Abstract
Multidrug resistance (MDR) in clinical isolates of Escherichia coli can be associated with overexpression of marA, a transcription factor that upregulates multidrug efflux and downregulates membrane permeability. Using random transposome mutagenesis, we found that many chromosomal genes and environmental stimuli affected MarA-mediated antibiotic resistance. Seven genes affected resistance mediated by MarA in an antibiotic-specific way; these were mostly genes encoding unrelated enzymes, transporters, and unknown proteins. Other genes affected MarA-mediated resistance to all antibiotics tested. These genes were acrA, acrB, and tolC (which encode the major MarA-regulated multidrug efflux pump AcrAB-TolC), crp, cyaA, hns, and pcnB (four genes involved in global regulation of gene expression), and the unknown gene damX. The last five genes affected MarA-mediated MDR by altering marA expression or MarA function specifically on acrA. These findings demonstrate that MarA-mediated MDR is regulated at multiple levels by different genes and stimuli, which makes it both complex and fine-tuned and interconnects it with global cell regulation and metabolism. Such a regulation could contribute to the adaptation and spread of MDR strains and may be targeted to treat antibiotic-resistant E. coli and related pathogens.
Historically, infectious diseases have been the leading cause of mortality and morbidity until the discovery of antibiotics in the 20th century. However, the overuse and misuse of antibiotics have favored the emergence and spread of bacteria that are resistant to multiple antibiotics, threatening the treatment of bacterial infections worldwide (26, 27).
Multidrug resistance (MDR) can occur through the acquisition of extrachromosomal DNA, such as plasmids or transposons, via chromosomal mutations in genes coding for proteins targeted by the drug (e.g., mutations in gyrA that confer resistance to fluoroquinolones), or via altered expression of intrinsic mechanisms, such as efflux pumps that expel multiple classes of antibiotics out of the cell (2). Although increased intrinsic resistance usually produces moderate resistance levels, it can generate resistance to a wide array of antibiotics and other toxic chemicals. Moreover, this type of resistance may be a “stepping stone” to higher levels of resistance, such as in MDR clinical isolates having a combination of mutations in target genes and increased drug efflux (1, 16).
The Escherichia coli marA gene, which forms part of the chromosomal marRAB operon, was the first global regulator of intrinsic MDR discovered (18, 22). It has been extensively studied because of the widespread interest in E. coli as a major source of intestinal infections in developing countries and because of outbreaks of food poisoning and extraintestinal infections in industrialized countries (23). Moreover, marA orthologs or marA-like regulators are widespread among bacteria, including many clinically relevant pathogens, such as Salmonella enterica, Shigella flexneri, Klebsiella pneumoniae, Yersinia pestis and other Enterobacteriaceae, Neisseria gonorrhoeae, and Staphylococcus aureus (1, 2, 16).
The product of marA, the transcription factor MarA, directly activates or represses many chromosomal genes. MarA produces intrinsic multiple antibiotic resistance (Mar) mainly by upregulating the expression of acrAB and tolC, which encode AcrAB-TolC, the major multidrug efflux pump in E. coli (28, 39), and by upregulating micF, a regulatory antisense RNA which downregulates ompF translation and thus the OmpF porin-mediated outer membrane permeability to antibiotics. MarA also upregulates superoxide resistance (acnA, fumC, fpr, sodA, and zwf) and DNA repair (nfo), among other effects (5, 6).
Overexpression of marA is often found in MDR clinical isolates and is the result of spontaneous mutations in the autorepressor marR or in the DNA-binding sites of MarR (16, 31). Overexpression of marA also occurs when MarR is inactivated by chemicals such as salicylate (10, 46) or by cellular proteins such as TktA (13). Moreover, transcription of marA is activated by MarA and by the MarA homologs SoxS and Rob, mostly in the presence of the helper protein Fis (32). The intracellular level of MarA depends also on the Lon protease, which constitutively degrades MarA (21).
At a single-gene level, the function of MarA on at least one operon, hdeAB, which is involved in acid resistance, depends also on growth phase, pH, and other regulators of hdeAB (42). However, little is known about how the function of MarA on MDR is regulated at a global level. Here, we report that many genes and environmental stimuli are involved in a fine-tuned modulation of MarA-mediated MDR by affecting marA expression or function.
MATERIALS AND METHODS
Growth conditions.
All cultures were grown in LB medium (10 g of tryptone, 5 g of yeast extract, and 10 g of NaCl per liter) at 37°C with agitation. Antibiotics were used at 100 μg ml−1 (ampicillin) and 25 μg ml−1 (chloramphenicol and kanamycin).
Strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. The ΔmarR knockout parental strain CR1000 was obtained using plasmid pCP20 as previously described (12) to remove the kanamycin resistance gene of strain JW5248 (E. coli BW25113 ΔmarR::kan; Keio collection [3]). The ΔmarRA knockout parental strain CR2000 was constructed using the λ Red recombinase method (12) and primers MarRAdelF2 (5′CCAGCGATCTGTTCAATGAAATTATTCCATTGGGTCGCTTAATCCATATGGTGTAGGCTGGAGCTGCTTC) and MarRAdelR2 (5′TTGCCTCAGTGACGTTGTCACGTTTTCAACTAGCTGTTGTAATGATTTAAATGGGAATTAGCCATGGTCC) (the sequences corresponding to plasmid pKD3 are in bold). Briefly, these primers were used to generate a PCR product of the cat gene of pKD3 with sequences flanking marRA at both ends. This product was used to replace wild-type marRA in E. coli BW25113, and the cat gene was then removed using plasmid pCP20. In-frame deletion of marR in CR1000 and marRA in CR2000 was confirmed by PCR amplification and sequencing using specific primers flanking the deleted genes.
TABLE 1.
Bacterial strains and plasmids used in this study
| Group | Genotype or relevant characteristic(s) | Reference or source |
|---|---|---|
| E. coli strains used for mutagenesis | ||
| HdeA100 | GC4468 zdd-239::Tn9 del1738 (Δ39 kb, including marRAB locus) (Chlr) hdeABp::lacZ (Ampr) on λ at λatt site/pMB102 | 42 |
| HdeA100 mutants | HdeA100 mutagenized using EZ-TN5 <R6Kγori/KAN-2> or EZ-TN5 <Kan-2> transposomes; Ampr Chlr Kanr | This study |
| E. coli strains used for multidrug resistance and gene expression experiments | ||
| BW25113 | Wild type; F− λ− Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) rph-1 Δ(rhaD-rhaB)568 hsdR514 | CGSC; 3 |
| JW5248 | BW25113 ΔmarR::kan; Kanr | CGSC; 3 |
| CR1000 | BW25113 ΔmarR | This study |
| CR2000 | BW25113 ΔmarRA | This study |
| CR1000/CR2000 inactivation mutants | Kanr | P1 HdeA100 mutant × CR1000/CR2000 |
| Complementation plasmids | Ptac lacIq; low copy number; Ampr | Mobile plasmid collection |
| pNTR-SD (control) | NBRP (E. coli) (NIG, | |
| pNT3-crp | Japan); 43 | |
| pNTR-SD-hns | ||
| pNTR-SD-cyaA | ||
| pNT3-pcnB | ||
| pNTR-SD-acrB | ||
| pNTR-SD-tolC | ||
| pNTR-SD-damX | ||
| Other plasmids | ||
| pCP20 | Plasmid for excision of kan and cat markers by FLP-mediated site-specific recombination; Ampr Chlr | 12 |
| pKD3 | Template for amplifying the cat gene; Chlr | 12 |
| pKD46 | λRed recombinase expression plasmid; Ampr | 12 |
| pMB102 | ori colE1 lacI lacZp::marA; Ampr | 40 |
| pNN608 | Single copy; acrAp-lacZ; Chlr | 30 |
Genetic procedures.
