Abstract
Isoniazid (INH) is an effective first-line antituberculosis drug. KatG, a catalase-peroxidase, converts INH to an active form in Mycobacterium tuberculosis, and katG mutations are major causes of INH resistance. In the present study, we sequenced katG of 108 INH-resistant M. tuberculosis clinical isolates. Consequently, 9 novel KatG mutants with a single-amino-acid substitution were found. All of these mutants had significantly lower INH oxidase activities than the wild type, and each mutant showed various levels of activity. Isolates having mutations with relatively low activities showed high-level INH resistance. On the basis of our results and known mutations associated with INH resistance, we developed a new hybridization-based line probe assay for rapid detection of INH-resistant M. tuberculosis isolates.
Isoniazid (INH) is an effective drug used in the treatment of tuberculosis and has been in common use to treat tuberculosis since its introduction in 1952 (4). However, the emergence of INH-resistant (Inhr) Mycobacterium tuberculosis is jeopardizing the continued utility of INH (10).
Drug resistance in M. tuberculosis is caused by mutations in restricted regions of the genome (36). Mutations in katG, the upstream region of the fabG1-inhA operon (PfabG1-inhA), and inhA are responsible for INH resistance (36). The katG gene encodes the bifunctional catalase-peroxidase enzyme that converts INH to an active form (35).
Previously, we developed a DNA sequencing-based method to detect mutations in regions associated with INH resistance in M. tuberculosis, including katG and PfabG1-inhA (28). Consequently, five novel mutations in katG associated with INH resistance were found (28). In the present study, we cloned 21 katG mutants, including 15 novel mutants, and compared their INH oxidase activities. Certain katG mutations were shown to cause high-level INH resistance, which suggests the possibility of determining the degree of INH resistance, such as high- or low-level resistance, by detecting these katG mutations. Furthermore, to detect these mutations in ordinary-scale clinical laboratories without sequencing, we developed a new hybridization-based line probe assay (LiPA) for INH resistance in M. tuberculosis isolates, which can be applied easily in clinical use.
MATERIALS AND METHODS
Bacterial strains and plasmids.
One hundred eight Inhr M. tuberculosis isolates were obtained from single patients at the International Medical Center of Japan and National Hospital Organization Tokyo National Hospital from 2003 to 2008. INH-susceptible (Inhs) M. tuberculosis strains H37Rv and IMCJ 2751 were used. The IMCJ 2751 isolate has a katG(G1388T) [KatG(R463L)] neutral mutation. The Escherichia coli strains and plasmids used in this study are listed in Table 1. E. coli TOP10F′ (Invitrogen, Carlsbad, CA) was used as the host for cloning. E. coli UM262 (17) was used as the host for expression of katG derived from clinical isolates and H37Rv.
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| E. coli strains | ||
| TOP10F′ | F′ [lacIq Tn10 (Tetr)] mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔΜ15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| UM262 | katG::Tn10 recA pro leu rpsL hsdM hsdR endl lacY | 17 |
| Plasmids | ||
| pTrcHis2-TOPO | TA cloning and expression vector; Apr Kmr | Invitrogen |
| pkatG-wt | pTrcHis2-TOPO carrying katG | This study |
| pkatG-1 | pkatG-wt carrying G1388T (neutral mutation) | This study |
| pkatG-2 | pkatG-1 carrying C379G | This study |
| pkatG-3 | pkatG-1 carrying C694T | This study |
| pkatG-4 | pkatG-wt carrying A398C | This study |
| pkatG-5 | pkatG-1 carrying T1147C | This study |
| pkatG-6 | pkatG-1 carrying 1297::C, Δ1305C | This study |
| pkatG-7 | pkatG-1 carrying a290g | This study |
| pkatG-8 | pkatG-1 carrying C1465A | This study |
| pkatG-9 | pkatG-wt carrying G944C | This study |
| pkatG-10 | pkatG-1 carrying T1259C | This study |
| pkatG-11 | pkatG-wt carrying G944C, G1159C | This study |
| pkatG-12 | pkatG-1 carrying G368A, G895A | This study |
| pkatG-13 | pkatG-1 carrying G1255C | This study |
| pkatG-14 | pkatG-1 carrying C195T (silent mutation), T527C | This study |
| pkatG-15 | pkatG-wt carrying Δ(478-479) | This study |
| pkatG-16 | pkatG-1 carrying G944C | This study |
| pkatG-17 | pkatG-wt carrying Δ371G | This study |
| pkatG-18 | pkatG-1 carrying C1894T | This study |
| pkatG-19 | pkatG-wt carrying C945A | This study |
| pkatG-20 | pkatG-1 carrying Δ(571-576) | This study |
| pkatG-21 | pkatG-1 carrying G1624C | This study |
Drug susceptibility testing.
All clinical isolates, H37Rv, and IMCJ 2751 were tested for drug susceptibility. Strains were analyzed by an agar proportion method with egg-based Ogawa medium (Vit Spectrum-SR [Kyokuto Pharmaceutical Industrial Co., Tokyo, Japan] or Wellpack [Japan BCG Laboratory, Tokyo, Japan]), which is based on a slightly modified WHO protocol (3) and is recommended by the Japanese Society of Tuberculosis (3, 12). The medium contained INH (0.2 μg/ml and 1.0 μg/ml), rifampin (RIF) (40 μg/ml), ethambutol (EB) (2.5 μg/ml), kanamycin (KM) (20 μg/ml), p-aminosalicylic acid (PAS) (0.5 μg/ml), streptomycin (SM) (10 μg/ml), ethionamide (TH) (20 μg/ml), enviomycin (EVM) (20 μg/ml), cycloserine (CS) (30 μg/ml), and levofloxacin (LVFX) (1.0 μg/ml). The results of drug susceptibility testing are shown in Table S1 in the supplemental material.
