaging the replication signal over hundreds of kilobases of DNA and many replication origins. So, although it is known that large regions on mammalian chromosomes replicate with reproducible timing, it is possible that replication timing is heterogeneous at higher resolution (Labit et al. 2008). Budding yeast experiments and recent mammalian microarray experiments, on the other hand, have high spatial resolution, in the kilobase range, but they assay the average behavior of individual origins over a population of cells. So, in these experiments, it is possible that replication timing is heterogeneous on a single-cell level. These analyses therefore do not demonstrate that individual origins fire are predetermined times, although they are often interpreted to do so.
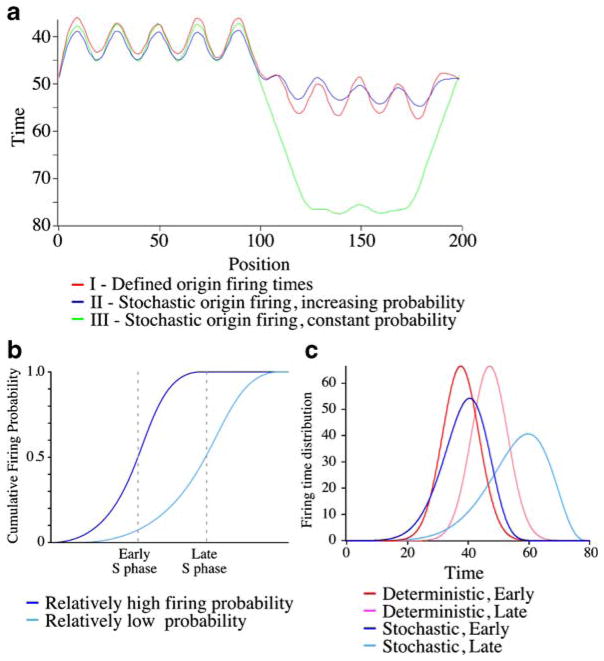
The fact that both the mammalian and budding yeast experiments average the behavior of many origins makes determining the timing of individual origin firing difficult. Moreover, the data from these experiments is usually presented and analyzed as trep replication profiles, that is, graphs of when during S phase each locus in the genome is, on average, half replicated (Fig. 1a). These replication profiles, in which origins are peaks and peak height correlates with origin timing and efficiency, are very useful for visualizing average replication kinetics. However, they discard much of the information from the experiment and cannot distinguish late, efficient origins from early, inefficient origins.
Fig. 1.
Stochastic origin firing can produce defined replication timing patterns. a Three simulated replication profiles. Each simulation covers ten origins spaced every 20 kb. The five origins on the right fire early, those on the left fire late. In these simulations, all of the early region is on average replicated before any of the late origins fire. Model I: Origins fire with a defined average time and variance. The early origins fire at 37.5±6 min, and the late origins fire at 47±6 min. Model II: The origins fire stochastically, with a probability that increases during S phase. The early-firing origins have a higher relative probability, and the late-firing origins have a lower relative firing probability. In this incarnation of the increasing-probability model, the increase in firing probability follows a power law that increases the firing probability of a late origin by tenfold between early S phase and late S phase. However, other kinetic schemes with a similar increase in firing probability would work just as well. Model III: The origins fire stochastically with a constant firing probability. b The cumulative firing probability of a representative early- and late-firing origin from Model II. An origin with relatively high firing probability is likely to fire in early S phase. An origin with relatively low firing probability is unlikely to fire in early S phase, but as its firing probability increases, it becomes likely to fire in late S phase. c The firing kinetics of a representative early- and late-firing origin from Models I and II
These uncertainties about the behavior of individual origins has allowed for two very different types of models of origin regulation. The first type, which we will call the deterministic models, assumes that each origin has an intrinsic firing time set by a mechanism that organizes origins within a predetermined replication timing program. In such models, origins are envisaged to fire at their pre-programmed time, plus or minus some small error. If they do not fire at their pre-programmed time, they will not fire at other times during S phase. Deterministic models predict homogeneous replication kinetics in a population of cells.