PCR, phage P1 transduction, and plasmid or transposome electroporation were performed according to standard procedures (45). DNA sequencing was performed at Tufts University Core Facility. β-Galactosidase assays for studying the expression of the acrA-lacZ transcriptional fusion in single-copy plasmid pNN608 were performed as previously described (42). Bacterial two-hybrid assays for studying interactions between MarA and other proteins were performed by the adenylate cyclase system/β-galactosidase selection method, as previously described (13).
Mutagenesis and identification of mutants with reduced MarA function.
E. coli HdeA100 (Table 1) (42) is a ΔmarRAB strain that has marA in a plasmid, cloned under the control of an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter, and bears a hdeABp-lacZ chromosomal transcriptional fusion. This strain produces blue colonies on LB X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates and white colonies on LB X-Gal-IPTG plates because MarA represses hdeAB expression. To find genes important for MarA function, HdeA100 cells were electroporated with the EZ-TN5 <Kan-2> or EZ-TN5 <R6Kγori/KAN-2> kanamycin resistance transposome (Epicentre) and plated on LB containing ampicillin, kanamycin, 0.5 mM IPTG, and 40 μg ml−1 X-Gal. Mutant colonies in which MarA failed to repress hdeAB (blue colonies) were selected and repurified, and their phenotype was reconfirmed.
Transposome insertion points were identified by arbitrarily primed PCR (AP-PCR) or by direct genomic sequencing and confirmed by PCR amplification and sequencing using specific primers flanking the transposome-disrupted genes. AP-PCR was performed essentially as described by Das et al. (11), using the arbitrary primers described by Gonin et al. (19) (Arb1 and Arb6 in round 1 and Arb2 in round 2) and the specific primers TNF3 (5′GGCAAAGCAAAAGTTCAAAATCACC) in round 1 and TN5F2 (5′GTCCACCTACAACAAAGCTCTCATCAACCGTGG) in round 2. The final AP-PCR products were purified using a QIAquick PCR purification kit (Qiagen) before sequencing. Direct genomic sequencing was performed at the Tufts University Core Facility, using the internal transposome primers included in the Epicentre transposome kit and genomic DNAs obtained using a Wizard Genomic DNA purification kit from Promega.
MIC determinations.
Mutations were transferred from strain HdeA100 (ΔmarRAB hdeABp-lacZ, with marA cloned in a plasmid) to the parental strains CR1000 (ΔmarR) and CR2000 (ΔmarRA) by P1 transduction and were reconfirmed by PCR before their effects on antibiotic susceptibility to cefoxitin, norfloxacin, chloramphenicol, and minocycline were studied. The different mutants were grown overnight in liquid cultures and their MICs determined using Mueller-Hinton (M-H) plates and Etest strips (AB Biodisk) according to the manufacturer's specifications. MIC readings were performed after incubation for 20 h at 37°C. M-H plates were supplemented with ampicillin and 0.5 mM IPTG in plasmid complementation experiments, with 1 mM cyclic AMP (cAMP) (44) in complementation experiments of cyaA mutants with cAMP, and with 300 mM NaCl or 0.4% d-glucose in the environmental stimuli experiments. Etest strips were a generous gift from AB Biodisk (Solno, Sweden).
Gene expression experiments.
Mutants showing reduced MarA-mediated MDR were studied by reverse transcription followed by real-time quantitative PCR (RT-qPCR) to determine the expression levels of marA, acrA, tolC, and micF. These genes comprise both class I (marA and acrA) and class II (tolC and micF) MarA-activated promoters (33). gapA, which encodes the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) enzyme, was used as an endogenous reference gene according to the method of Viveiros et al. (47). The specific primers for real-time qPCR of these genes are described elsewhere (47). These primers were also used to generate the PCR products used to perform qPCR standard plots for each gene.
Briefly, overnight cultures of the different CR1000 (ΔmarR)- and CR2000 (ΔmarRA)-derivative mutants were grown overnight, diluted 1:1,000 in fresh LB, and grown for 3 to 4 h to an optical density at 600 nm (OD600) of about 0.3. Then, total RNA in the cultures was stabilized using RNAprotect bacterial reagent (Qiagen) and isolated after lysozyme-proteinase K digestion of bacteria by using an RNeasy minikit (Qiagen) and On-Column DNase I digestion (Qiagen) according to the manufacturer's specifications. RNAs were then treated with RQ1 RNase-free DNase from Promega to remove any remaining DNA and repurified with the RNeasy minikit, and their purity from proteins and concentration was determined using a NanoDrop ND-1000 spectrophotometer. Reverse transcription was performed using the SuperScript III first-strand synthesis system from Invitrogen, the specific primers aforementioned, and 200 ng of RNA. RT-associated reactions without retrotranscriptase (RT− reactions) were used as controls to confirm the lack of contaminating DNA in the RNA samples. The cDNAs obtained from the RT+ and RT− reactions were quantified after 45 cycles using a Mx3000P detection system (Stratagene) with 25-μl reaction mixtures of primers (300 nM each), cDNA (4 μl of a 1:10 dilution of the RT+ or RT− reaction mixture), and 2× QuantiTect SYBR green qPCR master mix from Qiagen (12.5 μl). Absolute transcript numbers were calculated using standard plots obtained by qPCR of serial dilutions of gene-specific, gel-purified PCR products of known concentrations.
marA mRNA polyadenylation and half-life experiments in the parental strain and the pcnB mutant.
Polyadenylation of marA transcripts was determined by RT-PCR performed as described above, using the marA-specific forward primer and an oligo(dT)20 primer as the reverse primer, followed by agarose gel analysis of the amplification products. For the marA mRNA half-life experiments, overnight cultures of each strain were diluted 1:1,000 in fresh LB and grown for 3 to 4 h to an OD600 of about 0.3. Then, RNAs were extracted before (time zero) and after addition of rifampin (500-μg ml−1 final concentration, which stops RNA synthesis) and culture incubation at 37°C for 0.5, 1, 2, and 4 min. The amount of marA transcripts was determined by RT-qPCR as described above. Half-life was defined as elapsed time × log 2/log (beginning amount/final amount).
Statistical analysis.
Statistically significant differences in MDR or gene expression were determined by Student's t test (two independent samples with equal variance, with two-tailed distribution), using Microsoft Excel 2003 software.
RESULTS AND DISCUSSION
Identification of 24 different genes whose inactivation alters MarA function.
We used random transposome mutagenesis to find chromosomal genes affecting MarA-dependent MDR. We could not directly screen for mutants with altered MDR, since we would obtain many mutants that affected MDR by mechanisms unrelated to MarA. Therefore, we chose an alternative primary screening method which detected reduction of MarA function. As detailed in Materials and Methods, we used the HdeA100 strain (ΔmarRAB hdeABp-lacZ, with marA cloned in a plasmid) to select mutants in which MarA had lost its ability to repress the expression of the hdeABp-lacZ fusion (blue colonies). After screening 11,000 mutants (about 2.5 hits per gene), we found 24 different chromosomal genes whose inactivation prevented hdeAB repression by MarA. They encoded unrelated proteins, including transcriptional regulators (cspG, crp, hns, and ompR), enzymes (appA, cyaA, degP, maoC, metL, pcnB, recD, treC, and ynfE), transport proteins (acrA, acrB, mhpT, nikD, and tolC), and others (alx, damX, metV, yfdG, yibL, and yniD) (see Table 3).
TABLE 3.
