Abstract
Control of viral replication is a major therapeutic goal to reduce morbidity and mortality from chronic hepatitis B virus (HBV) infection. Recently, methylation has been identified as a novel host defense mechanism, and methylation of viral DNA leads to downregulation of HBV gene expression. To better understand the mechanisms of HBV methylation, cell lines were exposed to HBV using a model system that mimics natural infection and the expression of host DNA methyltransferase genes (DNMTs) was measured. DNMT1, DNMT2, and DNMT3 were all significantly upregulated in response to HBV. DNMT3 was further studied because of its known role in the de novo methylation of DNA. Cotransfection experiments with full-length HBV and DNMT3 led to the downregulation of viral protein and pregenomic RNA production. To investigate whether the upregulation of DNMTs could also have an effect on the methylation of host DNA, cell lines were exposed to HBV in two independent model systems, one that mimics natural infection and a second model with temporary transfection. Host DNA methylation was measured by DNA microarray analysis. Increased methylation of host CpG islands was detected in both experimental systems. Two CpG islands, corresponding to genes SUFU and TIRAP, were selected, and the downregulation of these genes in hepatocellular carcinomas was confirmed. In conclusion, hepatocytes respond to HBV infection by upregulating DNMTs. The DNMTs methylate viral DNA, leading to decreased viral gene expression and decreased viral replication. However, virus-induced overexpression of DNMTs also leads to methylation of host CpG islands.
With more than 400 million persons estimated to be chronically infected with hepatitis B virus (HBV) around the world, HBV infection is a major cause of morbidity and mortality, particularly when it results in hepatocellular carcinoma (HCC). Hepatocellular carcinomas develop because of a combination of factors, including viral integration, the development of cirrhosis, and the actions of the X protein on a wide variety of signaling and metabolic pathways. Recently, the overall amount of viral replication has also been strongly linked to the risk of carcinogenesis (1). Thus, understanding host and viral factors that regulate HBV replication may provide insights into preventing hepatocellular carcinoma. In this regard, recent studies have identified epigenetic modifications of HBV DNA as a novel mechanism for the control of viral gene expression (15, 22).
However, our understanding of the mechanism for methylation of viral DNA in the natural history of HBV infection remains incomplete. Liu et al. recently reported that DNA methyltransferase gene 1 (DNMT1) expression is upregulated in hepatocytes exposed to HBV DNA (9). Increased expression of DNMTs could give infected cells greater ability to methylate viral DNA and control viral replication. Relevant to this, cell lines exposed to HBV can methylate viral DNA (22) and the presence of methylated viral DNA has been confirmed in human tissues (21-23). However, overexpression of DNMTs in liver tissues could potentially have untoward consequences. In fact, marked overexpression of the viral X protein can lead to methylation of host DNA (14) and chronic viral hepatitis has also been linked to hepatocellular carcinomas with methylation of host tumor suppressor gene promoters, i.e., a “methylator phenotype” (24).
In this study, it is shown that HBV infection upregulates DNMTs, which in turn leads to the methylation of HBV DNA and decreased viral gene expression. However, in the same cells there is simultaneous methylation of host CpG islands, including CpG islands that overlap promoters for genes linked to carcinoma. These experiments identify and place into biological context the dual role of DNMT expression as having both a protective role and a potentially harmful effect within HBV-infected hepatocytes.
MATERIALS AND METHODS
Infection of cells with unmodified HBV particles.
To investigate which DNMTs are upregulated in human cells after they are exposed to native viral particles, a protocol was used that was derived from the work of Paran et al. (12), which utilizes dimethyl sulfoxide (DMSO) to directly infect cells without transfection. HepG2 and Huh7 cells were split into standard 24-well plates at 50% cell density in high-glucose Dulbecco's modified Eagle medium (Invitrogen) with 2% DMSO and 10% fetal bovine serum until confluent (typically day 3), at which time the medium was changed and the fetal bovine serum was reduced to 2%. DMSO was maintained at 2% throughout the experiment. A full medium change was performed at day 6, at which time human serum containing a total of 8 log copies of intact HBV virions was added. The HBV virions were from unmodified human serum. Control wells were treated with human serum that was negative for hepatitis C virus (HCV) and HBV infection (AccuRun 803; Boston Biomedica). At 14 h, the medium containing HBV was removed, the cells were washed, and fresh medium with 2% DMSO and 2% fetal bovine serum was placed on the cells. No additional medium changes took place for the remainder of the experiments, which went on for 15 days after the exposure to HBV.
In control experiments performed to better understand the performance of this model system, the levels of hepatitis B surface antigen (HBsAg) and HBeAg in the supernatant were found to quickly increase and peak by 3 days after HBV exposure and then to drop to very low but still detectable levels that plateaued by day 8 (data not shown).
As a separate control for the effect of viral DNA without viral replication, cells were transfected with two fragments of HBV DNA. Each HBV fragment contains over half the HBV genome, and together they contain more than the full length of the HBV genome, but because of their size and orientation they cannot be ligated within the cells to form intact covalently closed circular DNA (cccDNA). After 14 h, test and control cells were extensively washed with minimal essential medium and fresh medium with DMSO was added.
DNMT expression by real-time PCR.
For DNMT mRNA studies, total RNA was extracted from cells using TRIzol (Invitrogen) followed by precipitation with isopropyl alcohol per manufacturer's instructions. Each time point had at least six replicates. RNA extracts were DNase I treated (Invitrogen), and cDNA was synthesized with oligo(dT) primers and the Superscript First-Strand synthesis system for reverse transcriptase PCR (RT-PCR) (Invitrogen). Expression levels were normalized to that of beta-glucuronidase.
Real-time PCR was performed using the Fast Start SYBR green master mix (Roche). One microliter of cDNA was used as the input template for each real-time PCR (Table 1). PCR cycling conditions were 10 min at 95°C followed by 40 cycles of 95°C for 20 s, 55°C for 25 s, and 72°C for 25 s. The specificity was confirmed by melt curve analysis and band size in 1% agarose gels. Dilution studies demonstrated real-time PCR efficiencies of greater than 98% for each of the DNMT primer sets.
