Abstract
Cardioviruses (e.g., Theiler's murine encephalomyelitis virus [TMEV]) are members of the Picornaviridae family that cause myocarditis and encephalitis in rodents. Recently, several studies have identified human cardioviruses, including Saffold virus (SAFV) and a related virus named human TMEV-like cardiovirus (HTCV). At least eight cardiovirus genotypes are now recognized, with SAFV and most strains of HTCV belonging to genotypes 1 and 2, respectively; genotype 2 strains are the most common in the population. Although a genotype 3 cardiovirus has recently been cultured (SAFV-3), the genotype 1 and 2 cardioviruses have been difficult to propagate in vitro, hindering efforts to understand their seroprevalence and pathogenicity. Here we present the isolation and characterization of a genotype 2 human cardiovirus (HTCV-UC6). Notably, successful cultivation of HTCV-UC6 from stool required the addition of cytokine-blocking antibodies to interrupt downstream antiviral pathways. Unlike SAFV-3, HTCV-UC6 exhibited slow replication kinetics and demonstrated only a moderate cytopathic effect. Serologic assays revealed that 91% of U.S. adults carry antibodies to the genotype 2 cardioviruses, of which 80% generate neutralizing antibodies, in agreement with previous data showing that cardiovirus infection is widespread in humans. We also demonstrate an acute cardiovirus seroconversion event in a child with diarrhea and vomiting, thus reporting for the first time evidence linking cardiovirus infection to diarrheal disease in humans.
Cardioviruses are positive-strand RNA viruses in the Picornaviridae family that have been associated with myocarditis, encephalitis, and demyelinating disease in rodents (4, 12). The Cardiovirus genus consists of two viral species: Theilovirus, the prototype of which is Theiler's murine encephalomyelitis virus (TMEV), and Encephalomyocarditis virus (EMCV). Prior to 2007, the only known candidate human cardiovirus was the Vilyuisk virus, isolated from the cerebrospinal fluid of a patient with encephalitis (14). However, the Vilyuisk virus was isolated only after multiple passages in mouse cell culture and is highly related to known murine cardioviruses, raising concerns that it was in fact a contaminating animal cardiovirus (13).
In 2007, the complete sequence of the first human cardiovirus, named Saffold virus (SAFV), was recovered from an archived 1981 stool culture from an infant with a fever of unknown origin (11). Independently, we used a panviral microarray to identify a related human cardiovirus, designated HTCV (human TMEV-like cardiovirus), in respiratory secretions from a child with an influenza-like illness (isolate HTCV-UC1) (5). Subsequent analysis of a large cohort study of diarrheal transmission revealed similar viruses in six stool samples from U.S. children (HTCV-UC2 to -UC7) (5). Human cardioviruses have since been found in stool samples from both healthy children and children with gastroenteritis in Brazil, Germany, and China (7, 23) and in respiratory samples from children with influenza-like illnesses in Canada (1). They have also been found in stools from South Asian children who either had nonpolio acute flaccid paralysis or were healthy (3). Thus, infection with human cardioviruses appears to be worldwide, with PCR prevalence rates ranging from 0.5% to 12% (3, 5, 7, 23). The high diversity of the human cardioviruses is evidenced by the identification of at least eight VP1 genotypes (3, 12). The discovery of the existence of many human cardiovirus strains related to TMEV has raised a number of questions concerning the epidemiology and pathogenicity of these viruses.
Although a genotype 3 human cardiovirus has recently been cultured (24), related viruses of genotypes 1 and 2 have been difficult to propagate in vitro (7, 11, 24). Here we present the isolation, characterization, and seroepidemiology of a human cardiovirus (HTCV-UC6) of genotype 2, the most common of the eight known genotypes (3, 12). We demonstrate the utility of adding anti-cytokine antibodies to the culture medium to interrupt known downstream antiviral pathways and thus promote viral growth. We also provide the first direct evidence of seroconversion to cardiovirus infection in a child with diarrhea and confirm previous data showing that cardiovirus infection is widespread in humans.
MATERIALS AND METHODS
Cell lines.
The LLC-MK2 (rhesus monkey kidney epithelial), HeLa (human cervical carcinoma), A549 (human lung carcinoma), MRC-5 (human embryonic lung fibroblast), SK-N-BE2 (human neuroblastoma), Caco-2 (human intestinal carcinoma), BSC-1 (monkey kidney epithelial), Vero (monkey kidney epithelial), BHK-21 (baby hamster kidney), and CHO (Chinese hamster ovary) cell lines were obtained from the cell culture facility of the University of California, San Francisco (UCSF). The HDFK (human diploid fetal kidney) cell line was kindly provided by David Schnurr (California Department of Public Health, Richmond, CA). Most cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 1× nonessential amino acids (Invitrogen, Carlsbad, CA), 10% fetal bovine serum, 100 U of penicillin/ml, and 100 μg of streptomycin/ml, with the following exceptions: (i) for the A549 cell line, the medium used was a 1:1 mixture of DMEM and Ham's F12K medium (Invitrogen, Carlsbad, CA), and (ii) for the Caco-2 cell line, 20% fetal bovine serum instead of 10% was used.
Cultivation of HTCV-positive stool samples.
The SIFT (Stanford Infection and Familial Transmission) study is a large prospective study initiated in 1999 to investigate the association between Helicobacter pylori infection and diarrheal disease (16). Both stool and acute- and convalescent-phase serum samples (the convalescent phase is defined as >3 months after the acute phase) were collected from household members with gastroenteritis as well as from asymptomatic contacts. Previously, screening of the SIFT cohort revealed six stool samples harboring cardioviruses (designated HTCV-UC2 through HTCV-UC7) (5).
Approximately 200 mg of stool from each of these six HTCV-positive stool samples was aliquoted and mixed with 2 ml of phosphate-buffered saline (PBS). Stool-PBS mixtures were further diluted 1:10 in PBS and were clarified by centrifugation for 10 min at 4,000 × g and passage through a 0.2-μm-pore-size filter. Cell lines at 80% confluence were infected for 24 h at 37°C and were then washed seven times with PBS, and the medium was replaced with fresh culture medium at day 1. Viral replication was monitored over 14 days by blinded visual inspection under light microscopy for cytopathic effect (CPE) and serial quantitative reverse transcription-PCR (RT-PCR) assays of cell culture supernatants for HTCV at days 3, 7, 10, and 14 postinfection.
