Abstract
With an obligate intracellular lifestyle, Alphaproteobacteria of the order Rickettsiales have inextricably coevolved with their various eukaryotic hosts, resulting in small, reductive genomes and strict dependency on host resources. Unsurprisingly, large portions of Rickettsiales genomes encode proteins involved in transport and secretion. One particular transporter that has garnered recent attention from researchers is the type IV secretion system (T4SS). Homologous to the well-studied archetypal vir T4SS of Agrobacterium tumefaciens, the Rickettsiales vir homolog (rvh) T4SS is characterized primarily by duplication of several of its genes and scattered genomic distribution of all components in several conserved islets. Phylogeny estimation suggests a single event of ancestral acquirement of the rvh T4SS, likely from a nonalphaproteobacterial origin. Bioinformatics analysis of over 30 Rickettsiales genome sequences illustrates a conserved core rvh scaffold (lacking only a virB5 homolog), with lineage-specific diversification of several components (rvhB1, rvhB2, and rvhB9b), likely a result of modifications to cell envelope structure. This coevolution of the rvh T4SS and cell envelope morphology is probably driven by adaptations to various host cells, identifying the transporter as an important target for vaccine development. Despite the genetic intractability of Rickettsiales, recent advancements have been made in the characterization of several components of the rvh T4SS, as well as its putative regulators and substrates. While current data favor a role in effector translocation, functions in DNA uptake and release and/or conjugation cannot at present be ruled out, especially considering that a mechanism for plasmid transfer in Rickettsia spp. has yet to be proposed.
Type IV secretion systems (T4SSs) are macromolecular complexes that transport protein, DNA, and nucleoprotein across the bacterial cell envelope in both Gram-negative and Gram-positive species, as well as some wall-less bacteria and archaea (1, 32). Functioning in naked DNA uptake and release (60), conjugation (80), and the propagation of genomic islands (69), T4SSs are prominent factors in bacterial diversification and are responsible for the horizontal spread of antimicrobial resistance and virulence genes. T4SSs are also used by some species to deliver effector molecules (DNA and/or protein) into eukaryotic host cells (28), a process that facilitates infection and subsequent pathogenesis. It is assumed that all varieties of T4SSs form a channel that spans the cell envelope and culminates in a surface-exposed structure, such as a pilus (Fig. 1A). Despite this conserved architecture, genetic diversity in a multitude of features, including gene composition and organization, underlies the hundreds of T4SSs identified through genome sequencing. Recently, T4SSs have been classified into four groups: F, P, I, and GI (70). F-T4SSs and P-T4SSs (previously known as type IVA) are widespread systems represented by the archetypes encoded by the F plasmid of Escherichia coli (tra and trb) and the pTi plasmid of Agrobacterium tumefaciens (vir), respectively. I-T4SSs (previously known as type IVB) are typified by the icm/dot system of IncI plasmids, and examples in Legionella spp. and Coxiella burnetii are the best characterized. GI-T4SSs, distinct systems that function in transferring the genomic islands with which they are associated (70, 71), are also widespread and can be further classified into sublineages based on gene content and arrangement (73). The growing diversity of T4SSs will undoubtedly continue to challenge attempts at their classification and the unraveling of their evolutionary origins.
FIG. 1.
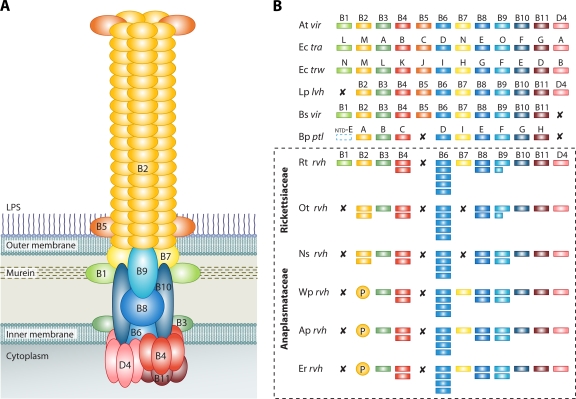
P-T4SSs. (A) Model of the vir P-T4SS encoded on the pTi plasmid of A. tumefaciens. B1 to B11, VirB1 to VirB11; D4, VirD4. (B) Comparison of the rvh P-T4SSs from Rickettsiales with similar P-T4SSs from other bacteria. At vir, A. tumefaciens Ti plasmid P-T4SS; Ec tra, E. coli IncN plasmid pKM101 P-T4SS; Ec trw, E. coli plasmid R388 P-T4SS; Lp lvh, L. pneumophila P-T4SS; Bs vir, Brucella suis P-T4SS; and Bp ptl, B. pertussis P-T4SS. VirB1 of B. pertussis is depicted with the N-terminal glycohydrolase domain of PtlE (ntd-E) (107). The rvh examples are shown within the dashed-line box: Rt rvh, R. typhi P-T4SS; Ot rvh, O. tsutsugamushi P-T4SS; Ns rvh, Neorickettsia sennetsu P-T4SS; Wp rvh, Wolbachia pipientis P-T4SS; Ap rvh, A. phagocytophilum P-T4SS; and Er rvh, Ehrlichia ruminantium P-T4SS. X indicates that no gene for the component has been annotated and no subjects were detectable using tblastn; P represents the proliferation of rvhB2 genes, putative VirB2-like encoding genes.
Alphaproteobacteria of the order Rickettsiales are diverse obligate intracellular species with a wide range of eukaryotic hosts (22, 23, 105, 125). Many species within the two well-characterized families, Anaplasmataceae and Rickettsiaceae, pose severe threats to livestock and human health. The agricultural and medical ramifications have resulted in the rapid accumulation of over 30 complete or nearly complete genome sequences from a diverse array of Rickettsiales taxa. Despite the common ancestry (127) and strictly intracellular lifestyles of Rickettsiales, the manner of genome reduction and reliance on host resources vary greatly across lineages (36, 63, 95). While few syntenic regions are found across Rickettsiales genomes (63), a conserved P-T4SS is a particularly definitive feature of these bacteria. Since the completion of sequencing of the first Rickettsiales genome, that of Rickettsia prowazekii (5), a reduced P-T4SS (lacking homologs of virB1, virB2, virB5, and virB7) has been uncovered in all subsequently sequenced genomes, with anomalous duplication of genes homologous to virB4, virB6, virB8, and virB9 suggesting rich functionality and with genes split into multiple islets across the genomes. We recently performed a detailed informatics analysis of the P-T4SS of Rickettsia spp. and concluded that, relative to the canonical vir P-T4SS of A. tumefaciens, this transporter lacks only a homolog of virB5, the gene encoding the minor pilus subunit (55).
In this review, we expand our prior analysis of the Rickettsia T4SS, in which we named this transporter rvh (Rickettsiales vir homolog), to encompass T4SSs of all Rickettsiales (Fig. 1B). An assumption is made that the acquisition of a P-T4SS was pivotal in the transition from an extracellular to an obligate intracellular lifestyle. We address the nature of duplication of rvh components (rvhB4, rvhB8, and rvhB9) and proliferation of another component (rvhB6) and draw special attention to the components that tend to elude automated genome annotation (rvhB1, rvhB2, and rvhB7). The translocated proteins encoded by the latter genes define the most plastic attributes of the rvh P-T4SS and, coupled with the deletion of a virB5 homolog, imply coevolution of the transporter and the bacterial cell envelope. Despite conservation of the rvh T4SS across the Rickettsiales, there is little information regarding rvh regulation and substrate transport. Learning more about the manner in which the various rvh T4SSs assemble and function in the bacterial outer membrane (OM) may present novel opportunities for vaccine development and drug targeting, and we discuss these possibilities in relation to rvh adaptations to host cell environments.
LATERAL ACQUISITION OF THE rvh P-T4SS
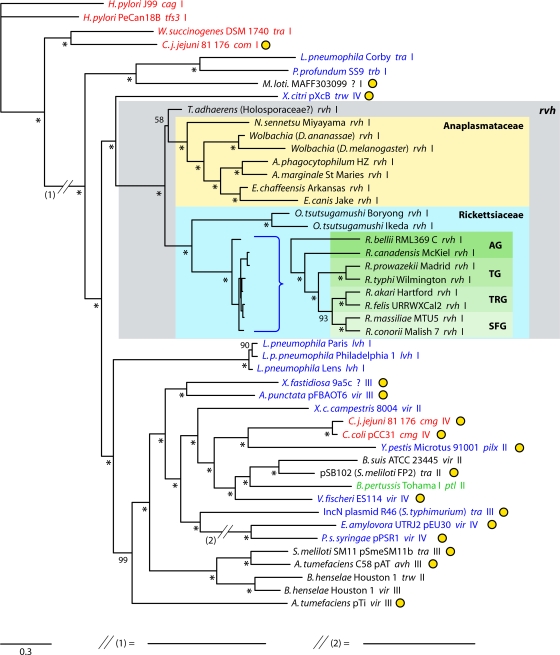
Previously, it was determined by phylogenetic analysis of P-T4SSs that lateral gene transfer (LGT) has spread these transporters across divergent bacterial lineages and that effector molecule translocation has evolved from the primitive function of conjugation (50). Consistent with this observation, it has been demonstrated previously that the rvh T4SS is related to P-T4SSs from certain Gammaproteobacteria (Legionella spp. and Photobacterium profundum) and Epsilonproteobacteria (Helicobacter pylori, Wolinella succinogenes, and Campylobacter jejuni) (26, 93). Our phylogeny estimation for 47 P-T4SSs based on half of the vir-like components (virB4, virB8 to virB11, and virD4) is in accord with this notion that the rvh P-T4SS was derived from a non-Alphaproteobacteria ancestor (Fig. 2). We recovered the trw P-T4SS, carried on plasmid pXcB of the Gammaproteobacteria species Xanthomonas citri, as the closest xenolog of rvh, as in a previous analysis based on 73 VirB4 and VirB4-like proteins (50). As demonstrated in other studies, the lvh P-T4SSs of Legionella spp., the trb P-T4SS of P. profundum, and the unnamed P-T4SS carried on plasmid pMLa of Mesorhizobium loti are the next closest xenologs of rvh, all having branched from the P-T4SSs of H. pylori, C. jejuni, and W. succinogenes that are involved in DNA uptake and release.
FIG. 2.
Estimated phylogeny for 47 diverse P-T4SSs. Taxa are colored according to proteobacterial class: red, Epsilonproteobacteria; blue, Gammaproteobacteria; black, Alphaproteobacteria; and green, Betaproteobacteria. Each taxon name is appended with its P-T4SS nomenclature and a number (I, II, III, or IV) referring to recently proposed categories of P-T4SSs (93). Plasmid-encoded P-T4SSs are indicated by yellow circles. The 18 sampled Rickettsiales taxa are within a gray box. Rickettsia spp. are shown as a cladogram due to limited sequence divergence relative to that in the remaining taxa, with the following abbreviations: AG, ancestral group; TG, typhus group; TRG, transitional group; and SFG, spotted fever group (56, 57). The tree is from two independent Bayesian analyses of six P-T4SS proteins (VirB4, VirB8 to VirB11, and VirD4). The topology of the sampled tree with the greatest likelihood (Lnl = −201,623.248) is shown, with branch support assessed from probabilities of clade occurrence in the posterior distribution of 1,906 sampled trees. Asterisks indicate probabilities of 100%. For Wolbachia species, the species in which they are symbionts are listed in parentheses (D. ananassae, Drosophila ananassae; D. melanogaster, Drosophila melanogaster). C. j. jejuni, C. jejuni subsp. jejuni; T. adhaerens, “Trichoplax adhaerens”; L. p. pneumophila, L. pneumophila subsp. pneumophila; X. fastidiosa, Xylella fastidiosa; A. punctata, Aeromonas punctata; X. c. campestris, Xanthomonas campestris pv. campestris; C. coli, Campylobacter coli; Y. pestis, Yersinia pestis; S. meliloti, Sinorhizobium meliloti; V. fischeri, Vibrio fischeri; S. typhimurium, Salmonella enterica serovar Typhimurium; E. amylovora, Erwinia amylovora; P. s. syringae, Pseudomonas syringae subsp. syringae; B. henselae, Bartonella henselae.