Isolation of genomic DNA.
Genomic DNA from M. tuberculosis was extracted as described previously (22).
DNA sequencing of INH resistance-related genes.
The furA-katG operon and its upstream region were amplified by PCR with primers −129furA (5′-GCTCATCGGAACATACGAAG-3′) and katG+50 (5′-GTGCTGCGGCGGGTTGTGGTTGATCGGCGG-3′). The fabG1-inhA operon and PfabG1-inhA were also amplified, using primers −200fabG1 (5′-TTCGTAGGGCGTCAATACAC-3′) and inhA+40 (5′-CCGAACGACAGCAGCAGGAC-3′). PCR products were used as templates for direct DNA sequencing. DNA sequences were compared with the H37Rv sequence using Genetyx-Mac, version 14.0.2 (Genetyx Corporation, Tokyo, Japan).
Construction of plasmids.
The coding regions of katG from H37Rv, IMCJ 2751, and Inhr clinical isolates with katG mutations were amplified by PCR with the primers katG-F-ccc (5′-CCCGAGCAACACCCACCCATTACAGAAAC-3′) and katG-R (5′-TCAGCGCACGTCGAACC-3′) and cloned into pTrcHis2-TOPO (Invitrogen) using the TA cloning method. The pTrcHis2-TOPO vector encodes a C-terminal peptide containing a c-myc epitope and a 6×His tag. However, the expressed recombinant KatG protein did not have any additional amino acid residues, such as the c-myc epitope and the 6×His tag, because the katG-R reverse primer included the native stop codon. The DNA sequences of all clones were confirmed by sequencing.
RFLP.
IS6110-probed restriction fragment length polymorphism (RFLP) was performed as described previously (22). Patterns with more than 70% similarity were postulated to form a cluster.
Immunoblotting.
Proteins separated by SDS-PAGE were transferred onto Immun-Blot polyvinylidene difluoride (PVDF) membranes (Bio-Rad, Hercules, CA). The proteins on the membranes were detected using primary antibodies specific for KatG (28). KatG was visualized with horseradish peroxidase-conjugated secondary antibodies.
Enzyme assays.
KatG mediates free-radical formation from INH oxidation in the presence of H2O2. The activities of KatG were detected spectrophotometrically by following the reduction of nitroblue tetrazolium (NBT) at A560 (28, 32). Peroxidase activity was monitored spectrophotometrically by following the oxidation of 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) at A405 (21). Catalase activity was measured spectrophotometrically by following the degradation of H2O2 at A240 (21). The catalase activity is shown as values subtracted from that of the vector control. All assays were carried out at 25°C. The absorbance was read 200 s after the initiation of the reaction.
LiPA.
The line probe assay (LiPA) was performed as described previously (1, 29). In brief, 41 oligonucleotide probes were designed to cover mutations in the furA-katG (35 probes for katG and 2 for furA), PfabG1-inhA (2 probes), and fabG1 (2 probes) regions (Table 2). These probes were immobilized on two strips. Six regions, located within PfabG1-inhA (477 bp), fabG1 (209 bp), furA (256 bp), and katG (612 bp, 698 bp, and 907 bp), were amplified by nested PCR. Immobilized probes on the two strips were hybridized with the biotinylated PCR products and then incubated with streptavidin labeled with alkaline phosphatase. The color development was performed by incubation with 5-bromo-4-chloro-3′-indolylphosphatase p-toluidine and NBT.
TABLE 2.
Locations of 41 oligonucleotide probes designed to cover a mutation(s) associated with INH resistance
| Probe | Amino acid (nucleotide) region covered by probe |
|---|---|
| inhA-1 | (−17 to −3)a |
| inhA-2 | 95-100 |
| fabG1-1 | 202-206 |
| fabG1-2 | 230-235 |
| furA-1 | 12-17 |
| furA-2 | 6-12 |
| katG-1 | 45-51 |
| katG-2 | 63-68 |
| katG-3 | 92-97 |
| katG-4 | 94-99 |
| katG-5 | 105-111 |
| katG-6 | 123-127 |
| katG-7 | 132-137 |
| katG-8 | 135-140 |
| katG-9 | 140-145 |
| katG-10 | 157-163 |
| katG-11 | 170-174 |
| katG-12 | 174-179 |
| katG-13 | 178-183 |
| katG-14 | 190-194 |
| katG-15 | 228-236 |
| katG-16 | 247-252 |
| katG-17 | 256-261 |
| katG-18 | 271-277 |
| katG-19 | 294-299 |
| katG-20 | 313-318 |
| katG-21 | 323-327 |
| katG-22 | 326-330 |
| katG-23 | 383-387 |
| katG-24 | 389-391 |
| katG-25 | 417-422 |
| katG-26 | 457-462 |
| katG-27 | 479-482 |
| katG-28 | 486-490 |
| katG-29 | 522-528 |
| katG-30 | 539-543 |
| katG-31 | 553-558 |
| katG-32 | 565-569 |
| katG-33 | 591-596 |
| katG-34 | 631-635 |
| katG-35 | 707-712 |
Nucleotide position relative to the initiation codon of fabG1.
RESULTS
Drug susceptibility profiles.
As shown in Table S1 in the supplemental material, among 108 Inhr isolates, 65 (60%) were resistant to INH at 0.2 μg/ml but susceptible to INH at 1.0 μg/ml. The remaining 43 (40%) were resistant to INH at 1.0 μg/ml. Among the 108 isolates, 44 (41%) were resistant to INH but susceptible to other antituberculosis drugs. Thirteen (12%) were multidrug-resistant (MDR) isolates and five (5%) were extensively drug resistant (XDR).