The overall details of the quantitative SYBR green real-time RT-PCR assay for HTCV have been described previously (5). Briefly, total RNA was extracted using the Zymo ZR viral RNA kit (Zymo Research, Orange, CA) according to the manufacturer's instructions. Quantitative RT-PCR was performed on a Stratagene MX3005P real-time PCR system with cardiovirus-specific primers CardioUTR-208F (5′-CATTTTCCGGCCCAGGCTAAG-3′) and CardioUTR-308R (5′-GTGAACAAGCGGCAAGGGAG-3′) using the Qiagen One-Step RT-PCR kit (Qiagen, Valencia, CA) with a 12.5-μl PCR mixture containing 2.75 μl H2O, 2.5 μl 5× PCR buffer, 2.5 μl Q solution, 0.5 μl deoxynucleoside triphosphates (dNTP), 0.5 μl RT/Taq solution, 0.75 μl of each primer, 0.25 μl SYBR green, and 2 μl of total RNA. Conditions for the PCR were as follows: 50°C for 30 min; 95°C for 15 min; 40 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s; and a final extension at 72°C for 7 min. An estimate of the viral titer in terms of viral genome equivalents was made using a standard curve derived from an in vitro-transcribed and quantitated cardiovirus mRNA standard (5). Melting curve analysis was employed to verify that the target amplicon was correctly amplified.
For cultivation in anti-cytokine antisera, mouse anti-human alpha interferon (IFN-α) (PBL Interferon Source, Piscataway, NJ) and/or anti-human tumor necrosis factor alpha (TNF-α) (R&D Systems, Minneapolis, MN) antibodies were added to the culture medium at a final concentration of 10 μg/ml. Anti-cytokine antibodies were added on day zero, at the time of infection, and on day 1, after the washes and the replacement of the medium with fresh culture medium.
Northern blot hybridization.
To generate the HTCV-UC6 RNA probe for Northern blotting, total RNA extracted from the HTCV-UC6 stool sample was amplified using 40 cycles of standard RT-PCR with primers CardioUTR-208F and CardioUTR-308R under the same conditions described above for quantitative RT-PCR, except that no SYBR green was used. To generate the poliovirus RNA probe, total RNA extracted from poliovirus type 1 (Sabin strain) was amplified using 40 cycles of standard RT-PCR with primers EV-1F (5′-CGGCCCCTGAATGCGGCTAA-3′) and EV-2R (5′-GAAACACGGACACCCAAAGTA-3′) (17). The ∼100-bp amplicons for cardiovirus and poliovirus were gel purified and cloned using a dual T7/Sp6 promoter TOPO TA cloning kit (Invitrogen, Carlsbad, CA). TOPO TA plasmids containing the ∼100-bp inserts were sequenced to ensure the presence of the correct sequence and to identify the insert orientation. Immediately prior to the hybridization step, radioactive positive- and negative-strand RNA probes were transcribed using the T7/Sp6-Maxiscript in vitro transcription kit (Ambion, Austin, TX) with the incorporation of [α-32P]UTP at 800 Ci/mmol. After synthesis of the RNA probe, the DNA template was removed by digestion with Turbo DNase at 37°C for 1 h by using the Turbo DNA-free kit (Ambion, Austin TX) according to the manufacturer's instructions.
RNA electrophoresis was performed on total RNA extracted from HTCV-UC6 cells at days 3, 7, 10, and 14 postinfection and from poliovirus cells at 4 h postinfection (5 μg/well), as well as on positive-strand in vitro-transcribed mRNA from HTCV-UC1 as a positive control (100 ng/well). The positive-control mRNA was generated by in vitro transcription of the cloned, nearly full length genome of HTCV-UC1 by using the Megascript T7 kit (Ambion, Austin, TX) according to the manufacturer's instructions. RNA samples were loaded onto 1.0% agarose denaturing gels prepared in 1× formaldehyde gel-running buffer, and electrophoresis was run at 100 V for 4 h. A 0.5- to 10-kb RNA ladder was used for size estimation (Invitrogen, Carlsbad, CA). After electrophoresis, gels were rinsed twice in 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and the capillary transfer of gels to Hybond N nylon membranes (Amersham Pharmacia, Sunnyvale, CA) was done overnight in 10× SSC. RNA was fixed to the capillary membranes by exposure to UV light at 254 nm using a Stratalinker 1800 UV cross-linker (Stratagene, La Jolla, CA). Capillary membranes were incubated with a radioactive probe (HTCV-UC6, poliovirus, or β-actin control) overnight at 65°C in a rolling bottle hybridization oven. After a wash, the hybridized probe was detected using a phosphorimager.
Growth curves.
A virus stock of HTCV-UC6 (passage 1 [P1]) was produced in LLC-MK2 cells, aliquoted, and titrated by quantitative RT-PCR. For growth curves, monolayer cells were infected with virus at a calculated titer of 108 viral genome equivalents/ml for 24 h at 37°C. After a 24-h incubation, cells were washed seven times with PBS, and the medium was replaced with fresh culture medium. Cells were then observed daily for evidence of CPE. At days zero, 1 (before and after washing), 3, 7, and 10 postinfection, 10% of the volume of the cell culture supernatant, or 50 μl, was removed for determination of viral titers by quantitative RT-PCR, as described above. Fresh medium was then added to replace the volume that was removed. For the anti-cytokine antibody and viral tropism experiments, cell lines were infected after they had reached 80 to 90% confluence. For the cell density experiments, LLC-MK2 cells were infected with virus at 100%, 90%, 70%, 50%, 40%, 20%, and 5% confluence, and viral titers were calculated at day 10 postinfection.
Direct immunofluorescence.
Direct immunofluorescent assays were performed on LLC-MK2 cells infected with HTCV-UC6 (P1) at days 3, 7, and 10 by using the Pan-Enterovirus diagnostic kit (Millipore, Temecula, CA) according to the manufacturer's instructions.
Genome sequencing.
Total RNA was extracted from stool suspensions harboring HTCV-UC6 using the Zymo ZR viral RNA kit (Zymo Research, Orange, CA) according to the manufacturer's instructions. Specific RT-PCR was done with the Qiagen One-Step RT-PCR kit (Qiagen, CA) using the same conditions given above for quantitative RT-PCR, except that no SYBR green was used and extension was performed at 72°C for 1 min/kb. Amplified cDNA libraries were generated from extracted HTCV-UC6 RNA and were subjected to deep sequencing on an Illumina genetic analyzer IIx instrument as previously described (20). From one deep-sequencing lane containing 433,522 raw sequence reads, 881 reads aligned to cardioviruses with high stringency by BLASTN (E value, <1 × 10−10) (2). These reads were used to design PCR primers for HTCV-UC6. The sequences of the 5′ and 3′ ends of HTCV-UC6 were recovered using a 5′/3′ RACE (rapid amplification of cDNA ends) kit (Invitrogen, Carlsbad, CA). The primers used for genome sequencing are available upon request. Amplified PCR fragments were cloned into TOPO TA vectors and were sequenced at 3× redundancy (Elim Biopharmaceuticals, Hayward, CA). Sequence analysis and assembly were performed using Geneious software (Biomatters Ltd., Auckland, New Zealand).
