Abstract
DNA damage is a source of carcinogenicity and is also the source of cytotoxicity of γ-radiolysis and antitumor agents, such as the enediynes. The dioxobutane lesion (DOB) is produced by a variety of DNA damaging agents, including the aforementioned. Repair of DOB is important for maintaining the integrity of the genome, as well as to counteract therapeutic agents that target DNA. We demonstrate that the DOB lesion efficiently and irreversibly inhibits repair by DNA polymerase β, an integral enzyme in base excision repair. Irreversible inhibition of Pol β by DOB suggests that this lesion provides a chemical explanation for the cytotoxicity of drugs that produce it, and explains previously unexplained observations in the literature concerning abasic lesions that are not repaired efficiently. Finally, these observations provide the impetus for the design of a new family of inhibitors of Pol β.
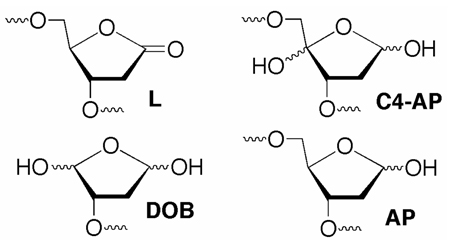
DNA oxidation is a source of carcinogenicity. It is also the source of cytotoxicity of γ-radiolysis and antitumor agents, such as the enediynes. Double-strand breaks (dsbs) are often thought of as an extremely challenging form of DNA damage for cells to overcome. However, few damaging agents produce dsbs in high yields.1 In contrast, oxidized abasic lesions (e.g. L, C4-AP, DOB) resulting