Abstract
An interaction network connecting mRNA capping enzymes, the RNA polymerase II (Pol II) carboxyl-terminal domain (CTD), elongation factor Spt5, and the Cdk7 and Cdk9 protein kinases is thought to comprise a transcription elongation checkpoint. A crux of this network is Spt5, which regulates early transcription elongation and has an imputed role in pre-mRNA processing via its physical association with capping enzymes. Schizosaccharomyces pombe Spt5 has a distinctive CTD composed of tandem nonapeptide repeats of the consensus sequence 1TPAWNSGSK9. The Spt5 CTD binds the capping enzymes and is a substrate for threonine phosphorylation by the Cdk9 kinase. Here we report that deletion of the S. pombe Spt5 CTD results in slow growth and aberrant cell morphology. The severity of the spt5-ΔCTD phenotype is exacerbated by truncation of the Pol II CTD and ameliorated by overexpression of the capping enzymes RNA triphosphatase and RNA guanylyltransferase. These results suggest that the Spt5 and Pol II CTDs play functionally overlapping roles in capping enzyme recruitment. We probed structure-activity relations of the Spt5 CTD by alanine scanning of the consensus nonapeptide. The T1A change abolished CTD phosphorylation by Cdk9 but did not affect CTD binding to the capping enzymes. The T1A and P2A mutations elicited cold-sensitive (cs) and temperature-sensitive (ts) growth defects and conferred sensitivity to growth inhibition by 6-azauracil that was exacerbated by partial truncations of the Pol II CTD. The T1A phenotypes were rescued by a phosphomimetic T1E change but not by capping enzyme overexpression. These results imply a positive role for Spt5 CTD phosphorylation in Pol Il transcription elongation in fission yeast, distinct from its capping enzyme interactions. Viability of yeast cells bearing both Spt5 CTD T1A and Pol II CTD S2A mutations heralds that the Cdk9 kinase has an essential target other than Spt5 and Pol II CTD-Ser2.
Eukaryal mRNA processing is linked physically and temporally to transcription elongation. The earliest processing step is mRNA capping, which can occur as soon as the 5′ triphosphate terminus of the nascent RNA extrudes from elongating RNA polymerase (6, 15). The cellular RNA capping enzymes are directed to nascent mRNAs by binding to the phosphorylated carboxyl-terminal domain (CTD) of the largest subunit of RNA polymerase II (Pol II) (7, 8, 18, 34, 29, 57). The Pol II CTD, consisting of tandem heptapeptide repeats of the consensus sequence Y1S2P3T4S5P6S7, functions as a landing pad for diverse cellular proteins that regulate the initiation, elongation, and termination steps of Pol II transcription, modify chromatin structure, and catalyze or regulate RNA capping, splicing, and polyadenylation (30, 37). The inherently plastic CTD structure is sculpted by cyclin-dependent kinases (Cdks) that have various positional specificities and act at different stages of the transcription cycle. The Cdk7 kinase (a component of transcription factor TFIIH) acts at or shortly after initiation to install Ser5-PO4 and Ser7-PO4 marks on the CTD (1), of which Ser5-PO4 is a critical determinant of capping enzyme recruitment and mRNA capping in vivo (20, 22, 49).
Capping enzymes may also access the transcription complex by binding to the Pol II elongation factor Spt5 (32, 52). Spt5 is a large polypeptide (∼1,000 to 1,200 amino acids [aa]) composed of multiple domain modules, including a distinctive C-terminal repeat domain (the “Spt5 CTD”) that directly binds RNA capping enzymes and is targeted for threonine phosphorylation by the Cdk9 protein kinase (33, 56). Spt5 exerts both negative and positive effects on transcription elongation (2, 17, 21, 39, 44, 50, 55). For example, metazoan Spt5 elicits an elongation arrest at promoter-proximal sites that is alleviated by the Cdk9 subunit of P-TEFb (positive transcription elongation factor b) (50, 51). Cdk9 phosphorylates the Pol II CTD and the Spt5 CTD. Studies with analog-sensitive kinase mutants implicate the Spt5 CTD as a bona fide substrate for Cdk9 (or its budding yeast ortholog Bur1) (26, 49, 58). Whereas interactions of the capping enzyme with the Spt5 CTD are independent of CTD phosphorylation (in contrast to the capping enzyme/Pol II CTD interactions) (32), the conversion of Spt5 from a negative to a positive elongation mode requires Spt5 CTD phosphorylation by Cdk9 (5, 56).
In the fission yeast Schizosaccharomyces pombe, the essential Cdk9 kinase exists as a stable heterotrimeric complex in vivo with its cyclin partner Pch1 and the mRNA cap methyltransferase Pcm1 (13, 33, 36). S. pombe Cdk9 also interacts with Pct1, the RNA triphosphatase component of the capping apparatus (35). In turn, Pct1 and Pce1 (the guanylyltransferase component of the fission yeast capping system) bind directly and independently to the unphosphorylated Spt5 CTD and the phosphorylated Pol II CTD (32, 34). These interactions and others noted above underlie the proposal of a transcription elongation checkpoint that ensures a temporal window for capping of nascent mRNAs (27, 35, 36, 45).
The checkpoint model is attractive insofar as there is evidence that positive and negative regulation of transcription can occur at the step of capping enzyme recruitment (6, 12) and that either diminished cap guanylylation activity or defective installation of the guanylyltransferase-recruiting Ser5-PO4 Pol II CTD mark can result in the production of uncapped transcripts that suffer premature 5′ exonucleolytic decay (20, 41). However, recent studies highlight that the putative checkpoint is either not enforced or not required on a large fraction of cellular transcription units. For example, genetic or pharmacological inhibition of Cdk9 or Spt5 exerts fairly narrow effects on the levels of certain mRNAs, rather than a global transcriptional dyscrasia (24, 25, 49). One explanation for the limited impact of such inhibition is that there is functional overlap built into the systems that coordinate transcription elongation and mRNA capping. One potential source of functional overlap is the independent interactions of the capping enzymes with the Pol II and Spt5 CTDs.
S. pombe Spt5 and its CTD are the focus of the present study. Spt5 is essential for cell growth in fission yeast (32), as it is in budding yeast and in human somatic cells (24, 47). The 990-aa S. pombe Spt5 protein consists of an acidic N-terminal domain, central NusG-like NGN and KOW domains, and an exceptionally regular CTD (aa 801 to 990) composed of tandem repeats of the consensus nonapeptide T1P2A3W4N5S6G7S8K9 (see Fig. 2). S. pombe Spt5 forms a stable heterodimeric complex with the 105-aa S. pombe Spt4 protein (42). The Spt4-docking site on Spt5 is localized to a trypsin-resistant segment from aa 218 to 381 (42). The Spt4-binding module is conserved in Saccharomyces cerevisiae and human Spt5, wherein it adopts a compact tertiary structure composed of a central antiparallel β-sheet flanked by three α-helices (14, 16, 53). A genetic analysis of Spt4 in S. pombe revealed it to be inessential for growth at 25 to 30°C but critical at 37°C (42). Thus, the unconditionally essential Spt5 protein must be performing Spt4-independent functions. Initial insights into functional compartmentalization of Spt5 emerged from studies of the effects of partial deletions of its CTD nonamer repeat array. As few as three nonamer repeats sufficed for normal S. pombe growth, but only when Spt4 was present (42). Synthetic lethality of an spt51-835 spt4Δ double mutant at 34°C suggested that interaction of Spt4 with the central domain of Spt5 overlaps functionally with the Spt5 CTD.
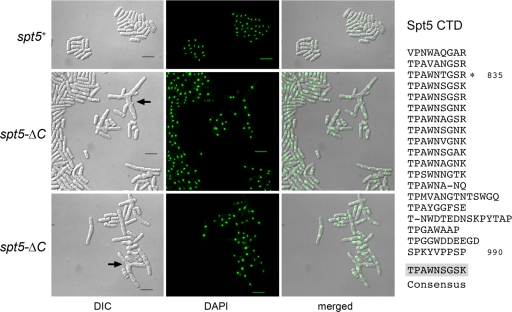
FIG. 2.
Deletion of the Spt5 CTD results in aberrant cell morphology. The amino acid sequence of the Spt5 CTD is displayed at right, with the nonamer repeats aligned vertically. The consensus sequence TPAWNSGSK is shown below the alignment. Previously it was shown that truncation of the CTD to amino acid 835, leaving three nonamer repeats, had no apparent impact on cells growth. In the present study, the entire CTD was deleted in the spt5-ΔC strains of S. pombe. Cell morphology and nuclear DNA localization were assessed by light microscopy. spt5+ and spt5-ΔC cells were cultured in YES medium at 30°C. Exponentially growing cells were fixed in 70% ethanol, treated with 4′,6-diamidino-2-phenylindole (DAPI), and then visualized by differential interference contrast (DIC) and fluorescence (DAPI) microscopy (Nikon Eclipse E600 microscope equipped with a Spot camera). The bars in the DIC images are 10 μm. The arrows denote branched cells.
Here we have studied the consequences of complete deletion of the Spt5 CTD repeat array, which results in slow growth and aberrant cell morphology. The spt5-ΔCTD phenotype is enhanced by truncating the Pol II CTD and suppressed by overexpressing capping enzymes. These results suggest that the Spt5 and Pol II CTDs play functionally overlapping roles in capping enzyme recruitment in fission yeast. We illuminated structure-activity relations in the Spt5 CTD by position-specific alanine scanning. The effects of Thr1 and Pro2 mutations on growth and 6-azauracil sensitivity suggest a positive role for Spt5 CTD phosphorylation in transcription elongation that overlaps elongation functions of the Pol II CTD.
