Abstract
Soluble, divalent cation-dependent oxaloacetate decarboxylases (ODx) catalyze the irreversible decarboxylation of oxaloacetate to pyruvate and CO2. Although these enzymes have been characterized in different microorganisms, the genes that encode them have not been identified, and their functions have been only poorly analyzed so far. In this study, we purified a soluble ODx from wild-type C. glutamicum about 65-fold and used matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis and peptide mass fingerprinting for identification of the corresponding odx gene. Inactivation and overexpression of odx led to an absence of ODx activity and to a 30-fold increase in ODx specific activity, respectively; these findings unequivocally confirmed that this gene encodes a soluble ODx. Transcriptional analysis of odx indicated that there is a leaderless transcript that is organized in an operon together with a putative S-adenosylmethionine-dependent methyltransferase gene. Biochemical analysis of ODx revealed that the molecular mass of the native enzyme is about 62 ± 1 kDa and that the enzyme is composed of two ∼29-kDa homodimeric subunits and has a Km for oxaloacetate of 1.4 mM and a Vmax of 201 μmol of oxaloacetate converted per min per mg of protein, resulting in a kcat of 104 s−1. Introduction of plasmid-borne odx into a pyruvate kinase-deficient C. glutamicum strain restored growth of this mutant on acetate, indicating that a high level of ODx activity redirects the carbon flux from oxaloacetate to pyruvate in vivo. Consistently, overexpression of the odx gene in an l-lysine-producing strain of C. glutamicum led to accumulation of less l-lysine. However, inactivation of the odx gene did not improve l-lysine production under the conditions tested.
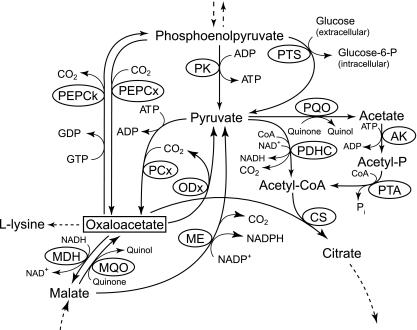
Corynebacterium glutamicum is a respirative, Gram-positive soil bacterium that is well suited to industrial amino acid production of, e.g., l-glutamate, l-lysine, and l-valine (4, 28). This organism possesses a rather complex phosphoenolpyruvate (PEP)-pyruvate-oxaloacetate node (Fig. 1) compared to model organisms, such as Escherichia coli and Bacillus subtilis (49). Due to the importance of this node for supply of precursors for amino acid synthesis and due to the fact that all enzymes of this node show significant activity in glucose-grown cells (13, 17, 20, 22, 42, 52), much attention has been focused on identifying targets for metabolic engineering (5, 18, 24, 26, 36, 39, 41, 46, 47, 56-58). Optimization of cellular oxaloacetate concentrations seems to be crucial, especially for improving l-lysine production. This possibility was proposed by Menkel et al. (29) and was indicated by overexpression of the pyruvate carboxylase gene (40), inactivation of PEP carboxykinase (46), inactivation of citrate and methylcitrate synthases (45), and disruption of the malate:quinone oxidoreductase gene (31). However, there have not been many studies addressing the role of oxaloacetate decarboxylase (ODx), an enzyme that has high levels of activity in different C. glutamicum strains (21) and catalyzes the irreversible decarboxylation of oxaloacetate (25), at this key branch point.
FIG. 1.
Phosphoenolpyruvate (PEP)-pyruvate-oxaloacetate node in C. glutamicum. Abbreviations: AK, acetate kinase; CS, citrate synthase; MDH, malate dehydrogenase; ME, malic enzyme; MQO, malate:quinone oxidoreductase; ODx, oxaloacetate decarboxylase; PCx, pyruvate carboxylase; PDHC, pyruvate dehydrogenase complex; PEPCk, PEP carboxykinase; PEPCx, PEP carboxylase; PK, pyruvate kinase; PQO, pyruvate:quinone oxidoreductase; PTA, phosphotransacetylase; PTS, phosphotransferase system; CoA, coenzyme A.
In general, oxaloacetate decarboxylases fall into two classes: (i) the sodium-dependent membrane-bound and biotin-containing oxaloacetate decarboxylases and (ii) the divalent cation-dependent soluble oxaloacetate decarboxylases. The mem-brane-bound oxaloacetate decarboxylases have been the subject of extensive studies; the genes encoding these enzymes have been identified, and the regulation, structure, function, and catalytic mechanisms of several of these enzymes have been elucidated (6, 7, 9-11). The other oxaloacetate decarboxylases are cytoplasmic enzymes that are absolutely dependent on the presence of divalent cations, such as Mn2+, Co2+, Mg2+, Ni2+, or Ca2+, and have been found in different microorganisms, including different species of Pseudomonas (19, 27, 35) and Acetobacter (2), C. glutamicum (21), Veillonella parvula (34), and Azotobacter vinelandii (43). Although some decarboxylases of the soluble type have been characterized, the genes that encode them have not been identified, and the functions of the enzymes in bacteria are not quite clear (49).
The oxaloacetate decarboxylase protein purified from C. glutamicum M2 and characterized by Jetten and Sinskey (20) has been reported to have an activity of 0.63 U/mg protein and a Km of 2.1 mM for the substrate oxaloacetate, and it was inhibited by Cu2+, Zn2+, ADP, GDP, coenzyme A, and succinate with Ki values of 0.2, 0.8, 1.2, 1.8, 2.4, and 2.8 mM, respectively. Nevertheless, from the data obtained no evidence concerning its function and role in growth and l-lysine production can be deduced. Metabolic network analysis addressing the actual in vivo fluxes and flux ratios at the PEP-pyruvate-oxaloacetate metabolic branch point produced contradictory results for the fluxes from oxaloacetate-malate to pyruvate. Whereas Petersen et al. (38) did not identify a direct carbon flux from oxaloacetate to pyruvate in wild-type C. glutamicum, Klapa et al. (24) observed high fluxes from oxaloacetate-malate to pyruvate in the l-lysine-producing strain C. glutamicum ATCC 21799. Since Klapa et al. (24) concluded that the malic enzyme was inactive under the conditions tested, the high fluxes from oxaloacetate-malate to pyruvate were attributed solely to high oxaloacetate decarboxylase activity and therefore indicated that oxaloacetate decarboxylase has a prominent role in l-lysine production.
The present study describes for the first time genetic and functional characterization of a soluble oxaloacetate decarboxylase from a prokaryotic organism. We isolated, analyzed, and expressed the odx gene encoding this enzyme in C. glutamicum and constructed and characterized C. glutamicum strains with altered oxaloacetate decarboxylase activities to clarify the significance of the oxaloacetate decarboxylation reaction for carbon distribution and l-lysine production.
MATERIALS AND METHODS
Bacteria, plasmids, oligonucleotides, and culture conditions.
