Abstract
Acidianus filamentous virus 1 (AFV1) (Lipothrixviridae) is an enveloped filamentous virus that was characterized from a crenarchaeal host. It infects Acidianus species that thrive in the acidic hot springs (>85°C and pH <3) of Yellowstone National Park, WY. The AFV1 20.8-kb, linear, double-stranded DNA genome encodes 40 putative open reading frames whose sequences generally show little similarity to other genes in the sequence databases. Because three-dimensional structures are more conserved than sequences and hence are more effective at revealing function, we set out to determine protein structures from putative AFV1 open reading frames (ORF). The crystal structure of ORF157 reveals an α+β protein with a novel fold that remotely resembles the nucleotidyltransferase topology. In vitro, AFV1-157 displays a nuclease activity on linear double-stranded DNA. Alanine substitution mutations demonstrated that E86 is essential to catalysis. AFV1-157 represents a novel class of nuclease, but its exact role in vivo remains to be determined.
The properties of double-stranded DNA (dsDNA) viruses that infect Crenarchaea living in acidic hot springs (pH 1.5 to 3 and 75 to 95°C) are radically different from those of the viruses that infect Bacteria and Eukarya. Not only are the shapes of these viruses distinct from those of all other known viruses, but ∼80% of their open reading frames (ORFs) do not share any sequence similarity with ORFs of other viruses or of cellular life forms apart from other archaeal viruses (44). Seven novel viral families have been created to categorize their unique characteristics: the spindle-shaped Fuselloviridae, the filamentous Rudiviridae and Lipothrixviridae, the bottle-shaped Ampullaviridae, the droplet-shaped Guttaviridae, the spherical Globuloviridae, the two-tailed Bicaudaviridae, and the unclassified Sulfolobus turreted icosahedral virus (STIV) (43). The study of archaeal viruses is still in its infancy compared to that of eukaryal and bacterial viruses, and little is known regarding crenarchaeal virus life cycles, virus-host relationships, genetics, or biochemistry. Further studies of these viruses are expected to provide genetic, biochemical, and evolutionary insight into their crenarchaeal hosts and the requirements for life in the harsh environments. Transcription cycles of the fusellovirus Sulfolobus spindle-shaped virus 1 (SSV1) (16) and the rudiviruses Sulfolobus islandicus rod-shaped virus 1 and 2 (SIRV1 and SIRV2) (24) have been analyzed. Also, recent results concerning the lytic viruses STIV and SIRV2 shed new light on their replication cycle and interaction with their hosts (7, 9, 38). Because structures generally are much better conserved than sequences, structural studies have aimed at uncovering functional and evolutionary relationships that are not apparent from the primary sequence. To date, 11 protein structures from crenarchaeal viruses have been reported (18, 19, 22, 23, 25-30, 33). Most of these proteins share structural similarity with proteins of known function: three winged-helix proteins are likely involved in transcriptional regulation (two from the fusellovirus SSV1 [27, 33] and one from STIV [28]); one glycosyltransferase from STIV displays the GT-A fold (30); one adaptor protein from SSV1 is similar to the repressor of primer (ROP) of Escherichia coli (26); and the major coat protein of STIV has revealed the first evolutionary relationship spanning the three domains of life (25). The structures of the highly conserved ORFs among these viruses, ORF109 of lipothrixvirus AFV3 (22) and ORFB116 from STIV (29), suggest that they are DNA-binding proteins that function in transcriptional regulation.
Filamentous viruses, the most abundant morphotype in these extreme environments, form the new viral order Ligamenvirales, which is divided into the Rudiviridae and Lipothrixviridae families (43). Lipothrixviruses (Acidianus filamentous virus 1 to 9, Thermoproteus tenax virus 1 to 3, and Sulfolobus islandicus filamentous virus) (8, 43), which were the first enveloped filamentous viruses with linear dsDNA genomes discovered, infect acidophilic and hyperthermophilic Crenarchaea. They are classified into α, β, γ, and δ genera based on their genomic properties and on the diversity of their terminal appendages, which are involved in host cell recognition. AFV1 is a γ-lipothrixvirus isolated from an acidic hot spring in Yellowstone National Park, where the temperature is above 85°C and the pH below 3 (6). The linear, double-stranded 20.8-kb DNA genome of AFV1 encodes 40 putative ORFs, 32% of which are homologous to viral ORFs from the lipothrixvirus SIFV and the rudiviruses SIRV1 and SIRV2. The predicted products generally are too dissimilar to the sequences in the public databases to allow functional assignment; only two glycosyltransferases, two CopG-like proteins, and one transcription regulator have been detected (6). It is highly unlikely that all of the encoded proteins consist entirely of unique protein folds serving novel functions. Therefore, to get insight into the biology of the Lipothrixviridae, we performed crystallographic studies of the AFV1 proteome. Although the solved structures of AFV1-102 (23) and the homologs AFV1-99 (19) and SIFV-014 (18) have not revealed any structural homologs, they have suggested that the proteins are involved in protein-protein interaction and are minor structural components, respectively.
Here, we report the crystal structure of AFV1-157 and its biochemical characterization. AFV1-157 is a 157-residue protein with one homolog in the fusellovirus Sulfolobus spindle-shaped virus Ragged Hills (SSVRH). It has a novel α+β fold that remotely resembles the nucleotidyltransferase topology. We demonstrated that (i) AFV1-157 exhibits in vitro nuclease activity that degrades linear dsDNA, and (ii) the E86 residue is essential for the nuclease activity.