IS6110-probed RFLP.
The results of IS6110-probed fingerprinting of the 108 Inhr isolates are shown in Fig. S1 in the supplemental material. Five clusters were detected, consisting of a total of 63 isolates (58%), including 12 (11%) in cluster I, 22 (20%) in cluster II, 12 (11%) in cluster III, 12 (11%) in cluster IV, and 5 (5%) in cluster V. These observations suggested that the majority of Inhr isolates in Japan expanded in a clonal manner.
Correlation between drug susceptibility and IS6110-probed RFLP.
With regard to the degree of INH resistance, the proportions of high-level Inhr isolates, i.e., isolates resistant to INH (1.0 μg/ml), were 1 (8%) in cluster I, 8 (36%) in cluster II, 4 (33%) in cluster III, 4 (33%) in cluster IV, and 5 (100%) in cluster V. These results indicated that the majority of isolates belonging to cluster I were resistant to INH (0.2 μg/ml) and susceptible to INH (1.0 μg/ml) and that those belonging to cluster V were highly resistant to INH. Six of 13 MDR isolates (46%) and 1 of 5 XDR isolates (20%) belonged to the clusters, but other MDR and XDR isolates did not belong to any clusters, indicating that they emerged sporadically in Japan.
Mutations in furA-katG, fabG1-inhA, and their upstream regions.
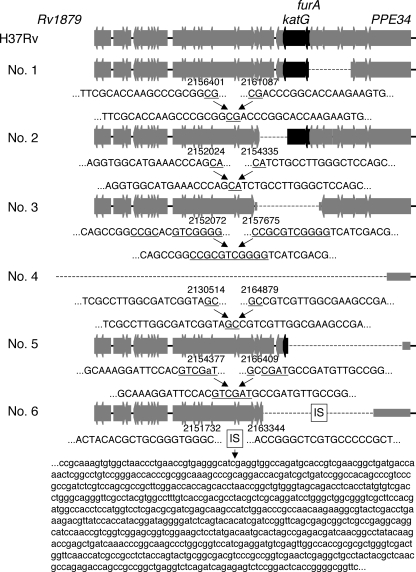
We sequenced the furA-katG operon, the fabG1-inhA operon, and their upstream regions in all Inhr isolates tested. Of the 108 isolates, 105 had at least one mutation (see Table S1 in the supplemental material), while the remaining 3 had no mutations in the regions sequenced. Of the 105 isolates with mutations, 64 had mutations in the furA-katG operon, 62 had mutations in fabG1-inhA operon, and 21 had mutations in both regions. Of the 64 with mutations in the furA-katG operon, six had a large-scale deletion adjacent to the furA-katG operon (Fig. 1; see also Table S1 in the supplemental material). As shown by genetic maps (Fig. 1), these isolates had large-scale deletions, ranging in size from 2.3 to 34.4 kb. The remaining 58 isolates did not have large-scale deletions.
FIG. 1.
Maps of large-scale deleted regions adjacent to katG in six Inhr M. tuberculosis isolates. Bold arrows indicate the open reading frames annotated in the H37Rv genome sequence (http://genolist.pasteur.fr/TubercuList/). The dotted lines correspond to the deleted regions, with the end sequences and H37Rv genome coordinates given below. Underlined sequences are possible substrates for recombination. The box labeled “IS” represents the 750-bp fragment of IS6110. Numbers 1 to 6 represent the names of the isolates and correspond to the numbers shown in Table S1 in the supplemental material. A nucleotide shown in lowercase in region 5 indicates a mutation.
Twenty-eight different mutations were found among the 58 isolates with mutations in the furA-katG operon (see Table S1 in the supplemental material). Twenty-three were in katG, two were in furA, and three were in the intergenic region. Seven different mutations were found among the 62 isolates with mutations in the fabG1-inhA operon (see Table S1 in the supplemental material). Three were in the upstream region, two were in fabG1, and two were in inhA. Of the 28 different mutations found in the furA-katG operon, 22 were novel (2 in furA, 3 in the intergenic region of the furA-katG operon, and 17 in katG). Of the seven different mutations found in the fabG1-inhA operon, four were novel: one in the upstream region of the fabG1-inhA operon, two in fabG1, and one in inhA (see Table S1 in the supplemental material).
Correlation between INH resistance and mutations.
We recently reported 5 novel mutations in katG (28). Including these mutations, 280 different mutations in katG were found in PubMed (http://www.ncbi.nlm.nih.gov/sites/entrez?db=pubmed) when articles were searched by the keywords “katG,” “mutation,” and “tuberculosis.” In addition, six mutations in the upstream region of the fabG1-inhA operon, including C−15T, and seven in inhA cause INH resistance (27, 28, 36). In this study, we found an additional 17 novel mutations in katG. One was a silent mutation (C195T [A65A]), while the other 16 caused amino acid substitutions. These mutations and amino acid substitutions are shown in Table 3. Furthermore, several novel mutations were detected in the present study: one in fabG1 (G609A [L203L]), one in furA (C41T [A14V]), and three in the intergenic region of the furA-katG operon (G−7A, A−10C, and G−12A).
