Abstract
The WalRK two-component system plays important roles in maintaining cell wall homeostasis and responding to antibiotic stress in low-GC Gram-positive bacteria. In the major human pathogen, Streptococcus pneumoniae, phosphorylated WalRSpn (VicR) response regulator positively controls the transcription of genes encoding the essential PcsB division protein and surface virulence factors. WalRSpn is phosphorylated by the WalKSpn (VicK) histidine kinase. Little is known about the signals sensed by WalK histidine kinases. To gain information about WalKSpn signal transduction, we performed a kinetic characterization of the WalRKSpn autophosphorylation, phosphoryltransferase, and phosphatase reactions. We were unable to purify soluble full-length WalKSpn. Consequently, these analyses were performed using two truncated versions of WalKSpn lacking its single transmembrane domain. The longer version (Δ35 amino acids) contained most of the HAMP domain and the PAS, DHp, and CA domains, whereas the shorter version (Δ195 amino acids) contained only the DHp and CA domains. The autophosphorylation kinetic parameters of Δ35 and Δ195 WalKSpn were similar [Km(ATP) ≈ 37 μM; kcat ≈ 0.10 min−1] and typical of those of other histidine kinases. The catalytic efficiency of the two versions of WalKSpn∼P were also similar in the phosphoryltransfer reaction to full-length WalRSpn. In contrast, absence of the HAMP-PAS domains significantly diminished the phosphatase activity of WalKSpn for WalRSpn∼P. Deletion and point mutations confirmed that optimal WalKSpn phosphatase activity depended on the PAS domain as well as residues in the DHp domain. In addition, these WalKSpn DHp domain and ΔPAS mutations led to attenuation of virulence in a murine pneumonia model.
The WalRK two-component regulatory system (TCS) plays pivotal roles in maintaining cell wall and surface homeostasis in low GC Gram-positive bacteria (14, 41, 93). Recent global transcription analyses suggest that it may also respond to cell wall stresses, such as antibiotic addition (22, 38, 68). In Bacillus and Staphylococcus species, both the WalR (YycF) response regulator and the WalK (YycG) histidine kinase are essential in that they cannot be depleted (20, 23, 53). In contrast, the WalR (VicR) response regulator of Streptococcus species is essential, whereas the WalK (VicK) histidine kinase is not essential under standard growth conditions, and the corresponding gene can be knocked out (18, 58, 72, 89). The WalRK TCS was initially discovered in Bacillus subtilis, where it was designated as YycFG (20), but it is widespread in other species, where it has other names. A recent proposal was made to unify this nomenclature (14, 15), and we refer to this TCS from S. pneumoniae as WalRKSpn, instead of VicRK as used previously (58, 72, 89).
WalRK regulons include genes that mediate peptidoglycan biosynthesis, cell division, and cell surface proteins (4, 9, 15, 54, 58, 59), but the specific genes regulated are dissimilar in different species (reviewed in references 14 and 93). In B. subtilis, WalRKBsu positively regulates several cell wall hydrolase genes and the ftsZ cell division gene and negatively regulates genes that modulate hydrolase activity (9, 23, 34). Likewise, the WalRKSau regulon of Staphylococcus aureus contains several murein hydrolases (15, 16). In these species, the essentiality of the WalRK TCS has been ascribed to misregulation of a combination of genes, since none of the hydrolase genes is individually essential (15), and ftsZ is transcribed from other promoters not regulated by WalRK (23). In S. pneumoniae, the essentiality of WalRSpn is due to its positive regulation of pcsB, which encodes a putative hydrolase that plays a critical role in cell wall biosynthesis and division (4, 27, 57, 58). Besides cell wall hydrolases and division proteins, the WalRK regulons of different Streptococcus species includes genes encoding surface virulence factors and enzymes of exopolysaccharide biosynthesis (1, 42, 51, 59, 72).
The WalK histidine kinases of Streptococcus species have sensing domains that are structurally different from those of Bacillus, Staphylococcus, and most other species (60, 84). WalKBsu, which is typical of one class, contains two transmembrane domains flanking an extracytoplasmic domain. The transmembrane domains of WalKBsu interact with the membrane domains of the ancillary WalHI (YycHI) proteins to negatively regulate phosphorylation levels of the WalRBsu response regulator (84-86). In addition, WalKBsu colocalizes with FtsZ at the septa of dividing B. subtilis cells (24). In contrast, WalKSpn, which exemplifies the other class, contains only a single transmembrane domain connected to an extracellular peptide of only 12 amino acids (Fig. 1, line 1) (47, 60, 89). Streptococcus species also lack homologues of WalHI. On the other hand, the cytoplasmic domains of both classes of WalK histidine kinases are highly similar and include HAMP (linker), PAS (potential signal binding made up of PAS and PAC motifs), DHp (dimerization and histidine phosphorylation), and CA (catalytic ATPase) domains typical of other histidine kinases (Fig. 1, line 1) (14, 25, 60, 93). The signals that are sensed by WalK histidine kinases are not yet known in any species, although it has been speculated that Lipid II derivatives may act as signals of the class that includes WalKBsu and WalKSau (14).
FIG. 1.
Domain organization of the protein constructs used in the present study. Full-length WalKSpn (VicK) (line 1) contains 449 amino acids organized into five architectural and functional domains based on the SMART database (smart.embl-heidelberg.de): TM (anchoring transmembrane domain; amino acids 13 to 32), HAMP linker domain (amino acids 16 to 84), PAS domain consisting of PAS and PAC motifs (amino acids 94 to 202), DHp (dimerization histidine phosphoryltransfer [HisKA]; amino acids 208 to 274), and CA (kinase catalytic domain [HATPase]; amino acids 323 to 435). Histidine residue 218 (H218) is phosphorylated in the autokinase reaction. Numbering of full-length WalKSpn was extended to the soluble, truncated WalKSpn derivatives purified and characterized in the present study (lines 2 to 10; Materials and Methods; see Fig. S1 in the supplemental material). The affinity tags on the constructs are indicated. Full-length WalRSpn (VicR) contains 234 amino acids organized into two domains: a receiver domain (amino acids 2 to 112) and an effector domain (amino acids 154 to 230). Aspartate residue 52 in the receiver domain is phosphorylated in the transferase reaction with WalKSpn∼P constructs, and the effector domain contains the helix-turn-helix DNA binding motif. See the text for further details.
Given their regulatory importance, relatively little enzymological characterization of WalK histidine kinases and WalR response regulators has been reported. Autophosphorylation of WalK and phosphoryl group transfer between WalK∼P and WalR was demonstrated for homologues from B. subtilis (34, 35, 91, 96) and Enterococcus faecalis (52). In addition, phosphoryl transfer that reflects physiologically relevant cross talk was detected between PhoRBsu∼P and WalRBsu (34, 35). A refolded, soluble construct of WalKSpn lacking the transmembrane domain was shown to have autokinase activity and to carry out phosphoryltransfer to purified WalRSpn (18). However, the only quantitative analysis of WalK autophosphorylation was reported for highly truncated versions of WalKSau and WalKSpn containing only the DHp and CA domains (12). We report here a comparison of the kinetics of the autokinase activity of a nearly full-length construct of WalKSpn with a highly truncated version and their activities in phosphoryltransfer to full-length WalRSpn and dephosphorylation of WalRSpn∼P. We also analyzed the effects of an internal PAS domain deletion and changes of key amino acids in the DHp domain of WalKSpn on these activities. We show that the autokinase and phosphoryltransfer reactions were largely unaffected by the absence of the HAMP and PAS domains but unexpectedly, optimal WalKSpn phosphatase activity for WalRSpn∼P depended on the PAS domain. We also show that the WalKSpn internal PAS deletion and the point mutations in the DHp domain characterized here biochemically are important for pneumococcal virulence in a mouse model of infection.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in the present study are listed in Tables S1 and S2 in the supplemental material. Genomic DNA used to construct protein expression plasmids was obtained from S. pneumoniae serotype 2 strains R6 and D39 (see reference 48). In most cases, deletion and point mutations in cloned walKSpn were constructed by fusion PCR (58, 59) using mutagenic primers and primers containing appropriate restriction sites (see Table S3 in the supplemental material). In three cases where mutations already existed, appropriate regions were simply amplified from the S. pneumoniae genome. PCR amplicons were cloned into the BamHI and BsaI or BsmFI sites of plasmid pSumo (LifeSensors, Inc.) and into NotI and XhoI sites of plasmid pET28a (Novagen, Inc.) to generate protein expression vectors (see Table S2 in the supplemental material). Recombinant expression plasmids were transformed into competent Escherichia coli strain DH5α and then into strain BL21(DE3)Rosetta/pLysS (see Table S1 in the supplemental material). All expression plasmids were verified by sequencing.
WalKSpn mutants in S. pneumoniae were constructed by the Janus method of allele replacement used previously (67, 82, 88). A ΔwalKSpn::[kanR-rpsL+] amplicon was transformed into strain IU1781 (D39 rpsL1 [resistant to 150 μg of streptomycin per ml]), resulting in strain IU1885 that is resistant to 250 μg of kanamycin per ml and sensitive to streptomycin. Markerless amplicons containing mutations in walKSpn (ΔwalKSpn), walKSpn (H218A), walKSpn (T222R), and walKSpn (ΔPAS [absence of amino acids 104 to 198])) were constructed by fusion PCR (58, 59) using the primers listed in Table S2 in the supplemental material that introduce the desired amino acid substitutions or deletions. The ΔwalKSpn deletion retained 60 bp at the 5′ and 3′ ends of walKSpn to maintain any transcription or translation signals that might affect the expression of the closely spaced adjacent walRSpn and walJSpn genes. Transformation of IU1885 with the markerless amplicons crossed out the walKSpn::[kanR-rpsL+] region, resulting in colonies resistant again to 150 μg of streptomycin per ml and sensitive to kanamycin. Mutants were checked for gene duplications by PCR, and mutations were confirmed by DNA sequencing of genomic DNA (48). The presence of capsule was confirmed in each transformant by the Quellung reaction (48).
Overexpression and purification of proteins.
E. coli strains were grown with shaking at 30°C in standard LB media (MP Biomedicals) supplemented with antibiotics required to maintain expression vectors (see Table S1 in the supplemental material) and other additives as indicated (see Table S4 in the supplemental material). After reaching an optical density at 620 nm (OD620) of ca. 0.2 to 0.6, cultures were induced by addition of IPTG at concentrations listed in Table S4.
Protein expression and solubility were estimated by SDS-PAGE (70). Cells from 1 ml of uninduced and induced cultures, adjusted to equal OD620 levels, were collected by centrifugation for 3.5 min at 16,100 × g, resuspended in 100 μl of Laemmli sample buffer (Bio-Rad) containing 5% (vol/vol) β-mercaptoethanol, and boiled for 5 min. Equal volumes (≈15 μl) of samples were resolved by 10% Tris-glycine SDS-PAGE. Gels were stained with Coomassie brilliant blue dye (70), and the levels of protein induction were estimated visually by comparing uninduced and induced samples relative to molecular weight markers (Invitrogen). To estimate the solubility of recombinant proteins, 5 to 10 ml of induced cultures were collected by centrifugation as described above and resuspended in 3 ml of cold buffer A (20 mM NaPO4, 0.5 M NaCl, 40 mM imidazole [pH 7.4]). Cells were lysed by passage through a chilled French pressure cell (20,000 lb/in2), and insoluble material was collected by centrifugation at 8,000 × g for 10 min at 4°C. Insoluble inclusion bodies in pellets were resuspended in 2 ml of buffer A. An equal volume of 2× Laemmli sample buffer was added to the supernatants and resuspended insoluble material. After boiling for 5 min, supernatant and pellet samples were loaded and analyzed by SDS-PAGE. If a band of the correct size was detected in the soluble fraction of the induced culture, then larger cultures were grown for protein purification.