MATERIALS AND METHODS
Cloning, expression, and purification.
The coding sequence of AFV1-157 was amplified by PCR from cDNA using two primers containing the attB sites of the Gateway recombination system (Invitrogen). The cDNA was cloned in the pDEST17 plasmid according to the Gateway cloning technology protocol (Invitrogen) (48). A tobacco etch virus (TEV) protease cleavage site was inserted between attB1 and the gene of interest. The resulting vector was transformed in Escherichia coli strain Rosetta(DE3)pLysS (Novagen) (18, 19). After overnight induction with 1 mM isopropyl 1-thio-β-d-galactopyranoside at 25°C, the cells were harvested by centrifugation for 10 min at 4,000 × g. The bacterial pellets were resuspended in lysis buffer (50 mM Tris, pH 8, 300 mM NaCl, 10 mM imidazole) supplemented with 0.25 mg/ml lysozyme, 1 μg/ml DNase, 20 mM MgSO4, and antiproteases (Complete EDTA-free antiproteases; Roche) and were frozen at −80°C. After thawing and sonication, the lysates were heated at 60°C for 15 min and clarified by centrifugation for 30 min at 12,000 × g. The overexpressed proteins were first purified by nickel column affinity chromatography (5 ml His-Trap; GE Healthcare) on a Pharmacia Äkta fast-performance liquid chromatograph (FPLC). The N-terminal hexahistidine tag was cleaved by the addition of the TEV protease with a mass ratio of TEV/AFV1-157 of 1/10 for 1 h at 20°C. The protein then was purified from the flowthrough of nickel affinity chromatography followed by a preparative Superdex 200 HR26/60 gel filtration in 10 mM HEPES, pH 7.5, 500 mM NaCl. Purified proteins were characterized by SDS-PAGE, matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF; Autoflex; Brüker), and trypsin peptide mass fingerprint and far-UV circular dichroism (Jasco J-810). The molecular weight was calculated using analytical size-exclusion chromatography (SEC) on a high-performance liquid chromatography (HPLC) system (Waters) with on-line multiangle laser light-scattering absorbance and refractive index detectors (MALLS/UV/RI) (Wyatt Technology, Santa Barbara, CA) (46). The SEC was performed on a Superose 12 10/30 column (Amersham Bioscineces) with 10 mM HEPES, pH 7.5, 500 mM NaCl as the eluent, and the molecular weight was calculated using ASTRA V software (Wyatt Technology) with a refractive index increment (dn/dc) of 0.180 ml/g.
E86A and K57A mutant production.
The single-amino-acid mutations E86A and K57A were generated using the QuikChange multisite-directed mutagenesis kit (Stratagene). The pDEST17/AFV1-157 vector was completely amplified using PfuTurbo DNA polymerase and mutated primers; the E86 and K57 codons were mutated to encode Ala using the primers E86A-F (5′-GACAAAGACGGAAAGATAGAAGCAAGAACAGTGTACAGCATTGAG-3′) and K57A-F (5′-CTGTTACCAGGCAATTACATCGCATTCGGTCTATACGTATTAAAAAAC- 3′), respectively (substituted nucleotides are in boldface type). The PCR amplification products were treated with the restriction enzyme DpnI (Fermentas) to eliminate the native plasmid. A one-microliter aliquot was transformed into XL10-Gold ultracompetent cells, which then were spread on an LB plate containing 100 μg/ml ampicillin. Mutations were verified by automated DNA sequencing (GATC Biotech, France). The expression and purification protocols were identical to those used for the native protein.
Crystallization and structure determination.
Crystals were obtained by the sitting-drop vapor diffusion method at 20°C using a nanodrop-dispensing robot (Honeybee 961; Cartesian Inc.) in 96-well Greiner crystallization plates. The drops contained 100 nl of the protein at 7.4 mg/ml mixed with 100 nl of crystallization reagent, 100 mM Tris, pH 8.5, 1.0 M LiCl2, 0.01 M NiCl2 (Structure Screens 2; Molecular Dimensions Ltd.). After optimization, one larger crystal was obtained by mixing 300 nl of the protein at 7.4 mg/ml with 100 nl of 100 mM Tris, pH 8, 0.9 M LiCl2, 0.01 M NiCl2. This crystal was soaked for 10 min in 5 mM mercury(II) chloride (Sigma Aldrich) for subsequent anomalous phasing, cryoprotected in mother liquor supplemented with 25% glycerol, and flash-frozen in liquid nitrogen at 100 K. One single-wavelength anomalous diffraction (SAD) data set was collected at the mercury peak wavelength (λ = 1.0005 Å) at beamline ID14-4 of the European Synchrotron Radiation Facility (ESRF; Grenoble, France) using an ADSC Q4r charge-coupled device (CCD) detector. Data extended to 2.0-Å resolution and were processed with MOSFLM/SCALA (11). The crystal was found to belong to the p3121 space group, with cell dimensions a = b = 65.64 Å and c = 85.57 Å, and contained one molecule in the asymmetric unit (VM = 2.8 Å3/Da; 56% solvent) (Table 1). The mercury substructure (two sites) was solved using SHELXD. Phase calculation and density modification were performed using SHELXE (47). ArpWarp (39) was used to autotrace ∼70% of the asymmetric unit, and the remaining part was manually built using COOT (14). The initial model was manually and iteratively completed by following the Fourier difference map through refinement cycles. The model was refined with PHENIX (1) at 2.0-Å resolution using TLS (translation/libration/screw) parameters (49) applied to three TLS groups (residues 5 to 105, 106 to 123, and 123 to 154), yielding R and Rfree values of 0.169 and 0.236, respectively. The stereochemistry of the final model was checked with PROCHECK (31). The atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) at RCSB as entry 3II2.