The second type of model, which we will call the stochastic models, posits that the firing time of an individual origin in a population is heterogeneous, firing early in some cells and late in others. Stochastic models also assume that the firing of neighboring origins is independent. Of course, when an origin fires, its replication forks will passively replicate neighboring origins, preventing them from firing; stochasticity simply means that in the time before they are passively replicated, the chance that one of the neighboring origins will itself fire is not affected.
The distinction between the two classes of models is not absolute; one can pass smoothly from one to the other. For example, even under a deterministic model, one expects small variations in the firing times of an origin. Thus, in a trivial sense, all models are stochastic. The real question concerns the degree of stochasticity and whether the stochasticity itself plays an important role in replication control. Thus, one might, more loosely, call a model deterministic if the variation in origin firing times is much less than the duration of S phase and stochastic if they are a substantial fraction. Therefore, in classifying a model as stochastic, one has to make a case that goes beyond the mere existence of stochastic aspects.
Stochastic models have been motivated by studies that demonstrate heterogeneous patterns of origin firing (Patel et al. 2006; Czajkowsky et al. 2008). The two central conclusions of these studies are that eukaryotic replication origins fire inefficiently and stochastically, instead of firing efficiently at defined times during S phase. It is well established that many eukaryotic origins are inefficient (Hamlin et al. 2008). The efficiency of yeast origins vary, with some being as high as 90% and other less than 10% (Raghuraman et al. 2001; Heichinger et al. 2006). Metazoan origins are less well characterized, but estimates of their efficiency ranges from 5% to 20% (Lebofsky et al. 2006; Hamlin et al. 2008). The inefficient nature of origins implies that they have some probability of firing which is balanced by the probability of being passively replicated by a fork from a neighboring origin. If, by chance, a nearby origin fires first, the origin is likely to get passively replicated; the longer an origin goes without being passively replicated, the better the chance of it firing itself.
The stochastic nature of origin firing has been difficult to address experimentally. It is most apparent in high-resolution, single-molecule analyses because bulk techniques, such as microarrays, average out the behavior of individual origins, obscuring stochastic effects. Nonetheless, in situations in which it has been possible to test the hypothesis, origins do fire stochastically (Lebofsky et al. 2006; Patel et al. 2006; Czajkowsky et al. 2008).
The inefficient, stochastic nature of origin firing was first observed in the rapid cell cycles of frog and fruit fly embryos. Frog embryos replicate their genome extremely quickly; embryonic S phase lasts about 20 min, as compared to 8 h for adult somatic cells. Furthermore, they initiate replication at random locations in the genome (Hyrien and Mechali 1993). The fact that the genomes of these embryonic cells are transcriptionally inactive allowed the possibility that they may be replicated differently from transcriptionally active cells. However, yeast and somatic mammalian cells show similar inefficient, stochastic origin firing, the major difference being that yeast and somatic metazoan cells use defined origin loci (Lebofsky et al. 2006; Patel et al. 2006; Czajkowsky et al. 2008).
The most compelling case for inefficient, stochastic firing is in budding yeast, where directly comparable bulk and single-molecule studies have been done. Czajkowsky et al. (2008) examined origin usage on Chromosome VI in over 100 different cells and found that no two used the same pattern of origin firing. Nonetheless, when they averaged the behavior of all of their chromosomes, they produced replication profiles strikingly similar to previous trep profiles obtained from microarray experiments (Raghuraman et al. 2001; Yabuki et al. 2002; Alvino et al. 2007). This comparison demonstrates that stochastic firing is compatible with defined replication timing. It also demonstrates the potential pitfalls of over-interpreting ensemble behavior, such as trep profiles; although each locus has a defined time at which it is half replicated on average, in any individual cell, the timing can vary greatly.
As we have suggested above, the distinction between deterministic timing and stochastic firing is something of a false dichotomy. All chemistry, and therefore all biology, is inherently stochastic. The question is not whether origin firing is stochastic, it is how important is the probabilistic nature of origin firing in the regulation of replication and, more importantly, how can stochastic origin firing be accommodated in realistic models that predict the patterns of replication timing seen in vivo.