Effect of genes whose inactivation prevented repression of hdeAB by MarA on MarA-mediated MDR
| Protein/function | Gene | Locusb | Level of MarA-mediated MDR (% of parental level)a |
|||
|---|---|---|---|---|---|---|
| FOX | NOR | CHL | MIN | |||
| Transcriptional regulators | ||||||
| Cold shock protein CspG | cspGpc | b0990 | 54 | 91 | 93 | 75 |
| Cyclic AMP receptor protein | crp | b3357 | 21 | 33 | 29 | 38 |
| DNA-binding protein H-NS | hns | b1237 | 19 | 23 | 20 | 35 |
| OmpR transcriptional dual regulator | ompR | b3405 | 67 | 77 | 93 | 100 |
| Enzymes | ||||||
| Acid phosphatase | appA | b0980 | 68 | 84 | 93 | 100 |
| Adenylate cyclase | cyaA | b3806 | 43 | 41 | 46 | 32 |
| Serine protease Do | degP | b0161 | 94 | 91 | 50 | 100 |
| Putative ring-cleavage enzyme of phenylacetate degradation | maoC | b1387 | 100 | 61 | 75 | 40 |
| Aspartate kinase/homoserine dehydrogenase | metL | b3940 | 120 | 91 | 100 | 100 |
| Poly(A) polymerase I; RNA modification | pcnB | b0143 | 10 | 23 | 33 | 30 |
| RecBCD DNA helicase/exonuclease | recD | b2819 | 80 | 77 | 50 | 65 |
| Trehalose-6-phosphate hydrolase | treC | b4239 | 94 | 46 | 85 | 100 |
| Oxidoreductase subunit paralog of DmsA | ynfE | b1587 | 94 | 68 | 93 | 100 |
| Transport | ||||||
| AcrAB-TolC multidrug efflux system | acrA | b0463 | 16 | 0 | 8 | 40 |
| AcrAB-TolC multidrug efflux system | acrB | b0462 | 6 | 9 | 8 | 40 |
| Putative propionic acid transporter | mhpT | b0353 | 68 | 91 | 100 | 100 |
| Subunit of nickel ABC transporter | nikD | b3479 | 68 | 68 | 50 | 125 |
| TolC outer membrane channel | tolC | b3035 | 0 | 0 | 0 | 40 |
| Others | ||||||
| Putative membrane protein | alx | b3088 | 120 | 68 | 100 | 65 |
| Predicted membrane-anchored protein | damX | b3388 | 40 | 46 | 50 | 30 |
| tRNA-methionine | metV | b2816 | 74 | 91 | 75 | 65 |
| CPS-53 (KpLE1) prophage; bactoprenol-linked glucose translocase (flippase) | yfdG | b2350 | 68 | 91 | 93 | 65 |
| Conserved protein | yibLpc | b3602 | 68 | 68 | 50 | 100 |
| Predicted unconserved protein | yniDpc | b4535 | 94 | 77 | 125 | 50 |
Abbreviations: FOX, cefoxitin; NOR, norfloxacin; CHL, chloramphenicol; MIN, minocycline. MarA-mediated MDR for each mutant was calculated as the fold increase in MIC for the ΔmarR background (CR1000-derivative mutant) compared to the MIC for the ΔmarRA background (CR2000-derivative mutant) by using the ΔmarR MIC/ΔmarRA MIC ratio. This ratio for the parental strains (the MIC for the ΔmarR parental strain CR1000 divided by the MIC for the ΔmarRA parental strain CR2000) for each antibiotic (shown in Table 2, footnote b) was considered to represent 100% MarA-mediated antibiotic resistance, whereas a ratio of 1 (with the MIC for the ΔmarR background being equal to the MIC for the ΔmarRA background) was considered to represent 0% MarA-mediated antibiotic resistance. These two reference points were used to calculate the percentage of MarA-mediated antibiotic resistance for each mutant according to its ΔmarR MIC/ΔmarRA MIC ratio. When inactivation of a gene significantly reduced MarA-mediated resistance to an antibiotic compared to the level for the parental strain (P < 0.01, associated with reductions of 50% or more), the corresponding value is highlighted in bold. For parental strains, n = 9; for mutants, n = 2 to 5.
The locus tags refer to accession numbers (http://biocyc.org/ecocyc/index.shtml).
The insertion of the transposome was in the promoter of the gene. All the other insertions were within the open reading frame (ORF) of the genes.
Our assumption was that some of these genes would also affect MarA function on genes other than hdeAB and, thus, would be important for MarA-mediated MDR. On the other hand, the effects of some of them on MarA may be specific for hdeAB or may be the result of an indirect effect on the complex regulatory network of hdeAB (see reference 42), or some might affect the expression or copy number of the IPTG-inducible, marA-containing plasmid in the strain HdeA100. Such genes, identified just because of the screening method used, were not expected to affect MarA-mediated MDR; however, some may still be relevant for MarA-mediated MDR if, independently, they were also involved in regulation of marA expression or MarA function (for example, global regulators and other genes with pleiotropic effects).
Since the goal of this work was to find genes important for MarA-mediated MDR, the identified genes were not studied with regard to how they affected hdeAB repression by MarA. Instead, we performed a second screen, involving antibiotic resistance, to find which of them affected MarA-mediated MDR. This second screen was not performed in the HdeA100 background because of the aforementioned limitations and because, in this strain, marA is cloned in a multicopy IPTG-inducible plasmid. In the HdeA100 strain, therefore, the promoter and copy number of marA are not physiological, and ampicillin is needed in the plates to maintain the plasmid, which may alter the effects of the antibiotics tested in the second screen.
For the second screen, therefore, we transferred the mutations in the identified genes to the parental strains CR1000 (E. coli BW25113 ΔmarR; overexpresses marA) and CR2000 (E. coli BW25113 ΔmarRA; MarA free). These two strains were chosen for several reasons. There is almost no phenotypic difference in antibiotic susceptibility between a wild-type strain and a ΔmarRA strain (Table 2) because the expression of marA in a wild-type strain (BW25113) is strongly repressed by MarR. A system in which marA expression is not repressed is necessary to study MarA-mediated MDR. In a wild-type strain, such derepression can be achieved by adding to the culture medium chemicals such as salicylate, known to induce marA expression by inactivating MarR (see the introduction). However, salicylate affects cell growth, MDR, and gene expression independently from MarA, which would complicate our studies and their interpretation. A more appropriate system for our screen involved the use of a ΔmarR parental strain, which constitutively overexpresses marA at physiological levels from its native location in the chromosome and, thus, is similar to MDR clinical isolates that overexpress marA because they have mutations in marR. As a comparing strain, we chose the ΔmarRA parental strain instead of the wild-type strain because both strains are physiologically similar (similar antibiotic susceptibilities) and because the ΔmarRA strain is completely MarA free so it is the best reference for antibiotic susceptibility and gene expression levels in the absence of MarA. Moreover, using this strain instead of the wild type prevents genes that indirectly affect marA expression via MarR, e.g., proteins that interact with MarR, from being found in our screen (13).
TABLE 2.
MICs and levels of MarA-mediated MDR in the wild-type and parental strains
| Strain | Description or genotype | MIC (μg/ml)a,b |
|||
|---|---|---|---|---|---|
| FOX | NOR | CHL | MIN | ||
| BW25113 | Wild type | 4.0 | 0.09 | 10.7 | 3.3 |
| CR2000 | BW25113 ΔmarRA (MarA-free parental strain) | 3.2 | 0.07 | 8.1 | 2.4 |
| CR1000 | BW25113 ΔmarR (MarA-overproducing parental strain) | 10.6 | 0.21 | 23.9 | 4.9 |
The results are presented as the average MICs (the standard errors of the means were 0 to 15% [n = 3 to 9]) for cefoxitin (FOX), norfloxacin (NOR), chloramphenicol (CHL), and minocycline (MIN). MarA mediated low-level resistance to all four antibiotics, as shown by the significant increase in MICs in the ΔmarR parental strain compared to the levels for the ΔmarRA parental and wild-type strains. MarA also mediated resistance to other antibiotics tested (ampicillin, cephalothin, nalidixic acid, rifampin, erythromycin, tetracycline, and doxycycline) but had no effect on susceptibility to aminoglycosides, trimethoprim-sulfamethoxazole, imipenem, or fosfomycin (data not shown).
