Abstract
Neurospora crassa dbf-2 encodes an NDR (nuclear Dbf2-related) protein kinase, homologous to LATS1, a core component of the Hippo pathway. This pathway plays important roles in restraining cell proliferation and promoting apoptosis in differentiating cells. Here, we demonstrate that DBF-2 is involved in three fundamental processes in a filamentous fungus: cell cycle regulation, glycogen biosynthesis, and conidiation. DBF-2 is predominantly localized to the nucleus, and most (approximately 60%) dbf-2 null mutant nuclei are delayed in mitosis, indicating that DBF-2 activity is required for properly completing the cell cycle. The dbf-2 mutant exhibits reduced basal hyphal extension rates accompanied by a carbon/nitrogen ratio-dependent bursting of hyphal tips, vast glycogen leakage, defects in aerial hypha formation, and impairment of all three asexual conidiation pathways in N. crassa. Our findings also indicate that DBF-2 is essential for sexual reproduction in a filamentous fungus. Defects in other Hippo and glycogen metabolism pathway components (mob-1, ccr-4, mst-1, and gsk-3) share similar phenotypes such as mitotic delay and decreased CDC-2 (cell division cycle 2) protein levels, massive hyphal swellings, hyphal tip bursting, glycogen leakage, and impaired conidiation. We propose that DBF-2 functions as a link between Hippo and glycogen metabolism pathways.
The nuclear Dbf2-related (NDR) protein kinases are essential components of signaling networks that control cellular processes in various organisms, including morphogenesis, exit from mitosis, cytokinesis, proliferation, differentiation, and apoptosis (23). Based on structural and functional conservation over long evolutionary distances, NDR kinases can be ascribed to one of two subgroups. One is comprised of mammalian NDR1/2, and the other is comprised of LATS1/2 (large tumor suppressor 1/2), as well as their orthologs and related kinases in different organisms.
The filamentous fungus Neurospora crassa has two NDR kinases, which representative both subgroups. These are encoded by cot-1 (colonial temperature sensitive 1; NCU07296.3) and dbf-2 (NCU09071.3). While cot-1 (an ortholog of human NDR1/2) is involved in apical hyphal cell elongation and polarity (71), the role of dbf-2 (an ortholog of human LATS1/2) is yet unknown. Additionally, significant sequence similarities (52.8%) between the catalytic domains of DBF-2 and COT-1 raise the question whether their functions or localizations overlap. COT-1 has been localized to several intracellular compartments, including the cytoplasm and nucleus, and in association with the plasma membrane and various proteins (18, 19, 54). The role that COT-1 plays in the formation of branched cellular structures as well as its cellular localization has been suggested to be preserved throughout evolution (54, 77). Similar conservation may also exist among DBF-2 and its orthologs, of which human LATS1 and Drosophila melanogaster WTS/LATS are, by far, the most extensively analyzed (13, 45, 53, 74, 75).
Both human LATS1 and NDR1 are widely expressed nuclear serine-threonine kinases that have been implicated in cell proliferation and/or tumor progression. LATS1 has been recently established as one of the key members operating at the core machinery of an emerging conserved signal transduction pathway yet to be elucidated in fungi—the Hippo pathway (13, 22, 75). In D. melanogaster as well as in higher eukaryotes, the Hippo pathway was found to participate in processes such as cell growth, proliferation, cell cycle, apoptosis, organ size tumorigenesis, and cell contact inhibition (13, 45, 74). As part of this novel mechanism of regulation, DBF2 orthologs act in close cooperation with other key members of the apparently conserved Hippo pathway such as serine/threonine sterile 20 (STE20)-like kinases, which have been implicated in the upstream activation of NDR kinases, and coactivators of the Mps-one binder (MOB) family, which have been found to associate with multiple NDR family kinases at the N terminus (23, 45). This mode of three-way interactions is well illustrated in Drosophila via the activation of the NDR kinase wts/lats by the STE20-like hippo (hpo) and its association with the coactivator mats (mob as a tumor suppressor) (22). Recent studies showing parallel interactions between the human homologs of wts/lats, hpo, and mats stress that this conserved pathway, harboring DBF2 orthologs, is important in development and is a vulnerable target for misregulation in cancer (45, 53).
At least three key members of the Hippo pathway can be identified in Saccharomyces cerevisiae, based on structural and functional conservation: the NDR kinase DBF2, the coactivator MOB1, and the Ste20-like kinase (CDC15). It was suggested that the binding of MOB1 to DBF2 enables CDC15 to phosphorylate DBF2 (35). Apart from its involvement in Hippo signaling, previous studies of yeasts also revealed the involvement of DBF2 in exit from mitosis and cytokinesis (66). Yeast DBF2 interacts genetically and physically with MOB1, an interaction imperative both for its translocation from the nucleus to the mother-bud neck during cell cycle (16) and for its phosphorylation and activation by CDC15 (35). Consistent with its late mitotic role, DBF2 protein kinase expression peaks after the metaphase-to-anaphase transition (66). These components (and others) are part of the mitotic exit network (MEN) and parallel septation initiation network (SIN) described in budding and fission yeasts, respectively, and whose counterparts have been identified in filamentous fungi (21, 28). In S. cerevisiae, DBF2 has also been identified as a component of the CCR4 (carbon catabolite repression 4) transcriptional complex, a general transcriptional regulator which affects expression of numerous genes both positively and negatively (32). In addition, DBF2 disruption in yeasts results in glycogen accumulation, indicative of the involvement of DBF2 in glycogen metabolism (69). DBF2 shares at least one essential function with a homolog, DBF20, as yeasts with either gene deleted are viable but deletion of both genes results in lethality (66).
In contrast to yeasts, filamentous fungi posses a significant number of additional, complex, and at times unique cellular processes such as hyphal elongation and conidiation, all of which require tight regulation. Thus, it is not surprising that in the filamentous ascomycete Aspergillus nidulans, the DBF2 ortholog SIDB has apparently gained additional functions beyond its conserved role in cell cycle control and cytokinesis. This is exemplified by its involvement in regulation of asexual reproduction, as disruption of SIDB results in abolishment of conidiation (26).
In this study, the involvement of the conserved N. crassa Hippo pathway component dbf-2 was identified in three fundamental processes: cell cycle regulation, glycogen biosynthesis, and conidiation. Furthermore, by studying the involvement of other genes encoding proteins whose activity is associated with dbf-2, we have established a functional link between Hippo signaling, glycogen metabolism, and conidiation in a filamentous fungus.
MATERIALS AND METHODS
Fungal strains, media, and growth conditions.
General procedures and media used for growth, preservation, and manipulation of N. crassa were as previously described (9, 10) or can be obtained through the Fungal Genetic Stock Center (FGSC; www.fgsc.net). The N. crassa strains used in this study (Table 1) were grown in either liquid or solid (supplemented with 1.5% agar) Vogel's minimal medium with 1.5% (wt/vol) sucrose (Vs), unless otherwise stated. The ccr-4, gsk-3, and mst-1 strains were obtained (from the FGSC) as heterokaryons and purified, by isolation of microconidia, to a homokaryon state. When required, the medium was supplemented with 100 μg/ml hygromycin B (Calbiochem, Riverside, CA), 20 μg/ml noursethricin (Werner Bioagents, Jena, Germany), 2.5 μg/ml benomyl, or 100 μg/ml histidine.
Table 1.
Neurospora crassa strains used in this study
| Strain | Relevant genotypea | Comment | Source or reference |
|---|---|---|---|
| 4200 | ORS-SL6a | Wild type, mat a | FGSCb catalog no. 4200 |
| ku-70 strain | Δmus-51::bar+; his-3; mat a | FGSC catalog no. 9717 | |
| rgb-1RIP strain | rgb-1RIP::hphr; mat A | NCU09377.3 | 69 |
| ppe-1 strain | Δppe-1::hphr; mat A | NCU03436.3 | S. Seiler (Georg-August-University, Germany) |
| mob-1 strain | Δmob-1::hphr; mat a | NCU01605.3 | S. Seiler (Georg-August-University, Germany) |
| mst-1 strain | Δmst-1::hphr; Δmus-51::bar+; mat a | NCU00772.3 | FGSC catalog no. 11523 |
| ccr-4 strain | Δccr-4::hphr; Δmus-51::bar+; mat a | NCU07779.3 (heterokaryon) | FGSC catalog no. 11498 |
| gsk-3 strain | Δgsk-3::hphr; Δmus-51::bar+; mat a | NCU04185.3 (heterokaryon) | FGSC catalog no. 11500 |
| dbf-2 strain | Δdbf-2::hphr; Δmus-51::bar+; his-3; mat a | NCU09071.3 | This study |
| G10 | his-3+::Pdbf-2-dbf-2+-sgfp+; Δmus-51::bar+; mat A | dbf-2 (A) pED4 transformant | This study |
| b7_1 | Pdbf-2-dbf-2+-sgfp+; natr; Δmob-1::hphr; mat a | mob-1 (a) pED4 and pBSIIgpdnatr cotransformant | This study |
| R54 | Δccr-4::hphr; Δmus-51::bar+; mat a | This study | |
| s1_d | Pdbf-2-dbf-2+-sgfp+; natr Δccr-4::hphr; Δmus-51::bar; mat a | R54 (a) pED4 and pBSIIgpdnatr cotransformant | This study |
| r4_6 | Pdbf-2-dbf-2+-sgfp+; natr; rgb-1RIP::hphr; mat A | rgb-1RIP (A) pED4 and pBSIIgpdnatr cotransformant | This study |
| t3_2 | Pdbf-2-dbf-2+-sgfp+; natr; Δsit-4::hphr; mat A | sit-4 (A) pED4 and pBSIIgpdnatr cotransformant | This study |
| 32 | Δgsk-3::hphr; Δmus-51::bar+; mat a | This study | |
| hex-1 strain | Δhex-1; pan-2−; mat A | NCU8332.3 | 25 |
| prk-9 strain | prk-9::hphr; Δmus-51::bar+; mat A/a | NCU04096.3 | 7 |
natr, nourseothricin resistant; hphr, hygromycin resistant.
