Abstract
Previously, it was shown that the CTCF paralogous gene, BORIS (brother of the regulator of imprinted sites) is expressed in male germ cells, but its function in spermatogenesis has not been defined. To develop an understanding of the functional activities of BORIS, we generated BORIS knockout (KO) mice. Mice homozygous for the null allele had a defect in spermatogenesis that resulted in small testes associated with increased cell death. The defect was evident as early as postnatal day 21 and was manifested by delayed production of haploid cells. By gene expression profiling, we found that transcript levels for Gal3st1 (also known as cerebroside sulfotransferase [CST]), known to play a crucial role in meiosis, were dramatically reduced in BORIS KO testes. We found that CST is expressed in testis as a novel testis-specific isoform, CST form FTS, that has a short exon 1f. We showed that BORIS bound to and activated the promoter of CST form FTS. Mutation of the BORIS binding site in the promoter reduced the ability of BORIS to activate the promoter. These findings define transcriptional regulation of CST expression as a critical role for BORIS in spermatogenesis.
CTCF is a ubiquitously expressed chromatin factor containing an 11-zinc-finger (11-ZF) DNA binding domain that is highly conserved from Drosophila melanogaster to humans (39, 43, 61). Identities among vertebrate orthologs of 90 to 99% imply critical functions for the protein now known to include regulation of gene expression and chromatin organization (10, 28, 41). CTCF was originally identified as a repressor of Myc but was later shown to function in regulation of transcription when bound to promoters of specific genes such as APP, Rb, p53, hTERT, and ARF (9, 11, 12, 29, 35, 46, 49, 54, 57). In addition, CTCF is also known to act as a chromatin insulator, which blocks inappropriate activation or inactivation of genes by flanking regulatory elements (39). A constitutive insulator function of CTCF was originally identified in a reporter construct based on the HS4 element cloned from the chicken β-globin locus (3, 21), while later studies revealed that CTCF also mediate methylated CpG-sensitive insulation of imprinted genes (24). CTCF was found to bind unmethylated sequences in the maternal allele of the H19 imprinting control region (H19 ICR) as a part of a complex three-dimensional loop structure that partitions the enhancer from IGF2, resulting in inhibition of expression. CTCF could not bind methylated H19 ICR sequences of paternal origin, resulting in loss of enhancer blocking activity and activation of IGF2 expression (14, 33). More recently, CTCF sites have been found to maintain epigenomic integrity of unstable repeats (34) and latency control of herpes simplex virus (HSV), Epstein-Barr virus (EBV), and Kaposi's sarcoma-associated herpesvirus (2, 5, 50).
The gene BORIS (brother of the regulator of imprinted sites), also known as CTCFL (CTCF-like) and CTCF-T (CTCF testis specific), has been identified as a mammalian paralog of CTCF (28, 37). Recent studies showed that a BORIS ortholog is present in the platypus, reptiles, and higher organisms (20). Evolutionarily, expression of BORIS became progressively restricted to the testis, whereas CTCF is ubiquitously expressed in all species tested. This implies that BORIS has acquired testis-specific functions in mammalian organisms.
In addition to its normal expression in the testis, recent studies revealed that various tumors and cancer cell lines are also BORIS positive, with frequent coexpression of cancer testis antigens (CTA) (7, 17, 48, 52, 55). In addition, it was shown that conditional expression of BORIS induced expression of a series of CTA genes, including MAGE-A1, NY-ESO-1 (17, 52), and SPANX (30). This suggests that BORIS may normally act to regulate expression of CTA genes, although some data suggest otherwise (25).
Recent genome-wide analyses of CTCF binding sites by chromatin immunoprecipitation (ChIP)-chip or ChIP-sequencing analysis identified approximately 14,000 or more CTCF binding sites throughout the genome (6, 27). Kim et al. (27) documented that almost 80% of the binding sites shared consensus recognition sequences with evolutionary conservation. It is important to recognize that in contrast to the 11-ZF domain, the N- and C-terminal regions of BORIS share no homology with similarly placed CTCF sequences. This suggests that BORIS may exhibit functions distinct from CTCF based upon partner interactions that are dependent on these unique domains. In fact, BAT3 and PRMT7 were found to interact with BORIS through the unique N-terminal domain, implying that BORIS is functionally distinct from CTCF despite sharing DNA recognition sequences (23, 40).
Production of seminolipid, a primary glycolipid in the testis, is catalyzed by cerebroside sulfotransferase (CST), which is encoded by Gal3st1 through sulfation of galactosylalkylacylglycerol (19, 53). The importance of CST and its product in spermatogenesis was revealed through analyses of CST knockout (KO) mice, which displayed male-specific sterility due to a spermatogenesis defect apparent before the first meiotic division (18, 60). Mouse CST has eight splicing variants, all of which have the same coding region with different 5′-untranslated region (5′-UTR) sequences (16). Although different tissues express various combinations of CST splicing variants, testis expresses only CST form F, implying that there are testis-specific transcription factors that activate the promoter of this unique isoform. In view of the crucial function of CST in spermatogenesis, regulation of the CST form F promoter should be a critical determinant for proper spermatogenesis.
In this study, we analyzed BORIS KO mice to develop a firm understanding of the physiological functions of BORIS. We found that BORIS−/− mice exhibited a spermatogenesis defect at meiosis. We identified a novel testis-specific CST splicing variant, CST form FTS, for which expression is dramatically suppressed by the loss of BORIS. Our findings suggest that BORIS plays an important function in spermatogenesis through regulation of CST expression.
MATERIALS AND METHODS
Antibodies.
A glutathione S-transferase (GST)-tagged mouse BORIS C-terminal fragment containing amino acid residues 538 to 636 was prepared from Escherichia coli and used to immunize two rabbits. Serum antibodies were affinity purified on a GST-BORIS C-terminal fragment column, and then GST-reactive antibodies were depleted with a GST column. Antibodies were produced and purified by GenScript (Piscataway, NJ).
Targeted disruption of the BORIS gene.
BORIS knockout mice were generated at Ozgene (Ozgene Pty. Ltd., Bentley, Western Australia, Australia). The targeting vector was designed to replace the BORIS gene from the first methionine to exon 8 with an eGFP reporter casette flanked by LoxP/Lox511 sequences and a PGK-neomycin cassette flanked by FLP recombinase target (FRT) sequences. Primers used for targeting vector construction, genotyping, and generation of probes for Southern blotting are listed in Table S1 of the supplemental material. The targeting vector was linearized and electroporated into 129S1/Sv-derived W9.5 embryonic stem (ES) cells. Targeted ES cells were injected into blastocysts from C57BL/6 (B6) mice. Chimeric male mice were crossed with C57BL/6 females, and the BORIS+/− mice obtained were mated to obtain BORIS null mice. The PGK-neomycin cassette was deleted by crossing with B6.Cg-Tg(ACTFLPe)9205Dym/J mice (The Jackson Laboratory, Bar Harbor, ME). The use of mice in this study was approved by the NIAID Animal Care and Use Committee under protocol LIP-5.
Histochemistry.
Testes were fixed with 10% formalin and embedded in paraffin. Five-micrometer sections were prepared for histological analysis. Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) staining was performed using the DeadEnd colorimetric TUNEL system (Promega, Madison, WI) according to the manufacturer's protocol. TUNEL-positive cells in a field, which corresponded to approximately 0.6 mm2, were counted to quantify apoptotic cells in testes. Three mice were examined for each genotype.
DNA flow cytometry.
DNA flow cytometry was performed as described previously with slight modifications (31). Briefly, testicular cells that were fixed with 70% ethanol at least overnight were treated with 0.5% pepsin solution for 75 min at 37°C followed by treatment with phosphate-buffered saline (PBS) supplemented with 80 μg/ml RNase and 1% fetal bovine serum (FBS) for 20 min at room temperature. Cells were then stained with PBS containing 5 μg/ml propidium iodide (PI) and 1% FBS for 30 min on ice. Stained cells were analyzed with a FACSCalibur (Becton Dickinson, Franklin Lakes, NJ).
Purification of spermatocytes and round spermatids.
Spermatocytes and round spermatids were purified by centrifugal elutriation (38) followed by flow cytometric sorting of cells stained with Vybrant DyeCycle Green (Invitrogen, Carlsbad, CA) to obtain cell fractions with high purity. Briefly, decapsulated testes were treated with collagenase followed by treatment with trypsin, and the dissociated cells were used for centrifugal elutriation. Partially purified spermatocytes and the round spermatids fractions were incubated with 10 μM Vybrant DyeCycle Green for 30 min at 32°C followed by 4,6-diamidino-2-phenylindole (DAPI) staining. Cells were then sorted on a FACSAria (Becton Dickinson) to purify spermatocytes and round spermatids. DAPI-positive dead cells were eliminated. The purity of cells was confirmed by flow cytometry analysis of DNA content using sorted cells fixed with ethanol (spermatocytes, 95.7% ± 2.7%; round spermatids, 92.2% ± 2.5% [means ± standard deviations]).