Proteins were purified as described previously (59) with the following modifications. Induced cultures (0.3 to 1 liter) were chilled on ice and centrifuged at 8,000 × g for 10 min, and cell pellets were resuspended in 20 to 40 ml of buffer A supplemented with protease cocktail inhibitor III (Calbiochem). All remaining steps were performed at 4°C. Cells were lysed by two passes through a French press cell (20,000 lb/in2). Lysates were centrifuged twice at 8,000 × g for 20 min and filtered in a 50-ml disposable manifold containing a 0.22-μm-pore-size membrane (Millipore) to remove debris. The filtrate was applied to a HisTrap HP column (GE Healthcare) preequilibrated with buffer A using a peristaltic pump at a flow rate of 0.5 ml per min. Loaded columns were attached to a Shimadzu 10A Biocompatible high-pressure liquid chromatography (HPLC) system, and proteins were eluted by using a linear 60-min gradient of 40 to 500 mM imidazole in buffer A at a flow rate of 0.5 ml per min. Proteins were detected by monitoring A220 and A280. Fractions containing recombinant proteins were checked for contaminants by SDS-PAGE and pooled. Purified protein samples were concentrated and exchanged into final optimized storage buffers (see Table S4 in the supplemental material) by using Amicon ultracentrifugal filters (Millipore) according to the manufacturer's instructions. Alternatively, overnight dialysis in Slide-A-Lyzer cassettes (Thermo Scientific) was used when fast exchange to storage buffers in Amicon filters caused protein aggregation (see Table S4 in the supplemental material). To improve protein solubility, the composition of storage buffers was optimized by testing for aggregation by centrifugation at 100,000 × g for 15 min and reassaying protein concentrations in supernatants. Protein purities were estimated visually on stained SDS gels to be >95% (see Fig. S1 in the supplemental material). Similar results were obtained in the assays described below for several different preparations of the purified WalKSpn constructs and WalRSpn.
Determination of protein concentration.
The concentrations of purified proteins were determined by using the DC protein assay kit (Bio-Rad) as instructed by the manufacturer using bovine serum albumin (Sigma Fraction V) dissolved in storage buffer as the standard (see Table S4 in the supplemental material). For CD measurements, protein concentrations were determined by using a MicroBCA protein assay kit (Pierce) as instructed by the manufacturer.
Determination of WalKSpn autophosphorylation kinetic parameters.
Autophosphorylation kinetic parameters were determined by using an SDS-PAGE method described previously (21) with the following modifications. Various WalKSpn constructs (1.1 to 1.7 μM; see Table S4 in the supplemental material) were preequilibrated in reaction buffer B (50 mM Tris-HCl [pH 7.8], 200 mM KCl, 5 mM MgCl2) for 10 min at 25°C. Reducing agents were omitted, because addition of 2 mM dithiothreitol diminished WalKSpn autophosphorylation activity. The autophosphorylation reactions were started by adding various concentrations (6, 12.5, 50, 100, and 225 μM) of [γ-32P]ATP (specific activity, 1.1 to 2.5 Ci/mmol; Perkin-Elmer, catalog no. BLU502Z). At designated times (15, 30, 45, and 60 s), 15-μl samples were removed, and reactions were stopped by adding the samples to 15 μl of 2× Laemmli sample buffer containing 5% (vol/vol) β-mercaptoethanol. Final samples (20 μl) were analyzed without heating by 10% Tris-glycine SDS-PAGE (21). After electrophoresis, gels were soaked for 20 min in 2% (vol/vol) glycerol and dried for 1 h at 80°C on a vacuum gel dryer (Bio-Rad). Dried gels were exposed to a storage phosphor screen (GE Healthcare) and analyzed by using a Typhoon Variable Mode Imager 9200 (Amersham) and ImageQuant 5.2 software (Molecular Dynamics). The amount of WalKSpn∼P in each lane was quantified by using a standard curve generated by spotting known concentrations of [γ-32P]ATP. Initial rates were calculated from linear regression plots of WalKSpn∼P formed versus time (Fig. 2), and Michaelis-Menten kinetic parameters (Km and kcat) (see references 12, 21, 28, 50, 61, and 78 for precedents) were obtained by fitting velocities to ATP concentrations using a nonlinear regression program (GraphPad Prism). The autophosphorylation of the H218A or T222R mutant WalKSpn (N)-Sumo construct was not detectable or very low, respectively. These constructs (2.4 to 3.3 μM; see Table S4 in the supplemental material) were preequilibrated in reaction buffer B for 5 min at 25°C, and reactions were initiated by adding 12.5 μM [γ-32P]ATP (specific activity, 5 to 10 Ci/mmol). Samples were removed at different times (1, 2.5, 5, 10, 15, and 20 min) and processed as described above.
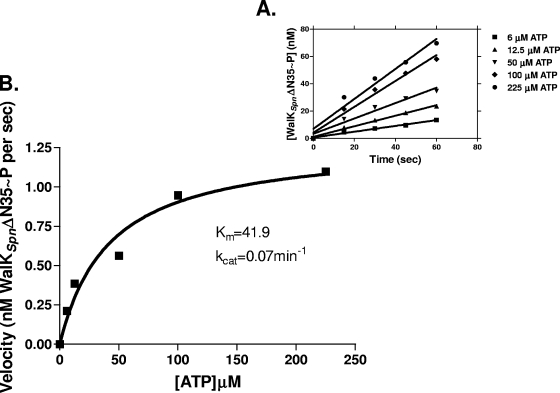
FIG. 2.
Progress curves of autophosphorylation reactions. Representative curves used to determine the kinetic parameters in Table 1 are shown. (A) Time course used to calculate the initial rates of WalKSpnΔN35 (C)-His autophosphorylation (Fig. 1, line 2) at different ATP concentrations. (B) Velocity versus [ATP] curve based on A used to calculate Km(ATP) and kcat for WalKSpnΔN35 (C)-His autophosphorylation. See Materials and Methods for details. These reactions contained 1.1 μM WalKSpnΔN35 (C)-His in a volume of 100 μl at 25°C.
Combined assay of WalKSpn autophosphorylation and phosphoryltransfer to WalRSpn.
Combined reactions were carried out based as described previously (12, 65) with the following modifications. WalKSpn constructs (2.2 to 3.4 μM; see Table S4 in the supplemental material) were autophosphorylated for 3 min in 100-μl reactions containing 50 mM Tris-HCl (pH 7.8), 200 mM KCl, 12.5 μM [γ-32P]ATP (5 Ci/mmol), 15 to 20% (vol/vol) glycerol (to maintain WalRSpn solubility later in the reaction), and either 5 mM MgCl2 or 3.8 mM CaCl2. The progression of WalKSpn autophosphorylation was monitored at 0.5, 1, and 3 min by removing 15-μl samples and stopping reactions as described above for the autophosphorylation assay. At 3 min, 9.6 μM WalRSpn (N)-His was added to the reaction mixtures containing WalKSpn∼P without removal of excess ATP. Samples (15 μl) were removed 1.5, 4.5, and 19.5 min after WalRSpn addition and processed and analyzed as described above. Amounts of WalKSpn∼P and WalRSpn∼P were calculated relative to the amount of WalKSpn∼P at the time of WalRSpn addition, which was set at 100%. To evaluate the effects of the purified PAS domain, we incubated WalKSpnΔPAS constructs [WalKSpnΔN195 (N)-Sumo (2.6 μM) or WalKSpnΔN195 (C)-His (2.9 μM)] with purified PAS domain [PAS (N)-Sumo (7.1 μM) or PAS (C)-His (5.6 μM)] for 10 min at 25°C prior to initiation of the autophosphorylation reaction.
Quantification of phosphoryltransfer efficiency between WalKSpn∼P constructs and WalRSpn.
WalKSpnΔN35 (C)-His (2.5 μM), WalKSpnΔN35 (N)-Sumo (2.5 μM), WalKSpnΔN195 (C)-His (3 μM), or WalKSpnΔN195 (N)-Sumo (2.5 μM) constructs were autophosphorylated for 20 min at 25°C in 100 μl of 50 mM Tris-HCl (pH 7.9), 200 mM KCl, either 5 mM MgCl2 or 5 mM CaCl2, 15 to 20% (vol/vol) glycerol (to maintain WalRSpn solubility later in the reaction), and 500 μM [γ-32P]ATP (0.5 Ci/mmol). Excess ATP was removed from reactions by using spin desalting columns (Pierce). The recovery of the proteins after desalting was tested by using the DC protein assay and 8% Tris-glycine SDS-PAGE. WalKSpn∼P concentrations after desalting ranged from 1.3 to 2.0 μM for different preparations. Then, 15-μl samples of desalted WalKSpn∼P were added to 2× Laemmli buffer to determine the amounts of WalKSpn∼P. At t = 0, 0.25 μM WalRSpn (N)-His was added to the remainder of the WalKSpn∼P sample to start the phosphoryltransfer reaction, and 15-μl samples were taken after 30, 60, 120, and 240 s and processed and analyzed as described above. The phosphoryltransfer efficiency between WalKSpn∼P and WalRSpn was evaluated by exponential decay plots of remaining WalKSpn∼P versus time after addition of WalRSpn (Fig. 3) (7, 78, 81) rather than measuring the rates of WalRSpn∼P formation, which is subject to WalKSpn phosphatase activity. Half-lives of WalKSpn∼P were corrected for the intrinsic stability of WalKSpn∼P in the absence of WalRSpn (average t1/2 ≈ 660 s).
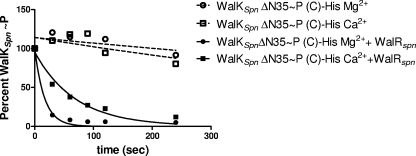
FIG. 3.
WalKSpn∼P disappearance due to phosphoryltransfer to added WalRSpn. A representative reaction progression curve used to determine half-lives in Table 2 is shown. Open symbols, WalKSpnΔN35 (C)-His∼P decay in the absence of WalRSpn in reaction mixtures containing Mg2+ or Ca2+; closed symbols, decay of WalKSpnΔN35 (C)-His∼P in parallel reaction mixtures containing Mg2+ or Ca2+ and WalRSpn added at t = 0. See Materials and Methods for details. These reactions contained 1.5 μM WalKSpnΔN35 (C)-His∼P and 0.25 μM WalRSpn in a reaction volume of 100 μl at 25°C.
Phosphorylation of WalRSpn by acetyl phosphate and quantification of WalRSpn∼P autodephosphorylation and WalKSpn-catalyzed dephosphorylation.