The CR1000 MIC/CR2000 MIC ratios were as follows: for FOX, 3.3; for NOR, 3.1; for CHL, 3.0; and for MIN, 2.0. These ratios represent 100% MarA-mediated resistance to each antibiotic (see Table 3), that is, the increase in the MIC resulting from physiological overexpression of marA in the ΔmarR parental strain (CR1000) compared to the MIC for the ΔmarRA parental strain (CR2000).
After transferring the mutations in the identified genes to the ΔmarR and ΔmarRA parental strains, we examined the susceptibilities of the resulting strains to four structurally and functionally unrelated bactericidal (cefoxitin and norfloxacin) or bacteriostatic (chloramphenicol and minocycline) antibiotics whose increased MICs were MarA dependent (their MICs were increased in the ΔmarR parental strain compared to the level for the ΔmarRA parental strain) (Table 2).
Fifteen identified genes affect MarA-mediated antibiotic resistance.
Compared to the levels for the ΔmarR and ΔmarRA parental strains, inactivation of 15 of the 24 identified genes reduced MarA-mediated resistance to at least one antibiotic by 50% or more; in contrast, inactivation of 9 of the 24 genes (cspG, ompR, appA, metL, yfnE, mhpT, alx, metV, and yfdG) did not significantly alter MarA-mediated antibiotic resistance (Table 3).
Inactivation of 7 of the 15 genes affecting resistance reduced MarA-mediated resistance to only one antibiotic: these genes were nikD, degP, recD, and yibL (chloramphenicol), maoC and yniD (minocycline), and treC (norfloxacin) (Table 3). This finding suggests that these genes do not affect marA expression or MarA function on genes essential for the Mar phenotype, such as acrAB or tolC. In contrast, these seven genes may have a role in some MarA-mediated cell responses that are important only for certain antibiotics or may be necessary for such responses to be effective. These genes were not further studied.
Inactivation of the other eight genes (crp, hns, cyaA, pcnB, acrA, acrB, tolC, and damX) decreased MarA-mediated MDR to all the antibiotics tested (Table 3). This was expected for acrA, acrB and tolC because of the major role of AcrAB-TolC in the Mar phenotype but was not known for the other five genes. Because of their general effect in MarA-mediated MDR, these genes were studied in more detail (see Fig. 1 to 4).
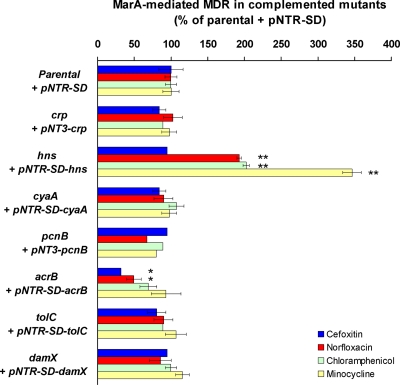
FIG. 1.
Complementation of genes whose inactivation reduced MarA-mediated MDR to all antibiotics tested. Those genes whose inactivation reduced MarA-mediated MDR to all antibiotics tested (Table 3) were added back on a plasmid to their respective mutants to study if MarA-mediated MDR was restored. All plasmids were derivatives of the control plasmid pNTR-SD (Table 1). The percentage of MarA-mediated MDR was calculated as explained in Table 2, footnote b, and Table 3, footnote a, with 100% MarA-mediated antibiotic resistance defined as the ratio resulting from dividing, for each antibiotic, the MIC for the ΔmarR parental strain CR1000 bearing the pNTR-SD plasmid by the MIC for the ΔmarRA parental strain CR2000 bearing pNTR-SD. The results are presented as the average ± the standard error of the mean (n = 4). Statistically significant differences are shown as * (P < 0.05) or ** (P < 0.01).
FIG. 4.
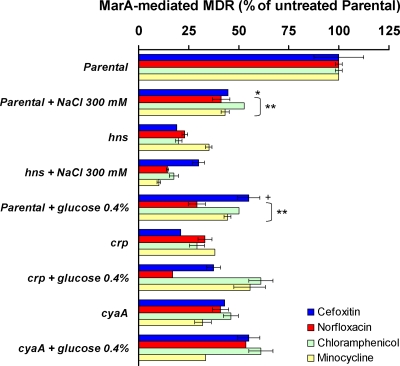
Effects of crp, cyaA, and hns on adaptation of MarA-mediated MDR to environmental stimuli. The percentage of MarA-mediated MDR was calculated as explained in Table 3, footnote a. The results are presented as the average ± the standard error of the mean (n = 3). When different conditions for the same strains were compared, addition of 0.4% glucose or 300 mM NaCl was found to produce statistically significant differences in MarA-mediated MDR in the parental strain (P was <0.01 for all antibiotics except for cefoxitin, in which case P was <0.05 for NaCl and P was <0.1 for glucose; these statistical significances are shown in the figure as “**,” “*,” and “+,” respectively), whereas these additions produced no differences in the mutants studied. Levels of MarA-mediated MDR were significantly different between the parental strain and the mutants in the absence of glucose and NaCl (P < 0.01 [see Table 3]; these statistical significances are not indicated in this figure), but no significant differences between them were found in the presence of glucose or NaCl.
crp and cyaA.
CRP is one of the major global transcriptional regulators in E. coli. It is involved in regulation of catabolic operons in response to the energetic status of the cell and in other functions. CRP directly activates or represses the transcription of about 200 genes, although it probably interacts with many other low-affinity binding sites in the chromosome. The activity of CRP is triggered by binding of the second messenger cAMP, whose conversion from ATP is catalyzed by the adenylate cyclase enzyme CyaA in response to glucose starvation and other stresses (references 20 and 35 and references therein).
Inactivation of crp or cyaA moderately reduced MarA-mediated MDR to all the antibiotics tested (Table 3). Complementation of these mutants by the crp or cyaA gene on a plasmid restored MarA-mediated MDR (Fig. 1). Also, addition of 1 mM cAMP to the culture medium complemented the cyaA inactivation, whereas it had no effect on the parental or crp strains (data not shown). Inactivation of crp or cyaA had previously been shown to increase MDR to oxacillin, macrolides, and crystal violet in a strain also inactivated for the major pump AcrAB-TolC because CRP-cAMP represses the expression of the accessory multidrug efflux pump MdtEF (38). In contrast, we show here that CRP-cAMP contributes to MDR in E. coli in the presence of MarA and the AcrAB-TolC pump.
