Abstract
Bacterial blight of onion (BBO) is an emerging disease that is present in many onion-producing areas. The causal agent, Xanthomonas axonopodis pv. allii, is seed transmitted. A reliable and sensitive diagnostic tool for testing seed health is needed. Detection of X. axonopodis pv. allii was achieved using a multiplex nested PCR assay developed using two randomly amplified polymorphic DNA (RAPD) and amplified fragment length polymorphism (AFLP) sequences corresponding to pilus assembly genes (pilW and pilX) and the avrRxv gene, respectively. The multiplex nested PCR was used with a large collection of X. axonopodis pv. allii strains pathogenic to onion and/or other Allium species isolated in different regions of the world. The internal primers used in the multiplex PCR assay directed amplification for all 86 X. axonopodis pv. allii strains tested, resulting in a 401-bp amplicon, a 444- to 447-bp amplicon, or both amplicons, depending on the strain. No amplification was obtained for 41 unrelated phytopathogenic bacteria and for 14 saprophytic bacteria commonly isolated from onion leaves and seeds. Most Xanthomonas strains also did not produce amplicons, except for nine strains classified in X. axonopodis genetic subgroup 9.1 or 9.2 and not pathogenic to onion. Nevertheless, sequence signatures distinguished most of these strains from X. axonopodis pv. allii. The assay detected X. axonopodis pv. allii in seed lots with contamination levels of 5 × 102 CFU g−1 or higher. The sensitivity threshold of the multiplex nested PCR assay was found to be 1 infected seed in 27,340 seeds. This PCR-based assay should be useful for certifying that commercial seed lots are free of this important seed-borne pathogen.
Onion (Allium cepa L.) is grown worldwide, covering a total area of roughly 6.7 million acres in at least 125 different countries. The total annual bulb production in 2007 was estimated to be 64.5 million tons (http://apps.fao.org/faostat/). Bulb production is threatened by bacterial blight of onion (BBO) caused by a xanthomonad classified as X. axonopodis pv. allii (41). First identified in Hawaii (4), this pathogen emerged in the 1990s and 2000s in several countries, including Brazil (25), Venezuela (55), the Caribbean Islands (29), different states of the continental United States (16, 28, 47, 50), Japan (17), the Republic of South Africa (52), and the Mascarene Archipelago (Mauritius and Réunion Islands) (33). Consequently, X. axonopodis pv. allii is now included on the European and Mediterranean Plant Protection Organization (EPPO) A1 list.
BBO leaf and scape symptoms start as small water-soaked spots that enlarge into chlorotic lesions and typically collapse at the point of initial infection. Leaf tip death and blight reduce the photosynthetic capacity of plants, leading to a reduction in bulb size. Yield losses ranging from 20 to 50% have been recorded under conditions conducive to efficient development of disease (28, 50).
Pathogenicity tests have indicated that the host range of X. axonopodis pv. allii includes several Allium species (garlic [A. sativum L.], Welsh onion [A. fistulosum L.], shallot [A. cepa var. ascalonicum Backer[, chive [A. schoenoprasum L.], and leek [A. porrum L.]) (41), as well as at least two Citrus species (11). Worldwide, most outbreaks have been reported on onion, but outbreaks have also affected leek and garlic in the Mascarene Archipelago (33) and Welsh onion in Japan (17). Compared to some Xanthomonas pathovars, X. axonopodis pv. allii is phenotypically and genetically diverse (11, 12, 33, 41).
Onion seeds originating from diseased fields were identified as a possible pathway for X. axonopodis pv. allii transmission (42). Inoculum associated with seeds contaminated at a rate of 4/10,000 has been used as the primary inoculum in field epidemiological studies (43). Recently, onion seeds were identified as the most probable pathway for the introduction of the pathogen into Réunion Island from the neighboring country Mauritius (33). Currently, the pathogen is isolated from onion seeds or from plant material using semiselective media (42, 51) and a subsequent identification step consisting of pathogenicity tests and/or molecular typing techniques (12, 33). This approach is time-consuming, and false-negative results can occur when population densities of the pathogen are low because of bacterial microbiota associated with seeds.
For several pathosystems involving xanthomonads, it has been shown that long-distance spread of pathogens can occur through contaminated seeds (53), and sensitive methods to certify that seed lots are pathogen free have been developed (20). Therefore, making highly specific and sensitive PCR-based diagnostic tools available for X. axonopodis pv. allii is a priority. PCR-based techniques have been reported to be highly efficient for detecting and identifying xanthomonads from seeds, such as X. campestris pv. carotae (23), X. oryzae pv. oryzae (46), Xanthomonas pathogens that cause cereal leaf streak (21), and X. axonopodis pv. manihotis (31). In the case of X. axonopodis pv. manihotis, a nested PCR (N-PCR) protocol was used to enhance the sensitivity and specificity of detection. In addition, multiplex PCR protocols have been developed in order to detect several pathogens (13) or genetically heterogeneous strains of a single pathovar (24, 56) simultaneously.
In this study we developed PCR primers specific to X. axonopodis pv. allii for sensitive and specific detection of all strains of this pathovar, and we evaluated the reliability of the method for detection of the pathogen in contaminated onion seed lots.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and DNA extraction.
The 86 X. axonopodis pv. allii strains used in this study and related information are shown in Table S1 in the supplemental material. Bacterial strains belonging to different Xanthomonas pathovars (60 strains) and species (25 strains) and to different genera (16 strains) and saprophytic bacteria isolated from onion (18 strains) were used for specificity analyses (see Table S2 in the supplemental material). The saprophytic bacteria were identified at the genus level using the Biolog system (6, 42). Strains were stored at −80°C on beads in cryovials (Microbank; Prolab Diagnostics, Austin, TX) and/or as lyophiles for long-term storage. Strains were routinely grown on YPGA (yeast extract, 7 g liter−1; peptone, 7 g liter−1; glucose, 7 g liter−1; agar, 18 g liter−1; pH 7.2) at 28°C. Strains that grew poorly on YPGA were cultivated on modified Wilbrink medium (40). One-day-old bacterial cultures were used for PCR assays and pathogenicity tests. DNA was extracted from 2-ml bacterial cultures (16 h of incubation with agitation at 28°C in YP broth [yeast extract, 7 g liter−1; peptone, 7 g liter−1; pH 7.2]) using a DNeasy blood and tissue kit (Qiagen, Courtaboeuf, France) and following the manufacturer's instructions. DNA concentrations were estimated by fluorometry (TKO 100 fluorometer; Hoefer, San Francisco, CA).
Selection of randomly amplified polymorphic DNA (RAPD) markers.