Phosphorylation of WalRSpn with acetyl phosphate was carried out as described earlier (59) with the following modifications. WalRSpn (N)-His (11.8 μM) was incubated with 40 mM acetyl phosphate (Fluka) in reaction buffer (50 mM Tris-HCl [pH 7.4], 200 mM KCl, 4 mM MgCl2, and 40% [vol/vol] glycerol) for 75 min at 37°C. Excess acetyl phosphate was removed by using spin desalting columns (Pierce). The recovery of WalRSpn∼P was ≈50% after desalting as determined by the DC protein assay. Desalted WalRSpn∼P was incubated at 25°C in the presence or absence of ADP (13.2 μM) (t = 0). At times ranging from 10 min to 22.5 h, samples were removed, and the amounts of WalRSpn∼P and WalRSpn were determined by reversed-phase HPLC using a Phenomenex Jupiter 300A C4 column and a Shimadzu 10A HPLC system (33, 59). Eluent A was composed of 20% (vol/vol) acetonitrile and 0.1% (vol/vol) trifluoroacetic acid in water, and eluent B was composed of 60% (vol/vol) acetonitrile and 0.1% (vol/vol) trifluoroacetic acid in water. A linear gradient from 50% eluent A plus 50% eluent B to 100% eluent B (no eluent A) was formed during a period of 18 min at a flow rate of 1 ml/min. Proteins were detected by monitoring the A220. The relative amounts of WalRSpn∼P and WalRSpn were calculated from peak areas and normalized to starting samples, which contained ≈85% WalRSpn∼P. The half-lives of WalRSpn∼P were calculated from exponential decay plots with time (GraphPad Prism), where the rate constant of autodephosphorylation, kauto, was ln2/t1/2 (98).
WalRSpn∼P dephosphorylation catalyzed by WalKSpn PAS+ constructs in the presence of nucleotide cofactors was determined as follows. Desalted WalRSpn∼P (5.9 μM) was incubated in the presence of WalKSpnΔN35 (C)-His (2.0 μM) at 25°C in 50 mM Tris-HCl (pH 7.4 or 7.8), 200 mM KCl, 2 mM MgCl2, and 40% glycerol. The following nucleotide cofactors were added to some of the reaction mixtures: ATP (13.2 μM), ATP-γS (13.2 μM), or ADP (13.2 or 120 μM). Samples were removed at time points ranging from 10 min to 5 h, and the extent of WalRSpn∼P dephosphorylation was determined by C4-HPLC as described above. The rate constant for dephosphorylation of WalRSpn∼P in the presence of WalKSpn was determined by the following formula: k = ln2/t1/2 − ln2/t1/2 auto, where t1/2auto is the half-life of WalRSpn∼P in the absence of histidine kinase (98).
WalRSpn∼P dephosphorylation catalyzed by WalKSpn DHp mutants was determined at 25°C as described above in reactions containing desalted WalRSpn∼P (4.3 to 5.8 μM), WalKSpnΔN35 (N)-Sumo (3.0 μM), WalKSpnΔN35 H218A (N)-Sumo (2.1 μM), or WalKSpnΔN35 T222R (N)-Sumo (1.7 μM), and ADP (9.5 to 12.8 μM). WalRSpn∼P dephosphorylation catalyzed by WalKSpn PAS domain mutants was determined at 25°C as described above in reactions containing desalted WalRSpn∼P (4.7 to 5.3 μM), WalKSpnΔN195 (N)-Sumo (2.1 μM), WalKSpnΔN195 (C)-His (2.2 μM), WalKSpnΔN35 D133N,N136Y,L140R (N)-Sumo (2.6 μM), or WalKSpnΔN35 ΔPAS[104-198] (N)-Sumo (1.8 μM), and ADP (11.0 to 11.9 μM).
CD spectroscopy of wild-type and mutant WalKSpn constructs.
Circular dichroism (CD) spectra were obtained for purified WalKSpnΔN35 (N)-Sumo, WalKSpnΔN35 H218A (N)-Sumo, WalKSpnΔN35 T222R (N)-Sumo, WalKSpnΔN35 D133N,N136Y,L140R (N)-Sumo, and WalKSpnΔN195 (N)-Sumo using a Jasco J-715 CD spectropolarimeter using a previously published protocol (30). Protein concentrations varied from 0.097 to 0.164 mg/ml in 10 mM potassium phosphate buffer (pH 7.4) in a 0.1-cm quartz cell (Starna). Proteins were exchanged into this buffer by using spin desalting columns (Pierce) to remove interfering components. Three independent spectra of each protein were recorded at 25°C by using a scanning speed of 100 nm per min with 0.5-nm intervals. The wavelength range was set from 190 to 240 nm with a bandwidth of 2 nm. Spectra were averaged, smoothed using a Savitsky-Golay filter with a smoothing window of 15 points (30), and corrected for buffer absorbance in the absence of proteins. The raw data was converted to the mean residue ellipticity: [θ] in degrees cm2 dmol−1 = (millidegrees × mean residue weight)/(path length in mm × concentration in mg ml−1), where the mean residue weight is the molecular weight of the protein divided by the number of amino acids minus 1. The [θ] values were used to perform secondary structure analyses with Selcon3 software from Dichroweb (see Table S5 in the supplemental material) (79, 80, 92).
Growth of S. pneumoniae strains.
Parent and walKSpn mutant strains were grown statically in 16-by-100-mm glass tubes at 37°C in an atmosphere of 5% CO2 as described previously (67, 88). Briefly, bacteria were inoculated from frozen stocks into 5.0 ml of brain heart infusion (BHI) broth (BD), serially diluted in BHI broth, and propagated overnight. Overnight cultures that were still in exponential growth phase (OD620 ≈ 0.2 to 0.4) were diluted to a OD620 of ≈0.1, and 50 μl of these diluted cultures was inoculated into 5.0 ml of BHI broth lacking antibiotic to give a starting OD620 of ≈0.001. Tubes were gently inverted before OD620 readings were obtained at approximately 1-h intervals using a Spectronic 20 spectrophotometer.
Murine pneumonia model of infection.
All procedures were approved in advance by the Institutional Animal Care and Use Committee and were performed according to recommendations of the National Research Council. Procedures were carried out as described previously (43) with the following changes. Male ICR (21 to 24 g; Harlan) mice were anesthetized by inhaling 4% isoflurane (Butler Animal Health Supply) delivered by an EZAnesthesia system (Euthanex Corp.) for 5 min. Nine or six mice in replicate experiments were inoculated intranasally with each bacterial strain to be tested. Aliquots (1 ml) of each strain growing exponentially in BHI broth (OD620 ≈ 0.240) were microcentrifuged for 10 min at 13,500 × g, and cell pellets were resuspended in 1 ml of phosphate-buffered saline (pH 7.4) solution. A 50-μl sample of this suspension (≈7.0 × 106 CFU) was used as the inoculum. Anesthetized mice were placed on their backs, and their mouths were gently closed to allow inhalation of the 50-μl inoculum, which was delivered in aliquots to the center of the noses. To ensure inhalation, mice were suspended vertically from their teeth after inoculation for ≈1 min until they started to awaken from the anesthesia. CFU in inocula were confirmed by serial dilution and plating. Mice were monitored at ≈6-h intervals. Death was not used as an endpoint. Moribund mice were euthanized by CO2 asphyxiation, and that time point was used as “time of death” in survival curves. Kaplan-Meier survival curves and log-rank tests were generated by using GraphPad Prism software.
RESULTS
Overexpression and purification of proteins.
We initially attempted to purify active full-length WalKSpn based on methods published for histidine kinases and other signal transducers (8, 83). Although we could overexpress sufficient amounts of protein, full-length WalKSpn was insoluble, even using these conditions. Based on extensive precedents from other histidine kinases (12, 21, 32, 61, 76), we turned to truncated versions of WalKSpn for these initial kinetic analyses. The longest active form of pneumococcal WalKSpn that retained autokinase activity was truncated for the first 35 amino acids specifying the transmembrane domain and a short section of the HAMP domain (Δ35 constructs, Fig. 1, lines 2 to 6). Several truncations that extended further into the HAMP domain were insoluble or inactive (data not shown). For comparison with the only published kinetic study of WalKSpn (12), we also characterized WalKSpn truncated for the transmembrane, HAMP, and PAS domains (Δ195 constructs, Fig. 1, lines 7 to 9). For each WalKSpn truncation, we added a Sumo or His6 tag to the amino or carboxyl terminus, respectively, and purified the proteins by affinity chromatography as described in Materials and Methods and Table S4 in the supplemental material. We also purified the PAS domain alone fused to the Sumo or His6 tag (Fig. 1, line 10). Full-length WalRSpn response regulator fused to an amino-terminal His10 tag (Fig. 1, line 11) was purified as before (59).
Attempts to remove the affinity tag from the WalKSpnΔN35 (N)-Sumo construct with Sumo protease were not successful, because the protein lost autokinase activity (data not shown). Therefore, to control for tag-specific effects, we characterized both the N-Sumo and C-His versions of each purified WalKSpn protein. The tag effects that we observed were generally small. We characterized amino acid substitutions in (N)-Sumo WalKSpn constructs, because they were generally more soluble than the corresponding (C)-His WalKSpn proteins (see Table S4 in the supplemental material). CD spectra confirmed that mutant WalKSpn proteins were not grossly misfolded compared to the wild-type protein (see Table S5 in the supplemental material). Many substitutions and small internal deletions in the PAS domain resulted in insoluble WalKSpn that could not be purified (see Fig. S2 and Table S4 in the supplemental material). We were able to improve the solubility of full-length WalRSpn (N)-His by increasing the glycerol and salt concentration in its storage and reaction buffers (see Materials and Methods and Table S4 in the supplemental material) (59).
Kinetic parameters of WalKSpn autophosphorylation do not depend on the presence of the HAMP and PAS domains.
The autokinase kinetic parameters of the WalKSpn Δ35 and Δ195 proteins fused to the (C)-His tag were nearly the same within experimental error (Fig. 2 and Table 1, lines 1 and 5). The (N)-Sumo constructs had comparable Km(ATP) values to their (C)-His counterparts (Table 1, lines, 1, 2, 5, and 6). There was some variation in the kcat values of the (N)-Sumo compared to the (C)-His constructs, where the kcat of WalKSpnΔ35 (N)-Sumo was ≈2.6-fold greater than that of the (C)-His version (Table 1, lines 1 and 2). However, taken together, the absence of the HAMP and PAS domains in the WalKSpn Δ195 constructs did not appreciably affect the autophosphorylation kinetic parameters, and tag-specific effects, although present for the (N)-Sumo constructs, tended to be marginal. The Km(ATP) of the WalKSpnΔ195 (N)-Sumo protein used here (28 μM; Table 1, line 6) was somewhat higher than that reported previously for a comparable WalKSpn Δ195 construct fused to an N-terminal avidin-His tag (3 μM), although the kcat rates were comparable for the two constructs (12). The autophosphorylation kinetic parameters of WalKSpn reported here are similar to those reported for other truncated histidine kinases, such as WalKSau, KinA, NarQ, and HpKA (12, 21, 31, 61).
TABLE 1.
Kinetic parameters of WalKSpn histidine kinase autophosphorylationa
| Enzyme constructb | Mean ± SEM (n) |
kcat/Km (M−1 min−1) | |
|---|---|---|---|
| Km for ATP (μM) | kcat (min−1) | ||
| WalKSpn PAS+ constructs | |||
| 1. WalKSpnΔN35 (C)-His | 42.0 ± 2.2 (3) | 0.084 ± 0.008 (3) | 2,000 |
| 2. WalKSpnΔN35 (N)-Sumo | 43.8 ± 8.3 (4) | 0.216 ± 0.02 (4) | 4,930 |
| 3. WalKSpnΔN35 H218A (N)-Sumo | NA | ||
| 4. WalKSpnΔN35 T222R (N)-Sumo | LA | ||
| WalKSpn PAS mutant constructs | |||
| 5. WalKSpnΔN195 (C)-His | 36.7 ± 1.9 (3) | 0.072 ± 0.004 (3) | 1,960 |
| 6. WalKSpnΔN195 (N)-Sumo | 28.4 ± 6.0 (4) | 0.030 ± 0.003 (4) | 1,060 |
| 7. WalKSpnΔN195 H218A (N)-Sumo | NA | ||
| 8. WalKSpnΔN195 T222R (N)-Sumo | LA | ||
| 9. WalKSpnΔN35 D133N, N136Y, L140R (N)-Sumo | 72.4 ± 29.1 (2) | 0.204 ± 0.04 (2) | 2,820 |
| 10. WalKSpnΔN35 ΔPAS[104-198] (N)-Sumo | 257 ± 17 (2) | 0.258 ± 0.03 (2) | 1,000 |
Kinetic parameters were determined at 25°C as described in Materials and Methods. Reaction mixtures contained 1.1 to 1.7 μM concentrations of the indicated WalKSpn constructs. The means are shown for the indicated number of independent experiments in parentheses (n). “NA” indicates no activity was detected in 20 min. “LA” indicates that autophosphorylation activity was not detected in 1 min and could be detected only in 5-min reactions, in which the relative amount of WalKSpn T222R∼P was <10% compared to the wild-type protein (data not shown).