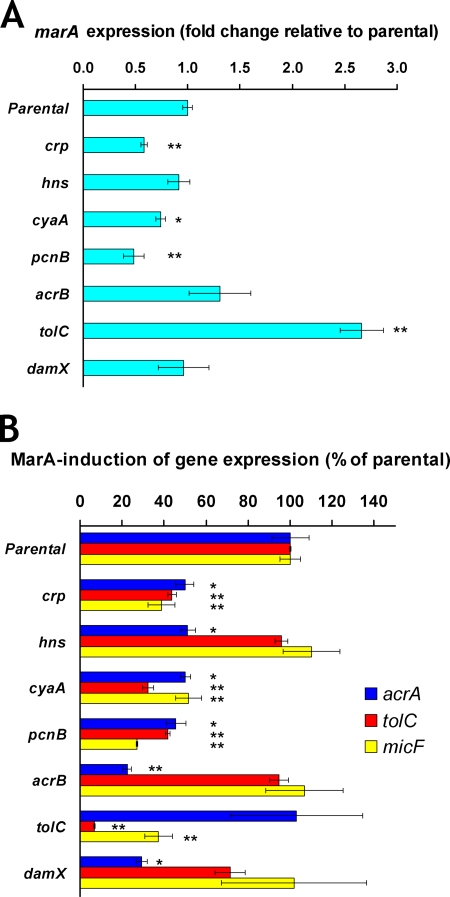
The reason for such a reduction in MarA-mediated MDR in the crp and cyaA mutants seems to be that the expression of marA (Fig. 2A) and, in consequence, the MarA induction of acrA, tolC, and micF (Fig. 2B) were significantly reduced in both mutants. Earlier studies had demonstrated that CRP-cAMP binds to and activates marRAB transcription in vitro but not in cells grown in minimal medium (20, 49). Our results agree with those in vitro results and suggest that activation of marA expression by CRP-cAMP does occur in cells grown in rich medium, which is important for full acrA, tolC, and micF activation by MarA and thus for MarA-mediated MDR. However, indirect effects of CRP-cAMP on regulation of marA expression cannot be ruled out, given the many genes regulated by CRP-cAMP.
FIG. 2.
Gene expression in mutants with reduced MarA-mediated MDR to all antibiotics tested. The effect of each inactivated gene on the expression of marA, acrA, tolC, micF, and gapA was measured by RT-qPCR (see Materials and Methods). The results are presented as the average ± the standard error of the mean (n = 3 to 5). Statistically significant differences for a mutant compared to the level for the parental strain are shown as * (P < 0.05) or ** (P < 0.01). (A) Fold change in marA expression in different ΔmarR-derivative mutants relative to the level for the ΔmarR parental strain CR1000. Fold 1 means no change in marA expression in a mutant compared to the level for the parental strain. gapA was used as an endogenous reference gene, and its expression was not affected by marA (gapA expression was the same in the ΔmarR and ΔmarRA parental strains; data not shown). Except for pcnB, none of the inactivated genes tested significantly affected the expression of gapA. The pcnB mutant showed reductions in both gapA and marA levels. However, such reductions seem to be an effect specific for these genes and not a general effect produced by pcnB inactivation on all RNAs, since inactivation of pcnB in the ΔmarRA background produced the same reduction in gapA levels but did not decrease the levels of the other genes studied (acrA, tolC, and micF) (data not shown). (B) Effect of inactivation of each gene on MarA induction of acrA, tolC, and micF expression. The percentage of MarA induction of gene expression for each mutant was calculated by comparing the ΔmarR/ΔmarRA ratio (in terms of the number of tran-scripts per ng of RNA) observed for the mutant to that observed for the parental strain, as explained for the percentage of MarA-mediated MDR in Table 3, footnote a.
Therefore, crp and cyaA play a role in MarA-mediated MDR by being involved in upregulation of marA expression and, in consequence, of MarA-regulated genes.
hns.
H-NS is a nucleoid-associated DNA-binding protein that plays a major role in the organization of the bacterial chromosome and in regulation of gene expression. It affects the transcription of over 5% of E. coli genes, usually acting as a repressor or as a gene silencer. H-NS plays a pleiotropic role in bacterial response to environmental stimuli such as temperature, osmolarity, pH, and starvation (17).
Inactivation of hns strongly reduced MarA-mediated MDR to all the antibiotics tested (Table 3). Addition of hns on a plasmid restored (cefoxitin) or significantly increased (norfloxacin, chloramphenicol, and minocycline) MarA-mediated MDR (Fig. 1). H-NS has previously been shown to decrease MDR in a strain inactivated for the major multidrug efflux pump AcrAB-TolC. This occurred because H-NS represses acrEF, mdtEF, and emrKY expression. These three operons are not expressed in a wild-type strain grown in LB and encode three TolC-dependent MDR efflux pumps whose activity is detected only in the absence of AcrAB-TolC (37). In contrast, we found that H-NS increases MDR in E. coli in the presence of MarA and an intact AcrAB-TolC pump.
When we studied the hns mutant at the level of gene expression, we found that marA expression and MarA-dependent induction of tolC and micF expression were unaffected (Fig. 2). However, the level of acrA induction by MarA was half that in the parental strain (Fig. 2B). This finding indicates that the reduced MarA-mediated MDR observed in the hns mutant is caused by a decrease in the AcrAB component of the AcrAB-TolC multidrug efflux pump. Such a specific contribution by H-NS to MarA induction of acrA, but not the other MarA-regulated genes tested, is of interest because H-NS is known to have no effect on regulation of acrA per se (37). Moreover, such an effect also occurs on hdeAB, which is regulated by MarA and H-NS both independently and synergistically (42). Decreased induction of acrA expression by MarA in the hns mutant (Fig. 2B) did not involve alteration of marA expression (Fig. 2A), and no protein-protein interaction between H-NS and MarA was found in two-hybrid experiments (data not shown). Thus, this effect is either indirectly mediated by other H-NS-regulated genes or attributable to synergic MarA binding or function at the acrA promoter caused by H-NS effects on DNA topology. An effect of DNA topology changes on acrA expression has not been reported before; however, DNA looping or changes in DNA supercoiling produced by H-NS, even when bound far from a promoter, can alter the DNA affinity of other transcription factors (reference 17 and references therein).
Therefore, hns plays an essential role in MarA-mediated MDR by being necessary for full MarA induction of acrA expression.
pcnB.
The pcnB gene encodes the enzyme poly(A) polymerase I (PAP I), which, together with polynucleotide phosphorylase and other proteins, is involved in polyadenylation, processing, and degradation of the vast majority of E. coli mRNA transcripts in exponential phase (reference 36 and references therein).
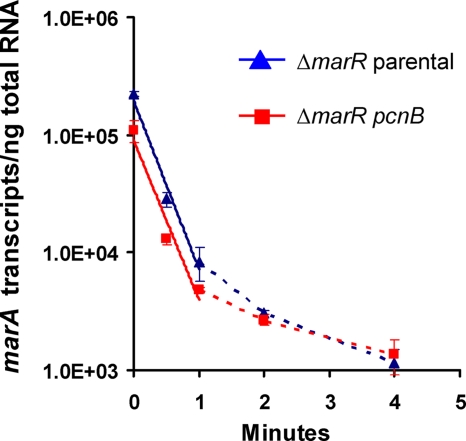
Inactivation of pcnB strongly reduced MarA-mediated MDR to all the antibiotics tested (Table 3); addition of pcnB on a plasmid to this mutant restored MarA-mediated MDR (Fig. 1). The reason for such a reduction in MarA-mediated MDR in the pcnB mutant seems to be that the levels of marA transcripts (Fig. 2A) and, by consequence, MarA induction of acrA, tolC, and micF (Fig. 2B) were strongly reduced in this mutant compared to the levels for the parental strain. Further experiments showed that marA transcripts were polyadenylated both in the parental strain and in the pcnB mutant (RT-PCR products of the same size were obtained; data not shown). Moreover, we found no significant differences in the half-lives of the marA mRNA transcripts between the parental strain and the pcnB mutant (Fig. 3). These findings suggest that the reduction in marA transcript levels found in the pcnB mutant compared to the level for the parental strain is not the result of a decrease in marA mRNA stability but rather the result of a decrease in marA transcription. Considering that a role for PAP I in transcription has not been reported, the effect of PAP I on marA expression may be indirectly mediated by other proteins, such as SoxS and Rob, two activators of marA transcription whose mRNA stabilities have been found to be increased after overproduction of PAP I (36).
FIG. 3.
