Abstract
Ste12 and Ste12-like proteins are transcription factors found exclusively in the fungal kingdom. In the yeast model Saccharomyces cerevisiae, where the first member was identified, Ste12p was shown to regulate mating and invasive/pseudohyphal growth. In recent literature, there have been several reports of Ste12-like factors in multiple fungal systems, yeasts or filamentous fungi, with saprophytic or parasitic life-styles. In all these models, Ste12 and Ste12-like factors are involved in the regulation of fungal development and pathogenicity. In this review, we discuss the features, the regulation, and the role of Ste12 and Ste12-like factors by highlighting the similarities and dissimilarities that occur within this group.
Fungi, like any other living organisms, are capable of perceiving changes in their environment and inducing appropriate adaptative responses. Morphogenetic programs initiated by environmental constraints include pseudohyphal and invasive growth, dimorphic transitions, mating, or other specialized cell differentiations. Understanding these processes at a molecular level has been the subject of intensive investigation to gain insights into the underlying mechanisms. A specific class of fungal transcription factors, grouped under the term Ste12-like factors, has emerged as major players in these adaptation schemes. The term Ste12 came from the identification of a yeast sterile mutant. The corresponding Ste12 protein was originally found to be a target of the Fus3 mitogen-activated protein kinase (MAPK) cascade regulating mating (14). Ste12p was then also identified as a major regulator of yeast invasive growth and pseudohyphal development, as a target of the Kss1p MAPK cascade (18, 29). Ste12p works as a homodimer within the mating signaling cascade, whereas it acts in tandem with the Tec1p transcription factor in the invasive/pseudohyphal growth cascade. In true filamentous fungi, Ste12-like proteins play essential roles in sexual development and pathogenicity. They are designated Ste12-like factors since they contain two C-terminally located tightly linked C2H2 zinc fingers, which are absent in yeast Ste12p. The recent identification of an STE12-like gene from the arbuscular mycorrhizal fungus Glomus intraradices suggests a very ancient origin of this gene family in the fungal kingdom (46). It also supports the intriguing line of thought that these proteins have been recruited to serve as molecular switches to allow the environmental adaptation of various fungi with a wide range of developmental modes and ecological niches.
Ste12 TRANSCRIPTION FACTORS: FUNCTIONAL DOMAINS AND DNA BINDING SPECIFICITIES
The first Ste12 gene was isolated from the yeast Saccharomyces cerevisiae (14) as a key regulator of two major developmental fates: the differentiation of yeast cells into a mating-competent cell type and the transition to filamentous growth occurring in response to nutrient deprivation. Since then, transcription factors showing similarities to yeast Ste12p were identified exclusively in fungi with saprophytic or parasitic life-styles (Table 1).
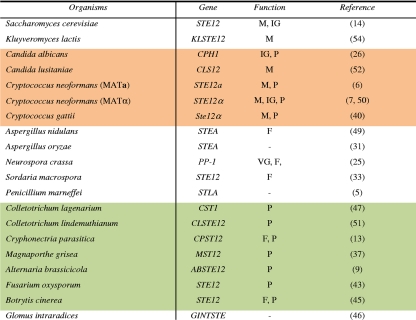
Table 1.
Identification and role of STE12 and STE12-like genesa
The impact of gene inactivation was assessed with regard to vegetative growth (VG), mating (M), fertility (F), conidiation (C), invasive growth (IG), and pathogenicity (P). Animal and plant pathogens are indicated by orange and green backgrounds, respectively.
Ste12 proteins are all characterized by the presence of an N-terminally located homeodomain-like motif named Ste (InterPro identification number IPR003120), which is involved in DNA binding (53). The homeodomain was originally identified in several Drosophila melanogaster homeotic proteins containing a helix-turn-helix structure that is involved in DNA binding (19). The fungal Ste homeodomain is highly divergent from other characterized homeodomains, but some residues involved in DNA binding are conserved (53). A crucial region for DNA binding is the KQKVFFWFSVA sequence located in the third helix. Very few studies aimed at identifying the DNA binding site of fungal Ste12p have been reported. However, this 11-amino-acid region is highly conserved in all fungal Ste12 proteins analyzed so far, suggesting that DNA sequences recognized by Ste12 proteins are similar. In yeast, the most extensively studied DNA sequence that is able to interact with an Ste12 protein is called the pheromone-responsive element (PRE), containing the consensus sequence TGAAACA, which was initially identified by footprinting and gel mobility shift assays (53). In response to pheromones, homodimers of Ste12p bind to a pair of adjacent PREs (21, 34, 38).
