Abstract
Poly(ADP-ribose) polymerase (PARP) is an abundant, chromatin-associated, NAD-dependent enzyme that functions in multiple chromosomal processes, including DNA replication and chromatin remodeling. The Epstein-Barr virus (EBV) origin of plasmid replication (OriP) is a dynamic genetic element that confers stable episome maintenance, DNA replication initiation, and chromatin organization functions. OriP function depends on the EBV-encoded origin binding protein EBNA1. We have previously shown that EBNA1 is subject to negative regulation by poly(ADP-ribosyl)ation (PARylation). We now show that PARP1 physically associates with OriP in latently EBV-infected B cells. Short hairpin RNA depletion of PARP1 enhances OriP replication activity and increases EBNA1, origin recognition complex 2 (ORC2), and minichromosome maintenance complex (MCM) association with OriP. Pharmacological inhibitors of PARP1 enhance OriP plasmid maintenance and increase EBNA1, ORC2, and MCM3 occupancy at OriP. PARylation in vitro inhibits ORC2 recruitment and remodels telomere repeat factor (TRF) binding at the dyad symmetry (DS) element of OriP. Purified PARP1 can ribosylate EBNA1 at multiple sites throughout its amino terminus but not in the carboxy-terminal DNA binding domain. We also show that EBNA1 linking regions (LR1 and LR2) can bind directly to oligomers of PAR. We propose that PARP1-dependent PARylation of EBNA1 and adjacently bound TRF2 induces structural changes at the DS element that reduce EBNA1 DNA binding affinity and functional recruitment of ORC.
Epstein-Barr Virus (EBV) is a human gammaherpesvirus that exists predominantly as a chromatinized episome in latently infected B lymphocytes (20, 33a). The latent infection is associated with B-cell immortalization in vitro and various B-cell and epithelial cell malignancies (46). Latent cycle DNA replication depends on the interaction of virus-encoded EBNA1 protein binding to the viral origin of plasmid replication (OriP) (19, 24). The minimal replicator of OriP consists of the dyad symmetry (DS) element that contains two pairs of EBNA1 binding sites (6, 45). The pairs of EBNA1 binding sites are flanked by telomere repeat factor (TRF) binding sites (33) that bind to TRF2 or TRF1 with different levels of occupancy depending on the stage of the cell cycle (11, 13). The TRF binding sites contribute to DNA replication efficiency and episome maintenance and have been implicated in the RNA-dependent recruitment of the origin recognition complex (ORC) (5, 30). The TRFs can also recruit cellular factors involved in DNA recombination and repair (33), and their complete role in regulating OriP function is not fully elucidated.
Poly(ADP-ribose) polymerase 1 (PARP1) is a highly abundant chromatin-associated enzyme that catalyzes the covalent attachment of ADP-ribose polymers (PAR) to various protein substrates (4, 21, 39, 44). PARP1 is a founding member of an extended family of related enzymes that catalyze PAR from NAD substrates (4). PARP1 binds to chromatin through a high-mobility group (HMG)-like DNA binding domain, and its PAR activity is allosterically activated through binding to single-stranded or nicked DNA substrates (21). PARP1 plays a critical role in various DNA processing activities, including DNA damage recognition and repair, transcription, DNA replication, and telomere end protection (10, 21, 44). PARP1 has also been found to function as a chromatin-organizing factor and can substitute for histone H1 as a linker histone at some chromatin locations (22, 41). Not surprisingly, PARP1 has also been implicated in regulating viral DNA replication and gene expression (16, 31, 42).
PARP1 and another PARP enzyme, tankyrase, have been implicated in the regulation of EBV latent cycle replication and episome maintenance (12, 13). Both PARP1 and tankyrase 1 were identified as proteins that bind to the DS element in biochemical assays. Tankyrase 1 was shown to bind to two regions of EBNA1 and to downregulate the EBNA1 replication function in a PAR-dependent manner. The biochemical effects of EBNA1 poly(ADP-ribosyl)ation (PARylation) were not completely clear, and the additional role of PARP1 was not investigated in detail (12). In this study, we investigate the role of PARP1 in regulation of OriP and the mechanistic effects of PARylation of EBNA1 by PARP1.
MATERIALS AND METHODS
Cells.
EBV-negative adherent 293 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and antibiotics in a 5% CO2 incubator at 37°C. EBV-positive Burkitt lymphoma (BL) cell line (Raji and Mutu I) and EBV-immortalized human B lymphoblastoid cell line (LCL3457) cells were cultured in suspension in RPMI 1640 medium supplemented with 10% fetal bovine serum and antibiotics in a 5% CO2 incubator at 37°C. 293-ZKO cells have been described previously (15) and were cultured as adherent cells in DMEM supplemented with 10% fetal bovine serum and 100 μg/liter hygromycin B in a 5% CO2 incubator at 37°C. 293-ZKO cells carry a bacmid containing the EBV genome lacking the BZFL1 open reading frame (Zta knockout [ZKO]) and express a green fluorescent protein (GFP) and hygromycin B drug resistance gene.
Plasmids and recombinant proteins.
OriP plasmid (N503) is a derivative of pREP10 (Invitrogen), with the enhanced GFP (eGFP) gene inserted into the NheI-Asp718 site (13). Cloning vector pBKSII+ (BKS) is commercially available from Agilent/Strategene, Inc. Short hairpin RNA (shRNA) expression vectors were generated as described previously (32). Briefly, short hairpins of 27 to 29 nucleotides for PARP1 were expressed by the U6 promoter in the pENTR/D-Topo vector (Invitrogen). PGEM1 plasmid containing the U6 promoter was used as the template for PCR with the Sp6 primer (CACCGATTTAGGTGACACTATAG) and PARP1 primer (AAAAAAAGGACAAGACGTACGCTAAGAACAACCCCCAAGCTTCGGAGTTGTTCTTAGCGCACATCTTGTCCCGGTGT TTCGTCCTTTCCACAA). Glutathione S-transferase (GST)-EBNA1 derivatives were cloned into pGEX-4T. EBNA1 derivatives GST-LR1 (where LR1 is linking region 1) and GST-LR2 fusion proteins were described previously (30). Purified proteins for EBNA1 and TRF1 were generated using baculovirus expression systems and purified using an amino-terminal hexahistidine tag. PARP1 protein was purchased from Trevigen, Inc.
Western blotting.