PCR assays were performed using a PE9600 cycler (Applied Biosystems, Courtaboeuf, France). Total DNA from 22 strains of X. axonopodis pv. allii isolated from various geographical locations and six other xanthomonads were used (see Tables S1 and S2 in the supplemental material). PCR amplification, extraction of fragments from agarose gels, and cloning were performed under conditions described elsewhere (38). Sequence data were obtained for X. axonopodis pv. allii strains CFBP 6364, CFBP 6366, CFBP 6369, and CFBP 6379 using the double-strand single-pass sequence method (Genome Express, Meylan, France) and universal primers T7 and SP6. Cloned DNA sequences were submitted as queries using the nucleotide BLAST program (BLASTN) and a nucleotide collection (nr/nt) available online (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi; accessed November 2009). In all experiments, sequence alignment and sequence analysis were performed using the Geneious software (v4.7; Biomatters Ltd., Auckland, New Zealand).
Selection of AFLP markers.
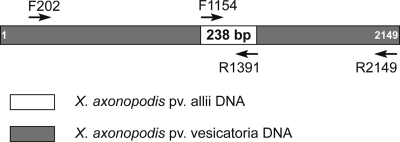
Preamplification and selective amplification of amplified fragment length polymorphism (AFLP) sequences were performed with a GeneAmp PCR system 9700 thermocycler (Applied Biosystems, Courtaboeuf, France). Preliminary data (33; I. Robène-Soustrade, unpublished data) allowed identification of putative specific markers of X. axonopodis pv. allii. In the present study, X. axonopodis pv. allii strains CFBP 6369, CFBP 6380, CFBP 6384, JX36-1, CFBP 6386, and CFBP 6382 representative of X. axonopodis pv. allii strains, which did not produce amplicons using the RAPD 80-21 primer, were used. One strain of X. citri pv. citri (IAPAR 306) and one strain of X. campestris pv. campestris (CFBP 5251) were used as controls. AFLP experiments were performed as previously described (2), except that unlabeled MspI+A, MspI+C, MspI+T, or MspI+G primer and the SacI+C or SacI+CT primer were used for selective amplification. All adaptors and AFLP primers (Applied Biosystems, Courtaboeuf, France) used are listed in Table 1. After migration on a 5% acrylamide gel as described elsewhere (37), DNA was visualized using a silver staining method (34). Potentially interesting AFLP fragments were removed from acrylamide gels using a scalpel and incubated at 60°C for 2 h in 50 μl 1× Goldstar Red Taq polymerase buffer (Eurogentec, Seraing, Belgium). Five microliters was used as the template for amplification with the corresponding unlabeled selective MspI+N/SacI+CT primer pair under selective amplification conditions, as mentioned above. The DNA fragments were cloned and sequenced as described above. The F1154 and R1391 primers (Table 1) were designed using a 270-bp fragment isolated from a CFBP 6382 gel and were used for PCR amplification with 20 ng of total DNA from strains CFBP 6366, CFBP 6380, CFBP 6384, JX36-1, CFBP 6386, CFBP 6357, and CFBP 6382 as follows. PCRs were performed with 25-μl reaction mixtures containing 3 mM MgCl2, 100 μM each deoxynucleoside triphosphate (dNTP), 0.2 μM each primer, 1.25 U Dap Goldstar polymerase (Eurogentec, Seraing, Belgium), and 0.5 μl template DNA in 10× opti-buffer. The amplification program included denaturation at 94°C for 5 min, 40 cycles consisting of denaturation at 95°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 2 min, and a final extension step at 72°C for 5 min. The 238-bp amplicons obtained were cloned and sequenced for all strains as described above. DNA sequences were submitted as queries using the BLAST nucleotide program (BLASTN) as described above. PCR amplification was performed again for the same strains under the same PCR conditions except for the annealing temperature (65°C instead of 60°C) using two sets of primers: primers F1154 and R2149 and primers F202 and R1391. These primers were designed using either the sequence of the 238-bp amplicon or the avrRxv gene sequence of X. axonopodis pv. vesicatoria, as shown in Fig. 1. Amplicons were cloned and sequenced as described above, and the sequences were submitted as queries using the BLAST nucleotide program (BLASTN) as described above.
TABLE 1.
Primers and adaptors used in this study
| Adaptor or primer | Sequence (5′ → 3′) |
|---|---|
| AFLP | |
| Ligation adaptors | |
| A1MSP | GACGATGAGTCCTGAG |
| A2MSP | CGCTCAGGACTCATC |
| A1SAC | CTCGTAGACTGCGTACAAGCT |
| A2SAC | TGTACGCAGTCTACG |
| Preamplification primers | |
| PMSP | GATGAGTCCTGAGCGG |
| PSAC | TAGACTGCGTACAAGCTC |
| Selective unlabeled amplification primers | |
| SacI+C | TAGACTGCGTACAAGCTCC |
| SacI+CT | TAGACTGCGTACAAGCTCCT |
| MspI + 1 selective nucleotide | GATGAGTCCTGAGCGG plus A, C, T, or G |
| Primers for amplification of avrRxv | |
| F1154 | GACCATATGCCTGGTTTCG |
| R1391 | CCTTAAGGTGCTGACTTTCG |
| F202 | ATTATCCGCGCATTGTCG |
| R2149 | GTTGACGGATCTGGCGTTG |
| Primers for multiplex nested PCR | |
| Pxaa1U | GGCTCTAATACGACGTTGACGAT |
| Pxaa1L | AAATTCATGCGCGTTTTCAATAG |
| Pxaa2U | CTCAAGCAGCAGTCGTTTTCA |
| Pxaa2L | ATGCTTCGATTGACATGCTGT |
| Nxaa1U | TTACGTCGCAAACAATCCAGATA |
| Nxaa1L | GGGCACCATTGACATTATCAGTT |
| Nxaa2U | ATGCCTGGTTTCGTGAA |
| Nxaa2L | CTACGGCTCAGCGACTC |
FIG. 1.
Amplification of a DNA fragment similar to the avrRxv gene in X. axonopodis pv. allii. The arrows indicate the positions and directions of priming of the primers chosen in the X. axonopodis pv. vesicatoria avrRxv gene and the X. axonopodis pv. allii AFLP fragment.
Multiplex PCR and multiplex nested PCR.
PCRs were performed with a GeneAmp PCR system 9700 thermocycler (Applied Biosystems, Courtaboeuf, France). Different templates were tested, including purified bacterial genomic DNA (20 ng μl−1), bacterial suspensions from cultures (a single colony in 1 ml of sterile deionized water, boiled for 2 min and chilled on ice), and seed extracts prepared as described below.