All bacterial strains and plasmids and their relevant characteristics are described in Table 1. The oligonucleotides used and their sequences are also listed in Table 1. Strain C. glutamicum DM1729 was constructed from C. glutamicum WT (= ATCC 13032) by sequentially introducing point mutations into the chromosomal pyc, hom, and lysC genes (16). The resulting pyc(P458S), lysC(T311I), and hom(V59A) alleles encode deregulated variants of pyruvate carboxylase and aspartate kinase and a homoserine dehydrogenase with lower activity (36). For construction of C. glutamicum DM1730 the chromosomal pck gene in C. glutamicum DM1729 was inactivated exactly as described previously, using the suicide vector pK19mobsacB-Δpck (46). A complex tryptone-yeast extract medium (2× TY) (48) was used for C. glutamicum and E. coli cultures, except for precultures of the l-lysine production strain C. glutamicum DM1730, which were grown in 3.7% (wt/vol) brain heart infusion (BHI) (Difco). For growth on 4-hydroxyphenylacetate (HPA) (2 mM) or gentisate (2 mM or 8 mM) as the sole carbon and energy source, M9 minimal medium (48) (pH 8.4) supplemented with 0.25 g MgSO4·7H2O per liter, 0.01 g CaCl2 per liter, 16.4 mg FeSO4·7H2O per liter, 10 mg MnSO4·H2O per liter, 0.2 mg CuSO4·5H2O per liter, 1.0 mg ZnSO4·7H2O per liter, 0.02 mg NiCl2·6H2O per liter, 0.2 mg biotin per liter, and 0.01% (wt/vol) yeast extract (Difco) was used. For growth on glucose, acetate, or lactate (at the concentrations indicated in the Results) CGXII minimal medium (14) with a higher (NH4)2SO4 concentration (20 g/liter) was used in studies of amino acid production. C. glutamicum was grown aerobically at 30°C and E. coli was grown at 37°C in 60-ml cultures in 500-ml baffled Erlenmeyer flasks on a rotary shaker (B. Braun certomat sII) at 120 rpm. Plasmids were maintained by supplementing the media with kanamycin (50 μg ml−1) or spectinomycin (100 μg ml−1 for E. coli and 250 μg ml−1 for C. glutamicum).
TABLE 1.
Strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Relevant characteristic(s) or sequence | Source, reference, or use |
|---|---|---|
| E. coli DH5α | F− φ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rk− mk+) phoA supE44 thi gyrA96 relA1 λ− | Invitrogen |
| C. glutamicum strains | ||
| WT | Wild-type strain ATCC 13032 | American Type Culture Collection |
| WT(pEKEx5) | WT carrying plasmid pEKEx5 | This study |
| WT(pEKEx5_odx) | WT carrying odx on plasmid pEKEx5 | This study |
| WT(pEKEx2_odx) | WT carrying odx on plasmid pEKEx2 | This study |
| WT Δodx | odx deletion mutant of WT | This study |
| WT Δpyk | pyk deletion mutant of WT | 33 |
| WT Δpyk(pEKEx5) | pyk deletion mutant of WT carrying plasmid pEKEx5 | This study |
| WT Δpyk(pEKEx5_odx) | pyk deletion mutant of WT carrying odx on plasmid pEKEx5 | This study |
| DM1729 | l-Lysine-producing strain, pyc(P458S) hom(V59A) lysC(T311I) | 16 |
| DM1730 | l-Lysine-producing strain, pyc(P458S) hom(V59A) lysC(T311I) Δpck | B. Bathe, Evonik Degussa AG |
| DM1730(pEKEx5) | DM1730 carrying plasmid pEKEx5 | This study |
| DM1730(pEKEx5_odx) | DM1730 carrying odx on plasmid pEKEx5 | This study |
| DM1730 Δodx | odx deletion mutant of DM1730 | This study |
| Plasmids | ||
| pK19mobsacB | Vector for allelic exchange in C. glutamicum, KmroriV oriT sacB lacZα | 50 |
| pK19mobsacB_Δodx | pK19mobsacB carrying the odx gene with internal 679-bp deletion | This study |
| pEKEx5 | Expression vector for use in E. coli and C. glutamicum, Spr Ptac His6 | 30 |
| pEKEx5_odx | pEKEx5 carrying structural odx gene | This study |
| pEKEx2 | Expression vector used in E. coli and C. glutamicum, Kmr Ptac | 15 |
| pEKEx2_odx | pEKEx5 carrying structural odx gene with gap ribosomal binding site | This study |
| Oligonucleotides (primers) | ||
| cg1458DelFrag1For | 5′-GCGGATCCCACCGGCATCAAATTGTGTC-3′ | Deletion in odx, BamHI site (underlined) |
| cg1458DelFrag1Rev | 5′-GGAATGGAGTATGGAAGTTGGCACGGGCGGTGAGGTTAG-3′ | Deletion in odx, artificial crossover overlap (underlined) |
| cg1458DelFrag2For | 5′-CCAACTTCCATACTCCATTCCCATCGGCAAGCTGGGCAAC-3′ | Deletion in odx, artificial crossover overlap (underlined) |
| cg1458DelFrag2Rev | 5′-GCGGATCCTTGCCTTGAGCACAATGTC-3′ | Deletion in odx, BamHI site (underlined) |
| cg1458_oe_for | 5′-CGGGATCCATGCGTTTTGGACGA-3′ | Amplification of odx, BamHI site (underlined) |
| cg1458_oe_rev | 5′-GCGTCGACTTAGGCGTCCACAAC-3′ | Amplification of odx, SalI site (underlined) |
| cg1458ForGapPEx2 | 5′-GCGTCGACAGGAGAACAAACATGCGTTTTGGACGAATTGC-3′ | Amplification of odx, SalI site (underlined) and gap ribosome binding site (bold type) |
| cg1458RevEx2 | 5′-CGGGATCCTTAGGCGTCCACAACTG-3′ | Amplification of odx, BamHI site (underlined) |
| Opcg1458For | 5′-CGAAGGTGAGCTCGCAGTAG-3′ | Analysis of transcriptional organization of odx |
| Opcg1459Rev | 5′-GATGCCGGTGGTGACGTATC-3′ | Analysis of transcriptional organization of odx |
| Opcg1459ForPos | 5′-ATGGATCACCACCATCAC-3′ | Analysis of transcriptional organization of odx |
| Opcg1459RevPos | 5′-TTTCCGACAGCCACTTTG-3′ | Analysis of transcriptional organization of odx |
| Opcg1459RT | 5′-TCGTTGCCTTGAGCACAATG-3′ | Analysis of transcriptional organization of odx |
| SP1_odx | 5′-AAGGAAGGGATTCGGATTGG-3′ | Analysis of transcriptional start site of odx |
| SP2_odx | 5′-AGTGATTCTGCGGACTTC-3′ | Analysis of transcriptional start site of odx |
| SP3_odx | 5′-ATCGGCGTAGTTACGGCCAATC-3′ | Analysis of transcriptional start site of odx |
DNA preparation and transformation.
Isolation of chromosomal DNA and plasmids from C. glutamicum was performed as described previously (15). Preparation of plasmids from E. coli was carried out either essentially as described by Birnboim (3) or, for sequence validation, by MWG Biotech AG with an Amersham GFX preparation kit used as described by the manufacturer. Electroporation of E. coli was performed with competent cells using the method of Dower et al. (12). Transfer of DNA into C. glutamicum was performed as described by Molenaar et al. (32).
RNA preparation.
RNA was isolated from C. glutamicum as described previously (52). To ensure that a DNA-free RNA preparation was obtained, a PCR with RNA as the template and primer pair OpCg1459ForPos/OpCg1459RevPos was performed. As a positive control, chromosomal DNA was used instead of RNA. Since no amplicon was obtained with RNA as the template, the possibility of DNA contamination could be excluded.
PCR techniques and 5′ rapid amplification of cDNA ends (5′-RACE).
Amplification of desired DNA fragments was performed with a Biometra personal cycler by using Taq DNA polymerase, buffers, and deoxynucleotide triphosphates purchased from Fermentas and oligonucleotides obtained from biomers.net as the primers. The PCR conditions used were based on the primer constitution and on the fragment length, both computed by CloneManager Professional Suite (version 7.11; Sci Ed Central). PCR products were purified using a Nucleospin extract kit (Macherey-Nagel).