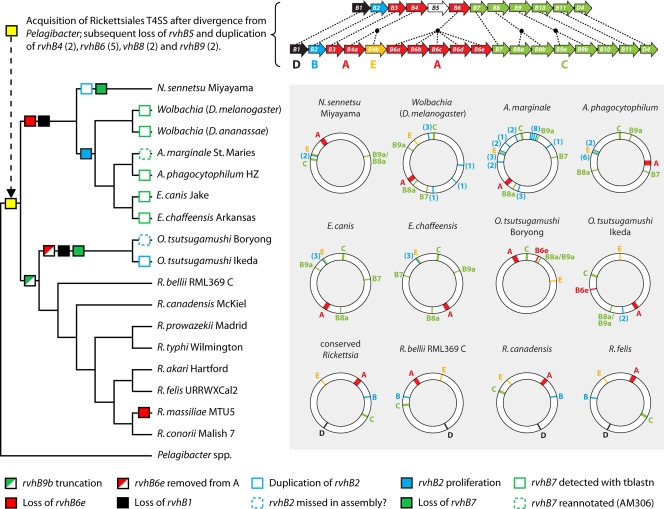
Our analysis, as well as the findings of prior studies (55, 106, 108), supports the vertical transmission of the rvh T4SS after the split of “Candidatus Pelagibacter” from the Rickettsiales ancestor. The most parsimonious recreation of an LGT event that equipped this ancestor with a P-T4SS involves a transporter containing 12 genes (rvhB1 to rvhB11 and rvhD4), with ancestral deletion, recombination, and duplication events creating a modified transporter comprising 18 genes (rvhB1 to rvhB3, rvhB4a and rvhB4b, rvhB6a to rvhB6e, rvhB7, rvhB8a and rvhB8b, rvhB9a and rvhB9b, rvhB10, rvhB11, and rvhD4). The modern genomic distribution of this ancestral operon is fragmented across all sequenced Rickettsiales genomes (Fig. 3; see also Fig. S1 in the supplemental material) and, coupled with lineage-specific gene deletion, proliferation, and truncation events, defines a variable rvh P-T4SS.
FIG. 3.
Diversification in rvh T4SS architecture across the major lineages of Rickettsiales. The schematic at the top depicts the deletion and duplication events that possibly occurred prior to the split of the major Rickettsiales lineages (corresponding to the yellow box at the root of the cladogram). The color scheme for the rvh components is consistent throughout the figure and reflects the five islets (A to E) of the Rickettsia rvh P-T4SS described previously (55). The cladogram is simplified from data obtained by phylogeny estimation across 31 rvh P-T4SSs (see Fig. S1 in the supplemental material). Colored boxes in the cladogram depict deviations from the ancestral rvh P-T4SS and are explained at the bottom. Synteny maps (shaded inset) are shown for select taxa. Note that in Rickettsiales other than Rickettsia spp., rvhB9a, rvhB8a, and rvhB7 are removed from islet C. Also, multiple rvhB2 orthologs per genome are depicted, with corresponding numbers in parentheses.
NATURE OF rvh DUPLICATION
For Rickettsia spp., we reported previously that only one ortholog of each of the singly duplicated genes (rvhB4a, rvhB8b, and rvhB9a) adhered to the conserved properties of similar genes in other P-T4SSs (55). Both rvhB4b and rvhB8a encode proteins with similar defects in Rickettsia spp. and the other Rickettsiales, as observed in our prior informatics analysis (see Table S1 in the supplemental material). In contrast, the product of rvhB9b, which in Rickettsia spp. is missing the C-terminal domain, demonstrated previously via nuclear magnetic resonance (NMR) (16), cryo-electron microscopy (52), and crystallography (29) to bind VirB7 in other systems, is full-length in the Anaplasmataceae (see Table S1 in the supplemental material). We had hypothesized (55), given the mirrored arrangements of the rvhB9a-rvhB8a locus (antisense) and the rvhB8b-rvhB9b locus (sense), with rvhB7 at the center of these loci (Fig. 3), that an intrachromosomal recombination event truncated rvhB9b in Rickettsia spp.; however, this pattern does hold throughout the Rickettsiales order. rvhB9b is also truncated in the genomes of Orientia tsutsugamushi strains, yet the rvhB8b-rvhB9b locus is well separated from the rvhB9a-rvhB8a locus in O. tsutsugamushi genomes, as it is in the Anaplasmataceae genomes. The exact manner in which Rickettsiaceae rvhB9b became truncated is unclear; either the entire C islet (rvhB9a rvhB8a rvhB7 rvhB8b rvhB9b rvhB10 rvhB11 rvhD4) exhibits the ancestral gene order and the rvhB9a-rvhB8a-rvhB7 segment became separated outside Rickettsia spp. or a recombination event coupled the duplicate rvhB9 and rvhB8 genes in Rickettsia spp. to the remaining genes within islet C. Nonetheless, a major difference between the T4SSs of Anaplasmataceae and Rickettsiaceae is the presence of two full-length copies of rvhB9 in the former and a truncated rvhB9b in the latter.
The proliferated rvhB6 genes, encoding proteins RvhB6a to RvhB6d, are present in most sequenced Rickettsiales genomes and are arrayed contiguously within one predicted operon (Fig. 3). A fifth gene, rvhB6e, is found only in Rickettsiaceae genomes, and while arrayed with rvhB6a to rvhB6d in the Rickettsia genomes, the gene is well separated from the rvhB6 operon in O. tsutsugamushi genomes. The proteins encoded by rvhB6 are perhaps the most intriguing features of the rvh P-T4SS. While all the proteins contain a complete VirB6/TrbL (PF04610) domain that is comparable to full-length VirB6 and VirB6-like orthologs in other species harboring P-T4SSs, the Rickettsiales open reading frames (ORFs) encode additional flanking regions that lack similarity to any proteins in public databases (55). The nature of these “gangly arms” flanking the VirB6/TrbL domain, coupled with the unusual proliferation of the corresponding genes in the highly reduced Rickettsiales genomes, in which gene duplication is atypical, brings attention to these curious proteins. VirB6/TrbL proteins resemble ComEC channel proteins, which are involved in the uptake of environmental DNA (42). It is possible that RvhB6 proteins play a role in DNA import/export, with the various duplications forming multiple diverse channels that maximize the potential for LGT in environments (such as those in protozoa and macrophages) with high rates of congener contact. Given that VirB6 in A. tumefaciens has been demonstrated previously to facilitate substrate transfer (66) yet is not a component of the core P-T4SS channel (52), the multiple RvhB6 proteins may equip Rickettsiales with substrate selectivity in the various host environments encountered by these obligate intracellular bacteria. However, the strong selection keeping the rvhB6 loci contiguous, presumably within tightly regulated operons (55), suggests that posttranscriptional regulation of rvhB6 genes would be necessary for environment-specific utilization of the various duplicate proteins in the bacteria. This idea is supported by the rvhB6e gene of O. tsutsugamushi strain Ikeda, which is truncated and well separated from the remaining rvhB6 genes in islet A (Fig. 3). However, all four rvhB6 genes of Ehrlichia chaffeensis are coexpressed in tick and human cells, and various RvhB6-RvhB6 and RvhB6-RvhB9 interactions suggest more than one RvhB6 protein may assemble at the inner membrane (IM) channel region of the rvh T4SS (8). Nonetheless, given that two rickettsial RvhB6-encoding genes (rvhB6d in R. bellii strain OSU 85 389 and rvhB6e in R. massiliae) (see Table S2 in the supplemental material) have undergone pseudogenization, not all copies of these genes may be functional despite the near global conservation across the Rickettsiales.
THE ENIGMATIC RvhB2 PROTEINS
VirB2 and related proteins constitute the major subunit of the Agrobacterium T-pilus and related structures (77, 79). The signal sequences of these proteins are processed in the IM, with subsequent species-specific processing of the C and/or N termini (44, 45, 72, 78, 80, 92, 112). This role in T-pilus formation is supported by findings from various studies demonstrating the presence of VirB2 in complexes with VirB5 (the minor subunit of the T-pilus) and VirB7 (17, 67, 76, 86, 115, 132). In particular, the VirB2-VirB5 pilus complex is dependent on the periplasmic interaction between VirB4 and VirB8, which promotes the formation of extracellular pili (132). Unlike pTi-encoded VirB5 (VirB5Ti), VirB2Ti is critical for substrate transfer through the P-T4SS scaffold (27, 64). Polymers (115) of these proteins probably span the entire periplasm (32), as made evident by the presence of at least two predicted transmembrane-spanning regions in nearly all available sequences (55). Recently, it was hypothesized that VirB2 polymers might “snake” along the VirB10 antenna domain, lining the outside of the entire P-T4SS channel from the IM to the OM (65) and binding VirB5 at the OM, with continual polymerization as a T-pilus in species that protract these extracellular appendages. However, it cannot be ruled out that VirB2 polymerizes within the chamber of the VirB7-VirB9-VirB10 core complex prior to binding VirB5 and extending extracellularly as the T-pilus (51), a hypothesis supported by the observation that the transmembrane helices within the TraF/VirB10 crystal appear to be caved in (in the absence of VirB2) (29).
Several prior reviews of T4SSs reported a lack of genes encoding either the major or the minor pilin subunit within Rickettsiales genomes. However, our recent bioinformatics analysis of 13 Rickettsia genomes identified a candidate VirB2-encoding ORF, rvhB2, predicted to be a single transcriptional unit well separated from other T4SS genes (55). In certain Anaplasmataceae genomes, a duplicated ORF previously named orfX (24, 94), associated with the major surface protein 2 (msp2) superfamily, has also recently been designated a putative virB2 homolog (23, 99). Here, we draw attention to recently identified ORFs in the O. tsutsugamushi and Anaplasmataceae genomes which suggest that VirB2 proteins are likely to be an essential component of the rvh P-T4SS (see references 35 and 114 and Fig. S2 in the supplemental material). Interestingly, unlike the genomes of Rickettsia spp., the remaining Rickettsiales genomes have rvhB2 present at least twice and the genomes of Wolbachia, Anaplasma, and Ehrlichia species show proliferation of this gene (yielding three or more paralogs) (Fig. 3). Together with a closely related ORF named orfY, a total of 22 candidate rvhB2 sequences in the Anaplasma marginale genome were identified (data not shown), exemplifying the operation of selection on the retention of numerous rvhB2 paralogs in the derived Anaplasmataceae. Like that of other duplicate rvh genes in Rickettsiales, the expression of rvhB2 paralogs may be specific to the host environment. Supporting this hypothesis, the rvhB2 paralogs of Anaplasma phagocytophilum are differentially expressed in tick and mammalian cell cultures (99). Alternatively, based on its probable secretion to the OM and likely exposure to the host immune system, the major pilin component may have become co-opted into a diverse antigen family that increases the chance of host immune avoidance, a role other duplicate genes and functional pseudogenes in the msp2 superfamily have (23). Supporting this possibility, the majority of rvhB2 genes in A. marginale are arrayed with msp2 and msp3 genes and their associated functional pseudogenes (25). Regardless of the provisional role that RvhB2 plays in rvh assembly and function, experimental evidence suggests surface exposure of the protein. For example, in A. marginale, RvhB2 induces a T-cell response in cattle as part of a protective bacterial membrane vaccine (89, 119), and the two orthologs in Neorickettsia risticii are coexpressed by an operon and localized predominantly at the poles, where they form focal complexes (85).
RvhB7: THE NEEDLE IN THE HAYSTACK
T4SSs typically encode small lipoproteins under 100 amino acids that are secreted to the periplasm and are essential for substrate transfer. In A. tumefaciens, the small lipoprotein VirB7 primarily binds and stabilizes VirB9 in the OM (13, 47, 118) via an essential disulfide bond (4, 11, 62, 132). Structural studies of this interaction in the P-T4SS encoded by the plasmid pKM101 of E. coli illustrate that a disulfide bond is not universal (16, 52), suggesting that other protein interactions in VirB7-VirB9 heterodimer formation can suffice (9). Along with VirB10 and VirB9, VirB7 is a component of the core T4SS complex, and it is inserted into the OM with the C-terminal domains of VirB9 and VirB10 (52). Prior comprehensive studies of type IV secretion have noted that VirB7 and related proteins are not always encoded within T4SSs (26, 93). However, given their small size, it is likely that many ORFs encoding these lipoproteins are not annotated by automated gene prediction methods, especially if the genomic positions of these ORFs are not arrayed with those of other T4SS genes.