MATERIALS AND METHODS
Deletion of the Spt5 CTD in fission yeast.
pUC19-based plasmids used for integration of ura4+ or kanMX markers downstream of spt5-(1-800) were constructed as follows. An spt5 gene fragment extending from an internal NdeI site (at nucleotide +1736) to nucleotide +2400 of the spt5 open reading frame (ORF) was amplified by PCR using a reverse primer that introduced a stop codon at +2401 and a flanking BamHI site. The DNA fragment was restricted and inserted upstream of the ura4 and kanMX genes in plasmids pUC19-ura4-spt53′ and pUC19-kanMX-spt53′ (42). The spt5ΔC5′-ura4-spt53′ and spt5ΔC5′-kanMX-spt53′ integration cassettes (which contain the flanking 534-bp genomic DNA fragment 3′ of the native spt5 stop codon) were excised and transformed into a diploid S. pombe strain. Ura+ or Geneticin-resistant transformants were selected, and diagnostic Southern blotting was used to verify targeted insertion into the chromosomal spt5 locus. The heterozygous diploids were then sporulated, and tetrads were dissected to obtain spt5-(1-800)::ura4+ or spt5-(1-800)::kanMX haploids, referred to herein as spt5-ΔC strains.
Missense mutations of the Spt5 CTD array.
CTD cassettes composed of tandem repeats of the wild-type nonamer consensus peptide or a T1A, P2A, W4A, N5A, K9A, or T1E variant were constructed as follows. Pairs of complementary 27-mer 5′-phosphorylated DNA oligonucleotides were designed that, when annealed, consisted of a 23-bp duplex with 5′ GATC overhangs, the sense strand of which encodes either the wild-type peptide GSKTPAWNS or the mutant peptides GSKAPAWNSGS (T1A), GSKTAAWNSGS (P2A), etc. (oligonucleotide sequences are available on request). Reactions of the annealed oligonucleotides with DNA ligase generated concatameric arrays of the repeating units, either in a tail-to-head orientation encoding a tandem nonapeptide repeat or in unfruitful tail-to-tail and head-to-head orientations. The tail-to-tail ligation events generate a BamHI cleavage site (5′-GGATCC) at the junctions, while the head-to-head junctions generate a BglII site (5′-AGATCT). In contrast, the in-frame junctions (5′-GGATCT) are resistant to BamHI and BglII. The ligation products were digested with BamHI and BglII and then resolved by PAGE. Resistant DNA fragments of ∼200 bp were isolated from the gels and then ligated into the BamHI site of pHis10Smt3, which had been modified to contain a translation stop codon immediately downstream of the BamHI site. DNA sequencing established the continuity of the ORFs, the number of nonapeptide repeats (either 7 or 8 in the constructs used here), and the desired wild-type or mutated amino acid sequences of each repeat.
S. pombe strains with Spt5 CTD missense mutations.
The wild-type and mutated Spt5 CTD cassettes were PCR amplified from the respective pHis10Smt3-(CTD)7-8 plasmid templates with primers designed to introduce BglII and SmaI sites at the 5′ and 3′ ends, respectively. The PCR products were digested with BglII and SmaI and then fused to the spt5-(1-800) ORF of a pDS474-based plasmid (10) by ligation between an engineered BglII site and a filled-in BamHI site that flanked the stop codon. The resulting plasmids were named pDS472-Spt5-(CTD)7, pDS472-Spt5-(T1A)7, etc. Cassettes for integration of the mutated spt5 alleles into S. pombe were constructed by restricting these plasmids with NdeI (at nucleotide +1736 of the spt5 ORF) and SmaI (in the pDS472 vector) and inserting the ∼860-bp CTD-encoding fragments upstream of the ura4 gene in the pUC19-ura4-spt53′ plasmid. The spt5CTD5′-ura4-spt53′ integration cassettes were excised and transformed into S. pombe cells. Ura+ transformants were selected and analyzed by diagnostic Southern blotting to verify correct integration.
Truncations of the S. pombe Rpb1 CTD array.
To generate C-terminal truncations of Rpb1, we first constructed pUC19-natMX-rpb13′, a plasmid containing the natMX gene upstream of a 552-bp segment of S. pombe genomic DNA 3′ of the rpb1+ stop codon. We then inserted upstream of natMX a series of rpb1 gene fragments extending from an EcoRI site (at nucleotide +4086 in the rpb1+ ORF) to a newly created stop codon at position +4816, +4858, +4879, +4900, +4921, +4984, +5086, or +5194 that truncated the Rpb1 CTD to 8, 10, 11, 12, 13, 16, 20, or 26 heptads, respectively. Excised rpb1ΔCTD5′-natMX-rpb13′ DNA fragments were introduced into diploid S. pombe cells. Transformants were selected on yeast extract with supplements (YES) agar medium containing 0.1 mg/ml nourseothricin (clonNAT; Werner Bioagents), and the targeted insertions were verified by diagnostic Southern blotting. The heterozygous rpb1+/rpb1-ΔCTD diploids were then sporulated, and tetrads were dissected.
Combining Spt5 CTD deletion with Rpb1 CTD truncations.
spt5-(1-800)::ura4+ (h+) cells were mixed with each of the rpb1-ΔCTD::natMX (h−) strains (rpb1-26, rpb1-20, rpb1-16, rpb1-13, and rpb1-12) on mating/sporulation agar (11), and the plates were incubated for 2 to 3 days at 30°C. We then dissected 5 to 20 tetrads for each cross and germinated the haploids on YES medium at 30°C. Their genotypes were determined to identify ura+ natR double mutants. Whereas tetrad dissection did yield the spt5-ΔC rpb1-26 and spt5-ΔC rpb1-20 strains, no viable ura+ natR haploids were isolated during tetrad dissections for the other crosses. To screen a larger number of haploid progeny, we performed random spore analysis (11). After plating ∼500 spores (determined by counting) on YES agar to determine the percentage that were viable, we plated 3,000 to 8,000 viable spores to selective medium lacking uracil and containing nourseothricin.
Rpb1 CTD-S2A mutant.
The S. pombe rpb1-12xS2ACTD strain was kindly provided by Jim Karagiannis, University of Western Ontario (23). Genomic DNA was isolated, and a 950-bp C-terminal segment of the rpb1-12xS2ACTD strain was amplified by PCR using a forward primer that annealed upstream of the EcoRI site at position +4086 within the rpb1 ORF and a reverse primer that introduced an XbaI site downstream of the stop codon in the rpb1-12xS2ACTD strain. The gene segment was restricted and inserted into pUC19. To expand the S2A repeat domain, we restricted pUC19-rpb1(12xS2A)5′ with AarI (at the site corresponding to nucleotide position +4703 within the rpb1+ ORF) and AvaI (at a site 16 nucleotides downstream of the AarI site in pUC19-rpb1(12xS2A)5′) and inserted therein a short duplex DNA that was generated by annealing two oligonucleotides (5′-ATG CCC TCT TCC CCA TCC TAC GCA CCA ACT TCA CCA TCT TAT GCT CCG ACT TCC and 5′-T CGG GGA AGT CGG AGC ATA AGA TGG TGA AGT TGG TGC GTA GGA TGG GGA AGA GG). The resulting plasmid was named pUC19-rpb1(14xS2A)5′. The 990-bp EcoRI/XbaI fragment encoding the modified CTD was excised from pUC19-rpb1(14xS2A)5′ and cloned into pUC19-term-natMX-rpb13′, a modified version of pUC19-natMX-rpb13′, in which a 320-bp DNA segment from the S. cerevisiae TPI1 transcription termination/polyadenylation signal had been inserted upstream of the natMX gene. In parallel, the pUC19-based plasmid for integration of rpb1-16 was modified to contain the TPI1 termination/polyadenylation signal. The integration cassettes were excised and transformed into haploid S. pombe strains to obtain nourseothricin-resistant rpb1-S2A and rpb1-16t cells.
Combining the Rpb1-S2A and Rpb1-16 variants with Spt5 CTD mutations.
The linear rpb1(14xS2A)5′-term-natMX-rpb13′ integration cassette was introduced into homozygous spt5-(CTD)7/spt5-(CTD)7, spt5-T1A/spt5-T1A, and spt5-T1E/spt5-T1E diploids, and nourseothricin-resistant transformants were selected. The resulting rpb1+/rpb1-S2A diploid strains were then sporulated, and tetrads were dissected to obtain the rpb1-S2A spt5-(CTD)7, rpb1-S2A spt5-T1A, and rpb1-S2A spt5-T1E haploid strains that were Ura+ and nourseothricin resistant. A set of control strains containing the same spt5-CTD alleles in the rpb1-16 background (which has a truncated Rbp1 CTD array composed of wild-type heptads) was generated by introducing the linear rpb1(1-16)5′-term-natMX-rpb13′ integration cassette into the spt5-(CTD)7, spt5-T1A, and spt5-T1E haploids and selecting for nourseothricin-resistant transformants. Correct targeting of the rpb1 locus in each case was confirmed by colony PCR and diagnostic Southern blotting.
Recombinant Pce1 and Pct1 proteins.