Serum samples.
For the HTCV seroprevalence study, a total of 57 serum samples were collected anonymously from adult blood donors presenting to the Blood Systems Research Institute in San Francisco, CA. The vast majority of donors were male (90%), with an average age of 40.0 ± 17.6 years (range, 16 to 83 years). These samples were tested in a blinded fashion for seroreactivity to genotype 2 cardioviruses by a virus neutralization assay and a VP1 enzyme-linked immunosorbent assay (ELISA). Six deidentified serum samples consisting of 3 paired acute- and convalescent-phase sera were also tested for seroreactivity to cardioviruses. These sera were obtained from household contacts of a patient with cardiovirus-associated diarrhea identified in a prior study (5, 15). All serum samples were analyzed under protocols approved by the UCSF Institutional Review Board.
Virus neutralization assay.
Diluted serum samples were preincubated with HTCV-UC6 (P1) virus stock (at a calculated titer of 108 viral genome equivalents/ml) for 1 h at 37°C. Serum-virus mixtures were then used to inoculate LLC-MK2 cells, and growth curves were generated as described above. Since viral CPE was not always apparent with the slow-growing HTCV-UC6 (P1) strain, virus neutralization was also assessed by quantitative RT-PCR titers at day 10 postinfection (see below for the definition of neutralization). In the seroprevalence study, a 1:20 dilution was used for the virus neutralization assay. For the six serum samples from the family of the patient for whom cardiovirus infection was identified, three 16-fold serum dilution steps were used, starting at a 1:20 dilution.
Expression and purification of HTCV-UC1 VP1 capsid protein.
The HTCV-UC1 VP1 capsid gene, flanked with NdeI and XhoI restriction sites, was amplified from the HTCV-UC1 genomic plasmid by two-step sticky-end PCR with the following primers: cardioVP1-NdeI-long (5′-TATGGGAGTGGATAACGCAGAAAAAGGCAAGG-3′), cardioVP1-XhoI-short (5′-GTTGAAGCTCCAAAACAGGGGTCGGGC-3′), cardioVP1-NdeI-short (5′-TGGGAGTGGATAACGCAGAAAAAGGCAAGG-3′), and cardioVP1-XhoI-long (5′-TCGAGTTGAAGCTCCAAAACAGGGGTCGGGC-3′). The resulting PCR products were purified, mixed together in equal ratios, and ligated into a NdeI/XhoI-digested pET15b vector (Novagen), which contained a 6×His tag on the N terminus (His6). The sequence-confirmed pEt15b vector containing the HTCV-UC1 VP1 gene was transformed into Escherichia coli BL21(DE3)/pLysS/pRIL, and recombinant His6-VP1 protein expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 37°C for 5 h. His6-VP1 was purified as previously described (22). Briefly, cells were lysed in TEN buffer (50 mM Tris-HCl [pH 8.0], 1 mM EDTA, and 100 mM NaCl) with 0.5 mg/ml lysozyme for 30 min in ice, followed by 3 min of sonication at 60% power. The cell pellet was washed three times by sonication with 0.5% deoxycholate in TEN buffer and three times with TEN buffer alone. His6-VP1 protein was solubilized in 20 mM Tris-HCl (pH 8.0)-500 mM NaCl-7 M urea, and the solubilized pellet fraction was then purified by nickel-nitrilotriacetic acid (NTA) column chromatography (Qiagen, Valencia, CA). The resulting eluate was loaded onto a 25-ml HiTrap desalting column (GE Life Sciences, Piscataway NJ) and was then eluted in TEN buffer. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using 4 to 12% Bis-Tris NuPage gels (Invitrogen, Carlsbad, CA) and Coomassie staining. Two milligrams of purified His6-VP1 was used for polyclonal antibody generation in rabbits (Pacific Immunology, Ramona, CA).
VP1 ELISA.
Maxisorp 96-well microtiter plates (Nunc, Naperville, IL) were coated overnight with purified HTCV-UC1 VP1 protein (0.15 μg/well) at 4°C in 50 μl PBS. Wells were washed twice in PBS containing 0.05% Tween 20 (PBST) and were blocked in blocking buffer (PBST containing 1% bovine serum albumin) for 2 h at room temperature. Well were washed again twice in PBST and were then incubated with serum samples, diluted in blocking buffer, in triplicate overnight at 4°C. For the seroprevalence study, serum samples were diluted 1:100 in blocking buffer. For the six serum samples from the family of the patient for whom cardiovirus infection was identified, eight 2-fold dilutions were used, starting at a 1:20 dilution. Diluted serum samples were preincubated with 0.6 μg protein overnight at 4°C. Plates were washed four times with PBST and were then incubated with a horseradish peroxidase (HRP)-conjugated anti-human IgG antibody (Invitrogen, Carlsbad, CA) diluted 1:1,000 in 50 μl blocking buffer for 2 h at room temperature. Plates were then washed four times in PBST and were incubated with 50 μl Ultra-TMB (Sigma) for 30 min at room temperature. An equal volume of 2 M H2SO4 was added to stop the reaction, and endpoint absorbance values (optical density at 450 nm [OD450]) were read immediately on a SpectraMax plate reader.
Determination of antibody seroprevalence.
We set a threshold for seroprevalence according to the results of the VP1 ELISA on nonneutralizing control human sera. Among the 57 random donor sera tested, the five serum samples with the smallest absorbance values (OD, ≤0.43) were all nonneutralizing both by examination for CPE and by quantitative viral titers at day 10. Thus, seroprevalence was defined as the percentage of human sera with OD values above 0.43. For antibody neutralization, we defined a serum sample as neutralizing if it was capable of reducing the viral titer at day 10 postinfection by 3 log units from that with a nonneutralizing inoculum (i.e., inoculation with virus alone).
Nucleotide sequence accession number.
The whole-genome sequence of HTCV-UC6 has been deposited in the GenBank database (accession number GU595289).
RESULTS
Growth of a genotype 2 human cardiovirus.