An increasing-probability model reconciles stochastic origin firing with defined replication timing
It is possible to create models in which stochastic firing of origins produces defined patterns of replication timing (Rhind 2006; Lygeros et al. 2008; de Moura et al. submitted for publication; Yang et al. submitted for publication). Here, we present a simple version meant only to illustrate the essential features of such a model. We describe a technically more sophisticated version elsewhere (Yang et al. submitted for publication).
It is clear that uniformly stochastic origin firing is incompatible with defined patterns of replication timing; such firing would lead to great heterogeneity between cells but uniform replication timing across the genome when averaged over a population. However, stochastic firing need not be uniform. The stochastic firing of an origin is characterized by a parameter, its firing probability, that describes the chance of it firing during any given time span (See Nomenclature for definitions). This parameter determines the average time it would take an origin to fire if it was never passively replicated. Firing probability can vary between origins and can explain why some origins fire earlier than others.
Nomenclature
| Origin | A site in the genome where replication can initiate; in some organisms and cell types, such sites may be well-defined by cis-acting sequence features; in other organisms and cell types, many, or even all, sites in the genome may act as origins |
| Origin firing or origin initiation | The irreversible conversion of a licensed origin into bidirectional replication forks |
| Efficiency | The fraction of cells in which an origin fires during S phase |
| Firing probability | The probability that an as-yet-unfired origin will fire during a specific time period |
| Relative firing probability | The firing probability of an origin measured relative to the firing probability of all other origins in the genome, irrespective of its absolute firing probabilities |
Variation in firing probability between origins easily explains why an origin with a relatively high firing probability would fire early and one with a relatively low probability would not. However, in the simplest version of stochastic firing models, an origin with a relatively low firing probability is unlikely to fire at any time during S phase. Therefore, this version of the model fails to fire origins in late S phase and suffers from the so-called random gap problem; the problem that if origins fire stochastically, at some frequency, origins will fail to fire across a large genomic region, leading to inefficient replication (Laskey 1985; Lucas et al. 2000; Hyrien et al. 2003; Lygeros et al. 2008).
To efficiently replicate late-replicating parts of the genome, stochastic-firing models need to include some mechanism to ensure that origins with relatively low firing probability, which are unlikely to fire early in S phase, are nonetheless able to fire later in S phase. One such mechanism is to have the probability of origins firing increase as S phase progresses (Lucas et al. 2000; Yang and Bechhoefer 2008). Thus, any origin that does not fire or get passively replicated in early S phase will become much more likely to fire in late S phase (Fig. 1b). Such an increasing probability of firing has been observed in all genome-wide replication-kinetics data sets that have been examined (Goldar et al. 2009). Since the firing probability of all origins starts off very low in such models and increases throughout S phase, the important parameter is relative firing probability; origins with high relative probability tend to fire early, and those with low relative firing probability tend to be passively replicated or fire late. A model that incorporates both varying firing probability and increasing firing probability as S phase progresses captures the essential behavior of in vivo replication kinetics. Possible mechanisms underlying such a model are discussed below.
The power of the increasing-probability model is demonstrated in the simulations presented in Fig. 1a. In this figure, we compare two models of origin firing, both of which lead to a pattern of replication with early- and late-firing origins. Both models fit an idealized pattern of chromosomal replication in which the chromosome is divided into two regions, an early-replicating region and a late-replicating region; each region has five origins spaced 20 kb apart, and the average replication times of the regions is such that all of the DNA in the early region replicates, on average, before any of the origins in the late region fire. The first is a model based on defined origin-firing times. Each origin has a characteristic firing time and fires at that time plus or minus a deviation drawn from a Gaussian distribution. The second is the increasing-probability model. Each origin has a defined relative firing probability, but the firing probability of all origins increases over time, making them more likely to fire later in S phase (Fig. 1b). In this increasing-probability model, the origins with high relative probability are more likely to fire and therefore, on average, replicate early, whereas those with low relative probability are unlikely to replicate early. However, as S phase progresses, the firing probability of all origins increases, so eventually, all origins reach a point by which if they have not been passively replicated, they are likely to fire. Thus, low-probability origins distant from high-probability origins are likely to fire in later S phase; exactly when they fire, on average, depends on their relative firing probability.