FGSC, Fungal Genetic Stock Center.
For the carbon/nitrogen (C/N) ratio experiments, glucose and ammonium nitrate were used as carbon and nitrogen sources, respectively. Briefly, nitrogen limitation was imposed by a stepwise decrease in the ammonium nitrate concentration while the glucose level was kept constant (1.5%) with C/N ratios ranging from 0.1 to 10 (wt/wt).
For determining the effect of benomyl on mitotic delay, strains were first cultured overnight and subsequently transferred to a fresh medium containing the drug for 4 h prior to DAPI (4′,6-diamidino-2-phenylindole) staining.
Electroporation was performed according to the method of Margolin et al. (38). The transformants were screened by their resistance to either hygromycin B or nourseothricin and by phenotypic complementation of his-3 (ability to grow without histidine supplementation).
Nucleic acid extraction and analysis.
Recombinant DNA methods and N. crassa total RNA isolation were performed according to standard protocols (52) and as previously described (76). PCR was performed according to standard protocols (52), using SuperTerm JMR801 polymerase (JMR Holdings, Saint Louis, MO). Primers used in this study are listed in Table 2. Sequencing was performed at the Center for Genomic Technologies, the Hebrew University of Jerusalem, Israel.
Table 2.
Primers used in this study
| Name | Sequence | Use |
|---|---|---|
| dbf-2 1247F | TCGCTTCTACATTGCCGAGA | PCR |
| dbf-2 2058R | TTGCTGCTTCTCATGCACCT | PCR |
| dbf-2gf13F | TCTAGATGCTGATGTCCTCTGCCTTG | PCR |
| dbf-2gf4208R | GATATCCATCGTACCAAAATTGTTGCTCTC | PCR |
| PED4NF3066 | GTCCTCCTTGAAGTCGATGC | PCR |
| PED4NR3864 | GCGTAGATGCTAACCAGCGTA | PCR |
| HPH565F | CGGTGTCGTCCATCACAGTT | PCR |
| HPH1139R | TCGCCCTTCCTCCCTTTATT | PCR |
| cdc-2rtF | GATACTGCCACAGCCACCGT | Real-time RT-PCR |
| cdc-2rtRN | ATGCCTTGAATGCGGGATAGG | Real-time RT-PCR |
| actrt575604F | TCCATCATGAAGTGCGATGTC | Real-time RT-PCR |
| actrt575437R | TTCTGCATACGGTCGGAGAGA | Real-time RT-PCR |
For reverse transcription-PCR (RT-PCR), RNA samples were treated with RQ1 RNase-free DNase (Promega, Madison, WI) and then purified with the RNeasy kit (Qiagen) according to the manufacturer's protocol. Purified RNA (5 μg) was used for the RT procedures using SuperScript II RNase H reverse transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Relative quantification of gene expression was performed using SYBR green real-time RT-PCR on an ABI Prism 5700 sequence detection system (Applied Biosystems, Foster City, CA). RT-PCR mixtures were composed of a 12-pmol concentration of each primer, 12.5 μl of SYBR green PCR master mix (Applied Biosystems), 5 μl of cDNA (a 1:100 dilution of the cDNA product produced as described above), and nuclease-free water to a final volume of 25 μl. Amplification conditions were as follows: 30 min at 48°C, 10 min at 95°C, and then 40 cycles that consisted of 15 s at 95°C and 1 min at 60°C. Total cDNA abundance in the samples was normalized using the act gene (NCU04173.3), which encodes actin, as a control. In all experiments, samples were amplified in triplicate, and the average cycle threshold was then calculated and used to determine the relative expression of each gene. Three independent experiments were carried out in the same manner, and the final average and standard error of the relative expression values were calculated.
Disruption of dbf-2.
Disruption of dbf-2 was performed using the Neurospora genome project gene knockout kit (obtained from the FGSC) according to the method of Colot et al. (8). Briefly, the construct for homologous recombination of NCU09071.3 was provided by the Neurospora knockout project. The cassette was transformed into the N. crassa ku-70 strain (FGSC catalog no. 9717), which facilitates high-frequency gene replacement events of the construct at the target locus by preventing nonhomologous recombination due to the disruption of a gene encoding a protein required for nonhomologous end joining of double-stranded DNA breaks (40). Several transformants were obtained and screened for dbf-2 gene replacement by PCR using primers dbf-2 1247F and dbf-2 2058R (for the dbf-2 open reading frame [ORF]) and HPH565F and HPH1139R (to identify homologous recombination events) and were verified by Southern blot analysis. One of the candidate dbf-2 deletion transformants was crossed with an N. crassa wild-type (wt) strain (FGSC catalog no. 4200) to obtain the dbf-2 deletion in a wild-type background. Since we were not able to obtain viable dbf-2 ascospores, dbf-2 deletion transformants in a ku-70 background were used throughout this study, while both ku-70 and wt strains were used as control strains.
Determining growth rate.
For growth rate measurements, 10 μl of a conidial suspension (1 × 107 conidia/ml) was inoculated in race tubes (9) containing Vogel's minimal medium. The race tubes were incubated for several days at 34°C, and the radial growth was measured twice daily.
DAPI and calcofluor white staining.
A DAPI stock solution (14.3 mM) was prepared by dissolving 5 mg of DAPI in 1 ml of dimethyl formamide (DMF) and stored at −20°C. In order to prepare a DAPI working solution (300 nM), 1 μl of DAPI stock solution was added to 50 ml of phosphate-buffered saline (PBS; 15 mM Na2HPO4, 5 mM NaH2PO4, 32 mM NaCl, pH 7.2). Neurospora cultures, 1 to 3 days old, grown in Vs liquid medium were dyed with the DAPI working solution and immediately analyzed by fluorescent microscopy. The fluorescent digital images acquired were used for the quantification of nuclear morphotypes in each strain. The prevalence of elongated nuclei was calculated based on the average of three experiments, in which 10 fields were counted for each strain. For calcofluor white staining, samples were treated with a solution of 10-μg/ml calcofluor prior to observation by fluorescent microscopy.
Production of a DBF-2::GFP fusion protein construct.
The putative dbf-2 (NCU09071.3) promoter and ORF (approximately 4.2 kb) were amplified, without the stop codon, as an EcoRV/XbaI fragment (4.2 kb) utilizing primers dbf-2gf13F and dbf-2gf4208R (Table 2) and cloned into a pDrive vector (Qiagen) to yield pED3. The pED3 insert, retrieved by an EcoRV/XbaI double digestion, was ligated with SmaI/XbaI-double-digested pMF267 (a promoterless sgfp plasmid used for green fluorescent protein [GFP] tagging, targeted to the his-3 locus of N. crassa [15]). This yielded pED4, an expression plasmid used for DBF-2 localization and for complementation of the dbf-2 mutation by expressing a full-length N. crassa DBF-2 protein with a C-terminal GFP tag, under the regulation of the dbf-2 promoter. pED4 was linearized by NdeI and transformed into dbf-2. PCR amplification and sequencing of a pED4 sequence (798 bp) located at the dbf-2 and sgfp ORF intersection, utilizing primers PED4NF3066 and PED4NR3864 (Table 2), were used to verify full insertion of the pED4 plasmid into dbf-2 transformants exhibiting full complementation of the dbf-2 and his-3 mutations.
Glycogen analysis.
Iodine vapor staining was conducted based on the work of Wilson et al. (69), with some modifications. Liquid medium used for growth of 2-day-old mycelial cultures was filtrated and vacuum concentrated with a Speed-Vac (Savant, Farmingdale, NY). From each sample, a droplet of concentrated medium was placed at the center of a Whatman filter paper (no. 1). Similarly, mycelium-colonized agar plugs (0.5 cm in diameter) obtained from the expanding margins of 4-day-old cultures grown in solid media were first placed at the center of a Whatman filter paper and then quickly removed, so that the mycelial strands were “printed” on the filter paper. The filter papers on which both liquid and solid medium-grown samples had been printed were then sealed with 3 to 4 granules of crystalline iodine (BDH Chemicals, Poole, United Kingdom) in 50-ml tubes for approximately 5 min and subsequently removed and visualized by light microscopy.
For glycogen identification by 1H nuclear magnetic resonance (NMR) analysis, hyphal tip cytoplasmic leakages observed at the expanding margins of dbf-2 cells were collected (utilizing a dissecting microscope) from 50 (5-cm-diameter) petri dishes of 3-day-old dbf-2 cultures. After lyophilization, the sample (2.5 mg), dissolved in D2O (0.5 ml), was analyzed on a Bruker Avance 400 spectrometer (operating at 400.17 MHz for 1H), and spectra were compared to those of a glycogen standard (Sigma-Aldrich).