To determine whether human cardioviruses are culturable, six HTCV-positive stool samples (HTCV-UC2 through HTCV-UC7) obtained from a diarrheal cohort from Northern California were used to infect rhesus monkey kidney (LLC-MK2), human diploid fetal kidney (HDFK), and HeLa cell lines. Replication was monitored over 14 days for visual evidence of a cytopathic effect (CPE). Viral titers were estimated in terms of viral genome equivalents using serial quantitative SYBR green real-time reverse transcription-PCR (RT-PCR) of cell culture supernatants. CPE was not observed in any of the cell cultures, and quantitative RT-PCR assays were nearly all negative. However, for the stool sample containing the genotype 2 cardiovirus HTCV-UC6, a transient rise in viral titers at day 3 was observed in supernatants from infected LLC-MK2 cells, followed by a continuous decline in titers to day 14 (Fig. 1A, squares). Multiple attempts to passage the supernatant from the HTCV-UC6-infected LLC-MK2 cells were unsuccessful (data not shown).
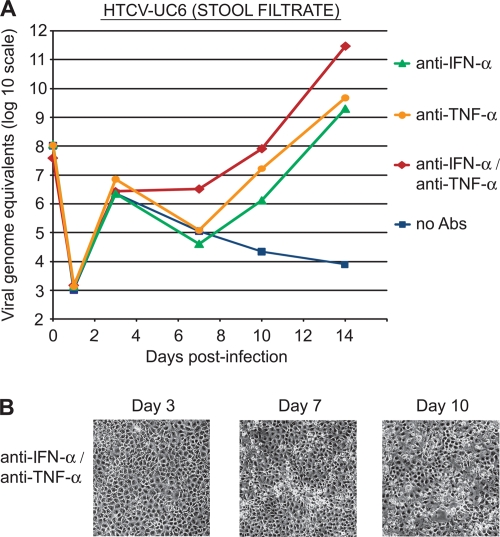
FIG. 1.
Replication of HTCV-UC6 in an LLC-MK2 cell line. (A) Growth kinetics of HTCV-UC6 (stool filtrate) over 10 days with the addition of an anti-IFN-α antibody (triangles), an anti-TNF-α antibody (circles), both antibodies (diamonds), or no antibody (squares) to the culture medium. Viral titers are calculated as viral genome equivalents/ml. (B) A moderate cytopathic effect (CPE) was first visible at day 7 after infection with HTCV-UC6 and after addition of anti-IFN-α/anti-TNF-α antibodies to the culture medium. After infection with HTCV-UC6, a similar CPE was observed with the addition of the anti-IFN-α or anti-TNF-α antibody alone (data not shown).
Cultivation with anti-cytokine antibodies.
We hypothesized that the addition of monoclonal antibodies against the cytokines alpha interferon (IFN-α) and tumor necrosis factor alpha (TNF-α) to the cell culture medium would promote viral growth by countering the antiviral effects of these two cytokines in both human and monkey cell lines (18, 19). Anti-IFN-α/anti-TNF-α antibodies were added to the cell culture medium upon infection with HTCV-UC6 on day zero, and after the washes on day 1, at a final concentration of 10 μg/ml. In LLC-MK2 cells, the addition of both antibodies produced a >8-log rise in viral titers by day 14 (Fig. 1A, diamonds), whereas addition of either antibody alone resulted in a less-pronounced, 6- to 7-log increase in viral titers (Fig. 1A, circles and triangles). This finding of viral growth with the addition of either anti-IFN-α alone, anti-TNF-α alone, or both antibodies to the cell culture medium was correlated with the observation of a weak CPE that was first visible at day 7 after infection (Fig. 1B), Control infections performed using only antibody or stool harboring HTCV-UC6 virus alone did not produce any visible CPE (data not shown).
Virus replication characteristics.
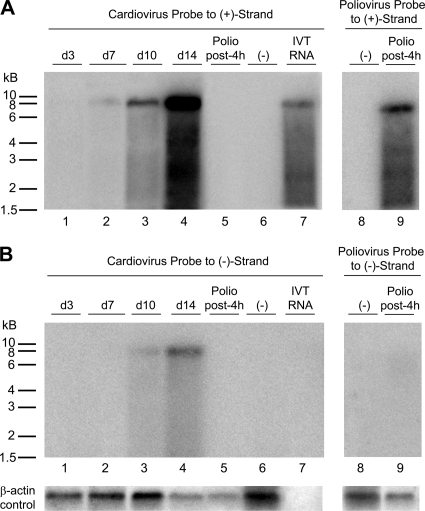
To confirm that viral replication was indeed occurring in HTCV-UC6-infected LLC-MK2 cells supplemented with anti-IFN-α/anti-TNF-α antibodies, Northern blot hybridization of extracted RNA at days 3, 7, 10, and 14 postinfection was performed using radioactively labeled positive-strand and negative-strand RNA probes directed against the 5′ untranslated region (5′ UTR) of HTCV-UC6 (Fig. 2). As positive controls, we also performed Northern blot analyses of in vitro-transcribed mRNA from a plasmid-encoded nearly full length genome of HTCV-UC1 (a related genotype 2 cardiovirus) and of poliovirus cultured in HeLa cells and harvested at 4 h postinfection. Northern blot analysis demonstrated that both positive- and negative-strand viral mRNAs were being synthesized in HTCV-UC6-infected LLC-MK2 cells. Production of positive-strand mRNA was first visible at day 7 postinfection (Fig. 2A, lane 2), whereas production of negative-strand mRNA was not observed until day 10 (Fig. 2B, lane 3). In HTCV-UC6-infected cells, a prominent RNA species corresponding to the full-length 8-kb genome and a lower-molecular-weight smear were observed (Fig. 2A, lanes 2 to 4, and B, lanes 3 and 4); these Northern blots were similar in appearance to those from the 7.5-kb poliovirus at 4 h postinfection (Fig. 2A, lane 9). As expected, positive-strand mRNA was substantially more abundant than negative-strand viral RNA for both HTCV-UC6 and poliovirus; poliovirus negative-strand mRNA was not even visible by Northern blot analysis.
FIG. 2.
Northern blot analysis of extracted RNA from virus-infected cells. Radioactive Northern blot analysis was performed using HTCV-UC6-infected LLC-MK2 cells, poliovirus-infected HeLa cells, and HTCV-UC1 in vitro-transcribed (IVT) genomic mRNA with cardiovirus- or poliovirus-specific probes to positive-strand (A) or negative-strand (B) RNA. β-Actin served as the loading control. Lane 6, marked (−), refers to the control uninfected LLC-MK2 cell line.