Effect of pcnB inactivation on marA mRNA stability. The absolute numbers of marA transcripts per ng of total RNA before addition of rifampin (time zero) were 2.3·105 for the ΔmarR parental strain (shown in blue), 1.1·105 for the ΔmarR pcnB mutant (shown in red), and 6.3·103 for the wild-type strain BW25113 (not shown). The half-life of marA mRNA was calculated as detailed in Materials and Methods, using the linear part of the ΔmarR parental strain and ΔmarR pcnB mutant curves (shown as a continuous line; data within the first minute). No significant differences were found: the marA mRNA half-lives were 0.21 min for the ΔmarR parental strain and 0.22 min for its derivative pcnB mutant.
Therefore, pcnB-encoded PAP I plays an important role in MarA-mediated MDR by being necessary, probably indirectly, for full marA expression.
acrA, acrB, and tolC.
Upregulation of the expression of the AcrAB-TolC multidrug efflux pump is the main basis for MarA-mediated MDR (28, 39), which explains why inactivation of acrA, acrB, or tolC dramatically reduced MarA-mediated MDR to all the antibiotics tested (Table 3). Such reduction was similar for each of these three genes (Table 3). This fact suggests that other minor multidrug efflux pumps in which AcrA or TolC are involved (28) do not play a role in MarA-mediated MDR under the conditions tested here.
These three genes may have additional effects on the MarA system besides their role in antibiotic efflux, since we found in our first screen that their inactivation also reduced MarA repression of hdeAB by an unknown mechanism. Therefore, we studied them in more detail to determine if they have other effects on MarA-mediated MDR.
To study additional effects of the AcrAB component of the AcrAB-TolC pump, we focused on the acrB mutant. As expected, complementation of this mutant with the acrB gene on a plasmid restored MarA-mediated MDR (Fig. 1). However, such a complementation was only partial for two antibiotics (cefoxitin and norfloxacin), which suggested that the inactivation of acrB might be also affecting the expression of the upstream acrA gene. In fact, when we studied the acrB mutant at the level of gene expression (RT-qPCR), we found that acrA transcriptional activation by MarA was indeed significantly reduced (Fig. 2B). However, marA expression (Fig. 2A) and tolC and micF activation by MarA (Fig. 2B) were not affected. To study whether this reduction in acrA activation by MarA was produced by the insertion of the transposome in acrB or by the absence of the AcrB protein, we added a single-copy plasmid bearing an acrA-lacZ transcriptional fusion to the parental and acrB mutant strains. In the parental strain, MarA activation of acrA-lacZ expression obtained by β-galactosidase assays (not shown) was similar to MarA activation of acrA expression found by RT-qPCR, showing the equivalence of both assays; however, in the acrB mutant, acrA-lacZ activation by MarA was no longer reduced compared to the level for the parental strain (not shown), in contrast to the RT-qPCR results. This finding shows that the reduced acrA activation by MarA in the acrB mutant compared to the level for the parental strain observed by RT-qPCR was produced artifactually by the cis transposome insertion in acrB and not by the absence of the AcrB protein.
Complementation of the tolC mutant with tolC on a plasmid restored MarA-mediated MDR, as expected (Fig. 1). When we studied this mutant at the level of gene expression, we found that marA expression was significantly increased (2.7-fold) compared to the level for the parental strain (Fig. 2A). This result agrees with a recent publication by Rosner and Martin (41). Using a different method (lacZ fusions) to study gene expression, they found that marA and soxS expression, Rob activity, and the expression of several genes belonging to the mar regulon were increased in a tolC mutant (41). They did not find the mechanism by which inactivation of tolC affects marA expression, but they propose that TolC is involved in one or more pumps responsible for effluxing cellular metabolites; in a tolC mutant, these metabolites would not be eliminated and would trigger the activation of the MarA/SoxS/Rob system in order to upregulate TolC-mediated efflux and restore homeostasis (41). The metabolites and pump(s) involved are still unknown (41). However, they found that the mechanism by which tolC inactivation alters marA expression is independent of the major AcrAB-TolC pump, since inactivation of acrAB did not produce the changes in marA expression and other changes found when tolC is inactivated (41). Our findings agree with their result, since we also found no alteration of marA expression in the acrB mutant (Fig. 2A).
Interestingly, despite marA expression being higher in the tolC mutant (Fig. 2A), the degree of activation of acrA and micF expression by MarA in this mutant was equal to or less than that observed in the parental strain (Fig. 2B). However, in both the ΔmarR and the ΔmarRA backgrounds, the absolute numbers of acrA and micF transcripts per ng of RNA in the tolC mutant were 2- to 4-fold higher than those observed in the parental strain (data not shown). These findings may be explained by the increased soxS expression and Rob activity also found in a tolC mutant (41). SoxS and Rob are two MarA homologs that are functionally similar to MarA with respect to their binding sites and the genes that they regulate. Thus, their increased amount/activity in the tolC mutant (41) would explain the higher levels of acrA and micF expression in absolute numbers that we found in the tolC mutant compared to the level for the parental strain. Moreover, the functional overlapping of these regulators with MarA may explain why the level of marA expression was higher in the tolC mutant than in the parental strain but the level of MarA induction of acrA and micF was not higher. Induction of the expression of acrA and micF by additional amounts/activity of SoxS and/or Rob (41) would impede additional induction of these genes by MarA, since all three regulators have the same binding site. These findings may be also explained by the different response that some genes have in vivo to changes in the concentration of MarA (34).
Therefore, acrA, acrB, and tolC are essential for MarA-mediated MDR because of the role of the AcrAB-TolC pump in antibiotic efflux. Moreover, tolC is involved in regulation of marA expression by an unknown and probably indirect mechanism.
damX.
The damX gene encodes a predicted membrane-anchored protein conserved among many Enterobacteriaceae. Little is known about its regulation and function. Lyngstadaas et al. (29) found that overproduction of DamX interfered with cell division. However, they suggested that such an effect could be indirect or nonspecifically produced by nonphysiological amounts of DamX, since they found no differences between a wild-type and a damX-inactivated strain. Leclerc et al. (25) found that the damX homolog in Salmonella might be involved in adherence to and invasion of human intestinal cells.
We found that inactivation of damX moderately decreased MarA-mediated MDR to all the antibiotics tested (Table 3). This is the first report of a role for damX in E. coli when expressed at physiological levels. Addition of damX on a plasmid restored MarA-induced MDR (Fig. 1). Inactivation of damX had no effect on marA expression or on MarA induction of tolC and micF (Fig. 2). However, it dramatically decreased MarA induction of acrA expression (Fig. 2B). This finding indicates that the reduction in MarA-mediated MDR observed in this mutant was the result of a decrease in the AcrAB-TolC multidrug efflux pump. Considering that marA expression was unaffected (Fig. 2A), that no protein-protein interaction between DamX and MarA was found in two-hybrid experiments (data not shown), and that DamX is a predicted membrane protein, the effect of DamX on acrA induction by MarA is likely to be indirect.
Therefore, damX contributes to MarA-mediated MDR by specifically enhancing, probably indirectly, MarA induction of acrA expression.
The identified genes can mediate the adaptation of MarA-mediated MDR to environmental stimuli.
Understanding how the genes found here affect MarA-mediated MDR provides a better knowledge of how marA functions in E. coli. Since the expression/activity of many of these genes/proteins are known to be regulated by different environmental stimuli, we studied whether these genes played a role in adaptation of MarA-mediated MDR to different environmental conditions. As examples, we focused on crp, cyaA, and hns (Fig. 4).
As mentioned above, CRP-cAMP regulates bacterial response to glucose starvation and other stress conditions. In the presence of glucose, CyaA does not catalyze the conversion of ATP to cAMP and CRP is not activated. As shown in Fig. 4, we found that addition of glucose (0.4%, wt/vol) to the medium indeed reduced MarA-mediated MDR to all the antibiotics tested in the parental strain. Such a reduction was similar to that observed for the crp and cyaA mutants in the absence of glucose. On the contrary, addition of glucose did not significantly alter MarA-mediated MDR in the crp and cyaA mutants (Fig. 4). Thus, MarA-mediated MDR is greater under glucose starvation conditions than in the presence of glucose, and such an effect is dependent on crp and cyaA.