TABLE 3.
katG mutations found in Inhr isolates
| Clone | Mutation(s) |
|
|---|---|---|
| Nucleotide | Amino acid | |
| katG-1a | G1388T | R463L |
| katG-2c | C379Gb | Q127Eb |
| katG-3c | C694Tb | P232Sb |
| katG-4 | A398Cb | N133Tb |
| katG-5c | T1147Cb | S383Pb |
| katG-6c | 1297::Cb, Δ1305Cb | KQT433-435QADb |
| katG-7c | A290G | H97R |
| katG-8c | C1465Ab | R489Sb |
| katG-9 | G944C | S315T |
| katG-10c | T1259Cb | M420Tb |
| katG-11 | G944C, G1159Cb | S315T, D387Hb |
| katG-12c | G368Ab, G895A | G123Eb, G299S |
| katG-13c | G1255C | D419H |
| katG-14c | C195Tb, T527Cb | A65Ab, M176Tb |
| katG-15 | Δ(478-479)b | Frame shiftb |
| katG-16c | G944C | S315T |
| katG-17 | Δ371Gb | Frame shiftb |
| katG-18c | C1894tb | R632Cb |
| katG-19 | C945A | S315R |
| katG-20c | Δ(571-576)b | Δ(191W-192E)b |
| katG-21c | G1624Cb | D542Hb |
katG-1 carrying a G1388T (R463L) neutral mutation was cloned from the Inhs strain IMCJ 2751.
These mutations have not previously been reported. Other mutations were previously reported in references 36 (G1388T), 7 (A290G), 36 (G944C), 7 (G895A), 6 (G1255C), and 36 (C945A).
This clone also had a G1388T neutral mutation.
We will report elsewhere that these mutations in furA and the intergenic region are associated with INH resistance induced by downregulation of katG expression (H. Ando and T. Kirikae, unpublished results), and those in fabG1 are also associated with INH resistance induced by upregulation of inhA expression (Ando et al., unpublished). In the present study, we examined whether novel mutations in katG are associated with INH resistance.
Correlation between mutations and IS6110-probed RFLP.
As shown in Fig. S1 and Table S1 in the supplemental material, all isolates belonging to cluster I detected in the IS6110-probed RFLP analysis, 11 (50%) in cluster II, and 8 (67%) in cluster III had a C−15T mutation in the inhA promoter region. All isolates in cluster IV had a C41T mutation in furA. All isolates in cluster V had a G944C/G945A (S315T/R) mutation. Isolates harboring katG mutations, except those with the G944C/G945A (S315T/R) mutation, did not cluster in the IS6110-probed RFLP.
Enzymatic activity of the novel KatG mutants.
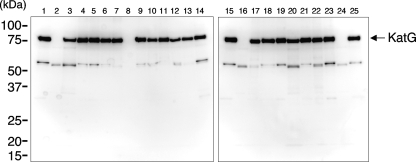
We cloned a wild-type (WT) katG gene (pkatG-wt) from H37Rv, a katG gene carrying a G1388T neutral mutation (pkatG-1) from IMCJ 2751, and 20 katG genes harboring mutations causing amino acid substitutions (pkatG-2 to -21) from Inhr isolates (Tables 1 and 3). Among the mutants, 15 were novel and 6 had been reported previously (the katG-1, -7, -9, -13, -16, and -19 mutants) (Table 3). These katG genes were expressed in katG-deficient E. coli UM262. As shown in Fig. 2, E. coli isolates with katG-wt expressed KatG (lanes 1 and 15), whereas E. coli isolates with an empty vector did not (lanes 2 and 16). E. coli isolates carrying katG mutants other than the katG-15 (lane 8) and katG-17 (lane 24) mutants expressed KatG proteins at levels similar to those observed for E. coli isolates carrying pkatG-wt. E. coli isolates with katG-15 (lane 8) and katG-17 (lane 24), which had a frame shift mutation (Table 3), did not express katG.
FIG. 2.
Western blot of whole-cell extracts from katG-deficient E. coli strain UM262 transformed with the empty vector, pTrcHis2-TOPO, or recombinant plasmids expressing various KatG mutations as follows: lanes 1 and 15, WT; lanes 2 and 16, empty vector; lane 3, R463L and D542H; lane 4, S315T and R463L; lane 5, Q127E and R463L; lane 6, P232S and R463L; lane 7, G123E, G299S, and R463L; lane 8, frame shift mutation from position 160; lane 9, S315T and D387H; lane 10, R463L and R489S; lane 11, S315R; lane 12, M420T and R463L; lane 13, A65A, M176T, and R463L; lane 14, H97R and R463L; lane 17, Δ(191W-192E) and R463L; lane 18, N133T; lane 19, R463L; lane 20, R463L and R632C; lane 21, S315T; lane 22, D419H and R463L; lane 23, S383P and R463L; lane 24, frame shift mutation from position 124; lane 25, in-frame insertion and deletion and R463L. The positions of molecular mass markers are shown on the left.
INH oxidase, peroxidase, and catalase activities were assessed using these clones (Table 4). Of the cloned mutants, one with KatG(R463L) from IMCJ 2751 showed levels of these activities similar to those observed for the wild type, and the KatG(R463L) mutation was not associated with INH resistance (Table 4). With regard to INH oxidase activity, E. coli isolates with katG-2 to -8 showed 1/3 to 1/17 less activity than those with katG-wt. E. coli isolates carrying katG-9 to -13 showed reduced activity compared to those carrying katG-2 to -8. E. coli isolates carrying katG-14 to -21 showed no activity (i.e., levels similar to those observed for vector controls). These results indicated that the degree of INH oxidase activity is correlated with that of INH resistance. E. coli isolates with katG-wt and katG-1 showed the highest levels of INH oxidase activity, and M. tuberculosis isolates with these genes were sensitive to INH. E. coli isolates carrying katG-2 to -8 showed slightly weaker activities, and M. tuberculosis isolates with these genes were resistant to INH at 0.2 μg/ml but susceptible to INH at 1.0 μg/ml. E. coli isolates with katG-9 to -21 showed weak or no activity, and M. tuberculosis isolates with these genes were resistant to INH at 1.0 μg/ml.