Specific primers were designed with Primer3 software (44) and were synthesized by Genecust (Evry, France). The sequences of the primers used for the first and second rounds of amplification are shown in Table 1. The first round of PCR was performed using 25-μl reaction mixtures containing 3 mM MgCl2, 100 μM each dNTP, 0.2 μM primer Pxaa1U, 0.2 μM primer Pxaa1L, 0.2 μM primer Pxaa2U, 0.2 μM primer Pxaa2L, 1.25 U Goldstar Red Taq polymerase (Eurogentec, Seraing, Belgium), and 0.5 to 5 μl template DNA (see below) in 75 mM Tris-HCl-20 mM (NH4)2SO4-0.01% Tween 20 buffer (pH 8.8). The amplification program included denaturation at 94°C for 5 min, 40 cycles consisting of denaturation at 95°C for 1 min, annealing at 63°C for 1 min, and extension at 72°C for 2 min, and a final extension step at 72°C for 5 min. The second round of PCR was performed with 1 to 5 μl of amplicons obtained from the first DNA reaction (see below) in 25-μl mixtures as described above for the first round, except that 0.2 μM primer Nxaa1U, 0.2 μM primer Nxaa1L, 0.2 μM primer Nxaa2U, and 0.2 μM primer Nxaa2L were used. The amplification program consisted of denaturation at 95°C for 5 min, followed by 30 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 40 s and a final extension at 72°C for 5 min. PCR products were separated by electrophoresis in 1% (amplicons from the first-round PCR) or 3% (amplicons from the nested PCR) Seakem LE agarose (FMC Bioproducts, Philadelphia, PA) and stained with ethidium bromide.
Specificity and sensitivity of multiplex nested PCR.
DNA fragments amplified with the Pxaa1U/Pxaa1L primer pair were sequenced for 27 X. axonopodis pv. allii strains (see Table S1 in the supplemental material) and 18 strains identified as members of other Xanthomonas pathovars (see Table S2 in the supplemental material). The fragments amplified with the Pxaa2U and Pxaa2L primers were sequenced for 28 X. axonopodis pv. allii strains (see Table S1 in the supplemental material) and 17 strains identified as members of other Xanthomonas pathovars (see Table S2 in the supplemental material). For these experiments, PCRs were performed as described above, except that Dap Goldstar polymerase (Eurogentec, Seraing, Belgium) was used, as described above, with bacterial suspensions as the templates. Amplicons were sequenced using Macrogen (Seoul, South Korea), as described previously. All strains listed in Tables S1 and S2 in the supplemental material were tested with the multiplex nested PCR assay (see above) using 0.5-μl bacterial suspensions as the templates. Amplicons were diluted 1/100 in deionized water for the second round of PCR to prevent inhibition of the PCR due to high template DNA concentrations.
The sensitivity of the multiplex nested PCR was determined using bacterial dilution series, one-half of which were mixed with seed samples. Suspensions prepared from overnight cultures on YPGA plates of X. axonopodis pv. allii strains CFBP 6385, CFBP 6366, and CFBP 6367 were adjusted spectrophotometrically to obtain a concentration of 1 × 108 CFU ml−1 (optical density at 600 nm [OD600], 0.05) and serially 10-fold diluted in 0.01 M sterile Sigma 7-9 buffer (pH 7.2; Sigma, Saint-Quentin Fallavier, France). Samples containing 10 g of healthy onion seeds (A. cepa cv. Véronique, harvested from plants growing in an area free of X. axonopodis pv. allii) were soaked in 50 ml of 0.01 M sterile Sigma 7-9 buffer (pH 7.2) and inoculated with bacterial suspensions at final concentrations ranging from 1 × 100 CFU ml−1 to 1 × 107 CFU ml−1. Negative controls were inoculated with 0.01 M sterile Sigma 7-9 buffer (pH 7.2). Similar dilution series not mixed with seed samples were analyzed concomitantly. After 48 h of maceration at 4°C, samples were plated in duplicate on NCTM1 semiselective medium (42) with a spiral device (Interscience, Saint Nom La Bretèche, France), and bacterial genomic DNA was extracted using a quick alkaline DNA extraction method (5). For each experiment, two (for suspensions containing ≥1 × 104 CFU ml−1) or three (for suspensions containing <1 × 104 CFU ml−1) aliquots (5 μl each) were used as templates. The experiment was replicated once.
Detection of X. axonopodis pv. allii in seed samples collected from a diseased field.
A seed lot collected from onion plants (A. cepa cv. Véronique) growing in experimental diseased plot P2 (15) was checked for the presence of X. axonopodis pv. allii. The number of samples (35 samples per experiment, each consisting of 10 g of seeds) was determined based on the hypergeometric distribution in order to detect at least one contaminated seed in 30,000 seeds (P = 0.05). The experiment was replicated once. Seed samples were soaked in 50 ml of 0.01 M sterile Sigma 7-9 buffer (pH 7.2; Sigma, Saint-Quentin Fallavier, France) for 48 h. Undiluted macerates and macerates diluted 1:10 were plated on the semiselective NCTM1 medium with a spiral device (Interscience, Saint Nom La Bretèche, France). All macerates were analyzed at least twice by multiplex nested PCR after alkaline DNA extraction, as described above. Negative controls in which water was used as the template were included in all experiments. Nested PCR is sometimes known to produce false-positive results. Therefore, only samples in which the expected DNA fragments were detected at least twice were scored as positive. Multiplex nested PCR was also used to confirm the identity of xanthomonad-like colonies recovered from seed macerates on the semiselective medium. Seed contamination rates derived from plate counts and from multiplex nested PCR analyses were calculated as described elsewhere (22). The same seed lot was mixed with healthy onion seeds (A. cepa cv. Véronique) at ratios of 1:1.1, 1:2.0, and 1:12.8, and two, two, and three independent samples, respectively, from these mixtures were analyzed as described above.
The exact distribution of positive samples in 35 samples for a contamination rate of 0.01% was calculated using Bayes' formula and the binomial distribution of the number of positive seeds. The correlation between the contamination rates calculated from plate counts or in multiplex nested PCR assays and the dilution ratios of infected seeds to healthy seeds was analyzed with the Pearson's product moment correlation coefficient (32). All statistical analyses were performed using R statistical software (version 2.8.1; R Development Core Team, Vienna, Austria).
Pathogenicity tests.
The pathogenicity of all strains of X. axonopodis pv. allii (see Table S1 in the supplemental material) was confirmed by verifying Koch's postulates using the host species from which the strain originated. The pathogenicity of some genetically related strains not assigned to this pathovar (see Table S2 in the supplemental material) was checked using onion cultivar Red Creole as described previously (42).
Nucleotide sequence accession numbers.
The DNA sequences of the AFLP-derived fragment (1,948 bp) and the 80-21 RAPD fragment (987 bp), both obtained from strain CFBP6366, have been deposited in the GenBank database under accession numbers GU084403 and GU084404, respectively. The GenBank accessions for the DNA sequences amplified with primers Pxaa1U and Pxaa1L and with primers Pxaa2U and Pxaa2L are GU736580 to GU736624 and GU736625 to GU736669, respectively.
RESULTS
Selection of RAPD and AFLP markers specific for X. axonopodis pv. allii.