For analysis of the transcriptional organization of odx (cg1458) in C. glutamicum, 1.4 μg of total RNA from C. glutamicum WT was used for reverse transcriptase PCR (RT-PCR)) with primer pairs Opcg1458For/Opcg1459Rev and Opcg1459ForPos/Opcg1459RevPos. Synthesis of cDNA was performed by using a Fermentas first-strand cDNA synthesis kit and the gene-specific primer Opcg1459RT.
For identification of the transcription start site of the odx (cg1458) gene, a 5′-RACE experiment was carried out using a Roche second-generation 5′/3′-RACE kit according to the manufacturer's instructions. In short, first-strand cDNA was generated using gene-specific primer SP1_odx. After poly(A) tailing of the purified cDNA, the tailed cDNA was amplified in a first PCR with an oligo(dT) anchor primer and gene-specific primer SP2_odx. To increase the output, a second nested PCR was conducted with the PCR anchor primer and primer SP3_odx.
Enzyme assays.
In order to determine the oxaloacetate decarboxylase enzyme activity in cell extracts, C. glutamicum cells were harvested in mid-exponential growth phase by centrifugation for 10 min in an Eppendorf 5804 R centrifuge at 4,500 × g and 4°C. Cells were washed once with 20 ml of 200 mM Tris-HCl (pH 7.3) and resuspended in an appropriate volume of the same buffer (approximately 2 μl buffer per mg cell pellet). The cell suspension was transferred into 2-ml screw-cap vials together with 250 mg of glass beads (diameter, 0.1 mm; Roth) and subjected to mechanical disruption three times for 34 s at speed 6.5 with a RiboLyser (Hybaid) at 4°C with intermittent cooling on ice for 5 min. After disruption, the glass beads and cellular debris were removed by centrifugation for 20 min in an Eppendorf 5804 R centrifuge at 20,000 × g and 4°C. The supernatant was used to assay the oxaloacetate decarboxylase activity. The protein concentration was determined by using a bicinchoninic acid (BCA) protein assay reagent kit from Pierce with bovine serum albumin as the standard.
Oxaloacetate decarboxylase activity was assayed spectrophotometrically using a discontinuous system essentially as described by Benziman et al. (2). The decarboxylation mixture contained 100 mM Tris-HCl (pH 7.3), 10 mM MgCl2, 2.5 mM EDTA, 10 mM oxaloacetate, and between 30 and 60 μl of cell extract (corresponding to 0.45 mg and 0.90 mg of protein, respectively). After preincubation of the reaction mixture at 30°C for 3 min, the decarboxylation reaction was started by addition of oxaloacetate, and it was terminated after 3 min of incubation by adding 2% (vol/vol) trichloroacetic acid and cooling on ice. After neutralization with 2 M KOH and removal of the precipitate by centrifugation (14,000 × g; 4°C; 2 min; Sigma 202 MK centrifuge), the pyruvate formed was measured by using l-lactate dehydrogenase (l-LDH) from pig heart (Roche) and monitoring the decrease in absorbance at 340 nm of NADH (ɛ = 6.22 mM−1 cm−1) with an Amersham Ultrospec 2100 pro spectrophotometer. The assay mixture contained 100 mM Tris-HCl (pH 7.3), 0.4 mM NADH, 10 U l-LDH, and an appropriate amount of decarboxylase reaction supernatant. Control assays without crude extract were used to correct for nonenzymatic decarboxylation of oxaloacetate. One unit of oxaloacetate decarboxylase activity was defined as the amount of enzyme that converted 1 μmol of oxaloacetate per min.
Malic enzyme activity was determined spectrophotometrically essentially as described by Gourdon et al. (17). The reaction mixture used for oxidative decarboxylation of malate contained Tris-HCl buffer (100 mM, pH 8.4), MgCl2 (5 mM), NADP+ (2 mM), l(−)-malate (40 mM), and between 20 and 40 μl of crude extract (corresponding to 0.3 and 0.6 mg of protein, respectively).
Pyruvate kinase activity was measured spectrophotometrically essentially as described by Gubler et al. (18). The assay mixture contained Tris-HCl buffer (pH 7.3), MgCl2 (15 mM), ADP (1 mM), 0.4 mM NADH, 12 mM PEP, 5 U l-LDH, and between 5 and 10 μl of crude extract (corresponding to 0.075 and 0.15 mg of protein, respectively).
Purification of oxaloacetate decarboxylase.
For purification of oxaloacetate decarboxylase from cell extract of C. glutamicum WT, cells were grown to an optical density at 600 nm (OD600) of about 5 in 5 liters of 2× TY complex medium in a 10-liter B. Braun Biostat B fermentation system, washed once in 900 ml buffer A (50 mM Tris-HCl, pH 7.0), resuspended in 50 ml of the same buffer, and disrupted mechanically with a French pressure cell (SLM Aminco) at 1,800 lb/in2 (40 K cell) five times with intermittent cooling on ice. After removal of cellular debris by centrifugation (Eppendorf 5804 R centrifuge; 20,000 × g; 4°C; 20 min), the salt concentration in the supernatant was adjusted to 1 M (NH4)2SO4. Subsequently, centrifugation for 1.5 h in a Beckman L8-M ultracentrifuge at 150,000 × g and 4°C removed the membrane and precipitated the proteins, and the supernatant was applied to a HiPrep 16/10 Phenyl FF (Amersham Biosciences) column (1.6 by 10 cm) previously equilibrated with 1 M (NH4)2SO4 in buffer A for hydrophobic interaction chromatography. Absorbed proteins were eluted with a stepwise gradient consisting of 600, 300, and 0 mM (NH4)2SO4. Fractions with oxaloacetate decarboxylase activity collected at 0 mM (NH4)2SO4 were applied to a Mono Q HR 5/5 (Pharmacia Biotech) column (5 by 0.5 cm) preequilibrated with buffer A for concentration and stabilization of the enzyme. Absorbed proteins were eluted with a stepwise gradient consisting of 100, 300, 500, and 1,000 mM NaCl in buffer A. A single fraction with oxaloacetate decarboxylase activity collected at 300 mM NaCl was then applied to a HiLoad 16/60 Superdex-200 (Amersham Bioscience) prep-grade column (60 by 2.6 cm) preequilibrated with 100 mM NaCl in buffer A. Oxaloacetate decarboxylase activity was collected in three fractions with elution volumes between 72 and 75.7 ml. These fractions were combined, diluted with buffer A, and applied again to the MonoQ HR 5/5 column that had been preequilibrated with buffer A. Absorbed proteins were eluted using a 15-ml gradient consisting of 100 to 500 mM NaCl. Oxaloacetate decarboxylase was collected in a single fraction at 318 mM NaCl.
Kinetic parameters.
Km and Vmax values for oxaloacetate decarboxylase were determined by creating substrate saturation curves by varying the oxaloacetate concentration over at least a 100-fold range with a constant amount of purified enzyme (2.58 μg). Velocity data were fitted to the Michaelis-Menten equation (V = Vmax·[S]/Km + [S]) by nonlinear least-square regression using SigmaPlot (Systat Sofware Inc.).
Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis.
Analysis of protein bands cut out of colloidal Coomassie blue-stained acrylamide gels was performed at IBT-1, Forschungszentrum Jülich, with a Voyager-DE STR biospectrometry workstation (Applied Biosystems) essentially as described previously (51).
Analytics.