TABLE 4.
Enzymatic activities of KatG mutants detected in this study
| Plasmid | Amino acid mutation(s) |
Mean activity ± SDa |
Additional mutation associated with INH resistance | INH resistance levelb | |||
|---|---|---|---|---|---|---|---|
| Not previously reported | Previously reported | INH oxidase (103A560 units) | Peroxidase (102A405 units) | Catalase (102A240 units) | |||
| pTrcHis2-TOPOc | 4.84 ± 0.17 | 6.89 ± 0.70 | 0.00 ± 0.24 | ||||
| pkatG-wt | 177.16 ± 18.50 | 286.08 ± 0.43 | 142.26 ± 0.16 | S | |||
| pkatG-1 | R463L | 162.00 ± 11.31 | 289.62 ± 1.40 | 141.85 ± 0.13 | S | ||
| pkatG-2 | Q127E | R463L | 60.18 ± 0.95 | 256.07 ± 7.80 | 143.21 ± 0.35 | PfabG1-inhAC−15T | 0.2 |
| pkatG-3 | P232S | R463L | 54.47 ± 0.36 | 62.25 ± 0.05 | 76.63 ± 0.52 | 0.2 | |
| pkatG-4 | N133T | 40.67 ± 6.31 | 36.00 ± 0.26 | 100.61 ± 5.55 | 0.2 | ||
| pkatG-5 | S383P | R463L | 38.49 ± 0.04 | 42.24 ± 3.64 | 107.65 ± 4.13 | PfabG1-inhAC−15T | 0.2 |
| pkatG-6 | KQT433-435QADd | R463L | 20.02 ± 0.48 | 106.47 ± 1.17 | 142.81 ± 0.11 | PfabG1-inhAC−15T | 0.2 |
| pkatG-7 | H97R, R463L | 17.42 ± 0.35 | 26.80 ± 0.44 | 27.86 ± 2.01 | 0.2 | ||
| pkatG-8 | R489S | R463L | 10.40 ± 0.16 | 27.34 ± 0.27 | 14.83 ± 0.93 | PfabG1-inhAC−15T | 0.2 |
| pkatG-9 | S315T | 8.83 ± 0.04 | 102.00 ± 2.54 | 71.26 ± 1.71 | 1.0 | ||
| pkatG-10 | M420T | R463L | 8.42 ± 0.14 | 21.02 ± 0.37 | 46.45 ± 0.20 | 1.0 | |
| pkatG-11 | D387H | S315T | 7.93 ± 0.08 | 34.75 ± 0.61 | 35.71 ± 0.41 | 1.0 | |
| pkatG-12 | G123E | G299S, R463L | 6.87 ± 0.66 | 6.02 ± 0.17 | −0.70 ± 1.42 | PfabG1-inhAT−8C | 1.0 |
| pkatG-13 | D419H, R463L | 6.30 ± 0.52 | 7.67 ± 0.01 | 4.49 ± 0.39 | 1.0 | ||
| pkatG-14 | M176Te | R463L | 5.14 ± 0.01 | 4.67 ± 0.07 | 1.06 ± 0.30 | PfabG1-inhAC−15T | 1.0 |
| pkatG-15 | Frame shiftf | 5.02 ± 0.24 | 4.01 ± 0.57 | −1.75 ± 1.16 | 1.0 | ||
| pkatG-16 | S315T, R463L | 3.83 ± 0.18 | 84.41 ± 0.17 | 117.07 ± 7.56 | 1.0 | ||
| pkatG-17 | Frame shiftg | 3.30 ± 0.69 | 4.59 ± 0.09 | 2.07 ± 1.51 | 1.0 | ||
| pkatG-18 | R632C | R463L | 3.26 ± 0.13 | 1.56 ± 0.08 | −7.41 ± 0.76 | 1.0 | |
| pkatG-19 | S315R | 3.19 ± 0.76 | 3.24 ± 0.02 | −2.36 ± 0.71 | 1.0 | ||
| pkatG-20 | Δ(191W-192E)h | R463L | 2.78 ± 0.09 | 2.09 ± 0.04 | 2.61 ± 1.86 | 1.0 | |
| pkatG-21 | D542H | R463L | 1.63 ± 0.49 | 0.32 ± 0.17 | −7.00 ± 0.69 | 1.0 | |
Mean (n = 3) ± SD.
The INH susceptibility levels for clinical isolates with katG mutations are shown, as follows: S, INH sensitive; 0.2, resistant to INH (0.2 μg/ml) and susceptible to INH (1.0 μg/ml); and 1.0, resistant to INH (1.0 μg/ml).
A vector control.
1297::C and Δ1305C.
This isolate had an additional A65A silent mutation.
Δ(478-479).
Δ371G.
Δ(571-576).
The peroxidase and catalase activities of E. coli isolates with mutations were correlated well with each other and also with INH oxidase activity (Table 4). However, in E. coli isolates carrying some clones, peroxidase/catalase activities were different from INH oxidase activity, i.e., E. coli isolates with katG-16 and -9 showed weak activity.
Development of a LiPA for detection of INH resistance.
To detect novel mutations associated with INH resistance, we developed a new LiPA based on the reverse hybridization principle (25). Forty-one oligonucleotide probes were designed for the LiPA to detect mutations containing the furA-katG operon, the fabG1-inhA operon, PfabG1-inhA, and fabG1 (Table 2). As shown in Fig. S2 in the supplemental material, the LiPA could detect all mutations found in this study.