TABLE 1.
Data collection and refinement statistics
| Parameter | Mercury | Native | K57A | E86A |
|---|---|---|---|---|
| Data collection statistics | ||||
| Space group | P3121 | P3121 | P3121 | P3121 |
| Cell dimensions (Å, °) | a = b = 65.64, c = 85.57 | a = b = 65.44, c = 85.60 | a = b = 64.76, c = 85.74 | a = b = 63.88, c = 85.35 |
| Wavelength (Å) | 1.005 | 0.93 | 0.93 | 0.93 |
| Resolutiona (Å) | 50-2.0 (2.1-2.0) | 40-2.7 (2.85-2.7) | 47-3.1 (3.27-3.1) | 45-3.3 (3.48-3.3) |
| No. of unique reflections | 14,251 | 6,151 | 4,047 | 3,286 |
| Rsyma,b | 0.098 (0.17) | 0.084 (0.46) | 0.07 (0.40) | 0.09 (0.41) |
| I/σIa | 10.7 (6.3) | 10.3 (3.1) | 29.2 (6.8) | 21.2 (5.4) |
| Completnessa (%) | 96.3 (96.7) | 100 (100) | 100 (100) | 99.9 (100) |
| Multiplicitya | 6 (6.2) | 5.2 (5.3) | 11.6 (12) | 10.3 (10.6) |
| Refinement statistics | ||||
| Resolutiona (Å) | 47-2.0 (2.12-2.0) | 25-2.7 (2.77-2.7) | 22-3.10 (3.18-3.1) | 32-3.3 (3.38-3.3) |
| No. of reflectionsa | 14,181 (2,175) | 5,764 (356) | 3,788 (227) | 3,088 (221) |
| Completnessa (%) | 95.3 (95) | 99.8 (97.9) | 100 (100) | 99.8 (98.3) |
| Rwork/Rfreea,c | 0.169/0.236 (0.15/0.20) | 0.27/0.28 (0.32/0.47) | 0.27/0.31 (0.36/0.44) | 0.26/0.31 (0.27/0.31) |
| No. of nonhydrogen atoms | ||||
| Protein | 1,317 | 1,299 | 1,268 | 1,286 |
| Heteroatoms | 8 | 2 | 1 | 1 |
| Water | 120 | |||
| RMSD bond (Å) | 0.010 | 0.015 | 0.012 | 0.013 |
| RMSD angles (°) | 1.14 | 1.55 | 1.33 | 1.42 |
| Ramachandran plot (%) | ||||
| Most favored regions | 100 | 98.6 | 99.3 | 99.3 |
| Additional allowed | 1.4 | 0.7 | 0.7 | |
| Avg B values (Å2) | ||||
| Protein | 32.1 | 76.4 | 87.1 | 105 |
| Heteroatoms | 34.7 | 59.4 | 47.6 | 42.4 |
| Water | 43.2 |
Values in parentheses refer to the highest-resolution bin.
Rsym = ΣhΣi|Ihi − (Ih)|/ΣhΣI(Ih), where Ii is the ith observation of reflection h and (Ih) is the weighted average intensity for all observations l of reflection h.
Calculated from randomly chosen reflections (6%).
The native structure was solved at 2.7-Å resolution and refined with final R and Rfree values of 0.27 and 0.28, respectively. The data set was collected at λ = 0.93 Å (ID14-3; ESRF, Grenoble, France) using an ADSC Q4r CCD detector. Data were processed with MOSFLM/SCALA (11) and reindexed with POINTLESS (15) in the p3121 space group relative to the Hg-SAD data set, applying the operators −h, −k, and l. Refinement cycles were applied to the Hg-SAD structure with REFMAC5 (35) using TLS parameters (49) applied to the entire chain (Table 1). Atomic coordinates and structure factors of the native protein have been deposited in the Protein Data Bank at RCSB as entry 3II3.
The mutants K57A and E86A were crystallized at 7.4 mg/ml in 20% P8000, 200 mM MgCl2, 100 mM Tris, pH 8.5, and in 100 mM Tris, pH 8.5, 1.0 M LiCl2, 0.01 M NiCl2, respectively. The crystal for mutant K57A was cryoprotected in mother liquor supplemented with 20% ethylene glycol. The data were collected at λ = 0.93 Å at ESRF beamline ID14-2 using an ADSC Q4r CCD detector. Resolution extended to 3.1 and 3.3 Å, respectively. The data were processed with XDS (20) (for the mutant K57A) and with MOSFLM (for the mutant E86A), reindexed with POINTLESS (15) applying the operators −h, −k, and l in the p3121 space group, and scaled with SCALA (11). Refinement cycles were applied to the Hg-SAD structure with REFMAC5 (35) using TLS parameters (49) applied to the whole chain. The final K57A mutant structure was refined at 3.1-Å resolution with R and Rfree values of 0.27 and 0.31, respectively. The E86A mutant was refined at 3.3-Å resolution with R and Rfree values of 0.26 and 0.31, respectively (Table 1). The atomic coordinates and structure factors of the K57A and E86A mutants have been deposited in the Protein Data Bank at RCSB as entries 3ILD and 3ILE, respectively.