from hydrogen atom abstraction from the 2'-deoxyribose ring are produced in greater yields by a variety of DNA damaging agents.2 These lesions exhibit potentially important reactivity and effects on repair enzymes. For instance, 2-deoxyribonolactone (L) cross-links base excision repair enzymes, while C4-AP and DOB form DNA interstrand cross-links.3–6 Furthermore, a cross-link involving C4-AP is converted into a dsb by a bacterial nucleotide excision repair system.7 We report here that the DOB lesion efficiently and irreversibly inhibits repair by DNA polymerase β (Pol β).
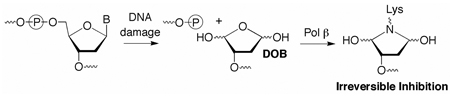
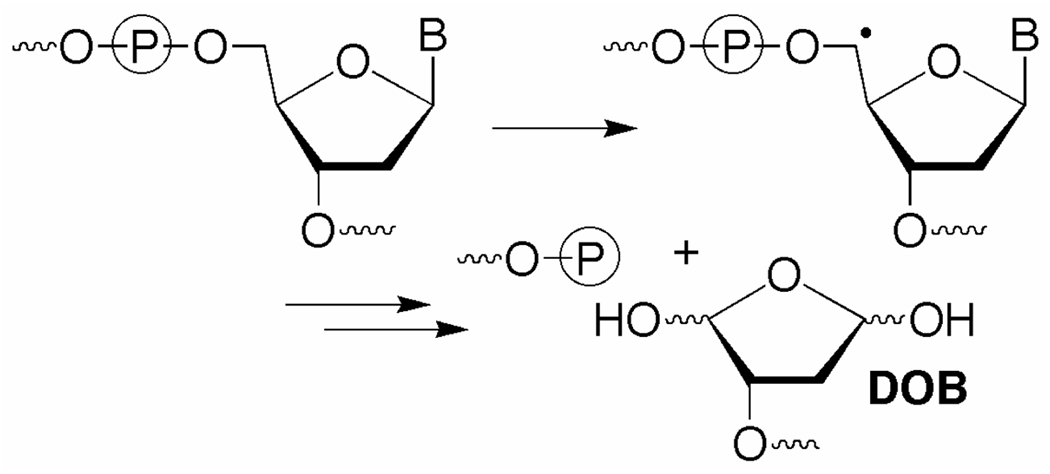
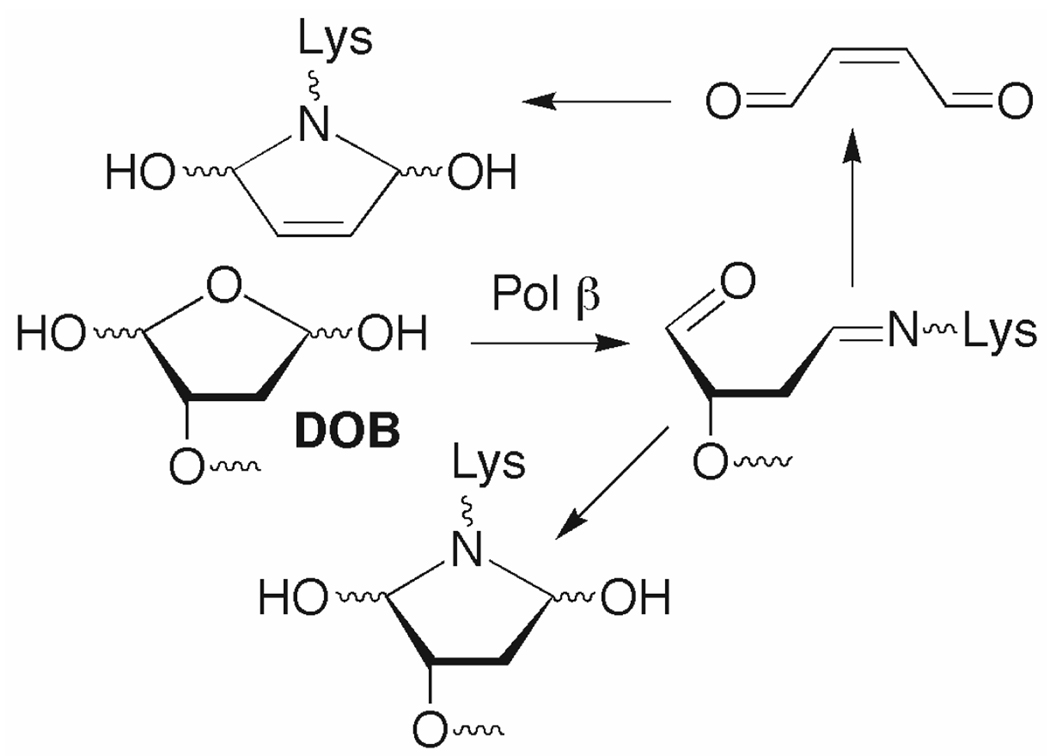
The DOB lesion is produced concomitantly with a strand break following C5'-hydrogen atom abstraction (Scheme 1).8 It has been detected in DNA exposed to antitumor agents as well as ionizing radiation, and accounts for ~5% of the deoxyribose oxidation products from the latter.9 DOB eliminates butenedial, which reacts to yield likely promutagenic exocyclic adducts with dC and dA.10 DOB also yields DNA interstrand cross-links by reacting selectively with dA opposite a 3'-adjacent thymidine.5 Given the potential detrimental effects of these secondary products, efficient repair of DOB should be important.
Scheme 1.