Primary antibodies to EBNA1 (Advanced Biotechnologies, Inc.), PARP1 (Trevigen), PCNA (Santa Cruz Biotechnology), TRF2 and TNKS1 (Imgenex), TRF1 and minichromosome maintenance complex 3 (MCM3) (Abcam), ORC2 (MBL International Corporation), and PAR (monoclonal; Trevigen) were used according to manufacturer's specifications. Rabbit polyclonal antibodies to EBNA1 and human Rap1 (hRap1) were generated against recombinant protein and affinity purified.
In vitro poly(ADP-ribosyl)ation assay.
One microgram of purified GST-EBNA1 protein was incubated in 50 μl of reaction buffer (50 mM Tris-HCl [pH 8] 5 mM KCl, 25 mM MgCl2, and 200 ng of sonicated salmon sperm DNA) with 0.5 units of human PARP1 high specific activity (Trevigen) in the presence or absence of 1 mM NAD+ (Sigma) for 1 h at 25°C. The reaction was terminated by adding 950 μl of ice-cold 25% trichloroacetic acid (TCA; Sigma). The samples were vortexed and then centrifuged at 14,000 × g at 4°C for 10 min. The sediments were washed three times with 20% ice-cold TCA, vortexed, and centrifuged, and the TCA was removed. The precipitates were resuspended in 30 μl of 2× SDS-PAGE loading buffer and subjected to SDS-PAGE and Western blot analysis. For the in vitro poly(ADP-ribosyl)ation of DNA affinity-purified proteins, the samples were incubated in 20 μl of reaction buffer for 1 h at 25°C, and the reaction was terminated by the addition of 2× SDS-PAGE loading buffer.
DNA replication and plasmid maintenance assay.
A DNA replication assay was performed as described previously (11, 13). For replication assays involving shRNAs, the OriP plasmid (3 μg) and a plasmid expressing small interfering RNA (siRNA; 3 μg) were cotransfected into 293 cells using Lipofectamine 2000 reagent (Invitrogen). At 24 h posttransfection, cells were detached by trypsin (0.05%), washed twice with phosphate-buffered saline (PBS), replated onto 10-cm plates, and cultured for another 2 days. Plasmid DNA was extracted by a modified Hirt method (Invitrogen). The nucleic acid pellets were dissolved in 30 μl of Tris-EDTA, and DNA was subjected to restriction digestion with DpnI and BamHI or with only BamHI. DNA samples were analyzed on 0.7% agarose gels in the absence of ethidium bromide and transferred to Zeta membranes (Bio-Rad) for Southern blotting. Radiographic images were quantified by PhosphorImager analysis.
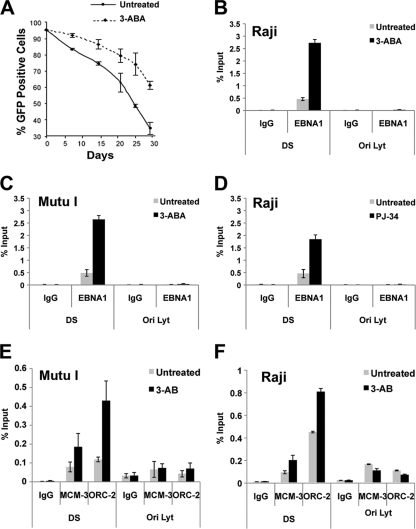
For maintenance of the EBV genome, 293-ZKO cells were cultured in the absence or presence of 3-aminobenzamide ([3-AB] 3 mM) without hygromycin selection. The percentage of GFP-positive cells was measured by fluorescence-activated cell sorter (FACS) analysis every 5 or 10 days.
DNA affinity purification assays.
Soluble nuclear extract fractions were obtained from Raji cells by the Dignam extraction method. Nuclear extract binding with biotinylated DNA bound to streptavidin-labeled magnetic Dynabeads (Dynal Biotech) was described previously (13). Recombinant EBNA1 was prebound to DS element or BKS DNA coupled to magnetic beads for 45 min at room temperature (RT). The bound material was washed at least three times with 1 ml of D150 (20 mM HEPES, pH 7.9, 0.2 mM EDTA, 20% glycerol, 150 mM KCl, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM dithiothreitol [DTT], and protease inhibitor cocktail) containing 0.05% Ipegal (Sigma). The bound proteins were then either eluted from the beads by boiling for 15 min in 2× SDS loading buffer or incubated with NAD+ (1 mM) for in vitro poly(ADP-ribosyl)ation, as described above.
ChIP assay.
A chromatin immunoprecipitation (ChIP) assay followed the protocol provided by Upstate Biotechnology, Inc., with minor modifications as previously described (11). Additional modifications are as follows. DNAs were sonicated to between 200- and 350-bp DNA fragments on a Diagenode Bioruptor according to the manufacturer's protocol, and real-time PCR was performed with a SYBR green probe in an ABI Prism 7000 using 1/100 to 1/2,500 of the ChIP DNA, according to the manufacturer's specified parameters. The rabbit polyclonal antibodies used for the ChIP were as follows: anti-PARP1 (Trevigen), anti-Orc2 (Pharmigen), anti-MCM3 (Abcam), and anti-EBNA1, which was raised against full-length EBNA1. The primers for amplification were as follows: for the DS element, ATGTAAATAAAACCGTGACAGCTCAT and TTACCCAACGGGAAGCATATG; for OriLyt (lytic origin of replication), GCCCGTTGGGTTTCATTAAG and CCAAATCTCGCGGACCTCTA. For ChIP assays involving inhibition of PARP1, EBV-positive cells were cultured in the absence or presence of 3 mM 3-aminobenzamide (Alexis) or 5 μM PJ34 for 72 h prior to analysis by ChIP. For the ChIP assay involving PARP1 depletion, EBV-positive 293-ZKO cells were transfected with a plasmid expressing shRNA against PARP1 (shPARP1) or an empty vector as a negative control (shControl) as described above. Samples were prepared as for the ChIP for EBV-positive cell lines.
EMSA.
Electrophoretic mobility shift assays (EMSAs) were described previously for EBNA1 LR1 and LR2 RNA binding (30). PAR oligomers were purchased from Biomol, Inc., and radiolabeled with polynucleotide kinase and[γ-32P]ATP. Unincorporated nucleotides were removed on a Microspin G50 column (Amersham Biosciences). In a 20-μl reaction mixture, purified GST proteins (200 ng) were added to a reaction mixture containing 0.5 μg of poly(dI-dC), 5% glycerol, 5 mM MgCl2, and 10,000 cpm of 32P-labeled DNA probe. Reaction mixtures were incubated for 30 min at 25°C, electrophoresed in a 1.5% agarose horizontal gel in 0.5× Tris-borate buffer (35) at 60V for 2 h, and visualized by PhosphorImager.