Figure 1a also demonstrates the importance of increasing firing probability in the stochastic model. We included a third simulation with stochastic origin firing but with constant firing probability. Without some mechanism to ensure that origins with low firing probability fire eventually, the late-replicating region of the chromosome is primarily passively replicated, obscuring late origins in the trep replication profile.
The simulations in Fig. 1 not only demonstrate that the increasing-probability model can account for defined replication times as successfully as a deterministic model but also show that the two types of models are experimentally distinguishable. In particular, the behavior of late-firing origins in the two models is significantly different (Fig. 1c). Although the trep for the late origins in the two models is similar (Fig. 1a), the actual firing times of the origins in the stochastic models is much later (Fig. 1c). Furthermore, the distribution of firing times of the late origins in the stochastic model is greater than that expected in a deterministic model (Fig. 1c and Yang et al. submitted for publication). Often, the kinetic data necessary to distinguish the models is discarded in the creation of trep replication profiles. However, a kinetic analysis has been done for fission yeast replication and is consistent with increasing firing probability in later S phase (Eshaghi et al. 2007). Furthermore, our kinetic analysis of published budding yeast microarray data supports a stochastic, increasing-probability model and is incompatible with simple deterministic models (Yang et al. submitted for publication).
Potential biochemical mechanisms for the increasing-probability model
For the increasing-probability model to be able to explain replication kinetics in vivo, there need to be plausible biochemical mechanisms for its two main functions: the increase in firing probability during S phase and the difference in firing probability between origins.
Mechanisms for increasing firing probability during S phase
Several mechanisms for increasing the probability of origin firing as S phase progresses have been proposed (Lucas et al. 2000; Hyrien et al. 2003; Rhind 2006; Goldar et al. 2008; Lygeros et al. 2008; Gauthier and Bechhoefer 2009). They can be grouped into three broad categories: polymerase recycling, limiting activator, and increasing activator. Polymerase-recycling models posit that there is a limiting member of the replication fork, perhaps the replicative polymerase itself, and that once all of this factor is incorporated into forks, no more forks can be established (Hyrien et al. 2003; Goldar et al. 2008; Rhind 2008; Gauthier and Bechhoefer 2009). Technically, polymerase recycling limits fork establishment not origin firing, per se, but the effect on replication kinetics is the same. Polymerase-recycling models lead to a constant number of forks replicating the genome at all times, the number of which is set by the number of molecules of the limiting factor. However, since the amount of unreplicated DNA decreases as S phase progresses, the ratio of forks to unreplicated DNA increases during S phase. Thus, later in S phase, the number of forks being established, relative to the amount of unreplicated DNA, goes up. This effect is the equivalent of having origins fire with higher probability during later S phase. The simplest polymerase-recycling models are not consistent with the suggestion that the number of replication forks increases during replication stress (Ge et al. 2007; Blow and Ge 2009). However, they can explain the observation that slowing fork progression also slows origin firing to the same extent (Rhind 2008).
In the limiting-activator models, an activator, for example, the Dbf4-dependent replication kinase (DDK), is sufficient to fire only a certain number of origins each minute (Rhind 2006; Lygeros et al. 2008). However, as in the polymerase-recycling model, as S phase progresses, that number of origins is a larger fraction of the remaining unfired origins, and so the firing probability of the remaining origins increases. Limiting-activator models have the advantage that they do not explicitly restrict the number of active forks and therefore are compatible with models in which fork density increases during replication stress (Blow and Ge 2009). Although a limiting-activator model can produce realistic S-phase completion times (Lygeros et al. 2008), published implementations do not fit experimental replication-kinetics data (Goldar et al. 2008).
DDK in fission yeast appears to be a diffusible, catalytic, rate-limiting activator—the requisite characteristics to satisfy the limiting-activator model (Patel et al. 2008). However, there are multiple regulated steps in origin activation, and there is no reason that the limiting step need be the same in every organism. For example, although DDK and Cdc45 seem to be rate limiting for origin firing in fission yeast (Patel et al. 2008; Wu and Nurse 2009), Cdk1 and Cdk2 have been suggested to be rate limiting in vertebrates (Krasinska et al. 2008; Katsuno et al. 2009), and Cdc28-Clb5 seems to regulate origin firing in budding yeast (Donaldson et al. 1998; McCune et al. 2008).