PRE sequences are also recognized by the homeodomain present in Clste12p, an Ste12-like protein isolated from the plant pathogen Colletotrichum lindemuthianum (51). However, the interaction of Ste12 proteins with their cis-acting sequences in filamentous fungi is poorly understood. Whether Ste12-like proteins bind solely or as homo- or heterodimers remains to be clarified. Further studies on the precise determination of cis-acting sequences binding to Ste12-like proteins in filamentous fungi will certainly help to predict target genes of this class of transcriptional factors. The presence of a C2H2 domain is a distinguishing feature of Ste12-like factors isolated from true filamentous fungi. However, its role is still unclear. DNA binding is not impaired by the deletion of this domain in vitro (8, 51), but the integrity of the C2H2 domain is required for the function of the protein in vivo, as was shown for the plant pathogens C. lindemuthianum and Magnaporthe grisea (36, 51).
ROLE OF Ste12 TRANSCRIPTION FACTORS: MATING AND CELL FUSION
In yeast, Ste12p regulates sexual development by binding to the PRE motif present in pheromone-responsive genes. Ste12p activity is controlled by the Fus3 MAPK pathway leading to the activation of genes involved in mating. Several Ste12 mutants impaired in sexual reproduction in filamentous fungi were characterized (Table 1). The STEA gene from Aspergillus nidulans codes for an Ste12-like protein. STEA mutant strains are sterile and can differentiate into neither ascogenous tissue nor fruiting bodies (cleistothecia) (49). The Neurospora crassa pp-1 mutant, disrupted in an STE12 ortholog, shows a severe reduction of the growth rate and fails to develop protoperithecia (25). A similar phenotype has been observed for the mak-2 mutant disrupted in a FUS3 homolog gene, suggesting that pp-1 in N. crassa is controlled by this MAPK pathway. A complete loss of female fertility was also observed for Cryphonectria parasitica (13). However, the function of Ste12p in female fertility is not a general feature. The deletion of the STE12-like gene from the homothallic ascomycete Sordaria macrospora affects neither hyphal development nor fruiting-body formation but causes highly impaired ascus and ascospore formation (33). In contrast, the Ste12 mutation does not have any effect on sexual development in other models such as the phytopathogen M. grisea (37). From these studies, a specific role of Ste12p and Ste12-like proteins in sexual development cannot be ascribed for all fungal species.
Conidia, conidial germ tubes, and hyphae in close vicinity to each other commonly undergo fusion to produce an interconnected network. It is widely assumed that hyphal fusion is important for communication, organelle translocation, and nutrient transport. Hyphal fusion during vegetative growth in filamentous fungi is somewhat comparable to other cell fusion events, such as the fusion of opposite mating-type cells, and might involve the same molecular basis. From this point of view, an N. crassa mutant disrupted in a FUS3/KSS1 ortholog gene, MAK-2, showed a loss of vegetative hyphal fusion (35). However, all three N. crassa MAPK pathways are essential for hyphal fusion. A decrease in conidial anastomosis tube formation has also been observed for the C. lindemuthianum Ste12 mutant (our unpublished results).
ROLE OF Ste12 TRANSCRIPTION FACTORS: INVASIVE GROWTH AND PATHOGENICITY
Fungi generally display either of two forms: yeast or filamentous. In contrast to unicellular yeasts, the growth mode of filamentous fungi involves polarized cell growth that results from apical extension. Dimorphic fungi share the remarkable ability to switch between the yeast-like and the filamentous forms. In baker's yeast, invasive growth is characterized by the formation of pseudohyphae, which are chains of yeast-phase diploid cells. These cells display elongation, a mitotic delay, symmetric cell division, and unipolar budding (20, 24). Pathogenic fungi are able to differentiate into a series of highly specialized infection cells, notably the appressorium, to penetrate their host. Interestingly, Ste12 and Ste12-like factors are important for pathogenesis in all animal and plant pathogens tested so far (Table 1), and further functional analyses revealed their importance in the setting up of a pathogenicity genetic program specific to the host. This indicates that Ste12-like factors are required for these developmental processes, which accompany the invasive colonization of a new environment.
A switch between yeast and filamentous morphotypes is required for pathogenicity in the human pathogen Candida albicans. This transition is controlled in part by Cph1p, the C. albicans Ste12p homolog, and Efg1p, a homolog of the yeast protein Pdh1p that is involved in pseudohyphal development. The double mutant efg1 cph1 displays severe morphological defects, with a nonfilamentation phenotype, and is avirulent (28). However, filaments were observed under a certain number of conditions, notably in vivo, emphasizing the role of independent signaling pathways in regulating these processes (42).