For quantification of glucose consumption and l-lysine formation in the culture fluid, 1-ml samples were removed from the cultures and centrifuged at room temperature for 10 min at 13,500 × g. Each supernatant was used for determination of the glucose concentration with an enzymatic test kit from Roche Diagnostics and for measuring the amino acid concentration by reverse-phase high-pressure liquid chromatography as described by Blombach et al. (4).
Computational analysis.
Databank searches and alignment studies were carried out by using BLAST, KEGG, and ERGO (1, 23, 37).
RESULTS
Oxaloacetate decarboxylase activity in C. glutamicum.
To determine whether a functional oxaloacetate decarboxylase is present in C. glutamicum, specific activities were determined for cell extracts of C. glutamicum WT grown in complex medium or in minimal medium containing 2% (wt/vol) glucose, 1% (wt/vol) potassium acetate, or 2% (wt/vol) sodium l-lactate as the sole carbon and energy source. Cells were harvested at the early, mid-, and late exponential and early and late stationary growth phases. The highest specific activities (about 0.66 U/mg protein) were found at the early and mid-exponential growth phases in cells grown in complex medium (Table 2). When cells were harvested at other growth phases during growth in complex medium (data not shown) or at any growth phase during growth in minimal medium containing glucose, acetate, or lactate, the oxaloacetate decarboxylase activities were about 1.5-fold lower and nearly identical (Table 2). These results show that C. glutamicum WT contains a functional oxaloacetate decarboxylase and indicate that this enzyme is only weakly regulated or not regulated by the carbon source and that its activity only partially depends or does not depend on the growth phase.
TABLE 2.
Specific activities of oxaloacetate decarboxylase in cell extracts of different C. glutamicum strains in complex medium and minimal medium containing different carbon sourcesa
| C. glutamicum strain | Medium | ODx sp act (U/mg protein) |
|---|---|---|
| WT | TY | 0.658 ± 0.116 |
| MM + glucose | 0.426 ± 0.078 | |
| MM + acetate | 0.463 ± 0.036 | |
| MM + lactate | 0.534 ± 0.076 | |
| TY + 4-hydroxyphenylacetate | 0.534 ± 0.068 | |
| TY + gentisate | 0.583 ± 0.096 | |
| WT Δodx | TY | 0.007 ± 0.007 |
| TY + 4-hydroxyphenylacetate | 0.016 ± 0.008 | |
| TY + gentisate | 0.023 ± 0.012 | |
| MM + gentisate | 0.045 ± 0.009 | |
| WT(pEKEx5) | TY | 0.495 ± 0.027 |
| WT(pEKEx5_odx) | TY | 18.736 ± 1.668 |
| DM1730 | MM + glucose | 0.437 ± 0.011 |
| DM1730 Δodx | MM + glucose | 0.009 ± 0.004 |
| DM1730(pEKEx5) | MM + glucose | 0.427 ± 0.063 |
| DM1730(pEKEx5_odx) | MM + glucose | 34.245 ± 8.143 |
The activities in complex medium (TY) and minimal medium (MM) are expressed as means ± standard deviations of at least three independent cultures and three determinations per experiment.
Purification of oxaloacetate decarboxylase and identification of open reading frame (ORF) cg1458.
Using cell extracts of C. glutamicum WT, oxaloacetate decarboxylase was purified 65-fold by using the five-step protocol outlined in Materials and Methods. After separation by SDS-PAGE and subsequent Coomassie blue staining (Fig. 2), one strong protein band detected at a relative molecular mass of about 32 kDa was cut out, digested with trypsin, and used for MALDI-TOF analysis. Matching peptide fragments accounted for 29% (78/268) of the deduced amino acids in the sequence encoded by ORF cg1458, and this is the first identification of a potential gene encoding a soluble oxaloacetate decarboxylase.
FIG. 2.
SDS-PAGE of oxaloacetate decarboxylase (ODx) from C. glutamicum after each step of the purification procedure. Lanes M, prestained molecular mass standard; lane 1, crude extract; lane 2, extract after ultracentrifugation; lane 3, pooled fractions with ODx activity after hydrophobic interaction chromatography; lane 4, pooled fractions with ODx activity after anion-exchange chromatography (stepwise elution); lanes 5 and 6, single fractions with ODx activity after size exclusion chromatography; lane 7, single fraction with ODx activity after anion-exchange chromatography (gradient elution).
Analysis of ORF cg1458 and its chromosomal organization.
ORF cg1458 is 807 bp long, and the predicted gene product consists of 268 amino acids. The computed molecular mass of this product (29 kDa) corresponds well to the experimentally determined molecular mass. ORFs that are directly upstream of and in the same direction as cg1458 putatively code for a signal transduction protein containing cyclic AMP-binding and cystathione beta-synthase (CBS) domains (cg1456) and a DNA polymerase III epsilon subunit (cg1457). Downstream of cg1458 and separated by only 1 bp, cg1459 codes for a putative S-adenosylmethionine-dependent methyltransferase.
A database search revealed two different annotations for the cg1458 gene product, which acts either as a 2-hydroxyhepta-2,4-diene-1,7-dioate (HHDD) isomerase in tyrosine metabolism (KEGG) or as a fumarylpyruvate hydrolase in gentisate metabolism (ERGO). Alignment studies with functionally analyzed enzymes having one of these catalytic functions revealed 35% and 28% identity to the C-terminal decarboxylase and N-terminal isomerase domain, respectively, of HpcE of E. coli, which acts as a 4-hydroxyphenylacetate degradation bifunctional isomerase/decarboxylase (55), and 32% identity to the fumarylpyruvate hydrolase of C. glutamicum encoded by ORF cg3350 (53). Furthermore, cg1458 has been annotated to be paralogous to cg3350 (KEGG).
Transcriptional analysis of ORF cg1458.
RT-PCR experiments were performed to analyze the organization of the C. glutamicum cg1458 transcript. RT-PCR using a primer covering the 3′ end of cg1458 and the 5′ end of cg1459 and total RNA of C. glutamicum resulted in an amplification product about 700 bp long, indicating that cg1458 and cg1459 are organized in an operon.
To identify the transcriptional initiation site of the C. glutamicum cg1458 gene, 5′-RACE experiments were performed with three individually isolated RNAs from three separate cultures of C. glutamicum WT. With all RNAs, a single signal was obtained, corresponding to the T residue of the annotated start codon ATG, indicating a leaderless transcript. A second start codon together with preceding sequences resembling ribosomal binding sites was found 33 bp downstream of the annotated translation initiation site in the sequence of cg1458. N-terminal sequencing of the purified protein was not successful. However, the identification of peptide fragments covering the area surrounding and in front of the second ATG by MALDI-TOF analysis of the purified enzyme clearly ruled out the alternative start codon.
Inactivation and overexpression of ORF cg1458.
To test whether cg1458 in fact codes for oxaloacetate decarboxylase, the chromosomal cg1458 gene was deleted, and it was also overexpressed from pEKEx5 in C. glutamicum WT. The resulting strains, designated C. glutamicum WT Δodx and C. glutamicum WT(pEKEx5_odx), were tested to determine whether they grew on different media and to determine their specific oxaloacetate decarboxylase activities.
The C. glutamicum WT and WT Δodx strains grew equally well (based on doubling time and final optical density) in complex medium and in minimal medium containing 2% (wt/vol) glucose or 2% (wt/vol) potassium acetate. There was also no difference in the growth characteristics of the two strains when cells were grown in minimal medium containing gentisate (data not shown). Neither C. glutamicum WT nor the WT Δodx mutant grew in minimal medium containing 4-hydroxyphenylacetate, an intermediate in tyrosine metabolism. These results indicate that cg1458 is not essential for growth of C. glutamicum under the conditions tested and that involvement of the cg1458 product in degradation of aromatic compounds is unlikely.