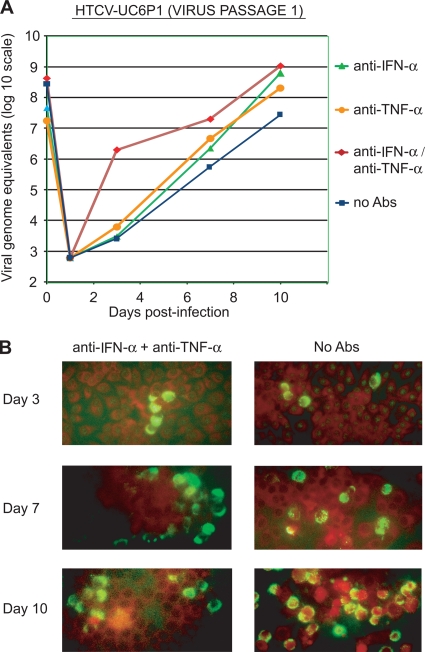
To further characterize the effect of adding anti-cytokine antibodies to the culture medium on viral propagation, the replication kinetics of singly passaged HTCV-UC6 (HTCV-UC6 [P1]) with or without anti-cytokine antibodies were examined by quantitative RT-PCR of cell culture supernatants in LLC-MK cells (Fig. 3). HTCV-UC6 (P1) was capable of replicating independently of antibody (Fig. 3A, squares), suggesting that it had acquired mutations after a single passage that facilitated adaptation to culture. There was a further enhancement in viral replication with the addition of anti-IFN-α alone, anti-TNF-α alone, or both antibodies; the most-pronounced difference was seen by day 3 postinfection with the addition of both antibodies (Fig. 3A, diamonds).
FIG. 3.
Effects of anti-cytokine antibodies on the replication of singly passaged HTCV-UC6 in LLC-MK2 cells. (A) Growth kinetics of HTCV-UC6 (P1) over 10 days with the addition of an anti-IFN-α antibody (triangles), an anti-TNF-α antibody (circles), both antibodies (diamonds), or no antibody (squares) to the culture medium. (B) Direct immunofluorescence assay for detection of cardioviruses in cells infected with HTCV-UC6 and no antibodies (right) or HTCV-UC6 with the addition of anti-IFN-α/anti-TNF-α antibodies to the culture medium (left) at days 3, 7, and 10 postinfection.
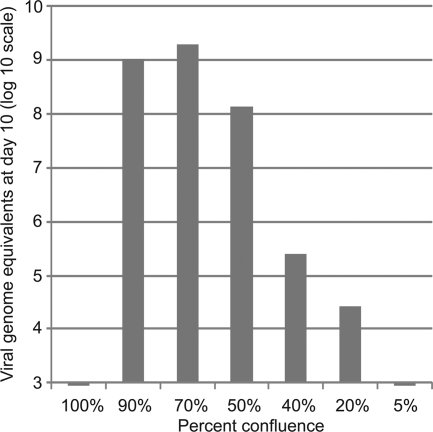
To observe virus propagation in LLC-MK2 cells with or without anti-IFN-α/anti-TNF-α antibodies, we performed direct immunofluorescent assays using the Pan-Enterovirus diagnostic kit (Millipore, Temecula, CA). This assay, although designed for panenterovirus detection, had been shown previously to be reactive against cardiovirus isolates (1). The percentage of infected cells was very low (<5%) at day 3, increasing to 10 to 15% by day 7 and peaking at 30 to 50% by day 10 (Fig. 3B). To confirm antibody-independent growth of passaged virus, HTCV-UC6 was passaged six times serially in LLC-MK2 cells, exhibiting identical replication kinetics for each passage (data not shown). HTCV passaging was also critically dependent on the growth state of the LLC-MK2 cells; successful infection and passaging did not occur when the cells were 100% confluent but required infection of fresh LLC-MK2 cells that were 50 to 90% confluent (Fig. 4).
FIG. 4.
Replication of HTCV-UC6 (P1) at various cell confluences. Viral stocks were inoculated into LLC-MK2 cells at cell confluences of 100%, 90%, 70%, 50%, 40%, 20%, and 5%. The y axis shows the viral titer (viral genome equivalents/ml) in the cell culture supernatant at day 10 postinfection.
Viral tropism in vitro.
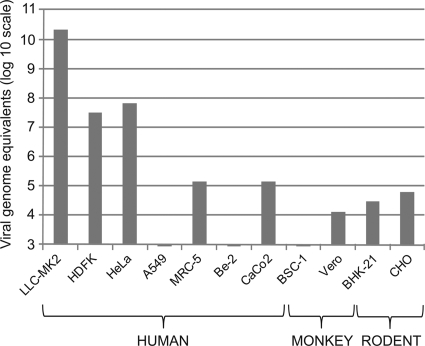
HTCV-UC6 (P1) was used to infect the following cell lines: LLC-MK2 (rhesus monkey kidney epithelial), HDFK (human diploid fetal kidney), HeLa (human cervical carcinoma), A549 (human lung carcinoma), MRC-5 (human embryonic lung fibroblast), SK-N-BE2 (human neuroblastoma), Caco-2 (human intestinal carcinoma), BSC-1 (monkey kidney epithelial), Vero (monkey kidney epithelial), BHK-21 (baby hamster kidney), and CHO (Chinese hamster ovary). In all cases, infection was monitored by quantitative RT-PCR for viral RNA in the culture supernatant (Fig. 5). The Caco-2 and SK-N-BE2 cell lines were selected given the presumed fecal-oral route of transmission for cardioviruses and the known association of rodent cardioviruses with neurological disease (4). The data show that virus replication was most efficient in LLC-MK2 cells, with a calculated titer of >1011 viral genome equivalents per ml at day 10 postinfection. Intermediate levels of viral replication were observed in HDFK and HeLa cells, and low-level replication proceeded in MRC-5, Caco-2, Vero, BHK-21, and CHO cell lines. No evidence of HTCV replication was observed in the A549, SK-N-BE2, and BSC-1 cell lines.
FIG. 5.
Replication of HTCV-UC6 (P1) in human, monkey, and rodent cell lines. The y axis shows the viral titer (viral genome equivalents/ml) in the cell culture supernatant at day 10 postinfection.
Complete genome sequencing and comparison with other human cardioviruses.