Contrasting results on the role of Ste12-like proteins have been obtained with Cryptococcus species. In Cryptococcus neoformans, two Ste12-like genes, STE12α and STE12a, correspond to the α and a mating types, respectively. Ste12α and Ste12a contain the typical Ste12-like domains, i.e., a homeodomain and a couple of zinc fingers, but share only 45% sequence similarity to each other (6). The deletion of STE12α in serotype A revealed that this gene is involved in haploid filamentation yet is dispensable for mating and virulence (55), whereas the deletion of STE12α in serotype D led to a decrease in virulence and a downregulation of capsule-associated genes (7). The deletion of STE12a further confirmed the importance of the Ste12 factor for pathogenicity (6). Recently, the disruption of an STE12α gene in Cryptococcus gattii led to a marked decrease of virulence in mouse models as well as a decrease in melanin production (40). Together, these results confirm the essential role of STE12 genes in Cryptococcus pathogenicity.
In plant pathogens, the phenotype of Ste12-like mutants is characterized by a defect in appressorium-mediated penetration (37, 47, 51). Upon germination on plant surfaces, some phytopathogenic fungi (Magnaporthe spp. and Colleotrichum spp.) differentiate into a melanized, dome-shaped cell, within which high turgor pressure is generated through the accumulation of glycerol, enabling the mechanical breaching of the plant cuticle. A penetration peg is emitted at that stage, allowing penetration into the first host epidermal cell and the subsequent differentiation of invasive hyphae by the fungus. A detailed analysis of the M. grisea Ste12 mutant (mst12) revealed the absence of penetration peg formation, although normal melanized appressoria were observed (36). However, a defect in microtubule reorganization, which occurs normally during penetration peg formation, was observed for the mutant, leading to a misorientation of the physical forces exerted by the appressorium turgor (36).
The role of Ste12-like factors in pathogenesis is not restricted to fungi that develop highly melanized appressoria since Ste12-like mutants with impaired or reduced virulence were identified for C. parasitica, Botrytis cinerea, Alternaria brassicicola, and Fusarium oxysporum (9, 13, 17, 43, 45). The virulence of Ste12 mutants of the chestnut blight fungus C. parasitica is reduced, and transcriptional changes induced by the mutation share significant similarity to those induced by hypovirus infection (13). An STE12 gene in the root pathogen F. oxysporum was recently studied (43). The corresponding mutant is impaired in pathogenesis on tomato tissues and in the penetration of cellophane membranes. Interestingly, these mutants also exhibit a reduced production of amylase and cellulase extracellular activities, which might explain, at least in part, the reduced penetration of plant tissues. Since these pathogens do not differentiate into specialized infectious structures, the phenotype of these Ste12-like mutants might be explained by an altered ability to produce extracellular pathogenicity effectors.
REGULATION OF Ste12 ACTIVITY IN YEAST
Yeast Ste12p is phosphorylated by the Fus3p and Kss1p MAPKs, involved in the pheromone response and in invasive/pseudohyphal growth, respectively (16, 39). The phosphorylation of Ste12p by either Fus3p or Kss1p was established as a key step for inducing the activity of Ste12p (Fig. 1). Two regulatory proteins, Dig1p and Dig2p, interact with Ste12p and block its activity. This complex is dissociated after the phosphorylation of Ste12p, allowing the derepression of target genes (3, 12). The involvement of Ste12p in two distinct responses raises the question of signaling specificity. In each of the two responses, Ste12p will regulate a different set of genes and will bind on specific promoter elements as a homodimer in the case of pheromone-responsive genes or as a heterodimer in association with the transcription factor Tec1p in the case of filamentation-induced genes. As mentioned above, Ste12p recognizes a DNA motif called the PRE, and a pair of PREs is present in the promoter of pheromone-regulated genes. In a cells, Ste12p activates a-specific genes together with the MADS box transcription factor Mcm1p (23). The physical interaction of Ste12p with Mcm1p and Matα1p homologs was shown by yeast two-hybrid analysis of the homothallic ascomycete S. macrospora (33).
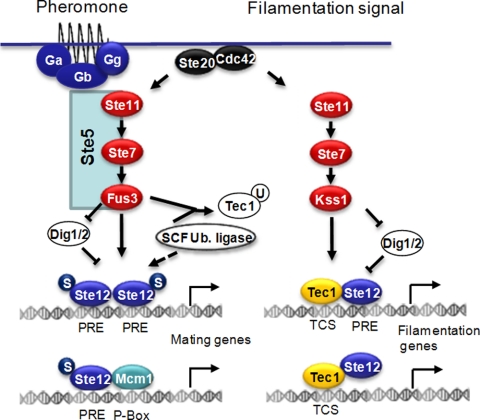
Fig. 1.