TABLE 1.
Primers used in this study
| Target | Primer, 5′ to 3′ | Product size (bp) |
|---|---|---|
| DNMT primers | ||
| DNMT1 mRNA, F (13) | GGTTCTTCCTCCTGGAGAATGTC | 146 |
| DNMT1 mRNA, R | GTCTGGGCCACGCCGTACTG | |
| DNMT2 mRNA, F | ACAGACTGCAGAGGATGTGC | 167 |
| DNMT2 mRNA, R | TCTTCTCAGGAAATCCGAACTC | |
| DNMT3a mRNA, F | TAAGCTGGAGCTGCAGGAGT | 179 |
| DNMT3a mRNA, R | GGAAACCAAATACCCTTTCCA | |
| DNMT3a v3 mRNA, F | ACTTGGAGAAGCGGAGTGAG | 195 |
| DNMT3a v3 mRNA, R | TGGTCTCCTTCTGTTCTTTGC | |
| DNMT3a v4 mRNA, F | AAGCCTCAAGAGCAGTGGAA | 190 |
| DNMT3a v4 mRNA, R | AAGCAGACCTTTAGCCACGA | |
| DNMT3b mRNA, F (13) | ACCACCTGCTGAATTACTCACGC | 146 |
| DNMT3b mRNA, R | GATGGCATCAATCATCACTGGATT | |
| HBV primers | ||
| HBV pregenomic RNA, F (7) | CACCTCTGCCTAATCATC | 148 |
| HBV pregenomic RNA, R | GGAAAGAAGTCAGAAGGCAA | |
| HBV precore RNA, F (7) | GGTCTGCGCACCAGCACC | 178 |
| HBV precore RNA, R | GGAAAGAAGTCAGAAGGCAA | |
| HBV cccDNA, F (7) | GTGCCTTCTCATCTGCCGG | 400 |
| HBV cccDNA, R | GGAAAGAAGTCAGAAGGCAA | |
| Bisulfite PCR primers | ||
| ONECUT BISULF 1F | TTTAAAGAGAAGATGTTTTGTTAAAGG | 218 |
| ONECUT BISULF 1R | AATTCCAACCCAAAAAAAACC | |
| PINX1 BISULF 1F | AGTAGGGGGAATTTTTATAGTGGTT | 204 |
| PINX1 BISULF 1R | CCTACTACAAACTCCTTCCTTAACTC | |
| LSM10 BISULF 1F | GGAGAAATTATAATTTTTAGAATGTTT | 147 |
| LSM10 BISULF 1R | AAAAAAAACCTAACCCAAATC | |
| RAI1 BISULF 1F | GTGGTATATAGGGGATATTTGAGG | 177 |
| RAI1 BISULF 1R | AACTATAAATCTCAACTACAACAAAC | |
| CENTG2 BISULF 1F | TTTGTAGTAATATATTTTTTTAGGGATAAT | 132 |
| CENTG2 BISULF 1R | TCTACAACAAATAACAAAAACAAAAAAC | |
| TIRAP BISULF 1F | AATAAAATTTAGGTTTTAGGTTGTTA | 171 |
| TIRAP BISULF 1R | AAACAACCCCTATATCCACTAC | |
| TRIM8 BISULF 1F | TTAGGGAGGTGGAGTTTTAGTTTG | 297 |
| TRIM8 BISULF 1R | AAAATAATATCCCTACATCAAAACC | |
| NUP62 BISULF 1F | TGGTTTTTTTTAATTTAGGTGGTGT | 236 |
| NUP62 BISULF 1R | CAACCTTCTTCAAATCCCACTACTA | |
| POLR2J2 BISULF 1F | GATTAAGTAAAAGTTGTTTTTTTT | 232 |
| POLR2J2 BISULF 1R | AAACCTCTTATTAAAACTCTAATAAC | |
| Gene expression primers | ||
| ONECUT RT 1F | AACCCTGGAGCAAACTCAAA | 172 |
| ONECUT RT 1R | AAGACCAACCTGGGCTTTTT | |
| PINX1 RT 1F | TCCTCGGACAAGAAGGAAAA | 171 |
| PINX1 RT 1R | GCCCTCGGGAGTCTTCTTAC | |
| LSM10 RT 1F | GCTACGTCCACATCCCAGAT | 154 |
| LSM10 RT 1R | GCATGGGCAACATAGTGAGA | |
| RAI1 1F | GTGTTCCAGCTGCCAAGAAG | 166 |
| RAI1 1R | CGTTGGGGTGGATTACTACG | |
| CENTG2 1F | GACGAGGTGAACGAGACCTG | 150 |
| CENTG2 1R | CGTAGGCCAGAGCTGTGTT | |
| TIRAP 1F | CTTCACCAATGCCTGGTCTC | 152 |
| TIRAP 1R | TCTTCTTGGGCTTCTTCAGC | |
| TRIM8 RT 1F | CCTGCAGCTGCTCTTTGATA | 152 |
| TRIM8 RT 1R | CGTTCAGGAAGAGCTTGGAG | |
| NUP62 RT 1F | GCCCGAAGACATCTACCAGA | 175 |
| NUP62 RT 1R | ATCCTCGGACATCACGTCTC | |
| SUFU RT 1F | ACTGAGGAGCATCCTTACGC | 141 |
| SUFU RT 1R | AGGCCAGCTGTACTCTTTGG | |
| POLR2J2 RT 1F | ATCTCACACTGAGGGCCAAC | 164 |
| POLR2J2 RT 1R | TTCGTCATTCATCACCAGGA |
Cotransfections with HBV and DNMTs.