DISCUSSION
The results of RFLP and sequence analysis in the present study indicated that there are several predominant strains of Inhr M. tuberculosis with different genetic backgrounds in Japan (see Fig. S1 and Table S1 in the supplemental material). These strains had katG(G944C) (S315T), an inhA promoter mutation, fabG1(G609A) (L203L), and furA(C41T) (A14V) (see Table S1 in the supplemental material). Inhr isolates were reported to expand clonally in several regions, including northwestern Russia (20), the Netherlands (30), San Francisco, CA (13), Venezuela (2), and Sierra Leone (15). These clonal Inhr strains had a KatG(S315T) or inhA promoter mutation. Gagneux et al. (13) reported that the strains carrying the KatG(S315T) or inhA promoter mutation were more likely to spread than those carrying other mutations; our results were consistent with these previous findings. In addition, strains with fabG1(G609A) (L203L) and furA(C41T) (A14V) mutations were also more likely to spread in Japan.
Of Inhr isolates, a smaller number (22%) had S315T/R mutations in Japan (Table S1). The prevalences of the KatG(S315T) mutation in M. tuberculosis strains from around the world differ, especially with regard to the prevalence of tuberculosis. In regions where the prevalence of tuberculosis is low or intermediate, the mutation has been reported relatively infrequently: it occurred in 26% to 30% of 95 isolates from Singapore (16) and Madrid (23) and rarely in isolates from Scotland (11) and Finland (19). In contrast, the S315T mutation accounted for INH resistance in 52% to 64% of strains in Africa (8, 14, 31), 79% in Peru (9), 91% in Russia (18), and 58% in New York, NY. (23).
We found four KatG mutations (D419H, M420T, D542H, and R632C) that are associated with high-level INH resistance, and we also found three KatG mutations (H97R, N133T, and P232S) that are associated with low-level INH resistance (Table 4). The S315 mutation is known to confer high-level INH resistance (24, 26, 33). KatG is a functional homodimer, and each monomer is composed of two domains that are mainly α-helical. The N-terminal domain contains a heme binding site, whereas the C-terminal domain lacks this feature (34). The high-level INH resistance-associated mutations D419H and M420T are located in the region connecting the N-terminal and C-terminal domains (5). The interdomain interactions between the N-terminal and C-terminal domains of the two monomers are essential for forming the functional homodimer (5). The changes in the interdomain interactions due to the D419H and M420T mutations may result in loss of enzymatic activities of KatG. D542H and R632C are located in the 16th and 19th α-helices in the C-terminal domain, respectively, and showed no enzymatic activities, although the functional role of the C-terminal domain in KatG remains unclear (5, 34). The mutations associated with low-level INH resistance, H97R, N133T, and P232S, are located adjacent to the INH binding pocket (5). They may weakly affect the binding affinity of INH. The S315T mutation located at the INH binding pocket could block binding of INH without interfering with catalysis (5).
The new LiPA was able to distinguish high-level INH resistance (resistant to 1.0 μg/ml) from low-level INH resistance (resistant to 0.2 μg/ml and sensitive to 1.0 μg/ml) in clinical isolates without sequencing. Thus, we were able to determine the degree of INH resistance using this LiPA. This assay would be useful in clinical application in combination with culture-based drug susceptibility tests. We have recently developed a LiPA to detect a pncA mutation(s) for rapid detection of pyrazinamide-resistant M. tuberculosis (29), which was shown to be readily usable in clinical applications (1). The whole procedure takes only 9 h, and the estimated cost per sample is $35. The clinical trials for in vitro diagnosis are in progress (from April 2009 to March 2010) in Japan. The trials will reveal the specificity of the LiPA. It will be beneficial especially in developing countries where the laboratories are scarcely equipped because of the high cost of setting them up.
Assessment of INH oxidase activities of M. tuberculosis isolates may provide useful information about INH resistance. The INH oxidase activities of KatG mutants showed good correlations with the degree of INH resistance (Table 4). Other enzymatic activities of KatG mutants, i.e., peroxidase and catalase activities, were also correlated with the degree of INH resistance (Table 4). However, the activities of the S315T mutant were not, i.e., this mutant showed catalase-peroxidase activities but no INH oxidase activity (Table 4). Other S315 mutants, such as the S315R (Table 4) and S315N (32) mutants, have lost all three kinds of enzymatic activity. Thus, the Inhr isolates with KatG(S315T), retaining catalase-peroxidase activities, may have a survival advantage, and this may explain the global spread of strains with the KatG(S315T) mutation.
Supplementary Material
Acknowledgments
We thank Akiko Seshimo for excellent technical assistance.
This work was supported by Health Sciences Research grants (H21-SHINKO-IPPAN-016) and a Grant for International Health Research (21A-105) from the Ministry of Health, Labor and Welfare of Japan.
Footnotes
Published ahead of print on 8 March 2010.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Ando, H., S. Mitarai, Y. Kondo, T. Suetake, J. I. Sekiguchi, S. Kato, T. Mori, and T. Kirikae. 2009. Pyrazinamide resistance in multidrug-resistant Mycobacterium tuberculosis isolates in Japan. Clin. Microbiol. Infect. [Epub ahead of print.] doi: 10.1111/j.1469-0691.2009.03078.x. [DOI] [PubMed]
- 2.Aristimuno, L., R. Armengol, A. Cebollada, M. Espana, A. Guilarte, C. Lafoz, M. A. Lezcano, M. J. Revillo, C. Martin, C. Ramirez, N. Rastogi, J. Rojas, A. V. de Salas, C. Sola, and S. Samper. 2006. Molecular characterisation of Mycobacterium tuberculosis isolates in the First National Survey of Anti-tuberculosis Drug Resistance from Venezuela. BMC Microbiol. 6:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aziz, M., A. Laszlo, M. Raviglione, H. Rieder, M. Espinal, and A. Wright. 2003. Guidelines for surveillance of drug resistance in tuberculosis, Second edition ed. World Health Organization, Geneva, Switzerland.