Regulation of Ste12p activity in S. cerevisiae. In response to pheromone, the Fus3 MAPK pathway (Ste11-Ste7-Fus3) relays a phosphorylation signal to reach the Dig1p/2p complex, leading to dissociation and Ste12p phosphorylation. Ste12p homodimerization allows the transcription of mating genes containing PRE sequences. Stimulation by the mating pheromone promotes the SUMOylation of Ste12p. In the case of prolonged pheromone treatment, Ste12p is subjected to degradation through a proteasome-mediated mechanism. Upon phosphorylation by Fus3p, Tec1p is rapidly degraded via a ubiquitin-mediated mechanism (SCF Ub. Ligase). In response to filamentation signals, a phosphorylation cascade is transduced via the Kss1 MAPK pathway (Ste11p-Ste7p-Kss1p), leading to the phosphorylation of Ste12p and the dissociation of the Dig1p/2p complex from FRE sequences. The dimerization of phosphorylated Ste12p with Tec1p allows the transcription of filamentation responsive genes. P, phosphorylation; S, SUMOylation; U, ubiquitination.
Tec1p associates with a motif called a TEA/ATTS consensus sequence (TCS). The association of a PRE and a TCS has been designated a filamentous responsive element (FRE) and should allow the specific expression of the filamentous gene upon the binding of a Ste12p-Tec1p heterodimer. However, many filamentation genes do not contain PRE motifs, suggesting that their regulation is mediated mainly by the TCS. Recent results suggest that in the case of filamentation genes, Tec1p is able to tether Ste12p to activate gene expression (11).
The maintenance of signaling specificity during the pheromone response relies on the phosphorylation of Tec1p by the mating-specific MAPK Fus3. Tec1p Fus3-dependent phosphorylation induces the degradation of Tec1p by the proteasome machinery (2, 10). Signaling specificity is reinforced by the stabilization of Ste12p by the ubiquitin-related modifier SUMO upon the pheromone response. However, prolonged exposure to the pheromone leads to ubiquitin-mediated destabilization and decreased amounts of Ste12p (15).
REGULATION OF Ste12-LIKE FACTOR ACTIVITY IN FILAMENTOUS FUNGI
Much less is known about the regulation of Ste12-like proteins in filamentous fungi. Importantly, a direct interaction between an Ste12-like protein and a MAPK has not been reported. Accordingly, the yeast Ste12 protein domain interacting with the MAPK Kss1p is not conserved in Ste12-like factors from euascomycetous fungi. Ste12-like factors also lack domains interacting with potential homologs of Tec1p, Dig1p, and Dig2p. This finding suggests that Ste12-like proteins could be regulated in these species by a different set of proteins. In C. neoformans, two-hybrid assays failed to identify any interacting proteins when Ste12αp was used as bait (8). The same assays carried out with M. grisea identified only a weak interaction between the Kss1p and the Ste12p homologs, named Pmk1p and Mst12p, respectively (37). Moreover, the Pmk1 and Mst12 mutants display different phenotypes: the Pmk1 mutant does not form appressoria, whereas mst12 forms melanized appressoria with normal morphology but which are unable to penetrate plant tissues (37). The regulation of Ste12-like factors by ubiquitination or SUMOylation has not been identified so far.
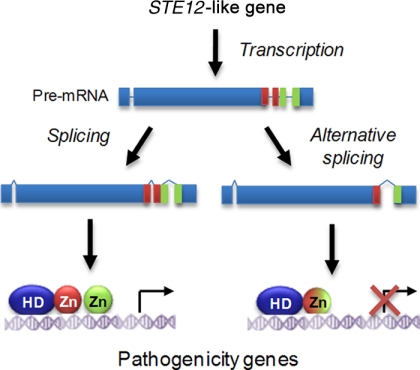
Besides posttranslational modifications, the activity of C. lindemuthianum and B. cinerea STE12-like genes can be modulated at the transcriptional level through the production of an alternatively spliced transcript (Fig. 2) (45, 51). The spliced variant encodes a protein with a single zinc finger, which can exert dominant negative activity on pathogenesis when overexpressed in transgenic strains (45, 51). These results suggest a novel role for the zinc finger domain, absent in yeasts, allowing the modulation of Ste12-like protein activity through the control of a ratio between a repressive form and an inductive form. Interestingly, the same exon-intron structure occurs for STE12-like genes from other filamentous fungi, opening the possibility that a similar regulation takes place in other species.
Fig. 2.
Regulation of STE12-like genes by alternative splicing. STE12-like genes from filamentous fungi display a conserved intron-exon organization at the 3′ ends of their coding sequences. This part encodes two zinc finger domains (red and green circles). In the plant pathogens C. lindemuthianum and B. cinerea, an alternative splicing event corresponding to exon skipping has been identified. This results in the production of a protein with only one new zinc finger displaying an inhibitory effect on pathogenesis when overexpressed in wild-type strains (45, 51). HD, homeodomain; Zn, zinc finger.