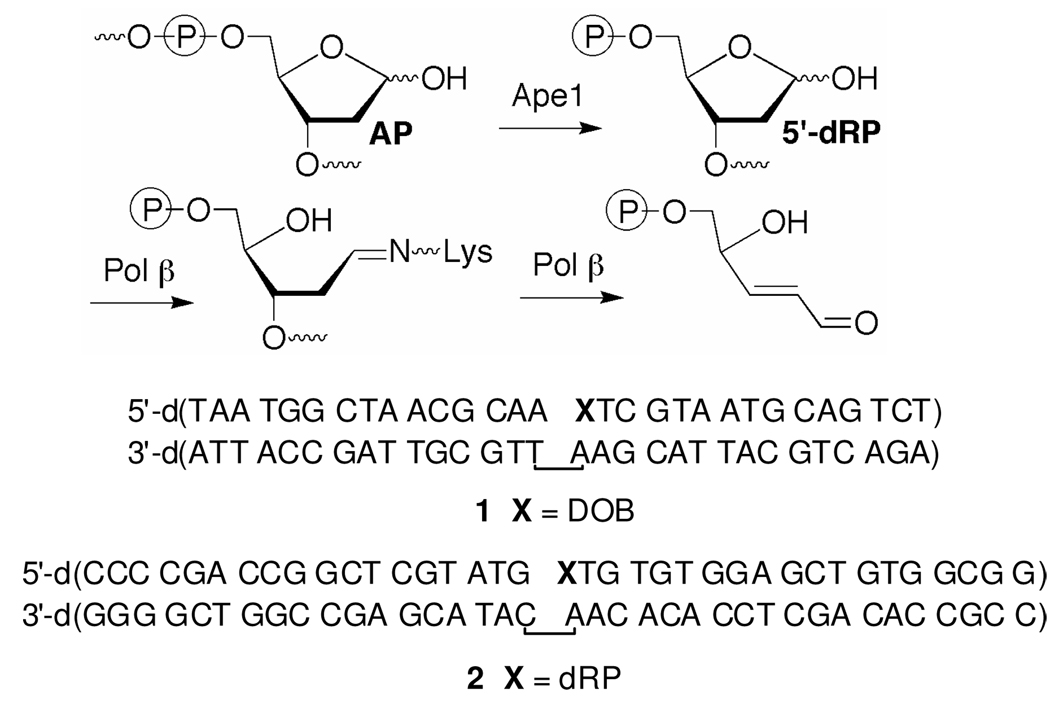
Abasic sites (AP) are typically removed by a series of enzymes in the base excision repair pathway (BER, Scheme 2). In mammalian cells the lesion (5'-dRP) is excised by Pol β following AP incision at its 5'-phosphate by apurinic endonuclease (Ape1).11,12 Excision is achieved via a lyase mechanism in which Pol β forms an intermediate Schiff base with the 5'-dRP intermediate.13 Pol β then fills the gap using an appropriate dNTP, and repair is completed by an ATP dependent DNA ligase. Following this paradigm, we expected that Pol β excision would be the first step in BER of DOB because its formation in conjunction with a direct strand break obviates the need for Ape1.
Scheme 2.
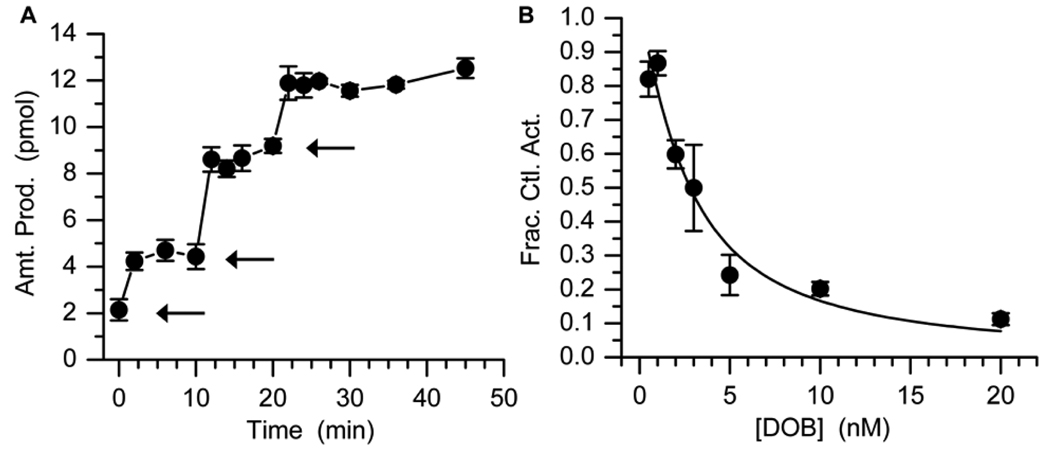
DOB excision by Pol β was examined using a ternary complex (1) in which the oligonucleotide containing the lesion was prepared via solid phase synthesis as previously reported.14 Subjection of 3'-32P-1 (200 nM, strand containing DOB is labeled) to Pol β (5 nM) rapidly produced a small amount of the expected oligonucleotide product, but the reaction stopped after a few minutes (Figure 1A). Additional aliquots of Pol β yielded the same observation. Approximately 4 equivalents of product were produced from each aliquot of enzyme. In contrast, a comparable substrate containing AP (3'-32P-2, strand containing dRP is labeled) was completely consumed by Pol β when the substrate was present in 40-fold excess.15 These observations suggested that Pol β was inhibited by DOB containing DNA. Additional evidence for inhibiton by DOB was gleaned by preincubating (3 min, the amount of time to inactivate Pol β in Figure 1A) Pol β with varying concentrations of 1 prior to adding the enzyme to 3'-32P-2 and measuring the amount of product produced over 10 min at 37 °C (Figure 1B). Inhibition depended upon the concentration of 1 and the IC50 value was 2.8 ± 0.3 nM in the presence of 2.5 nM Pol β.