- 4.Bernstein, J., W. A. Lott, B. A. Steinberg, and H. L. Yale. 1952. Chemotherapy of experimental tuberculosis. V. Isonicotinic acid hydrazide (nydrazid) and related compounds. Am. Rev. Tuberc. 65:357-364. [DOI] [PubMed] [Google Scholar]
- 5.Bertrand, T., N. A. Eady, J. N. Jones, Jesmin, J. M. Nagy, B. Jamart-Gregoire, E. L. Raven, and K. A. Brown. 2004. Crystal structure of Mycobacterium tuberculosis catalase-peroxidase. J. Biol. Chem. 279:38991-38999. [DOI] [PubMed] [Google Scholar]
- 6.Brossier, F., N. Veziris, C. Truffot-Pernot, V. Jarlier, and W. Sougakoff. 2006. Performance of the genotype MTBDR line probe assay for detection of resistance to rifampin and isoniazid in strains of Mycobacterium tuberculosis with low- and high-level resistance. J. Clin. Microbiol. 44:3659-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cardoso, R. F., R. C. Cooksey, G. P. Morlock, P. Barco, L. Cecon, F. Forestiero, C. Q. Leite, D. N. Sato, M. de Lourdes Shikama, E. M. Mamizuka, R. D. Hirata, and M. H. Hirata. 2004. Screening and characterization of mutations in isoniazid-resistant Mycobacterium tuberculosis isolates obtained in Brazil. Antimicrob. Agents Chemother. 48:3373-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dobner, P., S. Rusch-Gerdes, G. Bretzel, K. Feldmann, M. Rifai, T. Loscher, and H. Rinder. 1997. Usefulness of Mycobacterium tuberculosis genomic mutations in the genes katG and inhA for the prediction of isoniazid resistance. Int. J. Tuberc. Lung Dis. 1:365-369. [PubMed] [Google Scholar]
- 9.Escalante, P., S. Ramaswamy, H. Sanabria, H. Soini, X. Pan, O. Valiente-Castillo, and J. M. Musser. 1998. Genotypic characterization of drug-resistant Mycobacterium tuberculosis isolates from Peru. Tuber. Lung Dis. 79:111-118. [DOI] [PubMed] [Google Scholar]
- 10.Espinal, M. A., A. Laszlo, L. Simonsen, F. Boulahbal, S. J. Kim, A. Reniero, S. Hoffner, H. L. Rieder, N. Binkin, C. Dye, R. Williams, and M. C. Raviglione. 2001. Global trends in resistance to antituberculosis drugs. World Health Organization-International Union against Tuberculosis and Lung Disease Working Group on Anti-Tuberculosis Drug Resistance Surveillance. N. Engl. J. Med. 344:1294-1303. [DOI] [PubMed] [Google Scholar]
- 11.Fang, Z., C. Doig, A. Rayner, D. T. Kenna, B. Watt, and K. J. Forbes. 1999. Molecular evidence for heterogeneity of the multiple-drug-resistant Mycobacterium tuberculosis population in Scotland (1990 to 1997). J. Clin. Microbiol. 37:998-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujiki, A. 2001. TB bacteriology examination to stop TB. The Research Institute of Tuberculosis, Tokyo, Japan.
- 13.Gagneux, S., M. V. Burgos, K. DeRiemer, A. Encisco, S. Munoz, P. C. Hopewell, P. M. Small, and A. S. Pym. 2006. Impact of bacterial genetics on the transmission of isoniazid-resistant Mycobacterium tuberculosis. PLoS Pathog. 2:e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haas, W. H., K. Schilke, J. Brand, B. Amthor, K. Weyer, P. B. Fourie, G. Bretzel, V. Sticht-Groh, and H. J. Bremer. 1997. Molecular analysis of katG gene mutations in strains of Mycobacterium tuberculosis complex from Africa. Antimicrob. Agents Chemother. 41:1601-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Homolka, S., E. Post, B. Oberhauser, A. G. George, L. Westman, F. Dafae, S. Rusch-Gerdes, and S. Niemann. 2008. High genetic diversity among Mycobacterium tuberculosis complex strains from Sierra Leone. BMC Microbiol. 8:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, A. S., I. H. Lim, L. L. Tang, A. Telenti, and S. Y. Wong. 1999. Contribution of kasA analysis to detection of isoniazid-resistant Mycobacterium tuberculosis in Singapore. Antimicrob. Agents Chemother. 43:2087-2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loewen, P. C., J. Switala, M. Smolenski, and B. L. Triggs-Raine. 1990. Molecular characterization of three mutations in katG affecting the activity of hydroperoxidase I of Escherichia coli. Biochem. Cell Biol. 68:1037-1044. [DOI] [PubMed] [Google Scholar]
- 18.Marttila, H. J., H. Soini, E. Eerola, E. Vyshnevskaya, B. I. Vyshnevskiy, T. F. Otten, A. V. Vasilyef, and M. K. Viljanen. 1998. A Ser315Thr substitution in KatG is predominant in genetically heterogeneous multidrug-resistant Mycobacterium tuberculosis isolates originating from the St. Petersburg area in Russia. Antimicrob. Agents Chemother. 42:2443-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marttila, H. J., H. Soini, P. Huovinen, and M. K. Viljanen. 1996. katG mutations in isoniazid-resistant Mycobacterium tuberculosis isolates recovered from Finnish patients. Antimicrob. Agents Chemother. 40:2187-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mokrousov, I., O. Narvskaya, T. Otten, E. Limeschenko, L. Steklova, and B. Vyshnevskiy. 2002. High prevalence of KatG Ser315Thr substitution among isoniazid-resistant Mycobacterium tuberculosis clinical isolates from northwestern Russia, 1996 to 2001. Antimicrob. Agents Chemother. 46:1417-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagy, J. M., A. E. Cass, and K. A. Brown. 1997. Purification and characterization of recombinant catalase-peroxidase, which confers isoniazid sensitivity in Mycobacterium tuberculosis. J. Biol. Chem. 272:31265-31271. [DOI] [PubMed] [Google Scholar]