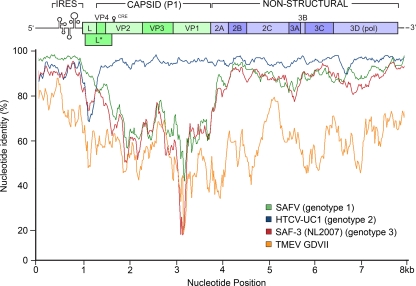
The whole-genome sequence of HTCV-UC6 was recovered directly from extracted stool RNA using PCR primers designed from the previously published sequence of HTCV-UC1 (GenBank accession number NC_010810), which generated amplified, overlapping fragments of 1 to 1.5 kb. Alignment of the full sequence of HTCV-UC6 with TMEV and one representative member each from human cardiovirus genotypes 1, 2, and 3 revealed the highest sequence similarity with HTCV-UC1 (genotype 2), with ∼90% nucleotide identity across the genome (Fig. 6). For the genotype 1 and 3 human cardioviruses, the nucleotide identity was considerably lower, especially in the capsid coding region. Based on whole-genome phylogenetic analysis, we confirmed that the HTCV-UC6 isolate is a genotype 2 cardiovirus, as previously shown by partial sequencing of the VP1 gene (5).
FIG. 6.
Nucleotide similarity plots of full-length cardiovirus sequences using HTCV-UC6 as the query sequence. Analysis was performed using a 200-nucleotide sliding window and a step size of 20 nucleotides. Data show the percentages of nucleotide identity between each cardiovirus sequence and the HTCV-UC6 sequence.
Seroprevalence by virus neutralization and VP1 ELISA.
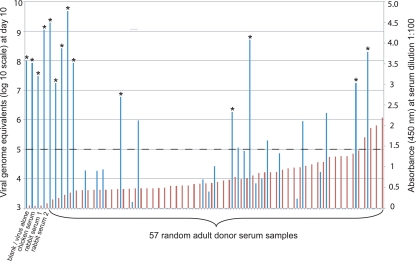
Successful propagation of HTCV-UC6 made it possible to perform a survey of seroprevalence among 57 randomly chosen, deidentified adult blood donors from California by virus neutralization. In parallel, an enzyme linked-immunosorbent assay (ELISA) for the detection of seroreactivity to cardiovirus VP1 was designed by expression and purification of the HTCV-UC1 VP1 protein. (Given the high nucleotide identity [95%] and the even greater amino acid identity (99%) in the VP1 region between HTCV-UC1 and HTCV-UC6, we reasoned that an ELISA based on the HTCV-UC1 VP1 protein would be cross-reactive with both members of the genotype 2 human cardioviruses.) Antibodies were found in 91% (52/57) of the adult donors by ELISA (Fig. 7). By use of a 3-log decrease in viral titers as a cutoff for neutralization, approximately 80% (42/52) of the seropositive donors, or 74% (42/57) of adult donors overall, produced neutralizing antibodies by the virus neutralization assay.
FIG. 7.
Seroprevalence of neutralizing antibodies to HTCV-UC6 among adult blood donors. The dual-axis column chart shows the virus titers (viral genome equivalents/ml) at day 10 postinfection by a cardiovirus neutralization assay (blue lines, left axis) and the absorbances at 450 nm measured by a cardiovirus VP1 ELISA (red lines, right axis) for 57 random adult-donor serum samples and negative controls. Asterisks indicate serum samples for which there was overt CPE at day 10. The horizontal dashed line indicates the 3-log-unit cutoff threshold for virus neutralization.
To verify the specificity of our serological assays and to investigate whether HTCV-UC6 antibodies were present in animals, we also analyzed serum specimens from rabbits, chickens, and monkeys. No antibodies were observed in serum samples from rabbits or chickens (Fig. 7), but antibodies to HTCV-UC6 were detected in 3 out of 3 rhesus monkey samples (data not shown), suggesting that cardioviruses related to HTCV may also circulate in nonhuman primates.
Cardiovirus seroconversion in a child with diarrhea.
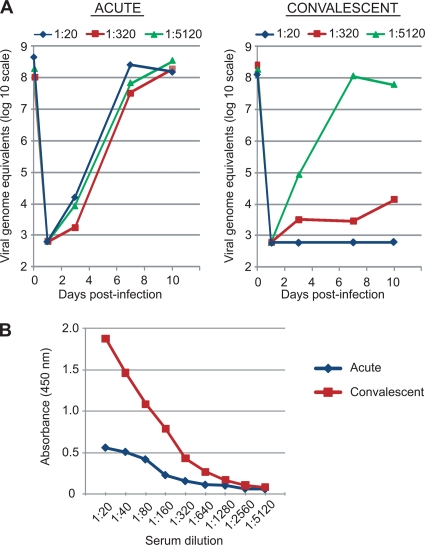
In serum samples collected from a cohort study of diarrheal transmission within households in Northern California (16), we identified a family for which a cardiovirus was detected in stool from the index case, an 8-month-old infant with diarrhea and vomiting. Although sera from this infant were unavailable, paired acute-phase and convalescent-phase (taken >3 months later) sera from three other members of the household, one child (age, 16 months) and two young adults (ages, 21 and 32 years), were available for analysis. Among these three individuals, the child had diarrhea and vomiting, whereas the adults were asymptomatic. The presence of neutralizing antibodies to cardioviruses was detected only in sera from the child with diarrhea and vomiting, and during the convalescent stage but not the acute stage (Fig. 8A). For this symptomatic child, a >8-fold rise in the cardiovirus-specific antibody titer was also observed by VP1 ELISA from the acute to the convalescent stage (Fig. 8B). Cardiovirus neutralization assays and VP1 ELISAs for the two adults were negative during both the acute and convalescent stages (data not shown). The only stool sample available for testing was a convalescent-stage sample from the symptomatic child, which, as expected, was negative for cardioviruses by RT-PCR. The serological data are consistent with an acute seroconversion event to cardiovirus infection occurring in a child with diarrhea and vomiting, but no evidence of seroconversion or even seropositivity in two other members of the family.
FIG. 8.
Acute seroconversion to cardiovirus infection in a child with diarrhea and vomiting. (A) Cardiovirus neutralization assay. Shown are the growth kinetics of HTCV-UC6 (P1) in LLC-MK2 cells over 10 days after a 24-h incubation of the virus with acute-phase (left) and convalescent-phase (right) sera at dilutions of 1:20 (diamonds), 1:320 (squares), and 1:5,120 (triangles). (B) Cardiovirus VP1 ELISA. Shown are the measured absorbances at 450 nm (OD450) for acute- and convalescent-phase sera at serial 2-fold dilutions from 1:20 to 1:5,120.