Surface spread staining.
Surface spreads of spermatocytes were prepared as described previously (42). Samples were stained with anti-SCP3 (Novus Biologicals, Littleton, CO) and anti-γH2AX antibodies (Millipore, Billerica, MA) for 1 h at room temperature followed by staining with Alexa Fluor 488- or Cy3-conjugated secondary antibody for 1 h at room temperature. Samples were counterstained with DAPI.
Microarray analysis.
Microarray chips printed by the NIAID Microarray Research Facility comprised approximately 18,000 genes represented by 70-mer oligonucleotides. We studied five sets of wild-type and BORIS−/− testes prepared from postnatal day 14 (P14) mice. After the raw data were normalized with the Lowess smoothing function, the significant genes were identified with significance analysis of microarrays (51) with a false discovery rate of 3%, followed by a selection of genes with changes greater than twofold. Expression of genes with significant changes by microarray analyses was validated by quantitative PCR (qPCR). Microarray data were deposited to the NCBI Gene Expression Omnibus database (accession number GSE19162).
RT-PCR and qPCR.
Total RNA was prepared using an RNeasy minikit (Qiagen, Valencia, CA). cDNA was prepared using the SuperScript III first-strand synthesis system (Invitrogen) according to the manufacturer's protocol. Quantiative PCR (qPCR) was performed using the SYBR green PCR master mix (Applied Biosystems, Foster City, CA) and the 7900HT sequence detection system (Applied Biosystems). Primers used for conventional reverse transcription-PCR (RT-PCR) and quantitative PCR are listed in Table S1 in the supplemental material. Primers for qPCR were designed using the Primer Express software (Applied Biosystems). Expression levels were normalized against the housekeeping gene Gapdh. For ΔCt calculations, a Ct value of 40 was used for samples that had Ct values over 40. Absolute quantification was performed to analyze the transcription level of CST form FTS and FSS in CST form FTotal by using a plasmid that contained the partial cDNA sequence of CST form FSS cloned from stomach cDNA as a standard. The numbers of samples prepared from individual mice for qPCR are described in the figure legends. Each sample subjected to qPCR was analyzed in triplicate. Student's t test was performed to evaluate statistical significance. Data shown are means ± standard deviations.
5′-RACE.
Total RNA prepared from 3-month-old mouse testes was subjected to 5′ rapid amplification of cDNA ends (5′-RACE) analysis utilizing the GeneRacer kit (Invitrogen), which ensures amplification of only full-length transcripts by eliminating truncated messages from the amplification process, according to the manufacturer's instructions. Reverse transcription was performed using a CST-specific reverse primer, 5′-CCATTGGGGAAAGCGAACTTGAG-3′, followed by nested PCR using a reverse primer specific for CST, 5′-AGTGTGCTGCTGGCGGTCTTGTG-3′. Both CST-specific primers were designed on the open reading frame of CST, which is common for all isoforms of CST. Purified PCR fragments were cloned in a pGEM-T vector (Promega), sequenced (Genomics Research Facility, Rocky Mountain Laboratories, NIAID, NIH, MT), and analyzed.
Bisulfite sequencing.
Bisulfite modification of genomic DNA prepared from kidney or spermatocytes was performed using an Imprint DNA modification kit (Sigma-Aldrich, St. Louis, MO). A 314-bp sequence was amplified using 5′-GGGAAAGTTTTTGTATTATTGTATGAT-3′ and 5′-AACTAACTCTCTACTAAACCTAAAAACC-3′ as primers with PCR Platinum Taq polymerase under the following conditions: 94°C for 2 min; 35 cycles of 94°C for 1 min, 52°C for 30 s, and 72°C for 1 min; and 72°C for 5 min. The amplicon was ligated into the pGEM-T vector. Following transformation, plasmids from individual bacterial colonies were isolated and subjected to sequencing.
EMSA and methylation interference assay.
An electrophoretic mobility shift assay (EMSA) and methylation interference assay were performed as described previously (13, 44). Briefly, proteins for the assays were synthesized using plasmid pET-16b (Novagen, Madison, WI) with inserts of BORIS, CTCF, CTCF zinc-finger domain (11-ZF) sequences, or luciferase T7 control DNA with the TnT in vitro transcription-translation system (Promega). One primer for each DNA fragment was labeled with [γ-32P]ATP and used for PCR to prepare DNA fragments. Amplicons were gel purified and used for the assays. For supershift assays, proteins and DNA fragments were preincubated for 30 min at room temperature for complex formation followed by incubation with antibodies for 20 min. A mixture of two rabbit anti-mouse BORIS antibodies and a mixture of mouse monoclonal antibodies against CTCF (44) were used for supershift assays. Samples were loaded onto 5% native polyacrylamide gels in 0.5×Tris-borate-EDTA. The supershifted band of BORIS was found at the top of the gel due to the polyclonality of the anti-mouse BORIS antibodies.
Luciferase assay, cells, and transfection.
Fragments containing the entire 5′-UTR sequences of CST form FTS and its promoter sequences starting from promoter position −179, −359, or −515 were cloned into pGL3-basic vector (Promega). NIH 3T3 cells cultured in Dulbecco's modified Eagle's medium supplemented with 10% FBS and penicillin-streptomycin were transfected using Fugene 6 (Roche, Indianapolis, IN) according to the manufacturer's protocol with luciferase constructs together with internal control vector and BORIS or CTCF expression vectors cloned into pCIneo where indicated. Cells were cultured for 48 h at 37°C with 5% CO2. Luciferase assays were performed using a Dual-Luciferase reporter assay system kit (Promega) according to the manufacturer's protocol. Three independent samples were analyzed for each experiment, and the numbers of experiments are described in the figure legends. Student's t test was performed to evaluate statistical significance. Luciferase activity data shown are the means ± standard errors of the means.
ChIP assay.
ChIP assays were performed based on the Upstate Biotechnology protocol. Briefly, decapsulated testes were washed with PBS and minced with a scalpel. Samples were fixed with 1% formaldehyde for 15 min with rocking followed by two washes with PBS. Fixed samples were homogenized with a Dounce homogenizer, and pelleted samples were suspended in SDS lysis buffer (20 mM Tris-HCl [pH 8], 0.1% SDS, 2 mM EDTA, 150 mM NaCl, 1% Triton X-100) supplemented with a protease inhibitor cocktail. Lysates corresponding to 30 mg of tissue were used for a ChIP assay. A mixture of rabbit anti-mouse BORIS antibodies was used for ChIP. Preimmune sera were used as a control. Primers used for quantitative PCR are listed in Table S1 of the supplemental material.
RESULTS
Targeted disruption of the BORIS gene.
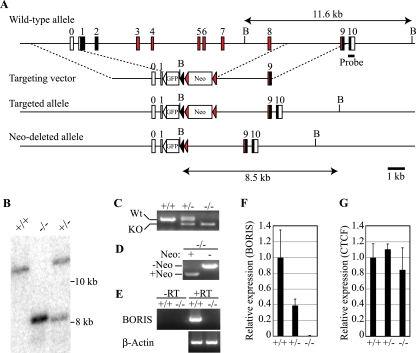
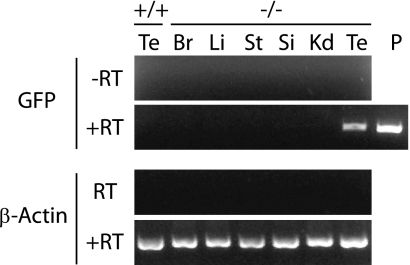
To identify the physiological function of BORIS and its target genes, we generated BORIS knockout mice. The BORIS gene, from the first methionine to exon 8, which includes ZF 1 to 10 and the N-terminal half of ZF11, was replaced with GFP and a neomycin cassette (Fig. 1A). We confirmed the recombination by Southern blotting (Fig. 1B), genomic PCR (Fig. 1C), RT-PCR (Fig. 1E), and quantitative RT-PCR (Fig. 1F), as well as the generation of mice with the PGK-neomycin cassette removed [BORIS−/− (-neo)] (Fig. 1D). Expression of CTCF did not change with the deletion of BORIS (Fig. 1G). By RT-PCR, we detected expression of GFP only in testis (Fig. 2), consistent with the normal expression pattern of BORIS transcripts; however, we could not detect GFP expression by fluorescence microscopy or flow cytometry (data not shown), probably due to low expression of GFP.