The Pce1 and Pct1 ORFs were PCR amplified from templates p132-PCE1 and pG1-PCT1 (34) with oligonucleotide primers that introduced BamHI and XhoI sites adjacent to the start and stop codons, respectively. The PCR fragments were restricted and inserted into plasmid pGEX-2TKN (Pharmacia) to yield expression plasmids encoding Pce1 and Pct1 fused to an N-terminal GST (glutathione-S-transferase) domain. The pGEX-Pct1 and pGEX-Pce1 plasmids were transformed into Escherichia coli BL21(DE3) Codon Plus (Novagen). Cultures derived from single transformants were maintained in logarithmic growth in LB medium containing 100 μg/ml ampicillin until the A600 of a 250-ml culture reached ∼0.7. The cultures were chilled on ice for 30 min, adjusted to 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and 2% (vol/vol) ethanol, and then incubated for 16 h at 17°C with constant shaking. Cells were harvested by centrifugation and stored at −80°C. All subsequent procedures were performed at 4°C. Thawed bacteria were resuspended in 12.5 ml of PBS (50 mM potassium phosphate, pH 7.2, 150 mM NaCl). The suspensions were adjusted to 0.2 mg/ml lysozyme and incubated for 45 min. Triton X-100 was added to a final concentration of 0.1%, and incubation was continued for 15 min. The cell suspensions were sonicated to reduce viscosity, and insoluble material was removed by centrifugation for 30 min at 13,000 rpm in a Sorvall SS34 rotor. The soluble extracts were mixed for 1 h with 0.5 ml of glutathione-Sepharose 4B resin (Pharmacia Biotech) that had been equilibrated in PBS. The resins were recovered by centrifugation, suspended in PBS, and poured into a column. The columns were washed twice with 10-ml aliquots of PBS and then eluted with 5 ml of 10 mM glutathione in PBS. The eluates were dialyzed against buffer D (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM dithiothreitol [DTT], 10% glycerol, 0.05% Triton X-100) and then stored at −80°C. The protein concentrations were determined by SDS-PAGE analysis of aliquots of the GST-Pce1 and GST-Pct1 polypeptides in parallel with increasing amounts of a bovine serum albumin (BSA) standard solution of known concentration. The gel was stained with Coomassie blue dye, and the staining intensities of GST-Pce1, GST-Pct1, and BSA were quantified with a Molecular Imager ChemiDoc densitometry system. The concentrations of the GST-Pce1 and GST-Pct1 polypeptides were determined by interpolation to the BSA standard curve. The yields of GST-Pce1 and GST-Pct1 from 250-ml bacterial cultures were ∼7 mg of each protein.
Recombinant Spt5-CTD proteins.
The wild-type and mutated His10-Smt3-Spt5-(CTD)7-8 plasmids (see above) were transformed into Escherichia coli BL21-Codon Plus (DE3). Cultures derived from single colonies were maintained in logarithmic growth in LB medium with 50 μg/ml kanamycin until the A600 of a 250-ml culture reached 0.6 to 0.8. The cultures were then placed on ice for 30 min and adjusted to 0.4 mM IPTG and 2% (vol/vol) ethanol before incubation was continued for 16 h at 17°C with constant shaking. Cells were harvested by centrifugation and stored at −80°C. All subsequent procedures were carried out at 4°C. The thawed cell pellets were resuspended in 12.5 ml of buffer A (50 mM Tris-HCl, pH 7.4, 10% sucrose, 250 mM NaCl). The cells were lysed, and soluble extracts were prepared as described above for the recombinant capping enzymes. The soluble lysates were mixed for 1 h with 1 ml of nickel-nitrilotriacetic acid (Ni-NTA)-agarose (Qiagen) that had been equilibrated with buffer A. The resins were recovered by centrifugation, suspended in 10 ml of buffer E (50 mM Tris HCl, pH 7.4, 250 mM NaCl, 10% glycerol) containing 25 mM imidazole, and then poured into columns. The columns were washed twice with 5-ml aliquots of the same buffer, and the bound proteins were then eluted stepwise with 3-ml aliquots of buffer E containing 300 and 500 mM imidazole. The elution profiles were monitored by SDS-PAGE. The 300 mM imidazole eluates containing the His10-Smt3-CTD polypeptides were dialyzed against buffer D. Protein concentrations were determined by SDS-PAGE and quantification of staining intensities as described above for the capping enzymes. The yields of His10-Smt3-CTD polypeptides recovered from 250 ml bacterial cultures were as follows: 8 mg (wild type), 6 mg (T1A variant), 10 mg (W4A variant), 6 mg (N5A variant), and 4.8 mg (K9A variant).
Binding of Pce1 to Spt5-CTD.
His10-Smt3-Spt5-CTD proteins (8 μg) were mixed with 8 μg of GST-Pce1 in 50 μl of buffer F (50 mM Tris-HCl [pH 7.4], 10% glycerol) containing 25 mM imidazole. Aliquots were removed to assess “input” material, and the samples were then mixed with 50 μl of a 5% slurry of magnetic Ni-NTA-agarose resin (Qiagen) in buffer F with 25 mM imidazole. The suspensions were incubated for 1 h at 4°C, after which the resins were collected and held at the bottoms of the tubes in a magnetic separator (Qiagen) while the supernatants containing unbound proteins were removed. The resins were washed twice with 500 μl of buffer F with 25 mM imidazole. The bound proteins were then eluted in 30 μl of 500 mM imidazole in buffer F.
Binding of Pct1 to Spt5-CTD.
His10-Smt3-Spt5-CTD proteins (8 μg) were mixed with 8 μg of GST-Pct1 in 50 μl of buffer E with 25 mM imidazole. Aliquots were removed to assess input material, and the samples were then mixed with 50 μl of a 5% slurry of magnetic Ni-NTA-agarose resin in buffer E with 25 mM imidazole. The suspensions were incubated for 1 h at 4°C, after which the resins were collected and held at the bottoms of the tubes in a magnetic separator while the supernatants containing unbound proteins were removed. The resins were washed twice with 500 μl of buffer E with 25 mM imidazole. The bound proteins were then eluted in 30 μl of 500 mM imidazole in buffer E.
Microscopy.
Microscopy was performed using live cells from exponentially growing liquid cultures of S. pombe. Cells were harvested by centrifugation, washed in water, and resuspended in water at ∼1 A600 unit/ml. Aliquots (5 μl) were applied to positively charged slides (Unifrost Plus; Azer Scientific). The yeast cells were visualized and photographed with a Nikon Eclipse E600 microscope (100× objective) coupled to an RT Slider Spot camera. The lengths of individual cells were measured by drawing a bar along the axis of each cell using the Spot Advanced software program (version 4.5).
Western blot analysis.
The haploid spt5+, spt5-ΔC, and various spt5-(1-800)-(CTD)7-8 strains were maintained in exponential growth in YES medium at 30°C. Aliquots of cells from exponentially growing cultures (containing equivalent A600 units) were harvested, and whole-cell extracts were prepared and subjected to SDS-PAGE and immunoblotting with affinity-purified rabbit anti-Spt5 antibodies as described previously (42). The immune complexes were visualized with horseradish peroxidase-conjugated anti-rabbit immunoglobulin using an enhanced chemiluminescence system. Where specified, the blot was stripped and reprobed with anti-PSTAIRE (Cdc2p34) antibody (Santa Cruz Biotechnology), which served as a cell extract loading control.
RESULTS
The Spt5 CTD is important for normal cell growth.
The CTD of S. pombe Spt5 comprises 18 nonamer repeats of the consensus sequence TPAWNSGSK (Fig. 2). To determine whether the CTD is important for spt5+ function in vivo, we introduced spt5-(1-800)::ura4+ into diploid cells so as to replace one copy of spt5+ with an allele encoding Spt5-ΔC, a variant that lacks the entire CTD. Ura+ haploids were recovered upon sporulation and dissection of spt5+/spt5-(1-800)::ura4+ cells, indicating that the Spt5 CTD is not essential for viability (Fig. 1A). However, the CTD is important for normal cell growth, insofar as spt5-ΔC cells formed smaller colonies than spt5+ sisters on YES agar at 30°C (Fig. 1A). The growth defect of spt5-ΔC cells on agar medium was also evident at 18°C, 20°C, 25°C, 32°C, 34°C, and 37°C (Fig. 1D and E; see also Fig. 4D). The doubling time of spt5-ΔC cells in YES liquid medium at 30°C was 3.5 h, versus 2.1 h for spt5+ cells (not shown).
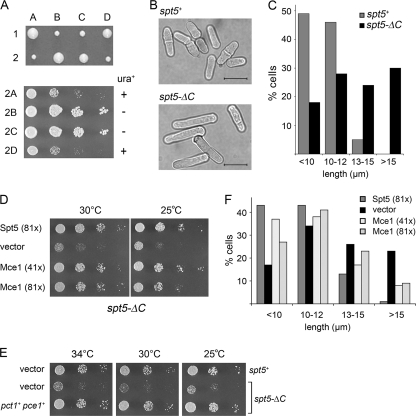
FIG. 1.