Nuclease assays.
Time-course nuclease assays were performed by using linear dsDNA from lambda phage and AFV1 virus as substrates. The DNA was treated either with wild-type AFV1-157 or the mutants E86A and K57A. Wild-type or mutant proteins (250 ng/μl) were incubated with the DNA substrates (25 ng/μl) at a DNA:protein mass ratio of 1:10 at 42°C for different time points in the presence of 25 mM MnCl2. Control reactions were performed with the DNA substrate in the presence of 1 μg of bovine serum albumin (BSA). The reactions were stopped by the addition of 50 mM EDTA. The reaction products were analyzed by 1% agarose gel electrophoresis and visualized by ethidium bromide (EtBr) staining. By following the same procedure, SnaBI-digested AFV1 genome (amplified by PCR) was incubated with wild-type and mutant proteins for 30 and 60 min.
Protein structure accession numbers.
The atomic coordinates and structure factors determined in the course of this work have been deposited in the Protein Data Bank at RCSB as entries 3II2, 3II3, 3ILD, and 3ILE.
RESULTS
Structure of AFV1-157.
The AFV1-157 crystal structure was solved by Hg-SAD (see Materials and Methods). The final model, from Ser5 to Ile154, was refined at 2.0-Å resolution with PHENIX, yielding final Rfactor and Rfree values of 0.169 and 0.236, respectively, and a good geometry (Table 1). Because the electron density of residues 106 to 123 was poorly defined, the occupancy of residues 107 to 110 was set at 0.5 during refinement. One mercury atom, refined with a partial occupancy of 0.25, was bound to Cys108-S, and a second mercury atom, refined with partial occupancy of 0.45, was bound to His77-Nδ. The native structure, which was refined at 2.7-Å resolution, superimposed onto the Hg-SAD structure with a root mean square deviation (RMSD) of 0.29 Å for the Cα atoms. Because the structure solved by Hg-SAD was at higher resolution than the native protein, it was used for the rest of the analysis.
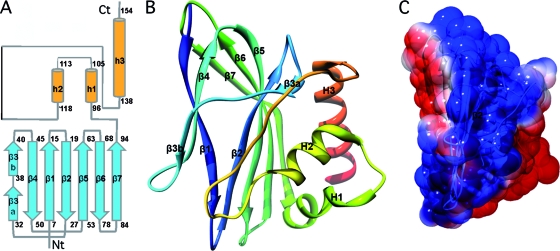
AFV1-157 is an α+β protein with the α and β parts clearly distinguishable in its topological diagram and three-dimensional (3D) structure (Fig. 1A and B). From the N terminus to Tyr95, two antiparallel β-sheets are formed by three and five strands, respectively (Fig. 1B). β-Sheet S1 involves the N terminus of β1 (residues 7 to 12), the C terminus of β3, and strand β4. β-Sheet S2 involves the C terminus of β1, strand β2, the N terminus of β3, and strands β5, β6, and β7. Strands β1 and β3 (β3 is composed of two stretches, β3a and β3b) are involved in both β-sheets S1 and S2. From Trp96 to the C terminus, three helices are packed against one S2 β-sheet side: H1 (residues 96 to 105), H2 (residues 109 to 118), and H3 (residues 138 to 154). Helix H2 is roughly perpendicular to the others and is connected to the longest helix, H3, via a 40-residue loop edging the S2 β-sheet. A 25-Å-width positively charged shallow cleft is delimited by this loop on one side, and the β4 strand and the turn between β6 and β7 are on the other. This surface, surrounded by negative charges, might be involved in nucleic acid interactions (Fig. 1C). Searching with the Dali server for similar structures in the PDB produced many nonsignificant hits with low Z values (3.9 or below), relatively high RMSD values (3 Å at best), and essentially overlapping results on three to four β-strands. This indicates that AFV1-157 has a new α+β fold.
FIG. 1.
Topological diagram and crystal structure of AFV1-157. (A) Topological diagram of the AFV1 ORF157 fold. The β strands and α helices are represented as broad arrows and cylinders, respectively, and are labeled with the residue number. (B) Ribbon representation of the AFV1-157 crystal structure. The color varies from blue to red from the N to the C terminus (rainbow color scheme). The β strands and helices are labeled β1 to β7 and H1 to H3, respectively. (C) The electrostatic potential calculated with APBS (4) (amber force field, 298 K) is represented on the solvent-accessible surface. Blue and red indicate positive (+52 mV) and negative (−52 mV) potential, respectively. Images were generated using Chimera (http://www.cgl.ucsf.edu/chimera/) (40).