DNase I footprinting analysis.
DNase I footprinting was performed as described previously (13). A 5′ end-labeled DS element probe was generated using 30 μCi of [α-32P]dATP (3,000 Ci/mmol; Perkin-Elmer) and 2 U of Klenow fragment (Roche) for 30 min at 25°C. Purified proteins were incubated in a reaction mixture containing 1× PBS, 5 mM MgCl2, 0.1 mM ZnSO4, 1 mM dithiothreitol, 0.1% NP-40, 10% glycerol, 1 μg of bovine serum albumin, 0.4 μg of poly(dI-dC), and 10,000 cpm of 32P-labeled probe. NAD or 3-AB was added into the reaction mixture, as indicated in the figures, in a final concentration of 1 mM or 3 mM, respectively. The protected probe was digested with different dilutions of DNase I (Sigma) and purified by phenol-chloroform extraction following proteinase K digestion. The DNA samples were then electrophoresed on a 7% denaturing, polyacrylamide sequencing gel at 33 mA and visualized by PhosphorImager.
RESULTS
PARP1 associates with the DS region of OriP in vivo.
Previous studies have shown that PARP1 can be isolated with DS region binding proteins in vitro (13). To determine if PARP1 could associate with the DS element in vivo, we used the ChIP assay to measure its binding to OriP (Fig. 1). We found that ChIP with PARP1 antibodies resulted in a ∼70- to 80-fold enrichment relative to an IgG control at the DS element in latently EBV-infected lymphoblastoid cell lines (LCL) or the Burkitt lymphoma-derived cell line Mutu I. No enrichment of PARP1 was detected at EBV OriLyt, which is largely inactive during latent infection. These findings indicate that PARP1 binds specifically to the DS element in two different cell lines latently infected with EBV.
FIG. 1.
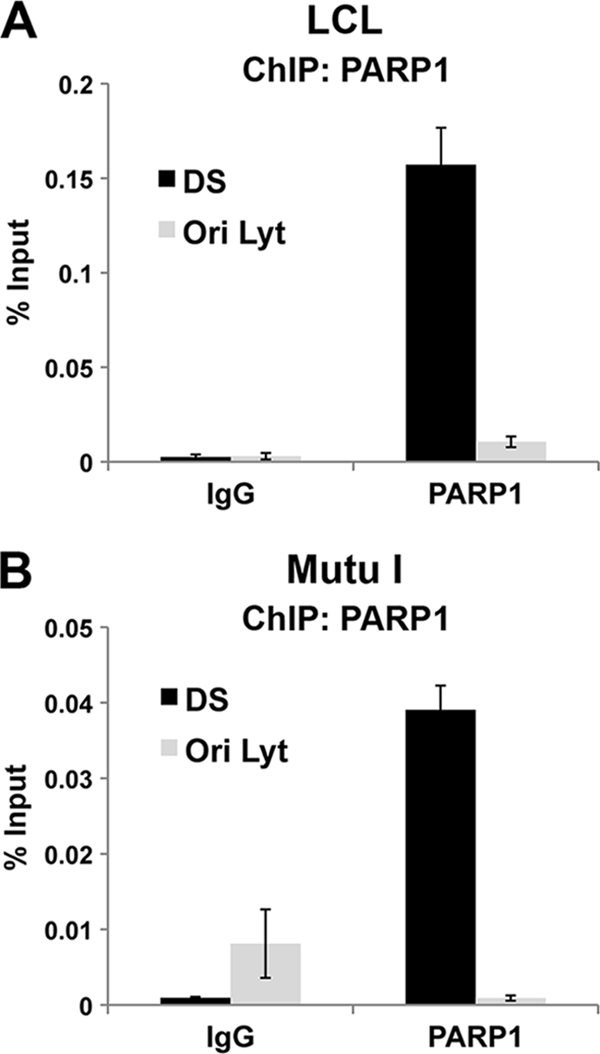
PARP1 interacts with the DS region of OriP in vivo. ChIP assays were performed with anti-PARP1 antibody or an IgG control using EBV-positive LCL or Mutu I cells. ChIP DNA was analyzed by quantitative PCR with primers specific for EBV regions DS or OriLyt.
PARP1 is a negative regulator of OriP-dependent DNA replication.
The functional significance of PARP1 binding to the DS element was assessed using shRNA depletion of PARP1 in 293 cells (Fig. 2). Western blot analysis indicated that PARP1 was efficiently depleted in the shPARP1-transfected cells while control protein PCNA levels were similar in shPARP1- and shControl-transfected cells (Fig. 2A). OriP replication was analyzed by transfection of an OriP-containing plasmid that also expresses EBNA1. DNA replication was assessed by comparing the DpnI-resistant form to the total DNA recovered and linearized by BamHI. In shControl-transfected 293 cells, OriP replication was readily detectable at levels commonly observed (Fig. 2B). In shPARP1-transfected cells, OriP replication was 3.7-fold greater than that in shControl-transfected cells. These findings indicate that PARP1 depletion enhances OriP replication efficiency in 293 cells.
FIG. 2.