Deletion of the Spt5 CTD affects cells growth and morphology. (A) Heterozygous spt5+/spt5-(1-800)::ura4+ diploids were sporulated, and asci were dissected. Individual spores (A, B, C, and D) from two tetrads (1 and 2) were germinated on YES agar at 30°C for 4 days (upper panel). The four haploid progeny of tetrad 2 were grown in liquid culture. The cultures were diluted to attain an A600 of 0.1, and aliquots (3 μl) of serial 10-fold dilutions were spotted on YES agar medium. The plates were photographed after incubation for 3 days at 30°C (lower panel). The spt5-ΔC haploids 2A and 2D are ura+; the spt5+ haploids 2B and 2C are ura−. (B) spt5+ and spt5-ΔC cells grown in YES medium at 30°C were examined by light microscopy. Bars, 10 μm. (C) The lengths of 270 spt5+ cells and 330 spt5-ΔC cells were measured and sorted into length bins as specified. The percentage of cells in each bin is represented in a bar graph. (D) Dosage suppression of the spt5-ΔC phenotype by mammalian capping enzyme Mce1. spt5-ΔC[spt5-(1-800)::ura4+] cells were transformed with LEU2 plasmids as specified. Serial dilutions of spt5-ΔC strains harboring a pREP81x-Spt5 plasmid (positive control), an empty vector plasmid (negative control), or plasmids for expression of Mce1 under the transcriptional control of an intermediate-strength (41x) or a low-strength (81x) nmt promoter were spotted on Leu− agar medium. The plates were photographed after incubation for 6 days at 25°C or 3 days at 30°C. (E) Dosage suppression of spt5-ΔC by fission yeast capping enzymes. The spt5+ and spt5-ΔC [spt5-(1-800)::kanMX] strains harbored two plasmids marked with ura4+ and LEU2, respectively. These were either empty vector plasmids or plasmids for expression of S. pombe pct1+ (RNA triphosphatase) and pce1+ (RNA guanylyltransferase) under the transcriptional control of the low-strength nmt (81x) promoter. Cultures were grown at 30°C in minimal medium lacking uracil and leucine. Aliquots (3 μl) of serial 10-fold dilutions were spotted on Ura− Leu− agar medium and incubated at 25°C (4 days), 30°C (3 days), or 34°C (3 days) as specified. (F) Overexpression of Mce1 partially rescues the elongated phenotype of spt5-ΔC cells. The lengths of 300 to 500 individual cells were measured. The percentages of cells in each of the four size categories are represented by vertical bars.
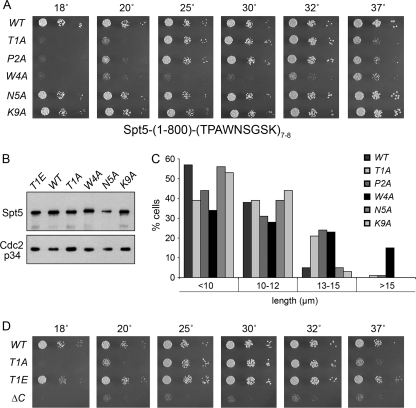
FIG. 4.
Effects of alanine substitutions in the Spt5 CTD. (A) S. pombe strains with the indicated chromosomal spt5-(1-800)-CTD alleles—in which 7 or 8 wild-type or mutated CTD nonamer repeats were fused to Spt5-(1-800)—were grown in liquid medium until the A600 reached 0.3 to 0.5. The cultures were adjusted to equalize the A600, and aliquots of serial 5-fold dilutions were spotted on YES agar medium. The plates were photographed after incubation for 8 days at 18°C, 5 days at 20°C, 3 days at 25 and 37°C, or 2 days at 30 and 32°C. WT, wild type. (B) Western blot analysis of Spt5. Whole-cell extracts of the indicated spt5-CTD strains were resolved by SDS-PAGE. The polypeptides were transferred to a membrane and probed by serial Western blotting with affinity-purified polyclonal anti-Spt5 antibody (top panel) and then with anti-Cdc2-p34 (PSTAIRE) antibody as a loading control (bottom panel). (C) Morphological phenotypes of spt5-CTD mutants. Cultures of the indicated spt5-CTD mutant strains were grown to mid-logarithmic phase at 30°C, and 300 to 500 individual cells were measured and sorted into the length bins specified. The percentage of cells in each bin is represented in the bar graph. (D) Effects of a phosphomimetic T1E change. Aliquots (3 μl) of serial 5-fold dilutions from exponentially growing cultures of the indicated strains were spotted onto YES agar medium. The plates were photographed after incubation for 8 days at 18°C, 6 days at 20°C, or 2.5 days at 30, 34, and 37°C.
We observed that cells lacking the Spt5 CTD were elongated compared to wild-type cells (Fig. 1B and C and Fig. 2). Whereas 49% and 46% of spt5+ cells grown in YES medium at 30°C were <10 μm and 10 to 12 μm, respectively, only 18% and 28% of the spt5-ΔC cells fell in these two categories. Thirty percent of spt5-ΔC cells were >15 μm (Fig. 1C), including hyperelongated cells (>20 μm) and cells with branches and multiple septa (Fig. 2). We conclude that the absence of the CTD results in slow growth and abnormal cell morphology.
Overexpression of capping enzymes alleviates the growth phenotype of spt5-ΔC cells.
The fission yeast mRNA capping enzymes Pct1 (RNA triphosphatase) and Pce1 (RNA guanylyltransferase) bind directly to the Spt5 CTD (32). If the recruitment of capping enzymes to the transcription complex is impaired in the absence of the Spt5 CTD, then overexpression of capping enzymes might suppress some of the spt5-ΔC phenotypes. We introduced into spt5-ΔC cells a multicopy plasmid for expression of the mammalian capping enzyme Mce1—a bifunctional triphosphatase-guanylyltransferase that binds avidly to the phosphorylated Pol II CTD (18)—under the control of the medium-strength (41x) and low-strength (81x) nmt1 promoters (9). Growth was assessed by spotting aliquots of serial dilutions of the cell cultures onto agar medium and incubating the plates at 25°C and 30°C. spt5-ΔC cells that had been transformed with a plasmid expressing Spt5 under the control of the 81x promoter served as a positive control. We found that cells harboring MCE1 plasmids grew as well as the Spt5 control cells, as gauged by colony size (Fig. 1D). nmt1-driven overexpression of both fission yeast capping enzymes (Pct1 plus Pce1) on multicopy plasmids also suppressed the slow-growth phenotype of spt5-ΔC cells (Fig. 1E). Furthermore, overexpression of Mce1 partially reversed the morphological defects of spt5-ΔC cells, insofar as only 8 to 9% of MCE1 cells were >15 μm long, compared to 23% of spt5-ΔC cells harboring the vector plasmid (Fig. 1F). In the suppressed strains, no branched or multiseptated cells were observed (not shown). These findings suggest that recruitment of the capping enzymes to the transcription complex is compromised in the absence of the Spt5 CTD, and we infer that dosage suppression is a consequence of increased binding of the capping enzymes to Pol II, likely via the Rpb1 CTD.
Functional overlap of the Spt5 and Pol II CTDs.
The CTD of the S. pombe Pol II large subunit Rpb1, which consists of 29 heptad repeats of the consensus sequence YSPTSPS (4) (Fig. 3), binds directly to Pct1 and Pce1 (34) and thus provides an alternative docking site for the capping enzymes on the Pol II elongation complex, potentially buffering the impact of deleting the Spt5 CTD. If so, we reasoned that deleting a portion of the Rpb1 CTD might exacerbate the growth defect of spt5-ΔC. The effects of incremental shortening of the Pol II CTD have been studied extensively in budding yeast (31, 54), yielding powerful genetic insights into transcriptional control mechanisms (48). The role of Pol II CTD length in S. pombe has received comparatively little attention (23).
FIG. 3.
Genetic interactions of rpb1-ΔCTD mutants with spt5-ΔC. The amino acid sequence of the S. pombe Rpb1 CTD (aa 1551 to 1752) is shown at right; the 29 heptapeptide repeats are stacked vertically. The C termini of the rpb1-ΔCTD mutants are indicated by asterisks. (A) Truncating the Rpb1 CTD impairs growth. S. pombe strains bearing the indicated rpb1 alleles were tested for growth on YES agar. Aliquots of serial 5-fold dilutions of cultures that had been adjusted to an A600 of 0.1 were spotted onto YES agar and incubated at the indicated temperatures. The plates were photographed after incubation for 7 days at 20°C, 4 days at 25°C, 3 days at 30°C, or 2 days at 37°C. rpb1 alleles are named according to the length of the Rpb1 protein variants; the numbers of heptad repeats are indicated. (B) Phenotypic enhancement of rpb1-ΔCTD mutations by spt5-ΔC. Viable haploid double mutants harboring spt5-ΔC and the indicated CTD truncations of rpb1 were analyzed by spotting on YES agar. The plates were photographed after incubation for 3 days at 30, 34, or 37°C. (C) Aberrant morphology of spt5-ΔC rpb1-ΔCTD double mutants. Aliquots of exponentially growing cultures were spotted on slides, and the cells were visualized by differential interference contrast microscopy. All images, including the highlighted inserts, were taken at the same magnification. Bars, 10 μm.
Here we integrated a series of rpb1-ΔCTD::natMX alleles into the rpb1+ locus in diploid cells, so as to express Rbp1 variants with 26, 20, 16, 13, 12, 11, 10, or 8 heptad repeats (named rpb1-26, rpb1-20, etc.) (Fig. 3). By sporulating the diploids and characterizing the haploid progeny, we established a clear hierarchy of CTD length effects on cell growth. For example, eight heptad repeats were lethal, i.e., we never recovered nourseothricin-resistant haploids after sporulation of rpb1+/rpb1-8 diploids. rpb1-10 cells were viable, but they grew poorly at 30°C and failed to form colonies at 20°C or 37°C (Fig. 3A). rpb1-11, -12, and -13 cells showed gradually improved growth at the low and high temperatures. The rpb1-16, rpb1-20, and rpb1-26 strains grew as well as the rpb1+ strain at all temperatures tested (Fig. 3A). These results indicate that 16 heptad repeats comprise a fully active Pol II CTD in fission yeast. Overexpression of Mce1 had no salutary effect on the conditional growth defects of the rpb1-10, -11, -12, and -13 strains (not shown), suggesting that additional Pol II functions were perturbed by these CTD truncations.