Expression vectors with full-length DNMT3a were used for cotransfection with full-length HBV DNA. Two splice variants were used for DNMT3a: splice variant 3 and splice variant 4 (Invitrogen). Pilot studies showed that both splice variants had similar abilities to reduce HBsAg and HBeAg production in cotransfection experiments (data not shown), and subsequent transfection studies used equal proportions of both variants. Full-length HBV DNA for transfection was generated essentially as originally reported by Gunther et al. (2). Briefly, the full-length HBV genome was cloned from the serum of an individual with chronic HBV infection, genotype D, who was HBsAg and HBeAg positive. The cloned virus was digested with SapI to release the HBV genome from the vector, the viral DNA was gel purified, and cells were cotransfected (Lipofectamine; Invitrogen) with 7 to 9 log copies of HBV DNA and equivalent copies of the DNMT3a vectors.
Cotransfection experiments were performed with the following cotransfectants: (i) HBV plus empty expression vector, (ii) HBV plus DNMT3a that had been inactivated by methylation of the DNMT3a vectors with CpG methyltransferase (New England Biolabs), (iii) HBV plus DNMT3a vectors, and (iv) empty vector plus HBV that had been inactivated by methylation of viral DNA with CpG methyltransferase (New England Biolabs). The enzyme CpG methyltransferase methylates all CpG dinucleotides.
In all experiments, the total amount of vector DNA and the total amount of viral DNA were the same across experimental conditions. In all experiments, the cells were washed and the growth medium was replaced at 24 h after transfection. Supernatant and cells were then harvested at 72 h. Each transfection was performed in triplicate with at least two replicates, providing at least six wells per experimental condition.
Detection of HBV proteins in cell culture supernatant and in human serum.
HBsAg and HBeAg enzyme-linked immunosorbent assays (ELISAs) (ETI-MAK-2 plus and ETI-EBK plus) were performed on 100 μl of supernatant per the manufacturer's instructions. Samples outside the linear range were diluted as necessary.
HBV RNA detection.
For HBV mRNA studies, at least three separate wells were studied from each experimental condition. Total RNA was extracted from cells using TRIzol (Invitrogen). Following DNase I treatment (Invitrogen), cDNA was synthesized with oligo(dT) primers using the Superscript First-Strand synthesis system (Invitrogen). The transfection model systems use large amounts of HBV DNA, and DNase treatment does not always remove all viral DNA. However, the mRNA signal was at least three cycles (approximately 10 times) greater than the residual DNA signal, showing that residual DNA contributed less than 10% to the HBV RNA quantification. Primers were used that detect only pregenomic RNA as well as primers that detect the combined total of precore and pregenomic RNA (Table 1). HBV RNA transcript levels were normalized to total cccDNA levels. Student t tests were used to test for differences between groups.
Microarray studies.
To investigate whether the DNMTs that are upregulated by HBV infection could also lead to new methylation of CpG islands in the human genome, we used HepG2 cells and compared the number of methylated CpG islands before and after infection with HBV. Cells were exposed to intact viral particles for up to 15 days as described above using the Paran et al. system. A second independent model system was then used to confirm the findings of increased methylation of host DNA following HBV infection. The model system of Gunther et al., which transfects full-length HBV DNA, was used as described above.
Quantification of the extent of methylation was performed in the Johns Hopkins Microarray core facility using the HCGI12K array (University Health Network, Toronto, Ontario, Canada), which contains 12,192 CpG island clones derived from the Sanger Center, United Kingdom (http://data.microarrays.ca/cpg/index.htm). After HBV infection, HepG2 cells were harvested at day 15. As controls, HepG2 cells at day 0 and at day 15 after mock infection with HBV-negative serum were used. Each experiment was performed in duplicate. Data analysis was performed at the Microarray core facility in a standard fashion as previously described (19). Briefly, data were analyzed after Lowess normalization using a limmaGUI package within R. Within-array normalization was performed using the print tip group (Lowess normalization), while between-array normalization was performed using quantiles. Comparative analysis (treated versus untreated) was performed using the least squares linear model. Genes with P values less than 0.001 were considered differently methylated between test and control when normalized log 2 ratios were ≤−0.6 or ≥0.6.
To confirm the methylation findings, 10 CpG island clones with increased methylation following HBV infection were chosen for additional studies. Bisulfite primers were designed (Methprimer) to a segment of each CpG island. Because the entire CpG islands were often substantially larger than can be confirmed in a bisulfite PCR amplicon, one segment of the CpG island was chosen for further study. After amplification and cloning, five to seven clones were sequenced. Real-time PCR primers were also designed to measure mRNA levels.
Human tissues.
We have previously reported that HCC with the methylator phenotype is associated with viral hepatitis as the underlying cause of liver disease (24). This same set of deidentified fresh frozen liver tissue was used as proof of principle to confirm that methylation on these CpG islands could be found in human liver tissues. A total of 16 tissues with chronic viral infection (8 HBV infections, 8 HCV infections) were tested, along with 8 paired tumor and nontumor tissues from livers with no known underlying liver disease.
RESULTS
All DNMTs are upregulated by HBV.
While methylation of HBV DNA in human serum and liver tissues has been previously described (4, 21-23), the specific methyltransferase(s) that plays a role in this process is not defined. Generally ascribed roles for the major methyltransferases include the following (16): DNMT1 methylates hemimethylated CpG dinucleotides and plays a key role in maintenance of methylation during cell division; DNMT2 (also known as TRDMT1) shares homologies to DNMT1 but lacks DNA methyltransferase capabilities and appears to play a role in methylation of structural RNA; DNMT3a and DNMT3b can methylate both unmethylated and hemimethylated CpG dinucleotides.