Determining conidial numbers.
In order to quantify the abundance of different conidial types, conidia were harvested from 15-day-old cultures and their total number was determined using a hemocytometer. Arthroconidia were counted separately based on their distinguishable rectangular shape. The total conidial suspension was filtrated via a Millipore Durapore Millex-SV 0.5-μm filter for separation of microconidia (∼3 μm [36]), and their number was then determined. The number of macrconidia was calculated by subtracting the number of arthroconidia and microconidia from the total conidial count. Conidial chains were counted as a single unit. Two independent experiments were carried out in the same manner, in which two conidial counts were conducted for each strain.
Western blot analysis.
Total protein isolation was conducted based on the work of Gorovits and Yarden (20). Cell lysate protein concentrations were quantified by the Bradford protein assay (5). Western blotting was performed by standard procedures (52) and based on that previously described by Gorovits and Yarden (20). Antibodies used throughout this study included anti-Cdc2 p34 (PSTAIRE) sc-53 (rabbit) primary antibody and the goat anti-rabbit peroxidase-coupled secondary antibody (Amersham Biosciences, Freiburg, Germany). General protein staining for demonstration of equal loading was performed with the Memcode reversible protein stain kit for nitrocellulose membranes (Pierce, Rockford, IL). Western blot analyses were repeated several times, using protein extracts from two independent experiments.
Microscopy.
Light microscopy was performed with a Zeiss Axioscope microscope equipped with a Nikon DXM1200F digital camera. Fluorescence microscopy was performed with two Zeiss filter sets (excitation filter, 365 nm, and emission filter, 420 nm; excitation filter, 395 to 440 nm, and emission filter, 470 nm) for visualizing DAPI staining and GFP, as well as calcofluor white, respectively.
For scanning electron microscopy (SEM), samples were fixed for 4 h with 5% (vol/vol) glutaraldehyde in 0.1 M phosphate buffer, pH 7.2. The samples were washed five times with the same buffer and then dehydrated in a series of 25 to 100% ethanol washes. The fixed samples were dried for 1 h in a CPD7 50 drier (Bio-Rad) and gold coated in an E5150 Polaron SEM coating system apparatus (Bio-Rad). The samples were observed under a JEOL (Tokyo, Japan) JSM 35 microscope.
RESULTS
DBF-2 is required for proper asexual development and sexual reproduction in N. crassa.
To analyze the role of DBF-2 in N. crassa, a dbf-2 deletion strain was produced using a cassette designed for dbf-2 homologous recombination (i.e., harboring the bacterial hph gene between the 5′ and 3′ flanking regions of dbf-2). The ku-70 strain was used to facilitate the homologous recombination (8). However, sexual reproduction was severely affected by the loss of dbf-2, as dbf-2 ascospores obtained from crosses between the dbf-2 mutant (in a ku-70 background) and the wt strain were not viable. Therefore, in order to obtain homokaryon deletion strains, microconidia harboring the dbf-2 deletion nuclei were isolated (12). The dbf-2 deletion strains grew significantly slower than the wild-type and ku-70 control strains. Specifically, when grown on Vs medium, the dbf-2 deletion strain exhibited reduced basal hyphal extension rates (17% of the rates of the wt and ku-70 control strains), as measured in race tubes, accompanied by a defect in septation (see Fig. S1 in the supplemental material) similar to that observed in the Aspergillus nidulans homolog sidB (26). In addition, the dbf-2 strain exhibited severe impairment of different stages of asexual reproduction. In particular, reduced aerial hypha formation and a conidial separation defect of macroconidia along with overproduction of micro- and arthroconidia were observed (detailed analysis is provided below). Overall, the pleiotropic nature of the dbf-2 mutant indicates that DBF-2 is a kinase involved in several processes in both vegetative and sexual developmental growth phases of N. crassa.
DBF-2 is a nuclear protein involved in mitosis.
Since in S. cerevisiae DBF2 was shown to be associated with anaphase and/or telophase progression (16, 65), we examined whether the deletion of the kinase would also affect cell cycle progression in N. crassa. Utilizing DAPI staining, we found that while nuclei in hyphae and conidia of the control strains were round, a distinctive elongated nuclear phenotype was evident in the dbf-2 mutant (Fig. 1). This phenotype resembles that described by Somers et al. (57), showing that N. crassa telophase is different from similar stages in other organisms, as toward telophase completion, the daughter nucleus appears elongated rather than having the regular, round shape regained during entrance to interphase. In order to provide a quantitative assessment of the phenotype observed, the percentage of elongated nuclei within the population was determined. This revealed a 6-fold-higher ratio of elongated nuclei in the dbf-2 mutant than that found in the control strains (Fig. 1). Even though a majority of the nuclei were delayed in mitosis, hyphal growth (albeit slower) was still maintained.
Fig. 1.
Mitosis is impaired in the dbf-2 mutant. In contrast to the round nuclear morphology in the wt and ku-70 control strains, the dbf-2 mutant nuclei are elongated, as determined by DAPI staining of hyphae (top panels). Round nuclei are also observed in conidia of the wt (middle panels; fluorescence and fluorescence with bright field, respectively), whereas nuclei in constricted proconidial chains of the dbf-2 strain are elongated. Scale bars represent 50 μm (wt and dbf-2 strains), 25 μm (ku-70 strain), and 10 μm (middle panels). The prevalence of elongated nuclei in the different strains is shown (bottom left panel). Data shown are based on the averages of three experiments, in which nuclei in 10 fields (of each strain) were counted. Bars indicate standard errors. DBF-2::GFP expressed in the nucleus of a pED4 dbf-2 transformant (G10) fully complements the dbf-2 mutant phenotype (bottom right panel).
Cellular localization of DBF-2 was determined by following the expression of a DBF-2::GFP fusion protein, expressed under the control of its natural (dbf-2) promoter. The G10 strain (Table 1), a dbf-2 mutant transformant expressing the DBF-2::GFP chimera, exhibited full phenotypic complementation of the dbf-2 mutant, indicative of proper activity of the fusion protein. Thus, the G10 strain exhibited normal hyphal morphology and growth rates, produced aerial hyphae, and conidiated in a manner indistinguishable from that of the wild type. Furthermore, no cytoplasmic leakage foci, characteristic of the parental strain (see below) were observed. Fluorescence microscopy of the G10 strain revealed that, in N. crassa, DBF-2 localizes to the nucleus (Fig. 1), with no evidence (at the resolution and fluorescent intensity used) for its presence in the cytoplasm.
Defects in DBF-2 function affect glycogen metabolism.
When cells were grown on either solid or liquid Vs medium, significant cytoplasmic leakage foci from hyphal tips were observed in cultures of dbf-2 mutants (Fig. 2A and B). As DBF2 was listed in a report on a systematic approach to identifying genes affecting glycogen storage in S. cerevisiae (69), we suspected that the phenotype observed at the hyphal tips of the dbf-2 strain may result from glycogen accumulation, even though no glycogen leakage has been associated with S. cerevisiae dbf2 mutants. This possibility was supported by results obtained following the exposure of cultures to iodine vapor, where staining (indicative of the presence of glycogen) was found to be markedly more intense in the dbf-2 mutant than in the wt (Fig. 2B). The identity of the polymer leaking from the dbf-2 mutant was further confirmed by 1H NMR analysis of the droplet material collected from growing hyphal tips, as the sample presented signals at δH 5.27 (broad singlet, 1H) and δH 3.00 to 4.00 (m, 6H), identical in all respects with the glycogen standard (Fig. 2C). Results of this analysis confirmed that the major substance within the droplets was, in fact, glycogen and established the fact that dbf-2 is involved in glycogen metabolism in a filamentous fungus.
Fig. 2.
The dbf-2 mutant is characterized by massive glycogen leakage. (A) Intact hyphal cells are characteristic of wt growth on standard media, while growth of the dbf-2 strain either on solid or in liquid media (middle and right panels, respectively) is accompanied by large cytoplasmic leakage foci at the hyphal tips (arrows). (B) Minimal iodine vapor staining is observed in the wt whereas intense iodine vapor staining is observed in the dbf-2 mutant grown in liquid (upper panel) or on solid (lower panel) media. Arrows indicate iodine vapor staining of leakage foci from hyphal tips of the dbf-2 strain. Images were obtained by light microscopy. (C) The predominant content of the cytoplasmic leakage is glycogen, as determined by 1H NMR analysis of the droplet material collected from growing dbf-2 mutant hyphal tips, compared with a glycogen standard (upper and lower spectra, respectively).
Given that reserve carbohydrate metabolism can be affected by the carbon/nitrogen (C/N) ratio in the medium (31), we cultured the dbf-2 strain on media with different C/N ratios. The sizes and localizations of glycogen leakages from dbf-2 mutant hyphae showed a clear correlation with variations in the C/N ratio. During growth on a high (10)-C/N-ratio medium (for comparison, Vs has a C/N ratio of approximately 11), the droplets observed were large and located mainly at hyphal tips (Fig. 3). When cells were cultured on media with decreasing C/N ratios, the size of the droplets was reduced and the location of leakage foci shifted to subapical regions of the hyphal extension fronts. A complete block in leakage was observed at a C/N ratio of 0.1. The observed phenotypic suppression of the glycogen leakage by alteration of C/N ratios is indicative of a regulatory link between dbf-2, glycogen metabolism, and environmental sensing.