To illuminate genetic interactions between the Pol II and Spt5 CTDs, we analyzed haploid progeny from genetic crosses between the spt5-ΔC strain and several of the rpb1-ΔCTD mutants. The rpb1-26 spt5-ΔC and rpb1-20 spt5-ΔC double mutants were recovered at the expected frequencies (20 to 25% of all spores analyzed). However, these strains were sicker than the spt5-ΔC single mutant at 30 to 37°C (Fig. 3B) and failed to form colonies at lower temperatures (data not shown). Light microscopy inspection of rpb1-26 spt5-ΔC and rpb1-20 spt5-ΔC cells grown in liquid culture at 30°C revealed a high percentage with elongated and aberrant shapes (Fig. 3C).
Genetic crosses between the spt5-ΔC strain and the rpb1-16 or rpb1-13 strain yielded double mutants that comprised ∼9% and ∼0.15%, respectively, of all haploid progeny, signifying progressive synthetic interactions with CTD-less Spt5 as the Pol II CTD was shortened below a critical threshold. Indeed, the rare surviving rpb1-13 spt5-ΔC strains could not be propagated when restreaked, and the rpb1-16 spt5-ΔC strain formed microscopic colonies only (data not shown). Finally, we failed to recover any rpb1-12 spt5-ΔC double mutants among ∼4,500 haploid progeny screened. These findings of synthetic lethality and synthetic sickness suggest that the C-terminal repeat domains of Rpb1 and Spt5 overlap functionally.
Alanine scanning of the Spt5 TPAWNSGSK nonamer: effects on growth and morphology.
To dissect the requirements for individual residues within the consensus repeat sequence T1P2A3W4N5S6G7S8K9, we mutated the chromosomal spt5 locus by replacing Thr1, Pro2, Trp4, Asn5, or Lys9 with alanine in each of 7 or 8 consecutive nonamer repeats that were fused to the C terminus of Spt5-(1-800) (Fig. 4A). Adding seven repeats of the wild type consensus sequence TPAWNSGSK to Spt5-(1-800) served as the positive control, insofar as the resulting spt5-(CTD)7 “wild-type” strain grew as well as spt5+ at all temperatures tested (Fig. 4; see also Fig. 7). Analysis of the CTD-Ala mutants showed that spt5-(N5A)8 and spt5-(K9A)7 formed wild-type-size colonies at all temperatures (Fig. 4A). In contrast, the spt5-(W4A)8 phenotype mirrored that of spt5-ΔC with respect to slow growth and cold sensitivity (Fig. 4A and D), signifying that single Trp-to-Ala mutations eliminated virtually all beneficial effects of the CTD on Spt5 function in vivo. spt5-(T1A)7 and spt5-(P2A)7 cells grew as well as wild-type cells at 25, 30, and 32°C but were slower growing at 37°C and formed only tiny colonies at 18 to 20°C (Fig. 4A and D). Thus, the T1A and P2A mutations were much milder than ΔC and W4A with respect to cell growth. Thr1 is the site of phosphorylation of the Spt5 CTD by the Cdk9/Pch1 kinase (33); the adjacent Pro2 completes the (S/T)P recognition motif for cyclin-dependent kinases. The finding of concordantly diminished CTD function at restrictive temperatures in T1A and P2A mutants suggests that defective CTD phosphorylation might underlie the observed growth defects. Note that the Spt5-CTD mutant variants were expressed to comparable levels as judged by Western blot analysis (Fig. 4B and other data not shown).
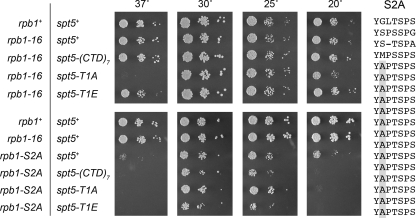
FIG. 7.
Effects of spt5-CTD mutations in combination with rpb1-S2A. The amino acid sequence of the S. pombe Rpb1 CTD-S2A variant is shown at right. The heptapeptide repeats are stacked vertically with the mutated Ser2 positions shaded in gray. The indicated fission yeast strains were maintained in logarithmic growth at 30°C in YES medium. The cultures were adjusted to an A600 of 0.01, and aliquots of 10-fold serial dilutions were spotted on YES agar medium. The genotypes of the strains with respect to the rpb1 and spt5 loci are indicated on the left. The plates shown in the upper panels were incubated for either 2 days at 37 and 30°C, 3 days at 25°C, or 5 days at 20°C. The plates shown in the lower panels were incubated for either 2 days at 37 and 30°C, 4 days at 25°C, or 7 days at 20°C.
We compared the cell size distributions of spt5-CTD-Ala mutants grown at 30°C in rich medium (Fig. 4C). Whereas spt5-(N5A)8 and spt5-(K9A)7 cells resembled spt5-(CTD)7 and only 3 to 5% were 13 to 15 μm long, the fractions of spt5-(T1A)7, spt5-(P2A)7, and spt5-(W4A)8 cells that were 13 to 15 μm long were 21, 24, and 23%, respectively (Fig. 4C). Only spt5-(W4A)8 had a significant population of cells that were >15 μm long (Fig. 4C) and aberrantly branched cells (data not shown). Overexpression of Mce1, which suppressed the growth defect of spt5-ΔC cells, also reversed the slow-growth phenotype of spt4-(W4A)8 cells at 25, 30, and 32°C (not shown). However, Mce1 overexpression did not relieve the cold sensitivity of spt5-(T1A)7 cells (not shown).
Effects of Spt5-CTD-Ala mutations on interaction with fission yeast capping enzymes.
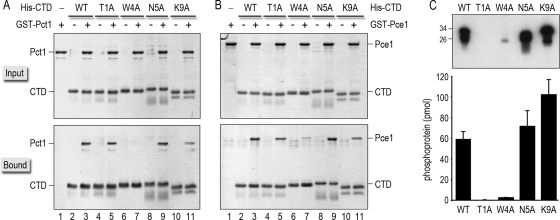
To illuminate the basis for the phenotypes of spt5-CTD-Ala mutants, we queried their effects on binding to Pct1 and Pce1. Recombinant GST-Pct1 and GST-Pce1 proteins were produced in bacteria and purified from soluble lysates by affinity chromatography. The Spt5-CTD proteins were produced in bacteria as N-terminal His10-Smt3 fusions. Whereas the recombinant wild-type, T1A, W4A, N5A, and K9A CTDs were readily isolated from soluble lysates by Ni2+ affinity chromatography, the P2A mutant was intractably insoluble and therefore not amenable to biochemical study. To assay CTD-capping enzyme interactions, the recombinant Pct1 and Pce1 proteins were mixed with the wild-type or mutant His10-Smt3-Spt5-CTD proteins and then adsorbed to Ni-agarose beads. The beads were recovered and washed extensively before the bound proteins were eluted with 0.5 M imidazole. The input and bound polypeptides were then analyzed by SDS-PAGE (Fig. 5A and B). Whereas each of the His10-Smt3-Spt5-CTD proteins was absorbed to the Ni-agarose resin via the His10 tag (lanes 2 to 11), the GST-tagged Pct1 and Pce1 proteins per se did not bind to Ni-agarose (lanes 1). However, when mixed with wild-type His10-Smt3-Spt5-CTD, Pct1 and Pce1 were recovered in the bound fraction (lanes 3). Pct1 and Pce1 bound to the T1A and N5A CTD mutants (lanes 5 and 9). They also bound to the K9A mutant, though their extents of binding to the K9A mutant were consistently lower than those to the other CTDs (lane 11). The instructive findings were as follows: (i) no Pct1 was recovered in the bound fraction when incubated with the W4A mutant (Fig. 5A, lane 7), and (ii) only a trace amount of Pce1 bound to the W4A mutant CTD versus the wild-type CTD (Fig, 5B, lane 7). These findings, together with those in the preceding section, suggest the following: (i) that impaired Spt5-capping enzyme interactions account for, or contribute to, the observed growth defect of spt5-(W4A)8 cells (an idea consistent with the rescue of the spt5-(W4A)8 growth defect by Mce1 overexpression) and (ii) the conditional growth defects of spt5-(T1A)7 and spt5-(P2A)7 cells have other causes.
FIG. 5.
Effects of Spt5 CTD-Ala mutations on binding to capping enzymes and on CTD phosphorylation by Cdk9. (A and B) Binding of GST-tagged Pct1 (A) or Pce1 (B) to wild-type and Ala-substituted His10-Smt3-Spt5-CTD proteins was assessed by Ni-agarose affinity chromatography as described in Materials and Methods. The input proteins are specified above the lanes by “+.” Aliquots comprising 10% of the input material (top panels) and 30% of the bead-bound material (bottom panels) were analyzed by SDS-PAGE. Polypeptides were visualized by staining the gels with Coomassie blue dye. (C) Kinase reaction mixtures (20 μl) containing 50 mM Tris acetate (pH 6.0), 1 mM DTT, 2.5 mM MnCl2, 50 μM [γ32P]ATP, ∼100 ng of recombinant Cdk9T212E/Pch1 kinase (36), and 1 μg of recombinant His10-Smt3-Spt5-CTD phosphoacceptor as specified were incubated for 1 h at 20°C. The reactions were quenched by adding SDS to a 1% final concentration. Aliquots (3 μl) of the reaction mixtures were then analyzed by 12% SDS-PAGE. The 32P-labeled proteins were visualized by autoradiography of the dried gel (top panel). The positions and sizes (kDa) of marker polypeptides are indicated on the left. The extents of label transfer from [γ32P]ATP to His10-Smt3-Spt5-CTD were quantified by scanning the gel with a Molecular Dynamics Typhoon PhosphorImager. The data were normalized to the initial reaction volume and are plotted as a bar graph in the bottom panel. Each datum is the average of results from three or four separate experiments ± SD.