H-NS regulates bacterial response to osmolarity and other stresses. H-NS activity strongly decreases at high osmolarity (e.g., 300 mM NaCl) because changes in H-NS protein structure and/or DNA curvature reduce H-NS affinity for its DNA-binding sites (reference 14 and references therein). We found that addition of 300 mM NaCl to the medium reduced MarA-mediated MDR to all the antibiotics tested in the parental strain (Fig. 4). This reduction was similar to that observed in the hns mutant without added NaCl. On the contrary, addition of NaCl did not significantly alter MarA-mediated MDR in the hns mutant (Fig. 4). Thus, high osmolarity reduces MarA-mediated MDR in a hns-dependent way.
Additional comments.
The results presented here demonstrate the importance of 15 chromosomal genes in modulating antibiotic resistance mediated by the transcriptional regulator MarA in E. coli. These genes encode unrelated transcriptional regulators, enzymes, transporters, and unknown proteins and can affect MarA response in general (by altering marA expression or MarA function) or specifically only for some antibiotics. Moreover, some of these genes can mediate the adaptation of MarA-mediated MDR to different environmental stimuli.
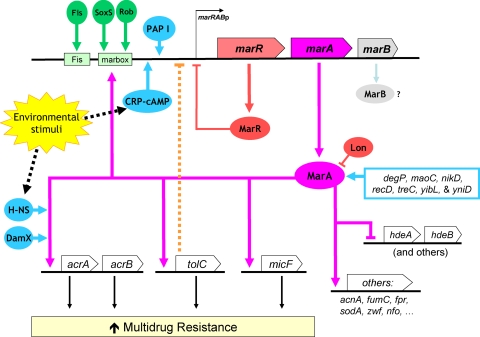
These and previous findings show that MarA-mediated MDR is regulated at multiple levels, including marA transcription, MarA protein stability, and MarA function on specific promoters (Fig. 5). They also show an interconnection of the MarA system with global regulation and cell metabolism. Why does the MarA system have such a multifaceted regulation? It may be a consequence of the complexity of antibiotic action and cell responses to antibiotics, including secondary targets, signaling effects, and indirect effects, such as antibiotic-induced generation of hydroxyl radicals or changes in gene expression and cell metabolism (8, 15, 24). On the other hand, such a regulation would allow integration of different signals as well as transitory and fine-tuned adaptation of the MarA-mediated response to different conditions, which would ensure that such a response is always adjusted to the cell needs. This idea agrees with reports showing that marA expression changes when cells are grown in different media or growth phases (4).
FIG. 5.
Regulation of MarA-mediated MDR. The figure was produced using data from the literature (see main text) and the results obtained here. Functional interactions, either direct or indirect, are represented as arrows for activation/induction and as “⊣” for repression. New genes found here to affect MarA-mediated MDR are in blue.
This complex regulation has additional implications. Modification of marA expression (directly or indirectly, e.g., via crp, cyaA, pcnB, or tolC) is a mechanism of generally altering the expression of the MarA regulon, which may thus affect other MarA-regulated functions such as virulence (1, 6, 9). On the other hand, altering MarA activity only on some genes allows a specific modification of only certain MarA functions. Here, we found that hns and damX specifically alter MarA activation of acrA expression and, thus, MarA-mediated MDR. We had previously found that other regulators of hdeAB were able to modify MarA activity on hdeAB (42). Therefore, gene-specific alteration of MarA activity by other proteins seems not unusual, which may explain why some genes are differently activated in vivo by the same concentrations of MarA, especially when these differences are poorly correlated with the in vitro affinity of MarA for their promoters (34).
The complexity and adaptability of the MarA response may favor the spread of MDR strains with mutations in the autorepressor marR. Also, the different levels of MDR found among different clinical isolates mutated in marR (e.g., reference 31) may be explained by additional mutations in the genes found here to modulate MarA-mediated MDR. Moreover, these findings raise the question of whether the effect of other MDR regulators, such as AcrR, a repressor of acrA, or the MarA-homologs SoxS and Rob, as well as marA orthologs or marA-like regulators in other clinically relevant pathogens, are also subject to this kind of fine-tuned regulation. Such a complex regulation of MDR may exist also in other bacteria, especially among Enterobacteriaceae. However, the specific regulatory details may be different since other species have different genes and regulatory networks, including additional MarA homologs and additional MDR regulators, and the genes found here to alter MarA-mediated MDR may have different effects. For example, in contrast with what was found for E. coli, inactivation of tolC in S. enterica does not affect marA expression but results in overexpression of the marA homolog ramA (48).
Finally, inhibitors of multidrug efflux pumps and inhibitors of MDR and virulence regulators, including MarA, have been proposed to enhance the effectiveness of antibiotics (7, 28). A potential alternative would involve the use of inhibitors of the expression or activity of genes that enhance marA expression or function. This approach would offer additional advantages, such as the possibility of inhibiting only some MarA functions or the possibility of synergistic effects, since some of these genes also have a role in MDR or virulence.
Acknowledgments
We thank Francis Domain for performing the two-hybrid experiments and Laura McMurry for stimulating discussion.
This work was supported by U.S. Public Health Service grant AI56021 from the National Institutes of Health.
Footnotes
Published ahead of print on 8 March 2010.
REFERENCES
- 1.Alekshun, M. N., and S. B. Levy. 1997. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob. Agents Chemother. 41:2067-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alekshun, M. N., and S. B. Levy. 2007. Molecular mechanisms of antibacterial multidrug resistance. Cell 128:1037-1050. [DOI] [PubMed] [Google Scholar]
- 3.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey, A. M., M. A. Webber, and L. J. Piddock. 2006. Medium plays a role in determining expression of acrB, marA, and soxS in Escherichia coli. Antimicrob. Agents Chemother. 50:1071-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbosa, T. M., and S. B. Levy. 2000. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J. Bacteriol. 182:3467-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbosa, T. M., and P. J. Pomposiello. 2005. The mar regulon, p. 209-223. In D. G. White, M. N. Alekshun, and P. F. McDermott (ed.), Frontiers in antimicrobial resistance: a tribute to Stuart. B. Levy. ASM Press, Washington, DC.
- 7.Bowser, T. E., V. J. Bartlett, M. C. Grier, A. K. Verma, T. Warchol, S. B. Levy, and M. N. Alekshun. 2007. Novel anti-infection agents: small-molecule inhibitors of bacterial transcription factors. Bioorg. Med. Chem. Lett. 17:5652-5655. [DOI] [PubMed] [Google Scholar]
- 8.Brazas, M. D., and R. E. Hancock. 2005. Using microarray gene signatures to elucidate mechanisms of antibiotic action and resistance. Drug Discov. Today 10:1245-1252. [DOI] [PubMed] [Google Scholar]
- 9.Casaz, P., L. K. Garrity-Ryan, D. McKenney, C. Jackson, S. B. Levy, S. K. Tanaka, and M. N. Alekshun. 2006. MarA, SoxS and Rob function as virulence factors in an Escherichia coli murine model of ascending pyelonephritis. Microbiology 152:3643-3650. [DOI] [PubMed] [Google Scholar]
- 10.Cohen, S. P., S. B. Levy, J. Foulds, and J. L. Rosner. 1993. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J. Bacteriol. 175:7856-7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das, S., J. C. Noe, S. Paik, and T. Kitten. 2005. An improved arbitrary primed PCR method for rapid characterization of transposon insertion sites. J. Microbiol. Methods 63:89-94. [DOI] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Domain, F., X. R. Bina, and S. B. Levy. 2007. Transketolase A, an enzyme in central metabolism, derepresses the marRAB multiple antibiotic resistance operon of Escherichia coli by interaction with MarR. Mol. Microbiol. 66:383-394. [DOI] [PubMed] [Google Scholar]
- 14.Dorman, C. J. 2004. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2:391-400. [DOI] [PubMed] [Google Scholar]
- 15.Dwyer, D. J., M. A. Kohanski, B. Hayete, and J. J. Collins. 2007. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol. Syst. Biol. 3:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dzink-Fox, J., and M. Oethinger. 2005. Identification of Mar mutants among clinical bacterial isolates, p. 224-234. In D. G. White, M. N. Alekshun, and P. F. McDermott (ed.), Frontiers in antimicrobial resistance: a tribute to Stuart. B. Levy. ASM Press, Washington, DC.