Figure 1.
Inhibition of Pol β lyase reaction by DOB. (A) Amount of product obtained from 3'-32P-1 (200 nM) upon addition of Pol β. Each arrow indicates the addition of 1 pmol Pol β. (B) Fraction of 5'-dRP (3'-32P-2, 100 nM) converted following preincubation with DOB (1).
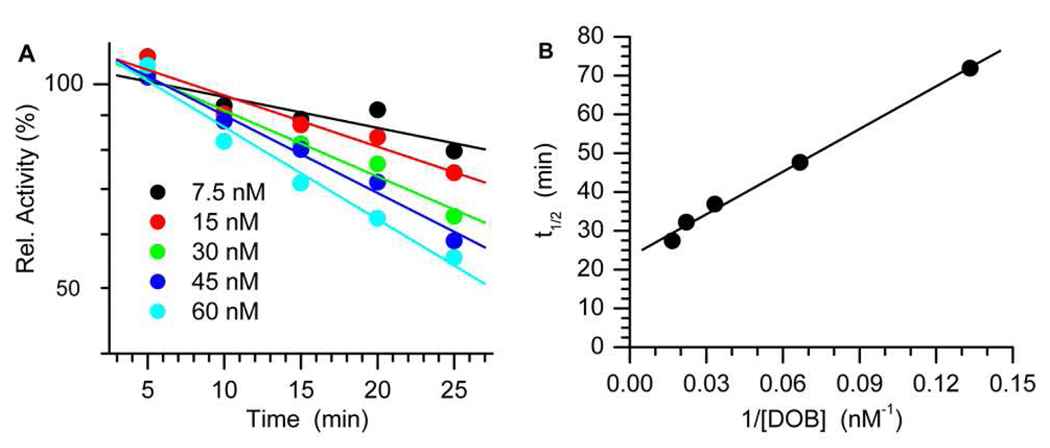
The rapid conversion of a small number of equivalents of 3'-32P-1 and the abrupt cessation of the reaction suggested that DOB irreversibly inhibited Pol β. Evidence for this mode of inhibition was obtained kinetically (Figure 2).15,16 The ratio of product produced from 3'-32P-2 (AP) in the presence of inhibitor relative to that formed in the absence of DOB was measured as a function of time for a series of inhibitor (1) concentrations (0–60 nM). A plot of the logarithm of the relative activities decayed linearly as a function of time (Figure 2A) and the pace at which inactivation occurred was proportional to the concentration of 1. The KI (12.8 ± 5.2 nM) and kinact (4.2 ± 0.9 × 10−4 s−1) were subsequently determined by plotting the half-life for inactivation versus the reciprocal of the concentration of 1 (DOB) (Figure 2B). Irreversible inhibition of Pol β by DOB is more efficient than that by 2-deoxyribonolactone (L), which cross-links Pol β following Ape1 incision, presumably because of greater electrophilicity of the aldehyde groups in the former.3,17
Figure 2.