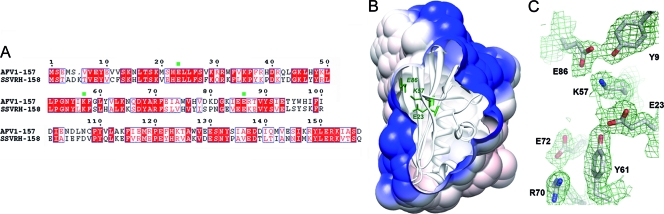
A sequence similarity search using the program PSI-BLAST returned a unique homolog sharing 49% identity, ORF158 from the archaeal fusellovirus Sulfolobus spindle-shaped virus Ragged Hills (SSVRH-158) (Fig. 2A). These proteins do not share sequence motifs or sequence similarities with any other proteins of known function. However, the CATHEDRAL server, which typically is used to access the CATH database by structure (37), classified AFV1-157 into the class alpha and beta proteins with the architecture of a two-layer sandwich. This server proposed the topology nucleotidyltransferase as a remote protein fold group (e-value of 0.69 and SSAP score of 69, the threshold value for the latter being 70), which is represented by the catalytic domain of the avian sarcoma virus (ASV) integrase (PDB no. 1ASV; class, architecture, topology, and homology [CATH] no. 1asvA00). Other members of the polynucleotidyltransferase superfamily include HIV-1 (13) and ASV (10) integrases, RNase H type 1 and 2 (12, 21, 34), the transposase MuA of the bacteriophage Mu (45), and the most distantly related member of this family, the Holliday junction resolvase RuvC from E. coli (3). These enzymes share the well-known RNase H fold, a central five-stranded mixed parallel and antiparallel β-sheet with α-helices on either side. AFV1-157 possesses a different topology and superimposes poorly onto the catalytic core domain of the ASV integrase (RMSD of 3.7 Å for 60 Cα atoms pairs without significant sequence similarity). Nevertheless, due to this slight resemblance and due to the properties of the AFV1-157 electrostatic surface, we tested whether AFV1-157 had any nuclease activity.
FIG. 2.
Putative catalytic residues. (A) Sequence alignment of AFV1-157 and SSVRH-158. Conserved residues are highlighted in red boxes. The three putative catalytic residues, E23, K57, and E86, are labeled with a green point. Sequence alignments were performed with ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/) and are represented with ESPript (17). (B) Visualization of the E23, E86, and K57 side chains as green sticks within the AFV1-157 surface (residues labeled in italics were substituted for alanine). The positive electrostatic potential is represented in blue. The image was generated with Chimera. (C) Close-up view of the putative catalytic residues. The side chains of E23, K57, E86, and surrounding residues are represented by gray sticks in the 2Fo−Fc Fourier map contoured at 1σ. The image was generated with Pymol (http://pymol.sourceforge.net/).
Putative catalytic residues.
Nucleases contain conserved minimal active-site motifs composed of typical acidic and basic residues. These residues may be aspartate, glutamate, histidine, asparagine, or lysine, depending on the nuclease family. These active-site residues coordinate catalytically essential divalent metal cofactors, most commonly magnesium or manganese. In our crystal structure we could assign some Ni2+ ions from the crystallization buffer to the highest peaks in the anomalous difference Fourier map of the native and Hg2+-soaked proteins (f″Ni2+, 0.93 Å = 1.7 e− and f″Ni2+, 1.0005 Å = 1.97 e−). These ions, however, are buried at the interfaces between the monomers and are coordinated by residues from symmetry-related molecules and are therefore likely not coordinated to putative catalytic residues. Moreover, AFV1-157 is a monomer in solution. We did not detect any other divalent metal ions coordinated by carboxylates. A search in the Catalytic Site Atlas (42) for putative catalytic residues, using the structure of AFV1-157 as the query, did not return hits relevant to a nuclease active site. In an attempt to identify the catalytic residues, we mapped all of the acidic and basic residues on the AFV1-157 crystal structure. Putative catalytic residues, E23-E86-K57 in the S1 β-sheet, were identified near the positively charged channel and were considered for functional characterization (Fig. 2B and C).
AFV1-157 nuclease activity.
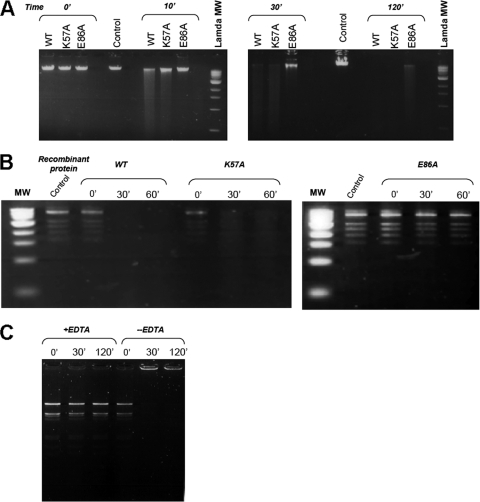
The purity of all analyzed protein samples was assessed after gel filtration by SDS-PAGE, and the structural integrity of the mutants K57A and E86A was verified by solving their crystal structures at 3.1 and 3.3 Å, respectively (Table 1). We performed nuclease assays with the wild-type protein (AFV1-157) and the mutants E86A and K57A on linear dsDNA substrates of phage lambda (Fig. 3A) and virus AFV1 (Fig. 3B). In control experiments performed without any recombinant protein, no nuclease activity could be detected (Fig. 3A and B). Upon incubation with AFV1-157 for 10 min, lambda DNA disappeared with the concomitant formation of a diffuse band, and all DNA was completely degraded after 30 min (Fig. 3A). The result was further confirmed by the degradation of fragments of AFV1 DNA produced by the SnaBI restriction enzyme: all restriction fragments were digested at the same rate (Fig. 3A). The AFV1-157 nuclease activity required the presence of a divalent cation; activity was detected with Mg2+, Zn2+, and Mn2+ and was completely abolished by the addition of EDTA (Fig. 3C).