The increasing-activator models are based on the idea that activity of a limiting activator increases as S phase progresses (Lucas et al. 2000; Goldar et al. 2008; Gauthier and Bechhoefer 2009). One explicit mechanism for increasing the activity of an activator is to have an excess of the activator in the cytoplasm that gets progressively concentrated in the nucleus during S phase. Other variants posit increased accumulation of an activator due to increased expression or stabilization. This class of models suffers from proposing a replication-independent timing mechanism. Therefore, to keep the timer synchronized with replication during perturbations, such a mechanism would require an active feedback loop that could monitor the progression of replication. Furthermore, as discussed above, because there are fewer potential origins to fire later in S phase, increasing the activity of a limiting activator may not be required to produce increasing firing probability.
Although simple versions of the increasing-probability model predict that firing probability continues to rise throughout S phase, in reality, firing probability seems to rise for most of S phase and then decline in late S phase (Goldar et al. 2008, 2009; Yang et al. submitted for publication). Such a decline does not interfere with the efficient completion of replication provided by increasing-probability models; in fact, it is consistent with the expected biochemical constraints on a diffusible activator (Gauthier and Bechhoefer 2009).
Mechanisms for varying relative firing probability
The relative firing probability of origins could be regulated in several ways. One factor that surely affects the probability of an origin firing is origin recognition complex (ORC) occupancy. If ORC binds to an origin in only 50% of cells, that origin can fire no more that 50% of the time. However, since ORC cannot license origins during S phase, this mechanism of regulating firing probability cannot lead to increasing firing probability later in S phase.
Chromatin structure is a plausible mechanism by which firing probability could be regulated. If chromatin structure restricts access of origin activators, it would decrease the efficiency with origins fire. This possibility is consistent with the observation that heterochromatin replicates late (Goren and Cedar 2003). In fact, in one example of early replicating heterochromatin in fission yeast, DDK is specifically recruited to the heterochromatin to overcome the late replication that the heterochromatin otherwise causes (Kim et al. 2003; Hayashi et al. 2009). Furthermore, regulation of firing probability by chromatin structure is consistent with the correlation seen between early replication and transcription. In fruit flies, this correlation is not seen at the level of individual genes, but only when averaged over 200 kb regions, suggesting the replication timing is not correlated with the transcriptional activity of any particular gene, but rather is affected by the general accessibility of large regions of chromatin (MacAlpine et al. 2004).
Another mechanism that could affect firing probability is the number of mini-chromosome maintenance (MCM) complexes loaded at origins (Yang et al. submitted for publication). MCM, the presumptive replicative helicase, is present at up to a 30-fold excess over the number needed to replicate the genome (Lei et al. 1996; Donovan et al. 1997; Hyrien et al. 2003). Some of that excess MCM is loaded at origins that will not fire, but some of the excess is loaded as multiple MCM complexes at individual origins (Edwards et al. 2002; Bowers et al. 2004). If each MCM has an intrinsic probability of initiating replication, an origin with ten pairs of MCM complexes will be ten times more likely to fire than an origin with one pair of MCM complexes (Yang et al. submitted for publication). Thus, the efficiency with which ORC loads MCM at a given origin, or the amount of time ORC is bound to an origin and able to load MCM, could determine an origins firing probability. Although multiple-loaded MCMs would need to move away from the loading site, they would presumably remain local, sufficiently close to the loading site to appear as a single origin in the replication profiles. This mechanism could also account for the observed increase in origin efficiency caused by lengthening mitosis (Wu and Nurse 2009); more time to load MCM during mitosis could lead to more efficient firing during S.