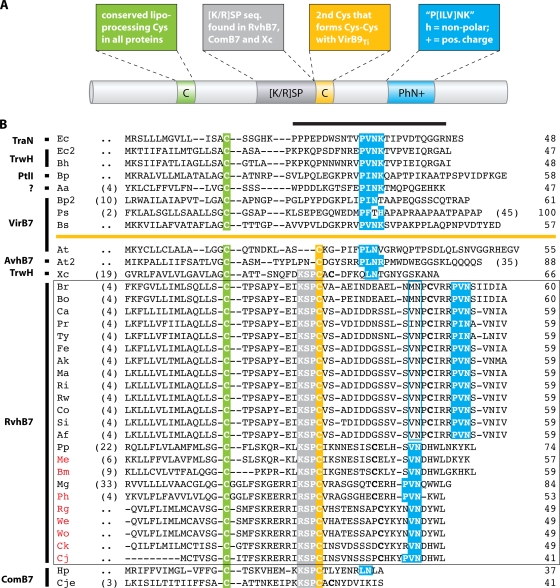
We recently identified a putative VirB7 homolog, RvhB7, encoded within all sequenced genomes of Rickettsia spp. (55). rvhB7 is located upstream of the rvhB8b-rvhB9b-rvhB10-rvhB11-rvhD4 cluster and is flanked by the rvhB9a-rvhB8a locus, which is carried on the opposite strand (Fig. 3). Bioinformatics analysis revealed that RvhB7 contains at least one Cys residue in addition to the conserved lipoprocessing Cys that is characteristic of all VirB7-like proteins, suggesting that RvhB7 is similar to VirB7Ti and other T4SS lipoproteins that may bind the C-terminal domain of VirB9 via a disulfide bridge. Other than these conserved features, RvhB7 proteins contain a candidate P(ILV)NK motif in the C-terminal region that is typical of most VirB7-like sequences (16). Additionally, a conserved sequence, (KI)KSP, directly flanking the second conserved Cys on the N-terminal side was found to be a feature shared only by the RvhB7 proteins of the Rickettsia spp. and the ComB7 proteins of H. pylori and C. jejuni, illustrating the possibility that rvh T4SSs descended from DNA competence systems (55).
Herein, we expand our analysis of putative rickettsial T4SS lipoproteins and illustrate that these molecules are likely to be part of the conserved rvh core as in other systems (Fig. 4). RvhB7 sequences of Rickettsia spp. were used to identify hypothetical proteins encoded in the genomes of A. marginale (hypothetical protein AM306) and the Wolbachia symbiont of Culex quinquefasciatus (hypothetical protein WPa_0823) that share the conserved characteristics of RvhB7 proteins. More refined tools revealed putative ORFs in most of the remaining sequenced Anaplasmataceae genomes that are currently not annotated as genes. Each of the protein sequences analyzed contains a predicted lipoprocessing site (Cys), a second conserved Cys, and several candidate P(ILV)NK motifs. The latter feature continues to become less conserved with the addition of more diverse VirB7-like sequences, and even simplifying the motif to PhN+, where h and + represent nonpolar and positively charged residues, respectively, still does not encompass the diversity in the C-terminal regions of these sequences or facilitate approaches to multiple-sequence alignment. The addition of these putative Anaplasmataceae sequences refines the conserved region shared by RvhB7 and the ComB7 proteins of H. pylori and C. jejuni to (K/R)SP, further supporting the evolution of rvh from competence systems. Additionally, we identified the (K/R)SP motif in the TrwH protein of X. citri, which is consistent with the phylogenetic position of the trw T4SS of X. citri as the closest xenolog of rvh (Fig. 2). Importantly, a recent study reports that the putative RvhB7 protein in A. marginale OM vaccine preparations is immunogenic for cattle vaccinees expressing several common major histocompatibility complex class II haplotypes (119). Altogether, these data strongly imply that RvhB7 is part of the conserved core rvh T4SS (RvhB7, RvhB9, and RvhB10) and is possibly surface exposed at the OM.
FIG. 4.
P-T4SS lipoproteins. (A) Schematic depiction of typical characteristics of VirB7 and related proteins. The color scheme is used to identify distinct features in the alignment (see the text for explanation). (B) Manual alignment of 13 diverse VirB7 and VirB7-like lipoproteins with 23 RvhB7 lipoproteins (the latter are boxed). Coordinates for each sequence are shown to the right, with numbers in parentheses to the left referring to flanking residues not shown in the alignment. Protein annotation is shown to the left. The black horizontal bar over the sequences of the predicted processed lipoproteins identifies the region of the NMR structure for the interaction of the TraO C-terminal domain (VirB9-like protein) and TraN (VirB7), both encoded by plasmid pKM101 in E. coli. Sequences above the orange line depict lipoproteins without a predicted Cys-Cys interaction with VirB9 and VirB9-like proteins: Ec, E. coli TraN, encoded by plasmid pKM101 (accession no. NP_511194); Ec2, E. coli TrwH protein (accession no. FAA00034); Bh, B. henselae TrwH-like protein (accession no. AAM82208); Bp, B. pertussis TraI protein, encoded by plasmid pSB102 (accession no. NP_361043); Aa, Aggregatibacter actinomycetemcomitans lipoprotein (accession no. NP_067577); Bp2, B. pertussis putative bacterial secretion system protein (accession no. NP_882291); Ps, P. syringae subsp. syringae VirB7 (accession no. NP_940729); and Bs, B. suis VirB7 (accession no. AAN33275). Sequences below the orange line represent proteins predicted to interact with VirB9 and VirB9-like proteins via a Cys-Cys bond: At, A. tumefaciens VirB7 (accession no. NP_536291); At2, A. tumefaciens AvhB7 (accession no. NP_396098); Xc, X. citri VirB7 (accession no. NP_942612); Br, R. bellii strain RML 369-C RvhB7 (accession no. YP_538183); Bo, R. bellii strain OSU 85 389 RvhB7 (accession no. YP_001495873); Ca, R. canadensis strain McKiel RvhB7 (accession no. YP_001492545); Pr, R. prowazekii strain Madrid E RvhB7 (accession no. NP_220672); Ty, R. typhi strain Wilmington RvhB7 (accession no. YP_067241); Fe, R. felis strain URRWXCal2 RvhB7 (accession no. YP_246480); Ak, R. akari strain Hartford RvhB7 (accession no. YP_001493229); Ma, R. massiliae strain MTU5 RvhB7 (accession no. YP_001499186); Ri, R. rickettsii strain Sheila Smith RvhB7 (accession no. YP_001494506); Rw, R. rickettsii strain Iowa RvhB7 (accession no. YP_001649753); Co, R. conorii strain Malish 7 RvhB7 (accession no. NP_360023); Si, R. sibirica strain 246 RvhB7 (accession no. ZP_00142155); Af, R. africae strain ESF-5 RvhB7 (accession no. ZP_02336216); Pp, protein of the Wolbachia symbiont of C. quinquefasciatus (accession no. YP_001975581); Me, protein of the Wolbachia symbiont of D. melanogaster; Bm, protein of the Wolbachia symbiont of Brugia malayi; Mg, A. marginale protein (accession no. YP_153652); Ph, A. phagocytophilum protein; Rg, E. ruminantium strain Gardel protein; We, E. ruminantium strain Welgevonden (Erwe) protein; Wo, E. ruminantium strain Welgevonden (Erum) protein; Ck, E. chaffeensis strain Arkansas protein; Cj, E. canis strain Jake protein; Hp, H. pylori protein (accession no. CAA10654); and Cje, C. jejuni protein (accession no. NP_863349). RvhB7 taxon codes colored red indicate that sequences corresponding to putative unannotated ORFs were recovered from tblastn searches of the Anaplasmataceae database (sequence coordinates are listed in Table S2 in the supplemental material). Additional conserved Cys residues in the RvhB7 sequences are in bold. A second putative PhN+ motif in the Rickettsia RvhB7 sequences is shaded in blue.
LOCALIZED MUREIN DEGRADATION IS SPECIFIC TO RICKETTSIA
Efficient T4SS transporter assembly across the cell envelope typically is associated with local disruption of peptidoglycan (PG) (Fig. 1). Descending from the free-living “Candidatus Pelagibacter,” Rickettsiales have not been found to encode a complete pathway for the synthesis of PG that is independent of host resources (see Fig. S3 in the supplemental material). PG from R. prowazekii similar to PGs from other Gram-negative species has been purified and demonstrated to incorporate d-alanine (103), and it is likely that a murein layer is synthesized in all Rickettsia spp., starting from host reserves of fructose-6-phosphate and/or glucosamine-6-phosphate. Prior studies have failed to detect PG in O. tsutsugamushi (3, 117), and a lack of enzymes able to convert fructose-6-phosphate to UDP-N-acetylglucosamine suggests the absence of PG in the cell envelope. Furthermore, alr, which encodes the enzyme that converts l-alanine to d-alanine, is deleted from O. tsutsugamushi genomes, as it is from all Anaplasmataceae genomes. Despite this finding, genes for most of the enzymes involved in amino sugar metabolism are present in the O. tsutsugamushi genomes (31, 98), as well as in A. marginale and Wolbachia genomes (49, 130). The same is true for genes encoding the enzymes responsible for the synthesis of lipid I and lipid II and the transport of anhydromuropeptides to the periplasm via the MurJ flippase (111). Genes encoding enzymes responsible for modification (i.e., transpeptidation and transglycosylation) of PG in the periplasm are highly conserved in Rickettsia spp. but found sporadically in the remaining Rickettsiales genomes. Outside the Rickettsiaceae, only A. marginale contains slt, the gene encoding the soluble lytic transglycosylase (LT) responsible for the excision of PG subunits from the murein layer. Rickettsia spp. encode the most complete pathway for recycling PG, with atypical proliferation of the IM AmpG permeases involved in PG subunit import to the cytoplasm (discussed below). Thus, from a genomics perspective, Rickettsia spp. and A. marginale are the only Rickettsiales that likely incorporate PG into their cell envelopes. The lack of rigid cell envelopes in Anaplasmataceae aside from A. marginale supports this viewpoint (33, 84, 109).
It is common for type II, III, and IV secretion systems, as well as DNA competence systems and bacteriophages, to encode LTs that hydrolyze PG (14, 40, 74, 81, 96). These specialized LTs (75) facilitate the local disruption of PG, allowing for efficient transporter assembly across the entire cell envelope or for host cell penetration in the case of bacteriophages. While typically much smaller than Slt and related PG autolysins, specialized LTs contain similar lysozyme-like folds and belong to an ancient glycohydrolase superfamily that includes plant chitinases, bacterial chitosanases, goose and hen g-type lysozymes, and phage T4 lysozymes (110). One of the best-characterized specialized LTs is VirB1Ti, which aside from its N-terminal lysozyme domain contains a processed C-terminal region that is secreted extracellularly and possibly involved in T-pilus formation (10, 133). While VirB1Ti is critical for T-pilus biogenesis, VirB1Ti mutants weaken but do not entirely abolish substrate transfer (15, 18, 53, 77), suggesting that Slt and/or other PG autolysins suffice in degrading PG for transporter assembly. VirB1-like proteins are also nonessential in other systems (14, 38, 39, 129); however, in the cag P-T4SS of H. pylori, the specialized LT is critical for both CagA translocation/phosphorylation and interleukin-8 induction in host cells (48).
We recently identified a virB1 homolog (rvhB1) that is conserved in all sequenced genomes of Rickettsia (55), supporting the likelihood that PG is a component of Rickettsia species cell envelopes. While rvhB1 is located independently of the other rvh genes in these genomes (Fig. 3), there is strong in silico evidence supporting RvhB1 as a specialized LT (55). Informatics data suggest that no RvhB1 homologs are encoded in the remaining Rickettsiales genomes (data not shown). Thus, if PG is synthesized in A. marginale and/or Wolbachia spp., the manner in which the T4SS scaffold assembles and spans the periplasm is likely to be different from that in Rickettsia spp.