- 17.Fang, F. C., and S. Rimsky. 2008. New insights into transcriptional regulation by H-NS. Curr. Opin. Microbiol. 11:113-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.George, A. M., and S. B. Levy. 1983. Gene in the major cotransduction gap of the Escherichia coli K-12 linkage map required for the expression of chromosomal resistance to tetracycline and other antibiotics. J. Bacteriol. 155:541-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonin, M., E. M. Quardokus, D. O'Donnol, J. Maddock, and Y. V. Brun. 2000. Regulation of stalk elongation by phosphate in Caulobacter crescentus. J. Bacteriol. 182:337-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grainger, D. C., D. Hurd, M. Harrison, J. Holdstock, and S. J. Busby. 2005. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc. Natl. Acad. Sci. U. S. A. 102:17693-17698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffith, K. L., I. M. Shah, and R. E. Wolf, Jr. 2004. Proteolytic degradation of Escherichia coli transcription activators SoxS and MarA as the mechanism for reversing the induction of the superoxide (SoxRS) and multiple antibiotic resistance (Mar) regulons. Mol. Microbiol. 51:1801-1816. [DOI] [PubMed] [Google Scholar]
- 22.Hächler, H., S. P. Cohen, and S. B. Levy. 1991. marA, a regulated locus which controls expression of chromosomal multiple antibiotic resistance in Escherichia coli. J. Bacteriol. 173:5532-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaper, J. B., J. P. Nataro, and H. L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123-140. [DOI] [PubMed] [Google Scholar]
- 24.Kohanski, M. A., D. J. Dwyer, B. Hayete, C. A. Lawrence, and J. J. Collins. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797-810. [DOI] [PubMed] [Google Scholar]
- 25.Leclerc, G. J., C. Tartera, and E. S. Metcalf. 1998. Environmental regulation of Salmonella typhi invasion-defective mutants. Infect. Immun. 66:682-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levy, S. B. 2002. The antibiotic paradox: how the misuse of antibiotics destroys their curative powers, 2nd ed. Perseus Publishing, Cambridge, MA.
- 27.Levy, S. B., and B. Marshall. 2004. Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10:S122-S129. [DOI] [PubMed] [Google Scholar]
- 28.Li, X. Z., and H. Nikaido. 2004. Efflux-mediated drug resistance in bacteria. Drugs 64:159-204. [DOI] [PubMed] [Google Scholar]
- 29.Lyngstadaas, A., A. Lobner-Olesen, and E. Boye. 1995. Characterization of three genes in the dam-containing operon of Escherichia coli. Mol. Gen. Genet. 247:546-554. [DOI] [PubMed] [Google Scholar]
- 30.Ma, D., M. Alberti, C. Lynch, H. Nikaido, and J. E. Hearst. 1996. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol. Microbiol. 19:101-112. [DOI] [PubMed] [Google Scholar]
- 31.Maneewannakul, K., and S. B. Levy. 1996. Identification for mar mutants among quinolone-resistant clinical isolates of Escherichia coli. Antimicrob. Agents Chemother. 40:1695-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin, R. G., and J. L. Rosner. 1997. Fis, an accessorial factor for transcriptional activation of the mar (multiple antibiotic resistance) promoter of Escherichia coli in the presence of the activator MarA, SoxS, or Rob. J. Bacteriol. 179:7410-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin, R. G., W. K. Gillette, S. Rhee, and J. L. Rosner. 1999. Structural requirements for marbox function in transcriptional activation of mar/sox/rob regulon promoters in Escherichia coli: sequence, orientation and spatial relationship to the core promoter. Mol. Microbiol. 34:431-441. [DOI] [PubMed] [Google Scholar]
- 34.Martin, R. G., E. S. Bartlett, J. L. Rosner, and M. E. Wall. 2008. Activation of the Escherichia coli marA/soxS/rob regulon in response to transcriptional activator concentration. J. Mol. Biol. 380:278-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinez-Antonio, A., and J. Collado-Vides. 2003. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 6:482-489. [DOI] [PubMed] [Google Scholar]
- 36.Mohanty, B. K., and S. R. Kushner. 2006. The majority of Escherichia coli mRNAs undergo post-transcriptional modification in exponentially growing cells. Nucleic Acids Res. 34:5695-5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishino, K., and A. Yamaguchi. 2004. Role of histone-like protein H-NS in multidrug resistance of Escherichia coli. J. Bacteriol. 186:1423-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishino, K., Y. Senda, and A. Yamaguchi. 2008. CRP regulator modulates multidrug resistance of Escherichia coli by repressing the mdtEF multidrug efflux genes. J. Antibiot. (Tokyo) 61:120-127. [DOI] [PubMed] [Google Scholar]
- 39.Okusu, H., D. Ma, and H. Nikaido. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178:306-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pomposiello, P. J., M. H. Bennik, and B. Demple. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183:3890-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosner, J. L., and R. G. Martin. 2009. An excretory function for the Escherichia coli outer membrane pore TolC: upregulation of marA and soxS transcription and Rob activity due to metabolites accumulated in tolC mutants. J. Bacteriol. 191:5283-5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruiz, C., L. M. McMurry, and S. B. Levy. 2008. Role of the multidrug resistance regulator MarA in global regulation of the hdeAB acid resistance operon in Escherichia coli. J. Bacteriol. 190:1290-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saka, K., M. Tadenuma, S. Nakade, N. Tanaka, H. Sugawara, K. Nishikawa, N. Ichiyoshi, M. Kitagawa, H. Mori, N. Ogasawara, and A. Nishimura. 2005. A complete set of Escherichia coli open reading frames in mobile plasmids facilitating genetic studies. DNA Res. 12:63-68. [DOI] [PubMed] [Google Scholar]
- 44.Sakamoto, Y., S. Furukawa, H. Ogihara, and M. Yamasaki. 2003. Fosmidomycin resistance in adenylate cyclase deficient (cya) mutants of Escherichia coli. Biosci. Biotechnol. Biochem. 67:2030-2033. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Seoane, A. S., and S. B. Levy. 1995. Characterization of MarR, the repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli. J. Bacteriol. 177:3414-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Viveiros, M., M. Dupont, L. Rodrigues, I. Couto, A. Davin-Regli, M. Martins, J. M. Pages, and L. Amaral. 2007. Antibiotic stress, genetic response and altered permeability of E. coli. PLoS One 2:e365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Webber, M. A., A. M. Bailey, J. M. Blair, E. Morgan, M. P. Stevens, J. C. Hinton, A. Ivens, J. Wain, and L. J. Piddock. 2009. The global consequence of disruption of the AcrAB-TolC efflux pump in Salmonella enterica includes reduced expression of SPI-1 and other attributes required to infect the host. J. Bacteriol. 191:4276-4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng, D., C. Constantinidou, J. L. Hobman, and S. D. Minchin. 2004. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 32:5874-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]