Fig. 3.
The extent of glycogen leakage in the dbf-2 strain is dependent on the carbon/nitrogen (C/N) ratio in the growth medium. The strain was cultured on minimal glucose (1.5%) medium with various concentrations of NH4NO3. Arrows mark the locations of glycogen droplets.
To date, the function of DBF-2 in signal transduction pathways regulating glycogen metabolism has not been characterized. Moreover, as it is a Hippo pathway component, the involvement of DBF-2 in glycogen metabolism implies a functional link between Hippo signaling and glycogen metabolism pathways. In order to further elucidate this possibility, we analyzed mutants of several genes (Table 1): mst-1 (a sterile-20-related kinase), mob-1 (Mps one binder), ccr-4 (carbon catabolite repression 4), and gsk-3 (glycogen synthase kinase 3), whose orthologs in different organisms have been shown to be involved in either Hippo signaling (mst-1 and mob-1) or glycogen biosynthesis (gsk-3 and ccr-4) pathways through interaction with DBF-2 orthologs (see the Discussion). This included, for the first time, a comparative morphological characterization of knockout mutants of these genes and of the dbf-2 mutant. We found that, with significant resemblance to the dbf-2 mutant, three noticeable defects were displayed by the strains harboring the mentioned mutations: cytoplasmic leakage, massive hyphal swellings, and hyphal tip bursting (Fig. 4). In addition, the mob-1 mutant also displayed a defect in septation (see Fig. S1 in the supplemental material). It was notable that the phenotypic severity ranged between the different mutants, as large cytoplasmic leakages (approximately 400 μm in diameter) were observed in the dbf-2 and mob-1 mutants while smaller ones (approximately 100 μm in diameter) were observed in the mst-1, gsk-3, and ccr-4 mutants. The striking similarities between the different phenotypic effects caused by disruption of these genes strongly indicate that the mutants share defects in a common pathway.
Fig. 4.
Defects in the putative N. crassa dbf-2 (Hippo) and glycogen metabolism pathway components share common morphological consequences. Typically, all the genetic defects are accompanied by hyphal swelling, bursting of hyphal tips (arrows in the center column), and cytoplasmic leakage (arrows in the right column).
Altered conidiation in strains harboring defects in dbf-2 and glycogen pathway components.
Aerial hypha formation is the initial step in macroconidial development (11, 60, 61). As the dbf-2 mutant does not produce proper aerial hyphae, we examined if defects in other Hippo and glycogen pathway components also confer similar morphological consequences. Our results show that in mutant strains lacking mst-1, mob-1, or ccr-4, reduced aerial hypha formation was evident (Fig. 5). The ccr-4 mutant, in particular, showed the most prominent resemblance to the dbf-2 mutant, with a fully arrested aerial hypha formation defect.
Fig. 5.
Various dbf-2 (Hippo) and glycogen metabolism pathway mutants exhibit altered aerial hypha production (as evident from the presence of fungal biomass on flask walls). All strains were photographed 2 weeks after inoculation.
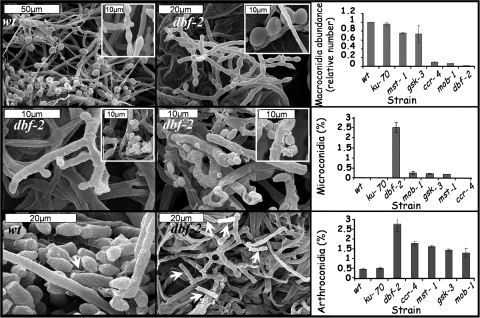
The involvement of Hippo and glycogen pathway components in asexual conidiation in N. crassa was further established by performing scanning electron microscopy analysis of the different mutants, at late stages of asexual development. As N. crassa produces three types of asexual conidia (61), we wanted to determine whether any or all of these pathways were affected. The qualitative morphological analysis was accompanied by quantification of the abundance of each conidial type produced by the individual mutants. In the different mutants harboring defects in the Hippo and glycogen pathways, the macroconidiation pathway is blocked prior to the formation of minor constrictions, resulting in conidiophore structures arrested at an immature stage. These results correlate well with a major decrease (up to 90%) in macroconidial abundance, which is due to the conidial separation defect (conidial chains were counted as single reproductive units). In contrast, microconidiation and arthroconidiation pathways were markedly upregulated, possibly as a bypass of the arrest in macroconidiation. This was expressed by a significant increase (orders of magnitude) in the percentage of microconidia produced as well as a higher percentage (3- to 5-fold) of arthroconidia in comparison to the control strains, in which almost no microconidia were formed and arthroconidia were only about 0.5% of the total conidia produced (Fig. 6).
Fig. 6.
Conidial production patterns in the dbf-2 mutant resemble those of other Hippo and glycogen metabolism pathway component mutants. The abundances of macroconidia (top panel), microconidia (middle panel), and arthroconidia (bottom panel) were determined in strains with mutations in genes involved in dbf-2 (Hippo) and glycogen signaling. While typical conidial separation can be observed in the wt strain, the process is impaired in the dbf-2 mutant (top panel). Microconidia are produced in abundance in the dbf-2 mutant and are at times engulfed by curling hyphae (middle panel). Typical production of arthroconidia in the wt strain is markedly lower than that observed in the dbf-2 mutant (bottom panel). Comparative quantification of the abundance of different conidial types is presented on the right side of each row.
Mitotic delay typifies defects in dbf-2 and glycogen pathway components in N. crassa.
As the morphological defects in the different dbf-2 and glycogen metabolism pathway mutants highly resembled those observed in the dbf-2 mutant, we examined if the defects imposed also include impairment of cell cycle progression. DAPI staining of nuclei revealed that, with remarkable similarity to the dbf-2 strain, the mutants analyzed (mst-1, ccr-4, gsk-3, and mob-1) exhibited a significantly higher number of intensively stained elongated nuclei (indicative of mitotic delay) than that found in the control strains (Fig. 7). Specifically, the dbf-2 and ccr-4 mutants showed the highest percentage of elongated nuclei within the nuclear population (6-fold higher than the control strains), followed by mob-1 and gsk-3 (5- and 4-fold higher than the control strains, respectively) (Fig. 7). In order to verify that the common phenotypic feature of these mutants is, in fact, a delay in cell cycle progression, we determined the effect of benomyl, a beta-tubulin depolymerizing agent, on the elongated nuclei (49). In all cases, the use of a sublethal concentration of the drug (2.5 μg/ml) resulted in the alleviation of the high abundance of elongated nuclei, indicative of a true defect in the mitotic cycle (see Fig. S2 in the supplemental material).
Fig. 7.
Impaired mitosis accompanies defects in dbf-2 (Hippo) and glycogen pathway components. (A) Elongated nuclei (as visualized by DAPI staining) can be observed in the mst-1, ccr-4, mob-1, and gsk-3 strains. (B) Comparative quantification of the percentage of elongated nuclei in strains defective in putative components of N. crassa dbf-2 (Hippo) and glycogen metabolism pathways in comparison to the wt and ku-70 control strains. Bars indicate standard errors.
Altered cdc-2 expression in mutants impaired in dbf-2 and glycogen pathway components.
Biochemical and genetic analyses have shown that the human Hippo pathway component DBF2 homolog, LATS1, negatively regulates CDC2 (cell division cycle 2), probably by physical interaction (63). It is also known that inhibition of CDC2 triggers a phosphorylation-dephosphorylation cascade regulating glycogen metabolism through activation of glycogen synthase kinase 3β (GSK3β), a known inhibitor of glycogen biosynthesis (29, 39, 63). On the basis of these findings, we assumed that CDC2 expression would be affected by defects in different components of the dbf-2 (Hippo) and glycogen metabolism pathways in N. crassa. When probed with anti-CDC2 (34-kDa) antibodies, a significant decrease in CDC-2 protein levels could be observed in the different mutants, with the exception of gsk-3 (Fig. 8). The most striking decrease in CDC-2 levels was detected in the dbf-2, ccr-4, and mob-1 mutants. In most of the strains analyzed, two protein bands, approximately 1 kDa apart, were evident, suggesting that the antibodies used may have detected both the phosphorylated (35-kDa) and the unphosphorylated (34-kDa) forms of CDC-2 in the fungal protein extract. If this is the case, it is possible that the putative phosphorylation pattern may be altered in the ccr-4 mutant, which exhibited only the upper, 35-kDa band.
Fig. 8.