Rvh REGULATION, SUBSTRATES, AND PUTATIVE FUNCTION
The vir P-T4SS of A. tumefaciens and the icm/dot I-T4SS of Legionella pneumophila are regulated by two-component systems (54, 128, 134), suggesting tight correlation of T4SS gene expression and function. Other single genes have been demonstrated previously to control the expression of the vir loci of Brucella spp. (37, 41, 116). Regarding several intracellular species, it has been demonstrated that host conditions upregulate T4SS operons (21, 30, 113). Clustering of T4SS genes into one or a few operons undoubtedly enhances their coordinated regulation (32, 82). However, the conserved rvh T4SS is scattered in small islets throughout the genome in all lineages (Fig. 3), and in Rickettsia genomes, the 18 genes can be grouped into five islets (55). Counterbalancing this architectural anomaly, a single T4SS transcriptional regulator, ecxR (ECH_0795), has been identified in E. chaffeensis and induces coordinate expression of several of the rvh islets (30). Informatics data indicate that ecxR homologs are present in all genomes of the Anaplasmataceae but are not present in the Rickettsiaceae (see Table S3 in the supplemental material). Previously, we identified two conserved genes in Rickettsia spp. that are carried within predicted rvh operons. A RelA/SpoT-encoding gene adjacent to rvhD4 is of interest given that a stringent response protein similar to RelA/SpoT, Rsh, regulates the vir loci of Brucella spp. (41). A second gene encoding a conserved hypothetical protein is immediately downstream of the rvhB1 gene in all sequenced genomes, possibly having a role associated with type IV secretion. Undoubtedly, there will be further characterized rvh regulators given the need to coordinately express the scattered rvh islets.
Despite the rvh P-T4SS's being one of the features most conserved across Rickettsiales genomes, the function of the rvh P-T4SS is poorly characterized (55). The AnkA protein of A. phagocytophilum, which penetrates neutrophil nuclei and is essential for host infection (83, 104), has been described as the first rvh T4SS substrate and is able to be secreted via the vir P-T4SS of A. tumefaciens (83). However, no other protein or DNA molecules have been identified as substrates of the rvh T4SS, and the exact manner in which substrates are presented to and secreted by the transporter are unknown. For Rickettsia spp., two genes have garnered attention for possibly encoding rvh substrates. ralF, a gene encoding a Sec7 domain-containing protein, is known in prokaryotes only from Rickettsia spp. and Legionella spp. (34), and in L. pneumophila the protein is an I-T4SS effector that functions as a guanine nucleotide exchange factor in the recruitment of the ADP-ribosylation factor to occupied phagosomes (97). While the precise role of RalF in Legionella pathogenesis is unknown, an analogous function associated with phagosomal modification in Rickettsia spp. is unlikely given the immediate lysis of the phagosome upon host cell invasion. Furthermore, pseudogenization has eliminated nearly the entire ralF ORF in all sequenced genomes of spotted fever group rickettsiae. The second putative rvh effector is RickA, a protein considered to activate host Arp2/Arp3 complexes, resulting in actin nucleation (58, 68), which permits intercellular spread of some rickettsiae (61). Based on informatics data, which predict neither membrane nor periplasmic association of RickA (data not shown), and the fact that the protein is localized to the bacterial surface (59), it was hypothesized that RickA is secreted via the T4SS (58). Like ralF, the RickA gene is not present in all sequenced genomes of Rickettsia spp.; hence, if either or both of these candidates are true T4SS effectors, the rvh T4SS would secrete different substrates in various species/strains of Rickettsia, possibly contributing to lineage-specific pathogenicity.
It was speculated previously that the rvh T4SS must function in virulence factor secretion versus plasmid transfer because not all sequenced rickettsia genomes contain plasmids (102). Interestingly, the same investigation not only uncovered the first case of a plasmid system in a rickettsia (R. felis) but also provided electron microscopy images of pilus-like structures that were assumed to be associated with conjugation (102). As there is no complete set of genes in the R. felis genome encoding type II or type IV pili (data not shown) and the absence of a virB5 homolog would presumably prevent the formation of a T-pilus (as discussed above), the exact genetic architecture underlying these extracellular appendages in R. felis remains unknown. Similar pilus-like structures were observed in R. bellii strain RML 369-C (101) and R. massiliae (19), which both carry a nearly full set of genes related to the tra-trb operon of the F plasmid of E. coli, despite only the latter's harboring a plasmid system. Other rickettsiae harboring plasmids (e.g., R. africae, “R. monacensis,” and R. peacockii) do not carry full tra-trb-like operons, and no pilus-like structures have been reported. Thus, the significance of a tra-trb operon in relation to extracellular appendages and their possible role in the conjugation of rickettsial plasmids remains quite nebulous. What is clear is that a large mobile genetic element, which carries an F-T4SS highly similar to the F plasmid tra-trb operon, is detectable as products of pseudogenization across the sequenced Rickettsiales genomes (data not shown) and is highly proliferated in the genomes of O. tsutsugamushi strains (31, 98). Similar conjugation genes have also been detected in various rickettsiae associated with arthropods with no known association with vertebrates (124). Recent sequencing of the rickettsial endosymbiont of Ixodes scapularis (REIS) has revealed the largest number of conjugation genes within a Rickettsia genome, suggesting that ancient conjugative plasmids have propagated mobile elements across the Rickettsiales. Strong reductive evolution has eliminated the majority of these elements in most genomes, in particular a complete F-T4SS likely to be essential for conjugative transfer of plasmids. The ability of the rvh P-T4SS to function in conjugation cannot be ruled out, but the absence of any identified plasmids or pilus-like structures in the Anaplasmataceae argues strongly against a role in conjugation. However, a function in naked DNA uptake and release from host environments, particularly given the close phylogenetic relatedness of rvh to T4SSs that are involved in DNA competence (Fig. 2), cannot be overlooked (55). Rickettsiaceae may benefit from scavenging nucleotides upon DNA uptake since genes involved in de novo synthesis of nucleotides have been deleted relative to the genomes of Anaplasmataceae, which due to their vacuole-enclosed lifestyle do not have access to cytoplasmic nucleotides and have thus retained genes involved in purine and pyrimidine biosynthesis (23).
RICKETTSIALES: DIVERSE SCAFFOLDS CORRELATED WITH CELL ENVELOPE ARCHITECTURE?
Hundreds of sequenced bacterial genomes provide a foundation for understanding the diversity of T4SSs that define the lifestyles of many pathogens, as well as contribute to the continuum of speciation through the transfer of components of the bacterial mobile gene pool. While informatics tools and laboratory studies are useful for unveiling T4SS architecture and function, evolution has done the crucial experiments, and many T4SSs can be seen as the end products of millions of years of bacterial coevolution with various host and vector cells. This information, combined with a wealth of experimental data that define the archetypal vir P-T4SS of A. tumefaciens (Fig. 5A), allows for the observation of structural and functional diversification across a wide range of bacterial species. For example, in relation to the vir P-T4SS, the ptl P-T4SS of Bordetella pertussis has undergone three innovations that likely correlate with the secretion of its sole substrate, the pertussis holotoxin (PT) (Fig. 5B). First, a virB1 homolog is absent in the ptl system, yet given the presence of PG in B. pertussis, a mechanism for local degradation of PG is required. This task is accomplished by the fusion of a glycohydrolase domain to the N-terminal region of PtlE (a VirB8 homolog) that has demonstrated peptidoglycanase activity in both B. pertussis and E. coli (107). Thus, because VirB8 proteins are bitopic in the IM, this N-terminal glycohydrolase domain is located in the periplasm, allowing for simultaneous degradation of PG and T4SS channel assembly. Second, a homolog of the type IV coupling protein (T4CP), VirD4, is deleted in the ptl system. This is explained by the sec-dependent secretion of PT subunits to the periplasm (88, 100), with holotoxin assembly likely driving the formation of the core Ptl scaffold around PT prior to secretion (121). Thus, a T4CP would not be needed for entry of substrates from the cytoplasm to the periplasm, with the energetics generated from the remaining two IM ATPases, VirB4 (PtlC) and VirB11 (PtlH), enough to drive translocation of PT out of the bacterial cell. Lastly, the lack of a virB5 homolog in the ptl system correlates with the lack of an observed T4SS pilus in B. pertussis (126), as host cell contact is not required for secretion of PT.
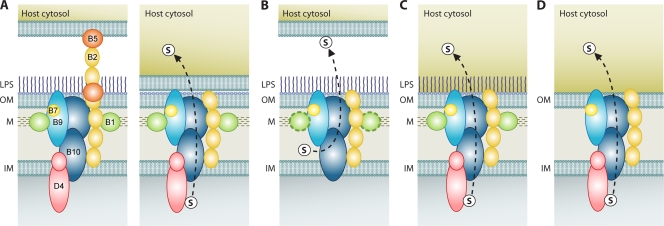
FIG. 5.
P-T4SS structural and functional diversification. The dashed-line arrows illustrate the mode of secretion, with substrates depicted by an encircled “s.” The color scheme of P-T4SS components VirB1 (green), VirB2 (light orange), VirB5 (dark orange), VirB7 (yellow), VirB9 (blue), VirB10 (dark blue), and VirD4 (pink) across four secretion systems implies homology. Distinct N and C termini of VirB10 and VirD4 are depicted. M, murein layer. (A) Model of transport for the A. tumefaciens vir P-T4SS. The two-step process of substrate attachment (left) and substrate transfer upon sloughing off of the T-pilus (right) is shown (51). (B) Model of transport for the B. pertussis ptl P-T4SS. VirB1 is distinguished to depict the N-terminal glycohydrolase domain of PtlE, a VirB8 homolog (107). (C) Model of transport for the Rickettsia rvh P-T4SS. (D) General model of transport for the O. tsutsugamushi rvh P-T4SS and for P-T4SSs in species of Anaplasmataceae that are not predicted to completely synthesize PG and LPS.
Similar comparisons between the T4SSs of members of the Rickettsiales and the vir system of A. tumefaciens can be made for inferring structural and functional diversification of the rvh transporter (Fig. 5C and D). Like the B. pertussis genome, all Rickettsiales genomes lack a virB5 homolog, suggesting that rvh-mediated secretion occurs in the absence of a T-pilus-like structure. As recent studies identify VirB5 as an adhesin involved in host cell recognition (2, 7, 131), a minor pilin homolog (and a T-pilus) would be unnecessary in Rickettsiales, as substrates would be directly secreted to and/or imported from the host environment (55). Thus, RvhB2 polymerization across the periplasm should terminate near the OM in all Rickettsiales species. As Rickettsia spp. are the only Rickettsiales lineage to synthesize lipopolysaccharide (LPS) (data not shown), RvhB2 may be highly surface exposed in the remaining Rickettsiales. This suggestion is supported by an RvhB2-induced T-cell response in cattle immunized with an A. marginale OM fraction (89). The lack of LPS may also expose a portion of the outer cap of the core T4SS, as antibodies specific for RvhB9 have been detected in dogs infected with Ehrlichia canis (46) and cattle infected with A. marginale (6, 122). Furthermore, both RvhB9 and RvhB10 elicit robust antibody and T-cell responses from cattle immunized with a protective A. marginale OM fraction (89-91). The loss of LPS synthesis in the Anaplasmataceae may have lead to greater exposure of the T4SS at the OM and may account for RvhB2 duplication and proliferation as a consequence of host cell immune system avoidance. In contrast to Rickettsiaceae genomes, all sequenced Anaplasmataceae genomes have full-length RvhB9 gene homologs, which may add to antigenic complexity in the OM portion of the T4SS. While the cell envelope composition of O. tsutsugamushi differs greatly from that of Rickettsia spp. and does not contain LPS (117), prediction of surface exposure of the T4SS is difficult since only one genome (that of strain Ikeda) contains the VirB2 paralog genes (Fig. 3). Nonetheless, as VirB9 is predicted to be surface exposed in other systems (16, 64), it is probable that some regions of the rvh transporter are surface exposed at the OM, thus making it worthy for exploration as a vaccine target.