FIG. 1.
(A) Targeting scheme for generation of BORIS (Ctcfl) KO mice. Deletion of the BORIS gene proceeds from the first methionine to exon 8, which includes ZF-1 to -10, and the N-terminal half of ZF-11 is replaced with GFP and a neomycin cassette. The probe used for Southern blotting is indicated. Filled boxes indicate open reading frames; red boxes indicate the coding regions of the 11-ZF domain; open boxes indicate untranslated regions; the open triangle indicates a LoxP site; the filled triangle indicates a Lox511 site. FRT sequences are shown with red triangles. B, BglII. (B) Southern blotting. Genomic DNA was digested with BglII and hybridized with a probe flanking the targeting construct at the 3′ end as diagramed in panel A. (C) Genomic PCR typing of BORIS−/− mice. (D) Genomic PCR typing of BORIS−/− (-neo) mice. (E) cDNAs prepared from testes of 3-month-old mice were subjected to RT-PCR. (F and G) cDNAs prepared from testes of 3-month-old mice were quantified by qPCR for expression of transcripts for BORIS (F) and CTCF (G) (n = 3). Expression levels are shown as the ratio to wild type.
FIG. 2.
Expression of GFP was analyzed by RT-PCR. cDNAs prepared from each tissue are indicated: Te, testis; Br, brain; Li, liver; St, stomach; Si, small intestine; Kd, kidney; P, positive control.
Spermatogenesis defect in BORIS−/− mice.
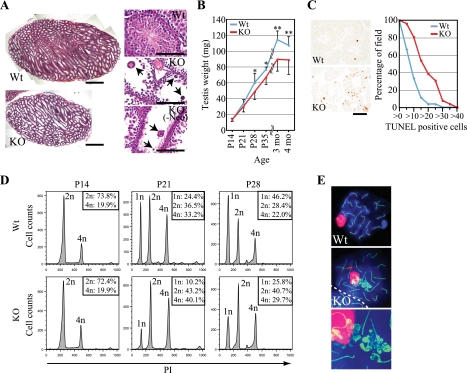
Matings of BORIS+/− mice yielded progeny at the expected Mendelian ratios (data not shown). As BORIS is exclusively expressed in testis (37) (Fig. 2), we focused our studies on testicular development and function. Although the BORIS−/− male mice were fertile (Table 1), we found that BORIS−/− testes were significantly smaller than those of their wild-type counterparts (Fig. 3A and B), indicating that BORIS had important functions in testicular development. A reduction in testicular size for BORIS−/− mice could be detected as early as P28 (Fig. 3B). To determine the basis for this difference, we performed histologic studies of testes from P28 BORIS−/− mice. Testes from BORIS−/− mice had reduced cellularity compared to testes of wild-type mice (Fig. 3A). Furthermore, testes from BORIS−/− mice had multinucleated cells (Fig. 3A, arrows), a feature found in a number of spermatogenesis-defective mice. We confirmed that BORIS−/− (-neo) mice also displayed a similar defect (Fig. 3A, right column, lower panel) and that they were fertile as well (data not shown). These results indicate that the spermatogenesis defect was due specifically to the loss of BORIS and was not affected by the presence of the PGK-neomycin cassette. To analyze whether the reduced cellularity of BORIS−/− testis might be due to increased cell death, we performed TUNEL staining and found that the frequency of apoptotic cells was greatly increased in BORIS−/− testis compared to wild-type testis (Fig. 3C). This result suggests that the smaller testes of BORIS−/− mice could be ascribed at least in part to increased cell death with effects on spermatogenesis.
TABLE 1.
Fertility analysisa
| Breeder (female × male) (n) | No. of litters (mean ± SD) | Litter size (mean ± SD) |
|---|---|---|
| B6 × BORIS+/+ (4) | 2.75 ± 0.50 | 6.90 ± 3.48 |
| B6 × BORIS−/− (4) | 2.00 ± 0.82 | 8.38 ± 0.74 |
| BORIS+/+ × B6 (3) | 3.67 ± 0.58 | 8.45 ± 2.16 |
| BORIS−/− × B6 (6) | 3.17 ± 0.41 | 9.74 ± 2.62 |
Two- to 3-month-old BORIS+/+ or BORIS−/− mice were mated with 2- to 3-month-old B6 mice for 3 months, and mean litter numbers and litter sizes were analyzed. All breeders produced offspring, and no significant difference was found in litter number and litter size between BORIS+/+ and BORIS−/− breedings.
FIG. 3.
(A) Hematoxylin and eosin staining of testes prepared from P28 wild-type, BORIS−/−, and BORIS−/− (-neo) mice. Arrows indicate multinucleated cells. Bars, 1 mm (left column); 100 μm (right column). (B) Sizes of wild-type (blue) and BORIS−/− (red) testes at various stages. *, P < 0.05; **, P < 0.005. (C) TUNEL staining of testes from P28 wild-type (upper panel) and BORIS−/− (lower panel) mice. Bar, 100 μm. TUNEL-positive cells in each field were counted in testes of wild-type (blue) and BORIS−/− (red) mice. (D) Representative results of DNA flow cytometry performed on testicular cells prepared from testes of P14, P21, and P28 of wild-type (upper row) and BORIS−/− (lower row) mice. The percentage of each population is shown in the inset. At least three mice were analyzed for each analysis. (E) Spermatocytes of wild-type (upper panel) and BORIS−/− (middle panel) mice were stained with SCP3 (green), γH2AX (red), and DAPI (blue). A dotted line indicates a boundary of adjacent cells. The lower panel shows a high-magnification image of aberrantly accumulated SCP3 in BORIS−/− spermatocytes.
To further analyze the effects of BORIS deficiency on the progression of spermatogenesis, we performed DNA flow cytometry on testicular cells. This allowed us to quantify subpopulations of testicular cells: tetraploid cells (primarily spermatocytes), diploid cells (a mixture of spermatogonia, secondary spermatocytes, and somatic cells), and haploid cells (spermatids). We found a deficiency in production of haploid cells in BORIS−/− testis compared to wild-type testis as early as P21 (17.8% ± 5.2% [wild type] versus 9.5% ± 5.2% [BORIS−/−]; P < 0.05) as well as at P28 (48.8% ± 2.3% [wild type] versus 28.9% ± 6.6% [BORIS−/−]; P < 0.05) (Fig. 3D). The fact that there was no significant difference in testicular weight at P21 was probably due to the scarcity of haploid cells at this stage of development, the beginning step in haploid cell production. To analyze the effect of BORIS on synapsis, we performed surface spread staining of spermatocytes. Although XY bodies positive for γH2AX were formed in spermatocytes of both BORIS−/− and wild-type mice, around 20% of BORIS−/− spermatocytes showed abnormal aggregation of the synaptonemal complex protein, SCP3, which does not coincide with γH2AX (Fig. 3E). This finding is similar to that of the abnormal structure reported in Dnmt3L-deficient spermatocytes (4). These results indicate that abnormalities in spermatogenesis in BORIS−/− mice begin around P21 and are evidenced by a significant delay and defect in meiosis.
Loss of BORIS is associated with reduced expression of CST.
We reasoned that alterations in gene expression responsible for the spermatogenesis defect found in BORIS−/− mice would be evident before day 21, when the defect could first be detected. To examine this possibility, we performed gene expression profiling using oligonucleotide microarrays to study testes prepared from P14 mice, an age at which we found no significant changes in the population of spermatogenic cells (wild type 2n, 70.6% ± 4.8%; wild type 4n, 20.7% ± 5.7%; BORIS−/− 2n, 74.7% ± 4.2%; BORIS−/− 4n, 18.7% ± 3.6%) (Fig. 3D). We identified 24 genes showing statistically significant differences in expression, and 4 of these genes showed more-than-2-fold differences in expression levels (see Fig. S2 in the supplemental material). To validate the differential expression, we employed qRT-PCR analyses and found three out of the four genes are truly differentially expressed between wild-type and BORIS−/− testis (Table 2). 1700019B21Rik is a gene with no known function that has been identified in mice and rats but not in humans. TSP50 is a testis-specific protease which is known to be a cancer testis antigen (47, 59). The expression is regulated in part by methylation of the promoter (22), is negatively regulated by p53 (56), and is abnormally activated in a high proportion of breast cancers (59). The third gene, Gal3st1, which encodes CST, showed the greatest reduction in transcripts in BORIS−/− testis (Table 2). Importantly, CST−/− mice exhibit male infertility due to an arrest of spermatogenesis prior to the metaphase of the first meiosis (18), paralleling the defect identified in BORIS−/− mice.