None of the RAPD markers tested were present in all strains of X. axonopodis pv. allii, nor were they present in strains classified as members of other pathovars. Nevertheless, several fragments were amplified from most X. axonopodis pv. allii strains and were not amplified from any unrelated bacterial strains except X. axonopodis pv. vesicatoria strains. The 80-21 primer (5′ACGCGCCAGG) produced an approximately 940-bp amplicon for 70% of the X. axonopodis pv. allii strains tested. DNA sequences derived from this fragment from strains CFBP 6364, CFBP 6366, CFBP 6369, and CFBP 6379 were highly similar (99% identity over 937 bp). Comparison of the target amplified sequence to sequences stored in the GenBank database (BLASTN) revealed a high level of similarity (98% over 933 bp) with two portions of contiguous genes (831 bp and 102 bp) encoding the type IV pilus assembly proteins PilW and PilX of X. axonopodis pv. vesicatoria, respectively. Other RAPD markers were analyzed in the same way, although no other RAPD marker was shared by the strains not amplified when the 80-21 primer was used.
Two AFLP fragments present in a majority of X. axonopodis pv. allii strains, including the strains for which no amplicons were obtained with the 80-21 RAPD primer, were identified. Due to the complexity of the AFLP profiles obtained when two selective bases were used in the selective amplification step, only simplified AFLP profiles using three selective bases were used to recover the target DNA fragments from gels. Even under these more stringent conditions, DNA sequences ligated into pGEM vectors suggested that contamination during excision of the fragments or comigration of amplification products that were the same size may have occurred. Interestingly, a clone obtained in the AFLP analysis using three selective nucleotides was obtained from strain CFBP 6382. This clone had a 270-bp sequence showing 90% similarity to a portion of the avrRxv avirulence gene of X. axonopodis pv. vesicatoria (GenBank accession number L20423). This fragment was amplified in all strains analyzed (88 to 100% identity with avrRxv for 238-bp amplicons depending on the strain). Primers used for PCR amplification and sequencing were designed using the combined chimeric sequence of this fragment and the sequence of avrRxv from X. axonopodis pv. vesicatoria. Using these primers, DNA sequences covering 1,948 bp were obtained for X. axonopodis pv. allii strains CFBP6357, CFBP 6366, CFBP 6382, and CFBP 6386 and DNA sequences covering 1,950 bp were obtained for strains CFBP 6384 and JX36-1. For strain CFBP 6380, a 1,504-bp partial sequence was obtained. All these sequences matched the avrRxv sequence from X. axonopodis pv. vesicatoria with levels of similarity ranging from 87 to 99%.
Specificity and sensitivity of the multiplex nested PCR assay.
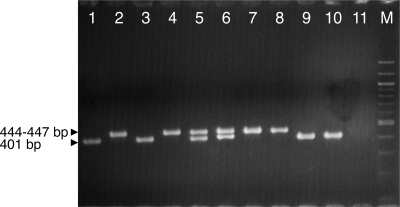
A multiplex nested PCR assay based on the two described DNA markers (referred to below as PIL and AVR) was performed by selecting compatible external and internal primers from the most conserved regions of sequences recovered for X. axonopodis pv. allii. The theoretical sizes of AVR amplicons were 995 bp (first round) and 401 bp (nested round) for all sequences analyzed. For the PIL marker, theoretical sizes of the amplicons were 697 bp (first round) and 447 bp (nested round) for all sequences analyzed except the strain CFBP 6364 sequence, which had a 3-bp deletion (694-bp and 444-bp amplicons). When the multiplex PCR assay was performed with all X. axonopodis pv. allii strains (n = 86), three categories of responses were observed on gels after the first round of amplification. Two amplicons of the expected sizes, about 700 bp (corresponding to the PIL marker) and 990 bp (corresponding to the AVR marker), were obtained for all strains from Cuba (n = 2), Japan (n = 2), Mauritius (n = 15), Réunion Island (n = 17), and Hawaii (n = 13) and for four strains from Brazil. One 700-bp amplicon (PIL) was amplified from all strains from Georgia (n = 3) and Venezuela (n = 7), from four strains from Colorado, and from two strains from South Africa. One 990-bp amplicon (AVR) was observed for all strains from Barbados (n = 5) and Texas (n = 7), three strains from South Africa, one strain from Brazil, and one strain from Colorado. In the second round of PCR, the corresponding internal amplicons were observed for the different strains, and they included two DNA fragments that were approximately 450 bp (PIL) and 400 bp (AVR) long for the first group of strains and single 450-bp and 400-bp amplicons for the second and third groups of strains, respectively (Fig. 2; see Table S1 in the supplemental material).
FIG. 2.
Multiplex nested PCR detection of X. axonopodis pv. allii strains. Lanes 1 to 10, strains CFBP 6384, CFBP 6385, CFBP 6380, CFBP 6379, CFBP 6369, CFBP 6107, JY 276, CFBP 6387, CFBP 6368, and CFBP 6382, respectively; lane 11 negative PCR control; lane M, 100-bp ladder (Invitrogen, Merelbeke, Belgium).
No amplification product was observed when the multiplex PCR assay was performed for saprophytic strains isolated from onion or for bacteria belonging to other bacterial genera (see Table S2 in the supplemental material). Most Xanthomonas strains also did not produce amplicons, except for a few strains belonging to X. axonopodis genetic group 9.2 sensu Rademaker et al., X. axonopodis pv. vesicatoria, X. axonopodis pv. citrumelo, X. axonopodis pv. cassavae, X. axonopodis pv. desmodii, X. axonopodis pv. desmodiigangetici, X. axonopodis pv. phyllanthi, X. axonopodis pv. tamarindi, and X. axonopodis pv. lespedezae. Depending on the pathovar, one or both approximately 700-bp and 990-bp DNA fragments (and the corresponding nested 450-bp and 400-bp amplicons) were observed. Amplicons were also observed for the AVR marker for strains of X. axonopodis pv. begoniae (genetic group 9.1 sensu Rademaker et al.). Pathogenicity tests showed that none of the strains positive for one or both markers in the multiplex assay were pathogenic to onion.
Sequencing of the amplicons from the first round of PCR confirmed the theoretical sizes of nested amplicons and showed that for the PIL marker the same 3-bp deletion occurred in the strains that originated from Cuba (n = 2) and Japan (n = 2) and in all of the strains belonging to X. axonopodis pv. vesicatoria. For strain LMG 955 a 3-bp insertion was found at the same site (450-bp nested amplicon). When these sequences were compared, specific signature sequences permitted us to distinguish X. axonopodis pv. allii from all nontarget strains except X. axonopodis pv. lespedezae strain NCPPB 938 (see the GenBank sequences deposited under accession numbers GU736580 to GU736624 and GU736625 to GU736669). Enzymatic restriction signatures that allowed us to distinguish X. axonopodis pv. allii from X. axonopodis pv. vesicatoria and X. axonopodis pv. begoniae were found (data not shown).