To test whether the cg1458 gene product functions as an oxaloacetate decarboxylase, oxaloacetate decarboxylase specific activities were determined for the C. glutamicum WT Δodx and C. glutamicum WT(pEKEx5_odx) strains and their ancestors. As shown in Table 2, the Δodx mutant did not exhibit any detectable oxaloacetate decarboxylase activity after growth in complex medium, whereas C. glutamicum WT(pEKEx5_odx) exhibited about 30-fold-higher specific oxaloacetate decarboxylase activity than the corresponding WT(pEKEx5) strain.
Only low levels of activity were observed in the Δodx mutant after growth in minimal medium containing 8 mM gentisate as a carbon source. These results show that the source of the oxaloacetate-decarboxylating activity in C. glutamicum WT is the product of the cg1458 gene; thus, this ORF was designated the odx gene.
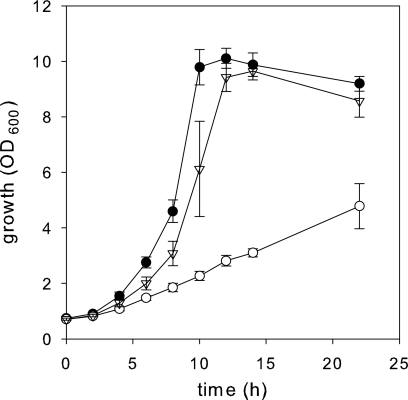
To test whether oxaloacetate decarboxylase directs carbon flux in the direction of pyruvate in vivo, growth experiments were performed with pyruvate kinase-deficient C. glutamicum strains. Netzer et al. (33) observed previously that pyruvate kinase appears to be essential for growth of C. glutamicum on acetate. Under gluconeogenic conditions, pyruvate availability for biomass synthesis is maintained by the pyruvate kinase reaction that converts a fraction of the PEP generated from oxaloacetate into pyruvate. Netzer et al. (33) found that in the absence of pyruvate kinase, increased malic enzyme (ME) activity is sufficient to redirect the carbon flux in the direction of pyruvate and to restore growth of a pyruvate kinase deletion mutant on acetate. In order to determine whether increased oxaloacetate decarboxylase activity also allows growth of the pyruvate kinase-deficient strain C. glutamicum WT Δpyk on acetate as a sole carbon source, the C. glutamicum WT and WT Δpyk strains harboring the odx expression plasmid pEKEx5_odx or the control plasmid pEKEx5 were constructed and analyzed. As shown in Fig. 3, the growth of C. glutamicum WT Δpyk(pEKEx5_odx) was similar to the growth of WT(pEKEx5) as these strains had growth rates of 0.27 ± 0.06 h−1 and 0.31 ± 0.01 h−1, respectively, and the final optical densities were 9.6 ± 0.3 and 10.1 ± 0.4, respectively, in minimal medium containing potassium acetate. However, the growth of C. glutamicum WT Δpyk(pEKEx5) was severely impaired, and the final optical density was only 4.8 after 20 h of cultivation (Fig. 3). C. glutamicum WT Δpyk(pEKEx5_odx) exhibited 17-fold-higher oxaloacetate decarboxylase activity (5.07 ± 1.42 U/mg protein) than C. glutamicum WT(pEKEx5) and WT Δpyk(pEKEx5), which had oxaloacetate decarboxylase activities of 0.30 ± 0.06 and 0.27 ± 0.04 U/mg protein, respectively. All three strains exhibited the same level of malic enzyme activity, and no pyruvate kinase activity was detected in the Δpyk mutants compared to strain C. glutamicum WT(pEKEx5) (data not shown). Thus, overexpression of the odx gene allows C. glutamicum WT Δpyk to grow on acetate as a sole carbon source, whereas the basal oxaloacetate decarboxylase and malic enzyme levels did not provide enough pyruvate for growth under these conditions. These results corroborate the conclusion that odx in fact codes for an enzyme that mediates decarboxylation of oxaloacetate to produce pyruvate in C. glutamicum.
FIG. 3.
Growth of C. glutamicum WT(pEKEx5) (•), WT Δpyk(pEKEx5) (○), and WT Δpyk(pEKEx5_odx) (▿) in CGXII minimal medium containing 1% (wt/vol) potassium acetate as the sole carbon source. Isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM) was added to the cultures after 4 h of growth.
Characteristics of the purified enzyme.
To determine the subunit structure and steady-state kinetic constants for purified oxaloacetate decarboxylase, strain C. glutamicum WT(pEKEx2_odx) was constructed for homologous overproduction of the native oxaloacetate decarboxylase lacking the N-terminal His tag, since the activity of purified His-tagged versions of oxaloacetate decarboxylase was unstable (about 70% of the activity was lost in 1 week).
The molecular mass of the native oxaloacetate decarboxylase was estimated by gel filtration using Superdex 200 10/300 GL and appeared to be 62.1 ± 1.3 kDa. We propose that the native enzyme has a homodimeric subunit structure and that the relative molecular mass is 29 kDa per subunit.
The enzyme showed Michaelis-Menten kinetics with oxaloacetate as the substrate. The Km was 1.39 ± 0.09 mM, and the Vmax was 201.1 ± 3.9 U/mg protein. Assuming that the molecular mass is about 62 kDa, this results in a kcat of 104 s−1 for each catalytic unit.
Different intermediates of the central metabolism were tested to determine their abilities to affect oxaloacetate decarboxylase activity; however, no effect was observed after addition of ADP (2.4 mM), GDP (3.6 mM), or succinate (5.6 mM) or after addition of PEP, fructose-6-phosphate, fructose-1,6-bisphosphate, malate, acetate, citrate, fumarate, aspartate, or acetyl coenzyme A (5 mM each).
Significance of oxaloacetate decarboxylase for amino acid production.
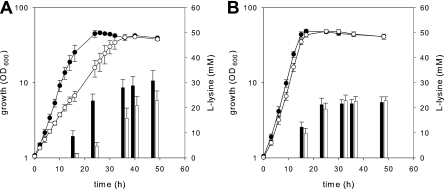
To analyze the influence of oxaloacetate decarboxylase activity on l-lysine production by C. glutamicum, an odx overexpresser and an oxaloacetate decarboxylase-negative derivative of the l-lysine producer C. glutamicum DM1730 were constructed. In minimal medium containing glucose, the odx-overexpressing strain DM1730(pEKEx5_odx) grew significantly slower than the corresponding strain DM1730(pEKEx5) (growth rates, 0.11 ± 0.01 h−1 and 0.20 ± 0.02 h−1, respectively; P < 0.001, Student's t test), but the final optical densities were the same (Fig. 4A). In cultures of both strains glucose was fully depleted when the stationary growth phase was reached. Amino acid analysis of the culture supernatants (Fig. 4A) revealed that for strain DM1730(pEKEx5_odx) the start of l-lysine production was delayed compared with strain DM1730(pEKEx5) and that significantly smaller amounts of this amino acid were produced (22.89 ± 3.84 mM and 30.78 ± 4.10 mM, respectively; P < 0.05, Student's t test) after 48 h of incubation. However, no differences in growth behavior and l-lysine production were observed when C. glutamicum DM1730 and the odx deletion mutant DM1730 Δodx were compared (Fig. 4B). These results indicate that strong overproduction of oxaloacetate decarboxylase negatively affects the growth of and l-lysine production by C. glutamicum DM1730; however, an absence of oxaloacetate decarboxylase does not have the desired beneficial effect on l-lysine production.
FIG. 4.