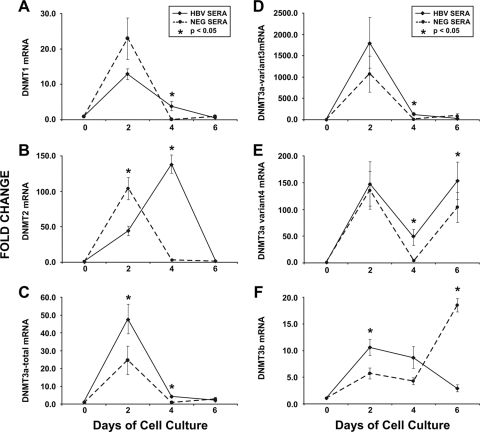
In cells without HBV exposure, the DNMT expression levels quickly increased, peaked at day 4, and decreased just as rapidly, returning to near-normal levels by day 6 (Fig. 1). Additional experiments showed that control cells without a fresh medium change or exposure to HBV showed a slight decrease in DNMT3a expression levels at days 2, 4, and 6 (data not shown), suggesting that fresh medium can cause an upregulation of DNMT activity and serves as a useful internal control. However, after cells were exposed to intact HBV virions, DNMT expression levels were significantly greater than those in control cells (Fig. 1). Overall, DNMT2 showed the greatest increase, followed by DNMT3a and DNTM1 (Fig. 1). These observations were confirmed in repeat experiments with HepG2 cells (data not shown).
FIG. 1.
DNMTs are upregulated following HBV exposure. Huh7 cell lines were exposed to human serum containing intact HBV particles (solid lines) or human serum negative for viral particles (dotted lines). Levels of mRNA were measured at 2-day intervals and normalized to levels at time zero. Each time point has at least six replicates. All data are shown as means ± standard errors of means. Groups were compared using Student t tests. (A) DNMT1 mRNA was significantly higher at day 4 in cells exposed to HBV (P = 0.049) but there was no difference at day 2 (P = 0.16) or 6 (P = 0.2). (B) DNMT2 mRNA expression in HBV-exposed cells was delayed compared to that in control cells, with significant differences at both days 2 (P = 0.01) and 4 (P > 0.001) between the two groups. No difference was seen on day 6 (P = 0.31). (C) DNMT3a total mRNA was significantly higher in cells exposed to HBV at day 2 (P = 0.04) and day 4 (P = 0.005) and borderline higher on day 6 (P = 0.051). (D) DNMT3a variant 3 mRNA levels were higher at day 4 (P = 0.02) but did not differ between control and HBV-exposed cells at day 2 (P = 0.33) or 6 (P = 0.12). (E) DNMT3a variant 4 mRNA levels showed a bimodal pattern, with separate peaks at days 2 and 6. The mRNA levels at days 4 and 6 were higher in HBV-infected cells (both P = 0.03), but there was no difference at day 2 (P = 0.8). (F) DNMT3b mRNA levels were higher in HBV-infected cells at day 2 (P = 0.03), no different at day 4 (P = 0.8), and lower at day 6 (P > 0.001).
DNMT3a expression reduces HBV viral protein production.
The next experiments focused on DNMT3 because of its recognized ability to methylate unmethylated DNA (16) and because DNMT3a can methylate stably expressed episomal DNA (17). While DNMT2 showed the greatest overall increase, its role in methylation of DNA remains unclear. Likewise, DNMT1 is generally considered to be mainly responsible for methylation of hemimethylated DNA.
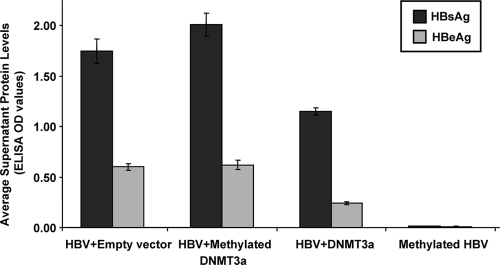
After cotransfection of Huh7 cells with HBV and an empty vector, both HBsAg and HBeAg were detectable in the supernatant (Fig. 2). When HBV was cotransfected with DNMT3a vectors that were inactivated by methylation, there was no decrease in HBsAg or HBeAg production. However, when HBV was transfected with fully active DNMT3a vectors, both HBsAg and HBeAg production significantly decreased (Fig. 2). As expected, HBV that was methylated prior to transfection showed markedly decreased expression of HBsAg or HBeAg (Fig. 2). Methylation of HBV DNA was detected by bisulfite PCR, cloning and sequencing, in experiments of cotransfection with DNMT3 but not with empty vector (data not shown).
FIG. 2.
DNMT3a can downregulate viral protein production. Huh7 cells were cotransfected with full-length HBV and empty vector, full-length HBV and methylated (inactivated) DNMT3a vectors, full-length HBV and DNMT3a vectors, and fully methylated HBV plus empty vector. All data are shown as means ± standard errors of means. Transfection with the methylated DNMT3a vectors did not affect the levels of HBsAg or HBeAg production (P = 0.14 and 0.57, respectively, by Student t tests). However, transfection with DNMT3a vectors significantly reduced both HBsAg and HBeAg secretion (P < 0.001 for both). As expected, methylation of HBV prior to transfection also significantly reduced both HBsAg and HBeAg secretion (P < 0.001 for both).
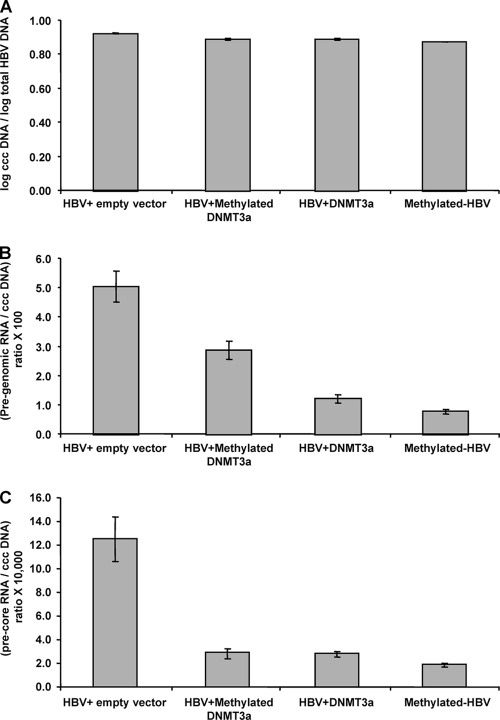
Methylation of HBV cccDNA reduces pregenomic viral RNA.