It has been hypothesized previously that species of Rickettsiales that invade vertebrate immune cells would benefit from a lack of PG synthesis (63). This proposal is consistent with evidence from other systems that host receptors of PG, nucleotide-binding oligomerization domain 1 (Nod1) and Nod2 proteins, detect by-products of PG degradation via inefficient anhydromuropeptide recycling (20, 120). As all members of the Anaplasmataceae replicate within intracellular vacuoles (43), discarded PG fragments may still be undetected by host cells in species that potentially synthesize PG. For free-living Rickettsiaceae, either PG synthesis does not occur (as in the case of O. tsutsugamushi) or a strategy may exist for “hiding” PG fragment release from immune cells. As discussed above, Rickettsia spp. contain many genes involved in the degradation and recycling of PG (see Fig. S3 in the supplemental material). Exceptionally, all sequenced genomes of Rickettsia spp. contain three to four copies of ampG, which encodes a permease involved in the import of PG monomers from the periplasm to the cytoplasm. As such genes are typically present only once in bacterial genomes, ampG proliferation in Rickettsia spp. may hint at increased necessity to recycle PG subunits rather then shed them freely into the host cytoplasm. The presence of a virB1 homolog in only the genomes of Rickettsia spp. suggests that PG fragments are likely to be released during the assembly of the rvh T4SS and that this process of cell envelope rearrangement may make various components of the T4SS scaffold vulnerable to the host immune response. Furthermore, the processing of the C-terminal region of VirB1Ti (VirB1*) (10) and its subsequent extracellular secretion and role in T-pilus formation (87, 133) are likely not specific to the A. tumefaciens vir T4SS. Processed VirB1 products in the cell lysate from Brucella abortus were identified previously (39), and informatics data suggest that a conserved Ala is the likely cleavage site in many VirB1 homologs, with all predicted VirB1* sequences containing tracts of repeated residues, particularly Pro-rich tracts (55). Across 13 Rickettsia spp., rvhB1 has the highest average number of codons evolving under positive selection among the 18 rvh genes (55), suggesting possible coevolution with host cell components. Nonetheless, RvhB1, as well as a putative RvhB1* form, may pose novel vaccine targets specific for species of Rickettsia.
CONCLUSIONS
Despite the genetic intractability of Rickettsiales as obligate intracellular bacteria, advances in understanding the mechanisms involved in the pathogenicity of these species are being made (12, 36, 123). This work outlines major achievements in the identification and characterization of components of the rvh T4SS, as well as its putative regulators and substrates. Past and present bioinformatics approaches have greatly facilitated our understanding of the genetic architecture of the rvh scaffold, and experimental evidence has identified several promising vaccine targets. Our synopsis here suggests a single event of inheritance of the rvh T4SS in the Rickettsiales progenitor, with lineage-specific diversification of rvh components likely a result of modifications to cell envelope structure. This coevolution of the rvh T4SS and cell envelope structure is likely driven by adaptations to various host cells and thus identifies the transporter as an important target for vaccine development. While current data favor a role in effector translocation, functions in DNA uptake and release and/or conjugation cannot at present be ruled out, especially considering that no mechanism for plasmid transfer in Rickettsia spp. has yet been proposed. Furthermore, the genomes of several attenuated strains of Rickettsia (R. prowazekii Madrid E and R. rickettsii Iowa) and species with no known pathogenicity in arthropod or vertebrate cells (REIS and R. peacockii Rustic) carry most or all of the rvh genes. Taking this finding into consideration, we expect major advances in the near future regarding knowledge about the rvh T4SS and its potential involvement in host disease.
ADDENDUM IN PROOF
Studies by Y. Rikihisa and M. Lin (Curr. Opin. Microbiol. 13:59-66, 2010) and H. Niu, V. Kozjak-Pavlovic, T. Rudel, and Y. Rikihisa (PLoS Pathog. 19:e1000774, 2010) were published during the production of this paper. They add substantial information regarding rvh substrates of the Anaplasmataceae.
Supplementary Material
Acknowledgments
The project described herein was supported by award numbers R01AI017828 and R01AI59118 (to A.F.A.) and R01AI053692 (to W.C.B.) from the National Institute of Allergy and Infectious Diseases (NIAID) and by funding through NIAID contract HHSN266200400035C to B.W.S.
The content of this paper is the responsibility solely of the authors and does not necessarily represent the official views of the NIAID or the National Institutes of Health.
We are grateful to Jim Kaper (University of Maryland) for the invitation to contribute this review. We thank Nick Carbonetti (University of Maryland) for helpful discussion on B. pertussis type IV secretion, Julie Hotopp (University of Maryland) for insight on Anaplasmataceae genomics, and Guy Palmer (Washington State University) and Steve Dumler (Johns Hopkins) for discussions on Anaplasmataceae biology. We are appreciative to members of the Azad lab and the Cyberinfrastructure Group (VBI) for critical advice during the completion of this work.
Biography
 Joseph J. Gillespie was born and raised in suburban Philadelphia, PA. In 1998 he obtained his B.S. degree from Widener University, studying the effects of controlled fire on arthropod populations in the New Jersey Pine Barrens. He continued to study arthropods in his M.S. (University of Delaware) and Ph.D. (Texas A&M University) programs, specializing in molecular evolution and phylogenetics. In 2006 he was hired by the Virginia Bioinformatics Institute (VBI) at Virginia Tech to study the bioinformatics of arthropod-borne bacteria. Since then he has developed an interest in the biology of obligate intracellular bacteria, especially rickettsiae. Of particular interest is the manner in which these bacteria coevolve with their eukaryotic hosts. He has remained with VBI as a senior research scientist and is also a visiting scientist in the Department of Microbiology and Immunology at the University of Maryland School of Medicine. He resides with his family in Maryland.
Joseph J. Gillespie was born and raised in suburban Philadelphia, PA. In 1998 he obtained his B.S. degree from Widener University, studying the effects of controlled fire on arthropod populations in the New Jersey Pine Barrens. He continued to study arthropods in his M.S. (University of Delaware) and Ph.D. (Texas A&M University) programs, specializing in molecular evolution and phylogenetics. In 2006 he was hired by the Virginia Bioinformatics Institute (VBI) at Virginia Tech to study the bioinformatics of arthropod-borne bacteria. Since then he has developed an interest in the biology of obligate intracellular bacteria, especially rickettsiae. Of particular interest is the manner in which these bacteria coevolve with their eukaryotic hosts. He has remained with VBI as a senior research scientist and is also a visiting scientist in the Department of Microbiology and Immunology at the University of Maryland School of Medicine. He resides with his family in Maryland.
 Kelly A. Brayton was born in Akron, OH, and grew up in India, Malaysia, Turkey, Greece, Lebanon, and Texas. She holds a B.A. in biology from Texas A&M and a Ph.D. in biochemistry from Purdue University. She started studying hemoparasitic diseases during her postdoctoral stint at the Onderstepoort Veterinary Institute in South Africa. She is currently an associate professor of microbial genomics in the Department of Veterinary Microbiology and Pathology, School for Global Animal Health, Washington State University, where she has developed a genomics program for veterinary pathogens. She led the sequencing efforts that resulted in genome sequences for the cattle pathogens A. marginale and Babesia bovis, among others. The availability of these sequences has catalyzed research on these organisms—allowing Kelly and her colleagues to pursue research on mechanisms of immune evasion, persistence, virulence, and transmission and vaccine development.
Kelly A. Brayton was born in Akron, OH, and grew up in India, Malaysia, Turkey, Greece, Lebanon, and Texas. She holds a B.A. in biology from Texas A&M and a Ph.D. in biochemistry from Purdue University. She started studying hemoparasitic diseases during her postdoctoral stint at the Onderstepoort Veterinary Institute in South Africa. She is currently an associate professor of microbial genomics in the Department of Veterinary Microbiology and Pathology, School for Global Animal Health, Washington State University, where she has developed a genomics program for veterinary pathogens. She led the sequencing efforts that resulted in genome sequences for the cattle pathogens A. marginale and Babesia bovis, among others. The availability of these sequences has catalyzed research on these organisms—allowing Kelly and her colleagues to pursue research on mechanisms of immune evasion, persistence, virulence, and transmission and vaccine development.
 Kelly P. Williams (Virginia Bioinformatics Institute) has broad interest in bacterial molecular biology and evolution, with special interests in RNAs, genomic islands, and phylogeny. His Ph.D. work (at the University of California, San Diego) was on the biochemistry of bacteriophage transcription. Postdoctoral research during appointments at the Salk Institute, the Consiglio Nazionale delle Ricerche (Rome, Italy), and the Whitehead Institute employed in vitro selection to explore diverse RNA functions. His laboratory at Indiana University focused on the mechanism and evolution of bacterial tmRNA.
Kelly P. Williams (Virginia Bioinformatics Institute) has broad interest in bacterial molecular biology and evolution, with special interests in RNAs, genomic islands, and phylogeny. His Ph.D. work (at the University of California, San Diego) was on the biochemistry of bacteriophage transcription. Postdoctoral research during appointments at the Salk Institute, the Consiglio Nazionale delle Ricerche (Rome, Italy), and the Whitehead Institute employed in vitro selection to explore diverse RNA functions. His laboratory at Indiana University focused on the mechanism and evolution of bacterial tmRNA.
 Marco A. Quevedo Diaz was born in Lima, Peru. He gained his M.S. degree in microbiology from the Faculty of Natural Sciences, Comenius University, in Bratislava, Slovakia, studying pdr genes in Saccharomyces cerevisiae. From 1998 to 2003, he was enrolled in the Ph.D. program in the Department of Rickettsiology at the Institute of Virology, Slovak Academy of Science, studying Coxiella burnetii phase variation and genetic transformation. Since that time, he has been fascinated with the biology of obligate intracellular bacteria, particularly rickettsiae and their interaction with eukaryotic hosts. Currently, he is at the Department of Microbiology and Immunology, School of Medicine, University of Maryland.
Marco A. Quevedo Diaz was born in Lima, Peru. He gained his M.S. degree in microbiology from the Faculty of Natural Sciences, Comenius University, in Bratislava, Slovakia, studying pdr genes in Saccharomyces cerevisiae. From 1998 to 2003, he was enrolled in the Ph.D. program in the Department of Rickettsiology at the Institute of Virology, Slovak Academy of Science, studying Coxiella burnetii phase variation and genetic transformation. Since that time, he has been fascinated with the biology of obligate intracellular bacteria, particularly rickettsiae and their interaction with eukaryotic hosts. Currently, he is at the Department of Microbiology and Immunology, School of Medicine, University of Maryland.
 Wendy C. Brown was born and raised in upstate New York. She obtained her B.A. degree in microbiology from Smith College and M.P.H. and Ph.D. degrees from Yale University, studying infectious disease epidemiology and T-cell immunology. She transitioned to research on tick-borne pathogens at the International Laboratory for Research on Animal Diseases in Nairobi, Kenya, where she studied vaccine development for theileriosis, and then went to Texas A&M University (as an associate professor), where she studied T-cell responses to B. bovis, and finally to Washington State University, where she has also worked on T-cell immunity to the rickettsial pathogen A. marginale. There, she is a Regents Professor in the Department of Veterinary Microbiology and Pathology and the School for Global Animal Health. Of particular interest is targeting outer membrane protein complexes, specifically the bacterial type IV secretion system, for vaccine development. She resides with her family in the Palouse.
Wendy C. Brown was born and raised in upstate New York. She obtained her B.A. degree in microbiology from Smith College and M.P.H. and Ph.D. degrees from Yale University, studying infectious disease epidemiology and T-cell immunology. She transitioned to research on tick-borne pathogens at the International Laboratory for Research on Animal Diseases in Nairobi, Kenya, where she studied vaccine development for theileriosis, and then went to Texas A&M University (as an associate professor), where she studied T-cell responses to B. bovis, and finally to Washington State University, where she has also worked on T-cell immunity to the rickettsial pathogen A. marginale. There, she is a Regents Professor in the Department of Veterinary Microbiology and Pathology and the School for Global Animal Health. Of particular interest is targeting outer membrane protein complexes, specifically the bacterial type IV secretion system, for vaccine development. She resides with her family in the Palouse.
 Abdu F. Azad is a professor of microbiology and immunology at the University of Maryland School of Medicine, Baltimore. He obtained his Ph.D. from the Johns Hopkins University School of Public Health, investigating the biology of a liver nematode, Capillaria hepatica, in Norway rats. His diverse scientific interests started with the epidemiology of human intestinal parasites, flea taxonomy (he described five new species), and the ecology and natural history of mammal-borne pathogens in Asia and Africa. His current research is focused primarily on the biology of arthropod-borne rickettsial pathogens, particularly understanding how these bacterial agents with reduced genomes cause infection and disease. Additionally, he has continued his long-term interest in investigating the genetic and molecular bases of preerythrocytic stages of malaria parasites in protective immunity. Aside from his research, he considers training as his major professional endeavor and is very proud of the scientific accomplishments of his former students and postdoctoral fellows.