FIG. 3.
Nuclease activity of AFV1-157 (A) Activity on linear DNA of the lambda phage. DNA was treated with wild-type AFV1-157 or K57A and E86A mutants for 10, 30, and 120 min. Control reactions were carried out for 10 and 120 min. (B) Activity on the AFV1 genome. The control reaction mixture contains no enzyme. The AFV1 DNA restriction fragments were treated with wild-type AFV1-157 or K57A and E86A mutants for 10, 30, and 60 min. (C) Activity in the presence of a divalent cation quencher. Lambda DNA digested with EcoRI-HindIII restriction enzymes was incubated with AFV1-157 and 25 mM MnCl2 either in the presence or absence of 50 mM EDTA for 30 and 120 min.
The nuclease activity of the E86A mutant was highly impaired. Lambda DNA and restriction fragments from AFV1 DNA were unaffected by the mutant, even at longer incubation times (Fig. 3A and B). These results strongly indicate that E86 is involved in the catalysis and is essential for the nuclease activity. The nuclease activity of the K57A mutant was comparable to that of the wild type, as gauged by its ability to degrade DNA within 120 min (Fig. 3).
DISCUSSION
Like the majority of the ORFs in the AFV1 genome, the sequence of AFV1-157 provides no insight into its role in the viral cycle. We purified the recombinant version of this protein to solve its structure and test the ensuing hypothesis of its biochemical function. Its structure reveals a novel α+β fold without structural homologs in the PDB. However, a slight similarity to the nucleotidyltransferase CATH topology prompted us to test for nuclease activity. We demonstrated that in vitro AFV1-157 is able to degrade linear dsDNA. Moreover, E86 was identified as an essential catalytic residue for nuclease activity.
Nucleases often have poorly conserved primary sequences, except for the motifs related to the catalytic site. They can be involved in various biological processes and often display broad substrate specificity, which can complicate the identification of their cellular targets. Sequence-based and structure-based classifications of nucleases have been described (2, 36). Structure databases, such as CATH or SCOP, can provide more relevant functional information than purely sequence-based predictions. From our structural and biochemical data, we propose that AFV1-157 is a representative of a novel class of nucleases.
Nucleases utilize one, two, or three metal cofactors for the hydrolytic reaction. We have demonstrated that the protein is active only in the presence of divalent cations such as Zn2+, Mg2+, and Mn2+. We ignore, however, the identity of the physiological activating metal, the binding stoichiometry, and the nature of the metal-binding sites. Catalytic metals in nucleases fulfill a combination of roles, such as activating the nucleophile (generally a water molecule), neutralizing the negative charge in the transition state, or facilitating the departure of the leaving group (36). It has been reported that the catalytically relevant binding of metal ions may occur only in the presence of the substrate or at ion concentrations much higher than those that are physiologically relevant (41, 50). We showed that E86 is essential for nuclease activity, and we hypothesize that it is involved in metal binding. We further propose that K57, which is positioned between E86 and E23 and is not essential for catalysis, could bind to and orient the substrate rather than stabilize the transition state, as reported for the nuclease MutH (32).
Although we have established that AFV1-157 displays nuclease activity in vitro, its exact role in the viral cycle remains speculative. Nucleases can be involved in ubiquitous viral functions of genome replication and transcription. Moreover, it recently has been demonstrated that AFV1 infection and propagation leads to the loss of the circular form, but not of the integrated form, of pAH1, a conjugative plasmid-like element in Acidanius hospitalis (5). This loss correlates positively with the increase in the intracellular amounts of AFV1 DNA. AFV1-157 therefore also could be involved in the regulation of the existence of pAH1 in its episomal state. AFV1-157 could be part of an in vivo a complex with other viral or cellular proteins such as DNA-binding domains. The identification and characterization of these complexes will be mandatory to unravelling the cellular function of this new nuclease.
Acknowledgments
This work was supported in part by the Marseille-Nice Génopole and by grant ARCHAEALVIR (NT05-2_41674) from the Agence Nationale de Recherche.
Footnotes
Published ahead of print on 3 March 2010.