Each line of data is preceded by a “line number” (“1.”, “2.”, etc.) in the first column. These lines are referenced by number in the text at the corresponding in-text table callouts.
Autophosphorylation of the WalKSpn ΔN35 and ΔN195 decreased in the presence of 2 mM DTT (data not shown). This result contrasts with a recent report that addition of reducing reagent increased the autokinase activity of full-length WalKEfa from E. faecalis (52). Different numbers and locations of cysteine residues in WalKSpn and WalKEfa may underlie this difference. WalKEfa contains three cysteine residues, one in the PAS domain and two in the CA domain. In contrast, WalKSpn contains a single cysteine (C240) in the DHp domain near the phosphorylated histidine (H218). Whether disulfide bond formation and covalent dimerization modulate WalKSpn function remains to be investigated, especially in the context of the unusual production of high levels of hydrogen peroxide by S. pneumoniae (see reference 67).
H218A, T222R, and ΔPAS mutant WalKSpn have abolished or reduced autokinase activity.
We determined the autophosphorylation parameters for several mutant WalKSpn proteins that were soluble. As expected, WalKSpn H218A mutants (Fig. 1, lines 4 and 9) lacked autokinase activity (Table 1, lines 3 and 7), because they are missing the histidine residue that is phosphorylated. The T222R substitution in the DHp domain (Fig. 1, lines 3 and 8) was tested, because a comparable change in some histidine kinases, such as EnvZ, results in increased autokinase activity, while abolishing phosphatase activity for the cognate phosphorylated response regulator (2, 17). However, the WalKSpn T222R substitution greatly reduced autokinase activity (Table 1, lines 4 and 8). A refolded WalKSpn containing a L100R substitution in the PAS domain was also reported to lack autokinase activity (18), although proper folding of this mutant protein was not confirmed. In this first study, we did not introduce other changes at this or other positions in the WalKSpn DHp domain.
Several amino acid substitutions and small internal deletions in the predicted β strands of the WalKSpn PAS domain resulted in insoluble protein (see Fig. S2 and Table S4 in the supplemental material). Inability to obtain soluble histidine kinases containing amino acid substitutions in their PAS domains has been reported before (e.g., E. coli NtrB [63]). Mutant WalKSpn containing three substitutions (D133N, N136Y, and L140R) in a predicted α-helical region of PAS (Fig. 1, line 5) had the same autokinase Km(ATP) and kcat as the wild-type protein within experimental error (Table 1, lines 2 and 9). In contrast, an internal deletion of PAS (ΔPAS[104-198]; Fig. 1, line 6) in the one construct that was soluble increased the WalKSpn autokinase Km(ATP) by ≈6-fold without affecting the kcat (Table 1, lines 2 and 10). Therefore, internal deletion of the WalKSpn PAS domain reduced the relative catalytic efficiency of the autokinase reaction.
Absence of the HAMP and PAS domains of WalKSpn has a minimal effect on the kinetic preference of the phosphoryltransfer reaction.
We determined half-lives of WalKSpn∼P during the phosphoryltransfer reaction to WalRSpn as described in Materials and Methods (see Fig. 3 and Table 2). Phosphorylation of WalKSpn was carried out in reaction mixtures containing either Mg2+ or Ca2+, and the same divalent cation was present in the subsequent phosphoryltransfer reactions. As observed for other TCS pairs (32, 81), the initial rates of this reaction were too rapid to measure by steady-state methods in reactions containing excess WalRSpn substrate. Therefore, we determined the half-lives of WalKSpn∼P in reactions containing an excess of WalKSpn over WalRSpn, which was present at a concentration lower than the typical Km for TCS pairs (see references 10 and 77). The resulting half-lives of WalKSpn∼P should reflect the kinetic preference (kcat/Km) of the phosphoryltransfer reaction (see references 13, 76, and 77).
TABLE 2.
Half-lives of WalKSpn∼P in phosphoryltransfer reactions to WalRSpna
| Enzyme constructb | Mean ± SEM |
|
|---|---|---|
| WalKSpn∼P half-life(s) + Mg2+ | WalKSpn∼P half-life(s) + Ca2+ | |
| WalKSpn PAS+ constructs | ||
| 1. WalKSpnΔN35 (C)-His | 12.5 ± 0.7 | 60.6 ± 7.1 |
| 2. WalKSpnΔN35 (N)-Sumo | 26.3 ± 3.1 | 501 ± 122 |
| WalKSpn ΔPAS mutant constructs | ||
| 3. WalKSpnΔN195 (C)-His | 23.0 ± 3.6 | 114 ± 31 |
| 4. WalKSpnΔN195 (N)-Sumo | 15.9 ± 2.2 | 64.1 ± 13.2 |
Reactions were performed at 25°C in buffers containing Mg2+ or Ca2+ as described in Materials and Methods. Reaction mixtures contained 1.3 to 2.0 μM concentrations of the indicated WalKSpn constructs and 0.25 μM WalRSpn (N)-His. The experiment was performed independently twice. The average intrinsic half-life of WalKSpn∼P was ≈660 s in the presence of either cation.
See Table 1, footnote b.
The kinetic preference of phosphoryltransfer was similar for the WalKSpn Δ35 and Δ195 constructs in reaction mixtures containing Mg2+ ion (Table 2, lines 1 to 4). Although there may be some minor variation due to tag effects, these data indicate that the absence of the HAMP and PAS domains had minimal effect on the kinetic preference of the phosphoryltransfer reaction. The rate of decrease of WalKSpn∼P amount during phosphoryltransfer depended strongly on Mg2+ ion and was reduced severalfold when Ca2+ replaced Mg2+ in reaction mixtures (Fig. 3 and Table 2). We do not know why substitution of Mg2+ with Ca2+ had a much more pronounced effect on WalKSpn Δ35 (N)-Sumo compared to the other constructs (Table 2, line 2).
WalKSpn phosphatase activity depends on the PAS domain.
Many histidine kinases possess a phosphatase activity that plays a role in preventing unwanted cross talk (3, 49, 74). However, there are several notable exceptions of histidine kinases that lack phosphatase activity, such as KinA and PhoR of B. subtilis (19, 73, 90). Gel-based combined phosphoryltransferase assays revealed a significant phosphatase activity of the WalKSpn Δ35 constructs (Fig. 4A and B and see Fig. S3 in the supplemental material). In the presence of Mg2+ ion, low amounts of WalRSpn∼P were detected, and there was a clear loss of labeled phosphate from the amount present in the starting WalKSpn∼P. Previously, it was shown that the phosphatase activity of the EnvZ histidine kinase was strongly reduced in reaction mixtures containing Ca2+ instead of Mg2+ ion (17, 99). Similar to results with EnvZ, considerably more WalRSpn∼P was detected in phosphoryltransfer reactions containing Ca2+ instead of Mg2+ (Fig. 4A and B and see Fig. S3 in the supplemental material), a finding consistent with a WalKSpn phosphatase activity.
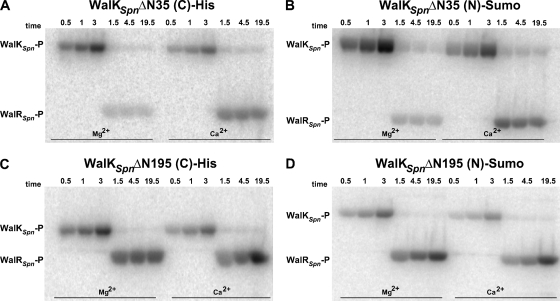
FIG. 4.
Autoradiographs showing autophosphorylation of WalKSpn Δ35 and Δ195 constructs, phosphoryltransfer to WalRSpn, and WalKSpn phosphatase of WalRSpn∼P. Representative time courses are shown and quantitated in Fig. S3 and S4 in the supplemental material. Combined reactions of WalKSpn autophosphorylation and WalRSpn phosphoryltransfer were performed at 25°C in reaction mixtures containing Mg2+ or Ca2+ as described in Materials and Methods. WalKSpn autophosphorylation reactions proceeded for 3 min before WalRSpn was added without removal of ATP (t = 0). Reactions contained the following concentrations of proteins: 2.2 μM WalKSpnΔN35 (C)-His (A), 2.9 μM WalKSpnΔN195 (C)-His (B), 3.4 μM WalKSpnΔN35 (N)-Sumo (C), and 2.6 μM WalKSpnΔN195 (N)-Sumo (D). Each reaction contained 9.6 μM WalRSpn.
Unexpectedly, similar amounts of WalRSpn∼P were detected in combined phosphoryltransfer reactions containing WalKSpn Δ195 and either Mg2+ or Ca2+ ion (Fig. 4C and D and see Fig. S4 in the supplemental material). Since the kinetic parameters for the autokinase and phosphoryltransfer reactions were similar for the WalKSpn Δ35 and Δ195 constructs (Tables 1 and 2), these data imply that the WalKSpn phosphatase activity was significantly reduced in the absence of the HAMP, PAS, or both domains. Consistent with this interpretation, WalRSpn∼P continued to accumulate in reactions containing WalKSpn Δ195 (19.5 min, Fig. 4C and D and Fig. S4), but not WalKSpn Δ35 (19.5 min, Fig. 4A and B and Fig. S3). In the former case, autophosphorylation of WalKSpn Δ195 and phosphoryltransfer to WalRSpn continued to occur, because the WalKSpn phosphatase activity was significantly reduced. In the latter case, the WalKSpn phosphatase acted on WalRSpn∼P, even in reaction mixtures containing Ca2+.
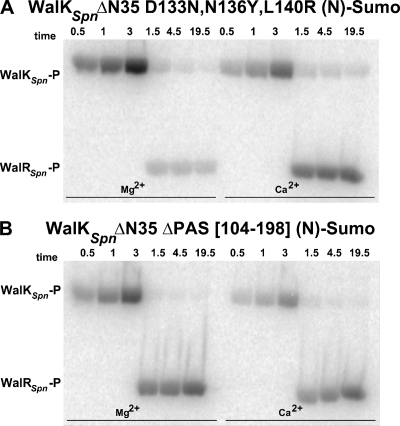
The conclusion that the phosphatase activity depends on the WalKSpn PAS domain was supported by the finding that the WalKSpn internal ΔPAS[104-198] mutant protein had reduced WalRSpn∼P phosphatase activity in combined phosphoryltransferase assays. Similar to the results for the WalKSpn Δ195 construct, WalRSpn∼P continued to accumulate in reactions containing WalKSpn(ΔPAS[104-198]) and either Mg2+ or Ca2+ (Fig. 5B and see Fig. S5 in the supplemental material). In contrast, the mutant WalKSpn with the triple (D133N, N136Y, and L140R) substitutions in the PAS domain served as a control and did not show diminished WalRSpn∼P phosphatase activity (Fig. 5A and Fig. S5). Finally, since the phosphatase activity depended on the WalKSpn PAS domain, we attempted to restore the phosphatase activity of WalKSpn Δ195 by adding back purified PAS domain in trans (Fig. 1, line 10). Added purified PAS domain did not change the autokinase and phosphoryltransferase activities of the WalKSpn Δ195 constructs, nor was WalKSpn phosphatase activity restored (data not shown). However, we do not know whether the purified PAS domain folded correctly.