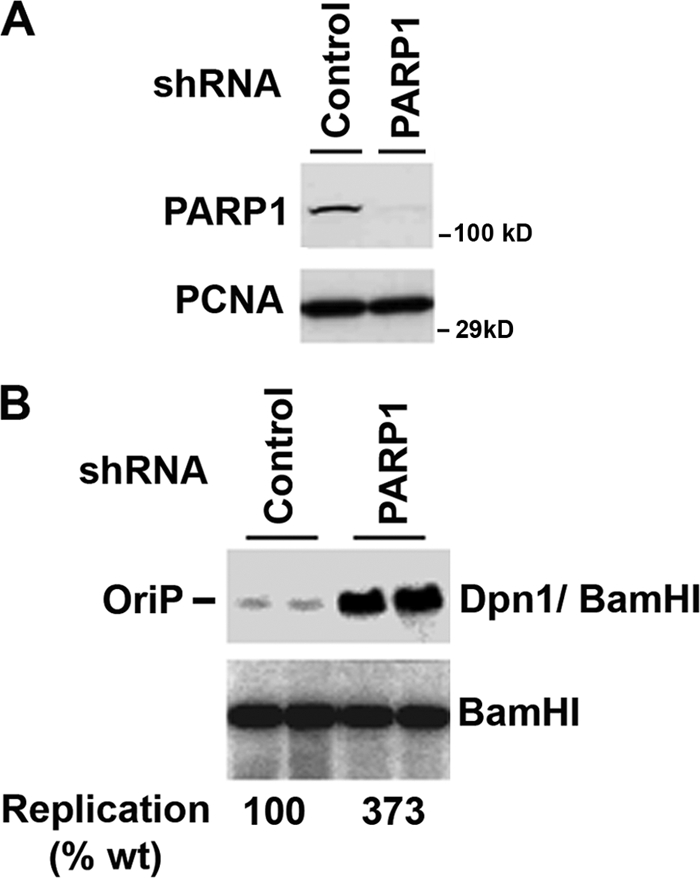
PARP1 depletion enhances OriP-dependent plasmid DNA replication. (A) 293 cells were cotransfected with either shRNA targeting PARP1 or a nontargeting control and with OriP-EBNA1 containing plasmid and subjected to Western blotting. (B) Cells were assayed for transient DNA replication at 72 h posttransfection using DpnI resistance assays. DpnI/BamHI-resistant DNA (top panel) was compared to the total recovered DNA linearized by BamHI (lower panel). The ratio of the DpnI/BamHI to BamHI DNA is indicated below as the percent wild-type control replication (% wt). Experiments are shown in duplicate, and the average of four independent replicates (data not shown) was quantified.
PARP inhibitors enhance EBNA1 binding in vivo.
Previous studies have shown that the PARP inhibitor 3-AB can enhance the episome maintenance of OriP-containing plasmids and stabilize EBV genomes in latently infected B lymphocytes (13). Consistent with these findings, EBV genomes were lost at a lower rate in 293-ZKO cells with 3-AB treatment than in cells without treatment (Fig. 3A). To further investigate the mechanistic role of PARP enzymatic activity on protein interactions at the DS element, we examined the effect of the PARP inhibitors 3-AB or PJ34 on EBNA1 binding to the DS element using ChIP assays. We found that 3 mM 3-AB treatment increases EBNA1 binding ∼8-fold in Raji cells (Fig. 3B) and ∼4-fold in Mutu I cells (Fig. 3C) as measured by ChIP assay. Similarly, the highly specific PJ34 PARP1 inhibitor caused a ∼4-fold increase in EBNA1 binding in Raji cell ChIP assays (Fig. 3D). EBNA1 was not detected at the control region OriLyt, indicating that the ChIP assay is specific for EBNA1-DS element binding.
FIG. 3.
PARP inhibitors increase EBNA1 occupancy at the DS site. (A) 293-ZKO cells were cultured in the presence or absence of 3 mM 3-AB without hygromycin selection. EBV episome maintenance was assayed by FACS analysis of the percentage of GFP-positive cells on the indicated days. The curve reflects the rate of episome loss derived from at least three independent experiments. (B to D) ChIP assays were performed with antibodies specific for EBNA1 or control IgG and analyzed using primers specific for the DS or OriLyt region in Raji cells treated with or without 3-AB (3 mM for 72 h) (B), Mutu I cells treated with or without 3-AB (3 mM for 72 h) (C), or Raji cells treated with or without PJ34 (5 μM for 72 h) (D). (E and F) ChIP assays were performed with antibodies specific for MCM3, ORC2, or control IgG and analyzed using primers specific for the DS or OriLyt region in Mutu I or Raji cells.
Replication initiation at OriP requires the association of the cellular origin recognition complex (ORC) and minichromomosome maintenance complex (MCM) components (9, 14, 34, 37). To determine if the PARP inhibitor 3-AB altered ORC and MCM occupancy at OriP, we assayed ChIP binding of ORC2 and MCM3 in Mutu I (Fig. 3E) or Raji cells (Fig. 3F) after treatment with or without 3-AB. As expected, both MCM3 and ORC2 were enriched at the DS region of OriP relative to control IgG in untreated cells. Addition of 3-AB caused a ∼2.5- to 4-fold increase in both MCM3 and ORC2 in Mutu I and Raji cells. No increase in MCM3 or ORC2 binding was observed at the control OriLyt DNA regions. These findings suggest that inhibitors of PARP1 increase EBNA1 occupancy, as well as cellular prereplication complexes, at the DS element in latently infected Burkitt lymphoma cell lines.
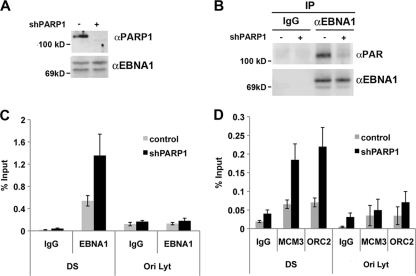
To determine whether PARP1 was responsible for the changes in EBNA1 and prereplication complex binding, we assayed the effects of shRNA depletion of PARP1 in EBV-positive 293-ZKO cells. Cells were transfected with shRNA control or shPARP1 and first assayed by Western blotting to confirm that PARP1 protein was depleted relative to EBNA1, which served as a loading control for this experiment (Fig. 4A). EBNA1 proteins were then subjected to immunoprecipitation using antibodies specific for EBNA1 or control IgG (Fig. 4B). The immunoprecipitated proteins were then assayed by Western blotting with anti-PAR or anti-EBNA1 antibodies. We found that equal quantities of EBNA1 protein were immunoprecipitated from PARP1-depleted cells (Fig. 4B, lower panel) but that this form of EBNA1 had reduced reactivity to PAR antibody (Fig. 4B, top panel). This suggests that PARP1 is required for PARylation of EBNA1 in vivo. The effect of PARP1 depletion on EBNA1 binding to OriP was monitored by ChIP assay (Fig. 4C). We found that PARP1 depletion caused a 2- to 3-fold increase in EBNA1 binding at the DS element but not at the control OriLyt. Similarly, we found that PARP1 depletion caused a ∼3-fold increase in MCM3 and ORC2 binding at the DS element relative to the IgG control ChIP and relative to the control OriLyt DNA (Fig. 4D). The shRNA depletion of PARP1 had results similar to those of pharmacological inhibition of PARP enzymes, suggesting that PARP1 is a primary target of the chemical inhibitors and largely responsible for the PARylation of EBNA1 in vivo.