Ste12 TARGETS AND DOWNSTREAM EFFECTORS
S. cerevisiae Ste12p regulates the expression of a wide array of genes. From a study using yeast cells treated with appropriate nutrient concentrations to induce the invasive/pseudohyphal switch, two major Ste12p target genes emerged: FLO11 and PGU1. FLO11 encodes a cell surface-associated protein that promotes yeast flocculation, pseudohyphal growth, and adhesion to plastic surfaces (27, 41). Microarray analyses further identified a number of putative Ste12p targets differentially regulated in response to mating factors or upon the induction of invasive growth (30, 44). Among them are genes involved in cell cycle arrest, cell fusion, nuclear fusion, and membrane and cell wall biosynthesis. Genome-wide approaches combining chromatin immunoprecipitation (ChIP) and DNA microarray technology were carried out to search for target genes in the mating pathway and in the invasive/pseudohyphal growth pathway (56). This large-scale study revealed that Ste12p binds to different sets of genes during these two processes and that each set of genes specifies a developmental program (56). However, most of the filamentation gene promoters do not contain PREs, the Ste12p DNA binding sequence, raising the question of their regulation by Ste12p. In that case, it was suggested that Tec1p bound to TCS is able to tether Ste12p, allowing the transcriptional activation of the target genes (11).
Several genes encoding virulence traits in animal parasites are regulated by Ste12-like factors. These include adhesin and protease genes in C. albicans (1, 32), which play an important role in biofilm establishment. In C. neoformans, capsule-associated laccase and superoxide dismutase genes were found to act downstream of Ste12p (6). In phytopathogenic fungi, the deletion of the STE12 homolog led to a severe decrease in levels of pectinase production as well as a downregulation of a gene coding for a cell surface protein in C. lindemuthianum (51). The effect of an Ste12-like mutation on pectinase production is consistent with the identification of Ste12p binding site-like cis-acting elements in a pectinase gene (22). The regulation of pectinase genes by STE12-like genes is correlated with the finding that PGU1, a yeast pectinase gene, is induced during invasive growth (30). Interestingly, the overexpression of STEA, the STE12 homolog from Aspergillus oryzae, led to the overproduction of secreted cell wall-degrading enzymes (31).
A microarray analysis of C. parasitica led to the identification of 152 genes that were underexpressed in an Ste12 mutant (13). A significant number of these genes were also downregulated in strains infected by a virulence-attenuating hypovirus, suggesting that the STE12-like gene may be a major target of virus infection (13). Together, these results for different systems show that a part of Ste12-like protein target genes encodes extracellular proteins involved in the degradation of organic compounds present in the environment or playing a role in cell adhesion. However, a more complete view of genes regulated by Ste12-like proteins in filamentous fungi remains to be obtained.
CONCLUDING REMARKS AND OUTLOOK
The colonization of new environments by fungi requires the expression of a specific genetic program that leads to the differentiation of new cellular structures allowing the dissemination, adhesion, and production of extracellular proteins involved in the degradation of complex substrates. From the studies described above, Ste12 and Ste12-like proteins emerge as regulators of these adaptation requirements, which can act in concert with other transcription factors. Recently, an STE12 homolog, GINSTE, was identified in the mycorrhizal fungus G. intraradices. GINSTE complements the yeast Ste12 mutant and allows the restoration of infectivity of an Ste12-like mutant of the plant pathogen C. lindemuthianum (46). While the precise role of GinSte in symbiosis remains to be elucidated, these findings suggest that Ste12-regulated mechanisms are very ancient and conserved in distantly related fungi. This also opens the possibility that a diversity of responses controlled by Ste12 and Ste12-like proteins has been generated, allowing the adaptation of each fungal species to their ecological niche. Divergence at the level of Ste12p binding sites might explain this variability. This point was recently investigated by comparing Ste12p binding sites in three yeast species during pseudohyphal growth by using chromatin immunoprecipitation and DNA microarray analysis (4). More than 300 binding sites were detected, most of which diverged among species (4), reflecting the regulation of a specific genetic network in each strain by Ste12p as a common regulator. A similar result emerged from a study focused on the yeast transcriptional regulator Mcm1p, which regulates hundreds of genes (48). Less than 20% of common Mcm1p-target gene connections are found in three yeast species, S. cerevisiae, Kluyveromyces lactis, and C. albicans. Further work is certainly needed to explore the potential of fungal adaptation at the gene regulatory sequence level. Having several dozen pathogenic fungal genomes in hand, this objective can now be achieved and will probably lead to the identification of new effectors of pathogenicity controlled by Ste12 factors.