FIG. 5.
Autoradiographs showing autophosphorylation of WalKSpn PAS domain mutant constructs, phosphoryltransfer to WalRSpn, and WalKSpn phosphatase of WalRSpn∼P. Representative time courses are shown and quantitated in Fig. S5. Combined reactions of WalKSpn autophosphorylation and WalRSpn phosphoryltransfer were performed at 25°C in reaction mixtures containing Mg2+ or Ca2+ as described in Materials and Methods. WalKSpn autophosphorylation reactions proceeded for 3 min before WalRSpn was added without removal of ATP (t = 0). Reactions contained the following concentrations of proteins: 3.4 μM WalKSpnΔN35 D133N,N136Y,L140R (N)-Sumo (A) and 2.5 μM WalKSpnΔN35 ΔPAS[104-198] (N)-Sumo (B). Each reaction contained 9.6 μM WalRSpn.
WalRSpn∼P autophosphatase activity is extremely low.
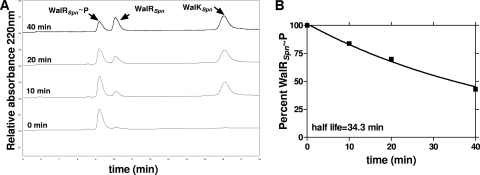
We previously showed that WalRSpn could be phosphorylated with ≈85% efficiency by incubation with acetyl phosphate (59). We used WalRSpn∼P from this reaction in HPLC-based assays for WalKSpn phosphatase activity as described in Materials and Methods (Fig. 6). Amino acid alignment predicted that WalRSpn∼P was likely to have a low rate of autophosphatase activity (see Table S6 in the supplemental material) (87). This prediction was confirmed by phosphatase assays showing that the half-life of WalRSpn∼P was ≈23 h at 25°C (Table 3, line 1). The WalRSpn∼P autophosphatase activity was not affected by addition of ADP, similar to other response regulators, including VanR, PhoQ, and DrrA (29, 71, 94).
FIG. 6.
WalKSpn phosphatase activity of WalRSpn∼P. Representative reaction progression curves used to determine the rate constants and half-lives in Tables 4 and 5 are shown. Reactions containing 13.2 μM ADP were carried out at 25°C as described in Materials and Methods. (A) Reversed-phase HPLC chromatograms showing dephosphorylation of WalRSpn∼P by WalKSpnΔN35 (C)-His with time, where t = 0 was the addition of the WalKSpnΔ35. Reactions contained 5.9 μM WalRSpn∼P and 2.0 μM WalKSpnΔN35 (C)-His. (B) Percent of WalRSpn∼P remaining with time was calculated from the areas under the WalRSpn∼P and WalRSpn, where 100% at t = 0 corresponded to 85% WalRSpn∼P in the starting sample. The rates of WalRSpn∼P disappearance and the half-lives were calculated as described in Materials and Methods.
TABLE 3.
Rates of WalRSpn∼P autodephosphorylation and dephosphorylation in the presence of WalKSpn constructsa
| Enzyme constructb | Mean ± SEM |
|
|---|---|---|
| k (min−1) | WalRSpn∼P half-life (min) | |
| WalRSpn∼P autophosphatase activity | ||
| 1. WalRSpn (N)-His | 0.000554 ± 0.000113 | 1,370 ± 320 |
| WalKSpn PAS+ phosphatase activity | ||
| 2. WalKSpnΔN35 (C)-His | 0.0195 ± 0.0002 | 34.5 ± 0.3 |
| 3. WalKSpnΔN35 (N)-Sumo | 0.036 ± 0.0007 | 18.9 ± 0.4 |
| 4. WalKSpnΔN35 T222R (N)-Sumo | 0.0025 ± 0.00065 | 228 ± 47.2 |
| 5. WalKSpnΔN35 H218A (N)-Sumo | 0.0028 ± 0.0004 | 210 ± 22 |
| WalKSpn PAS domain mutant phosphatase activity | ||
| 6. WalKSpnΔN195 (C)-His | 0.0042 ± 0.0001 | 147 ± 3 |
| 7. WalKSpnΔN195 (N)-Sumo | 0.0023 ± 0.0007 | 249 ± 58.7 |
| 8. WalKSpnΔN35 D133N,N136Y,L140R (N)-Sumo | 0.0184 ± 0.0015 | 36.6 ± 2.9 |
| 9. WalKSpnΔN35 ΔPAS[104-198] (N)-Sumo | 0.0024 ± 0.0009 | 234 ± 71.5 |
Dephosphorylation rates and half-lives of WalRSpn∼P were determined at 25°C in reaction mixtures containing Mg2+, ADP, and the indicated WalKSpn constructs as described in Materials and Methods. Reaction mixtures contained 4.3 to 5.9 μM WalRSpn(N)-His∼P and 1.7 to 3.0 μM concentrations of the indicated WalKSpn constructs. Experiments were performed at least two times.
See Table 1, footnote b.
WalKSpn catalyzed dephosphorylation of WalRSpn∼P is significantly reduced by deletion of the PAS domain.
Addition of WalKSpn decreased the half-life of WalRSpn∼P by ≈40- or ≈70-fold for the Δ35 (C)-His or (N)-Sumo constructs, respectively (Fig. 6 and Table 3, lines 2 and 3). This result directly demonstrates a strong WalKSpn phosphatase activity and indicates a relatively small (<2-fold) tag-specific effect. This decrease in half-life was comparable to that of truncated EnvZ for OmpR∼P in similar reaction mixtures (98, 99). The H218A and T222R substitutions reduced the WalKSpn phosphatase activity by ≈12-fold (Table 3, lines 3, 4, and 5). Thus, these mutations abolished or greatly reduced both the autokinase and phosphatase activities of these WalKSpn Δ35 constructs (Tables 1 and 3). However, the H218A mutant WalKSpn still retained measurable phosphatase activity (Table 3, line 5), whereas it totally lacked autokinase activity (Table 1, lines 3 and 7). Therefore, the WalKSpn phosphatase activity does not occur by a reversal of the phosphoryltransfer reaction and likely proceeds by release of inorganic phosphate (see references 25, 36, 39, and 75). Similar to other histidine kinases (37, 39, 44, 98, 99), WalKSpn required ADP or ATP for optimal phosphatase activity (Table 4). In addition, nonhydrolyzable ATPγS stimulated the WalKSpn phosphatase activity to the same extent as ADP and ATP, a finding consistent with the conclusion that WalKSpn phosphatase activity is not a simple reversal of the phosphoryltransfer reaction.
TABLE 4.
Rates of WalRSpn∼P dephosphorylation catalyzed by WalKSpn in the presence of nucleoside phosphate cofactorsa
| Cofactor (concn [μM])b | Mean ± SEM |
|
|---|---|---|
| k (min−1) | WalRSpn∼P half-life (min) | |
| 1. None | 0.0044 ± 0.00036 | 140 ± 10 |
| 2. ADP (13.2) | 0.0195 ± 0.0002 | 34.5 ± 0.3 |
| 3. ADP, pH 7.8 (13.2) | 0.029 ± 0.002 | 23.3 ± 1.9 |
| 4. ADP (120) | 0.018 ± 0.0004 | 37.1 ± 0.9 |
| 5. ATP (13.2) | 0.032 ± 0.006 | 21.3 ± 3.8 |
| 6. ATPγS (13.2) | 0.019 ± 0.0001 | 35.4 ± 0.2 |
Dephosphorylation rates and half-lives of WalRSpn∼P were determined at 25°C in reaction mixtures containing Mg2+ at pH 7.4 (or pH 7.8 where indicated) in the presence or absence of cofactors as described in Materials and Methods. Reaction mixtures contained 5.9 μM WalRSpn (N)-His∼P and 2.0 μM WalKSpnΔN35 (C)-His. Experiments were performed independently twice.
See Table 1, footnote b.
Finally, deletion of the PAS domain decreased the phosphatase activity by ≈13-fold for the WalKSpn (N)-Sumo constructs (Table 3, lines 3, 7, and 9). A smaller decrease (≈5-fold) was detected for the WalKSpn (C)-His constructs (Table 3, lines 2 and 6). As a control, amino acid substitutions in the WalKSpn PAS domain minimally affected the phosphatase activity (Table 3, lines 3 and 8). Together, these results support the conclusion from the gel-based combined assays (Fig. 4 and 5 and see Fig. S3 to S5 in the supplemental material) that the PAS domain is required for optimal WalKSpn phosphatase activity. The relative rates of the phosphatase reaction appeared to be more rapid in the combined gel-based than the HPLC-based assays (Fig. 4, 5, and 6 and Table 3). This difference may reflect some inactivation or conformational changes that occur when WalRSpn is phosphorylated by acetyl phosphate, which requires an extended incubation and removal of unincorporated acetyl phosphate (Materials and Methods).
ΔPAS and DHp mutations reduce pneumococcal virulence.
To relate the biochemical properties described above to pneumococcus physiology, we tested whether the mutants affected virulence (Fig. 7). Markerless walKSpn(H218A), walKSpn(T222R), and walKSpn(ΔPAS[104-198]) mutations in full-length walKSpn and a ΔwalKSpn deletion were crossed into the chromosome of virulent parent strain D39 rpsL1 (Materials and Methods) (see Table S1 in the supplemental material). The rpsL1 mutation was used in the allele exchange procedure and does not affect virulence in this pneumonia model of infection (67). Western blot analyses (see reference 4) confirmed that the walKSpn(H218A), walKSpn(T222R), and walKSpn(ΔPAS[104-198]) mutants produced similar amounts of WalKSpn protein as the D39 parent strain (data not shown). Attempts to detect mutant WalKSpn deleted for its transmembrane domain (Fig. 1, line 2) were not successful, possibly due to degradation.
FIG. 7.
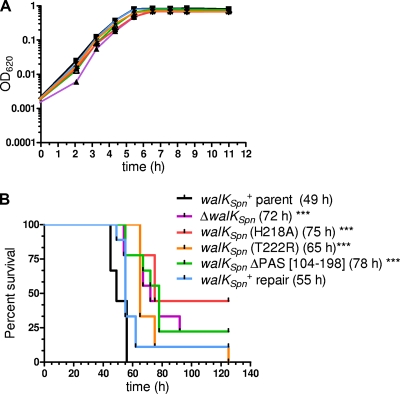
Growth and virulence properties of walKSpn+ and ΔwalKSpn mutant strains. Strain constructions, growths, and survival curve analyses were performed as described in Materials and Methods on the following strains: D39 rpsL1 parent (IU1781), D39 rpsL1 ΔwalKSpn (IU1896), D39 rpsL1 walKSpn (H218A) (IU3102), D39 rpsL1 walKSpn (T222R) (IU3104), D39 rpsL1 walKSpn ΔPAS[104-198] (IU2306), and D39 rpsL1 walKSpn+ repair (IU2193). (A) Representative growth curve of static BHI broth cultures at 37°C in an atmosphere of 5% CO2. The experiment was repeated numerous times for each strain. (B) Survival curve analysis of a murine pneumonia model using intranasal inoculation of nine mice for each bacterial strain. Median survival times are in parentheses, where “***” denotes P < 0.005 in log-rank (Mantel-Cox) tests. Similar results were obtained from an independent experiment using six mice per strain.