The first round of PCR performed with dilution series of strains CFBP 6366, CFBP 6367, and CFBP 6385 mixed with seed samples resulted in a detection sensitivity of 1 × 106 CFU ml−1, as determined by dilution plating on YPGA and NCTM1 medium. When multiplex nested PCR was used, the detection limit was 1 × 103 CFU ml−1. A signal was often obtained (for one or two of three replicates) when suspensions containing 1 × 102 CFU ml−1 were used as templates. The sensitivity of the assay was the same when the same dilution series that were not mixed with seed samples were tested.
Detection of X. axonopodis pv. allii in seed samples collected from a diseased field.
X. axonopodis pv. allii was detected in both experiments using the multiplex nested PCR assay with six and nine positive samples (Table 2). The values obtained corresponded to an average contamination rate (CR) of 0.01%. When calculating the exact distribution of positive samples for 35 samples for a CR of 0.01%, we found dispersion between 3 and 11 positive samples at a probability level of 96.6% and dispersion between 4 and 10 positive samples at a probability level of 89.6%, and the mean number of positive lots was 6.76. These data were consistent with the observed values for positive samples. Meanwhile, the pathogen was recovered on semiselective NCTM1 medium from four and one samples with an average CR of 0.0031%. Recovery of target bacterial colonies on NCTM1 medium proved to be difficult because of high population densities of saprophytic bacteria (>1 × 106 CFU per g of seeds) that were associated with onion seeds and rapidly colonized agar plates. No amplicon was obtained and no colony of X. axonopodis pv. allii was isolated on NCTM1 medium for two healthy seed lots analyzed as negative controls. The assay that was developed accurately estimated seed contamination rates, and there was a positive correlation (r = 0.90) between “theoretical” (as determined by seed dilution ratios) and experimentally determined seed contamination rates calculated from PCR data. In contrast, such a correlation could not be demonstrated when contamination rates derived from plate counts were used. At an experimentally determined contamination rate of 0.0037%, corresponding to one infected seed in 27,340 seeds, the multiplex nested PCR assay allowed detection of the bacterium in each nested PCR replicate. At a higher dilution (CR, 0.0012%; i.e., one contaminated seed in 82,000 seeds), only one of three replicates was positive.
TABLE 2.
Contamination rates and numbers of positive samples detected using seed macerates of infected seed lots
| Seeds | Assay | No. of seed samples | Multiplex nested PCR detection |
Semiselective isolation |
No. of lots detected by both methods | ||
|---|---|---|---|---|---|---|---|
| CR (%) | No. of positive lots | CR (%) | No. of positive lots | ||||
| Seed lot P2 (CR, 1/10,000)a | Assay 1 | 35 | 0.000077 | 6 | 0.000050 | 4 | 3 |
| Assay 2 | 35 | 0.000121 | 9 | 0.000012 | 1 | 1 | |
| Mean | 0.000099 | 0.000031 | |||||
| Seed lot P2 mixed with healthy seeds (1:1.1) (CR, 1/21,000)b | Assay 1 | 35 | 0.000050 | 4 | 0.000000 | 0 | 0 |
| Assay 2 | 35 | 0.000037 | 3 | 0.000024 | 2 | 0 | |
| Mean | 0.000043 | 0.000012 | |||||
| Seed lot P2 mixed with healthy seeds (1:2) (CR, 1/30,000)b | Assay 1 | 35 | 0.000037 | 3 | 0.000000 | 0 | 0 |
| Assay 2 | 35 | 0.000037 | 3 | 0,000012 | 1 | 0 | |
| Mean | 0.000037 | 0.000006 | |||||
| Seed lot P2 mixed with healthy seeds (1:12.8) (CR, 1/138.000)b | Assay 1 | 35 | 0.000000 | 0 | 0.000012 | 1 | 0 |
| Assay 2 | 35 | 0.000000 | 0 | 0.000000 | 0 | 0 | |
| Assay 3 | 35 | 0.000037 | 3 | 0.000012 | 1 | 0 | |
| Mean | 0.000012 | 0.000008 | |||||
| Healthy seeds | Assay 1 | 35 | 0.000000 | 0 | 0.000000 | 0 | |
| Assay 2 | 35 | 0.000000 | 0 | 0.000000 | 0 | ||
The seed contamination rate was determined by multiplex nested PCR analyses.
The theoretical seed contamination rate was obtained by diluting seed lot P2 with healthy seeds.
DISCUSSION
A multiplex nested PCR was developed for specific and sensitive detection of X. axonopodis pv. allii from onion seeds. Due to the high genetic diversity of this pathogen (11, 12, 33, 41), two PCR-based sequence-characterized amplified regions (SCARs) identified in RAPD and AFLP analyses were required to detect all isolates of this pathovar. The RAPD technique has been widely used for SCAR development to obtain PCR diagnostic tools for bacterial plant pathogens (23, 38, 54). Examples of SCARs derived from AFLP analysis are less common in molecular phytodiagnostics (10, 36), whereas this technique is capable of generating a large number of markers over a relatively short time without any prior sequence knowledge (48). In order to convert AFLP markers into SCAR markers, we reduced the complexity of AFLP fingerprints by increasing the number of selective bases. In spite of this precaution, the recovered DNAs were heterogeneous in terms of fragment composition (data not shown), which was probably the result of coisolation of amplification products that were of similar sizes, a situation previously documented for AFLP analysis (7). Nevertheless, here we succeeded in isolating an AFLP fragment of interest.
The target sequence identified by RAPD-PCR analyses displayed a high level of nucleotide similarity with sequences encoding the PilW and PilX proteins of X. axonopodis pv. vesicatoria. These proteins are involved in assembly of type IV pili (fimbriae), as demonstrated for Pseudomonas aeruginosa (3). Fimbriae are present in plant-pathogenic bacteria. They are involved in host colonization, plant cell adhesion, cell aggregate formation, or twitching motility. They are, therefore, important components of plant-bacterium interactions (9, 18, 19, 30). The target sequence identified in the AFLP analysis showed a high level of nucleotide similarity with the avrRxv avirulence gene of X. axonopodis pv. vesicatoria (57). This gene encodes a type III effector protein belonging to the AvrRxv/YopJ (C55) family of proteins, which presumably function as proteases and modulate the host response (14, 26). Thus, the two regions targeted in our multiplex PCR assay are potentially involved in pathogenic interactions with host plants.