(A) Growth of and l-lysine production by C. glutamicum DM1730(pEKEx5) (• and filled bars) and DM1730(pEKEx5_odx) (○ and open bars) and (B) growth of and l-lysine production by C. glutamicum DM1730 (• and filled bars) and DM1730 Δodx (○ and open bars) in CGXII minimal medium containing 4% (wt/vol) glucose as the sole carbon source.
DISCUSSION
This study describes for the first time identification of a gene encoding a soluble oxaloacetate decarboxylase. Due to the absence of sequence information for this type of enzyme, oxaloacetate decarboxylase from C. glutamicum WT was purified and subsequently analyzed by the MALDI-TOF method to determine the reading frame that encodes this enzyme.
The purified oxaloacetate decarboxylase of C. glutamicum WT (= ATCC 13032) has some similarities to the enzyme previously purified from a derivative of C. glutamicum ATCC 13059 by Jetten and Sinskey (20), but there are also some differences. The two enzymes exhibited the same behavior during separation with Q-Sepharose, eluting at 318 mM NaCl and 350 mM KCl, respectively. The activities were absolutely dependent on the presence of divalent cations, which is the outstanding characteristic of this class of soluble oxaloacetate decarboxylases. SDS-PAGE of the purified enzyme revealed a single ∼32-kDa subunit, similar to that observed by Jetten and Sinskey (31.7 kDa); however, determinations of the molecular masses of the native enzymes revealed differences in the subunit composition. Jetten and Sinskey (20) obtained a homotetramer, whereas the data obtained in this work favor a dimeric structure. Nevertheless, the values determined for oxaloacetate (a Michaelis-Menten constant of 1.4 mM, a Vmax of 201 U/mg protein, and a kcat of 104 s−1) for the single subunit were similar to those of the oxaloacetate decarboxylase characterized by Jetten and Sinskey (Km, 2.1 mM; Vmax, 158 U/mg protein; kcat, 78 s−1).
MALDI-TOF analysis and peptide mass fingerprinting of the purified oxaloacetate decarboxylase identified ORF cg1458 as the gene encoding this protein. This gene was formerly annotated the gene encoding 2-hydroxyhepta-2,4-diene-1,7-dioate (HHDD) isomerase (KEGG) and fumarylpyruvate hydrolase (ERGO), enzymes that are involved in the breakdown of 4-hydroxyphenylacetate (4-HPA) and gentisate, respectively. However, the oxaloacetate decarboxylase activities observed after overexpression and deletion of the cg1458 gene in C. glutamicum unequivocally showed that cg1458 in fact encodes the soluble oxaloacetate decarboxylase of C. glutamicum. To elucidate a possible additional role of cg1458 (odx) in degradation of the aromatic compounds mentioned above in C. glutamicum, growth experiments were carried out with 4-HPA and gentisate. However, since C. glutamicum WT, as well as the Δcg1458 (Δodx) mutant, did not grow in minimal medium containing 4-HPA, we doubt that there is a homoprotocatechuate pathway in C. glutamicum, like that described for various E. coli strains (8). Furthermore, the similarity in growth behavior between C. glutamicum WT and the Δcg1458 (Δodx) mutant during cultivation on gentisate indicates that involvement of cg1458 (odx) in degradation of this aromatic compound is very unlikely. Additionally, in a study of the changes in the corynebacterial proteome during growth on various aromatic compounds (44), clear differences in the expression pattern of cg3350 were observed, whereas cg1458 (odx) never appeared.
Due to the paralogous annotation of cg3350 and cg1458, it can be assumed that the enzyme encoded by cg3350 has oxaloacetate decarboxylase (side) activity. However, since only negligible oxaloacetate decarboxylase activity was observed in the Δcg1458 (Δodx) mutant cells after growth on gentisate compared to cells grown in complex medium, the possibility that the product of the formerly annotated paralogous gene cg3350 has oxaloacetate decarboxylase (side) activity can be excluded.
In addition to malic enzyme and PEP carboxykinase, oxaloacetate decarboxylase is one of the three enzymes at the PEP-pyruvate-oxaloacetate node that in principle is capable of catalyzing the initial reaction of gluconeogenesis (Fig. 1). However, the inability of a defined PEP carboxykinase-negative mutant to grow on acetate (46) indicated that PEP carboxykinase is the only enzyme responsible for PEP synthesis from tricarboxylic acid (TCA) cycle intermediates and that in C. glutamicum WT this enzyme cannot be functionally replaced by activities of malic enzyme and/or oxaloacetate decarboxylase together with PEP synthetase. Although the presence of a PEP synthetase in C. glutamicum has been proposed for some strains (21, 56), the results mentioned above and the inability of a pyruvate carboxylase-deficient strain to grow on lactate (39) argue against the presence of a functional PEP synthetase. Based on the results presented here, there is no direct evidence for involvement of oxaloacetate decarboxylase and/or malic enzyme in pyruvate formation from oxaloacetate. However, Netzer et al. (33) found that a pyruvate kinase-deficient strain of C. glutamicum grows very weakly on acetate, assuming that there is a lack of precursor for synthesis of amino acids derived from pyruvate. Moreover, overexpression of malE, encoding the malic enzyme, was sufficient to restore growth of the pyk deletion strain. In this work, we obtained evidence that the same is true for overexpression of the oxaloacetate decarboxylase gene. In accordance with the results of Netzer et al. (33), high oxaloacetate decarboxylase activity resulted in complementation of the previously observed growth defect of the pyk-deficient strain on acetate (Fig. 3) and proved that decarboxylation of oxaloacetate through oxaloacetate decarboxylase occurs in vivo.
Oxaloacetate is a key branch point intermediate between biosynthesis of amino acids belonging to the aspartate family and the central carbon and energy metabolism. Reactions that influence the synthesis and conversion of oxaloacetate are therefore considered important targets for optimization of amino acid production. Klapa et al. (24) suggested that high oxaloacetate decarboxylase activities occur under l-lysine-producing conditions based on their results obtained by metabolic flux analysis of C. glutamicum ATCC 21799 harboring a feedback-resistant aspartate kinase. To further elucidate the role of oxaloacetate decarboxylase in l-lysine production, odx was overexpressed and deleted in the l-lysine-producing strain C. glutamicum DM1730. Although overexpression of the odx gene led to accumulation of less l-lysine, inactivation of the odx gene did not improve l-lysine production, which disproved the proposal based on the results of Klapa et al. (24). Analysis of the kinetic properties revealed that the Km for oxaloacetate (1.4 mM) was higher than those of other enzymes catalyzing oxaloacetate conversion, such as citrate synthase, aspartate aminotransferase, and malate dehydrogenase (Km values, 0.0015 mM, 0.11 mM, and 0.25 mM, respectively) (15, 21, 54). The low affinity of oxaloacetate decarboxylase for oxaloacetate, a compound that occurs only at low intracellular concentrations but is involved in a multitude of reactions, is one possible explanation for the absence of a positive effect of the odx deletion on l-lysine production. It may be that the oxaloacetate decarboxylase with its relatively high Km represents an overflow system when cells encounter high intracellular oxaloacetate concentrations.
Acknowledgments
We thank Volker Wendisch for kindly providing the pyk deletion mutant of C. glutamicum and Lothar Eggeling for kindly providing plasmid pEKEx5. MALDI-TOF mass spectrometry was performed at Forschungszentrum Jülich under administration of Michael Bott, and we gratefully acknowledge his support.
This work was supported by Evonik Degussa GmbH (Feed Additives) and the BMBF (grants 0313704 “SysMAP” and 0313805G “GenoMik-Plus”).