Concluding remarks
The goal of this review is to make the point that there is no inherent conflict between stochastic origin firing and defined replication timing. To some extent, the difference between stochastic firing and deterministic firing is a semantic one, as we have suggested above. In the stochastic models presented here and elsewhere, origins fire—on average—at well-defined times. However, there are important mechanistic implications of favoring one class of models over the other. In deterministic-firing models, one must invoke a mechanism that measures the passage of S phase and fires origins at specific times. Plausible biochemical details for such mechanisms have yet to be proposed. In the stochastic, increasing-probability models, variations in replication timing are a natural, and in fact inevitable, consequence of variations in relative firing probability between origins. Furthermore, the increasing-probability models make general testable predictions, such as the differences in firing kinetic shown in Fig. 1c, and suggest specific biochemical mechanisms, which are also testable. Although the mechanisms described here can explain the function of the increasing-probability model, there may be other mechanisms that could do so as well. In particular, there is no need for the underlying mechanisms to be conserved; different mechanism could lead to increasing firing probability in different organisms. Nonetheless, whatever mechanisms turn out to operate in vivo, they must be able to reconcile the stochastic nature of origin firing with defined patterns of replication timing.
Methods
Monte Carlo simulations were performed in Igor Pro (www.wavemetrics.com) using custom scripts. The chromosome was simulated as an array of 200 loci, spaced 1 kb apart. Replication was simulated in 80, 1-min time-steps with replication fork rate of 1 kb/min. Each figure represents the results of 1000 simulations. The trep for each set of simulations was calculated and smoothed to produce the presented replication profiles. The replication profiles were normalized so that the early origins fired at the same time. Simulation code is available upon request.
Acknowledgments
We thank Sean Ryder, Lucienne Ronco, and Olivier Hyrien for critical reading of the manuscript and Olivier Hyrien and Arach Goldar for sharing their review in this volume before publication. We thank Alessandro de Moura and Conrad Nieduszynski for sharing their work prior to publication.
Abbreviations
- DDK
Dbf4-dependent kinase
- MCM
Mini-chromosome maintenance
- ORC
Origin recognition complex
Contributor Information
Nicholas Rhind, Email: Nick.Rhind@umassmed.edu, Department of Biochemistry and Molecular Pharmacology, University of Massachusetts Medical School, Worcester, MA 01605, USA.
Scott Cheng-Hsin Yang, Department of Physics, Simon Fraser University, Burnaby, BC V5A 1S6, Canada.
John Bechhoefer, Department of Physics, Simon Fraser University, Burnaby, BC V5A 1S6, Canada.
References
- Alvino GM, Collingwood D, Murphy JM, Delrow J, Brewer BJ, Raghuraman MK. Replication in hydroxyurea: it's a matter of time. Mol Cell Biol. 2007;27:6396–6406. doi: 10.1128/MCB.00719-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Ge XQ. A model for DNA replication showing how dormant origins safeguard against replication fork failure. EMBO Rep. 2009;10(4):406–412. doi: 10.1038/embor.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JL, Randell JC, Chen S, Bell SP. ATP hydrolysis by ORC catalyzes reiterative Mcm2–7 assembly at a defined origin of replication. Mol Cell. 2004;16:967–978. doi: 10.1016/j.molcel.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Cadoret JC, Meisch F, Hassan-Zadeh V, Luyten I, Guillet C, Duret L, Quesneville H, Prioleau MN. Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc Natl Acad Sci U S A. 2008;105:15837–15842. doi: 10.1073/pnas.0805208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czajkowsky DM, Liu J, Hamlin JL, Shao Z. DNA combing reveals intrinsic temporal disorder in the replication of yeast chromosome VI. J Mol Biol. 2008;375:12–19. doi: 10.1016/j.jmb.2007.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson AD, Raghuraman MK, Friedman KL, Cross FR, Brewer BJ, Fangman WL. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol Cell. 1998;2:173–182. doi: 10.1016/s1097-2765(00)80127-6. [DOI] [PubMed] [Google Scholar]