FIG. 4.
Increased EBNA1 binding and prereplication complex formation at OriP in PARP-depleted cells. (A) 293-ZKO cells were transfected with control or PARP1-targeting shRNA and assayed by Western blotting for depletion of PARP1 or EBNA1. (B) 293-ZKO cells transfected with control or shPARP1 expression vectors, as shown in panel A, were subjected to immunoprecipitation with anti-EBNA1 or control IgG. Immunoprecipitates were then analyzed by Western blotting with anti-PAR or anti-EBNA1. (C) ZKO-293 cells transfected with control or shPARP1 were assayed by ChIP with antibodies to EBNA1 or control IgG and analyzed by quantitative PCR at the DS and control OriLyt DNA regions. (D) The same experiment as in panel C, except that ChIP was performed with antibodies to MCM3, ORC2, or control IgG. α, anti.
PARylation alters protein composition at the DS element.
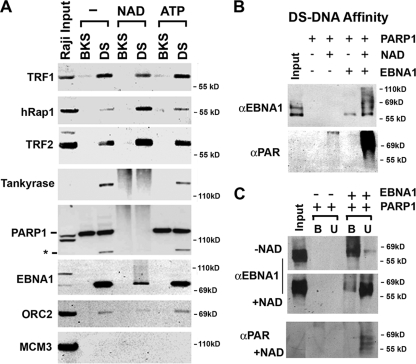
The effect of PARylation on the composition of proteins bound to the DS element was examined by DNA affinity purification followed by Western blot analysis (Fig. 5A). Raji nuclear extracts were incubated with magnetic beads linked to BKS control DNA or with DS element DNA. Bead-bound proteins were then incubated with either NAD, ATP, or buffer alone prior to analysis by Western blotting with antibodies to various known DS element-associated proteins. We found that TRF1 binding was slightly reduced by NAD while TRF2 and hRAP1 were generally increased by NAD treatment. Tankyrase and PARP1 proteins became heterogenous smears after NAD treatment, consistent with their known ability to be auto-ribosylated when they are incubated with NAD. EBNA1 protein also appeared as a heterogenous smear in NAD-treated reactions but to a lesser extent than PARP1 or tankyrase. ORC2 binding was slightly reduced by NAD treatment, but this apparent reduction may reflect a heterogeneous pattern caused by poly(ADP-ribosyl)ation, similar to that observed with EBNA1. MCM3 protein was not detected in any of the bound fractions. ATP treatment had no discernible effect on the bound proteins. We conclude that the NAD causes PARylation of several proteins bound to the DS element, including tankyrase, PARP1, and EBNA1, and alters the binding properties of TRFs and ORC that are thought to be important for DNA replication function.
FIG. 5.
Poly(ADP-ribosyl)ation alters protein binding to the DS element. (A) Raji nuclear extracts were subjected to affinity chromatography with biotinylated DS element or control BKS DNA. Purified complexes coupled with beads were then incubated with buffer control, NAD (1 mM), or ATP (1 mM). Proteins were then eluted and assayed by SDS-PAGE and Western blotting with antibodies for TRF1, hRap1, TRF2, tankyrase 1, PARP1, EBNA1, ORC2, or MCM3. The asterisk indicates a cleaved form of PARP1 that binds specifically to the DS site. (B) Purified recombinant PARP1 and EBNA1 proteins were bound to biotinylated DS element DNA immobilized on magnetic beads and then incubated with or without NAD+ (1 mM). Proteins were then eluted and analyzed by SDS-PAGE and Western blotting with anti-EBNA1 or anti-PAR antibody. (C) DS element DNA affinity-bound PARP1 and EBNA1 were incubated without or with NAD and then separated by centrifugation for bound (B) and unbound (U) proteins. Samples were then analyzed by SDS-PAGE and Western blotting with antibodies to EBNA1 (top panels) or anti-PAR (lower panel).
EBNA1 is subject to PARylation by purified PARP1 in vitro.
EBNA1 has been shown to be subject to PARylation in vivo and in vitro when incubated with purified tankyrase (10). We therefore examined whether PARP1 could also poly(ADP-ribosyl)ate (PARylate) EBNA1 using purified components in vitro (Fig. 5B). EBNA1 protein purified from baculovirus was incubated with purified PARP1 and then bound to the DS element using DNA affinity chromatography. The bound proteins were then incubated with or without NAD. Addition of PARP1 and NAD caused a significant increase in the mobility of EBNA1 protein on SDS-PAGE gels (Fig. 5B, top panel). The higher-mobility form of EBNA1 was also highly reactive to anti-PAR antibody (Fig. 5B, lower panel). These findings indicate that EBNA1 is subject to PARylation when incubated with purified PARP1 and DS element DNA in vitro.
PARylation of EBNA1 alters protein-DNA interactions with the DS element.
The effect of PARP1-mediated PARylation on EBNA1 DNA binding was examined further using a DNA affinity assay (Fig. 5C). Bound or unbound protein was measured by separating the DNA-bound protein from the supernatant. We found that in the absence of NAD, EBNA1 was mostly associated with the bound fraction (Fig. 5C, top panel, lane B). In contrast, addition of NAD caused the majority of EBNA1 to partition to the unbound fraction (Fig. 5C, middle panel, lane U). The PARylation of the unbound EBNA1 proteins was verified by Western blotting of the same samples with anti-PAR antibody (Fig. 5C, lower panel). These finding indicated that PARP1-dependent PARylation of EBNA1 destabilizes EBNA1 interaction with DS element DNA.
PARylation alters protein-DNA interactions at telomere repeat sites of the DS element.