HBV DNA replication takes place through the RNA intermediary of pregenomic RNA, which is transcribed from the viral cccDNA. Cotransfection of HBV with DNMT3a vectors led to a decrease in the production of pregenomic as well as precore RNA (Fig. 3). Together, these data indicate that methylation regulates production of both HBV protein and pregenomic RNA. Interestingly, the cotransfection with methylated DNMT3a expression vectors led to decreased precore RNA levels and to a lesser extent decreased pregenomic RNA levels. The reasons for this are not clear, and the reduction in viral RNA was not associated with decreased HBsAg or HBeAg in the supernatant. One possibility is that the methylation reactions did not go to completion, leaving some residual DNMT3a activity. In addition, there are at this time no data on the relative effects of methylation on various viral RNA transcripts, and it may be that some viral RNAs are more affected by methylation than others.
FIG. 3.
DNMT3a can downregulate viral RNA and pregenomic RNA production. All data are shown as means ± standard errors of means. (A) After cotransfections, the levels of HBV cccDNA did not differ between the various conditions (all P values not significant). (B) Pregenomic RNA levels were decreased under all conditions compared to the level of the control of HBV plus empty vector (all P < 0.003). Active DNTM3a reduced pregenomic RNA levels more than methylated DNMT3a vectors (P < 0.001). (C) Precore RNA levels were decreased under all conditions compared to the level of the control of HBV plus empty vector (all P < 0.003). Active DNTM3a did not reduce pregenomic RNA levels more than methylated DNMT3a vectors (P = 0.9). However, fully methylated HBV did have lower precore levels than both active DNMT3a (P < 0.001) and methylated DNMT3a (P = 0.04) cotransfections.
Increased DNMT expression leads to methylation of host CpG islands.
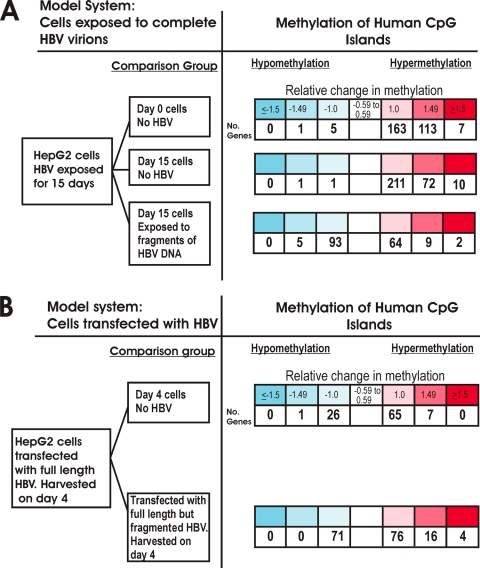
Using the same experimental conditions, a series of cell culture experiments were designed to investigate if exposure of HBV at physiological levels can lead to methylation of host genes. After HepG2 cells were exposed to HBV in the Paran model, 283 CpG islands showed increased methylation compared to that of baseline cells at day 0 (Fig. 4A). Interestingly, a very similar number (293 CpG islands) was found to be hypermethylated in day 15 HBV-infected cells compared to day 15 uninfected cells. A total of 209 hypermethylated CpG islands were found in common among these two experiments. In comparison to both controls, HBV-infected cells showed only a combined total of 8 CpG islands that were hypomethylated after HBV infection (Fig. 4A). These data indicate that HBV infection induces a significant degree of hypermethylation of the human genome. Exposure to HBV DNA alone, without any viral replication, also induces DNMT expression (9) and also led to methylation of host DNA, although to a lesser extent (Fig. 4). Interestingly, exposure to HBV DNA alone also led to a much larger number of CpG islands becoming hypomethylated (Fig. 4).
FIG. 4.
Exposure of HBV leads to increased methylation of host CpG islands. Microarray results are summarized for each experimental condition using heat maps. Log 2-transformed normalized fluorescence ratios are shown, with red indicating hypermethylation and blue indicating hypomethylation. (A) In the model system derived from the work of Paran et al., HepG2 cells were exposed to intact HBV virions in a model that mimics natural infection. Exposed cells showed increased methylation of host CpG islands. Very little hypomethylation was detected. HepG2 cells were also exposed to HBV DNA alone in a manner in which intact cccDNA could not be formed. This led to both hypo- and hypermethylation. (B) In the model system derived from the work of Gunther et al., full-length intact HBV genomes are directly transfected into cells. The same basic pattern emerged, with cells transfected with HBV showing increased methylation of host CpG islands. When cells were transfected with full-length HBV DNA in a manner in which intact cccDNA could not be formed, both hypo- and hypermethylation of host CpG islands were detected.
To confirm these important findings, a second model system was used, in which HBV DNA is directly transfected into cells. The key observations were confirmed: HBV replication induces host DNA methylation, and exposure to HBV DNA with viral replication can induce both hyper- and hypomethylation of host CpG islands (Fig. 4B). Further analysis of those CpG islands with the greatest degree of hypermethylation, with a ≥1.5 log 2 relative increase in methylation, showed that many of the same CpG islands were targeted in experiments with either full HBV virions or full-length HBV. In contrast, there was little gene overlap compared to the situation with cells exposed to full-length, but fragmented, HBV DNA (Fig. 5).
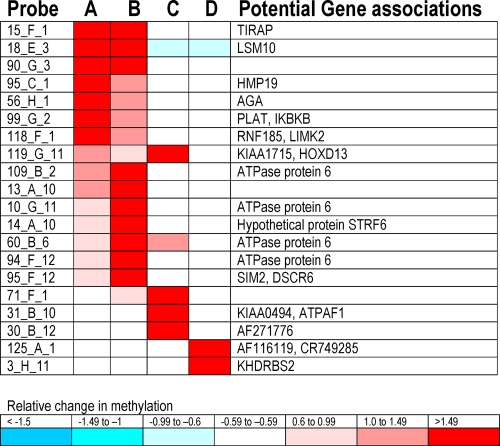
FIG. 5.