- Donovan S, Harwood J, Drury LS, Diffley JF. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci U S A. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MC, Tutter AV, Cvetic C, Gilbert CH, Prokhorova TA, Walter JC. MCM2–7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J Biol Chem. 2002;277:33049–33057. doi: 10.1074/jbc.M204438200. [DOI] [PubMed] [Google Scholar]
- Eshaghi M, Karuturi RK, Li J, Chu Z, Liu ET, Liu J. Global profiling of DNA replication timing and efficiency reveals that efficient replication/firing occurs late during S-phase in S. pombe. PLoS ONE. 2007;2:e722. doi: 10.1371/journal.pone.0000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangman WL, Hice RH, Chlebowicz-Sledziewska E. ARS replication during the yeast S phase. Cell. 1983;32:831–838. doi: 10.1016/0092-8674(83)90069-7. [DOI] [PubMed] [Google Scholar]
- Gauthier MG, Bechhoefer J. Control of DNA replication by anomalous reaction-diffusion kinetics. Phys Rev Lett. 2009;102:158104. doi: 10.1103/PhysRevLett.102.158104. [DOI] [PubMed] [Google Scholar]
- Ge XQ, Jackson DA, Blow JJ. Dormant origins licensed by excess Mcm2–7 are required for human cells to survive replicative stress. Genes Dev. 2007;21:3331–3341. doi: 10.1101/gad.457807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldar A, Labit H, Marheineke K, Hyrien O. A dynamic stochastic model for DNA replication initiation in early embryos. PLoS ONE. 2008;3:e2919. doi: 10.1371/journal.pone.0002919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldar A, Marsolier-Kergoat MC, Hyrien O. Universal temporal profile of replication origin activation in eukaryotes. PLoS ONE. 2009;4:e5899. doi: 10.1371/journal.pone.0005899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren A, Cedar H. Replicating by the clock. Nat Rev Mol Cell Biol. 2003;4:25–32. doi: 10.1038/nrm1008. [DOI] [PubMed] [Google Scholar]
- Hamlin JL, Mesner LD, Lar O, Torres R, Chodaparambil SV, Wang L. A revisionist replicon model for higher eukaryotic genomes. J Cell Biochem. 2008;105:321–329. doi: 10.1002/jcb.21828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi MT, Takahashi TS, Nakagawa T, Nakayama J, Masukata H. The heterochromatin protein Swi6/HP1 activates replication origins at the pericentromeric region and silent mating-type locus. Nat Cell Biol. 2009;11:357–362. doi: 10.1038/ncb1845. [DOI] [PubMed] [Google Scholar]
- Heichinger C, Penkett CJ, Bahler J, Nurse P. Genome-wide characterization of fission yeast DNA replication origins. EMBO J. 2006;25:5171–5179. doi: 10.1038/sj.emboj.7601390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmquist G, Gray M, Porter T, Jordan J. Characterization of Giemsa dark- and light-band DNA. Cell. 1982;31:121–129. doi: 10.1016/0092-8674(82)90411-1. [DOI] [PubMed] [Google Scholar]
- Hyrien O, Mechali M. Chromosomal replication initiates and terminates at random sequences but at regular intervals in the ribosomal DNA of Xenopus early embryos. EMBO J. 1993;12:4511–4520. doi: 10.1002/j.1460-2075.1993.tb06140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O, Marheineke K, Goldar A. Paradoxes of eukaryotic DNA replication: MCM proteins and the random completion problem. Bioessays. 2003;25:116–125. doi: 10.1002/bies.10208. [DOI] [PubMed] [Google Scholar]
- Katsuno Y, Suzuki A, Sugimura K, Okumura K, Zineldeen DH, Shimada M, Niida H, Mizuno T, Hanaoka F, Nakanishi M. Cyclin A-Cdk1 regulates the origin firing program in mammalian cells. Proc Natl Acad Sci U S A. 2009;106:3184–3189. doi: 10.1073/pnas.0809350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SM, Dubey DD, Huberman JA. Early-replicating heterochromatin. Genes Dev. 2003;17:330–335. doi: 10.1101/gad.1046203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasinska L, Besnard E, Cot E, Dohet C, Mechali M, Lemaitre JM, Fisher D. Cdk1 and Cdk2 activity levels determine the efficiency of replication origin firing in Xenopus. EMBO J. 2008;27:758–769. doi: 10.1038/emboj.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labit H, Perewoska I, Germe T, Hyrien O, Marheineke K. DNA replication timing is deterministic at the level of chromosomal domains but stochastic at the level of replicons in Xenopus egg extracts. Nucleic Acids Res. 2008;36:5623–5634. doi: 10.1093/nar/gkn533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskey RA. Chromosome replication in early development of Xenopus laevis. J Embryol Exp Morphol. 1985;89 (Suppl):285–296. [PubMed] [Google Scholar]