TABLE 2.
Differentially expressed genes in P14 BORIS−/− testis
| Gene | Accession no. | Fold changea based on: |
|
|---|---|---|---|
| Microarray | qRT-PCR | ||
| Gal3st1 | NM_016922 | −7.36 | −11.15 |
| 1700019B21Rik | XM_001473416 | −2.68 | −2.41 |
| Tsp50 | NM_146227 | −2.29 | −2.60 |
cDNAs prepared from P14 mice testes were used for qRT-PCR. qRT-PCR analysis of Gal3st1 showed a larger difference than microarray results, probably because qRT-PCR analysis has a greater dynamic range and returns more accurate data than microarrays.
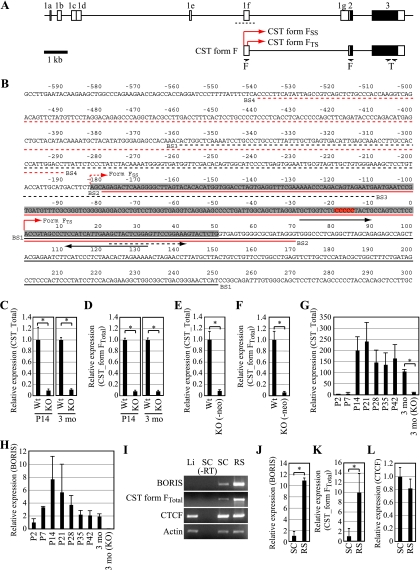
There are eight splice variants of CST expressed in different tissues (16), but only one, form F, is expressed in testis (Fig. 4A and B ). To confirm the microarray analyses, we used qRT-PCR to quantify total CST transcript levels and showed that CST transcript levels were reduced more than 10-fold in testis of BORIS−/− compared to wild-type mice at P14 (Fig. 4C). A comparable reduction of CST expression was also found in testes of mature mice (Fig. 4C). Furthermore, quantitation of transcripts specifically for CST form F showed comparable levels of reduction (Fig. 4D). We also confirmed that there was a comparable reduction in the expression of CST in BORIS−/− (-neo) testis (Fig. 4E and F). Analysis of CST transcript levels during testicular development showed an abrupt induction of expression at P14, when growing numbers of testicular cells differentiated into spermatocytes, with increased levels of CST transcripts being sustained thereafter (Fig. 4G). If BORIS regulates the expression of CST in testis, then BORIS should be expressed at or before the time that CST is induced. Indeed, BORIS transcripts were detectable from P2 and peaked at P14, with expression sustained at relatively high levels thereafter, consistent with the expression pattern of CST (Fig. 4H). Furthermore, we found that CST was highly expressed in round spermatids in correlation with the high expression of BORIS, while there was no correlation between the levels of expression of CST and CTCF (Fig. 4I to L). These results demonstrated that expression of CST is reduced in testes of BORIS−/− mice and suggest that BORIS might be directly involved in its transcriptional regulation.
FIG. 4.
(A) Genomic structure of the CST gene. Open and filled boxes represent UTRs and open reading frames, respectively. It has been reported that CST has eight splicing variants, all of which have the same coding region with different 5′-UTR sequences (16). CST form FTS is the testis-specific form among splicing variants expressed in other tissues. Arrows denote the positions of primers used to evaluate the expression of CST form FTotal (F) and total CST (T). Red arrows indicate the transcription start site of CST form FSS and CST form FTS. A dotted line indicates sequences shown in panel B. (B) Sequences surrounding exon 1f. The gray box represents the reported exon 1f sequences. The numbering is relative to the transcription start site of CST form FTS, which is marked by a red arrow. The reported transcription start site is marked by a dotted red arrow. The red box shows BORIS/CTCF-contacting residues as determined in a methylation interference assay. Black arrows indicate primers for CST form FSS. The dotted black arrow indicates a forward primer for CST form FTotal. DNA fragments used for EMSA are denoted by solid lines or dotted lines in black or red. (C) cDNAs prepared from wild-type or BORIS−/− mouse testis at P14 (n = 3) or at 3 months (n = 3) were subjected to qPCR to evaluate the expression of CST form F. Primers for total CST were used for the experiment. Expression levels are shown as the ratio to wild-type level. Asterisks denote statistical significance (P < 0.005). (D) cDNAs prepared from wild-type or BORIS−/− mouse testes at P14 (n = 3) or at 3 months (n = 3) of age were subjected to qPCR to evaluate the expression of CST form F. Primers for CST form FTotal were used for the experiment. Expression levels are shown as the ratio to the wild-type level. Asterisks denote statistical significance (P < 0.005). (E) cDNAs prepared from wild-type or BORIS−/− (-neo) mouse testis at P28 were subjected to qPCR to evaluate the expression of CST (n = 3). Primers for total CST were used for the experiment. Expression levels are shown as the ratio to wild type. The asterisk denotes statistical significance (P < 0.005). (F) cDNAs prepared from wild-type or BORIS−/− (-neo) mouse testis at P28 were subjected to qPCR to evaluate the expression of CST (n = 3). Primers for CST form FTotal were used for the experiment. Expression levels are shown as the ratio to wild type. The asterisk denotes statistical significance (P < 0.005). (G) cDNAs prepared from wild-type testes were subjected to qPCR analysis to evaluate the expression of CST during testis development. Primers for total CST were used for the experiment (n = 3). Expression levels are shown as the ratio to the P2 testis level. The asterisk denotes statistical significance (P < 0.005). (H) cDNAs prepared from wild-type testes were subjected to qPCR analysis to evaluate the expression of BORIS during testis development (n = 3). Expression levels are shown as the ratio to the P2 testis level. (I) Expression levels of BORIS, CST, and CTCF were analyzed by RT-PCR. cDNAs were prepared from liver (Li), spermatocytes (SC), and round spermatids (RS). (J to L) cDNAs prepared from SC and RS were subjected to qPCR analysis to evaluate the expression levels of BORIS (J), CST (K), and CTCF (L) (n = 3). Expression levels are shown as the ratio to the level in spermatocytes. Asterisks denote statistical significance (P < 0.05).
BORIS regulates expression of the testis-specific CST splicing variant CST form FTS.
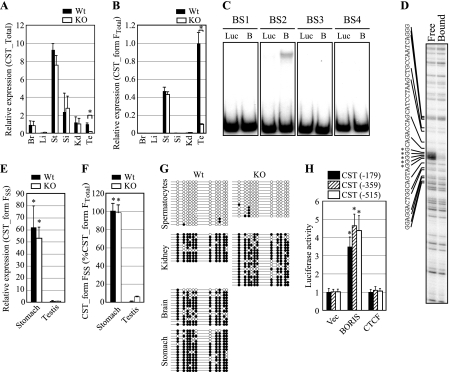
To analyze the expression pattern of CST, we performed qRT-PCR using cDNA prepared from various tissues. As reported previously, CST was widely expressed in multiple tissues including testis although expression of CST form F was restricted to stomach and testis (16) (Fig. 5A and B). Consistent with the known expression pattern of BORIS, expression of CST and CST form F was greatly diminished in BORIS−/− testis but not in the other tissues, including stomach, suggesting that CST form F promoter contains a potential BORIS binding site (Fig. 5A and B).
FIG. 5.
(A and B) Expression of CST and CST form F in various tissues of wild-type and BORIS−/− mice was analyzed by qPCR (n = 3). Primers for CST form FTotal were used in panel B. Expression levels are shown as the ratio to wild-type testis levels. Asterisks denote statistical significance (P < 0.005). (C) BORIS binding to the promoter region of exon 1f was analyzed by EMSA using fragments shown in Fig. 4B. Luciferase protein was used as a negative control. Luc, luciferase; B, BORIS. (D) Methylation interference assay of the BS2 fragment using 11-ZF. Partial sequences of the fragment are shown. Only the bottom strand is shown, as the top strand did not show any differences. Left lane, unbound fragments; right lane, bound fragments. Asterisks indicate contacting guanine residues. (E) Expression of CST form FSS in stomach and testis of wild-type and BORIS−/− mice was analyzed by qPCR (n = 3). Expression levels are shown as the ratio to wild-type testis. Asterisks denote statistical significance versus the testis sample (P < 0.005). (F) The ratio of CST form FSS in CST form FTotal was analyzed by qPCR (n = 3). Due to the massive and specific reduction of CST form FTS expression in BORIS−/− testis, the ratio of CST form FSS in CST form FTotal was slightly increased in BORIS−/− testis. Asterisks denote statistical significance versus the testis sample (P < 0.01). (G) Methylation status around exon 1f was analyzed by bisulfite sequencing. Open and filled circles represent unmethylated and methylated cytosines, respectively. (H) Activity of the CST form FTS promoter was analyzed by luciferase assay (n = 3). Empty vector (vec), BORIS, or CTCF expression vectors were cotransfected with each construct. Numbers indicate the 5′ end of each construct on the CST form FTS promoter region. Luciferase activities are shown as the ratio to the empty vector-transfected sample. Asterisks denote statistical significance versus empty vector-transfected sample (P < 0.05).