Heat map of the CpG islands with normalized fluorescence ratios of ≥1.5. The name of the probe on the chip is shown in the first column. Each probe identifies a specific area of the human genome that contains a CpG island that can be methylated. Each CpG island can have a specific linkage to potential genes, and these genes are shown in the last column. Column A shows the results for the experimental conditions of the HBV infection model compared to those of day 0 cells (row 1 in Fig. 4A); column B corresponds to the experimental conditions of the HBV infection model compared to those of day 15 cells (row 2 in Fig. 4A); column C corresponds to the experimental conditions of the HBV transfection model compared to those of untransfected cells (row 1 in Fig. 4B); column D corresponds to the experimental conditions of the HBV infection model compared to those of cells exposed to full-length but fragmented HBV (row 3 in Fig. 4A).
Ten genes were chosen for confirmation, and day 15 HBV-infected cells showed decreased expression of genes as well as increased methylation of CpG islands compared to those of the control (Table 2). Of these genes, SUFU and TIRAP were selected because of their marked reduction in expression in cells exposed to HBV, and their expression levels were measured in human HCC specimens. Compared to the average level of expression in liver tissue with no known underlying liver disease, cirrhotic liver specimens with HBV infection showed at least a 50% decrease in expression of TIRAP in one of eight cases. In livers with chronic HCV infection, two of eight cases showed at least a 50% decrease in expression of TIRAP. In contrast, for HCCs, four of seven tumors arising in the context of HBV had at least a 50% decrease in TIRAP expression compared to that for HCCs arising in livers with no known underlying liver disease. In addition, six of six cases had decreased SUFU expression, while two cases had no SUFU detected. For HCC arising in HCV cirrhosis, one of eight tumors showed decreased TIRAP and four of seven showed decreased SUFU expression compared to those of the control.
TABLE 2.
Probe designations for confirmation studiesa
| Closest gene | Methylation site designationb | Overlap with classic CpG islandc | Distance (bp) from methylation site to genef | CpG methylation (control:test)d | mRNA fold changee |
|---|---|---|---|---|---|
| POLR2J2 | 49_D_2 | Yes | 0 | 1/1:5/5 | 1.3 ± 0.6 |
| TRIM8g | 63_D_2 | Yes | 2,237 | 1/1:11/6 | 0.45 ± 0.01 |
| SUFUg | Yes | 0.14 ± 0.04 | |||
| LSM10 | 18_E_3 | Yes | 0 | 1/1:7/6 | 0.56 ± 0.2 |
| ONECUT1 | 13_F_3 | Yes | 0 | No difference | 0.75 ± 0.3 |
| TIRAP | 15_F_1 | Yes | 349 | No difference | 0.29 ± 0.01 |
| PINX1 | 15_G_3 | No | 51,937 | No difference | 0.38 ± 0.3 |
| RAI1 | 20_G_2 | No | 17,703 | 0/0:3/3 | 0.65 ± 0.11 |
| TOP3A | 84_H_7 | No | 0 | No difference | 0.73 ± 0.4 |
| NUP62 | 1_H_3 | Yes | 0 | 2/2:7/6 | 0.98 ± 0.07 |
Ten methylation sites were arbitrarily chosen for confirmation studies. The table shows the probe designations on the chip for each of the confirmed human CpG islands.
As defined in the array.
CpG islands as predicted by BLAT (http://genome.ucsc.edu/).
Number of methylated CpG dinucleotides/number of affected clones. DNA from two test and two control wells were pooled for amplification.
Average fold change compared to mRNA in day 0 cells. Two wells from both test and control were independently tested, and data are presented as means ± standard deviations.
0, methylation site is present within the gene.
Either gene may be relevant to this CpG location.
DISCUSSION
The results from this study show that DNMTs are upregulated in cells exposed to HBV and that, in turn, DNMT3a can methylate HBV DNA. This methylation leads to a reduction in viral protein production, viral mRNA production, and pregenomic RNA production. However, in the same experimental conditions, the DNMT upregulation also leads to methylation of host DNA. These findings are important because they show that methylation of human DNA can be induced by HBV infection in model systems where HBV DNA and protein levels closely mimic those of actual human infections. These experimental findings are also important in directly showing that in the same experimental conditions both viral DNA and host DNA are targets for methylation in cells exposed to HBV. These results provide a unifying model of the role of methylation in chronic hepatitis B infection and underscore the dual effects of methylation as potentially both protective and harmful.
Although the functions of individual DNMTs in human tissues vary as a group, they are responsible for maintaining the methylation status of DNA during cell division as well as for de novo CpG methylation. In this study, cells exposed to intact HBV particles responded by increasing the expression of DNMTs, typically within days 2 to 4. This rapid response suggests the possibility that DNMTs may serve as an important part of the cellular response to infection. In this regard, increased DNMT expression can also be triggered in human cells by viruses other than HBV. For example, acute HIV infection of lymphoid cells increases DNA methyltransferase expression (10). Taken together, these observations suggest that DNMT upregulation may be a nonspecific response to cellular infections, i.e., part of the innate immune system's response to HBV infection. In further support of this hypothesis, Huh7 cells express Toll-like receptors that can be triggered by the unmethylated CpG dinucleotides of HBV DNA, activating the NF-κB pathway (9). Activation of the NF-κB pathway has been shown to play a central role in the innate immune system's ability to inhibit HBV replication (3).