ACKNOWLEDGMENTS
This work was supported by CNRS and Université Paul Sabatier, Toulouse, France.
Footnotes
Published ahead of print on 5 February 2010.
REFERENCES
- 1.Argimon S., Wishart J. A., Leng R., Macaskill S., Mavor A., Alexandris T., Nicholls S., Knight A. W., Enjalbert B., Walmsley R., Odds F. C., Gow N. A., Brown A. J. 2007. Developmental regulation of an adhesin gene during cellular morphogenesis in the fungal pathogen Candida albicans. Eukaryot. Cell 6:682–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bao M. Z., Schwartz M. A., Cantin G. T., Yates J. R., III, Madhani H. D. 2004. Pheromone-dependent destruction of the Tec1 transcription factor is required for MAP kinase signaling specificity in yeast. Cell 119:991–1000 [DOI] [PubMed] [Google Scholar]
- 3.Bardwell L., Cook J. G., Voora D., Baggott D. M., Martinez A. R., Thorner J. 1998. Repression of yeast Ste12 transcription factor by direct binding of unphosphorylated Kss1 MAPK and its regulation by the Ste7 MEK. Genes Dev. 12:2887–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borneman A. R., Gianoulis T. A., Zhang Z. D., Yu H., Rozowsky J., Seringhaus M. R., Wang L. Y., Gerstein M., Snyder M. 2007. Divergence of transcription factor binding sites across related yeast species. Science 317:815–819 [DOI] [PubMed] [Google Scholar]
- 5.Borneman A. R., Hynes M. J., Andrianopoulos A. 2001. An STE12 homolog from the asexual, dimorphic fungus Penicillium marneffei complements the defect in sexual development of an Aspergillus nidulans steA mutant. Genetics 157:1003–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang Y. C., Penoyer L. A., Kwon-Chung K. J. 2001. The second STE12 homologue of Cryptococcus neoformans is MATa-specific and plays an important role in virulence. Proc. Natl. Acad. Sci. U. S. A. 98:3258–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang Y. C., Wickes B. L., Miller G. F., Penoyer L. A., Kwon-Chung K. J. 2000. Cryptococcus neoformans STE12α regulates virulence but is not essential for mating. J. Exp. Med. 191:871–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang Y. C., Wright L. C., Tscharke R. L., Sorrell T. C., Wilson C. F., Kwon-Chung K. J. 2004. Regulatory roles for the homeodomain and C2H2 zinc finger regions of Cryptococcus neoformans Ste12αp. Mol. Microbiol. 53:1385–1396 [DOI] [PubMed] [Google Scholar]
- 9.Cho Y., Kim K. H., La Rota M., Scott D., Santopietro G., Callihan M., Mitchell T. K., Lawrence C. B. 2009. Identification of novel virulence factors associated with signal transduction pathways in Alternaria brassicicola. Mol. Microbiol. 72:1316–1333 [DOI] [PubMed] [Google Scholar]
- 10.Chou S., Huang L., Liu H. 2004. Fus3-regulated Tec1 degradation through SCFCdc4 determines MAPK signaling specificity during mating in yeast. Cell 119:981–990 [DOI] [PubMed] [Google Scholar]
- 11.Chou S., Lane S., Liu H. P. 2006. Regulation of mating and filamentation genes by two distinct Ste12 complexes in Saccharomyces cerevisiae. Mol. Cell. Biol. 26:4794–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cook J. G., Bardwell L., Kron S. J., Thorner J. 1996. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes Dev. 10:2831–2848 [DOI] [PubMed] [Google Scholar]
- 13.Deng F., Allen T. D., Nuss D. L. 2007. Ste12 transcription factor homologue CpST12 is down-regulated by hypovirus infection and required for virulence and female fertility of the chestnut blight fungus Cryphonectria parasitica. Eukaryot. Cell 6:235–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Errede B., Ammerer G. 1989. STE12, a protein involved in cell-type-specific transcription and signal transduction in yeast, is part of protein-DNA complexes. Genes Dev. 3:1349–1361 [DOI] [PubMed] [Google Scholar]
- 15.Esch R. K., Wang Y., Errede B. 2006. Pheromone-induced degradation of Ste12 contributes to signal attenuation and the specificity of developmental fate. Eukaryot. Cell 5:2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gancedo J. M. 2001. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25:107–123 [DOI] [PubMed] [Google Scholar]
- 17.García-Sánchez M. A., Martín-Rodrigues N., Ramos B., de Vega-Bartol J. J., Perlin M. H., Díaz-Mínguez J. M. 23November2009, posting date fost12, the Fusarium oxysporum homolog of the transcription factor Ste12, is upregulated during plant infection and required for virulence. Fungal Genet. Biol doi:10.1016/j.fgb.2009.11.006 [DOI] [PubMed] [Google Scholar]