To determine if BORIS would indeed bind to the promoter region, we performed EMSAs using fragments derived from the promoter region of CST form F (Fig. 4B) and found that BORIS bound to fragment BS2 (Fig. 5C). This fragment was also capable of binding CTCF (Fig. 6C). Neither BORIS nor CTCF (data not shown) bound any of the other fragments tested. To identify the BORIS binding site in fragment BS2, we performed methylation interference assays of the fragment using the 11-ZF domain of CTCF, based on its known strong binding to CTCF target sequences in vitro and the shared recognition sequence of BORIS and CTCF (Fig. 5D). We found that 11-ZF bound to tandem guanine residues that are located within exon 1f (Fig. 4B).
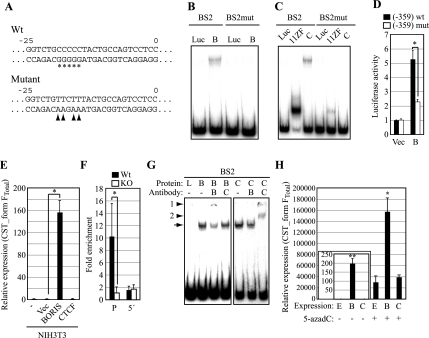
FIG. 6.
(A) Sequences of wild-type and mutants of the CST form FTS promoter. Asterisks indicate contacting guanine residues. Four contacting guanines were converted into adenines (arrowheads) in mutant constructs. Numbering shows the positions relative to the transcription start site of CST form FTS. (B) Binding of BORIS to the mutated BS2 fragment was analyzed by EMSA. Luc, luciferase; B, BORIS. (C) Binding of 11-ZF and CTCF to the mutated BS2 fragment was analyzed by EMSA as for panel B. C, CTCF. (D) Luciferase assays were performed using a mutated promoter of the CST form FTS (n = 3). Empty vector or BORIS expression vector were cotransfected with each construct. Luciferase activities are shown as the ratio to empty vector-transfected sample. The asterisk denotes statistical significance (P < 0.05). Vec, empty vector; B, BORIS. (E) Effect of BORIS on CST form FTotal expression was analyzed by transient transfection of a BORIS expression vector into NIH 3T3 cells. Expression of CST form FTotal was analyzed by qPCR (n = 3). Expression levels are shown as the ratio to mock-transfected sample. The asterisk denotes statistical significance versus empty vector-transfected sample (P < 0.005). Vec, empty vector. (F) Binding of BORIS on the CST form FTS promoter (P) and 1.6 kb upstream of the transcription start site (5′) in testis was analyzed in a ChIP-qPCR assay using anti-BORIS antibody (n = 4). The asterisk denotes statistical significance (P < 0.05). (G) Recognition of BORIS/DNA complexes by BORIS antibody was validated by supershift assay. Fragment BS2 was preincubated with BORIS, CTCF, or luciferase followed by incubation with antibody against BORIS or CTCF. The arrow indicates the shifted band. Arrowhead 1 indicates a supershifted band with BORIS antibody, and arrowhead 2 indicates a supershifted band with CTCF antibody. B, BORIS; L, luciferase; C, CTCF. (H) Effects of BORIS and 5-azadC on CST form FTotal expression were analyzed by transient transfection of a expression vector into NIH 3T3 cells either treated or not with 1 μM 5-azadC. Expression of CST form FTotal was analyzed by qPCR (n = 3). Cells were cultured with or without 5-azadC for 24 h followed by transient transfection. Cells were further cultured for 48 h and harvested for analysis. Expression levels are shown as the ratio to empty vector-transfected cells without 5-azadC. *, statistically significant versus empty vector-transfected cells without 5-azadC (P < 0.01); **, statistically significant versus empty vector-transfected cells with 5-azadC (P < 0.005). E, empty vector; B, BORIS; C, CTCF.
These results indicate that a BORIS/CTCF target site is located inside exon 1f. Although it was reported that BORIS/CTCF target sites could exist inside exonic sequences (52), most proteins that activate transcription bind upstream of the transcription start site. In view of the finding that the mechanisms governing activation of the CST form F promoter seemed to differ between testis and stomach (Fig. 5B), we hypothesized that there is a testis-specific transcription start site downstream of the BORIS/CTCF target site different from that previously described for exon 1f. To analyze this possibility, we performed 5′-RACE using testis RNA and found that all 10 CST transcripts analyzed started from exon 1f downstream from the BORIS/CTCF target site (Fig. 4B). Furthermore, we confirmed that CST form F transcripts highly expressed in stomach were initiated upstream from the BORIS/CTCF target site (Fig. 5E). Virtually all the CST form F transcripts expressed in testis were the short form, while the CST form F expressed in stomach was exclusively the longer form (Fig. 5F). Hereafter, we call the short form of the CST form F transcript CST form FTS, for testis specific, the originally reported longer form CST form FSS, for stomach specific, and both forms when measured together as CST form FTotal.
Methylation of CpG sequences in promoter regions is one of the major mechanisms responsible for suppression of gene expression. To analyze the methylation status of the CST form FTS promoter, we performed bisulfite sequencing and found that the region was highly methylated in tissues that didn't express CST form FTS—kidney, brain, and stomach—but was fully unmethylated in spermatocytes, even in BORIS−/− mice (Fig. 5G). This suggested that promoter methylation was responsible for suppression of CST form FTS expression, although BORIS being present or absent had no effect on the methylation status of its promoter.
Taken together, these results indicate that transcription of CST form FTS, which is initiated downstream from the BORIS/CTCF target site, is regulated by BORIS in testis, while transcription of CST form FSS is BORIS independent.
BORIS activates CST expression through binding to the BORIS/CTCF target site.
To further understand the role of BORIS in regulating the CST form FTS promoter, we performed luciferase assays using different fragments from the promoter region (Fig. 5H). All constructs cotransfected with a BORIS expression vector showed activation of the promoter, while cotransfection with a CTCF expression vector had no appreciable effect. This result suggests that BORIS activates the CST form FTS promoter by binding to the BORIS/CTCF target site while CTCF cannot.
As another approach to establishing that activation of the CST form FTS promoter was due to binding of BORIS to the BORIS/CTCF target site, we performed EMSA using fragments mutated on the residues recognized by the 11-ZF (Fig. 6A). BORIS did not induce a mobility shift with the mutated BS2 fragments, a finding common to the 11-ZF and full-length CTCF, indicating that BORIS as well as CTCF bound to the sequence (Fig. 6B and C). Furthermore, BORIS-induced activation of the promoter was diminished when the same mutation was introduced in the construct used for luciferase assays (Fig. 6D). We also confirmed that exogenous expression of BORIS, but not CTCF, induced expression of CST form FTotal in NIH 3T3 cells (Fig. 6E). To analyze binding of BORIS to the CST form FTS promoter region, we performed a ChIP-qPCR assay using whole testes derived from wild-type and BORIS−/− mice. Consistent with the in vitro findings, we found specific enrichment of BORIS on the CST form FTS promoter region from wild-type testis but not from BORIS−/− testis (Fig. 6F). The validity of using the rabbit anti-mouse BORIS antibodies for ChIP was confirmed by supershift assays (Fig. 6G).
Taken together, these results indicate that BORIS activates expression of CST form FTS through binding to the BORIS/CTCF target site.
DISCUSSION
To understand the physiological roles played by BORIS, we generated BORIS knockout mice and found that BORIS is critical to normal spermatogenesis (Fig. 3). Although there were no apparent abnormalities in the population of testicular cells at P14, we found delayed spermatogenesis as early as P21, suggesting that BORIS functions in meiosis. We conclude that the reduced size and the hypocellularity of testes in BORIS−/− mice are probably due to a failure of meiosis resulting in the apoptotic death found in BORIS−/− testis.