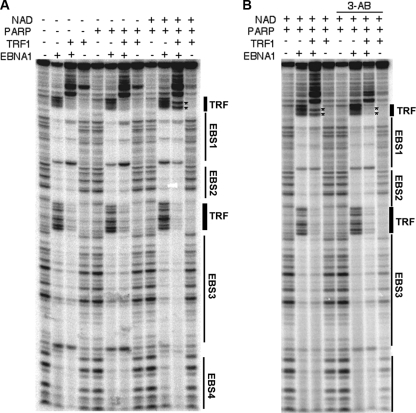
The effect of PARylation on DS element DNA structure was also investigated using a DNase I footprinting assay (Fig. 6). Previous studies had implicated EBNA1 and TRF1 as proteins subject to PARylation. We therefore examined the effect of PARylation on the DS element DNA when it was bound by EBNA1 with or without TRF1. TRF1 and EBNA1 bound to their respective sites on the DS element in the absence of PARP1 (Fig. 6A, lanes 2 to 4, counting from the left). Addition of PARP1 had little effect on the DNase I footprinting pattern (Fig. 6A, lanes 5 to 8). Addition of PARP1 plus NAD, however, caused a strong DNase I-hypersensitive site in the flanking TRF1 binding site (Fig. 6A, lane 11, asterisk). To determine if this DNase I-hypersensitive site was caused by PARP1-dependent PARylation, we tested whether the addition of 3-AB altered this footprinting pattern (Fig. 6B). We found that the PARP1- and NAD-dependent hypersensitive site induced at the TRF1 binding site could be eliminated by the addition of 3-AB (Fig. 6B, compare lanes 3 and 7, counting from the left). Although the precise structure of the protein bound to DNA is not known, we conclude that PARylation of either TRF1 or EBNA1 or both alters the DNase sensitivity at the outermost TRF1 binding sites in the DS element.
FIG. 6.
PARylation alters protein-DNA interactions at telomere repeat binding sites in the DS element. DNase I footprinting of DS element DNA using purified EBNA1, TRF1, and PARP1 with (+) or without (−) NAD, as indicated above each lane. EBNA1 binding sites (EBS) and TRF binding sites are indicated to the right. (B) Reaction mixtures were also incubated with or without 3-AB, as indicated. The asterisks indicate DNase I-hypersensitive sites induced by EBNA1, TRF1, PARP1, and NAD.
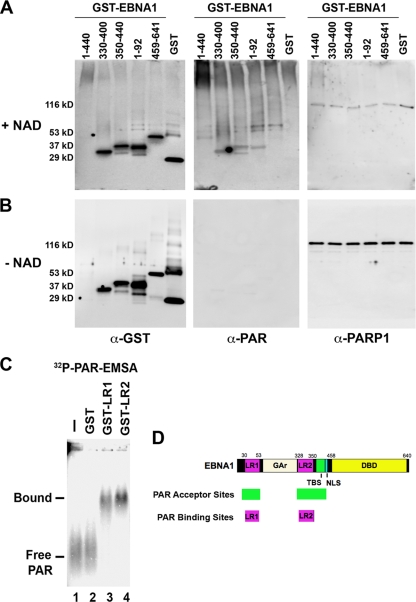
PARP1 can PARylate multiple sites in the EBNA1 amino terminal domain in vitro.
To identify potential PAR acceptor sites in EBNA1, we generated a series of GST-EBNA1 fusion proteins and assayed these for PAR reactivity after incubation with purified PARP1 and NAD in vitro (Fig. 7A and B). GST-EBNA1 comprised of EBNA1 residues 1 to 440 [GST-EBNA1(1-440)], GST-EBNA1(330-400), GST-EBNA1(350-440), GST-EBNA1(1-92), GST-EBNA1(459-641), and GST alone were assayed for NAD-dependent PAR reactivity by Western blotting. We found that GST-EBNA1(330-400), GST-EBNA1(350-440), and GST-EBNA1(1-92) were highly reactive with PAR antibody in the presence of NAD but not in reaction mixtures lacking NAD (Fig. 7A and B, compare middle panels). GST-EBNA1(1-440) was not expressed at high levels but could still be detected by Coomassie staining and Western blotting assays and was found to be PAR reactive. In contrast, GST-EBNA1(459-641) and GST alone were substantially less reactive to PAR antibody relative to their total input protein. Based on these studies, we conclude that PARP1 can PARylate multiple domains in the amino-terminal portion of EBNA1 but does not PARylate the carboxy-terminal DNA binding domain of EBNA1 (summarized in Fig. 7D).
FIG. 7.
EBNA1 domains involved in PARylation and PAR binding. (A) Purified GST-EBNA1 fusion proteins were assayed for in vitro poly(ADP-ribosyl)ation in the presence of NAD (1 mM) and then analyzed by SDS-PAGE and Western blotting with anti-PAR, anti-GST or anti-PARP1 antibody. (B) Purified GST-EBNA1 fusion proteins were assayed as above in the absence of NAD. (C) Purified GST, GST-LR1, or GST-LR2 protein was incubated with 32P-labeled PAR and assayed by EMSA. (D) Schematic of EBNA1 protein domains. Linking regions 1 and 2 (LR1 and LR2), glycine-alanine repeats (GAr), nuclear localization sequence (NLS), and the DNA binding domain (DBD) are indicated. PAR acceptor sites are indicated. (D) Schematic summary of EBNA1 domains involved in PARylation and PAR binding as determined in panels A to C.
EBNA1 can bind PAR through its linking regions (LR1 and LR2).
Previous studies have shown that EBNA1 LR1 and LR2 confer metaphase chromosome attachment function to EBNA1 as well as RNA binding activity (29). Since RNA binding domains may also bind PAR, we assayed the ability of GST-LR1 and GST-LR2 to bind radiolabeled PAR oligomers using EMSAs (Fig. 7C). We found that GST-LR1 and GST-LR2, but not GST alone, could efficiently bind radiolabeled PAR oligomers. These findings indicate that purified LR1 and LR2 can bind to PAR oligomers, suggesting that PAR may regulate EBNA1 functions by interaction with LR1 and LR2.