The internal primers of the multiplex PCR assay directed amplification for all strains of X. axonopodis pv. allii. Depending on the strain, one or two fragments were amplified. No amplification product was obtained for taxonomically unrelated bacteria (41 strains) or for saprophytic bacteria commonly isolated from onion leaves and seeds (14 strains). Amplicons were also obtained for some strains that are not classified in X. axonopodis pv. allii but belong to X. axonopodis genetic subgroup 9.1 or 9.2 sensu Rademaker et al. (35). Some of the pathovars are well documented as they cause diseases of economic concern (e.g., X. axonopodis pv. vesicatoria, X. axonopodis pv. citrumelo, X. axonopodis pv. cassavae, and X. axonopodis pv. begoniae), whereas no data or little data are available for the other pathovars. There have been no reports of detection of Xanthomonas pathovars other than X. axonopodis pv. allii in onion seeds. The probability of finding these pathovars on onion plants or seeds is negligible because the capacity to induce symptoms on onion plants is specific to X. axonopodis pv. allii strains, as demonstrated by inoculation of onion plants with different pathovars belonging to group 9.2 (11). In this study, we also verified that all strains not classified as X. axonopodis pv. allii that displayed a positive PCR signal were not pathogenic to onion. As these bacteria cannot multiply over a long period of time if they are in contact with onion tissue, it is very unlikely that they would be detected on onion plants and consequently in seeds. Thus, this molecular tool can be routinely used to specifically evaluate rates of seed contamination by X. axonopodis pv. allii. Nevertheless, the presence of signature sequences for most of the strains can be used to differentiate the other pathovars from X. axonopodis pv. allii if a false-positive signal is suspected. If necessary, AFLP or multilocus sequence analysis methods can also be used to confirm the diagnosis after isolation of the bacteria (8).
The PCR-based seed assay detected X. axonopodis pv. allii in artificially inoculated seed samples with contamination levels ranging from 5 × 102 to 5 × 107 CFU g−1. It is likely that the alkaline extraction procedure that was used prior to PCR concentrated the DNA and neutralized PCR inhibitors in onion seed extracts, as suggested by preliminary experiments (data not shown). The rate of contamination of the seed lot was estimated to be 0.01% by the multiplex nested PCR assay, consistent with previous data (15, 42). This rate is similar to those reported previously for other pathosystems involving xanthomonads (1). The sensitivity threshold of the multiplex nested PCR assay was found to be 1/27,340 when it was determined with the same naturally contaminated seed lot mixed with healthy seeds. This level of sensitivity is greater than the contamination rate reported for BBO outbreaks in a tropical environment (4.5/10,000) (43). This threshold is also consistent with the tolerance standards currently recommended in the International Rules of Seed Health Testing for phytopathogenic bacteria and with a lower threshold recently defined for X. campestris pv. campestris for avoiding spread among transplants in seed trays (39).
Our new multiplex nested PCR assay is less labor-intensive and time-consuming than isolation on semiselective media. The latter technique requires further evaluation of suspect colonies using other identification methods, including serological or molecular techniques and/or pathogenicity tests.
When used with naturally infected seed samples, the PCR assay was found to detect more positive samples than the traditional plating method. This was often due to high population densities of microbiota present on seeds, which were able to grow on the semiselective medium and inhibited the development of the target bacterium (42). Alternatively, it is possible that our PCR-based method detects free DNA from nonviable bacteria, as demonstrated for PCR detection of X. campestris pv. carotae (23), and thus results in false-positive responses. Nevertheless, detection of free target DNA in a seed lot represents a history of contamination, which indicates that there is a need for further analysis. We plan to compare the performance of biological enrichment followed by PCR (BIO-PCR) (49) and ethidium monoazide PCR (EMA-PCR) (27, 45) in terms of their capacities to determine the rates of occurrence of viable target cells in seed samples. We concluded that the multiplex nested PCR assay described here is a reliable and sensitive procedure for detecting and identifying X. axonopodis pv. allii, the BBO pathogen, in onion seeds and that it should be useful for international sanitary surveillance of seed exchanges. Additionally, this molecular tool could be useful for ecological and epidemiological investigations, especially investigations to determine the relationship between seed contamination and disease incidence or to understand better the life cycle of X. axonopodis pv. allii and assess the relative importance of the different stages of dissemination and conservation of this pathogen. A French patent concerning this detection technique and the target sequences is pending.
Supplementary Material
Acknowledgments
This work was funded by grants from the European Union (FEOGA), the Conseil Régional de La Réunion, and CIRAD.
We thank Philippe Roumagnac, Michel Roux-Cuvelier, and Emmanuel Jouen for helpful discussions; Christian Vernière and Philippe Roumagnac for reviewing the manuscript; and Anna Granet, Philippe Chiroleu, and Walter Grondin for technical assistance.
Footnotes
Published ahead of print on 5 March 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Agarwal, V. K., and J. B. Sinclair. 1997. Principles of seed pathology, 2nd ed. CRC Lewis Publishers, Boca Raton, FL.
- 2.Ah-You, N., L. Gagnevin, F. Chiroleu, E. Jouen, J. R. Neto, and O. Pruvost. 2007. Pathological variations within Xanthomonas campestris pv. mangiferaeindicae support its separation into three distinct pathovars that can be distinguished by amplified fragment length polymorphism. Phytopathology 97:1568-1577. [DOI] [PubMed] [Google Scholar]
- 3.Alm, R. A., J. P. Hallinan, A. A. Watson, and J. S. Mattick. 1996. Fimbrial biogenesis genes of Pseudomonas aeruginosa: pilW and pilX increase the similarity of type 4 fimbriae to the GSP protein-secretion systems and pWI encodes a gonococcal PilC homologue. Mol. Microbiol. 22:161-173. [DOI] [PubMed] [Google Scholar]