Abdu F. Azad is a professor of microbiology and immunology at the University of Maryland School of Medicine, Baltimore. He obtained his Ph.D. from the Johns Hopkins University School of Public Health, investigating the biology of a liver nematode, Capillaria hepatica, in Norway rats. His diverse scientific interests started with the epidemiology of human intestinal parasites, flea taxonomy (he described five new species), and the ecology and natural history of mammal-borne pathogens in Asia and Africa. His current research is focused primarily on the biology of arthropod-borne rickettsial pathogens, particularly understanding how these bacterial agents with reduced genomes cause infection and disease. Additionally, he has continued his long-term interest in investigating the genetic and molecular bases of preerythrocytic stages of malaria parasites in protective immunity. Aside from his research, he considers training as his major professional endeavor and is very proud of the scientific accomplishments of his former students and postdoctoral fellows.
 Bruno W. Sobral was born in Brazil. His undergraduate education was in agricultural engineering and his Ph.D. was in genetics at Iowa State University, with postdoctoral work in molecular evolution. After starting his own research group at the California Institute of Biological Research, La Jolla, in 1991, he went on to be the vice president of scientific programs at the National Center for Genome Resources in Santa Fe, NM. In 2000 he started the Virginia Bioinformatics Institute (VBI) at Virginia Tech as the founding executive and scientific director and as a professor in the Department of Plant Pathology, Physiology, and Weed Science. His Cyberinfrastructure Section at VBI has focused on bioinformatics, computational biology, and informatics-based approaches to infectious disease research. He is particularly interested in and focused on transdisciplinary approaches to research and development and currently is a professor and the director of the Cyberinfrastructure Section.
Bruno W. Sobral was born in Brazil. His undergraduate education was in agricultural engineering and his Ph.D. was in genetics at Iowa State University, with postdoctoral work in molecular evolution. After starting his own research group at the California Institute of Biological Research, La Jolla, in 1991, he went on to be the vice president of scientific programs at the National Center for Genome Resources in Santa Fe, NM. In 2000 he started the Virginia Bioinformatics Institute (VBI) at Virginia Tech as the founding executive and scientific director and as a professor in the Department of Plant Pathology, Physiology, and Weed Science. His Cyberinfrastructure Section at VBI has focused on bioinformatics, computational biology, and informatics-based approaches to infectious disease research. He is particularly interested in and focused on transdisciplinary approaches to research and development and currently is a professor and the director of the Cyberinfrastructure Section.
Editor: H. L. Andrews-Polymenis
Footnotes
Published ahead of print on 22 February 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Alvarez-Martinez, C. E., and P. J. Christie. 2009. Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. 73:775-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aly, K. A., and C. Baron. 2007. The VirB5 protein localizes to the T-pilus tips in Agrobacterium tumefaciens. Microbiology 153:3766-3775. [DOI] [PubMed] [Google Scholar]
- 3.Amano, K., A. Tamura, N. Ohashi, H. Urakami, S. Kaya, and K. Fukushi. 1987. Deficiency of peptidoglycan and lipopolysaccharide components in Rickettsia tsutsugamushi. Infect. Immun. 55:2290-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, L. B., A. V. Hertzel, and A. Das. 1996. Agrobacterium tumefaciens VirB7 and VirB9 form a disulfide-linked protein complex. Proc. Natl. Acad. Sci. U. S. A. 93:8889-8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersson, S. G., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Ponten, U. C. Alsmark, R. M. Podowski, A. K. Naslund, A. S. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-140. [DOI] [PubMed] [Google Scholar]
- 6.Araujo, F. R., C. M. Costa, C. A. Ramos, T. A. Farias, I. I. Souza, E. S. Melo, C. Elisei, G. M. Rosinha, C. O. Soares, S. P. Fragoso, and A. H. Fonseca. 2008. IgG and IgG2 antibodies from cattle naturally infected with Anaplasma marginale recognize the recombinant vaccine candidate antigens VirB9, VirB10, and elongation factor-Tu. Mem. Inst. Oswaldo Cruz 103:186-190. [DOI] [PubMed] [Google Scholar]
- 7.Backert, S., R. Fronzes, and G. Waksman. 2008. VirB2 and VirB5 proteins: specialized adhesins in bacterial type-IV secretion systems? Trends Microbiol. 16:409-413. [DOI] [PubMed] [Google Scholar]
- 8.Bao, W., Y. Kumagai, H. Niu, M. Yamaguchi, K. Miura, and Y. Rikihisa. 2009. Four VirB6 paralogs and VirB9 are expressed and interact in Ehrlichia chaffeensis-containing vacuoles. J. Bacteriol. 191:278-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baron, C. 2005. From bioremediation to biowarfare: on the impact and mechanism of type IV secretion systems. FEMS Microbiol. Lett. 253:163-170. [DOI] [PubMed] [Google Scholar]
- 10.Baron, C., M. Llosa, S. Zhou, and P. C. Zambryski. 1997. VirB1, a component of the T-complex transfer machinery of Agrobacterium tumefaciens, is processed to a C-terminal secreted product, VirB1*. J. Bacteriol. 179:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baron, C., Y. R. Thorstenson, and P. C. Zambryski. 1997. The lipoprotein VirB7 interacts with VirB9 in the membranes of Agrobacterium tumefaciens. J. Bacteriol. 179:1211-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Batut, J., S. G. Andersson, and D. O'Callaghan. 2004. The evolution of chronic infection strategies in the alpha-proteobacteria. Nat. Rev. Microbiol. 2:933-945. [DOI] [PubMed] [Google Scholar]
- 13.Bayan, N., I. Guilvout, and A. P. Pugsley. 2006. Secretins take shape. Mol. Microbiol. 60:1-4. [DOI] [PubMed] [Google Scholar]
- 14.Bayer, M., R. Eferl, G. Zellnig, K. Teferle, A. Dijkstra, G. Koraimann, and G. Hogenauer. 1995. Gene 19 of plasmid R1 is required for both efficient conjugative DNA transfer and bacteriophage R17 infection. J. Bacteriol. 177:4279-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bayer, M., R. Iberer, K. Bischof, E. Rassi, E. Stabentheiner, G. Zellnig, and G. Koraimann. 2001. Functional and mutational analysis of p19, a DNA transfer protein with muramidase activity. J. Bacteriol. 183:3176-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bayliss, R., R. Harris, L. Coutte, A. Monier, R. Fronzes, P. J. Christie, P. C. Driscoll, and G. Waksman. 2007. NMR structure of a complex between the VirB9/VirB7 interaction domains of the pKM101 type IV secretion system. Proc. Natl. Acad. Sci. U. S. A. 104:1673-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beaupre, C. E., J. Bohne, E. M. Dale, and A. N. Binns. 1997. Interactions between VirB9 and VirB10 membrane proteins involved in movement of DNA from Agrobacterium tumefaciens into plant cells. J. Bacteriol. 179:78-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berger, B. R., and P. J. Christie. 1994. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J. Bacteriol. 176:3646-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanc, G., H. Ogata, C. Robert, S. Audic, J. M. Claverie, and D. Raoult. 2007. Lateral gene transfer between obligate intracellular bacteria: evidence from the Rickettsia massiliae genome. Genome Res. 17:1657-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boneca, I. G. 2005. The role of peptidoglycan in pathogenesis. Curr. Opin. Microbiol. 8:46-53. [DOI] [PubMed] [Google Scholar]
- 21.Boschiroli, M. L., S. Ouahrani-Bettache, V. Foulongne, S. Michaux-Charachon, G. Bourg, A. Allardet-Servent, C. Cazevieille, J. P. Liautard, M. Ramuz, and D. O'Callaghan. 2002. The Brucella suis virB operon is induced intracellularly in macrophages. Proc. Natl. Acad. Sci. U. S. A. 99:1544-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Braig, H. R., B. D. Turner, and M. A. Perotti. 2008. Symbiotic rickettsia, p. 221-249. In K. Bourtzis and T. A. Miller (ed.), Insect symbiosis, vol. 3. CRC Press, Boca Raton, FL. [Google Scholar]
- 23.Brayton, K. A., M. J. Dark, and G. H. Palmer. 2009. Anaplasma, p. 85-116. In V. Nene and C. Kole (ed.), Genome mapping and genomics in animal-associated microbes. Springer, Berlin, Germany.
- 24.Brayton, K. A., L. S. Kappmeyer, D. R. Herndon, M. J. Dark, D. L. Tibbals, G. H. Palmer, T. C. McGuire, and D. P. Knowles, Jr. 2005. Complete genome sequencing of Anaplasma marginale reveals that the surface is skewed to two superfamilies of outer membrane proteins. Proc. Natl. Acad. Sci. U. S. A. 102:844-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brayton, K. A., D. P. Knowles, T. C. McGuire, and G. H. Palmer. 2001. Efficient use of a small genome to generate antigenic diversity in tick-borne ehrlichial pathogens. Proc. Natl. Acad. Sci. U. S. A. 98:4130-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao, T. B., and M. H. Saier, Jr. 2001. Conjugal type IV macromolecular transfer systems of Gram-negative bacteria: organismal distribution, structural constraints and evolutionary conclusions. Microbiology 147:3201-3214. [DOI] [PubMed] [Google Scholar]
- 27.Cascales, E., and P. J. Christie. 2004. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304:1170-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cascales, E., and P. J. Christie. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chandran, V., R. Fronzes, S. Duquerroy, N. Cronin, J. Navaza, and G. Waksman. 2009. Structure of the outer membrane complex of a type IV secretion system. Nature 462:1011-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng, Z., X. Wang, and Y. Rikihisa. 2008. Regulation of type IV secretion apparatus genes during Ehrlichia chaffeensis intracellular development by a previously unidentified protein. J. Bacteriol. 190:2096-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho, N. H., H. R. Kim, J. H. Lee, S. Y. Kim, J. Kim, S. Cha, S. Y. Kim, A. C. Darby, H. H. Fuxelius, J. Yin, J. H. Kim, J. Kim, S. J. Lee, Y. S. Koh, W. J. Jang, K. H. Park, S. G. Andersson, M. S. Choi, and I. S. Kim. 2007. The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host-cell interaction genes. Proc. Natl. Acad. Sci. U. S. A. 104:7981-7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christie, P. J., K. Atmakuri, V. Krishnamoorthy, S. Jakubowski, and E. Cascales. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59:451-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collins, N. E., J. Liebenberg, E. P. de Villiers, K. A. Brayton, E. Louw, A. Pretorius, F. E. Faber, H. van Heerden, A. Josemans, M. van Kleef, H. C. Steyn, M. F. van Strijp, E. Zweygarth, F. Jongejan, J. C. Maillard, D. Berthier, M. Botha, F. Joubert, C. H. Corton, N. R. Thomson, M. T. Allsopp, and B. A. Allsopp. 2005. The genome of the heartwater agent Ehrlichia ruminantium contains multiple tandem repeats of actively variable copy number. Proc. Natl. Acad. Sci. U. S. A. 102:838-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cox, R., R. J. Mason-Gamer, C. L. Jackson, and N. Segev. 2004. Phylogenetic analysis of Sec7-domain-containing Arf nucleotide exchangers. Mol. Biol. Cell 15:1487-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Darby, A. C., N. H. Cho, H. H. Fuxelius, J. Westberg, and S. G. Andersson. 2007. Intracellular pathogens go extreme: genome evolution in the Rickettsiales. Trends Genet. 23:511-520. [DOI] [PubMed] [Google Scholar]
- 37.Delrue, R. M., C. Deschamps, S. Leonard, C. Nijskens, I. Danese, J. M. Schaus, S. Bonnot, J. Ferooz, A. Tibor, X. De Bolle, and J. J. Letesson. 2005. A quorum-sensing regulator controls expression of both the type IV secretion system and the flagellar apparatus of Brucella melitensis. Cell. Microbiol. 7:1151-1161. [DOI] [PubMed] [Google Scholar]