Footnotes
Published ahead of print on 16 March 2010.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Benziman, M., A. Russo, S. Hochman, and H. Weinhouse. 1978. Purification and regulatory properties of the oxaloacetate decarboxylase of Acetobacter xylinum. J. Bacteriol. 134:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birnboim, H. C. 1983. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol. 100:243-255. [DOI] [PubMed] [Google Scholar]
- 4.Blombach, B., M. E. Schreiner, J. Holátko, T. Bartek, M. Oldiges, and B. J. Eikmanns. 2007. l-Valine production with pyruvate dehydrogenase complex-deficient Corynebacterium glutamicum. Appl. Environ. Microbiol. 73:2079-2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blombach, B., M. E. Schreiner, M. Moch, M. Oldiges, and B. J. Eikmanns. 2007. Effect of pyruvate dehydrogenase complex deficiency on l-lysine production with Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 76:615-623. [DOI] [PubMed] [Google Scholar]
- 6.Bott, M. 1997. Anaerobic citrate metabolism and its regulation in enterobacteria. Arch. Microbiol. 167:78-88. [PubMed] [Google Scholar]
- 7.Buckel, W. 2001. Sodium ion-translocating decarboxylases. Biochim. Biophys. Acta 1505:15-27. [DOI] [PubMed] [Google Scholar]
- 8.Cooper, R. A., and M. A. Skinner. 1980. Catabolism of 3- and 4-hydroxyphenylacetate by the 3,4-dihydroxyphenylacetate pathway in Escherichia coli. J. Bacteriol. 143:302-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimroth, P. 1997. Primary sodium ion translocating enzymes. Biochim. Biophys. Acta 1318:11-51. [DOI] [PubMed] [Google Scholar]
- 10.Dimroth, P., P. Jockel, and M. Schmid. 2001. Coupling mechanism of the oxaloacetate decarboxylase Na+ pump. Biochim. Biophys. Acta 1505:1-14. [DOI] [PubMed] [Google Scholar]
- 11.Dimroth, P., and B. Schink. 1998. Energy conservation in the decarboxylation of dicarboxylic acids by fermenting bacteria. Arch. Microbiol. 170: 69-77. [DOI] [PubMed] [Google Scholar]
- 12.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eikmanns, B. J. 1992. Identification, sequence analysis, and expression of a Corynebacterium glutamicum gene cluster encoding the three glycolytic enzymes glyceraldehyde-3-phosphate dehydrogenase, 3-phosphoglycerate kinase, and triosephosphate isomerase. J. Bacteriol. 174:6076-6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eikmanns, B. J., M. Metzger, D. Reinscheid, M. Kircher, and H. Sahm. 1991. Amplification of three threonine biosynthesis genes in Corynebacterium glutamicum and its influence on carbon flux in different strains. Appl. Microbiol. Biotechnol. 34:617-622. [DOI] [PubMed] [Google Scholar]
- 15.Eikmanns, B. J., N. Thum-Schmitz, L. Eggeling, K. U. Lüdtke, and H. Sahm. 1994. Nucleotide sequence, expression and transcriptional analysis of the Corynebacterium glutamicum gltA gene encoding citrate synthase. Microbiology 140:1817-1828. [DOI] [PubMed] [Google Scholar]
- 16.Georgi, T., D. Rittmann, and V. F. Wendisch. 2005. Lysine and glutamate production by Corynebacterium glutamicum on glucose, fructose and sucrose: roles of malic enzyme and fructose-1,6-bisphosphatase. Metab. Eng. 7: 291-301. [DOI] [PubMed] [Google Scholar]
- 17.Gourdon, P., M. F. Baucher, N. D. Lindley, and A. Guyonvarch. 2000. Cloning of the malic enzyme gene from Corynebacterium glutamicum and role of the enzyme in lactate metabolism. Appl. Environ. Microbiol. 66:2981-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gubler, M., M. Jetten, S. H. Lee, and A. J. Sinskey. 1994. Cloning of the pyruvate kinase gene (pyk) of Corynebacterium glutamicum and site-specific inactivation of pyk in a lysine-producing Corynebacterium lactofermentum strain. Appl. Environ. Microbiol. 60:2494-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horton, A. A., and H. L. Kornberg. 1964. Oxaloacetate 4-carboxy-lyase from Pseudomonas ovalis Chester. Biochim. Biophys. Acta 89:381-383. [DOI] [PubMed] [Google Scholar]
- 20.Jetten, M. S., and A. J. Sinskey. 1995. Purification and properties of oxaloacetate decarboxylase from Corynebacterium glutamicum. Antonie Van Leeuwenhoek 67:221-227. [DOI] [PubMed] [Google Scholar]
- 21.Jetten, M. S. M., G. A. Pitoc, M. T. Follettie, and A. J. Sinskey. 1994. Regulation of phospho(enol)-pyruvate- and oxaloacetate-converting enzymes in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 41: 47-52. [Google Scholar]
- 22.Jetten, M. S., and A. J. Sinskey. 1993. Characterization of phosphoenolpyruvate carboxykinase from Corynebacterium glutamicum. FEMS Microbiol. Lett. 111:183-188. [Google Scholar]
- 23.Kanehisa, M., and S. Goto. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klapa, M. I., J. Aon, and G. Stephanopoulos. 2003. Systematic quantification of complex metabolic flux networks using stable isotopes and mass spectrometry. Eur. J. Biochem. 270:3525-3542. [DOI] [PubMed] [Google Scholar]
- 25.Krampitz, L. O., and C. H. Werkman. 1941. The enzymic decarboxylation of oxaloacetate. Biochem. J. 35:595-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krömer, J. O., O. Sorgenfrei, K. Klopprogge, E. Heinzle, and C. Wittmann. 2004. In-depth profiling of lysine-producing Corynebacterium glutamicum by combined analysis of the transcriptome, metabolome, and fluxome. J. Bacteriol. 186:1769-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Labrou, N. E., and Y. D. Clonis. 1999. Oxaloacetate decarboxylase from Pseudomonas stutzeri: purification and characterization. Arch. Biochem. Biophys. 365:17-24. [DOI] [PubMed] [Google Scholar]
- 28.Liebl, W. 1991. The genus Corynebacterium—nonmedical, p. 1157-1171. In K. H. Schleifer, M. Dworkin, A. Balows, H. G. Trüper, and W. Harder (ed.), The procaryotes. Springer, New York, NY.