All strains grew at approximately the same rate (Fig. 7A), although the ΔwalKSpn and walKSpn(H218A) mutants, which lacked autokinase activity (Table 1, lines 3 and 7), consistently had lower growth yields (OD620 = 0.59 ± 0.03 and 0.60 ± 0.02, respectively) than the walKSpn+ parent and the walKSpn(T222R) and walKSpn(ΔPAS[104-198]) mutants (OD620 = 0.82 ± 0.02, 0.71 ± 0.07, and 0.79 ± 0.02, respectively). Repair of the ΔwalKSpn deletion back to wild-type restored the growth yield (OD620 = 0.80 ± 0.03). Finally, the walKSpn(H218A), walKSpn(T222R), walKSpn(ΔPAS[104-198]), and ΔwalKSpn mutants were all significantly attenuated for virulence to approximately the same extent (median survival time ≈ 73 h) in a murine pneumonia model compared to the walKSpn+ parent and repaired strains (median survival time ≈ 52 h) (Fig. 7B). Thus, WalKSpn containing an intact PAS domain is required for full virulence of serotype strain D39.
DISCUSSION
The data presented here show that the autokinase and phosphoryltransfer reactions of the WalRKSpn TCS do not depend strongly on the presence of the PAS domain under standard in vitro reaction conditions (Tables 1 and 2; Fig. 2 and 3). In contrast, the PAS domain is required for optimal WalKSpn phosphatase activity (Fig. 4, 5, and 6; Table 3). Prior to the present study, a WalKSpn phosphatase activity had been inferred by an inability to detect WalRSpn∼P in the presence of WalKSpn bound to membrane vesicles (89); however, a phosphatase activity was not demonstrated directly, although bioinformatic analysis had predicted this activity for WalKBsu and WalKSau (3). The data from WalRK TCSs in several bacterial species suggest that phosphorylated WalR∼P is required for positive activation of their regulons (18, 23, 58, 59). Since the WalRSpn∼P autophosphatase activity is extremely low (Table 3), the WalKSpn phosphatase system may play an important role in resetting the system back to the unphosphorylated WalR “off” state.
Signaling through the PAS domain may predominate in modulating WalR phosphorylation state in Streptococcus species, because WalKSpn lacks an extracytoplasmic domain and one of the transmembrane domains present in many other WalK homologues (47, 60, 66, 89, 93). In addition, the WalHI (YycHI) extracytoplasmic proteins that modulate WalK activity in B. subtilis and other species (84-86) are absent in Streptococcus species. The signals that impinge on the PAS domains of WalKSpn and its homologues in other bacteria are unknown (14, 93), and it remains to be determined whether binding of small molecules or proteins to the PAS domain modulates WalKSpn phosphatase activity in vivo. The mutant WalKSpn proteins containing the internal deletion of the PAS domain or amino acid replacements in the DHp domain characterized here (Tables 1 and 3) are stable in pneumococcal cells (see Results), and these mutational changes in WalKSpn attenuated pneumococcal virulence (Fig. 7 and below).
A substantial body of evidence supports a model in which the autokinase and phosphatase activities of EnvZ are balanced to regulate OmpR∼P amount (6, 36, 40, 69, 97). High osmolarity is thought to favor the EnvZ histidine autokinase activity that leads to phosphorylation of OmpR, whereas low osmolarity favors the EnvZ phosphatase activity that dephosphorylates OmpR∼P. In addition, a recently discovered modulator of EnvZ, called MzrA, leads to increased amounts of OmpR∼P, possibly by modulating the autokinase/phosphatase balance (26). However, EnvZ does not contain a cytoplasmic PAS domain, and the exact mechanisms of EnvZ signaling are still largely unknown. There are also some discrepancies between binding and kinetic data that remain to be resolved (45, 97). Modulation of the balance between autokinase and phosphatase activity has also been studied in detail for other histidine kinases that lack cytoplasmic PAS domains. For example, Mg2+ binding to an external sensing domain stimulates PhoQ phosphatase activity of serovar Typhimurium (11, 55).
There are a relatively limited number of precedents of cytoplasmic PAS domains regulating the phosphatase activity of histidine kinases. The phosphatase of the ResE histidine kinase is negatively regulated by anaerobiosis, and the ResE PAS domain may contribute to this process, although this has not yet been established experimentally (5, 56). A strong precedent is provided by the NtrB-NtrA TCS of E. coli. The CA catalytic domain of the NtrB histidine kinase inhibits the NtrA∼P phosphatase that resides in the DHp domain (39, 46). The DHp domain alone of NtrB (and EnvZ) is sufficient for phosphatase activity (39, 46, 99). Attempts to purify and test the DHp domain from WalKSpn were not successful, because the domain was insoluble (see Table S4 in the supplemental material). Inhibition of NtrB phosphatase activity in the DHp domain is relieved by the binding of the PII regulatory protein to the CA catalytic domain in the presence of cofactors, such as AMP-PNP (39, 62-64). The resulting conformational change allows activation of the NtrB phosphatase activity. The PAS domain has been proposed to act as an “anvil” that stabilizes the DHp phosphatase activity (39, 63). walRK operons do not encode a homologue to the gene that encodes PII. However, walRK operons do encode a β-lactamase-fold protein, called WalJ (YycJ; VicX), that can influence WalRK function under some conditions (58). The functions of WalJSpn remain to be determined.
Another important precedent appeared in a recent report of the structure of the complex formed between a PAS-containing ThkA histidine kinase and its TrrA cognate response regulator from Thermotoga maritima (95). This structural model has several features relevant to the work reported here. The PAS domain of the ThkA does not dimerize in the complete structure, but rather forms contacts with the CA catalytic domain. This interaction bends the DHp domain toward the CA catalytic domain. In addition, there were two distinct interactions between the DHp domain of ThkA and the TrrA response regulator and an unanticipated third interaction between the PAS domain and TrrA (95). These multiple interactions likely underlie the complex effects that analogous amino acid replacements in the DHp and CA domains have on the autokinase and phosphatase activities of different histidine kinases, such as EnvZ (17, 36, 98), NtrB (39, 63), and WalKSpn (see above). Similar to the findings presented here, an initial study of the autophosphatase, phosphoryltransferase, and phosphatase activities of the ThkA-TrrA TCS revealed that deletion of the PAS domain strongly reduced the ThkA phosphatase activity without significantly changing the autokinase or phosphoryltransferase activities (95). The interaction between the ThkA PAS domain and TrrA was invoked as a possible explanation for this large decrease in phosphatase activity. The detailed kinetic results in this report suggest a similar type of interaction for the WalKSpn-WalRSpn TCS and extend the dependence of the phosphatase activity on the PAS domain from a thermophilic to a mesophilic TCS.
The walKSpn(H218A) mutation that eliminated autokinase activity in truncated purified WalKSpn constructs (Table 1) caused reduced growth yields similar to the ΔwalKSpn deletion mutant (see Results). In contrast, the walKSpn(T222R) and walKSpn(ΔPAS[104-198]) mutants did not display reduced growth yields (Results). Microarray analysis of the ΔwalKSpn mutant compared to the walKSpn+ parent grown under these conditions revealed that significant changes in relative transcript amounts were confined to genes in the WalRKSpn regulon (unpublished result). Relative transcript amounts decreased in the ΔwalKSpn mutant by 2.0- to 4.4-fold for different genes in the regulon. Therefore, regulation by the WalKSpn histidine kinase was specific to its regulon under these condition.
All four walKSpn mutants were significantly attenuated to about the same extent in a pneumonia model of infection compared to the walKSpn+ parent and repaired strains (Fig. 7B). This result contrasts with a previous paper claiming that a walKSpn::kan insertion mutant was avirulent in a similar D39 strain (42). A major difference between the present study and the previous one is that we did not passage mutants through mice before characterization because of the possibility of selecting for additional mutations. In addition, the walKSpn(H218A) and walKSpn(T222R) mutants contain missense mutations that did not affect WalKSpn amounts (see Results). This result strongly argues against polarity effects on expression of downstream walJSpn as a cause for the reduced virulence. However, we cannot ascribe the reduced virulence of the walKSpn(H218A), walKSpn(T222R), and walKSpn(ΔPAS[104-198]) mutants solely to reduced WalRSpn∼P phosphatase activity (Table 3; Fig. 5). Purified WalKSpn(H218A) or WalKSpn(T222R) also lacked or had very low autokinase activity, respectively (Table 1), and even WalKSpn(ΔPAS[104-198]) had a moderately increased Km for ATP in the autokinase reaction, although its relative kcat was unchanged (Table 1). Therefore, if the pneumococcal ATP pool decreases during infection, then autophosphorylation of WalKSpn(ΔPAS[104-198]) could become kinetically limited. Nevertheless, taken together, these results indicate that the WalKSpn histidine kinase through its regulation of the WalKSpn regulon is required for normal growth in culture and that an intact WalKSpn PAS domain is required for full virulence in this pneumonia model of infection.
Supplementary Material
Acknowledgments
We thank Smirla Ramos for discussions and reading the manuscript, Tiffany Tsui for unpublished microarray data, and Jonathon Day for assistance with the CD data analysis.
This project was supported by grant AI060744 (to M.E.W.) from the National Institute of Allergy and Infectious Diseases.
The contents of this study are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Footnotes
Published ahead of print on 26 February 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ahn, S. J., Z. T. Wen, and R. A. Burne. 2007. Effects of oxygen on virulence traits of Streptococcus mutans. J. Bacteriol. 189:8519-8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiba, H., F. Nakasai, S. Mizushima, and T. Mizuno. 1989. Evidence for the physiological importance of the phosphotransfer between the two regulatory components, EnvZ and OmpR, in osmoregulation in Escherichia coli. J. Biol. Chem. 264:14090-14094. [PubMed] [Google Scholar]
- 3.Alves, R., and M. A. Savageau. 2003. Comparative analysis of prototype two-component systems with either bifunctional or monofunctional sensors: differences in molecular structure and physiological function. Mol. Microbiol. 48:25-51. [DOI] [PubMed] [Google Scholar]
- 4.Barendt, S. M., A. D. Land, L. T. Sham, W. L. Ng, H. C. Tsui, R. J. Arnold, and M. E. Winkler. 2009. Influences of capsule on the cell shape and chaining of wild-type and pcsB mutants of serotype 2 Streptococcus pneumoniae. J. Bacteriol. 191:3024-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baruah, A., B. Lindsey, Y. Zhu, and M. M. Nakano. 2004. Mutational analysis of the signal-sensing domain of ResE histidine kinase from Bacillus subtilis. J. Bacteriol. 186:1694-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batchelor, E., and M. Goulian. 2003. Robustness and the cycle of phosphorylation and dephosphorylation in a two-component regulatory system. Proc. Natl. Acad. Sci. U. S. A. 100:691-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belcheva, A., and D. Golemi-Kotra. 2008. A close-up view of the VraSR two-component system. A mediator of Staphylococcus aureus response to cell wall damage. J. Biol. Chem. 283:12354-12364. [DOI] [PubMed] [Google Scholar]
- 8.Bibikov, S. I., L. A. Barnes, Y. Gitin, and J. S. Parkinson. 2000. Domain organization and flavin adenine dinucleotide-binding determinants in the aerotaxis signal transducer Aer of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:5830-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bisicchia, P., D. Noone, E. Lioliou, A. Howell, S. Quigley, T. Jensen, H. Jarmer, and K. M. Devine. 2007. The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis. Mol. Microbiol. 65:180-200. [DOI] [PubMed] [Google Scholar]
- 10.Cai, S. J., and M. Inouye. 2002. EnvZ-OmpR interaction and osmoregulation in Escherichia coli. J. Biol. Chem. 277:24155-24161. [DOI] [PubMed] [Google Scholar]
- 11.Castelli, M. E., E. Garcia-Vescovi, and F. C. Soncini. 2000. The phosphatase activity is the target for Mg2+ regulation of the sensor protein PhoQ in Salmonella. J. Biol. Chem. 275:22948-22954. [DOI] [PubMed] [Google Scholar]
- 12.Clausen, V. A., W. Bae, J. Throup, M. K. Burnham, M. Rosenberg, and N. G. Wallis. 2003. Biochemical characterization of the first essential two-component signal transduction system from Staphylococcus aureus and Streptococcus pneumoniae. J. Mol. Microbiol. Biotechnol. 5:252-260. [DOI] [PubMed] [Google Scholar]
- 13.Copeland, R. A. 2000. Enzymes: a practical introduction to structure, mechanism, and data analysis, p. 136. Wiley-VCH, New York, NY.