DISCUSSION
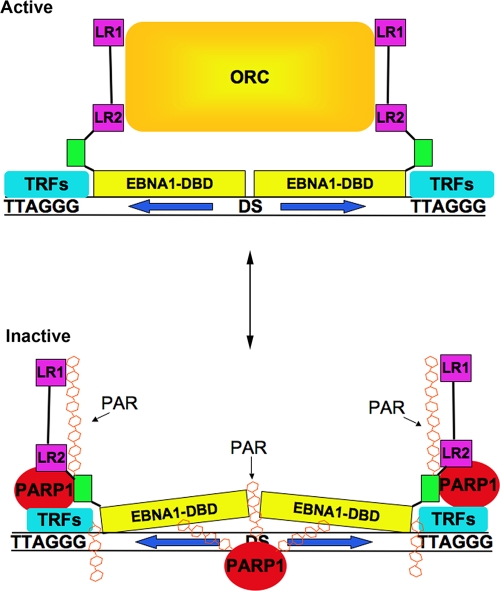
In earlier studies, we showed that OriP can be regulated by poly(ADP-ribosyl)ation (12, 13). We found that two proteins, tankyrase and PARP1, associate with the DS element. Tankyrase was found to bind directly to EBNA1, modify EBNA1 in vitro, and inhibit its DNA replication function (12). In the present study, we focused on the role of PARP1 in the regulation of OriP function. PARP1 localizes to the DS region of OriP in latently infected LCL and BL cells (Fig. 1). Similar to our findings with tankyrase, shRNA depletion of PARP1 enhanced transient replication of OriP (Fig. 2). Inhibitors of PARP, like 3-AB and PJ34, as well as shRNA depletion of PARP1, increased EBNA1, ORC2, and MCM3 occupancy at the DS element as measured by ChIP assay (Fig. 3). Addition of NAD to DS element-bound proteins in vitro resulted in the poly(ADP-ribosyl)ation of EBNA1, PARP1, and tankyrase as well as a remodeling of telomere repeat factors and a decrease in ORC2 binding (Fig. 5A). Poly(ADP-ribosyl)ation of EBNA1 promoted its dissociation from the DS element (Fig. 5B and C) and altered protein-DNA interactions at telomere repeat binding sites adjacent to EBNA1 binding sites in the DS region(Fig. 6). Poly(ADP-ribosyl)ation of EBNA1 was mapped to several subdomains in the EBNA1 amino terminus in vitro (Fig. 7B). Finally, we show that EBNA1 linking regions LR1 and LR2 were capable of binding directly to PAR oligomers in vitro (Fig. 7C). We conclude that PARP1 is a posttranslational modifier of EBNA1 that remodels DS protein-DNA structure to regulate OriP replication activity (Fig. 8).
FIG. 8.
Model of PAR-dependent nucleoprotein remodeling at the DS site. In the absence of PAR, EBNA1 can recruit ORC through interactions with LR1 and LR2, and telomere repeats are bound primarily by TRF2. PARylation induces changes in EBNA1 DNA binding and in LR1 and LR2 recruitment of ORC. PARylation also alters protein interactions with telomere repeats at the DS element. The precise binding sites of PARP1 with proteins and DNA are speculative.
PARP1 is an abundant enzyme that has multiple functions in chromatin organization (21, 41) and DNA damage response pathways (44). The PARP1 catalytic activity is stimulated through its allosteric binding to aberrant DNA structures, like double-strand breaks, single-strand nicks and bulges, and cruciforms (25). The DNA structure at OriP, and especially at the DS element bound by EBNA1, is known to have oxidizable thymine residues and is also capable of forming secondary structures, like cruciforms (7, 43). PARP1 can also interact with multiple cellular proteins, including several involved in DNA damage repair (e.g., DNA ligase IV and topoisomerase I), DNA replication (e.g., DNA polymerase alpha), and telomere length regulation (e.g., ATM, TRF2, and TRF1) (21). We found that PARP1 was highly enriched at the DS element DNA in several EBV-infected cell lines (Fig. 1). Therefore, it is likely that PARP1 associates with the DS element through its ability to recognize aberrant DNA structures and proteins associated with the DS region, including EBNA1, telomere repeat factors, and replication and repair factors known to assemble at the DS element.
Our data suggest that EBNA1 can be posttranslationally modified by PARP1 and that this can lead to a remodeling of protein interactions at the DS element. The regions of EBNA1 between amino acids (aa) 1 to 92 and aa 330 to 440 were efficient substrates for PARylation by PARP1 while the EBNA1 DNA binding domain (aa 459 to 641) was not subject to PARylation by purified PARP1 in vitro (Fig. 7A and B). Previous studies indicated that the EBNA1 regions of aa 58 to 93 and aa 419 to 440 were capable of binding and subject to PARylation by tankyrase (12). The region of aa 419 to 440 of EBNA1 has also been implicated in binding to herpesvirus-associated ubiquitin-specific protease (HAUSP), the ubiquitin protease that regulates MDM2 stability and p53 function (36). The region from aa 58 to 93 has been implicated in transcriptional activation of the Cp EBNA2 promoter (3). Thus, it is possible that PARylation of EBNA1 may regulate interactions of EBNA1 with other proteins, like HAUSP, that bind in close proximity to the PAR acceptor sites.
The linking regions of EBNA1 (LR1 and LR2) were both found to be capable of binding PAR oligomers in vitro (Fig. 7C). LR1 and LR2 have been implicated in the metaphase chromosome tethering function of EBNA1 (28) and are also essential for the DNA replication and transcription activation function of EBNA1 (27). LR1 and LR2 have also been implicated in AT-rich DNA binding (AT-hook activity) (38), as well as RNA binding to G-rich (26, 40) and G-quadruplex structures (29). The RNA binding activity of LR1 and LR2 was correlated with the ORC recruitment function of EBNA1 at OriP (30). PAR binding by these domains may promote interactions of EBNA1 with PARylated proteins, including PARP1. PAR binding may also alter EBNA1 interactions with AT-rich DNA or G-quadruplex RNA, which could disrupt EBNA1 function in metaphase chromosome tethering or ORC recruitment. It is not yet known whether the relative binding affinity for PAR is sufficient to compete with AT-rich DNA or G-quadruplex RNA, nor is it known whether EBNA1 binds PAR oligomers in vivo.
Although PARP1 appears to inhibit transient DNA replication efficiency of OriP, it is also possible that PARP1 provides a positive function to long-term maintenance of episomes containing OriP. PARP1 and PARylation may restrict DNA replication initiation at the DS element to prevent DNA damage and genetic instability. The presence of PARP1 at the DS site could represent a checkpoint for EBV replication. PARP1 is able to recognize DNA damage and recruits proteins involved in DNA repair at the site of the DNA breaks (1, 2). Moreover, PARP1 can detect stalled forks and recruit Mre11 at the replication block and promote replication restart (8). PARP1 may monitor DNA structure at the DS element and disable EBNA1-associated replication function when excessive DNA damage occurs at OriP. PARP1 may also coordinate cellular energy resources with the OriP origin activation function (17, 18). In this scenario, PARP1 may help regulate the relative copy number and replication efficiency of OriP and EBV episomes in response to the cellular metabolic state. In this respect, PARP1 may help preserve EBV genome integrity and stable maintenance.