DISCUSSION
Here we present the cultivation and serological analysis of HTCV-UC6, a human cardiovirus in the genotype 2 subgroup, the predominant subgroup to date in North America (1, 3, 5). A genotype 3 isolate, SAF-3 (NL2007), was recently found to grow efficiently in HeLa cells, with complete lysis of the monolayer by days 2 to 3 postinfection (24). However, propagation of genotype 1 and 2 human cardioviruses in cell culture has been difficult to date (1, 7, 11, 24), hampering detailed molecular and seroepidemiological analyses of these isolates. Consistent with earlier difficulties in growing genotype 1 and 2 cardioviruses, HTCV-UC6 replication was barely detectable during initial stool inoculation, and virus propagation could not be accomplished in the absence of cytokine-blocking antibodies. The cytokine blockade allowed rapid amplification of the virus copy number during the first passage, and HTCV-UC6 became completely independent of cytokine manipulation by passages 2 to 3.
Stocks of HTCV-UC6 passaged in LLC-MK2 cells are able to reach titers as high as 109 to 1010 viral genome equivalents/ml, implying a robust replicative cycle. However, HTCV-UC6 growth resulted in only moderate CPE, and a significant percentage of cells in the culture (>50%) failed to stain with a panenteroviral monoclonal antibody reactive for cardioviruses. One potential explanation for these findings may be abundant production of defective, noninfectious viral particles, resulting in actual titers of infectious virus significantly lower than those estimated. The marked difference in CPE between HTCV-UC6 and SAFV-3 (NL2007) may also reflect sequence divergence in the L protein (Fig. 6), which in TMEV has been found to mediate apoptosis in infected cells (8). Furthermore, even in the most susceptible lines, individual cells may differ in their permissivity for viral replication due to factors such as inherent heterogeneity of the cell line or cell culture density. Specifically, we found that the ability of LLC-MK2 cells to support HTCV propagation was sensitive to cell density. Successful infection did not occur at 100% confluence (Fig. 4), suggesting that active cell division may be necessary to support the growth of HTCV.
Northern blot analysis of both HTCV-UC6- and poliovirus-infected cells using virus-specific probes to positive- and negative-strand mRNA yielded a large mRNA species corresponding to the full-length genome, as well as a lower-molecular-weight smear. However, evidence of poliovirus replication was observed at a mere 4 h postinfection, whereas viral mRNA from HTCV-UC6 was not visible until day 7. The markedly slower replication kinetics of HTCV-UC6 relative to poliovirus may be related to inefficiency (or lack of processivity) in RNA replication, with the production of many incomplete and/or noninfectious RNA products. The weak cytolytic effect (Fig. 1B) and delayed replication kinetics (Fig. 2) of HTCV-UC6, a genotype 2 cardiovirus, are strikingly different from the virus replication characteristics of SAF-3 (a genotype 3 cardiovirus) (24), which may be due to the considerable sequence divergence between the two strains (Fig. 6). The growth characteristics of HTCV-UC6 are thus more similar to those seen previously in other slow-growing picornaviruses, such as hepatitis A (6).
Successful culturing of HTCV-UC6 from a primary stool sample required the addition of monoclonal antibodies to IFN-α and/or TNF-α to the cell culture medium, at least transiently (Fig. 1). The enhanced replication of HTCV-UC6 once IFN-α and TNF-α responses are attenuated suggests that the signaling pathways downstream of both IFN and TNF receptors express antiviral activity against human cardioviruses. The enhanced replication of HTCV-UC6 after attenuation of the cytokine response also raises the possibility that cardiovirus infection may exhibit greater pathogenicity in individuals who are immunocompromised and/or are taking medications that suppress these inflammatory cytokines, such as TNF inhibitors for autoimmune diseases (9). Consistent with our data suggesting that interferon may have antiviral activity against human cardioviruses is evidence that the leader (L) protein of TMEV inhibits immediate-early production of IFN-α/β in infected cells (21), suggesting that IFN has exerted selective pressure on the rodent cardiovirus that favors the blunting of the IFN-mediated antiviral response. Whether the L proteins of human cardioviruses can similarly suppress interferon production, and whether this plays a role in viral pathogenicity, is currently unknown.
Evidence that HTCV-like cardioviruses produce human infection is now overwhelming: the viruses have been repeatedly recovered from stool (3, 5, 7, 23), cluster phylogenetically in a distinct group clearly separate from other animal cardioviruses (3, 5, 7), and evoke serologic responses in a high proportion of humans (Fig. 6) (24). However, although the seroprevalence data show that cardiovirus infection is relatively common, they do not address its pathogenic potential. The studies reported here reveal both direct and indirect evidence of cardiovirus involvement in human gastroenteritis. Using a virus neutralization assay, we were able to demonstrate an acute seroconversion event to cardiovirus infection in a child with diarrhea and vomiting (Fig. 8). In addition, we found that cardioviruses were capable of replicating in intestinal cells (Caco-2), albeit at relatively low levels (Fig. 5).
Other, recently published studies also support an association between cardiovirus infection and diarrheal disease. An outbreak of enteric disease associated with a family gathering in Minnesota was reported in 2008, and despite extensive testing for a diarrheal pathogen, the only agent detected in multiple samples from the outbreak was an HTCV/Saffold-like virus (10). In a 9-month study of children with acute gastroenteritis in China, samples from all 12 of 373 children testing positive for cardioviruses were collected in a single month, suggesting a possible seasonal diarrheal outbreak (23). Further studies will be required to definitively ascertain the role of human cardioviruses in the etiology of diarrhea.
It is still unclear whether human cardioviruses can cause more-severe illnesses, such as myocarditis, encephalitis, or demyelinating disease (4, 12). Although we found that >90% of adults in California have antibodies to the genotype 2 cardioviruses (of which 80% are neutralizing) (Fig. 7), in close agreement with a previous study of worldwide seroprevalence for a genotype 3 cardiovirus (24), a proportion of the adult population (and most likely an even larger fraction of young children) is clearly still susceptible to cardiovirus infection. Neutralizing antibody levels may also wane with age, allowing for the possibility of reinfection in previously exposed individuals. In addition, at least eight cardiovirus genotypes have been characterized to date (3, 12), and given the broad sequence diversity among cardiovirus strains, it is possible that some genotypes are inherently more virulent than others. Strategies such as the cytokine blockade methods described here may be effective in facilitating cultivation of these other cardiovirus genotypes.
Acknowledgments
We thank Rachel Adams for assistance with the cell culturing and Northern blot experiments, Michael Busch at the Blood Systems Research Institute for providing donor serum samples, and Amy Kistler and Sanjay Chandriani for helpful advice.