- 4.Alvarez, A. M., I. W. Buddenhagen, E. S. Buddenhagen, and H. Y. Domen. 1978. Bacterial blight of onion, a new disease caused by Xanthomonas sp. Phytopathology 68:1132-1136. [Google Scholar]
- 5.Audy, P., C. E. Braat, G. Saindon, H. C. Huang, and A. Laroche. 1996. A rapid and sensitive PCR-based assay for concurrent detection of bacteria causing common and halo blights in bean seed. Phytopathology 86:361-366. [Google Scholar]
- 6.Bochner, B. R. 1989. Sleuthing out bacterial identities. Nature 339:157-158. [DOI] [PubMed] [Google Scholar]
- 7.Brugmans, B., G. M. Van der Hulst, R. G. F. Visser, P. Lindhout, and J. Van Eck. 2003. A new and versatile method for the successful conversion of AFLP markers into simple single locus markers. Nucleic Acids Res. 31:e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bui Thi Ngoc, L., C. Vernière, E. Jouen, N. Ah-You, P. Lefeuvre, F. Chiroleu, L. Gagnevin, and O. Pruvost. 2010. Amplified fragment length polymorphism and multilocus sequence analysis-based genotypic relatedness among pathogenic variants of Xanthomonas citri pv. citri and Xanthomonas campestris pv. bilvae. Int. J. Syst. Evol. Microbiol. 60:515-525. [DOI] [PubMed] [Google Scholar]
- 9.Cao, H., R. L. Baldini, and L. G. Rahme. 2001. Common mechanisms for pathogens of plants and animals. Annu. Rev. Phytopathol. 39:259-284. [DOI] [PubMed] [Google Scholar]
- 10.Esquibet, M., E. Grenier, O. Plantard, F. A. Andaloussi, and G. Caubel. 2003. DNA polymorphism in the stem nematode Ditylenchus dipsaci: development of diagnostic markers for normal and giant races. Genome 46:1077-1083. [DOI] [PubMed] [Google Scholar]
- 11.Gent, D. H., A. Al-Saadi, D. W. Gabriel, F. J. Louws, C. A. Ishimaru, and H. F. Schwartz. 2005. Pathogenic and genetic relatedness among Xanthomonas axonopodis pv. allii and other pathovars of X. axonopodis. Phytopathology 95:918-925. [DOI] [PubMed] [Google Scholar]
- 12.Gent, D. H., H. F. Schwartz, C. A. Ishimaru, F. J. Louws, R. A. Cramer, and C. B. Lawrence. 2004. Polyphasic characterization of Xanthomonas strains from onion. Phytopathology 94:184-195. [DOI] [PubMed] [Google Scholar]
- 13.Glick, D. L., C. M. Coffey, and M. A. Sulzinski. 2002. Simultaneous PCR detection of the two major bacterial pathogens of geranium. J. Phytopathol. 150:54-59. [Google Scholar]
- 14.Gürlebeck, D., F. Thieme, and U. Bonas. 2006. Type III effector proteins from the plant pathogen Xanthomonas and their role in the interaction with the host plant. J. Plant Physiol. 163:233-255. [DOI] [PubMed] [Google Scholar]
- 15.Humeau, L., P. Roumagnac, Y. Picard, I. Robène-Soustrade, F. Chiroleu, L. Gagnevin, and O. Pruvost. 2006. Quantitative and molecular epidemiology of bacterial blight of onion in seed production fields. Phytopathology 96:1345-1354. [DOI] [PubMed] [Google Scholar]
- 16.Isakeit, T., M. E. Miller, L. W. Barnes, E. R. Dickstein, and J. B. Jones. 2000. First report of leaf blight of onion caused by Xanthomonas campestris in the continental United States. Plant Dis. 84:201. [DOI] [PubMed] [Google Scholar]
- 17.Kadota, I., K. Uehara, H. Shinohara, and K. Nishiyama. 2000. Bacterial blight of Welsh onion: a new disease caused by Xanthomonas campestris pv. allii pv. nov. J. Gen. Plant Pathol. 66:310-315. [Google Scholar]
- 18.Li, Y., G. Hao, C. D. Galvani, Y. Meng, L. De La Fuente, H. C. Hoch, and T. J. Burr. 2007. Type I and type IV pili of Xylella fastidiosa affect twitching motility, biofilm formation and cell-cell aggregation. Microbiology 153:719-726. [DOI] [PubMed] [Google Scholar]
- 19.Liu, H. L., Y. W. Kang, S. Genin, M. A. Schell, and T. P. Denny. 2001. Twitching motility of Ralstonia solanacearum requires a type IV pilus system. Microbiology 147:3215-3229. [DOI] [PubMed] [Google Scholar]
- 20.Maddox, D. A. 1998. Implications of new technologies for seed health testing and the worldwide movement of seed. Seed Sci. Res. 8:277-284. [Google Scholar]
- 21.Maes, M., P. Garbeva, and O. Kamoen. 1996. Recognition and detection in seed of the Xanthomonas pathogens that cause cereal leaf streak using rDNA spacer sequences and polymerase chain reaction. Phytopathology 86:63-69. [Google Scholar]
- 22.Masmoudi, K., C. Duby, M. Suhas, J. Q. Guo, J. D. Taylor, and Y. Maury. 1994. Quality control of pea seed for pea seed-borne mosaic virus. Seed Sci. Technol. 22:407-414. [Google Scholar]
- 23.Meng, X. Q., K. C. Umesh, R. M. Davis, and R. L. Gilbertson. 2004. Development of PCR-based assays for detecting Xanthomonas campestris pv. carotae, the carrot bacterial leaf blight pathogen, from different substrates. Plant Dis. 88:1226-1234. [DOI] [PubMed] [Google Scholar]
- 24.Motiwala, A. S., M. Strother, A. Amonsin, B. Byrum, S. A. Naser, J. R. Stabel, W. P. Shulaw, J. P. Bannantine, V. Kapur, and S. Sreevatsan. 2003. Molecular epidemiology of Mycobacterium avium subsp. paratuberculosis: evidence for limited strain diversity, strain sharing, and identification of unique targets for diagnosis. J. Clin. Microbiol. 41:2015-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neto, J. R., V. A. Malavolta, Jr., M. A. Cardelli, and C. Sinigaglia. 1987. Ocorrencia de uma nova doença bacteriana em cebola, no estado de Sao Paulo. Summa Phytopathol. 13:10. [Google Scholar]
- 26.Noel, L., F. Thieme, J. Gabler, D. Buttner, and U. Bonas. 2003. XopC and XopJ, two novel type III effector proteins from Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 185:7092-7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nogva, H. K., S. M. Drømtorp, H. Nissen, and K. Rudi. 2003. Ethidium monoazide for DNA-based differentiation of viable and dead bacteria by 5′-nuclease PCR. Biotechniques 34:804-813. [DOI] [PubMed] [Google Scholar]
- 28.Nunez, J. J., R. L. Gilbertson, X. Meng, and R. M. Davis. 2002. First report of Xanthomonas leaf blight of onion in California. Plant Dis. 86:330. [DOI] [PubMed] [Google Scholar]
- 29.O'Garro, L. W., and L. P. Paulraj. 1997. Onion leaf blight caused by Xanthomonas campestris: alternative hosts and resistant onion genotypes. Plant Dis. 81:978-982. [DOI] [PubMed] [Google Scholar]
- 30.Ojanen-Reuhs, T., N. Kalkkinen, B. Westerlund-Wikstrom, J. Van Doorn, K. Haahtela, E. L. Nurmiaho-Lassila, K. Wengelnik, U. Bonas, and T. K. Korhonen. 1997. Characterization of the fimA gene encoding bundle-forming fimbriae of the plant pathogen Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 179:1280-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ojeda, S., and V. Verdier. 2000. Detecting Xanthomonas axonopodis pv. manihotis in cassava true seeds by nested polymerase chain reaction assay. Can. J. Plant Pathol. 22:241-247. [Google Scholar]