REFERENCES
- 1.Adams, P. D., R. W. Grosse-Kunstleve, L. W. Hung, T. R. Ioerger, A. J. McCoy, N. W. Moriarty, R. J. Read, J. C. Sacchettini, N. K. Sauter, and T. C. Terwilliger. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948-1954. [DOI] [PubMed] [Google Scholar]
- 2.Aravind, L., D. R. Walker, and E. V. Koonin. 1999. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 27:1223-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ariyoshi, M., D. G. Vassylyev, H. Iwasaki, H. Nakamura, H. Shinagawa, and K. Morikawa. 1994. Atomic structure of the RuvC resolvase: a Holliday junction-specific endonuclease from E. coli. Cell 78:1063-1072. [DOI] [PubMed] [Google Scholar]
- 4.Baker, N. A., D. Sept, S. Joseph, M. J. Holst, and J. A. McCammon. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 98:10037-10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basta, T., J. Smyth, P. Forterre, D. Prangishvili, and X. Peng. 2009. Novel archaeal plasmid pAH1 and its interactions with the lipothrixvirus AFV1. Mol. Microbiol. 71:23-34. [DOI] [PubMed] [Google Scholar]
- 6.Bettstetter, M., X. Peng, R. A. Garrett, and D. Prangishvili. 2003. AFV1, a novel virus infecting hyperthermophilic archaea of the genus acidianus. Virology 315:68-79. [DOI] [PubMed] [Google Scholar]
- 7.Bize, A., E. A. Karlsson, K. Ekefjard, T. E. Quax, M. Pina, M. C. Prevost, P. Forterre, O. Tenaillon, R. Bernander, and D. Prangishvili. 2009. A unique virus release mechanism in the Archaea. Proc. Natl. Acad. Sci. USA 106:11306-11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bize, A., X. Peng, M. Prokofeva, K. Maclellan, S. Lucas, P. Forterre, R. A. Garrett, E. A. Bonch-Osmolovskaya, and D. Prangishvili. 2008. Viruses in acidic geothermal environments of the Kamchatka Peninsula. Res. Microbiol. 159:358-366. [DOI] [PubMed] [Google Scholar]
- 9.Brumfield, S. K., A. C. Ortmann, V. Ruigrok, P. Suci, T. Douglas, and M. J. Young. 2009. Particle assembly and ultrastructural features associated with replication of the lytic archaeal virus sulfolobus turreted icosahedral virus. J. Virol. 83:5964-5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bujacz, G., M. Jaskolski, J. Alexandratos, A. Wlodawer, G. Merkel, R. A. Katz, and A. M. Skalka. 1995. High-resolution structure of the catalytic domain of avian sarcoma virus integrase. J. Mol. Biol. 253:333-346. [DOI] [PubMed] [Google Scholar]
- 11.Collaborative Computing Project No. 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 12.Davies, J. F., Jr., Z. Hostomska, Z. Hostomsky, S. R. Jordan, and D. A. Matthews. 1991. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science 252:88-95. [DOI] [PubMed] [Google Scholar]
- 13.Dyda, F., A. B. Hickman, T. M. Jenkins, A. Engelman, R. Craigie, and D. R. Davies. 1994. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science 266:1981-1986. [DOI] [PubMed] [Google Scholar]
- 14.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 15.Evans, P. 2006. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62:72-82. [DOI] [PubMed] [Google Scholar]
- 16.Fröls, S., P. M. Gordon, M. A. Panlilio, C. Schleper, and C. W. Sensen. 2007. Elucidating the transcription cycle of the UV-inducible hyperthermophilic archaeal virus SSV1 by DNA microarrays. Virology 365:48-59. [DOI] [PubMed] [Google Scholar]
- 17.Gouet, P., E. Courcelle, D. I. Stuart, and F. Metoz. 1999. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15:305-308. [DOI] [PubMed] [Google Scholar]
- 18.Goulet, A., S. Spinelli, S. Blangy, H. van Tilbeurgh, N. Leulliot, T. Basta, D. Prangishvili, C. Cambillau, and V. Campanacci. 2009. The crystal structure of ORF14 from Sulfolobus islandicus filamentous virus. Proteins 76:1020-1022. [DOI] [PubMed] [Google Scholar]
- 19.Goulet, A., S. Spinelli, S. Blangy, H. van Tilbeurgh, N. Leulliot, T. Basta, D. Prangishvili, C. Cambillau, and V. Campanacci. 2009. The thermo- and acido-stable ORF-99 from the archaeal virus AFV1. Protein Sci. 18:1316-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kabsch, W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Applied Crystallography. 26:795-800. [Google Scholar]
- 21.Katayanagi, K., M. Miyagawa, M. Matsushima, M. Ishikawa, S. Kanaya, M. Ikehara, T. Matsuzaki, and K. Morikawa. 1990. Three-dimensional structure of ribonuclease H from E. coli. Nature 347:306-309. [DOI] [PubMed] [Google Scholar]
- 22.Keller, J., N. Leulliot, C. Cambillau, V. Campanacci, S. Porciero, D. Prangishvilli, P. Forterre, D. Cortez, S. Quevillon-Cheruel, and H. van Tilbeurgh. 2007. Crystal structure of AFV3-109, a highly conserved protein from crenarchaeal viruses. Virol. J. 4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keller, J., N. Leulliot, B. Collinet, V. Campanacci, C. Cambillau, D. Pranghisvilli, and H. van Tilbeurgh. 2009. Crystal structure of AFV1-102, a protein from the Acidianus filamentous virus 1. Protein Sci. 18:845-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kessler, A., A. B. Brinkman, J. van der Oost, and D. Prangishvili. 2004. Transcription of the rod-shaped viruses SIRV1 and SIRV2 of the hyperthermophilic archaeon sulfolobus. J. Bacteriol. 186:7745-7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khayat, R., L. Tang, E. T. Larson, C. M. Lawrence, M. Young, and J. E. Johnson. 