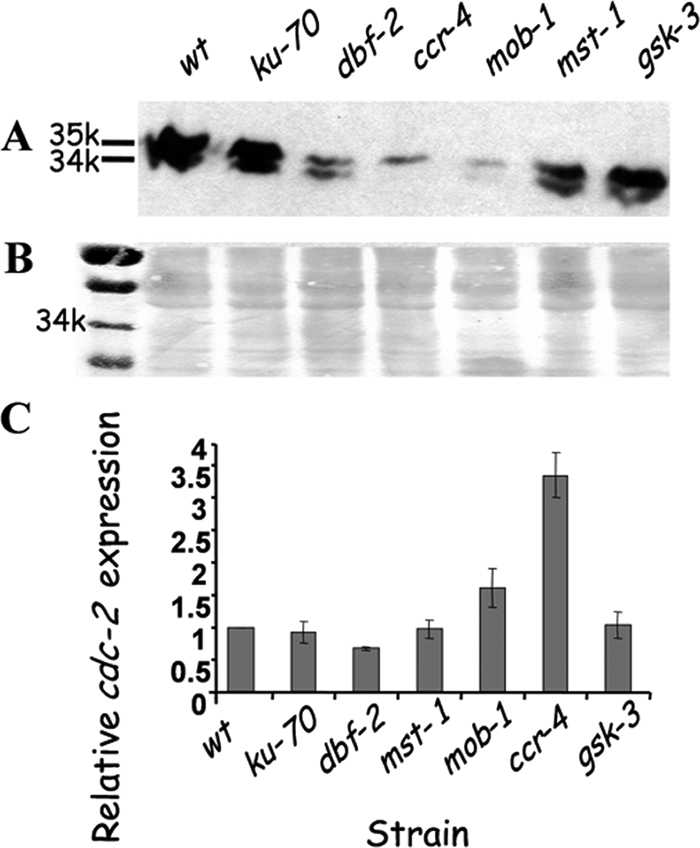
Altered cdc-2 expression in various mutants harboring defects in (dbf-2) Hippo and glycogen pathways. (A and B) Decreased cdc-2-encoded protein levels (detected at 34 and 35 kDa) in dbf-2, ccr-4, mob-1, and mst-1 mutants, compared to the wt and ku-70 control strains, as determined by Western blot analysis (A). A general protein stain demonstrating equal loading is shown as well (B). (C) Changes in cdc-2 expression in different mutants impaired in dbf-2 and glycogen pathway components as determined by real-time RT-PCR. The quantity of cdc-2 cDNA measured was normalized to that of actin cDNA, in extracts from each mutant. Data shown are the average expression levels based on three experiments. Bars indicate standard errors. The abundance of cDNA from the wt strain samples was assigned a value of 1.
The decrease in CDC-2 protein levels could be attributed to downregulation of cdc-2 transcription or, alternatively, to reduced protein stability. In order to examine whether cdc-2 transcription is decreased in the different mutants, real-time RT-PCR analysis was performed (utilizing primers cdc-2rtF and cdc-2rtRN and primers actrt575604F and actrt575473R [Table 2; Fig. 8C]). In contrast to the expected downregulation of cdc-2 transcript levels (which would explain the reduction in CDC-2 protein levels), no significant differences in cdc-2 transcript levels in dbf-2, mst-1, and gsk-3 mutants were observed. Moreover, 3- and 1.5-fold-higher cdc-2 transcription levels were observed in the ccr-4 and the mob-1 mutants, respectively. Thus, it is conceivable that the decrease in CDC-2 protein levels observed in the Hippo/glycogen pathway mutants is a consequence of reduced protein stability.
ccr-4 and rgb-1 are required for proper cellular localization of DBF-2.
The association of DBF2 with MOB1 and with CCR4 has been previously reported (23, 32). The fact that MOB1 has been shown to localize to the nucleus and the observation that CCR4 is cytoplasmic suggest that not all associations of the two proteins with DBF2 are simultaneous and that such associations may be involved in, or required for, proper localization of the kinase (16, 62, 64). Other proteins that may influence DBF2 localization are protein phosphatase 2A (PP2A) and sporulation-induced transcribed sequence (SIT4, a PP2A-related protein), as human PP2A inhibits LATS1 through inhibition of MST2 (STE20 homolog) and SIT4 is known to suppress the DBF2 mutation in yeasts (42, 46). In order to determine whether a deficiency of these proteins results in the altering of DBF-2's nuclear localization in N. crassa, the fused DBF-2::GFP protein was expressed in the ccr-4, mob-1, rgb-1RIP (in which a PP2A B regulatory subunit was inactivated by repeat-induced point mutations [72]), and ppe-1 (the N. crassa SIT4 homolog) mutants. No significant mislocalization of DBF-2 could be observed in the mob-1 and ppe-1 strains, where the protein was found to predominantly localize to the nucleus. However, in the ccr-4 and rgb-1RIP mutants, we observed a marked increase in cytoplasmic levels of DBF-2::GFP and its accumulation in large cytoplasmic organelles (Fig. 9). These findings link the intracellular localization of DBF-2 with CCR-4 as well as PP2A while at the same time demonstrating that MOB-1, though known to physically interact with DBF-2 (34), is not required for its nuclear localization. The differential localization of DBF-2 in the various mutants is an important initial step in determining the mechanistic nature of DBF-2 protein complex members.
Fig. 9.
Abnormal localization of DBF-2::GFP expressed in the mob-1, ppe-1, ccr-4, and rgb-1RIP strains (but not in the mob-1 or ppe-1 mutant) as determined by fluorescent microscopy. Accumulation of DBF-2 in the cytoplasm and in large cytoplasmic organelles is marked by arrows. For normal localization of DBF-2::GFP, see the G10 panel in Fig. 1.
DISCUSSION
In this study, we disrupted the N. crassa dbf-2 gene and studied the consequences of its inactivation. It was not surprising to identify, in addition to the conserved roles of dbf-2, novel (effect on sexual reproduction and dependency on ccr-4 and rgb-1 for localization), unique (involvement in three conidiation pathways), or altered (differential effect on CDC-2) functions of this kinase in N. crassa, as protein kinases have been shown to selectively adapt roles in different organisms (2).
DBF-2 is involved in multiple cellular functions in N. crassa.
Inactivation of dbf-2 in N. crassa results in a pleiotropic effect, and yet the mutant is viable. This is in contrast to the lethal consequence of inactivating both DBF2 and DBF20 in S. cerevisiae, indicating that in the filamentous fungus at least some functions of the protein are redundant. This is in line with previous observations pointing out the presence of differences in gene/protein function between yeasts and filamentous fungi (3, 17).
The lack of dbf-2 had a clear effect on hyphal growth, as evident from the fact that dbf-2 deletion strains exhibit reduced basal hyphal extension rates. The involvement of DBF-2 in this process may be a novel function gained in the course of evolution from yeasts, as additional inputs are likely to be required to modulate the complexity of filamentous growth. In agreement with our findings, mutants with mutations of the A. nidulans DBF2 homolog sidB have been reported to produce fluffy colonies which are significantly smaller than those of the wild type (26). The role of DBF2 in cellular growth has also been demonstrated in higher eukaryotes, as Lats1−/− mice exhibited significant growth retardation (70).
The dbf-2 strain exhibited reduced aerial hypha formation, and subsequently, macroconidiation was reduced. Other N. crassa mutants that exhibit defects in blastoconidiation which are accompanied by increased arthroconidium production include rgb-1RIP (72) and frost (which encodes a homolog of the yeast cdc1 gene [58]) mutants. In A. nidulans, sidB mutants fail to produce conidia altogether (26). Thus, even though both dbf-2 and sidB belong to the septation initiation network (for example, on the basis of septation defects; see Fig. S1 in the supplemental material), the roles of dbf-2 and sidB differ significantly, as our results clearly indicate that the involvement of DBF-2 in N. crassa conidiation occurs after commitment to this developmental process, in contrast to A. nidulans, where conidiation is blocked prior to initiation. This may well be linked to the divergent modes of A. nidulans and N. crassa conidiation (1), including the presence of multiple conidiation pathways in the latter. The crucial role of septation in growth and development is further emphasized by the fact that the dbf-2 mutant and the mob-1 mutant, both of which exhibit impaired septation, tend to accumulate suppressors in which septation is resumed. The nature of these suppressors has yet to be determined.
Our findings also indicate, for the first time, that DBF-2 is essential for sexual reproduction in a filamentous fungus. Interestingly, mice lacking the dbf-2 homolog (Lats1−/−) also showed infertility (70). How DBF-2 may affect the function of N. crassa mating type loci and interacting genes/gene products has yet to be elucidated. The fact that CDC15 (a DBF2-interacting protein in S. cerevisiae) is required for meiosis II exit, as well as for spore morphogenesis (44), suggests that DBF2 may be linked with sexual reproduction in yeasts as well.
We found that, in N. crassa, DBF-2 is predominantly localized in the nucleus, as has been shown in other organisms (41, 59, 66, 68, 73). Taking into consideration the role that DBF2 homologs have been shown to play in cell cycle progression (4, 62, 65–68), this is expected. Based on the elongated phenotype of the nuclei observed in this study, along with the effect of benomyl on nuclear morphology in the mutants, we concluded that in N. crassa, impairment of DBF-2 confers a block in cell cycle progression, as has been shown in other systems. Stretched/broken nuclear morphology has been observed in the N. crassa rho-4 (encoding a monomeric GTPase) mutant, which also exhibits septation defects and cytoplasmic leakage (50, 51). However, in addition to the morphological differences between rho-4 mutant nuclei and those observed in the dbf-2/glycogen pathway mutants, in the rho-4 mutant the altered nuclear morphology was accompanied by a variance in nuclear distribution along the hyphal cell which was attributed to aberrant actin and microtubule cytoskeleton assembly shown to occur in that mutant (51). The fact that we did not observe fragmented nuclei in the mutants studied here and that the benomyl treatment did not affect nuclear positioning is indicative that the elongated nuclear morphology was not due to a general defect in microtubule dynamics in these mutants.