Mutational effects on Spt5 CTD phosphorylation by Cdk9.
The Spt5 CTD is a substrate for threonine phosphorylation by S. pombe Cdk9/Pch1 (33, 49). The Cdk9 kinase is regulated by the S. pombe Cdk-activating kinase Csk1, which phosphorylates Cdk9 on Thr212 (36). The phosphomimetic mutant Cdk9T212E is activated constitutively (33, 36). Here we assessed the primary structure requirements for Spt5 phosphorylation by reacting recombinant wild-type and alanine-substituted His10-Smt3-Spt5-CTD proteins (1 μg; ∼50 pmol) with recombinant Cdk9T212E/Pch1 kinase and [γ32P]ATP. The reaction products were analyzed by SDS-PAGE, and the extents of 32P label transfer to the Spt5 CTD phosphoacceptors were quantified (Fig. 5C). Whereas 60 pmol of 32Pi was incorporated into the wild-type CTD, we detected no label transfer to the T1A CTD mutant (Fig. 5C). This result confirms previous inferences from phosphoamino acid analysis (33) that Thr1 is the direct target for Cdk9 phosphorylation. The N5A and K9A mutants were as good or better as substrates for Cdk9 than wild-type Spt5 CTD (Fig. 5C), consistent with the benign effect of the N5A and K9A changes on Spt5 activity in vivo. In contrast, the W4A mutation reduced the extent of CTD phosphorylation by a factor of 20 (Fig. 5C). The fact that Trp4 is critical for all aspects of Spt5 CTD function tested (cell growth and morphology, capping enzyme binding, and phosphorylation by Cdk9) implies that Trp4 is essential for proper folding of the Spt5 CTD, whether it be intrinsic to the CTD or templated by proteins that interact with the CTD.
Threonine-to-glutamate substitutions in the Spt5 CTD.
We replaced the chromosomal spt5+ locus with a T1E mutant in which Thr1 was changed to glutamate (a phosphomimetic) in each of 7 nonamer repeats fused to Spt5-(1-800). The spt5-(T1E)7 cells grew as well as wild-type spt5-(CTD)7 cells at all temperatures tested (Fig. 4D), and they also displayed wild-type morphology (not shown). These findings contrast with the cold-sensitive growth defect and modestly elongated shape of the spt5-(T1A)7 mutant (Fig. 4C and D). We surmise that Thr1 phosphorylation of the Spt5 CTD is important in vivo and there is no obvious penalty to a constitutive phosphomimetic state.
Spt5 CTD T1A and P2A mutations confer sensitivity to 6-azauracil.
The ribonucleotide-depleting drug 6-azauracil (6-AU) slows the growth of yeast strains carrying mutations in genes that encode proteins involved in transcription elongation (3, 28, 43, 46). To determine whether spt5-CTD-Ala mutant strains are sensitive to 6-AU, we tested their growth on agar plates containing 0, 200, or 300 μg/ml 6-AU (Fig. 6). In parallel, we assessed the effect of 6-AU on growth of cells carrying the spt5-CTD-Ala rpb1-16 and spt5-CTD-Ala rpb1-12 alleles (Fig. 6). The spt5-(N5A)8 and spt5-(T1E)7 mutants grew comparably to the spt5-(CTD)7 strain in the rpb1+, rpb1-16, and rpb1-12 strain backgrounds. In contrast, the CTD T1A and P2A mutations sensitized S. pombe to 300 μg/ml 6-AU in the rpb1+ background, resulting in slowed growth (Fig. 6, top panels). Whereas truncating the Rpb1 CTD to 16 and 12 heptad repeats had little or no effect on 6-AU sensitivity per se, the rpb1-16 and rpb1-12 alleles progressively exacerbated the 6-AU sensitivity of the spt5-(T1A)7 and spt5-(P2A)7 mutants (Fig. 6, middle and bottom panels). The distinctive effects of Thr1 and Pro2 mutations on 6-AU sensitivity imply a positive role for Spt5 CTD phosphorylation in transcription elongation in fission yeast.
FIG. 6.
Spt5 CTD T1A and P2A mutations sensitize fission yeast to growth inhibition by 6-azauracil. Exponentially growing cultures of S. pombe strains with the indicated rbp1 and spt5 genotypes were adjusted to an A600 of 0.1, and aliquots (3 μl) of serial 5-fold dilutions were spotted on synthetic agar medium lacking uracil and containing 6 mM NH4OH and 0, 200, or 300 μg/ml 6-azauracil (6-AU) as specified. The plates were incubated at 30°C for 4 days (no 6-AU), 5 days (200 μg/ml 6-AU), or 5.5 days (300 μg/ml 6-AU).
Combining Pol II CTD Ser2 and Spt5 CTD Thr1 mutations.
S. pombe Cdk9 catalyzes threonine phosphorylation of the Spt5 CTD and serine phosphorylation of the Pol II CTD (33). Probing of the Cdk9-phosphorylated Pol II CTD product with various antibodies suggested that Cdk9 acts on Ser2 and Ser5 of the YSPTSPS heptad (13, 36). Most of the bulk Ser2 phosphorylation of the fission yeast Rpb1 CTD in vivo depends on a different nonessential Cdk enzyme named Lsk1 (23). Nonetheless, a fraction of Rpb1 Ser2 phosphorylation in vivo appears to require Cdk9, as inferred from experiments with analog-sensitive kinase mutants (49). The finding that a mutated Rpb1 CTD in which Ser2 was replaced by alanine could sustain S. pombe viability, albeit with conditional defects in cytokinesis (23), had two important ramifications: (i) that Ser2 phosphorylation is not globally essential for Pol II transcription and (ii) that the essentiality of the S. pombe Cdk9 kinase (36) reflects its phosphorylation of a critical target other than Rpb1 CTD-Ser2. One obvious candidate is the Spt5 CTD Thr1, which, though also not essential, might overlap functionally with Rpb1 Ser2. To evaluate this scenario, we tested for mutational synergy between an Rpb1 CTD-S2A mutation and an Spt5 CTD T1A mutation.
We constructed an rpb1-S2A allele that encodes 14 tandem repeats of the mutant heptad YAPTSPS fused to the first 4 imperfect heptad variants of the wild-type Rpb1 subunit (Fig. 7). Our version of S2A has two more mutant heptads than that described previously (23). The haploid S. pombe rpb1-S2A strain grew as well as the rpb1+ and rpb1-16 (which contains a “wild type” CTD sequence of similar length) strains at 30°C but was slower growing at 25°C and extremely sick at 20 and 37°C (Fig. 7, bottom panels).
To assay mutational synergy, we combined the rpb1-S2A allele with spt5-(CTD)7, spt5-(T1A)7, and spt5-(T1E)7. The relevant findings were as follows: (i) truncating the Spt5 CTD to 7 “wild type” nonamer repeats (which had no effect in an rpb1-16 background) exacerbated the rpb1-S2A cold-sensitive phenotype (see results for 20°C); (ii) there was no synthetic growth defect for the rpb1-S2A spt5-(T1A)7 double mutant; (iii) the constitutive Spt5 phosphomimetic spt5-(T1E)7 mutation did not suppress the conditional growth defects of rpb1-S2A (Fig. 7). We surmise that phosphorylations of Rpb1 Ser2 and/or Spt5 Thr1 are not essential for viability of S. pombe and that Cdk9 phosphorylates another essential target site in vivo.
DISCUSSION
The Spt5 and Pol II CTDs perform overlapping essential functions.
We show here that the CTD nonamer repeat array of fission yeast Spt5 is important for normal physiology, insofar as its complete deletion elicits a constitutive slow-growth defect and aberrant cell morphology. The spt5-ΔC phenotype is mimicked by replacing Trp4 in each TPAWNSGSK nonapeptide with alanine, which attests to a likely structural role for this defining residue of the S. pombe Spt5 CTD. The Spt5 homologs of other eukarya also have CTD arrays with repeating (Thr/Ser)Pro motifs, albeit less regular in their spacing and primary structure than that of S. pombe (32). In particular, whereas the residue located two positions downstream of the Thr-Pro dipeptide in S. pombe Spt5 is a tryptophan in 12 of the CTD repeats (Fig. 2), it is never a tryptophan in the human, nematode, or zebra fish proteins. The Spt5 CTD of budding yeast S. cerevisiae is atypical in that it contains 15 tandem repeats of a proline-free hexapeptide motif of the consensus sequence S(A/T)WGG(A/Q), in which the first serine residue is a substrate for phosphorylation by the cyclin-dependent Bur1 kinase (26, 58). Budding yeast Bur1 is the ortholog of fission yeast Cdk9 (33, 35). Immediately upstream of this CTD hexapeptide array in S. cerevisiae Spt5 is a pair of Thr-Pro dipeptides, each of which is embedded in a short motif—905TPGWSS and 916TPAVNA—that bears some similarity to the S. pombe Spt5 CTD repeat unit. Probing of cell extracts with phosphoamino acid antibodies suggested that S. cerevisiae Spt5 contains phosphothreonine (58), though the threonine phosphorylation sites were not identified.
Notwithstanding these differences in CTD structure, there is an emerging consensus from genetic studies that the Spt5 CTD plays an important, but not strictly essential, role in Spt5 function in vivo. To wit, whereas knockdown of human Spt5 blocked proliferation of HeLa cells, cell growth was restored by a human Spt5 mutant lacking the CTD repeat module (24). This result prompted speculation of functional redundancy of the human Spt5 CTD. Deletion of the hexapeptide CTD repeat array of S. cerevisiae Spt5 had no effect on cell growth at 30°C but resulted in slow growth at 16°C (26) and sensitivity to 6-AU (58). (Note that the spt5-ΔCTD growth phenotype in budding yeast is less severe than that in S. pombe and appears more akin to the effects of the S. pombe spt5-T1A mutant.)