- 14.Dubrac, S., P. Bisicchia, K. M. Devine, and T. Msadek. 2008. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol. Microbiol. 70:1307-1322. [DOI] [PubMed] [Google Scholar]
- 15.Dubrac, S., I. G. Boneca, O. Poupel, and T. Msadek. 2007. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J. Bacteriol. 189:8257-8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubrac, S., and T. Msadek. 2004. Identification of genes controlled by the essential YycG/YycF two-component system of Staphylococcus aureus. J. Bacteriol. 186:1175-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dutta, R., T. Yoshida, and M. Inouye. 2000. The critical role of the conserved Thr247 residue in the functioning of the osmosensor EnvZ, a histidine kinase/phosphatase, in Escherichia coli. J. Biol. Chem. 275:38645-38653. [DOI] [PubMed] [Google Scholar]
- 18.Echenique, J. R., and M. C. Trombe. 2001. Competence repression under oxygen limitation through the two-component MicAB signal-transducing system in Streptococcus pneumoniae and involvement of the PAS domain of MicB. J. Bacteriol. 183:4599-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eldakak, A., and F. M. Hulett. 2007. Cys303 in the histidine kinase PhoR is crucial for the phosphotransfer reaction in the PhoPR two-component system in Bacillus subtilis. J. Bacteriol. 189:410-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fabret, C., and J. A. Hoch. 1998. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J. Bacteriol. 180:6375-6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foster, J. E., Q. Sheng, J. R. McClain, M. Bures, T. I. Nicas, K. Henry, M. E. Winkler, and R. Gilmour. 2004. Kinetic and mechanistic analyses of new classes of inhibitors of two-component signal transduction systems using a coupled assay containing HpkA-DrrA from Thermotoga maritima. Microbiology 150:885-896. [DOI] [PubMed] [Google Scholar]
- 22.Friedman, L., J. D. Alder, and J. A. Silverman. 2006. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 50:2137-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukuchi, K., Y. Kasahara, K. Asai, K. Kobayashi, S. Moriya, and N. Ogasawara. 2000. The essential two-component regulatory system encoded by yycF and yycG modulates expression of the ftsAZ operon in Bacillus subtilis. Microbiology 146:1573-1583. [DOI] [PubMed] [Google Scholar]
- 24.Fukushima, T., H. Szurmant, E. J. Kim, M. Perego, and J. A. Hoch. 2008. A sensor histidine kinase co-ordinates cell wall architecture with cell division in Bacillus subtilis. Mol. Microbiol. 69:621-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao, R., and A. M. Stock. 2009. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 63:133-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerken, H., E. S. Charlson, E. M. Cicirelli, L. J. Kenney, and R. Misra. 2009. MzrA: a novel modulator of the EnvZ/OmpR two-component regulon. Mol. Microbiol. 72:1408-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giefing, C., A. L. Meinke, M. Hanner, T. Henics, M. D. Bui, D. Gelbmann, U. Lundberg, B. M. Senn, M. Schunn, A. Habel, B. Henriques-Normark, A. Ortqvist, M. Kalin, A. von Gabain, and E. Nagy. 2008. Discovery of a novel class of highly conserved vaccine antigens using genomic scale antigenic fingerprinting of pneumococcus with human antibodies. J. Exp. Med. 205:117-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilles-Gonzalez, M. A., and G. Gonzalez. 1993. Regulation of the kinase activity of heme protein FixL from the two-component system FixL/FixJ of Rhizobium meliloti. J. Biol. Chem. 268:16293-16297. [PubMed] [Google Scholar]
- 29.Goudreau, P. N., P. J. Lee, and A. M. Stock. 1998. Stabilization of the phospho-aspartyl residue in a two-component signal transduction system in Thermotoga maritima. Biochemistry 37:14575-14584. [DOI] [PubMed] [Google Scholar]
- 30.Greenfield, N. J. 2007. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 1:2876-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grimshaw, C. E., S. Huang, C. G. Hanstein, M. A. Strauch, D. Burbulys, L. Wang, J. A. Hoch, and J. M. Whiteley. 1998. Synergistic kinetic interactions between components of the phosphorelay controlling sporulation in Bacillus subtilis. Biochemistry 37:1365-1375. [DOI] [PubMed] [Google Scholar]
- 32.Groban, E. S., E. J. Clarke, H. M. Salis, S. M. Miller, and C. A. Voigt. 2009. Kinetic buffering of cross talk between bacterial two-component sensors. J. Mol. Biol. 390:380-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Head, C. G., A. Tardy, and L. J. Kenney. 1998. Relative binding affinities of OmpR and OmpR-phosphate at the ompF and ompC regulatory sites. J. Mol. Biol. 281:857-870. [DOI] [PubMed] [Google Scholar]
- 34.Howell, A., S. Dubrac, K. K. Andersen, D. Noone, J. Fert, T. Msadek, and K. Devine. 2003. Genes controlled by the essential YycG/YycF two-component system of Bacillus subtilis revealed through a novel hybrid regulator approach. Mol. Microbiol. 49:1639-1655. [DOI] [PubMed] [Google Scholar]
- 35.Howell, A., S. Dubrac, D. Noone, K. I. Varughese, and K. Devine. 2006. Interactions between the YycFG and PhoPR two-component systems in Bacillus subtilis: the PhoR kinase phosphorylates the non-cognate YycF response regulator upon phosphate limitation. Mol. Microbiol. 59:1199-1215. [DOI] [PubMed] [Google Scholar]
- 36.Hsing, W., and T. J. Silhavy. 1997. Function of conserved histidine-243 in phosphatase activity of EnvZ, the sensor for porin osmoregulation in Escherichia coli. J. Bacteriol. 179:3729-3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Igo, M. M., A. J. Ninfa, J. B. Stock, and T. J. Silhavy. 1989. Phosphorylation and dephosphorylation of a bacterial transcriptional activator by a transmembrane receptor. Genes Dev. 3:1725-1734. [DOI] [PubMed] [Google Scholar]
- 38.Jansen, A., M. Turck, C. Szekat, M. Nagel, I. Clever, and G. Bierbaum. 2007. Role of insertion elements and yycFG in the development of decreased susceptibility to vancomycin in Staphylococcus aureus. Int. J. Med. Microbiol. 297:205-215. [DOI] [PubMed] [Google Scholar]
- 39.Jiang, P., M. R. Atkinson, C. Srisawat, Q. Sun, and A. J. Ninfa. 2000. Functional dissection of the dimerization and enzymatic activities of Escherichia coli nitrogen regulator II and their regulation by the PII protein. Biochemistry 39:13433-13449. [DOI] [PubMed] [Google Scholar]
- 40.Jin, T., and M. Inouye. 1993. Ligand binding to the receptor domain regulates the ratio of kinase to phosphatase activities of the signaling domain of the hybrid Escherichia coli transmembrane receptor, Taz1. J. Mol. Biol. 232:484-492. [DOI] [PubMed] [Google Scholar]
- 41.Jordan, S., M. I. Hutchings, and T. Mascher. 2008. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol. Rev. 32:107-146. [DOI] [PubMed] [Google Scholar]
- 42.Kadioglu, A., J. Echenique, S. Manco, M. C. Trombe, and P. W. Andrew. 2003. The MicAB two-component signaling system is involved in virulence of Streptococcus pneumoniae. Infect. Immun. 71:6676-6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kazmierczak, K. M., K. J. Wayne, A. Rechtsteiner, and M. E. Winkler. 2009. Roles of relSpn in stringent response, global regulation and virulence of serotype 2 Streptococcus pneumoniae D39. Mol. Microbiol. 72:590-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keener, J., and S. Kustu. 1988. Protein kinase and phosphoprotein phosphatase activities of nitrogen regulatory proteins NTRB and NTRC of enteric bacteria: roles of the conserved amino-terminal domain of NTRC. Proc. Natl. Acad. Sci. U. S. A. 85:4976-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.King, S. T., and L. J. Kenney. 2007. Application of fluorescence resonance energy transfer to examine EnvZ/OmpR interactions. Methods Enzymol. 422:352-360. [DOI] [PubMed] [Google Scholar]
- 46.Kramer, G., and V. Weiss. 1999. Functional dissection of the transmitter module of the histidine kinase NtrB in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 96:604-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lange, R., C. Wagner, A. de Saizieu, N. Flint, J. Molnos, M. Stieger, P. Caspers, M. Kamber, W. Keck, and K. E. Amrein. 1999. Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene 237:223-234. [DOI] [PubMed] [Google Scholar]
- 48.Lanie, J. A., W. L. Ng, K. M. Kazmierczak, T. M. Andrzejewski, T. M. Davidsen, K. J. Wayne, H. Tettelin, J. I. Glass, and M. E. Winkler. 2007. Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J. Bacteriol. 189:38-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laub, M. T., and M. Goulian. 2007. Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 41:121-145. [DOI] [PubMed] [Google Scholar]
- 50.Lesley, J. A., and C. D. Waldburger. 2003. Repression of Escherichia coli PhoP-PhoQ signaling by acetate reveals a regulatory role for acetyl coenzyme A. J. Bacteriol. 185:2563-2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu, M., T. S. Hanks, J. Zhang, M. J. McClure, D. W. Siemsen, J. L. Elser, M. T. Quinn, and B. Lei. 2006. Defects in ex vivo and in vivo growth and sensitivity to osmotic stress of group A Streptococcus caused by interruption of response regulator gene vicR. Microbiology 152:967-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma, P., H. M. Yuille, V. Blessie, N. Gohring, Z. Igloi, K. Nishiguchi, J. Nakayama, P. J. Henderson, and M. K. Phillips-Jones. 2008. Expression, purification and activities of the entire family of intact membrane sensor kinases from Enterococcus faecalis. Mol. Membr. Biol. 25:449-473. [DOI] [PubMed] [Google Scholar]
- 53.Martin, P. K., T. Li, D. Sun, D. P. Biek, and M. B. Schmid. 1999. Role in cell permeability of an essential two-component system in Staphylococcus aureus. J. Bacteriol. 181:3666-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mohedano, M. L., K. Overweg, A. de la Fuente, M. Reuter, S. Altabe, F. Mulholland, D. de Mendoza, P. Lopez, and J. M. Wells. 2005. Evidence that the essential response regulator YycF in Streptococcus pneumoniae modulates expression of fatty acid biosynthesis genes and alters membrane composition. J. Bacteriol. 187:2357-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Montagne, M., A. Martel, and H. Le Moual. 2001. Characterization of the catalytic activities of the PhoQ histidine protein kinase of Salmonella enterica serovar Typhimurium. J. Bacteriol. 183:1787-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakano, M. M., and Y. Zhu. 2001. Involvement of ResE phosphatase activity in down-regulation of ResD-controlled genes in Bacillus subtilis during aerobic growth. J. Bacteriol. 183:1938-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ng, W. L., K. M. Kazmierczak, and M. E. Winkler. 2004. Defective cell wall synthesis in Streptococcus pneumoniae R6 depleted for the essential PcsB putative murein hydrolase or the VicR (YycF) response regulator. Mol. Microbiol. 53:1161-1175. [DOI] [PubMed] [Google Scholar]