2005. Structure of an archaeal virus capsid protein reveals a common ancestry to eukaryotic and bacterial viruses. Proc. Natl. Acad. Sci. USA 102:18944-18949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kraft, P., D. Kummel, A. Oeckinghaus, G. H. Gauss, B. Wiedenheft, M. Young, and C. M. Lawrence. 2004. Structure of D-63 from sulfolobus spindle-shaped virus 1: surface properties of the dimeric four-helix bundle suggest an adaptor protein function. J. Virol. 78:7438-7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraft, P., A. Oeckinghaus, D. Kummel, G. H. Gauss, J. Gilmore, B. Wiedenheft, M. Young, and C. M. Lawrence. 2004. Crystal structure of F-93 from Sulfolobus spindle-shaped virus 1, a winged-helix DNA binding protein. J. Virol. 78:11544-11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larson, E. T., B. Eilers, S. Menon, D. Reiter, A. Ortmann, M. J. Young, and C. M. Lawrence. 2007. A winged-helix protein from Sulfolobus turreted icosahedral virus points toward stabilizing disulfide bonds in the intracellular proteins of a hyperthermophilic virus. Virology 368:249-261. [DOI] [PubMed] [Google Scholar]
- 29.Larson, E. T., B. J. Eilers, D. Reiter, A. C. Ortmann, M. J. Young, and C. M. Lawrence. 2007. A new DNA binding protein highly conserved in diverse crenarchaeal viruses. Virology 363:387-396. [DOI] [PubMed] [Google Scholar]
- 30.Larson, E. T., D. Reiter, M. Young, and C. M. Lawrence. 2006. Structure of A197 from Sulfolobus turreted icosahedral virus: a crenarchaeal viral glycosyltransferase exhibiting the GT-A fold. J. Virol. 80:7636-7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laskowski, R. A., D. S. Moss, and J. M. Thornton. 1993. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231:1049-1067. [DOI] [PubMed] [Google Scholar]
- 32.Lee, J. Y., J. Chang, N. Joseph, R. Ghirlando, D. N. Rao, and W. Yang. 2005. MutH complexed with hemi- and unmethylated DNAs: coupling base recognition and DNA cleavage. Mol. Cell 20:155-166. [DOI] [PubMed] [Google Scholar]
- 33.Menon, S. K., W. S. Maaty, G. J. Corn, S. C. Kwok, B. J. Eilers, P. Kraft, E. Gillitzer, M. J. Young, B. Bothner, and C. M. Lawrence. 2008. Cysteine usage in Sulfolobus spindle-shaped virus 1 and extension to hyperthermophilic viruses in general. Virology 376:270-278. [DOI] [PubMed] [Google Scholar]
- 34.Muroya, A., D. Tsuchiya, M. Ishikawa, M. Haruki, M. Morikawa, S. Kanaya, and K. Morikawa. 2001. Catalytic center of an archaeal type 2 ribonuclease H as revealed by X-ray crystallographic and mutational analyses. Protein Sci. 10:707-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 36.Nishino, T., and K. Morikawa. 2002. Structure and function of nucleases in DNA repair: shape, grip and blade of the DNA scissors. Oncogene 21:9022-9032. [DOI] [PubMed] [Google Scholar]
- 37.Orengo, C. A., A. D. Michie, S. Jones, D. T. Jones, M. B. Swindells, and J. M. Thornton. 1997. CATH—a hierarchic classification of protein domain structures. Structure 5:1093-1108. [DOI] [PubMed] [Google Scholar]
- 38.Ortmann, A. C., S. K. Brumfield, J. Walther, K. McInnerney, S. J. Brouns, H. J. van de Werken, B. Bothner, T. Douglas, J. van de Oost, and M. J. Young. 2008. Transcriptome analysis of infection of the archaeon Sulfolobus solfataricus with Sulfolobus turreted icosahedral virus. J. Virol. 82:4874-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perrakis, A., R. Morris, and V. S. Lamzin. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6:458-463. [DOI] [PubMed] [Google Scholar]
- 40.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 41.Pingoud, A., and A. Jeltsch. 2001. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 29:3705-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Porter, C. T., G. J. Bartlett, and J. M. Thornton. 2004. The catalytic site atlas: a resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Res. 32:D129-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prangishvili, D., P. Forterre, and R. A. Garrett. 2006. Viruses of the Archaea: a unifying view. Nat. Rev. Microbiol. 4:837-848. [DOI] [PubMed] [Google Scholar]
- 44.Prangishvili, D., R. A. Garrett, and E. V. Koonin. 2006. Evolutionary genomics of archaeal viruses: unique viral genomes in the third domain of life. Virus Res. 117:52-67. [DOI] [PubMed] [Google Scholar]
- 45.Rice, P., and K. Mizuuchi. 1995. Structure of the bacteriophage Mu transposase core: a common structural motif for DNA transposition and retroviral integration. Cell 82:209-220. [DOI] [PubMed] [Google Scholar]
- 46.Sciara, G., S. Blangy, M. Siponen, S. Mc Grath, D. van Sinderen, M. Tegoni, C. Cambillau, and V. Campanacci. 2008. A topological model of the baseplate of lactococcal phage Tuc2009. J. Biol. Chem. 283:2716-2723. [DOI] [PubMed] [Google Scholar]
- 47.Sheldrick, G. M. 2008. A short history of SHELX. Acta Crystallogr. A 64:112-122. [DOI] [PubMed] [Google Scholar]
- 48.Walhout, A. J., G. F. Temple, M. A. Brasch, J. L. Hartley, M. A. Lorson, S. van den Heuvel, and M. Vidal. 2000. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 328:575-592. [DOI] [PubMed] [Google Scholar]
- 49.Winn, M. D., M. N. Isupov, and G. N. Murshudov. 2001. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 57:122-133. [DOI] [PubMed] [Google Scholar]
- 50.Yang, W., J. Y. Lee, and M. Nowotny. 2006. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell 22:5-13. [DOI] [PubMed] [Google Scholar]