In humans, defects in LATS1 affect cell cycle progression as well, and yet this effect is not limited to a single specific stage (e.g., early prophase or early anaphase [63]). Taken together, it is clear that DBF2 has a role in cell cycle progression in a variety of organisms. Nonetheless, this is another example of functional deviation of this kinase across evolution. It is tempting to speculate that the presence of cytoplasmic continuity in the filamentous fungi may contribute to the higher uniformity of the effect that DBF2 has on the nuclear population in a given fungus. Even though a significant accumulation of nuclei delayed in mitosis was observed in the dbf-2 mutant, hyphal growth was only partially impaired. As not all nuclei appeared elongated, it is conceivable that an additional protein(s), yet to be identified, can enable progression (albeit partial or slower) through mitosis, parallel to DBF-2.
DBF-2 is involved in glycogen metabolism.
The dbf-2 mutant exhibited a C/N-ratio-dependent bursting of hyphal tips and vast cytoplasm leakage (Fig. 3). The cytoplasmic exudate was identified as glycogen, establishing a new role of DBF-2 in negative regulation of glycogen metabolism in N. crassa. As reserve carbohydrate metabolism can be affected by the C/N ratio in the medium (14, 31, 47), we propose that decreasing C/N ratios results in a dramatic reduction in glycogen accumulation and thus prevents the hyphal tip leakage phenomenon observed during growth of the dbf-2 mutant. The C/N-ratio-dependent bursting of hyphal tips and glycogen-rich exudates are typical of the dbf-2 strain and were not observed in an additional mutant (the hex-1 mutant [25]) exhibiting septation defects accompanied by cytoplasmic leakage, suggesting that this is not an effect common to all mutants exhibiting the mentioned phenotype. In yeasts, DBF2 has been shown to be required for the phosphorylation of the A and B subunits (Vma1p and Vma2pa, respectively) of the vacuolar H+-ATPase (37). As this ATPase has been suggested to affect glycogen degradation (69), the hyperaccumulation of glycogen that we have observed can perhaps be attributed to both the effect of DBF-2 on the glycogen synthesis pathway (see below) and its effect on glycogen degradation.
Mutants with defects in dbf-2 and glycogen metabolism pathway components share similar phenotypes.
In order to study the potential functional link between dbf-2 and glycogen metabolism, with an emphasis on the possibility of interactions between the tentative Neurospora Hippo and glycogen metabolism pathways, we analyzed mutants defective in several genes (mst-1 [a sterile-20-like kinase], mob-1 [Mps one binder], ccr-4, and gsk-3 [glycogen synthase kinase 3]) whose orthologs belong to the mentioned pathways. We found that, similarly to the dbf-2 strain, these mutants also display defects such as mitotic delay and decreased CDC-2 protein levels, massive hyphal swellings, hyphal tip bursting, glycogen leakage, and impaired conidiation. Even though mst-1 shows the highest structural similarity to the human Mst2 (shown to be the upstream activator of Lats1), N. crassa harbors an additional STE20-related kinase-encoding gene—prk-9 (NCU04096) (8)], which shows high structural similarity to the Schizosaccharomyces pombe SIN component sid1. The phenotypic defects observed in the prk-9 deletion strain also resemble those observed in the dbf-2 and glycogen pathway mutants (even though it appears that the presence of elongated nuclei in the mst-1 mutant exceeds that found in the prk-9 mutant, in a manner more resembling that seen in the dbf-2 mutant). Thus, the possibility that prk-9 may also feed into the mentioned pathways or even have functions overlapping with those of mst-1 should not be excluded. Our observations are supported by previous reports which have studied some of these components separately. For example, the fact that MOB1 and DBF2 can physically associate is well documented in Drosophila (27) and, recently, in N. crassa as well (34). The similar overlap in some of the phenotypic defects (including the typical SIN defect [see Fig. S1 in the supplemental material]) can thus be explained by the lack of one of the complex components. A similar argument can be made for CCR4, which has also been shown to complex with DBF2 (32). As both DBF2 and MOB1 have been shown to be regulated by STE20 (33, 48), the absence of this upstream component may well result in the lack of active DBF2/MOB1 and DBF2/CCR4 complexes.
On the basis of the reduction in CDC-2 levels in the dbf-2, ccr-4, and mob-1 mutant strains, we propose that DBF-2 may negatively regulate CDC-2 activity. This possibility can be derived on the basis of different reports concerning LATS1 which, once integrated, imply that a signal transduction pathway involving LATS1, CDC2, and PP1 (protein phosphatase 1) operates in cell cycle regulation and also, with the involvement of GSK3β (a glycogen synthase kinase), in glycogen biosynthesis in human cells (29, 39, 63).
In accordance with the described dependence of various cellular functions on DBF2, MOB1, and CCR4 (known to interact with each other or with other CCR4 transcriptional complex components [27]), the similarities in phenotypic consequences which are a result of impaired function of downstream components should be expected. This includes hyphal swelling, bursting, and glycogen leakage, which have been observed in all the Hippo/glycogen pathway components examined here (the most prominent being in the dbf-2, mob-1, and ccr-4 mutants). We suggest that due to impaired Hippo pathway signaling, the hyphae undergo glycogen hyperaccumulation which consequently causes the observed swelling and eventual bursting of hyphae. The different mutants also exhibited mitotic delay, implying a connection between glycogen accumulation and cell cycle progression. In yeast cells glycogen accumulated during G1 is metabolized at the bud emergence phase (43, 55). Thus, it is possible that due to the mitotic delay observed in the different N. crassa mutants, the cells continue to accumulate glycogen (a process initiated in G1). Stopping glycogen accumulation may depend on exit from mitosis.
One of the unique developmental attributes that can now be associated with both Hippo and glycogen metabolism pathways is asexual sporulation. In all the mutants analyzed, conidiation was significantly affected. However, the consequences of inactivation of the different genes were not uniform. The most striking deviation was in the ccr-4 mutant, where in contrast to the other mutants, the microconidiation pathway was not upregulated. One explanation for this is that microconidiation is a process regulated by a complex requiring only DBF-2 and MOB-1 (rather than a complex which includes CCR-4 as well).
DBF-2 as a link between Hippo and glycogen metabolism pathways.
To date, a functional link between the Hippo pathway and glycogen metabolism has not been proposed. On the basis of our results, along with those previously described, we suggest that at least some of the functions of the Hippo and glycogen biosynthesis pathways are coordinated.
It appears that the two pathways share a core factor—DBF2. This protein can associate with both Hippo pathway components (e.g., MOB1 and STE20 [6, 27, 34]), while it has also been shown to associate with CCR4, which is also involved in glycogen metabolism (via a transcriptional activator intermediate, identified in yeasts as MSN2/4 [30, 56]), or, possibly, by inhibition of CDC2, as implied by our results (Fig. 8). The striking similarities between the multiple phenotypic effects caused by disruption of these genes strongly suggest that they belong to common pathways. Further support for the interactions among at least some of the mentioned gene products was obtained by analyzing the effects of various mutant backgrounds on DBF-2 localization. The fact that DBF-2::GFP was improperly localized in the ccr-4 and rgb-1RIP (yet not in mob-1) mutants not only supports the functional link between some of these pathway components but also establishes, for the first time, a dependency of DBF-2 localization, in N. crassa, on CCR-4 and PP2A (protein phosphatase 2A; see below). In Drosophila CCR4 is mainly cytoplasmic (64). It is possible that small amounts of CCR-4 also function in the cytoplasm in N. crassa. If this be the case, the increased presence of DBF-2::GFP in the cytoplasm of the ccr-4 mutant can be explained as a means to compensate for the lack of a fungal cytoplasmic DBF2/CCR4 complex. The accumulation of DBF-2::GFP in large cytoplasmic organelles (Fig. 9) may serve to regulate the excess of DBF-2 in the cytoplasm.
The linking of these pathways can be based on the presence of an upstream activator—a member of the STE20 kinase family. Based on our results, MST-1 is a positive regulator of DBF-2 function and a negative regulator of glycogen biosynthesis. Other upstream elements include PP2A, which is a negative regulator of the human STE20 ortholog (42). It is likely that PP2A (and specifically the B regulatory subunit encoded by rgb-1) functions in a similar manner in N. crassa, as we have observed an increase in cytoplasmic DBF-2::GFP in the rgb-1RIP strain. The specificity of this interaction is supported by the fact that a defect in a structurally related phosphatase (ppe-1) did not result in mislocalization of DBF-2::GFP. The fact that a reduction in PP2A activity has been shown to correlate with a reduction in glycogen synthesis (7, 24) further supports the dbf-2 (Hippo) and glycogen metabolism pathway link. The presence of gsk-3, a major component of the glycogen synthesis pathway, downstream of DBF-2 has been established here, on the basis of the phenotypic consequences of its inactivation. We propose that its DBF-2-regulated function is conferred via the conserved glycogen biosynthesis pathway components cdc-2 and pp-1 (29, 39, 63).
Integrating our results led us to hypothesize that DBF-2 operates as part of different coexisting complexes with MOB-1 and CCR-4, which function in addition to other independent activities of the complex components. The possible association of DBF2 within these complexes may indicate that the protein serves as a linking unit between primary metabolism and developmental and growth processes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nir Osherov for his helpful comments on the manuscript and Stephen Osmani for his comments and suggestions concerning analysis of mitotic delay. We thank Stephan Seiler for the ppe-1 (SIT4-like) and mob-1 strains and Greg Jedd for the hex-1 strain.