Nevertheless, this potentially protective effect of DNMT upregulation may be offset over time either through viral manipulation of the host methylation machinery or through nonspecific methylation of host CpG islands as a result of chronic overexpression of DNMTs. For example, latent viruses, such as Epstein-Barr virus (EBV), are maintained in their latent state in part by methylation, suggesting that some viruses may have evolved strategies to manipulate host DNMTs to their advantage. Interestingly, a number of DNA viruses, including Kaposi's sarcoma-associated herpesvirus (20), HBV (14, 25), and EBV (5), encode viral proteins that can lead to upregulation of DNMTs and eventual methylation of host genes. In this study, we extend our understanding of this mechanism by showing that cells also increase the expression of DNMTs in response to viral replication. Thus, long-term virus-induced overexpression/dysregulation of DNMTs may have deleterious effects, including methylation of host genes. Further support for this possibility is seen in human livers with chronic viral infection or cirrhosis, which have increased levels of DNMTs compared to those in histologically normal livers (18). Hepatocellular carcinomas also show increased DNMT1 and -3 expression compared to those of paired nonneoplastic tissues (11, 18), and virus-related hepatocellular carcinomas tend to have more tumor suppressor gene methylation than those that arise in noncirrhotic livers (24). In the present study, we have directly confirmed these observations reported for human samples by using two separate HBV infection model systems and, in turn, showed that novel genes, such as SUFU and TIRAP, identified experimentally as being downregulated in response to HBV infection can be confirmed in human tissues. Also of interest is the finding that HBV can induce methylation of immunoregulatory genes, including some genes that are active against HBV. For example, HBV replication can cause de novo methylation and decreased expression of interleukin-4 (IL-4) (25), which benefits the virus because IL-4 expression inhibits HBV replication (8).
Taken together, these findings suggest a model (Fig. 6) in which infected cells respond by increasing DNMT expression, which may protect the cell by methylation of viral genomes, effectively turning off viral replication. However, long-term or marked upregulation of these same DNMTs may eventually lead to the inappropriate methylation of host immunoregulatory and tumor suppressor genes (Fig. 6). The HBV infection models used in this study cannot directly address questions of long-term HBV infection, and further studies will be needed to confirm the latter point. However, as previously noted, there is some support for this possibility from studies of human livers with chronic viral infection or cirrhosis, which have shown increased levels of DNMTs compared to those in histologically normal livers (18). Whether this model is applicable to chronic HCV infection is not clear. Hepatocellular carcinomas that arise in livers with chronic HCV hepatitis also have increased methylation of tumor suppressor genes, as seen in both this study and previous reports (24), presumably reflecting dysregulation of DNMT activity. However, the mechanism is unclear.
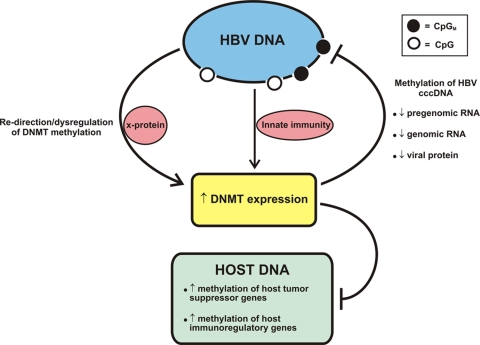
FIG. 6.
Model of HBV infection and its relationship to methylation. In this model, HBV replication is recognized by the infected cell, triggering activation of DNMTs. Once activated, DNMTs can methylate CpG dinucleotides in viral DNA, turning off viral RNA production. The presumed target is cccDNA. However, the increased expression of DNMTs is not exclusively to the benefit of the cell. DNMTs can also be co-opted/dysregulated by the viral X protein, diverting the DNMTs to methylate and downregulate genes that are disadvantageous to viral replication. Also, over time host tumor suppressor promoters can be methylated, leading to hepatocellular carcinomas with a methylator phenotype.
Previous work has shown that methylated nonintegrated HBV DNA can be found in human tissues (4, 22, 23) and that methylation of viral DNA reduces viral mRNA and protein production (23). Furthermore, methylation of CpG island 2 is greater in individuals who are HBeAg negative than in those who are HBeAg positive (4). In this study, these core observations are significantly extended by showing that pregenomic RNA, a key step in viral replication, is also reduced by methylation of cccDNA.
In conclusion, hepatocytes respond to HBV infection by upregulating DNMTs, which can methylate HBV viral DNA, leading to decreased viral replication and decreased viral gene expression. However, this protective effect is not without cost, as host genes can also be methylated at the same time. The factors affecting the ultimate outcome of the increased expression of DNMTs in chronic viral expression warrant further exploration.
Acknowledgments
This work was supported by NIDDK grant R01DK078686 (to M.T.).
Footnotes
Published ahead of print on 10 February 2010.
REFERENCES
- 1.Chen, C. J., H. I. Yang, J. Su, C. L. Jen, S. L. You, S. N. Lu, G. T. Huang, and U. H. Iloeje. 2006. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA 295:65-73. [DOI] [PubMed] [Google Scholar]
- 2.Gunther, S., B. C. Li, S. Miska, D. H. Kruger, H. Meisel, and H. Will. 1995. A novel method for efficient amplification of whole hepatitis B virus genomes permits rapid functional analysis and reveals deletion mutants in immunosuppressed patients. J. Virol. 69:5437-5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo, H., D. Jiang, D. Ma, J. Chang, A. M. Dougherty, A. Cuconati, T. M. Block, and J. T. Guo. 2009. Activation of pattern recognition receptor-mediated innate immunity inhibits the replication of hepatitis B virus in human hepatocyte-derived cells. J. Virol. 83:847-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo, Y., Y. Li, S. Mu, J. Zhang, and Z. Yan. 2009. Evidence that methylation of hepatitis B virus covalently closed circular DNA in liver tissues of patients with chronic hepatitis B modulates HBV replication. J. Med. Virol. 81:1177-1183. [DOI] [PubMed] [Google Scholar]
- 5.Hino, R., H. Uozaki, N. Murakami, T. Ushiku, A. Shinozaki, S. Ishikawa, T. Morikawa, T. Nakaya, T. Sakatani, K. Takada, and M. Fukayama. 2009. Activation of DNA methyltransferase 1 by EBV latent membrane protein 2A leads to promoter hypermethylation of PTEN gene in gastric carcinoma. Cancer Res. 69:2766-2774. [DOI] [PubMed] [Google Scholar]
- 6.Reference deleted.