- 18.Gavrias V., Andrianopoulos A., Gimeno C. J., Timberlake W. E. 1996. Saccharomyces cerevisiae TEC1 is required for pseudohyphal growth. Mol. Microbiol. 19:1255–1263 [DOI] [PubMed] [Google Scholar]
- 19.Gehring W. J., Muller M., Affolter M., Percival-Smith A., Billeter M., Qian Y. Q., Otting G., Wuthrich K. 1990. The structure of the homeodomain and its functional implications. Trends Genet. 6:323–329 [DOI] [PubMed] [Google Scholar]
- 20.Gimeno C. J., Ljungdahl P. O., Styles C. A., Fink G. R. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077–1090 [DOI] [PubMed] [Google Scholar]
- 21.Hagen D. C., McCaffrey G., Sprague G. F., Jr 1991. Pheromone response elements are necessary and sufficient for basal and pheromone-induced transcription of the FUS1 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 11:2952–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herbert C., Jacquet C., Borel C., Esquerré-Tugayé M. T., Dumas B. 2002. A cis-acting sequence homologous to the yeast filamentation and invasion response element regulates expression of a pectinase gene from the bean pathogen Colletotrichum lindemuthianum. J. Biol. Chem. 277:29125–29131 [DOI] [PubMed] [Google Scholar]
- 23.Johnson A. D. 1995. Molecular mechanisms of cell-type determination in budding yeast. Curr. Opin. Genet. Dev. 5:552–558 [DOI] [PubMed] [Google Scholar]
- 24.Kron S. J., Styles C. A., Fink G. R. 1994. Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 5:1003–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li D., Bobrowicz P., Wilkinson H. H., Ebbole D. J. 2005. A mitogen-activated protein kinase pathway essential for mating and contributing to vegetative growth in Neurospora crassa. Genetics 170:1091–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu H., Kohler J., Fink G. R. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723–1726 [DOI] [PubMed] [Google Scholar]
- 27.Lo W. S., Dranginis A. M. 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 9:161–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lo W. S., Raitses E. I., Dranginis A. M. 1997. Development of pseudohyphae by embedded haploid and diploid yeast. Curr. Genet. 32:197–202 [DOI] [PubMed] [Google Scholar]
- 29.Madhani H. D., Fink G. R. 1997. Combinatorial control required for the specificity of yeast MAPK signaling. Science 275:1314–1317 [DOI] [PubMed] [Google Scholar]
- 30.Madhani H. D., Galitski T., Lander E. S., Fink G. R. 1999. Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proc. Natl. Acad. Sci. U. S. A. 96:12530–12535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morita H., Hatamoto O., Masuda T., Sato T., Takeuchi M. 2007. Function analysis of steA homolog in Aspergillus oryzae. Fungal Genet. Biol. 44:330–338 [DOI] [PubMed] [Google Scholar]
- 32.Naglik J., Albrecht A., Bader O., Hube B. 2004. Candida albicans proteinases and host/pathogen interactions. Cell. Microbiol. 6:915–926 [DOI] [PubMed] [Google Scholar]
- 33.Nolting N., Poggeler S. 2006. A STE12 homologue of the homothallic ascomycete Sordaria macrospora interacts with the MADS box protein MCM1 and is required for ascosporogenesis. Mol. Microbiol. 62:853–868 [DOI] [PubMed] [Google Scholar]
- 34.Olson K. A., Nelson C., Tai G., Hung W., Yong C., Astell C., Sadowski I. 2000. Two regulators of Ste12p inhibit pheromone-responsive transcription by separate mechanisms. Mol. Cell. Biol. 20:4199–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pandey A., Roca M. G., Read N. D., Glass N. L. 2004. Role of a mitogen-activated protein kinase pathway during conidial germination and hyphal fusion in Neurospora crassa. Eukaryot. Cell 3:348–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park G., Bruno K. S., Staiger C. J., Talbot N. J., Xu J. R. 2004. Independent genetic mechanisms mediate turgor generation and penetration peg formation during plant infection in the rice blast fungus. Mol. Microbiol. 53:1695–1707 [DOI] [PubMed] [Google Scholar]
- 37.Park G., Xue C., Zheng L., Lam S., Xu J. R. 2002. MST12 regulates infectious growth but not appressorium formation in the rice blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact. 15:183–192 [DOI] [PubMed] [Google Scholar]