- 38.den Hartigh, A. B., H. G. Rolan, M. F. de Jong, and R. M. Tsolis. 2008. VirB3 to VirB6 and VirB8 to VirB11, but not VirB7, are essential for mediating persistence of Brucella in the reticuloendothelial system. J. Bacteriol. 190:4427-4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.den Hartigh, A. B., Y. H. Sun, D. Sondervan, N. Heuvelmans, M. O. Reinders, T. A. Ficht, and R. M. Tsolis. 2004. Differential requirements for VirB1 and VirB2 during Brucella abortus infection. Infect. Immun. 72:5143-5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dijkstra, A. J., and W. Keck. 1996. Peptidoglycan as a barrier to transenvelope transport. J. Bacteriol. 178:5555-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dozot, M., R. A. Boigegrain, R. M. Delrue, R. Hallez, S. Ouahrani-Bettache, I. Danese, J. J. Letesson, X. De Bolle, and S. Kohler. 2006. The stringent response mediator Rsh is required for Brucella melitensis and Brucella suis virulence, and for expression of the type IV secretion system virB. Cell. Microbiol. 8:1791-1802. [DOI] [PubMed] [Google Scholar]
- 42.Draskovic, I., and D. Dubnau. 2005. Biogenesis of a putative channel protein, ComEC, required for DNA uptake: membrane topology, oligomerization and formation of disulphide bonds. Mol. Microbiol. 55:881-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dumler, J. S., A. F. Barbet, C. P. Bekker, G. A. Dasch, G. H. Palmer, S. C. Ray, Y. Rikihisa, and F. R. Rurangirwa. 2001. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 51:2145-2165. [DOI] [PubMed] [Google Scholar]
- 44.Eisenbrandt, R., M. Kalkum, E. M. Lai, R. Lurz, C. I. Kado, and E. Lanka. 1999. Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J. Biol. Chem. 274:22548-22555. [DOI] [PubMed] [Google Scholar]
- 45.Eisenbrandt, R., M. Kalkum, R. Lurz, and E. Lanka. 2000. Maturation of IncP pilin precursors resembles the catalytic dyad-like mechanism of leader peptidases. J. Bacteriol. 182:6751-6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Felek, S., H. Huang, and Y. Rikihisa. 2003. Sequence and expression analysis of virB9 of the type IV secretion system of Ehrlichia canis strains in ticks, dogs, and cultured cells. Infect. Immun. 71:6063-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fernandez, D., T. A. Dang, G. M. Spudich, X. R. Zhou, B. R. Berger, and P. J. Christie. 1996. The Agrobacterium tumefaciens virB7 gene product, a proposed component of the T-complex transport apparatus, is a membrane-associated lipoprotein exposed at the periplasmic surface. J. Bacteriol. 178:3156-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fischer, W., J. Puls, R. Buhrdorf, B. Gebert, S. Odenbreit, and R. Haas. 2001. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol. Microbiol. 42:1337-1348. [DOI] [PubMed] [Google Scholar]
- 49.Foster, J., M. Ganatra, I. Kamal, J. Ware, K. Makarova, N. Ivanova, A. Bhattacharyya, V. Kapatral, S. Kumar, J. Posfai, T. Vincze, J. Ingram, L. Moran, A. Lapidus, M. Omelchenko, N. Kyrpides, E. Ghedin, S. Wang, E. Goltsman, V. Joukov, O. Ostrovskaya, K. Tsukerman, M. Mazur, D. Comb, E. Koonin, and B. Slatko. 2005. The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 3:e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frank, A. C., C. M. Alsmark, M. Thollesson, and S. G. Andersson. 2005. Functional divergence and horizontal transfer of type IV secretion systems. Mol. Biol. Evol. 22:1325-1336. [DOI] [PubMed] [Google Scholar]
- 51.Fronzes, R., P. J. Christie, and G. Waksman. 2009. The structural biology of type IV secretion systems. Nat. Rev. Microbiol. 7:703-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fronzes, R., E. Schafer, L. Wang, H. R. Saibil, E. V. Orlova, and G. Waksman. 2009. Structure of a type IV secretion system core complex. Science 323:266-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fullner, K. J., J. C. Lara, and E. W. Nester. 1996. Pilus assembly by Agrobacterium T-DNA transfer genes. Science 273:1107-1109. [DOI] [PubMed] [Google Scholar]
- 54.Gal-Mor, O., and G. Segal. 2003. Identification of CpxR as a positive regulator of icm and dot virulence genes of Legionella pneumophila. J. Bacteriol. 185:4908-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gillespie, J. J., N. C. Ammerman, S. M. Dreher-Lesnick, M. S. Rahman, M. J. Worley, J. C. Setubal, B. S. Sobral, and A. F. Azad. 2009. An anomalous type IV secretion system in Rickettsia is evolutionarily conserved. PLoS One 4:e4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gillespie, J. J., M. S. Beier, M. S. Rahman, N. C. Ammerman, J. M. Shallom, A. Purkayastha, B. S. Sobral, and A. F. Azad. 2007. Plasmids and rickettsial evolution: insight from Rickettsia felis. PLoS One 2:e266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gillespie, J. J., K. Williams, M. Shukla, E. E. Snyder, E. K. Nordberg, S. M. Ceraul, C. Dharmanolla, D. Rainey, J. Soneja, J. M. Shallom, N. D. Vishnubhat, R. Wattam, A. Purkayastha, M. Czar, O. Crasta, J. C. Setubal, A. F. Azad, and B. S. Sobral. 2008. Rickettsia phylogenomics: unwinding the intricacies of obligate intracellular life. PLoS One 3:e2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gouin, E., C. Egile, P. Dehoux, V. Villiers, J. Adams, F. Gertler, R. Li, and P. Cossart. 2004. The RickA protein of Rickettsia conorii activates the Arp2/3 complex. Nature 427:457-461. [DOI] [PubMed] [Google Scholar]
- 59.Gouin, E., H. Gantelet, C. Egile, I. Lasa, H. Ohayon, V. Villiers, P. Gounon, P. J. Sansonetti, and P. Cossart. 1999. A comparative study of the actin-based motilities of the pathogenic bacteria Listeria monocytogenes, Shigella flexneri and Rickettsia conorii. J. Cell Sci. 112(Pt. 11):1697-1708. [DOI] [PubMed] [Google Scholar]
- 60.Hamilton, H. L., N. M. Dominguez, K. J. Schwartz, K. T. Hackett, and J. P. Dillard. 2005. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol. Microbiol. 55:1704-1721. [DOI] [PubMed] [Google Scholar]
- 61.Heinzen, R. A., S. F. Hayes, M. G. Peacock, and T. Hackstadt. 1993. Directional actin polymerization associated with spotted fever group Rickettsia infection of Vero cells. Infect. Immun. 61:1926-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoppner, C., A. Carle, D. Sivanesan, S. Hoeppner, and C. Baron. 2005. The putative lytic transglycosylase VirB1 from Brucella suis interacts with the type IV secretion system core components VirB8, VirB9 and VirB11. Microbiology 151:3469-3482. [DOI] [PubMed] [Google Scholar]
- 63.Hotopp, J. C., M. Lin, R. Madupu, J. Crabtree, S. V. Angiuoli, J. A. Eisen, R. Seshadri, Q. Ren, M. Wu, T. R. Utterback, S. Smith, M. Lewis, H. Khouri, C. Zhang, H. Niu, Q. Lin, N. Ohashi, N. Zhi, W. Nelson, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, J. Sundaram, S. C. Daugherty, T. Davidsen, A. S. Durkin, M. Gwinn, D. H. Haft, J. D. Selengut, S. A. Sullivan, N. Zafar, L. Zhou, F. Benahmed, H. Forberger, R. Halpin, S. Mulligan, J. Robinson, O. White, Y. Rikihisa, and H. Tettelin. 2006. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jakubowski, S. J., E. Cascales, V. Krishnamoorthy, and P. J. Christie. 2005. Agrobacterium tumefaciens VirB9, an outer-membrane-associated component of a type IV secretion system, regulates substrate selection and T-pilus biogenesis. J. Bacteriol. 187:3486-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jakubowski, S. J., J. E. Kerr, I. Garza, V. Krishnamoorthy, R. Bayliss, G. Waksman, and P. J. Christie. 2009. Agrobacterium VirB10 domain requirements for type IV secretion and T pilus biogenesis. Mol. Microbiol. 71:779-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jakubowski, S. J., V. Krishnamoorthy, E. Cascales, and P. J. Christie. 2004. Agrobacterium tumefaciens VirB6 domains direct the ordered export of a DNA substrate through a type IV secretion system. J. Mol. Biol. 341:961-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jakubowski, S. J., V. Krishnamoorthy, and P. J. Christie. 2003. Agrobacterium tumefaciens VirB6 protein participates in formation of VirB7 and VirB9 complexes required for type IV secretion. J. Bacteriol. 185:2867-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jeng, R. L., E. D. Goley, J. A. D'Alessio, O. Y. Chaga, T. M. Svitkina, G. G. Borisy, R. A. Heinzen, and M. D. Welch. 2004. A Rickettsia WASP-like protein activates the Arp2/3 complex and mediates actin-based motility. Cell. Microbiol. 6:761-769. [DOI] [PubMed] [Google Scholar]
- 69.Juhas, M., D. W. Crook, I. D. Dimopoulou, G. Lunter, R. M. Harding, D. J. Ferguson, and D. W. Hood. 2007. Novel type IV secretion system involved in propagation of genomic islands. J. Bacteriol. 189:761-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Juhas, M., D. W. Crook, and D. W. Hood. 2008. Type IV secretion systems: tools of bacterial horizontal gene transfer and virulence. Cell. Microbiol. 10:2377-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Juhas, M., P. M. Power, R. M. Harding, D. J. Ferguson, I. D. Dimopoulou, A. R. Elamin, Z. Mohd-Zain, D. W. Hood, R. Adegbola, A. Erwin, A. Smith, R. S. Munson, A. Harrison, L. Mansfield, S. Bentley, and D. W. Crook. 2007. Sequence and functional analyses of Haemophilus spp. genomic islands. Genome Biol. 8:R237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kalkum, M., R. Eisenbrandt, and E. Lanka. 2004. Protein circlets as sex pilus subunits. Curr. Protein Pept. Sci. 5:417-424. [DOI] [PubMed] [Google Scholar]
- 73.Klockgether, J., D. Wurdemann, O. Reva, L. Wiehlmann, and B. Tummler. 2007. Diversity of the abundant pKLC102/PAGI-2 family of genomic islands in Pseudomonas aeruginosa. J. Bacteriol. 189:2443-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Koonin, E. V., and K. E. Rudd. 1994. A conserved domain in putative bacterial and bacteriophage transglycosylases. Trends Biochem. Sci. 19:106-107. [DOI] [PubMed] [Google Scholar]
- 75.Koraimann, G. 2003. Lytic transglycosylases in macromolecular transport systems of Gram-negative bacteria. Cell. Mol. Life Sci. 60:2371-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Krall, L., U. Wiedemann, G. Unsin, S. Weiss, N. Domke, and C. Baron. 2002. Detergent extraction identifies different VirB protein subassemblies of the type IV secretion machinery in the membranes of Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. U. S. A. 99:11405-11410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lai, E. M., O. Chesnokova, L. M. Banta, and C. I. Kado. 2000. Genetic and environmental factors affecting T-pilin export and T-pilus biogenesis in relation to flagellation of Agrobacterium tumefaciens. J. Bacteriol. 182:3705-3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lai, E. M., R. Eisenbrandt, M. Kalkum, E. Lanka, and C. I. Kado. 2002. Biogenesis of T pili in Agrobacterium tumefaciens requires precise VirB2 propilin cleavage and cyclization. J. Bacteriol. 184:327-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lai, E. M., and C. I. Kado. 2000. The T-pilus of Agrobacterium tumefaciens. Trends Microbiol. 8:361-369. [DOI] [PubMed] [Google Scholar]
- 80.Lawley, T. D., W. A. Klimke, M. J. Gubbins, and L. S. Frost. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 224:1-15. [DOI] [PubMed] [Google Scholar]
- 81.Lehnherr, H., A. M. Hansen, and T. Ilyina. 1998. Penetration of the bacterial cell wall: a family of lytic transglycosylases in bacteriophages and conjugative plasmids. Mol. Microbiol. 30:454-457. [DOI] [PubMed] [Google Scholar]
- 82.Lessl, M., and E. Lanka. 1994. Common mechanisms in bacterial conjugation and Ti-mediated T-DNA transfer to plant cells. Cell 77:321-324. [DOI] [PubMed] [Google Scholar]
- 83.Lin, M., A. den Dulk-Ras, P. J. Hooykaas, and Y. Rikihisa. 2007. Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell. Microbiol. 9:2644-2657. [DOI] [PubMed] [Google Scholar]