- 22.Otsuka, Y., P. Parniewski, Z. Zwolska, M. Kai, T. Fujino, F. Kirikae, E. Toyota, K. Kudo, T. Kuratsuji, and T. Kirikae. 2004. Characterization of a trinucleotide repeat sequence (CGG)5 and potential use in restriction fragment length polymorphism typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 42:3538-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piatek, A. S., A. Telenti, M. R. Murray, H. El-Hajj, W. R. Jacobs, Jr., F. R. Kramer, and D. Alland. 2000. Genotypic analysis of Mycobacterium tuberculosis in two distinct populations using molecular beacons: implications for rapid susceptibility testing. Antimicrob. Agents Chemother. 44:103-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pym, A. S., B. Saint-Joanis, and S. T. Cole. 2002. Effect of katG mutations on the virulence of Mycobacterium tuberculosis and the implication for transmission in humans. Infect. Immun. 70:4955-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossau, R., H. Traore, H. De Beenhouwer, W. Mijs, G. Jannes, P. De Rijk, and F. Portaels. 1997. Evaluation of the INNO-LiPA Rif. TB assay, a reverse hybridization assay for the simultaneous detection of Mycobacterium tuberculosis complex and its resistance to rifampin. Antimicrob. Agents Chemother. 41:2093-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saint-Joanis, B., H. Souchon, M. Wilming, K. Johnsson, P. M. Alzari, and S. T. Cole. 1999. Use of site-directed mutagenesis to probe the structure, function and isoniazid activation of the catalase/peroxidase, KatG, from Mycobacterium tuberculosis. Biochem. J. 338(3):753-760. [PMC free article] [PubMed] [Google Scholar]
- 27.Sala, C., F. Forti, E. Di Florio, F. Canneva, A. Milano, G. Riccardi, and D. Ghisotti. 2003. Mycobacterium tuberculosis FurA autoregulates its own expression. J. Bacteriol. 185:5357-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sekiguchi, J., T. Miyoshi-Akiyama, E. Augustynowicz-Kopec, Z. Zwolska, F. Kirikae, E. Toyota, I. Kobayashi, K. Morita, K. Kudo, S. Kato, T. Kuratsuji, T. Mori, and T. Kirikae. 2007. Detection of multidrug resistance in Mycobacterium tuberculosis. J. Clin. Microbiol. 45:179-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sekiguchi, J., T. Nakamura, T. Miyoshi-Akiyama, F. Kirikae, I. Kobayashi, E. Augustynowicz-Kopec, Z. Zwolska, K. Morita, T. Suetake, H. Yoshida, S. Kato, T. Mori, and T. Kirikae. 2007. Development and evaluation of a line probe assay for rapid identification of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis strains. J. Clin. Microbiol. 45:2802-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Doorn, H. R., P. E. de Haas, K. Kremer, C. M. Vandenbroucke-Grauls, M. W. Borgdorff, and D. van Soolingen. 2006. Public health impact of isoniazid-resistant Mycobacterium tuberculosis strains with a mutation at amino-acid position 315 of katG: a decade of experience in The Netherlands. Clin. Microbiol. Infect. 12:769-775. [DOI] [PubMed] [Google Scholar]
- 31.Victor, T. C., G. S. Pretorius, J. V. Felix, A. M. Jordaan, P. D. van Helden, and K. D. Eisenach. 1996. katG mutations in isoniazid-resistant strains of Mycobacterium tuberculosis are not infrequent. Antimicrob. Agents Chemother. 40:1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei, C. J., B. Lei, J. M. Musser, and S. C. Tu. 2003. Isoniazid activation defects in recombinant Mycobacterium tuberculosis catalase-peroxidase (KatG) mutants evident in InhA inhibitor production. Antimicrob. Agents Chemother. 47:670-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wengenack, N. L., J. R. Uhl, A. L. St. Amand, A. J. Tomlinson, L. M. Benson, S. Naylor, B. C. Kline, F. R. Cockerill III, and F. Rusnak. 1997. Recombinant Mycobacterium tuberculosis KatG(S315T) is a competent catalase-peroxidase with reduced activity toward isoniazid. J. Infect. Dis. 176:722-727. [DOI] [PubMed] [Google Scholar]
- 34.Wilming, M., and K. Johnsson. 2001. Inter- and intramolecular domain interactions of the catalase-peroxidase KatG from M. tuberculosis. FEBS Lett. 509:272-276. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, Y., B. Heym, B. Allen, D. Young, and S. Cole. 1992. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 358:591-593. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, Y., and A. Telenti. 2000. Genetics of drug resistance in Mycobacterium tuberculosis. ASM Press, Washington, DC.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.