Using microarray analyses, we found that BORIS regulated the expression of CST, as demonstrated by the greatly reduced levels of transcripts in BORIS−/− testis (Table 2). This alteration in CST expression was found in the testis but not the somatic tissues of BORIS−/− mice, keeping with the testis-specific expression of BORIS (Fig. 5A and B). The defect in spermatogenesis seen in BORIS-deficient mice at meiosis is a near replica of that seen in testes of CST knockout mice (18). On the other hand, although CST knockout mice showed complete sterility, BORIS knockout mice were still fertile. Since production of sperm far exceeds the number necessary for fertility, it is likely that residual expression of CST found in BORIS−/− testis is sufficient to maintain fertility even though it causes significant defect in spermatogenesis.
We found that CST was expressed in testis as a novel testis-specific isoform, CST form FTS, which has shorter exon 1f sequences (exon 1fS) than described previously (16). CST form FTS was the dominant form of CST expressed in testis even in BORIS KO mice (Fig. 5E and F), indicating that BORIS is important for activation of the CST form FTS promoter but not for defining the transcription start site of that form. Consistent with the testis-specific expression of CST form FTS, the promoter for this isoform was highly methylated in somatic tissues, including stomach, which expressed CST form FSS, while it was unmethylated in spermatocytes (Fig. 5G). This suggests that methylation of the promoter is responsible for the repressing expression of CST form FTS. The promoter for this isoform was unmethylated in spermatocytes not only in wild-type mice but also in BORIS−/− mice. The unmethylated status of the promoter may be responsible for the incomplete repression of CST expression in BORIS−/− testis, while also indicating that BORIS is not responsible for demethylation of the promoter. We confirmed that ectopic expression of BORIS in somatic cells treated with 5-aza-2′-deoxycytidine (5-azadC), which induces demethylation of CpG dinucleotides, synergistically activates the expression of CST form FTS (Fig. 6H), although ectopic expression of BORIS alone or treatment with 5-azadC alone could also induce expression. Taken together, these results suggest that demethylation of the promoter is a prerequisite for BORIS to fully activate CST form FTS expression.
We showed that BORIS binds the CST form FTS promoter. Methylation interference assays followed by EMSA with mutated fragments revealed that the exact BORIS binding site in the promoter is highly homologous to the TAGGGGG-containing CTCF/BORIS binding sequence mapped by DeJong and coworkers in the testis-specific ALF gene promoter (8, 26), which has been termed the consensus CTCF binding site as identified by genome-wide ChIP analysis (27). Consistent with the finding of a BORIS/CTCF target site in the promoter, ectopic expression of BORIS activated the CST form FTS promoter, but not the promoter mutated in the BORIS/CTCF target site. It is interesting that both BORIS and CTCF can recognize the BORIS/CTCF target site, but only BORIS can activate the promoter. The expression pattern of BORIS, CTCF, and CST in spermatogenic cells also suggests a BORIS-specific function in regulation of CST expression. These data provide a clear demonstration that BORIS is functionally different from CTCF, even though they have the same recognition sequences. Since the amino and carboxy termini of BORIS have no homology with the parallel sequences in CTCF, the distinct molecular functions of BORIS may be attributable to the unique protein-protein interactions specified by these sequences. Our observations on the role of BORIS binding site in activation of the CST form FTS transcription are remarkably consistent with results on regulation of testis-specific TFIIA α/β-like factor (ALF) gene promoter obtained by DeJong and coworkers, who showed that unlike CTCF, expression of BORIS had an activating effect on the ALF promoter-reporter construct (9, 26). However, ALF only starts to express at the time point we performed microarray analysis, and that may be why we missed it as a target (15).
Although orthologs of CST can be found in species from zebrafish to humans, the sequence of exon 1fS is found only in mammals. This suggests that the testis-specific regulation of CST expression by BORIS evolved after mammalians diverged. Interestingly, the sequence of exon 1fS is not well conserved among mammals, while the BORIS/CTCF target site as well as the splice site flanking exon 1fS are highly conserved (see Fig. S3 in the supplemental material). This suggests that the regulation of CST form FTS expression by BORIS, which depends on the BORIS/CTCF site, is important in mammalian species, although the 5′-UTR sequence of exon 1fS itself is not. It is noteworthy that BORIS is expressed in a variety of tissues in lower vertebrates, while expression is restricted to the testis in mammals (20). This suggests that the acquisition of BORIS-specific regulatory mechanisms developed in conjunction with the functional specialization of BORIS to testicular germ cells.
Aside from CST, we found a few other genes that are differentially expressed in testes of wild-type and BORIS-deficient mice. This is probably due in part to the fact that we analyzed testes from P14 mice to identify BORIS target genes that were responsible for initiating the spermatogenesis defect. This is the earliest stage of testicular development at which we can expect to identify differentially expressed genes. The fact that spermatogenesis is disturbed in BORIS KO mice makes the identification of potential BORIS target genes that function later in spermatogenesis difficult. Transcriptional profiles of purified testicular cell subsets would be certain to yield additional targets. However, we also found several genomic targets for BORIS binding that could not be correlated with transcriptional regulation, suggesting other BORIS functions. Further studies are required to examine these possibilities.
Although we found that BORIS is a major determinant of normal spermatogenesis, BORIS−/− male mice were still fertile, indicating that there are mechanisms that can compensate for the loss of BORIS. Since CTCF is the only known paralog for BORIS, CTCF may partially compensate for the loss of BORIS, enabling BORIS−/− male mice to remain fertile. To determine if this model is correct, it would be necessary to analyze BORIS/CTCF double mutant mice. In addition, we cannot rule out the possibility that there is a protein other than CTCF that can compensate for the loss of BORIS. Finally, the phenotypes associated with different gene knockout mice are notoriously strain dependent (1, 45). The mice evaluated in the current study had a mixed 129/B6 genetic background such that progressive introgression onto a pure background such as B6 could result in a strain with significant phenotypic differences.
Based on previous studies of BORIS and CTCF and their abilities to recognize common target sites, it is likely that BORIS will be found to have functions other than regulating spermatogenesis. Methylation-sensitive binding of CTCF to ICRs of H19/Igf2 (24), Rasgrf1 (58), and Kcnq1 (13) regulates allele-specific gene expression of these imprinted loci in mouse and human somatic cells. However, ICRs alone are neither necessary nor sufficient for resetting of imprinting upon germ line transmission (32). Additional ICR-targeting sequences have been mapped for the Rasgrf1 ICR (58) but remain unknown for H19/Igf2 and other ICRs. Therefore, remethylation may indeed occur upon replacement of BORIS to CTCF as suggested (37) and be targeted to paternal ICRs from a distant BORIS binding site that remains to be mapped in vivo (32). Although offspring of BORIS−/− mice develop normally, indicating that at least some sperm can develop normally without BORIS, there might be defective germ cells with aberrant imprinting destined to die in BORIS−/− testis, a suggestion consistent with the high rate of apoptosis seen in testes of BORIS-deficient mice.
Most promising, perhaps, is the finding that expression of Tsp50 is markedly altered in BORIS-deficient testes. TSP50 is known as a CTA (47, 59), is overexpressed in a high proportion of breast cancers (59), is a gene with promoter methylation associated with expression (22), and has a near-canonical CTCF target sequence in the promoter (data not shown). Further studies of this gene, its regulation, and its importance as a CTA may link it functionally to BORIS regulation of CTA and to BORIS as a CTA target for cancer treatment (36).
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the NIAID, NIH.