- 32.Pearson, K. 1926. On the coefficient of racial likeness. Biometrika 18:105-117. [Google Scholar]
- 33.Picard, Y., P. Roumagnac, D. Legrand, L. Humeau, I. Robène-Soustrade, F. Chiroleu, L. Gagnevin, and O. Pruvost. 2008. Polyphasic characterization of Xanthomonas axonopodis pv. allii associated with outbreaks of bacterial blight on three Allium species in the Mascarene archipelago. Phytopathology 98:919-925. [DOI] [PubMed] [Google Scholar]
- 34.Qu, L., X. Li, G. Wu, and N. Yang. 2005. Efficient and sensitive method of DNA silver staining in polyacrylamide gels. Electrophoresis 26:99-101. [DOI] [PubMed] [Google Scholar]
- 35.Rademaker, J. L. W., F. J. Louws, M. H. Schultz, U. Rossbach, L. Vauterin, J. Swings, and F. J. De Bruijn. 2005. A comprehensive species to strain taxonomic framework for Xanthomonas. Phytopathology 95:1098-1111. [DOI] [PubMed] [Google Scholar]
- 36.Radisek, S., J. Jakse, and B. Javornik. 2004. Development of pathotype-specific SCAR markers for detection of Verticillium albo-atrum isolates from hop. Plant Dis. 88:1115-1122. [DOI] [PubMed] [Google Scholar]
- 37.Risterucci, A. M., L. Grivet, J. A. K. N′Goran, I. Pieretti, M. H. Flament, and C. Lanaud. 2000. A high-density linkage map of Theobroma cacao L. Theor. Appl. Genet. 101:948-955. [DOI] [PubMed] [Google Scholar]
- 38.Robene-Soustrade, I., P. Laurent, L. Gagnevin, E. Jouen, and O. Pruvost. 2006. Specific detection of Xanthomonas axonopodis pv. dieffenbachiae in anthurium (Anthurium andreanum) tissues by nested PCR. Appl. Environ. Microbiol. 72:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts, S. J., J. Brough, and P. J. Hunter. 2007. Modelling the spread of Xanthomonas campestris pv. campestris in module-raised brassica transplants. Plant Pathol. 56:391-401. [Google Scholar]
- 40.Rott, P., M. Chatenet, M. Granier, and P. Baudin. 1988. L'échaudure des feuilles de canne à sucre provoquée par Xanthomonas albilineans (Ashby) Dowson. II. Diagnostic et spectre d'hotes de l'agent pathogene en Afrique tropicale. Agron. Trop. 43:244-251. [Google Scholar]
- 41.Roumagnac, P., L. Gagnevin, L. Gardan, L. Sutra, C. Manceau, E. R. Dickstein, J. B. Jones, P. Rott, and O. Pruvost. 2004. Polyphasic characterization of xanthomonads isolated from onion, garlic and Welsh onion (Allium spp.) and their relatedness to different Xanthomonas species. Int. J. Syst. Evol. Microbiol. 54:15-24. [DOI] [PubMed] [Google Scholar]
- 42.Roumagnac, P., L. Gagnevin, and O. Pruvost. 2000. Detection of Xanthomonas sp., the causal agent of onion bacterial blight, in onion seeds using a newly developed semi-selective isolation medium. Eur. J. Plant Pathol. 106:867-877. [Google Scholar]
- 43.Roumagnac, P., O. Pruvost, F. Chiroleu, and G. Hughes. 2004. Spatial and temporal analyses of bacterial blight of onion caused by Xanthomonas axonopodis pv. allii. Phytopathology 94:138-146. [DOI] [PubMed] [Google Scholar]
- 44.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers, p. 365-386. In S. Krawetz and S. Misener (ed.), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ. [DOI] [PubMed]
- 45.Rudi, K., B. Moen, S. M. Drømtorp, and A. L. Holck. 2005. Use of ethidium monoazide and PCR in combination for quantification of viable and dead cells in complex samples. Appl. Environ. Microbiol. 71:1018-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakthivel, N., C. N. Mortensen, and S. B. Mathur. 2001. Detection of Xanthomonas oryzae pv. oryzae in artificially inoculated and naturally infected rice seeds and plants by molecular techniques. Appl. Microbiol. Biotechnol. 56:435-441. [DOI] [PubMed] [Google Scholar]
- 47.Sanders, F. H., D. B. Langston, Jr., J. H. Brock, R. D. Gitaitis, D. E. Curry, and R. L. Torrance. 2003. First report of a leaf blight of onion caused by Xanthomonas spp. in Georgia. Plant Dis. 87:749. [DOI] [PubMed] [Google Scholar]
- 48.Savelkoul, P. H. M., H. J. M. Aarts, J. De Haas, L. Dijkshoorn, B. Duim, M. Otsen, J. L. W. Rademaker, L. Schouls, and J. A. Lenstra. 1999. Amplified-fragment length polymorphism analysis: the state of an art. J. Clin. Microbiol. 37:3083-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schaad, N. W., Y. Berthier-Schaad, A. Sechler, and D. Knorr. 1999. Detection of Clavibacter michiganensis subsp. sepedonicus in potato tubers by BIO-PCR and an automated real-time fluorescence detection system. Plant Dis. 83:1095-1100. [DOI] [PubMed] [Google Scholar]
- 50.Schwartz, H. F., and K. Otto. 2000. First report of leaf blight of onion caused by Xanthomonas campestris in Colorado. Plant Dis. 84:922. [DOI] [PubMed] [Google Scholar]
- 51.Schwartz, H. F., K. L. Otto, and D. H. Gent. 2003. Relation of temperature and rainfall to development of Xanthomonas and Pantoea leaf blights of onion in Colorado. Plant Dis. 87:11-14. [DOI] [PubMed] [Google Scholar]
- 52.Serfontein, J. J. 2001. Xanthomonas blight of onion in South Africa. Plant Dis. 85:442. [DOI] [PubMed] [Google Scholar]
- 53.Stall, R. E., T. R. Gottwald, M. Koizumi, and N. C. Schaad. 1993. Ecology of plant pathogenic xanthomonads, p. 265-299. In J. G. Swings and E. L. Civerolo (ed.), Xanthomonas. Chapman & Hall, London, United Kingdom.
- 54.Trebaol, G., C. Manceau, Y. Tirilly, and S. Boury. 2001. Assessment of the genetic diversity among strains of Xanthomonas cynarae by randomly amplified polymorphic DNA analysis and development of specific characterized amplified regions for the rapid identification of X. cynarae. Appl. Environ. Microbiol. 67:3379-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trujillo, G., and Y. Hernandez. 1999. Identificacion de bacterias fitopatogenas en semillas de plantas cultivadas, p. 10. In Proceedings of the XVI Congreso Venezolano de Fitopatologia. Sociedad Venezolano de Fitopatologia, Barquisimeto, Venezuela.
- 56.Weller, S. A., J. G. Elphinstone, N. C. Smith, N. Boonham, and D. E. Stead. 2000. Detection of Ralstonia solanacearum strains with a quantitative, multiplex, real-time, fluorogenic PCR (TaqMan) assay. Appl. Environ. Microbiol. 66:2853-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whalen, M. C., J. F. Wang, F. M. Carland, M. E. Heiskell, D. Dahlbeck, G. V. Minsavage, J. B. Jones, J. W. Scott, R. E. Stall, and B. J. Staskawicz. 1993. Avirulence gene Avrrxv from Xanthomonas campestris pv. vesicatoria specifies resistance on tomato line Hawaii 7998. Mol. Plant-Microbe Interact. 6:616-627. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.