To explore the potential functional redundancies of the S. pombe Spt5 CTD, we focused on the Rbp1 CTD heptad array, reasoning that because both CTDs bind proteins that act cotranscriptionally and receive inputs via phosphorylation of their component repeats, the presence of one of the CTDs might buffer the loss or truncation of the other CTD. Here we conducted the first incremental deletion analysis of the S. pombe Rpb1 CTD, which revealed the following: (i) 16 of the 29 heptad repeats are needed for wild-type growth on standard medium; (ii) less than 10 heptads is constitutively lethal; and (iii) serial trimmings between 13 and 10 heptads progressively worsen cell growth. Two prior studies had each documented a single viable S. pombe rpb1-ΔCTD allele, though their nomenclatures differed from each other and from ours with respect to heptad counting. By reference to the heptad alignment in Fig. 2, the viable but slow-growing rpb1-11 allele in the work of Schramke et al. (40) contained 12 repeats, and the rpb1-12xCTD allele of Karagiannis and Balasubramanian (23) contained 16 repeats.
A key finding of our study is that the otherwise viable CTD truncation mutations rpb1-11, -12, and -13 are synthetic lethal with spt5-ΔC, while rpb-16 and spt5-ΔC are synthetically very sick. Even a modest shortening of the Pol II CTD to 20 repeats exacerbated the spt5-ΔC growth defect. These results provide evidence for an essential overlapping function(s) of the Spt5 and Pol II CTDs in fission yeast. Similar inferences were drawn for budding yeast based on the observation that the viable spt5-ΔCTD allele was synthetic lethal with a viable ctk1Δ null allele that lacks the major yeast Rbp1 CTD-Ser2 kinase Ctk1 (26), a result taken to mean that Pol II Ser2 phosphorylation and Spt5 CTD phosphorylation are the functionally overlapping principles. However, this scenario cannot pertain in fission yeast, where we see no synergy between an spt5-T1A mutation that eliminates the Spt5 CTD phosphorylation site and an rpb1-S2A mutation.
Fortifying the CTD connection to capping enzymes in S. pombe.
The fission yeast capping enzymes RNA triphosphatase (Pct1) and RNA guanylyltransferase (Pce1) are separately encoded essential proteins that bind independently to the phosphorylated Rbp1 CTD heptad array and to the unphosphorylated S. pombe Spt5 CTD nonamer array (32, 34). The mammalian capping enzyme Mce1 is a single modular polypeptide composed of N-terminal triphosphatase and C-terminal guanylyltransferase domains. The Mce1 guanylyltransferase domain binds to the phosphorylated Pol II CTD and thereby ferries the covalently tethered triphosphatase module, which does not bind to the Pol II CTD on its own, to the mammalian Pol II elongation complex (18, 19). Both isolated domains of Mce1 can bind to human Spt5 via its CTD, and this interaction increases the efficiency of contranscriptional capping, especially when the Pol II CTD is in the dephosphorylated state (27, 52). It is noteworthy that Mce1 does not bind to the S. pombe Spt5 CTD (32). Our finding here that Mce1 is a dosage suppressor of the growth defect and elongated shape of the spt5-ΔC mutant attests that capping enzyme recruitment is likely a limiting factor in the absence of the Spt5 CTD. Overexpression of Mce1, or combined overexpression of Pct1 and Pce1, presumably drives their binding directly to the phosphorylated Rbp1 CTD, either by simple mass action or by competition with other cellular CTD-binding proteins. An appealing scenario, admittedly speculative at this stage, is that the fission yeast triphosphatase and guanylyltransferase bind initially to the unphosphorylated Spt5 CTD and are then handed off to the nearby Poll II CTD within the same transcription complex after Rbp1 is phosphorylated on Ser5. It is noteworthy that capping enzyme overexpression does not alleviate the slow growth of spt5-ΔC cells at low temperatures, indicating that the Spt5 CTD has other functions beyond capping.
A genetically separable contribution of Spt5 to transcription elongation relies on threonine phosphorylation.
Replacing the Spt5 CTD Thr1 side chain with alanine abolished its ability to serve as a phosphoacceptor substrate for Cdk9 without impacting the interactions with the capping enzymes Pct1 and Pce1. The T1A mutation elicited a milder phenotype in vivo than did complete deletion of the Spt5 CTD, said phenotype comprising cold-sensitive (cs) and temperature-sensitive (ts) growth (that was unaffected by Mce1 overexpression) and sensitivity to growth inhibition by 6-AU. The cs and ts growth and 6-AU sensitivity are both suppressed by the phosphomimetic T1E change, thereby implicating deficient Spt5 CTD phosphorylation as the culprit in a putative transcription elongation abnormality in T1A cells. 6-AU sensitivity of T1A cells was enhanced by shortening the Pol II CTD to 16 heptad repeats, though the rpb1-16 allele itself caused no 6-AU sensitivity. This trend progressed as spt5-T1A was combined with the rpb1-12 allele. We surmise from these results that the Pol II CTD and the threonine-phosphorylated Spt5 CTD play overlapping positive roles in elongation. However, as noted above, the overlap is not at the level of Ser2 phosphorylation of the Rpb1 CTD array, because there was no synergy between rpb1-S2A and spt5-T1A, as gauged by growth on standard medium. We thereby infer the existence of at least one additional critical target for the essential Cdk9 kinase. Conceivably, Cdk9 could phosphorylate a different position within the Rbp1 CTD heptad (Ser5 and/or Ser7) or yet another protein involved in transcriptional control in fission yeast. S. pombe Cdk9 can phosphorylate Rpb1 on Ser5 in vitro (36), and it is appealing to think that Cdk9 phosphorylation of Ser5, coordinated temporally (during the transcription cycle) and spatially (within the linear heptad array) with Cdk7's Ser5 phosphorylation function, might comprise an internal “CTD clock,” analogous to recent discussions of how Cdk7 Ser5 phosphorylation primes subsequent CTD phosphorylations of Ser2 (38, 49).
Acknowledgments
This work was supported by National Institutes of Health grant GM52470. S.S. is an American Cancer Society Research Professor.
Footnotes
Published ahead of print on 15 March 2010.
REFERENCES
- 1.Akhtar, M. S., M. Heidermann, J. R. Tietjen, D. W. Zhang, R. D. Chapman, D. Eick, and A. Z. Ansari. 2009. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol. Cell 34:387-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrulis, E. D., E. Guzman, P. Dumloring, J. Werner, and J. T. Lis. 2000. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 14:2635-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archambault, J., F. LaCroute, A. Ruet, and J. D. Friesen. 1992. Genetic interaction between transcription elongation factor TFIIS and RNA polymerase II. Mol. Cell. Biol. 12:4142-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azuma, Y., M. Yamagashi, R. Ueshima, and A. Ishihama. 1991. Cloning and sequence determination of the Schizosaccharomyces pombe rpb1 gene encoding the largest subunit of RNA polymerase II. Nucleic Acids Res. 19:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, Y., Y. Yamaguchi, Y. Tsugeno, J. Yamamoto, T. Yamada, M. Nakamura, K. Hisatake, and H. Handa. 2009. DISF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation. Genes Dev. 23:2765-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiu, Y. L., C. K. Ho, N. Saha, B. Schwer, S. Shuman, and T. M. Rana. 2002. Tat stimulates cotranscriptional capping of HIV mRNA. Mol. Cell 10:585-597. [DOI] [PubMed] [Google Scholar]
- 7.Cho, E., T. Takagi, C. R. Moore, and S. Buratowski. 1997. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxyl-terminal domain. Genes Dev. 11:3319-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fabrega, C., V. Shen, S. Shuman, and C. D. Lima. 2003. Structure of an mRNA capping enzyme bound to the phosphorylated carboxyl-terminal domain of RNA polymerase II. Mol. Cell 11:1549-1561. [DOI] [PubMed] [Google Scholar]
- 9.Forsburg, S. L. 1993. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 21:2955-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forsburg, S. L., and D. A. Sherman. 1997. General purpose tagging vectors for fission yeast. Gene 191:191-195. [DOI] [PubMed] [Google Scholar]
- 11.Forsburg, S. L., and N. Rhind. 2006. Basic methods for fission yeast. Yeast 23:173-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao, L., and D. S. Gross. 2008. Sir2 silences gene transcription by targeting the transition between RNA polymerase II initiation and elongation. Mol. Cell. Biol. 28:3979-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guiguen, A., J. Soutourine, M. Dewez, L. Tafforeau, M. Dieu, M. Raes, J. Vandenhaute, M. Werner, and D. Hermand. 2007. Recruitment of P-TEFb (Cdk9-Pch1) to chromatin by the cap-methyl transferase Pcm1 in fission yeast. EMBO J. 26:1552-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo, M., F. Xu, J. Yamada, T. Egelhofer, Y. Gao, G. A. Hartzog, M. Teng, and L. Niu. 2008. Core structure of the yeast Spt4-Spt5 complex: a conserved module for regulation of transcription elongation. Structure 16:1649-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagler, J., and S. Shuman. 1992. A freeze-frame view of eukaryotic transcription during elongation and capping of nascent mRNA. Science 255:983-986. [DOI] [PubMed] [Google Scholar]