Kinetic analysis of irreversible inhibition by DOB (1). (A) Effect of increasing [DOB] (1) on Pol β (7.5 nM) lyase reaction of AP (3'-32P-2, 500 nM). (B) Half-life of Pol β inactivation as a function of [DOB].
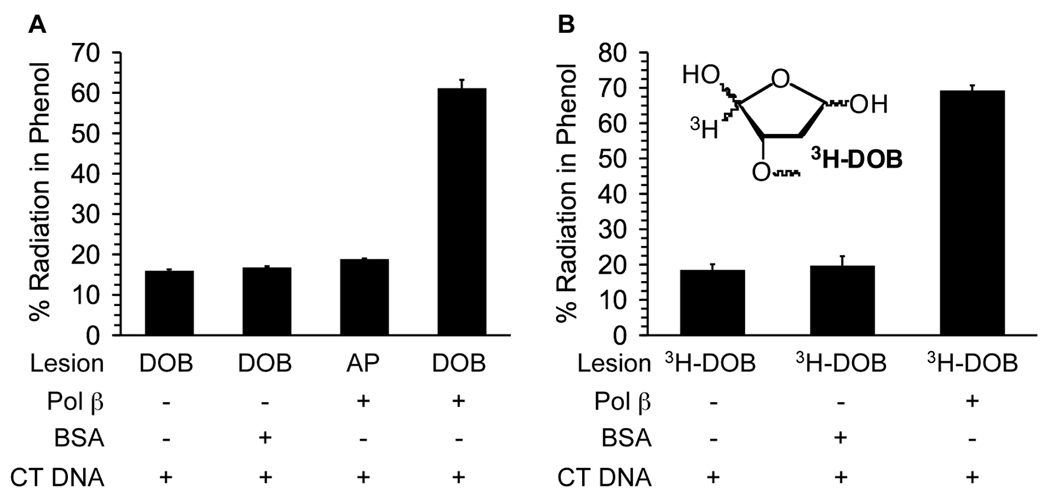
Two modes of irreversible inhibition of Pol β by DOB were envisioned. The DOB lesion could inactivate Pol β by trapping the nucleophilic lysine involved in Schiff base formation and/or the butenedial product could react with the enzyme. (Scheme 3 shows examples of possible products.) Attempts at isolating DNA-protein cross-linked product(s) by gel electrophoresis were unsuccessful. However, support for covalent protein modification was obtained from phenol extraction of reactions containing radiolabeled DOB substrate (Figure 3). Radiation in the phenol layer was quantified by liquid scintillation counting following reaction of 3'-32P-1 with excess Pol β. The amount of 32P retained in the phenol was 45 ± 2% after subtracting the background level of radiation extracted in the absence of protein (Figure 3A). The specificity for the covalent interaction between DOB and Pol β was supported by the absence of 32P above background in the phenol after incubation of 3'-32P-1 with bovine serum albumin or AP containing DNA (3'-32P-2) with Pol β (Figure 3A). Evidence for irreversible inhibition by butenedial released from DOB was obtained by preparing the ternary complex in which DOB was tritiated (3H-DOB, Figure 3).15 Extraction of 1 containing 3H-DOB reacted with excess Pol β resulted in 51 ± 2% of the radiation above background being retained in the phenol layer. Inactivation of Pol β by either pathway (Scheme 2) results in transfer of 3H from 1 to the phenol, but 32P transfer requires that DOB still be linked to the DNA. The difference between the percentage of 3H and 32P in the phenol layer suggests that ≥90% of the inhibition results from trapping by DOB, whereas butenedial contributes ≤10%.
Scheme 3.
Figure 3.
Determination of covalent trapping of Pol β (1.25 µM) via phenol extraction of reactions containing radiolabeled 1. (A) 3'-32P-1 (125 nM), (B) 3H-1 (125 nM).