Footnotes
Published ahead of print on 15 March 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Albrecht, K. H., M. Young, L. L. Washburn, and E. M. Eicher. 2003. Sry expression level and protein isoform differences play a role in abnormal testis development in C57BL/6J mice carrying certain Sry alleles. Genetics 164:277-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amelio, A. L., P. K. McAnany, and D. C. Bloom. 2006. A chromatin insulator-like element in the herpes simplex virus type 1 latency-associated transcript region binds CCCTC-binding factor and displays enhancer-blocking and silencing activities. J. Virol. 80:2358-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell, A. C., A. G. West, and G. Felsenfeld. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98:387-396. [DOI] [PubMed] [Google Scholar]
- 4.Bourc'his, D., and T. H. Bestor. 2004. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431:96-99. [DOI] [PubMed] [Google Scholar]
- 5.Chau, C. M., X. Y. Zhang, S. B. McMahon, and P. M. Lieberman. 2006. Regulation of Epstein-Barr virus latency type by the chromatin boundary factor CTCF. J. Virol. 80:5723-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuddapah, S., R. Jothi, D. E. Schones, T. Y. Roh, K. Cui, and K. Zhao. 2009. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 19:24-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Arcy, V., N. Pore, F. Docquier, Z. K. Abdullaev, I. Chernukhin, G. X. Kita, S. Rai, M. Smart, D. Farrar, S. Pack, V. Lobanenkov, and E. Klenova. 2008. BORIS, a paralogue of the transcription factor, CTCF, is aberrantly expressed in breast tumours. Br. J. Cancer 98:571-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeJong, J. 2006. Basic mechanisms for the control of germ cell gene expression. Gene 366:39-50. [DOI] [PubMed] [Google Scholar]
- 9.De La Rosa-Velazquez, I. A., H. Rincon-Arano, L. Benitez-Bribiesca, and F. Recillas-Targa. 2007. Epigenetic regulation of the human retinoblastoma tumor suppressor gene promoter by CTCF. Cancer Res. 67:2577-2585. [DOI] [PubMed] [Google Scholar]
- 10.Filippova, G. N. 2008. Genetics and epigenetics of the multifunctional protein CTCF. Curr. Top. Dev. Biol. 80:337-360. [DOI] [PubMed] [Google Scholar]
- 11.Filippova, G. N., S. Fagerlie, E. M. Klenova, C. Myers, Y. Dehner, G. Goodwin, P. E. Neiman, S. J. Collins, and V. V. Lobanenkov. 1996. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol. Cell. Biol. 16:2802-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filippova, G. N., C. F. Qi, J. E. Ulmer, J. M. Moore, M. D. Ward, Y. J. Hu, D. I. Loukinov, E. M. Pugacheva, E. M. Klenova, P. E. Grundy, A. P. Feinberg, A. M. Cleton-Jansen, E. W. Moerland, C. J. Cornelisse, H. Suzuki, A. Komiya, A. Lindblom, F. Dorion-Bonnet, P. E. Neiman, H. C. Morse III, S. J. Collins, and V. V. Lobanenkov. 2002. Tumor-associated zinc finger mutations in the CTCF transcription factor selectively alter its DNA-binding specificity. Cancer Res. 62:48-52. [PubMed] [Google Scholar]
- 13.Fitzpatrick, G. V., E. M. Pugacheva, J. Y. Shin, Z. Abdullaev, Y. Yang, K. Khatod, V. V. Lobanenkov, and M. J. Higgins. 2007. Allele-specific binding of CTCF to the multipartite imprinting control region KvDMR1. Mol. Cell. Biol. 27:2636-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han, L., D. H. Lee, and P. E. Szabo. 2008. CTCF is the master organizer of domain-wide allele-specific chromatin at the H19/Igf2 imprinted region. Mol. Cell. Biol. 28:1124-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han, S. Y., L. Zhou, A. Upadhyaya, S. H. Lee, K. L. Parker, and J. DeJong. 2001. TFIIAα/β-like factor is encoded by a germ cell-specific gene whose expression is up-regulated with other general transcription factors during spermatogenesis in the mouse. Biol. Reprod. 64:507-517. [DOI] [PubMed] [Google Scholar]
- 16.Hirahara, Y., M. Tsuda, Y. Wada, and K. Honke. 2000. cDNA cloning, genomic cloning, and tissue-specific regulation of mouse cerebroside sulfotransferase. Eur. J. Biochem. 267:1909-1917. [DOI] [PubMed] [Google Scholar]
- 17.Hong, J. A., Y. Kang, Z. Abdullaev, P. T. Flanagan, S. D. Pack, M. R. Fischette, M. T. Adnani, D. I. Loukinov, S. Vatolin, J. I. Risinger, M. Custer, G. A. Chen, M. Zhao, D. M. Nguyen, J. C. Barrett, V. V. Lobanenkov, and D. S. Schrump. 2005. Reciprocal binding of CTCF and BORIS to the NY-ESO-1 promoter coincides with derepression of this cancer-testis gene in lung cancer cells. Cancer Res. 65:7763-7774. [DOI] [PubMed] [Google Scholar]
- 18.Honke, K., Y. Hirahara, J. Dupree, K. Suzuki, B. Popko, K. Fukushima, J. Fukushima, T. Nagasawa, N. Yoshida, Y. Wada, and N. Taniguchi. 2002. Paranodal junction formation and spermatogenesis require sulfoglycolipids. Proc. Natl. Acad. Sci. U. S. A. 99:4227-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honke, K., Y. Zhang, X. Cheng, N. Kotani, and N. Taniguchi. 2004. Biological roles of sulfoglycolipids and pathophysiology of their deficiency. Glycoconj. J. 21:59-62. [DOI] [PubMed] [Google Scholar]
- 20.Hore, T. A., J. E. Deakin, and J. A. Marshall Graves. 2008. The evolution of epigenetic regulators CTCF and BORIS/CTCFL in amniotes. PLoS Genet. 4:e1000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou, C., H. Zhao, K. Tanimoto, and A. Dean. 2008. CTCF-dependent enhancer-blocking by alternative chromatin loop formation. Proc. Natl. Acad. Sci. U. S. A. 105:20398-20403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, Y., Y. Wang, M. Wang, B. Sun, Y. Li, Y. Bao, K. Tian, and H. Xu. 2008. Differential methylation of TSP50 and mTSP50 genes in different types of human tissues and mouse spermatic cells. Biochem. Biophys. Res. Commun. 374:658-661. [DOI] [PubMed] [Google Scholar]
- 23.Jelinic, P., J. C. Stehle, and P. Shaw. 2006. The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol. 4:e355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanduri, C., V. Pant, D. Loukinov, E. Pugacheva, C. F. Qi, A. Wolffe, R. Ohlsson, and V. V. Lobanenkov. 2000. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 10:853-856. [DOI] [PubMed] [Google Scholar]
- 25.Kholmanskikh, O., A. Loriot, F. Brasseur, E. De Plaen, and C. De Smet. 2008. Expression of BORIS in melanoma: lack of association with MAGE-A1 activation. Int. J. Cancer 122:777-784. [DOI] [PubMed] [Google Scholar]
- 26.Kim, M., D. Li, Y. Cui, K. Mueller, W. C. Chears, and J. DeJong. 2006. Regulatory factor interactions and somatic silencing of the germ cell-specific ALF gene. J. Biol. Chem. 281:34288-34298. [DOI] [PubMed] [Google Scholar]
- 27.Kim, T. H., Z. K. Abdullaev, A. D. Smith, K. A. Ching, D. I. Loukinov, R. D. Green, M. Q. Zhang, V. V. Lobanenkov, and B. Ren. 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128:1231-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klenova, E. M., H. C. Morse, 3rd, R. Ohlsson, and V. V. Lobanenkov. 2002. The novel BORIS + CTCF gene family is uniquely involved in the epigenetics of normal biology and cancer. Semin. Cancer Biol. 12:399-414. [DOI] [PubMed] [Google Scholar]
- 29.Klenova, E. M., R. H. Nicolas, H. F. Paterson, A. F. Carne, C. M. Heath, G. H. Goodwin, P. E. Neiman, and V. V. Lobanenkov. 1993. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol. Cell. Biol. 13:7612-7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kouprina, N., V. N. Noskov, A. Pavlicek, N. K. Collins, P. D. Schoppee Bortz, C. Ottolenghi, D. Loukinov, P. Goldsmith, J. I. Risinger, J. H. Kim, V. A. Westbrook, G. Solomon, H. Sounders, J. C. Herr, J. Jurka, V. Lobanenkov, D. Schlessinger, and V. Larionov. 2007. Evolutionary diversification of SPANX-N sperm protein gene structure and expression. PloS One 2:e359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krishnamurthy, H., G. C. Jagetia, and P. Jyothi. 1998. Radioprotective effect of zinc aspartate on mouse spermatogenesis: a flow cytometric evaluation. Mutat. Res. 401:111-120. [DOI] [PubMed] [Google Scholar]