- 29.Menkel, E., G. Thierbach, L. Eggeling, and H. Sahm. 1989. Influence of increased aspartate availability on lysine formation by a recombinant strain of Corynebacterium glutamicum and utilization of fumarate. Appl. Environ. Microbiol. 55:684-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Micklinghoff, J. C., K. J. Breitinger, M. Schmidt, R. Geffers, B. J. Eikmanns, and F. Bange. 2009. The role of the transcriptional regulator RamB (Rv0465c) in control of the glyoxylate cycle in Mycobacterium tuberculosis. J. Bacteriol. 191:7260-7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitsuhashi, S., M. Hayashi, J. Ohnishi, and M. Ikeda. 2006. Disruption of malate:quinone oxidoreductase increases l-lysine production by Corynebacterium glutamicum. Biosci. Biotechnol. Biochem. 70:2803-2806. [DOI] [PubMed] [Google Scholar]
- 32.Molenaar, D., M. E. van der Rest, and S. Petrović. 1998. Biochemical and genetic characterization of the membrane-associated malate dehydrogenase (acceptor) from Corynebacterium glutamicum. Eur. J. Biochem. 254:395-403. [DOI] [PubMed] [Google Scholar]
- 33.Netzer, R., M. Krause, D. Rittmann, P. G. Peters-Wendisch, L. Eggeling, V. F. Wendisch, and H. Sahm. 2004. Roles of pyruvate kinase and malic enzyme in Corynebacterium glutamicum for growth on carbon sources requiring gluconeogenesis. Arch. Microbiol. 182:354-363. [DOI] [PubMed] [Google Scholar]
- 34.Ng, S. K., M. Wong, and I. R. Hamilton. 1982. Properties of oxaloacetate decarboxylase from Veillonella parvula. J. Bacteriol. 150:1252-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Brien, R., D. T. Chuang, B. L. Taylor, and M. F. Utter. 1977. Novel enzymic machinery for the metabolism of oxalacetate, phosphoenolpyruvate, and pyruvate in Pseudomonas citronellolis. J. Biol. Chem. 252:1257-1263. [PubMed] [Google Scholar]
- 36.Ohnishi, J., S. Mitsuhashi, M. Hayashi, S. Ando, H. Yokoi, K. Ochiai, and M. Ikeda. 2002. A novel methodology employing Corynebacterium glutamicum genome information to generate a new l-lysine-producing mutant. Appl. Microbiol. Biotechnol. 58:217-223. [DOI] [PubMed] [Google Scholar]
- 37.Overbeek, R., N. Larsen, T. Walunas, M. D'Souza, G. Pusch, E. Selkov, K. Liolios, V. Joukov, D. Kaznadzey, I. Anderson, A. Bhattacharyya, H. Burd, W. Gardner, P. Hanke, V. Kapatral, N. Mikhailova, O. Vasieva, A. Osterman, V. Vonstein, M. Fonstein, N. Ivanova, and N. Kyrpides. 2003. The ERGO genome analysis and discovery system. Nucleic Acids Res. 31:164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petersen, S., A. A. de Graaf, L. Eggeling, M. Möllney, W. Wiechert, and H. Sahm. 2000. In vivo quantification of parallel and bidirectional fluxes in the anaplerosis of Corynebacterium glutamicum. J. Biol. Chem. 275:35932-35941. [DOI] [PubMed] [Google Scholar]
- 39.Peters-Wendisch, P. G., C. Kreutzer, J. Kalinowski, M. Pátek, H. Sahm, and B. J. Eikmanns. 1998. Pyruvate carboxylase from Corynebacterium glutamicum: characterization, expression and inactivation of the pyc gene. Microbiology 144:915-927. [DOI] [PubMed] [Google Scholar]
- 40.Peters-Wendisch, P. G., B. Schiel, V. F. Wendisch, E. Katsoulidis, B. Möckel, H. Sahm, and B. J. Eikmanns. 2001. Pyruvate carboxylase is a major bottleneck for glutamate and lysine production by Corynebacterium glutamicum. J. Mol. Microbiol. Biotechnol. 3:295-300. [PubMed] [Google Scholar]
- 41.Peters-Wendisch, P. G., B. J. Eikmanns, G. Thierbach, B. Bachmann, and H. Sahm. 1993. Phosphoenolpyruvate carboxylase in Corynebacterium glutamicum is dispensable for growth and lysine production. FEMS Microbiol. Lett. 112:269-274. [Google Scholar]
- 42.Peters-Wendisch, P. G., V. F. Wendisch, S. Paul, B. J. Eikmanns, and H. Sahm. 1997. Pyruvate carboxylase as an anaplerotic enzyme in Corynebacterium glutamicum. Microbiology 143:1095-1103. [DOI] [PubMed] [Google Scholar]
- 43.Plaut, G. W. E., and H. A. Lardy. 1949. The oxalacetate decarboxylase of Azotobacter vinelandii. J. Biol. Chem. 180:13-27. [PubMed] [Google Scholar]
- 44.Qi, S., M. T. Chaudhry, Y. Zhang, B. Meng, Y. Huang, K. Zhao, A. Poetsch, C. Jiang, S. Liu, and S. Liu. 2007. Comparative proteomes of Corynebacterium glutamicum grown on aromatic compounds revealed novel proteins involved in aromatic degradation and a clear link between aromatic catabolism and gluconeogenesis via fructose-1,6-bisphosphatase. Proteomics 7:3775-3787. [DOI] [PubMed] [Google Scholar]
- 45.Radmacher, E., and L. Eggeling. 2007. The three tricarboxylate synthase activities of Corynebacterium glutamicum and increase of l-lysine synthesis. Appl. Microbiol. Biotechnol. 76:587-595. [DOI] [PubMed] [Google Scholar]
- 46.Riedel, C., D. Rittmann, P. Dangel, B. Möckel, S. Petersen, H. Sahm, and B. J. Eikmanns. 2001. Characterization of the phosphoenolpyruvate carboxykinase gene from Corynebacterium glutamicum and significance of the enzyme for growth and amino acid production. J. Mol. Microbiol. Biotechnol. 3:573-583. [PubMed] [Google Scholar]
- 47.Sahm, H., L. Eggeling, and A. A. de Graaf. 2000. Pathway analysis and metabolic engineering in Corynebacterium glutamicum. Biol. Chem. 381:899-910. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 49.Sauer, U., and B. J. Eikmanns. 2005. The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol. Rev. 29:765-794. [DOI] [PubMed] [Google Scholar]
- 50.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 51.Schaffer, S., B. Weil, V. D. Nguyen, G. Dongmann, K. Günther, M. Nickolaus, T. Hermann, and M. Bott. 2001. A high-resolution reference map for cytoplasmic and membrane-associated proteins of Corynebacterium glutamicum. Electrophoresis 22:4404-4422. [DOI] [PubMed] [Google Scholar]
- 52.Schreiner, M. E., D. Fiur, J. Holátko, M. Pátek, and B. J. Eikmanns. 2005. E1 enzyme of the pyruvate dehydrogenase complex in Corynebacterium glutamicum: molecular analysis of the gene and phylogenetic aspects. J. Bacteriol. 187:6005-6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen, X., C. Jiang, Y. Huang, Z. Liu, and S. Liu. 2005. Functional identification of novel genes involved in the glutathione-independent gentisate pathway in Corynebacterium glutamicum. Appl. Environ. Microbiol. 71:3442-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shiio, I., M. Mori, and H. Ozaki. 1982. Amino acid aminotransferases in an amino acid-producing bacterium, Brevibacterium flavum. Agric. Biol. Chem. 46:2967-2977. [Google Scholar]
- 55.Tame, J. R. H., K. Namba, E. J. Dodson, and D. I. Roper. 2002. The crystal structure of HpcE, a bifunctional decarboxylase/isomerase with a multifunctional fold. Biochemistry 41:2982-2989. [DOI] [PubMed] [Google Scholar]
- 56.Vallino, J. J., and G. Stephanopoulos. 2000. Metabolic flux distributions in Corynebacterium glutamicum during growth and lysine overproduction. Reprinted from Biotechnology and Bioengineering, vol. 41, pp 633-646 (1993). Biotechnol. Bioeng. 67:872-885. [PubMed] [Google Scholar]
- 57.Wittmann, C., and E. Heinzle. 2001. Application of MALDI-TOF MS to lysine-producing Corynebacterium glutamicum: a novel approach for metabolic flux analysis. Eur. J. Biochem. 268:2441-2455. [DOI] [PubMed] [Google Scholar]
- 58.Wittmann, C., H. M. Kim, and E. Heinzle. 2004. Metabolic network analysis of lysine producing Corynebacterium glutamicum at a miniaturized scale. Biotechnol. Bioeng. 87:1-6. [DOI] [PubMed] [Google Scholar]