- Lebofsky R, Heilig R, Sonnleitner M, Weissenbach J, Bensimon A. DNA replication origin interference increases the spacing between initiation events in human cells. Mol Biol Cell. 2006;17:5337–5345. doi: 10.1091/mbc.E06-04-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Kawasaki Y, Tye BK. Physical interactions among Mcm proteins and effects of Mcm dosage on DNA replication in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5081–5090. doi: 10.1128/mcb.16.9.5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas I, Chevrier-Miller M, Sogo JM, Hyrien O. Mechanisms ensuring rapid and complete DNA replication despite random initiation in Xenopus early embryos. J Mol Biol. 2000;296:769–786. doi: 10.1006/jmbi.2000.3500. [DOI] [PubMed] [Google Scholar]
- Lygeros J, Koutroumpas K, Dimopoulos S, Legouras I, Kouretas P, Heichinger C, Nurse P, Lygerou Z. Stochastic hybrid modeling of DNA replication across a complete genome. Proc Natl Acad Sci U S A. 2008;105 (34):12295–12300. doi: 10.1073/pnas.0805549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine DM, Rodriguez HK, Bell SP. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev. 2004;18:3094–3105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll RM, Fangman WL. Time of replication of yeast centromeres and telomeres. Cell. 1988;54:505–513. doi: 10.1016/0092-8674(88)90072-4. [DOI] [PubMed] [Google Scholar]
- McCune HJ, Danielson LS, Alvino GM, Collingwood D, Delrow JJ, Fangman WL, Brewer BJ, Raghuraman MK. The temporal program of chromosome replication: genome-wide replication in clb5Δ Saccharomyces cerevisiae. Genetics. 2008;180 (4):1833–1847. doi: 10.1534/genetics.108.094359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel PK, Arcangioli B, Baker SP, Bensimon A, Rhind N. DNA replication origins fire stochastically in fission yeast. Mol Biol Cell. 2006;17:308–316. doi: 10.1091/mbc.E05-07-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel PK, Kommajosyula N, Rosebrock A, Bensimon A, Leatherwood J, Bechhoefer J, Rhind N. The Hsk1 (Cdc7) replication kinase regulates origin efficiency. Mol Biol Cell. 2008;19:5550–5558. doi: 10.1091/mbc.E08-06-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL. Replication dynamics of the yeast genome. Science. 2001;294:115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- Reynolds AE, McCarroll RM, Newlon CS, Fangman WL. Time of replication of ARS elements along yeast chromosome III. Mol Cell Biol. 1989;9:4488–4494. doi: 10.1128/mcb.9.10.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N. DNA replication timing: random thoughts about origin firing. Nat Cell Biol. 2006;8:1313–1316. doi: 10.1038/ncb1206-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N. An intrinsic checkpoint model for regulation of replication origins. Cell Cycle. 2008;7:2619–2620. doi: 10.4161/cc.7.17.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JH. DNA synthesis in chromosomes: implications of early experiments. Bioessays. 1989;10:121–124. doi: 10.1002/bies.950100407. [DOI] [PubMed] [Google Scholar]
- Wu PY, Nurse P. Establishing the program of origin firing during S phase in fission yeast. Cell. 2009;136:852–864. doi: 10.1016/j.cell.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki N, Terashima H, Kitada K. Mapping of early firing origins on a replication profile of budding yeast. Genes Cells. 2002;7:781–789. doi: 10.1046/j.1365-2443.2002.00559.x. [DOI] [PubMed] [Google Scholar]
- Yang SC, Bechhoefer J. How Xenopus laevis embryos replicate reliably: investigating the random-completion problem. Phys Rev E Stat Nonlin Soft Matter Phys. 2008;78:041917. doi: 10.1103/PhysRevE.78.041917. [DOI] [PubMed] [Google Scholar]