- 32.Lewis, A., and W. Reik. 2006. How imprinting centres work. Cytogenet. Genome Res. 113:81-89. [DOI] [PubMed] [Google Scholar]
- 33.Li, T., J. F. Hu, X. Qiu, J. Ling, H. Chen, S. Wang, A. Hou, T. H. Vu, and A. R. Hoffman. 2008. CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex 2 intrachromosomal loop. Mol. Cell. Biol. 28:6473-6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Libby, R. T., K. A. Hagerman, V. V. Pineda, R. Lau, D. H. Cho, S. L. Baccam, M. M. Axford, J. D. Cleary, J. M. Moore, B. L. Sopher, S. J. Tapscott, G. N. Filippova, C. E. Pearson, and A. R. La Spada. 2008. CTCF cis-regulates trinucleotide repeat instability in an epigenetic manner: a novel basis for mutational hot spot determination. PLoS Genet. 4:e1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lobanenkov, V. V., R. H. Nicolas, V. V. Adler, H. Paterson, E. M. Klenova, A. V. Polotskaja, and G. H. Goodwin. 1990. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene 5:1743-1753. [PubMed] [Google Scholar]
- 36.Loukinov, D., A. Ghochikyan, M. Mkrtichyan, T. E. Ichim, V. V. Lobanenkov, D. H. Cribbs, and M. G. Agadjanyan. 2006. Antitumor efficacy of DNA vaccination to the epigenetically acting tumor promoting transcription factor BORIS and CD80 molecular adjuvant. J. Cell. Biochem. 98:1037-1043. [DOI] [PubMed] [Google Scholar]
- 37.Loukinov, D. I., E. Pugacheva, S. Vatolin, S. D. Pack, H. Moon, I. Chernukhin, P. Mannan, E. Larsson, C. Kanduri, A. A. Vostrov, H. Cui, E. L. Niemitz, J. E. Rasko, F. M. Docquier, M. Kistler, J. J. Breen, Z. Zhuang, W. W. Quitschke, R. Renkawitz, E. M. Klenova, A. P. Feinberg, R. Ohlsson, H. C. Morse III, and V. V. Lobanenkov. 2002. BORIS, a novel male germ-line-specific protein associated with epigenetic reprogramming events, shares the same 11-zinc-finger domain with CTCF, the insulator protein involved in reading imprinting marks in the soma. Proc. Natl. Acad. Sci. U. S. A. 99:6806-6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meistrich, M. L. 1977. Separation of spermatogenic cells and nuclei from rodent testes. Methods Cell Biol. 15:15-54. [DOI] [PubMed] [Google Scholar]
- 39.Moon, H., G. Filippova, D. Loukinov, E. Pugacheva, Q. Chen, S. T. Smith, A. Munhall, B. Grewe, M. Bartkuhn, R. Arnold, L. J. Burke, R. Renkawitz-Pohl, R. Ohlsson, J. Zhou, R. Renkawitz, and V. Lobanenkov. 2005. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 6:165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen, P., G. Bar-Sela, L. Sun, K. S. Bisht, H. Cui, E. Kohn, A. P. Feinberg, and D. Gius. 2008. BAT3 and SET1A form a complex with CTCFL/BORIS to modulate H3K4 histone dimethylation and gene expression. Mol. Cell. Biol. 28:6720-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohlsson, R., R. Renkawitz, and V. Lobanenkov. 2001. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 17:520-527. [DOI] [PubMed] [Google Scholar]
- 42.Peters, A. H., A. W. Plug, M. J. van Vugt, and P. de Boer. 1997. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 5:66-68. [DOI] [PubMed] [Google Scholar]
- 43.Phillips, J. E., and V. G. Corces. 2009. CTCF: master weaver of the genome. Cell 137:1194-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pugacheva, E. M., V. K. Tiwari, Z. Abdullaev, A. A. Vostrov, P. T. Flanagan, W. W. Quitschke, D. I. Loukinov, R. Ohlsson, and V. V. Lobanenkov. 2005. Familial cases of point mutations in the XIST promoter reveal a correlation between CTCF binding and pre-emptive choices of X chromosome inactivation. Hum. Mol. Genet. 14:953-965. [DOI] [PubMed] [Google Scholar]
- 45.Raverot, G., J. Weiss, S. Y. Park, L. Hurley, and J. L. Jameson. 2005. Sox3 expression in undifferentiated spermatogonia is required for the progression of spermatogenesis. Dev. Biol. 283:215-225. [DOI] [PubMed] [Google Scholar]
- 46.Renaud, S., D. Loukinov, F. T. Bosman, V. Lobanenkov, and J. Benhattar. 2005. CTCF binds the proximal exonic region of hTERT and inhibits its transcription. Nucleic Acids Res. 33:6850-6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scanlan, M. J., A. J. Simpson, and L. J. Old. 2004. The cancer/testis genes: review, standardization, and commentary. Cancer Immun. 4:1. [PubMed] [Google Scholar]
- 48.Smith, I. M., C. A. Glazer, S. K. Mithani, M. F. Ochs, W. Sun, S. Bhan, A. Vostrov, Z. Abdullaev, V. Lobanenkov, A. Gray, C. Liu, S. S. Chang, K. L. Ostrow, W. H. Westra, S. Begum, M. Dhara, and J. Califano. 2009. Coordinated activation of candidate proto-oncogenes and cancer testes antigens via promoter demethylation in head and neck cancer and lung cancer. PloS One 4:e4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soto-Reyes, E., and F. Recillas-Targa. 25 January 2010, posting date. Epigenetic regulation of the human p53 gene promoter by the CTCF transcription factor in transformed cell lines. Oncogene. doi: 10.1038/onc.2009.509. [DOI] [PubMed]
- 50.Stedman, W., H. Kang, S. Lin, J. L. Kissil, M. S. Bartolomei, and P. M. Lieberman. 2008. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 27:654-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vatolin, S., Z. Abdullaev, S. D. Pack, P. T. Flanagan, M. Custer, D. I. Loukinov, E. Pugacheva, J. A. Hong, H. Morse III, D. S. Schrump, J. I. Risinger, J. C. Barrett, and V. V. Lobanenkov. 2005. Conditional expression of the CTCF-paralogous transcriptional factor BORIS in normal cells results in demethylation and derepression of MAGE-A1 and reactivation of other cancer-testis genes. Cancer Res. 65:7751-7762. [DOI] [PubMed] [Google Scholar]
- 53.Vos, J. P., M. Lopes-Cardozo, and B. M. Gadella. 1994. Metabolic and functional aspects of sulfogalactolipids. Biochim. Biophys. Acta 1211:125-149. [DOI] [PubMed] [Google Scholar]
- 54.Vostrov, A. A., and W. W. Quitschke. 1997. The zinc finger protein CTCF binds to the APBβ domain of the amyloid beta-protein precursor promoter. Evidence for a role in transcriptional activation. J. Biol. Chem. 272:33353-33359. [DOI] [PubMed] [Google Scholar]
- 55.Woloszynska-Read, A., S. R. James, P. A. Link, J. Yu, K. Odunsi, and A. R. Karpf. 2007. DNA methylation-dependent regulation of BORIS/CTCFL expression in ovarian cancer. Cancer Immun. 7:21. [PMC free article] [PubMed] [Google Scholar]
- 56.Xu, H., J. Shan, V. Jurukovski, L. Yuan, J. Li, and K. Tian. 2007. TSP50 encodes a testis-specific protease and is negatively regulated by p53. Cancer Res. 67:1239-1245. [DOI] [PubMed] [Google Scholar]
- 57.Yang, Y., W. W. Quitschke, A. A. Vostrov, and G. J. Brewer. 1999. CTCF is essential for up-regulating expression from the amyloid precursor protein promoter during differentiation of primary hippocampal neurons. J. Neurochem. 73:2286-2298. [DOI] [PubMed] [Google Scholar]
- 58.Yoon, B., H. Herman, B. Hu, Y. J. Park, A. Lindroth, A. Bell, A. G. West, Y. Chang, A. Stablewski, J. C. Piel, D. I. Loukinov, V. V. Lobanenkov, and P. D. Soloway. 2005. Rasgrf1 imprinting is regulated by a CTCF-dependent methylation-sensitive enhancer blocker. Mol. Cell. Biol. 25:11184-11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yuan, L., J. Shan, D. De Risi, J. Broome, J. Lovecchio, D. Gal, V. Vinciguerra, and H. P. Xu. 1999. Isolation of a novel gene, TSP50, by a hypomethylated DNA fragment in human breast cancer. Cancer Res. 59:3215-3221. [PubMed] [Google Scholar]
- 60.Zhang, Y., Y. Hayashi, X. Cheng, T. Watanabe, X. Wang, N. Taniguchi, and K. Honke. 2005. Testis-specific sulfoglycolipid, seminolipid, is essential for germ cell function in spermatogenesis. Glycobiology 15:649-654. [DOI] [PubMed] [Google Scholar]
- 61.Zlatanova, J., and P. Caiafa. 2009. CTCF and its protein partners: divide and rule? J. Cell Sci. 122:1275-1284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.