- 38.Pi H., Chien C. T., Fields S. 1997. Transcriptional activation upon pheromone stimulation mediated by a small domain of Saccharomyces cerevisiae Ste12p. Mol. Cell. Biol. 17:6410–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qi M., Elion E. A. 2005. MAP kinase pathways. J. Cell Sci. 118:3569–3572 [DOI] [PubMed] [Google Scholar]
- 40.Ren P., Springer D. J., Behr M. J., Samsonoff W. A., Chaturvedi S., Chaturvedi V. 2006. Transcription factor STE12α has distinct roles in morphogenesis, virulence, and ecological fitness of the primary pathogenic yeast Cryptococcus gattii. Eukaryot. Cell 5:1065–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reynolds T. B., Fink G. R. 2001. Bakers' yeast, a model for fungal biofilm formation. Science 291:878–881 [DOI] [PubMed] [Google Scholar]
- 42.Riggle P. J., Andrutis K. A., Chen X., Tzipori S. R., Kumamoto C. A. 1999. Invasive lesions containing filamentous forms produced by a Candida albicans mutant that is defective in filamentous growth in culture. Infect. Immun. 67:3649–3652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rispail N., Di Pietro A. 2009. Fusarium oxysporum Ste12 controls invasive growth and virulence downstream of the Fmk1 MAPK cascade. Mol. Plant Microbe Interact. 22:830–839 [DOI] [PubMed] [Google Scholar]
- 44.Roberts C. J., Nelson B., Marton M. J., Stoughton R., Meyer M. R., Bennett H. A., He Y. D., Dai H., Walker W. L., Hughes T. R., Tyers M., Boone C., Friend S. H. 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287:873–880 [DOI] [PubMed] [Google Scholar]
- 45.Schamber A., Leroch M., Diwo J., Mendgen K., Hahn M. 2010. The role of mitogen-activated protein (MAP) kinase signalling components and the Ste12 transcription factor in germination and pathogenicity of Botrytis cinerea. Mol. Plant Pathol. 11:105–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tollot M., Wong Sak Hoi J., van Tuinen D., Arnould C., Chatagnier O., Dumas B., Gianinazzi-Pearson V., Seddas P. M. 2009. An STE12 gene identified in the mycorrhizal fungus Glomus intraradices restores infectivity of a hemibiotrophic plant pathogen. New Phytol. 181:693–707 [DOI] [PubMed] [Google Scholar]
- 47.Tsuji G., Fujii S., Tsuge S., Shiraishi T., Kubo Y. 2003. The Colletotrichum lagenarium Ste12-like gene CST1 is essential for appressorium penetration. Mol. Plant Microbe Interact. 16:315–325 [DOI] [PubMed] [Google Scholar]
- 48.Tuch B. B., Galgoczy D. J., Hernday A. D., Li H., Johnson A. D. 2008. The evolution of combinatorial gene regulation in fungi. PLoS Biol. 6:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vallim M. A., Miller K. Y., Miller B. L. 2000. Aspergillus SteA (sterile12-like) is a homeodomain-C2/H2-Zn+2 finger transcription factor required for sexual reproduction. Mol. Microbiol. 36:290–301 [DOI] [PubMed] [Google Scholar]
- 50.Wickes B. L., Edman U., Edman J. C. 1997. The Cryptococcus neoformans STE12α gene: a putative Saccharomyces cerevisiae STE12 homologue that is mating type specific. Mol. Microbiol. 26:951–960 [DOI] [PubMed] [Google Scholar]
- 51.Wong Sak Hoi J. W., Herbert C., Bacha N., O'Connell R., Lafitte C., Borderies G., Rossignol M., Rougé P., Dumas B. 2007. Regulation and role of a STE12-like transcription factor from the plant pathogen Colletotrichum lindemuthianum. Mol. Microbiol. 64:68–82 [DOI] [PubMed] [Google Scholar]
- 52.Young L. Y., Lorenz M. C., Heitman J. 2000. A STE12 homolog is required for mating but dispensable for filamentation in Candida lusitaniae. Genetics 155:17–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan Y. L., Fields S. 1991. Properties of the DNA-binding domain of the Saccharomyces cerevisiae STE12 protein. Mol. Cell. Biol. 11:5910–5918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan Y. O., Stroke I. L., Fields S. 1993. Coupling of cell identity to signal response in yeast: interaction between the α 1 and STE12 proteins. Genes Dev. 7:1584–1597 [DOI] [PubMed] [Google Scholar]
- 55.Yue C., Cavallo L. M., Alspaugh J. A., Wang P., Cox G. M., Perfect J. R., Heitman J. 1999. The STE12α homolog is required for haploid filamentation but largely dispensable for mating and virulence in Cryptococcus neoformans. Genetics 153:1601–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zeitlinger J., Simon I., Harbison C. T., Hannett N. M., Volkert T. L., Fink G. R., Young R. A. 2003. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell 113:395–404 [DOI] [PubMed] [Google Scholar]