- 16.Hartzog, G. A., M. A. Basrai, S. L. Ricupero-Hovasse, P. Hieter, and F. Winston. 1996. Identification and analysis of a functional human homolog of the SPT4 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2848-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartzog, G. A., T. Wada, H. Handa, and F. Winston. 1998. Evidence that Spt4, Spt5 and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 12:357-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho, C. K., and S. Shuman. 1999. Distinct roles for CTD Ser2 and Ser5 phosphorylation in the recruitment and allosteric activation of mammalian capping enzyme. Mol. Cell 3:405-411. [DOI] [PubMed] [Google Scholar]
- 19.Ho, C. K., V. Sriskanda, S. McCracken, D. Bentley, B. Schwer, and S. Shuman. 1998. The guanylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 273:9577-9585. [DOI] [PubMed] [Google Scholar]
- 20.Hong, S. W., S. M. Hong, J. W. Yoo, Y. C. Lee, S. Kim, J. T. Lis, and D. Kee. 2009. Phosphorylation of the RNA polymerase II C-terminal domain by TFIIH kinase is not essential for transcription of Saccharomyces cerevisiae genome. Proc. Natl. Acad. Sci. U. S. A. 106:14276-14280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jennings, B. H., S. Shah, Y. Yamaguchi, M. Seki, R. G. Phillips, H. Handa, and D. Ish-Horowicz. 2004. Locus-specific requirements for Spt5 in transcriptional activation and repression in Drosophila. Curr. Biol. 14:1680-1684. [DOI] [PubMed] [Google Scholar]
- 22.Kanin, E. I., R. T. Kipp, C. Kung, M. Slattery, A. Viale, S. Hahn, K. M. Shokat, and A. Z. Ansari. 2007. Chemical inhibition of the THIIH-associated kinase Cdk7/Kin28 does not impair global mRNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 104:5812-5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karagiannis, J., and M. K. Balasubramanian. 2007. A cyclin-dependent kinase that promotes cytokinesis through modulating phosphorylation of the carboxy terminal domain of the RNA Pol II Rpb1p subunit. PLoS One 2:e433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komori, T., N. Inukai, T. Yamada, Y. Yamaguchi, and H. Handa. 2009. Role of human transcription elongation factor DSIF in the suppression of senescence and apoptosis. Genes Cells 14:343-354. [DOI] [PubMed] [Google Scholar]
- 25.Krishnan, K., N. Salomonis, and S. Guo. 2008. Identification of Spt5 target genes in zebrafish development reveals its dual activity in vivo. PLoS One 3:e3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, Y., L. Warfield, C. Zhang, J. Luo, J. Allen, W. H. Lang, J. Ranish, K. M. Shokat, and S. Hahn. 2009. Phosphorylation of the transcription factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol. Cell. Biol. 29:4852-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandal, S. S., C. Chu, T. Wada, H. Handa, A. J. Shatkin, and D. Reinberg. 2004. Functional interaction of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc. Natl. Acad. Sci. U. S. A. 101:7572-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mason, P. B., and K. Struhl. 2005. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell 17:831-840. [DOI] [PubMed] [Google Scholar]
- 29.McCracken, S., N. Fong, E. Rosonina, K. Yankulov, G. Brothers, D. Siderovski, A. Hessel, S. Foster, S. Shuman, and D. L. Bentley. 1997. 5′ Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated C-terminal domain of RNA polymerase II. Genes Dev. 11:3306-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meinhart, A., T. Kamenski, S. Hoeppner, S. Baumli, and P. Cramer. 2005. A structural perspective of CTD function. Genes Dev. 19:1401-1415. [DOI] [PubMed] [Google Scholar]
- 31.Nonet, M., D. Sweetser, and R. A. Young. 1987. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell 50:909-915. [DOI] [PubMed] [Google Scholar]
- 32.Pei, Y., and S. Shuman. 2002. Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. J. Biol. Chem. 277:19639-19648. [DOI] [PubMed] [Google Scholar]
- 33.Pei, Y., and S. Shuman. 2003. Characterization of the Schizosaccharomyces pombe Cdk9/Pch1 protein kinase: Spt5 phosphorylation, autophosphorylation and mutational analysis. J. Biol. Chem. 278:43346-43356. [DOI] [PubMed] [Google Scholar]
- 34.Pei, Y., S. Hausmann, C. K. Ho, B. Schwer, and S. Shuman. 2001. The length, phosphorylation state, and primary structure of the RNA polymerase II carboxyl-terminal domain dictate interactions with mRNA capping enzymes. J. Biol. Chem. 276:28075-28082. [DOI] [PubMed] [Google Scholar]
- 35.Pei, Y., B. Schwer, and S. Shuman. 2003. Interactions between fission yeast Cdk9, its cyclin partner Pch1, and mRNA capping enzyme Pct1 suggest an elongation checkpoint for mRNA quality control. J. Biol. Chem. 278:7180-7188. [DOI] [PubMed] [Google Scholar]
- 36.Pei, Y., H. Du, J. Singer, St. C. Amour, S. Granitto, S. Shuman, and R. P. Fisher. 2006. Cdk9 of fission yeast is activated by the CDK-activating kinase Csk1, overlaps functionally with the TFIIH-associated kinase Mcs6, and associates with the cap methyltransferase Pcm1 in vivo. Mol. Cell. Biol. 26:777-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phatnani, H. P., and A. L. Greenleaf. 2006. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 20:2922-2936. [DOI] [PubMed] [Google Scholar]
- 38.Qiu, H., C. Hu, and A. G. Hinnebusch. 2009. Phosphorylation of the Pol II CTD by KIN29 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol. Cell 33:752-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Renner, D. B., Y. Yamaguchi, T. Wada, H. Handa, and D. H. Price. 2001. A highly purified RNA polymerase II elongation control system. J. Biol. Chem. 276:42601-42609. [DOI] [PubMed] [Google Scholar]
- 40.Schramke, V., D. M. Sheedy, A. M. Denli, C. Bonila, K. Ekwall, G. J. Hannon, and R. C. Allshire. 2005. RNA-interference-directed chromatin modification coupled to RNA polymerase II transcription. Nature 435:1275-1279. [DOI] [PubMed] [Google Scholar]
- 41.Schwer, B., X. Mao, and S. Shuman. 1998. Accelerated mRNA decay in conditional mutants of yeast mRNA capping enzyme. Nucleic Acids Res. 26:2050-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwer, B., S. Schneider, Y. Pei, A. Aronova, and S. Shuman. 2009. Characterization of the Schizosaccharomyces pombe Spt5-Spt4 complex. RNA. 15:1241-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shaw, R. J., and D. Reines. 2000. Saccharomyces cerevisiae transcription elongation mutants are defective in PUR5 induction in response to nucleotide depletion. Mol. Cell. Biol. 20:7427-7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shim, E. Y., A. K. Walker, Y. Shi, and K. T. Blackwell. 2002. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev. 16:2135-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sims, R. J., R. Belotserkovskaya, and D. Reinberg. 2004. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 18:2437-2468. [DOI] [PubMed] [Google Scholar]
- 46.Squazzo, S. L., P. J. Costa, D. L. Lindstrom, K. E. Kumer, R. Simic, J. L. Jennings, A. J. Link, K. M. Arndt, and G. A. Hartzog. 2002. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21:1764-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swanson, M. S., E. A. Malone, and F. Winston. 1991. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol. Cell. Biol. 11:3009-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson, C. M., A. J. Koleske, D. M. Chao, and R. A. Young. 1993. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell 73:1361-1375. [DOI] [PubMed] [Google Scholar]
- 49.Viladevall, L., St. C. V. Amour, A. Rosebrock, S. Schneider, C. Zhang, J. J. Allen, K. M. Shokat, B. Schwer, J. K. Leatherwood, and R. P. Fisher. 2009. TFIIH and P-TEFb coordinate transcription with capping enzyme recruitment at specific genes in fission yeast. Mol. Cell 33:738-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. A. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12:343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wada, T., T. Takagi, Y. Yamaguchi, D. Watanabe, and H. Handa. 1998. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17:7395-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wen, Y., and A. J. Shatkin. 1999. Transcription elongation factor hSpt5 stimulates mRNA capping. Genes Dev. 13:1774-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wenzel, S., B. M. Martins, P. Rörsch, and B. M. Wöhrli. 2010. Crystal structure of the human transcription elongation factor DSIF hSpt4 subunit in complex with the hSpt5 dimerization interface. Biochem. J. 425:373-380. [DOI] [PubMed] [Google Scholar]
- 54.West, M. L., and J. L. Corden. 1995. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics 140:1223-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu, C. H., Y. Yamaguchi, L. R. Benjamin, M. Horvat-Gordon, J. Washinky, E. Enerly, J. Larsson, A. Lambertsson, H. Handa, and D. Gilmour. 2003. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 17:1402-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamada, T., Y. Yamaguchi, N. Inukai, S. Okamoto, T. Mura, and H. Handa. 2006. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell 21:227-237. [DOI] [PubMed] [Google Scholar]
- 57.Yue, Z., E. Maldonado, R. Pillutla, H. Cho, D. Reinberg, and A. J. Shatkin. 1997. Mammalian capping enzyme complements mutant S. cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc. Natl. Acad. Sci. U. S. A. 94:12898-12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou, K., W. H. W. Kuo, J. Fillingham, and J. F. Greenblatt. 2009. Control of transcriptional histone modification by the yeast BUR kinase substrate Spt5. Proc. Natl. Acad. Sci. U. S. A. 106:6956-6961. [DOI] [PMC free article] [PubMed] [Google Scholar]