This research was funded by the Israel Science Foundation and the Deutsche Forschungsgemeinschaft.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 4 December 2009.
REFERENCES
- 1.Bailey-Shrode L., Ebbole D. J. 2004. The fluffy gene of Neurospora crassa is necessary and sufficient to induce conidiophore development. Genetics 166:1741–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banuett F. 1998. Signalling in yeasts: an information cascade with links to filamentous fungi. Microbiol. Mol. Biol. Rev. 62:249–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borkovich K. A., Alex L. A., Yarden O., Freitag M., Turner G. E., Read N. D., Seiler S., Bell-Pedersen D., Paietta J., Plesofsky N., Plamann M., Goodrich-Tanrikulu M., Schulte U., Mannhaupt G., Nargang F. E., Radford A., Selitrennikoff C., Galagan J. E., Dunlap J. C., Loros J. J., Catcheside D., Inoue H., Aramayo R., Polymenis M., Selker E. U., Sachs M. S., Marzluf G. A., Paulsen I., Davis R., Ebbole D. J., Zelter A., Kalkman E. R., O'Rourke R., Bowring F., Yeadon J., Ishii C., Suzuki K., Sakai W., Pratt R. 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68:1–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bothos J., Tuttle R. L., Ottey M., Luca F. C., Halazonetis T. D. 2005. Human LATS1 is a mitotic exit network kinase. Cancer Res. 65:6568–6575 [DOI] [PubMed] [Google Scholar]
- 5.Bradford M. M. 1976. A rapid and sensitive method for the quantitation of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 6.Chan E. H., Nousiainen M., Chalamalasetty R. B., Schafer A., Nigg E. A., Sillie H. H. 2005. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene 24:2076–2086 [DOI] [PubMed] [Google Scholar]
- 7.Clotet J., Posas F., Hu G. Z., Ronne H., Arifio J. 1995. Role of protein phosphatase 2A in the control of glycogen metabolism in yeast. Eur. J. Biochem. 229:207–214 [PubMed] [Google Scholar]
- 8.Colot H. V., Park G., Turner G. E., Ringelberg C., Crew C. M., Litvinkova L., Weiss R. L., Borkovich K. A., Dunlap J. C. 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. U. S. A. 103:10352–10357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis R. H. 2000. Neurospora—contributions of a model organism Oxford University Press, New York, NY [Google Scholar]
- 10.Davis R. H., de Serres F. J. 1970. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 17A:79–143 [Google Scholar]
- 11.Ebbole D. J. 1996. Morphogenesis and vegetative differentiation in filamentous fungi. J. Genet. 75:361–374 [Google Scholar]
- 12.Ebbole D. J., Sachs M. S. 1990. A rapid and simple method for isolation of Neurospora crassa homokaryons using microconidia. Fungal Genet. Newsl. 37:17–18 [Google Scholar]
- 13.Edgar B. A. 2006. From cell structure to transcription: Hippo forges a new path. Cell 124:267–273 [DOI] [PubMed] [Google Scholar]
- 14.Francois J., Parrou J. L. 2001. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25:125–145 [DOI] [PubMed] [Google Scholar]
- 15.Freitag M., Hickey P. C., Raju N. B., Selker E. U., Read N. D. 2004. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet. Biol. 41:897–910 [DOI] [PubMed] [Google Scholar]
- 16.Frenz L. M., Lee S. E., Fesquet D., Johnston L. H. 2000. The budding yeast Dbf2 protein kinase localizes to the centrosomes and moves to the bud neck in late mitosis. J. Cell Sci. 113:3399–3408 [DOI] [PubMed] [Google Scholar]
- 17.Galagan J. E., Calvo S. E., Borkovich K. A., Selker E. U., Read N. D., Jaffe D., FitzHugh W., Ma L. J., Smirnov S., Purcell S., Rehman B., Elkins T., Engels R., Wang S., Nielsen C. B., Butler J., Endrizzi M., Qui D., Ianakiev P., Bell Pedersen D., Nelson M. A., Werner Washburne M., Selitrennikoff C. P., Kinsey J. A., Braun E. L., Zelter A., Schulte U., Kothe G. O., Jedd G., Mewes W., Staben C., Marcotte E., Greenberg D., Roy A., Foley K., Naylor J., Stange Thomann N., Barrett R., Gnerre S., Kamal M., Kamvysselis M., Mauceli E., Bielke C., Rudd S., Frishman D., Krystofova S., Rasmussen C., Metzenberg R. L., Perkins D. D., Kroken S., Cogoni C., Macino G., Catcheside D., Li W., Pratt R. J., Osmani S. A., DeSouza C. P., Glass L., Orbach M. J., Berglund J. A., Voelker R., Yarden O., Plamann M., Seiler S., Dunlap J., Radford A., Aramayo R., Natvig D. O., Alex L. A., Mannhaupt G., Ebbole D. J., Freitag M., Paulsen I., Sachs M. S., Lander E. S., Nusbaum C., Birren B. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422:859–868 [DOI] [PubMed] [Google Scholar]
- 18.Gorovits R., Propheta O., Kolot M., Dombradi V., Yarden O. 1999. A mutation within the catalytic domain of COT1 kinase confers changes in the presence of two COT1 isoforms and in Ser/Thr protein kinase and phosphatase activities in Neurospora crassa. Fungal Genet. Biol. 27:264–274 [DOI] [PubMed] [Google Scholar]
- 19.Gorovits R., Sjollema K. A., Sietsma J. H., Yarden O. 2000. Cellular distribution of COT1 kinase in Neurospora crassa. Fungal Genet. Biol. 30:63–70 [DOI] [PubMed] [Google Scholar]
- 20.Gorovits R., Yarden O. 2003. Environmental suppression of Neurospora crassa cot-1 hyperbranching: a link between COT1 kinase and stress sensing. Eukaryot. Cell 2:699–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris S. D. 2006. Cell polarity in filamentous fungi: shaping the mold. Int. Rev. Cytol. 251:41–77 [DOI] [PubMed] [Google Scholar]
- 22.Harvey K., Tapon N. 2007. The Salvador-Warts-Hippo pathway—an emerging tumour-suppressor network. Nat. Rev. Cancer 7:182–191 [DOI] [PubMed] [Google Scholar]
- 23.Hergovich A., Stegert M. R., Schmitz D., Hemmings B. A. 2006. NDR kinases regulate essential cell processes from yeast to humans. Nat. Rev. Mol. Cell Biol. 7:253–264 [DOI] [PubMed] [Google Scholar]
- 24.Jablonka W., Guzman S., Ramirez J., Montero-Lomeli M. 2006. Deviation of carbohydrate metabolism by the SIT4 phosphatase in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1760:1281–1291 [DOI] [PubMed] [Google Scholar]
- 25.Jedd G., Chua N.-H. 2000. A new self-assembled peroxisomal vesicle required for efficient resealing of the plasma membrane. Nat. Cell Biol. 2:226–231 [DOI] [PubMed] [Google Scholar]
- 26.Kim J. M., Lu L., Shao R., Chin J., Liu B. 2006. Isolation of mutations that bypass the requirement of the septation initiation network for septum formation and conidiation in Aspergillus nidulans. Genetics 173:685–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komarnitsky S. I., Chiang Y. C., Luca F. C., Chen J., Toyn J. H., Winey M., Johnston L. H., Denis C. L. 1998. DBF2 protein kinase binds to and acts through the cell cycle-regulated MOB1 protein. Mol. Cell. Biol. 18:2100–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krapp A., Simanis V. 2008. An overview of the fission yeast septation initiation network (SIN). Biochem. Soc. Trans. 36:411–415 [DOI] [PubMed] [Google Scholar]
- 29.Kwon Y. G., Lee S. Y., Choi Y., Greengard P., Nairn A. C. 1997. Cell cycle-dependent phosphorylation of mammalian protein phosphatase 1 by cdc2 kinase. Proc. Natl. Acad. Sci. U. S. A. 94:2168–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenssen E., James N., Pedruzzi I., Dubouloz F., Cameroni E., Bisig R., Maillet L., Werner M., Roosen J., Petrovic K., Winderickx J., Collart M. A., De Virgilio C. 2005. The Ccr4-Not complex independently controls both Msn2-dependent transcriptional activation—via a newly identified Glc7/Bud14 type I protein phosphatase module—and TFIID promoter distribution. Mol. Cell. Biol. 25:488–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lillie S., Pringle J. R. 1980. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J. Bacteriol. 143:1384–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu H.-Y., Toyn J. H., Chiang Y.-C., Draper M. P., Johnston L. H., Denis C. L. 1997. DBF2, a cell cycle-regulated protein kinase, is physically and functionally associated with the CCR4 transcriptional regulatory complex. EMBO J. 16:5289–5298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma C., Mitra A. 2002. Intrinsic direct repeats generate consistent post-transcriptional gene silencing in tobacco. Plant J. 31:37–49 [DOI] [PubMed] [Google Scholar]
- 34.Maerz S., Dettmann A., Ziv C., Liu Y., Valerius O., Yarden O., Seiler S. 2009. Two NDR kinase-MOB complexes function as distinct modules during septum formation and tip extension in Neurospora crassa. Mol. Microbiol. 74:707–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mah A. S., Jang J., Deshaies R. J. 2001. Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc. Natl. Acad. Sci. U. S. A. 98:7325–7330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maheshwari R. 1999. Microconidia of Neurospora crassa. Fungal Genet. Biol. 26:1–18 [DOI] [PubMed] [Google Scholar]