- 58.Ng, W. L., G. T. Robertson, K. M. Kazmierczak, J. Zhao, R. Gilmour, and M. E. Winkler. 2003. Constitutive expression of PcsB suppresses the requirement for the essential VicR (YycF) response regulator in Streptococcus pneumoniae R6. Mol. Microbiol. 50:1647-1663. [DOI] [PubMed] [Google Scholar]
- 59.Ng, W. L., H. C. Tsui, and M. E. Winkler. 2005. Regulation of the pspA virulence factor and essential pcsB murein biosynthetic genes by the phosphorylated VicR (YycF) response regulator in Streptococcus pneumoniae. J. Bacteriol. 187:7444-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ng, W. L., and M. E. Winkler. 2004. Singular structures and operon organizations of essential two-component systems in species of Streptococcus. Microbiology 150:3096-3098. [DOI] [PubMed] [Google Scholar]
- 61.Noriega, C. E., R. Schmidt, M. J. Gray, L. L. Chen, and V. Stewart. 2008. Autophosphorylation and dephosphorylation by soluble forms of the nitrate-responsive sensors NarX and NarQ from Escherichia coli K-12. J. Bacteriol. 190:3869-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pioszak, A. A., P. Jiang, and A. J. Ninfa. 2000. The Escherichia coli PII signal transduction protein regulates the activities of the two-component system transmitter protein NRII by direct interaction with the kinase domain of the transmitter module. Biochemistry 39:13450-13461. [DOI] [PubMed] [Google Scholar]
- 63.Pioszak, A. A., and A. J. Ninfa. 2003. Genetic and biochemical analysis of phosphatase activity of Escherichia coli NRII (NtrB) and its regulation by the PII signal transduction protein. J. Bacteriol. 185:1299-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pioszak, A. A., and A. J. Ninfa. 2003. Mechanism of the PII-activated phosphatase activity of Escherichia coli NRII (NtrB): how the different domains of NRII collaborate to act as a phosphatase. Biochemistry 42:8885-8899. [DOI] [PubMed] [Google Scholar]
- 65.Qin, L., R. Dutta, H. Kurokawa, M. Ikura, and M. Inouye. 2000. A monomeric histidine kinase derived from EnvZ, an Escherichia coli osmosensor. Mol. Microbiol. 36:24-32. [DOI] [PubMed] [Google Scholar]
- 66.Qin, Z., J. Zhang, B. Xu, L. Chen, Y. Wu, X. Yang, X. Shen, S. Molin, A. Danchin, H. Jiang, and D. Qu. 2006. Structure-based discovery of inhibitors of the YycG histidine kinase: new chemical leads to combat Staphylococcus epidermidis infections. BMC Microbiol. 6:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramos-Montanez, S., H. C. Tsui, K. J. Wayne, J. L. Morris, L. E. Peters, F. Zhang, K. M. Kazmierczak, L. T. Sham, and M. E. Winkler. 2008. Polymorphism and regulation of the spxB (pyruvate oxidase) virulence factor gene by a CBS-HotDog domain protein (SpxR) in serotype 2 Streptococcus pneumoniae. Mol. Microbiol. 67:729-746. [DOI] [PubMed] [Google Scholar]
- 68.Rogers, P. D., T. T. Liu, K. S. Barker, G. M. Hilliard, B. K. English, J. Thornton, E. Swiatlo, and L. S. McDaniel. 2007. Gene expression profiling of the response of Streptococcus pneumoniae to penicillin. J. Antimicrob. Chemother. 59:616-626. [DOI] [PubMed] [Google Scholar]
- 69.Russo, F. D., and T. J. Silhavy. 1991. EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J. Mol. Biol. 222:567-580. [DOI] [PubMed] [Google Scholar]
- 70.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 71.Sanowar, S., and H. Le Moual. 2005. Functional reconstitution of the Salmonella typhimurium PhoQ histidine kinase sensor in proteoliposomes. Biochem. J. 390:769-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Senadheera, M. D., B. Guggenheim, G. A. Spatafora, Y. C. Huang, J. Choi, D. C. Hung, J. S. Treglown, S. D. Goodman, R. P. Ellen, and D. G. Cvitkovitch. 2005. A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J. Bacteriol. 187:4064-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shi, L., W. Liu, and F. M. Hulett. 1999. Decay of activated Bacillus subtilis pho response regulator, PhoP approximately P, involves the PhoR approximately P intermediate. Biochemistry 38:10119-10125. [DOI] [PubMed] [Google Scholar]
- 74.Siryaporn, A., and M. Goulian. 2008. Cross-talk suppression between the CpxA-CpxR and EnvZ-OmpR two-component systems in E. coli. Mol. Microbiol. 70:494-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Skarphol, K., J. Waukau, and S. A. Forst. 1997. Role of His243 in the phosphatase activity of EnvZ in Escherichia coli. J. Bacteriol. 179:1413-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Skerker, J. M., B. S. Perchuk, A. Siryaporn, E. A. Lubin, O. Ashenberg, M. Goulian, and M. T. Laub. 2008. Rewiring the specificity of two-component signal transduction systems. Cell 133:1043-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Skerker, J. M., M. S. Prasol, B. S. Perchuk, E. G. Biondi, and M. T. Laub. 2005. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 3:e334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sourjik, V., and R. Schmitt. 1998. Phosphotransfer between CheA, CheY1, and CheY2 in the chemotaxis signal transduction chain of Rhizobium meliloti. Biochemistry 37:2327-2335. [DOI] [PubMed] [Google Scholar]
- 79.Sreerama, N., S. Y. Venyaminov, and R. W. Woody. 1999. Estimation of the number of alpha-helical and beta-strand segments in proteins using circular dichroism spectroscopy. Protein Sci. 8:370-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sreerama, N., and R. W. Woody. 1993. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal. Biochem. 209:32-44. [DOI] [PubMed] [Google Scholar]
- 81.Stewart, R. C. 1997. Kinetic characterization of phosphotransfer between CheA and CheY in the bacterial chemotaxis signal transduction pathway. Biochemistry 36:2030-2040. [DOI] [PubMed] [Google Scholar]
- 82.Sung, C. K., H. Li, J. P. Claverys, and D. A. Morrison. 2001. An rpsL cassette, Janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 67:5190-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Swem, L. R., X. Gong, C. A. Yu, and C. E. Bauer. 2006. Identification of a ubiquinone-binding site that affects autophosphorylation of the sensor kinase RegB. J. Biol. Chem. 281:6768-6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Szurmant, H., L. Bu, C. L. Brooks III, and J. A. Hoch. 2008. An essential sensor histidine kinase controlled by transmembrane helix interactions with its auxiliary proteins. Proc. Natl. Acad. Sci. U. S. A. 105:5891-5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Szurmant, H., M. A. Mohan, P. M. Imus, and J. A. Hoch. 2007. YycH and YycI interact to regulate the essential YycFG two-component system in Bacillus subtilis. J. Bacteriol. 189:3280-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Szurmant, H., K. Nelson, E. J. Kim, M. Perego, and J. A. Hoch. 2005. YycH regulates the activity of the essential YycFG two-component system in Bacillus subtilis. J. Bacteriol. 187:5419-5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thomas, S. A., J. A. Brewster, and R. B. Bourret. 2008. Two variable active site residues modulate response regulator phosphoryl group stability. Mol. Microbiol. 69:453-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tsui, H. C., D. Mukherjee, V. A. Ray, L.-T. Sham, A. L. Feig, and M. E. Winkler. 2010. Identification and characterization of noncoding small RNAs in Streptococcus pneumoniae serotype 2 strain D39. J. Bacteriol. 192:264-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wagner, C., A. Saizieu Ad, H. J. Schonfeld, M. Kamber, R. Lange, C. J. Thompson, and M. G. Page. 2002. Genetic analysis and functional characterization of the Streptococcus pneumoniae vic operon. Infect. Immun. 70:6121-6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang, L., C. Fabret, K. Kanamaru, K. Stephenson, V. Dartois, M. Perego, and J. A. Hoch. 2001. Dissection of the functional and structural domains of phosphorelay histidine kinase A of Bacillus subtilis. J. Bacteriol. 183:2795-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Watanabe, T., Y. Hashimoto, Y. Umemoto, D. Tatebe, E. Furuta, T. Fukamizo, K. Yamamoto, and R. Utsumi. 2003. Molecular characterization of the essential response regulator protein YycF in Bacillus subtilis. J. Mol. Microbiol. Biotechnol. 6:155-163. [DOI] [PubMed] [Google Scholar]
- 92.Whitmore, L., and B. A. Wallace. 2008. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers 89:392-400. [DOI] [PubMed] [Google Scholar]
- 93.Winkler, M. E., and J. A. Hoch. 2008. Essentiality, bypass, and targeting of the YycFG (VicRK) two-component regulatory system in gram-positive bacteria. J. Bacteriol. 190:2645-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wright, G. D., T. R. Holman, and C. T. Walsh. 1993. Purification and characterization of VanR and the cytosolic domain of VanS: a two-component regulatory system required for vancomycin resistance in Enterococcus faecium BM4147. Biochemistry 32:5057-5063. [DOI] [PubMed] [Google Scholar]
- 95.Yamada, S., H. Sugimoto, M. Kobayashi, A. Ohno, H. Nakamura, and Y. Shiro. 2009. Structure of PAS-linked histidine kinase and the response regulator complex. Structure 17:1333-1344. [DOI] [PubMed] [Google Scholar]
- 96.Yamamoto, K., T. Kitayama, S. Minagawa, T. Watanabe, S. Sawada, T. Okamoto, and R. Utsumi. 2001. Antibacterial agents that inhibit histidine protein kinase YycG of Bacillus subtilis. Biosci. Biotechnol. Biochem. 65:2306-2310. [DOI] [PubMed] [Google Scholar]
- 97.Yoshida, T., S. Cai, and M. Inouye. 2002. Interaction of EnvZ, a sensory histidine kinase, with phosphorylated OmpR, the cognate response regulator. Mol. Microbiol. 46:1283-1294. [DOI] [PubMed] [Google Scholar]
- 98.Zhu, Y., and M. Inouye. 2002. The role of the G2 box, a conserved motif in the histidine kinase superfamily, in modulating the function of EnvZ. Mol. Microbiol. 45:653-663. [DOI] [PubMed] [Google Scholar]
- 99.Zhu, Y., L. Qin, T. Yoshida, and M. Inouye. 2000. Phosphatase activity of histidine kinase EnvZ without kinase catalytic domain. Proc. Natl. Acad. Sci. U. S. A. 97:7808-7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.