- 7.Laras, A., J. Koskinas, E. Dimou, A. Kostamena, and S. J. Hadziyannis. 2006. Intrahepatic levels and replicative activity of covalently closed circular hepatitis B virus DNA in chronically infected patients. Hepatology 44:694-702. [DOI] [PubMed] [Google Scholar]
- 8.Lin, S. J., P. Y. Shu, C. Chang, A. K. Ng, and C. P. Hu. 2003. IL-4 suppresses the expression and the replication of hepatitis B virus in the hepatocellular carcinoma cell line Hep3B. J. Immunol. 171:4708-4716. [DOI] [PubMed] [Google Scholar]
- 9.Liu, X., Q. Xu, W. Chen, H. Cao, R. Zheng, and G. Li. 2009. Hepatitis B virus DNA-induced carcinogenesis of human normal liver cells by virtue of nonmethylated CpG DNA. Oncol. Rep. 21:941-947. [DOI] [PubMed] [Google Scholar]
- 10.Mikovits, J. A., H. A. Young, P. Vertino, J. P. Issa, P. M. Pitha, S. Turcoski-Corrales, D. D. Taub, C. L. Petrow, S. B. Baylin, and F. W. Ruscetti. 1998. Infection with human immunodeficiency virus type 1 upregulates DNA methyltransferase, resulting in de novo methylation of the gamma interferon (IFN-gamma) promoter and subsequent downregulation of IFN-gamma production. Mol. Cell. Biol. 18:5166-5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oh, B. K., H. Kim, H. J. Park, Y. H. Shim, J. Choi, C. Park, and Y. N. Park. 2007. DNA methyltransferase expression and DNA methylation in human hepatocellular carcinoma and their clinicopathological correlation. Int. J. Mol. Med. 20:65-73. [PubMed] [Google Scholar]
- 12.Paran, N., B. Geiger, and Y. Shaul. 2001. HBV infection of cell culture: evidence for multivalent and cooperative attachment. EMBO J. 20:4443-4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park, H. J., E. Yu, and Y. H. Shim. 2006. DNA methyltransferase expression and DNA hypermethylation in human hepatocellular carcinoma. Cancer Lett. 233:271-278. [DOI] [PubMed] [Google Scholar]
- 14.Park, I. Y., B. H. Sohn, E. Yu, D. J. Suh, Y. H. Chung, J. H. Lee, S. J. Surzycki, and Y. I. Lee. 2007. Aberrant epigenetic modifications in hepatocarcinogenesis induced by hepatitis B virus X protein. Gastroenterology 132:1476-1494. [DOI] [PubMed] [Google Scholar]
- 15.Pollicino, T., L. Belloni, G. Raffa, N. Pediconi, G. Squadrito, G. Raimondo, and M. Levrero. 2006. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology 130:823-837. [DOI] [PubMed] [Google Scholar]
- 16.Pradhan, S., and P. O. Esteve. 2003. Mammalian DNA (cytosine-5) methyltransferases and their expression. Clin. Immunol. 109:6-16. [DOI] [PubMed] [Google Scholar]
- 17.Ramsahoye, B. H., D. Biniszkiewicz, F. Lyko, V. Clark, A. P. Bird, and R. Jaenisch. 2000. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl. Acad. Sci. U. S. A. 97:5237-5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saito, Y., Y. Kanai, M. Sakamoto, H. Saito, H. Ishii, and S. Hirohashi. 2001. Expression of mRNA for DNA methyltransferases and methyl-CpG-binding proteins and DNA methylation status on CpG islands and pericentromeric satellite regions during human hepatocarcinogenesis. Hepatology 33:561-568. [DOI] [PubMed] [Google Scholar]
- 19.Schumacher, A., P. Kapranov, Z. Kaminsky, J. Flanagan, A. Assadzadeh, P. Yau, C. Virtanen, N. Winegarden, J. Cheng, T. Gingeras, and A. Petronis. 2006. Microarray-based DNA methylation profiling: technology and applications. Nucleic Acids Res. 34:528-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shamay, M., A. Krithivas, J. Zhang, and S. D. Hayward. 2006. Recruitment of the de novo DNA methyltransferase Dnmt3a by Kaposi's sarcoma-associated herpesvirus LANA. Proc. Natl. Acad. Sci. U. S. A. 103:14554-14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vivekanandan, P., R. Kannangai, S. C. Ray, D. L. Thomas, and M. Torbenson. 2008. Comprehensive genetic and epigenetic analysis of occult hepatitis B from liver tissue samples. Clin. Infect. Dis. 46:1227-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vivekanandan, P., D. Thomas, and M. Torbenson. 2008. Hepatitis B viral DNA is methylated in liver tissues. J. Viral Hepat. 15:103-107. [DOI] [PubMed] [Google Scholar]
- 23.Vivekanandan, P., D. Thomas, and M. Torbenson. 2009. Methylation regulates hepatitis B viral protein expression. J. Infect. Dis. 199:1286-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vivekanandan, P., and M. Torbenson. 2008. Epigenetic instability is rare in fibrolamellar carcinomas but common in viral-associated hepatocellular carcinomas. Mod. Pathol. 21:670-675. [DOI] [PubMed] [Google Scholar]
- 25.Zheng, D. L., L. Zhang, N. Cheng, X. Xu, Q. Deng, X. M. Teng, K. S. Wang, X. Zhang, J. Huang, and Z. G. Han. 2009. Epigenetic modification induced by hepatitis B virus X protein via interaction with de novo DNA methyltransferase DNMT3A. J. Hepatol. 50:377-387. [DOI] [PubMed] [Google Scholar]