- 37.Makrantoni V., Dennison P., Stark M. J. R., Coote P. J. 2007. A novel role for the yeast protein kinase Dbf2p in vacuolar H+-ATPase function and sorbic acid stress tolerance. Microbiology 153:4016–4026 [DOI] [PubMed] [Google Scholar]
- 38.Margolin B. S., Freitag M., Selker E. U. 1997. Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet. Newsl. 44:34–36 [Google Scholar]
- 39.Morfini G., Szebenyi G., Brown H., Pant H. C., Pigino G., DeBoer S., Beffert U., Brady S. T. 2004. A novel CDK5-dependent pathway for regulating GSK3 activity and kinesin-driven motility in neurons. EMBO J. 23:2235–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ninomiya Y., Suzuki K., Ishii C., Inoue H. 2004. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. U. S. A. 101:12248–12253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishiyama Y., Hirota T., Morisaki T., Hara T., Marumoto T., Iida S., Makino K., Yamamoto H., Hiraoka T., Kitamura N., Saya H. 1999. A human homolog of Drosophila warts tumor suppressor, h-warts, localized to mitotic apparatus and specifically phosphorylated during mitosis. FEBS Lett. 459:159–164 [DOI] [PubMed] [Google Scholar]
- 42.O'Neill E. E., Matallanas D., Kolch W. 2005. Mammalian sterile 20-like kinases in tumor suppression: an emerging pathway. Cancer Res. 65:5485–5487 [DOI] [PubMed] [Google Scholar]
- 43.Paalman J. W. G., Verwaal R., Slofstra S. H., Verkleij A. J., Boonstra J., Verrips C. T. 2003. Trehalose and glycogen accumulation is related to the duration of the G1 phase of Saccharomyces cerevisiae. FEMS Yeast Res. 3:261–268 [DOI] [PubMed] [Google Scholar]
- 44.Pablo-Hernando M. E., Arnaiz-Pita Y., Nakanishi H., Dawson D., del Rey F., Neiman A. M., Vazquez de Aldana C. R. 2007. Cdc15 is required for spore morphogenesis independently of Cdc14 in Saccharomyces cerevisiae. Genetics 177:281–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pan D. 2007. Hippo signaling in organ size control. Genes Dev. 21:886–897 [DOI] [PubMed] [Google Scholar]
- 46.Parkes V., Johnston L. H. 1992. SPO12 and SIT4 suppress mutations in DBF2, which encodes a protein kinase that is periodically expressed. Nucleic Acids Res. 20:5617–5623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parrou J. L., Enjalbert B., Plourde L., Bauche A., Gonzalez B., François J. 1999. Dynamic responses of reserve carbohydrate metabolism under carbon and nitrogen limitations in Saccharomyces cerevisiae. Yeast 15:191–203 [DOI] [PubMed] [Google Scholar]
- 48.Praskova M., Xia F., Avruch J. 2008. MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr. Biol. 18:311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prigozhina N. L., Oakley C. E., Lewis A. M., Nayak T., Osmani S. A., Oakley B. R. 2004. γ-Tubulin plays an essential role in the coordination of mitotic events. Mol. Biol. Cell 15:1374–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rasmussen C. G., Glass N. L. 2005. A Rho-type GTPase, rho-4, is required for septation in Neurospora crassa. Eukaryot. Cell 4:1913–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rasmussen C. G., Morgenstein R. M., Peck S., Glass N. L. 2008. Lack of the GTPase RHO-4 in Neurospora crassa causes a reduction in numbers and aberrant stabilization of microtubules at hyphal tips. Fungal Genet. Biol. 45:1027–1039 [DOI] [PubMed] [Google Scholar]
- 52.Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed.Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 53.Saucedo L. J., Edgar B. A. 2007. Filling out the Hippo pathway. Nat. Rev. Mol. Cell Biol. 8:613–621 [DOI] [PubMed] [Google Scholar]
- 54.Seiler S., Vogt N., Ziv C., Gorovits R., Yarden O. 2006. The STE20/germinal center kinase POD6 interacts with the NDR kinase COT1 and is involved in polar tip extension in Neurospora crassa. Mol. Biol. Cell 17:4080–4092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sillje H. H., ter Schure E. G., Rommens A. J., Huls P. G., Woldringh C. L., Verkleij A. J., Boonstra J., Verripset C. T. 1997. Effects of different carbon fluxes on G1 phase duration, cyclin expression, and reserve carbohydrate metabolism in Saccharomyces cerevisiae. J. Bacteriol. 179:6560–6565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith A., Ward M. P., Garrett S. 1998. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 17:3556–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Somers C. E., Wagner R. P., Hsu T. C. 1960. Mitosis in vegetative nuclei of Neurospora crassa. Genetics 45:801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sone T., Griffiths A. J. F. 1999. The frost gene of Neurospora crassa is a homolog of yeast cdc1 and affects hyphal branching via manganese homeostasis. Fungal Genet. Biol. 28:227–237 [DOI] [PubMed] [Google Scholar]
- 59.Sparks C. A., Morphew M., McCollum D. 1999. Sid2p, a spindle pole body kinase that regulates the onset of cytokinesis. J. Cell Biol. 146:777–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Springer M. L. 1993. Genetic control of fungal differentiation: the three sporulation pathways of Neurospora crassa. Bioessays 15:365–374 [DOI] [PubMed] [Google Scholar]
- 61.Springer M. L., Yanofsky C. 1989. A morphological and genetic analysis of conidiophore development in Neurospora crassa. Genes Dev. 3:559–571 [DOI] [PubMed] [Google Scholar]
- 62.Stoepel J., Ottey M. A., Kurischko C., Hieter P., Luca F. C. 2005. The mitotic exit network Mob1p-Dbf2p kinase complex localizes to the nucleus and regulates passenger protein localization. Mol. Biol. Cell 16:5465–5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tao W., Zhang S., Turenchalk G. S., Stewart R. A., St. John M. A., Chen W., Xu T. 1999. Human homologue of the Drosophila melanogaster lats tumour suppressor modulates CDC2 activity. Nat. Genet. 21:177–181 [DOI] [PubMed] [Google Scholar]
- 64.Temme C., Zaessinger S., Meyer S., Simonelig M., Wahle E. 2004. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J. 23:2862–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Toji S., Yabuta N., Hosomi T., Nishihara S., Kobayashi T., Suzuki S., Tamai K., Nojima H. 2004. The centrosomal protein Lats2 is a phosphorylation target of Aurora-A kinase. Genes Cells 9:383–397 [DOI] [PubMed] [Google Scholar]
- 66.Toyn J. H., Johnston L. H. 1994. The Dbf2 and Dbf20 protein kinases of budding yeast are activated after the metaphase to anaphase cell cycle transition. EMBO J. 13:1103–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Toyn J. H., Johnston L. H. 1993. Spol2 is a limiting factor that interacts with the cell cycle protein kinases Dbf2 and Dbf20, which are involved in mitotic chromatid disjunction. Genetics 135:963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Visintin R., Amon A. 2001. Regulation of the mitotic exit protein kinases Cdc15 and Dbf2. Mol. Biol. Cell 12:2961–2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilson W. A., Wang Z., Roach P. J. 2002. Systematic identification of the genes affecting glycogen storage in the yeast Saccharomyces cerevisiae: implication of the vacuole as a determinant of glycogen level. Mol. Cell. Proteomics 1:232–242 [DOI] [PubMed] [Google Scholar]
- 70.Yang X., Li D. M., Chen W., Xu T. 2001. Human homologue of Drosophila lats, LATS1, negatively regulates growth by inducing G2/M arrest or apoptosis. Oncogene 20:6516–6523 [DOI] [PubMed] [Google Scholar]
- 71.Yarden O., Plamann M., Ebbole D. J., Yanofsky C. 1992. cot-1, a gene required for hyphal elongation in Neurospora crassa, encodes a protein kinase. EMBO J. 11:2159–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yatzkan E., Yarden O. 1999. The B regulatory subunit of protein phosphatase 2A is required for completion of macroconidiation and other developmental processes in Neurospora crassa. Mol. Microbiol. 31:197–209 [DOI] [PubMed] [Google Scholar]
- 73.Yoshida S., Tohe A. 2001. Regulation of the localization of Dbf2 and Mob1 during cell division of Saccharomyces cerevisiae. Genes Genet. Syst. 76:141–147 [DOI] [PubMed] [Google Scholar]
- 74.Zeng Q., Hong W. 2008. The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell 13:188–192 [DOI] [PubMed] [Google Scholar]
- 75.Zhao B., Lei Q. Y., Guan K. L. 2008. The Hippo-Yap pathway: new connections between regulation of organ size and cancer. Curr. Opin. Cell Biol. 20:638–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ziv C., Gorovits R., Yarden O. 2008. Carbon source affects PKA-dependent polarity of Neurospora crassa in a CRE-1-dependent and independent manner. Fungal Genet. Biol. 45:103–116 [DOI] [PubMed] [Google Scholar]
- 77.Ziv C., Kra-Oz G., Gorovits R., Maerz S., Seiler S., Yarden O. 2009. Cell elongation and branching are regulated by differential phosphorylation states of the NDR kinase COT1 in Neurospora crassa. Mol. Microbiol. 74:974–989 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.