C.Y.C. was supported by NIH K08 grant AI074913 and an award from Abbott Diagnostics, Inc. D.G. and J.L.D. were supported by the Howard Hughes Medical Institute and a grant from the Doris Duke Foundation. J.P. was supported by NIH R01 grant AI042801.
Footnotes
Published ahead of print on 17 February 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Abed, Y., and G. Boivin. 2008. New Saffold cardioviruses in 3 children, Canada. Emerg. Infect. Dis. 14:834-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Blinkova, O., A. Kapoor, J. Victoria, M. Jones, N. Wolfe, A. Naeem, S. Shaukat, S. Sharif, M. M. Alam, M. Angez, S. Zaidi, and E. L. Delwart. 2009. Cardioviruses are genetically diverse and cause common enteric infections in South Asian children. J. Virol. 83:4631-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brahic, M., J. F. Bureau, and T. Michiels. 2005. The genetics of the persistent infection and demyelinating disease caused by Theiler's virus. Annu. Rev. Microbiol. 59:279-298. [DOI] [PubMed] [Google Scholar]
- 5.Chiu, C. Y., A. L. Greninger, K. Kanada, T. Kwok, K. F. Fischer, C. Runckel, J. K. Louie, C. A. Glaser, S. Yagi, D. P. Schnurr, T. D. Haggerty, J. Parsonnet, D. Ganem, and J. L. DeRisi. 2008. Identification of cardioviruses related to Theiler's murine encephalomyelitis virus in human infections. Proc. Natl. Acad. Sci. U. S. A. 105:14124-14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cromeans, T., H. A. Fields, and M. D. Sobsey. 1989. Replication kinetics and cytopathic effect of hepatitis A virus. J. Gen. Virol. 70(Pt 8):2051-2062. [DOI] [PubMed] [Google Scholar]
- 7.Drexler, J. F., L. K. Luna, A. Stocker, P. S. Almeida, T. C. Ribeiro, N. Petersen, P. Herzog, C. Pedroso, H. I. Huppertz, H. D. C. Ribeiro, Jr., S. Baumgarte, and C. Drosten. 2008. Circulation of 3 lineages of a novel Saffold cardiovirus in humans. Emerg. Infect. Dis. 14:1398-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan, J., K. N. Son, S. Y. Arslan, Z. Liang, and H. L. Lipton. 2009. Theiler's murine encephalomyelitis virus leader protein is the only nonstructural protein tested that induces apoptosis when transfected into mammalian cells. J. Virol. 83:6546-6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feldmann, M. 2009. Translating molecular insights in autoimmunity into effective therapy. Annu. Rev. Immunol. 27:1-27. [DOI] [PubMed] [Google Scholar]
- 10.Fuller, C., E. Cebelinski, J. Bartkus, B. Juni, K. Smith, and J. Besser. 2008. Enhanced laboratory testing of enteric disease outbreaks of unknown etiology in Minnesota, abstr. 89. Abstr. Int. Conf. Emerg. Infect. Dis. 2008.
- 11.Jones, M. S., V. V. Lukashov, R. D. Ganac, and D. P. Schnurr. 2007. Discovery of a novel human picornavirus in a stool sample from a pediatric patient presenting with fever of unknown origin. J. Clin. Microbiol. 45:2144-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang, Z., A. S. Kumar, M. S. Jones, N. J. Knowles, and H. L. Lipton. 2008. Phylogenetic analysis of the species Theilovirus: emerging murine and human pathogens. J. Virol. 82:11545-11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lipton, H. L. 2008. Human Vilyuisk encephalitis. Rev. Med. Virol. 18:347-352. [DOI] [PubMed] [Google Scholar]
- 14.Lipton, H. L., A. Friedmann, P. Sethi, and J. R. Crowther. 1983. Characterization of Vilyuisk virus as a picornavirus. J. Med. Virol. 12:195-203. [DOI] [PubMed] [Google Scholar]
- 15.Perry, S., M. de la Luz Sanchez, S. Yang, T. D. Haggerty, P. Hurst, G. Perez-Perez, and J. Parsonnet. 2006. Gastroenteritis and transmission of Helicobacter pylori infection in households. Emerg. Infect. Dis. 12:1701-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perry, S., L. Sanchez, S. Yang, T. D. Haggerty, P. Hurst, and J. Parsonnet. 2004. Helicobacter pylori and risk of gastroenteritis. J. Infect. Dis. 190:303-310. [DOI] [PubMed] [Google Scholar]
- 17.Rotbart, H. A. 1990. Enzymatic RNA amplification of the enteroviruses. J. Clin. Microbiol. 28:438-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruby, J., H. Bluethmann, and J. J. Peschon. 1997. Antiviral activity of tumor necrosis factor (TNF) is mediated via p55 and p75 TNF receptors. J. Exp. Med. 186:1591-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakuma, R., A. A. Mael, and Y. Ikeda. 2007. Alpha interferon enhances TRIM5α-mediated antiviral activities in human and rhesus monkey cells. J. Virol. 81:10201-10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sorber, K., C. Chiu, D. Webster, M. Dimon, J. G. Ruby, A. Hekele, and J. L. DeRisi. 2008. The long march: a sample preparation technique that enhances contig length and coverage by high-throughput short-read sequencing. PLoS One 3:e3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Pesch, V., O. van Eyll, and T. Michiels. 2001. The leader protein of Theiler's virus inhibits immediate-early alpha/beta interferon production. J. Virol. 75:7811-7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang, J. H., C. M. Liang, J. M. Peng, J. J. Shieh, M. H. Jong, Y. L. Lin, M. Sieber, and S. M. Liang. 2003. Induction of immunity in swine by purified recombinant VP1 of foot-and-mouth disease virus. Vaccine 21:3721-3729. [DOI] [PubMed] [Google Scholar]
- 23.Xu, Z. Q., W. X. Cheng, H. M. Qi, S. X. Cui, Y. Jin, and Z. J. Duan. 2009. New Saffold cardiovirus in children, China. Emerg. Infect. Dis. 15:993-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zoll, J., S. Erkens Hulshof, K. Lanke, F. Verduyn Lunel, W. J. Melchers, E. Schoondermark-van de Ven, M. Roivainen, J. M. Galama, and F. J. van Kuppeveld. 2009. Saffold virus, a human Theiler's-like cardiovirus, is ubiquitous and causes infection early in life. PLoS Pathog. 5:e1000416. [DOI] [PMC free article] [PubMed] [Google Scholar]