- 84.Lin, M., and Y. Rikihisa. 2003. Ehrlichia chaffeensis and Anaplasma phagocytophilum lack genes for lipid A biosynthesis and incorporate cholesterol for their survival. Infect. Immun. 71:5324-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lin, M., C. Zhang, K. Gibson, and Y. Rikihisa. 2009. Analysis of complete genome sequence of Neorickettsia risticii: causative agent of Potomac horse fever. Nucleic Acids Res. 37:6076-6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu, Z., and A. N. Binns. 2003. Functional subsets of the virB type IV transport complex proteins involved in the capacity of Agrobacterium tumefaciens to serve as a recipient in virB-mediated conjugal transfer of plasmid RSF1010. J. Bacteriol. 185:3259-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Llosa, M., J. Zupan, C. Baron, and P. Zambryski. 2000. The N- and C-terminal portions of the Agrobacterium VirB1 protein independently enhance tumorigenesis. J. Bacteriol. 182:3437-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Locht, C., and J. M. Keith. 1986. Pertussis toxin gene: nucleotide sequence and genetic organization. Science 232:1258-1264. [DOI] [PubMed] [Google Scholar]
- 89.Lopez, J. E., P. A. Beare, R. A. Heinzen, J. Norimine, K. K. Lahmers, G. H. Palmer, and W. C. Brown. 2008. High-throughput identification of T-lymphocyte antigens from Anaplasma marginale expressed using in vitro transcription and translation. J. Immunol. Methods 332:129-141. [DOI] [PubMed] [Google Scholar]
- 90.Lopez, J. E., G. H. Palmer, K. A. Brayton, M. J. Dark, S. E. Leach, and W. C. Brown. 2007. Immunogenicity of Anaplasma marginale type IV secretion system proteins in a protective outer membrane vaccine. Infect. Immun. 75:2333-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lopez, J. E., W. F. Siems, G. H. Palmer, K. A. Brayton, T. C. McGuire, J. Norimine, and W. C. Brown. 2005. Identification of novel antigenic proteins in a complex Anaplasma marginale outer membrane immunogen by mass spectrometry and genomic mapping. Infect. Immun. 73:8109-8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Maher, D., R. Sherburne, and D. E. Taylor. 1993. H-pilus assembly kinetics determined by electron microscopy. J. Bacteriol. 175:2175-2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Medini, D., A. Covacci, and C. Donati. 2006. Protein homology network families reveal step-wise diversification of Type III and Type IV secretion systems. PLoS Comput. Biol. 2:e173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Meeus, P. F., K. A. Brayton, G. H. Palmer, and A. F. Barbet. 2003. Conservation of a gene conversion mechanism in two distantly related paralogues of Anaplasma marginale. Mol. Microbiol. 47:633-643. [DOI] [PubMed] [Google Scholar]
- 95.Min, C. K., J. S. Yang, S. Kim, M. S. Choi, I. S. Kim, and N. H. Cho. 2008. Genome-based construction of the metabolic pathways of Orientia tsutsugamushi and comparative analysis within the Rickettsiales order. Comp. Funct. Genomics 2008:623145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mushegian, A. R., K. J. Fullner, E. V. Koonin, and E. W. Nester. 1996. A family of lysozyme-like virulence factors in bacterial pathogens of plants and animals. Proc. Natl. Acad. Sci. U. S. A. 93:7321-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nagai, H., J. C. Kagan, X. Zhu, R. A. Kahn, and C. R. Roy. 2002. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295:679-682. [DOI] [PubMed] [Google Scholar]
- 98.Nakayama, K., A. Yamashita, K. Kurokawa, T. Morimoto, M. Ogawa, M. Fukuhara, H. Urakami, M. Ohnishi, I. Uchiyama, Y. Ogura, T. Ooka, K. Oshima, A. Tamura, M. Hattori, and T. Hayashi. 2008. The whole-genome sequencing of the obligate intracellular bacterium Orientia tsutsugamushi revealed massive gene amplification during reductive genome evolution. DNA Res. 15:185-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nelson, C. M., M. J. Herron, R. F. Felsheim, B. R. Schloeder, S. M. Grindle, A. O. Chavez, T. J. Kurtti, and U. G. Munderloh. 2008. Whole genome transcription profiling of Anaplasma phagocytophilum in human and tick host cells by tiling array analysis. BMC Genomics 9:364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nicosia, A., M. Perugini, C. Franzini, M. C. Casagli, M. G. Borri, G. Antoni, M. Almoni, P. Neri, G. Ratti, and R. Rappuoli. 1986. Cloning and sequencing of the pertussis toxin genes: operon structure and gene duplication. Proc. Natl. Acad. Sci. U. S. A. 83:4631-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ogata, H., B. La Scola, S. Audic, P. Renesto, G. Blanc, C. Robert, P. E. Fournier, J. M. Claverie, and D. Raoult. 2006. Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLoS Genet. 2:e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ogata, H., P. Renesto, S. Audic, C. Robert, G. Blanc, P. E. Fournier, H. Parinello, J. M. Claverie, and D. Raoult. 2005. The genome sequence of Rickettsia felis identifies the first putative conjugative plasmid in an obligate intracellular parasite. PLoS Biol. 3:e248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pang, H., and H. H. Winkler. 1994. Analysis of the peptidoglycan of Rickettsia prowazekii. J. Bacteriol. 176:923-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Park, J., K. J. Kim, K. S. Choi, D. J. Grab, and J. S. Dumler. 2004. Anaplasma phagocytophilum AnkA binds to granulocyte DNA and nuclear proteins. Cell. Microbiol. 6:743-751. [DOI] [PubMed] [Google Scholar]
- 105.Perlman, S. J., M. S. Hunter, and E. Zchori-Fein. 2006. The emerging diversity of Rickettsia. Proc. Biol. Sci. 273:2097-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pichon, S., D. Bouchon, R. Cordaux, L. Chen, R. A. Garrett, and P. Greve. 2009. Conservation of the Type IV secretion system throughout Wolbachia evolution. Biochem. Biophys. Res. Commun. 385:557-562. [DOI] [PubMed] [Google Scholar]
- 107.Rambow-Larsen, A. A., and A. A. Weiss. 2002. The PtlE protein of Bordetella pertussis has peptidoglycanase activity required for Ptl-mediated pertussis toxin secretion. J. Bacteriol. 184:2863-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rances, E., D. Voronin, V. Tran-Van, and P. Mavingui. 2008. Genetic and functional characterization of the type IV secretion system in Wolbachia. J. Bacteriol. 190:5020-5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ristic, M., and J. P. Kreier. 1984. Genus I. Anaplasma Theiler 1970, 7AL, p. 720-722. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 110.Robertus, J. D., A. F. Monzingo, E. M. Marcotte, and P. J. Hart. 1998. Structural analysis shows five glycohydrolase families diverged from a common ancestor. J. Exp. Zool. 282:127-132. [PubMed] [Google Scholar]
- 111.Ruiz, N. 2008. Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 105:15553-15557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sagulenko, V., E. Sagulenko, S. Jakubowski, E. Spudich, and P. J. Christie. 2001. VirB7 lipoprotein is exocellular and associates with the Agrobacterium tumefaciens T pilus. J. Bacteriol. 183:3642-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schmiederer, M., R. Arcenas, R. Widen, N. Valkov, and B. Anderson. 2001. Intracellular induction of the Bartonella henselae virB operon by human endothelial cells. Infect. Immun. 69:6495-6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schneider, T. D., and R. M. Stephens. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shamaei-Tousi, A., R. Cahill, and G. Frankel. 2004. Interaction between protein subunits of the type IV secretion system of Bartonella henselae. J. Bacteriol. 186:4796-4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sieira, R., D. J. Comerci, L. I. Pietrasanta, and R. A. Ugalde. 2004. Integration host factor is involved in transcriptional regulation of the Brucella abortus virB operon. Mol. Microbiol. 54:808-822. [DOI] [PubMed] [Google Scholar]
- 117.Silverman, D. J., and C. L. Wisseman, Jr. 1978. Comparative ultrastructural study on the cell envelopes of Rickettsia prowazekii, Rickettsia rickettsii, and Rickettsia tsutsugamushi. Infect. Immun. 21:1020-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Spudich, G. M., D. Fernandez, X. R. Zhou, and P. J. Christie. 1996. Intermolecular disulfide bonds stabilize VirB7 homodimers and VirB7/VirB9 heterodimers during biogenesis of the Agrobacterium tumefaciens T-complex transport apparatus. Proc. Natl. Acad. Sci. U. S. A. 93:7512-7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sutten, E. L., J. Norimine, P. A. Beare, R. A. Heinzen, J. E. Lopez, K. Morse, K. A. Brayton, J. J. Gillespie, and W. C. Brown. 2010. Anaplasma marginale type IV secretion system proteins VirB2, VirB7, VirB11, and VirD4 are immunogenic components of a protective bacterial membrane vaccine. Infect. Immun. 78:1314-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Travassos, L. H., S. E. Girardin, D. J. Philpott, D. Blanot, M. A. Nahori, C. Werts, and I. G. Boneca. 2004. Toll-like receptor 2-dependent bacterial sensing does not occur via peptidoglycan recognition. EMBO Rep. 5:1000-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Verma, A., and D. L. Burns. 2007. Requirements for assembly of PtlH with the pertussis toxin transporter apparatus of Bordetella pertussis. Infect. Immun. 75:2297-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vidotto, M. C., E. J. Venancio, and O. Vidotto. 2008. Cloning, sequencing and antigenic characterization of rVirB9 of Anaplasma marginale isolated from Parana State, Brazil. Genet. Mol. Res. 7:460-466. [DOI] [PubMed] [Google Scholar]
- 123.Walker, D. H., and N. Ismail. 2008. Emerging and re-emerging rickettsioses: endothelial cell infection and early disease events. Nat. Rev. Microbiol. 6:375-386. [DOI] [PubMed] [Google Scholar]
- 124.Weinert, L. A., J. J. Welch, and F. M. Jiggins. 2009. Conjugation genes are common throughout the genus Rickettsia and are transmitted horizontally. Proc. Biol. Sci. 276:3619-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Weinert, L. A., J. H. Werren, A. Aebi, G. N. Stone, and F. M. Jiggins. 2009. Evolution and diversity of Rickettsia bacteria. BMC Biol. 7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Weiss, A. A., F. D. Johnson, and D. L. Burns. 1993. Molecular characterization of an operon required for pertussis toxin secretion. Proc. Natl. Acad. Sci. U. S. A. 90:2970-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Williams, K. P., B. W. Sobral, and A. W. Dickerman. 2007. A robust species tree for the alphaproteobacteria. J. Bacteriol. 189:4578-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Winans, S. C. 1992. Two-way chemical signaling in Agrobacterium-plant interactions. Microbiol. Rev. 56:12-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Winans, S. C., and G. C. Walker. 1985. Conjugal transfer system of the IncN plasmid pKM101. J. Bacteriol. 161:402-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wu, M., L. V. Sun, J. Vamathevan, M. Riegler, R. Deboy, J. C. Brownlie, E. A. McGraw, W. Martin, C. Esser, N. Ahmadinejad, C. Wiegand, R. Madupu, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, A. S. Durkin, J. F. Kolonay, W. C. Nelson, Y. Mohamoud, P. Lee, K. Berry, M. B. Young, T. Utterback, J. Weidman, W. C. Nierman, I. T. Paulsen, K. E. Nelson, H. Tettelin, S. L. O'Neill, and J. A. Eisen. 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2:E69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yeo, H. J., Q. Yuan, M. R. Beck, C. Baron, and G. Waksman. 2003. Structural and functional characterization of the VirB5 protein from the type IV secretion system encoded by the conjugative plasmid pKM101. Proc. Natl. Acad. Sci. U. S. A. 100:15947-15952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yuan, Q., A. Carle, C. Gao, D. Sivanesan, K. A. Aly, C. Hoppner, L. Krall, N. Domke, and C. Baron. 2005. Identification of the VirB4-VirB8-VirB5-VirB2 pilus assembly sequence of type IV secretion systems. J. Biol. Chem. 280:26349-26359. [DOI] [PubMed] [Google Scholar]
- 133.Zupan, J., C. A. Hackworth, J. Aguilar, D. Ward, and P. Zambryski. 2007. VirB1* promotes T-pilus formation in the vir-type IV secretion system of Agrobacterium tumefaciens. J. Bacteriol. 189:6551-6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zusman, T., G. Aloni, E. Halperin, H. Kotzer, E. Degtyar, M. Feldman, and G. Segal. 2007. The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol. Microbiol. 63:1508-1523. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.