Our data suggest that competing reactions in the Pol β active site result in inactivation in ~25% of the enzyme's encounters with DOB. Efficient Pol β inactivation is important for a number of reasons. For instance, these data suggest that DOB may be the 5'-abasic site of undetermined structure produced in cells upon exposure to H2O2 that are resistant to repair by Pol β.18 In addition, DOB's inhibition of Pol β may provide a chemical basis for the potent cytotoxicity of antitumor agents, such as neocarzinostatin. Finally, Pol β is a potentially important target for mechanism based inhibitors because it is overexpressed in tumor cells and is an attractive target for adjuvants to DNA targeted anticancer therapies.19 The reactivity of DOB suggests an approach for designing irreversible inhibitors that target the lyase site of this enzyme.
Supplementary Material
Acknowledgment
We are grateful to Aaron Jacobs and Jon Sczepanski for their careful reading of the manuscript, and for generous support from the National Institute of General Medical Sciences (GM-063028).
Footnotes
Supporting Information. Procedures for the synthesis of all compounds and all other experiments. Kinetic plots of irreversible inhibition by 1. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Kishikawa H, Jiang Y-P, Goodisman J, Dabrowiak JC. J. Am. Chem. Soc. 1991;113:5434–5440. [Google Scholar]
- 2.Gates KS. Chem. Res. Toxicol. 2009;22:1747–1760. doi: 10.1021/tx900242k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hashimoto M, Greenberg MM, Kow YW, Hwang J-T, Cunningham RP. J. Am. Chem. Soc. 2001;123:3161–3162. doi: 10.1021/ja003354z. [DOI] [PubMed] [Google Scholar]
- 4.Sczepanski JT, Jacobs AC, Majumdar A, Greenberg MM. J. Am. Chem. Soc. 2009;131:11132–11139. doi: 10.1021/ja903404v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan L, Greenberg MM. J. Am. Chem. Soc. 2009;131:15225–15231. doi: 10.1021/ja9061695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sczepanski JT, Jacobs AC, Greenberg MM. J. Am. Chem. Soc. 2008;130:9646–9647. doi: 10.1021/ja8030642. [DOI] [PubMed] [Google Scholar]
- 7.Sczepanski J, Jacobs AC, Van Houten B, Greenberg MM. Biochemistry. 2009;48:7565–7567. doi: 10.1021/bi901006b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldberg IH. Acc. Chem. Res. 1991;24:191–198. [Google Scholar]
- 9.Chen B, Bohnert T, Zhou X, Dedon PC. Chem. Res. Toxicol. 2004;17:1406–1413. doi: 10.1021/tx049818e. [DOI] [PubMed] [Google Scholar]
- 10.Chen B, Vu CC, Byrns MC, Dedon PC, Peterson LA. Chem. Res. Toxicol. 2006;19:982–985. doi: 10.1021/tx0601197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsumoto Y, Kim K. Science. 1995;269:699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- 12.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd ed. Washington, D.C.: ASM Press; 2006. [Google Scholar]
- 13.Prasad R, Beard WA, Strauss PS, Wilson SH. J. Biol. Chem. 1998;273:15263–15270. doi: 10.1074/jbc.273.24.15263. [DOI] [PubMed] [Google Scholar]
- 14.Kodama T, Greenberg MM. J. Org. Chem. 2005;70:9916–9924. doi: 10.1021/jo051666k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.See Supporting Information.
- 16.Silverman RB. The Organic Chemistry of Enzyme-Catalyzed Reactions. San Diego: Academic Press; 2000. [Google Scholar]
- 17.DeMott MS, Beyret E, Wong D, Bales BC, Hwang J-T, Greenberg MM, Demple B. J. Biol. Chem. 2002;277:7637–7640. doi: 10.1074/jbc.C100577200. [DOI] [PubMed] [Google Scholar]
- 18.Nakamura J, La DK, Swenberg JA. J. Biol. Chem. 2000;275:5323–5328. doi: 10.1074/jbc.275.8.5323. [DOI] [PubMed] [Google Scholar]
- 19.Gao Z, Maloney DJ, Dedkova LM, Hecht SM. Bioorganic & Medicinal Chemistry. 2008;16:4331–4340. doi: 10.1016/j.bmc.2008.02.071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.