Our data are also consistent with a role of PARP1 in a host antiviral response. Several studies have indicated that the host DNA repair machinery, of which PARP1 is a major component, can inhibit viral DNA replication (23). The finding that PARP1 can restrict the lytic cycle reactivation of Kaposi's sarcoma-associated herpesvirus (KSHV), another human gammaherpesvirus, strengthens the idea that PARylation could be a mechanism to control viral replication (31). However, as mentioned above, cellular DNA repair enzymes can also provide essential positive functions in viral replication and gene expression. Thus, PARP1 may function as a double agent that protects EBV from excess DNA damage while at the same time protecting the cell from too many EBV genomes. Since PARP1 is both abundant and ubiquitous, it may perform multiple and sometimes paradoxical roles in regulating virus infections.
Acknowledgments
We thank the Wistar Institute Cancer Center Core Facilities for Genomics and Protein Expression.
This work was supported from an NIH grant (CA117830) to P.M.L. and a postdoctoral fellowship from the Istituto Pasteur-Fondazione Cenci Bolognetti, Rome, Italy, to I.T.
Footnotes
Published ahead of print on 10 March 2010.
REFERENCES
- 1.Ahel, I., D. Ahel, T. Matsusaka, A. J. Clark, J. Pines, S. J. Boulton, and S. C. West. 2008. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature 451:81-85. [DOI] [PubMed] [Google Scholar]
- 2.Allinson, S. L., I. I. Dianova, and G. L. Dianov. 2003. Poly(ADP-ribose) polymerase in base excision repair: always engaged, but not essential for DNA damage processing. Acta Biochim. Pol. 50:169-179. [PubMed] [Google Scholar]
- 3.Altmann, M., D. Pich, R. Ruiss, J. Wang, B. Sugden, and W. Hammerschmidt. 2006. Transcriptional activation by EBV nuclear antigen 1 is essential for the expression of EBV's transforming genes. Proc. Natl. Acad. Sci. U. S. A. 103:14188-14193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ame, J. C., C. Spenlehauer, and G. De Murcia. 2004. The PARP superfamily. Bioessays 26:882-893. [DOI] [PubMed] [Google Scholar]
- 5.Atanasiu, C., Z. Deng, A. Wiedmer, J. Norseen, and P. M. Lieberman. 2006. ORC binding to TRF2 stimulates OriP replication. EMBO Rep. 7:716-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bashaw, J. M., and J. L. Yates. 2001. Replication from oriP of Epstein-Barr virus requires exact spacing of two bound dimers of EBNA1 which bend DNA. J. Virol. 75:10603-10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bochkarev, A., E. Bochkareva, L. Frappier, and A. M. Edwards. 1998. The 2.2 Å structure of a permanganate-sensitive DNA site bound by the Epstein-Barr virus origin binding protein, EBNA1. J. Mol. Biol. 284:1273-1278. [DOI] [PubMed] [Google Scholar]
- 8.Bryant, H. E., E. Petermann, N. Schultz, A. S. Jemth, O. Loseva, N. Issaeva, F. Johansson, S. Fernandez, P. McGlynn, and T. Helleday. 2009. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 28:2601-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaudhuri, B., H. Xu, I. Todorov, A. Dutta, and J. L. Yates. 2001. Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein-Barr virus. Proc. Natl. Acad. Sci., U. S. A. 98:10085-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.d'Adda di Fagagna, F., M. P. Hande, W. M. Tong, P. M. Lansdorp, Z. Q. Wang, and S. P. Jackson. 1999. Functions of poly(ADP-ribose) polymerase in controlling telomere length and chromosomal stability. Nat. Genet. 23:76-80. [DOI] [PubMed] [Google Scholar]
- 11.Deng, Z., C. Atanasiu, J. S. Burg, D. Broccoli, and P. M. Lieberman. 2003. Telomere repeat binding factors TRF1, TRF2, and hRAP1 modulate replication of Epstein-Barr virus OriP. J. Virol. 77:11992-12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng, Z., C. Atanasiu, K. Zhao, R. Marmorstein, J. I. Sbodio, N. W. Chi, and P. M. Lieberman. 2005. Inhibition of Epstein-Barr virus OriP function by tankyrase, a telomere-associated poly-ADP ribose polymerase that binds and modifies EBNA1. J. Virol. 79:4640-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng, Z., L. Lezina, C. J. Chen, S. Shtivelband, W. So, and P. M. Lieberman. 2002. Telomeric proteins regulate episomal maintenance of Epstein-Barr virus origin of plasmid replication. Mol. Cell 9:493-503. [DOI] [PubMed] [Google Scholar]
- 14.Dhar, S. K., K. Yoshida, Y. Machida, P. Khaira, B. Chaudhuri, J. A. Wohlschlegel, M. Leffak, J. Yates, and A. Dutta. 2001. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell 106:287-296. [DOI] [PubMed] [Google Scholar]
- 15.Feederle, R., M. Kost, M. Baumann, A. Janz, E. Drouet, W. Hammerschmidt, and H. J. Delecluse. 2000. The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 19:3080-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon-Shaag, A., Y. Yosef, M. Abd El-Latif, and A. Oppenheim. 2003. The abundant nuclear enzyme PARP participates in the life cycle of simian virus 40 and is stimulated by minor capsid protein VP3. J. Virol. 77:4273-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanai, M., W. M. Tong, E. Sugihara, Z. Q. Wang, K. Fukasawa, and M. Miwa. 2003. Involvement of poly(ADP-ribose) polymerase 1 and poly(ADP-ribosyl)ation in regulation of centrosome function. Mol. Cell. Biol. 23:2451-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanai, M., M. Uchida, S. Hanai, N. Uematsu, K. Uchida, and M. Miwa. 2000. Poly(ADP-ribose) polymerase localizes to the centrosomes and chromosomes. Biochem. Biophys. Res. Commun. 278:385-389. [DOI] [PubMed] [Google Scholar]
- 19.Kennedy, G., and B. Sugden. 2003. EBNA-1, a bifunctional transcriptional activator. Mol. Cell. Biol. 23:6901-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kieff, E., and A. B. Rickinson. 2007. Epstein-Barr virus and its replication, p. 2603-2654. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 21.Kraus, W. L. 2008. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr. Opin. Cell Biol. 20:294-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krishnakumar, R., M. J. Gamble, K. M. Frizzell, J. G. Berrocal, M. Kininis, and W. L. Kraus. 2008. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science 319:819-821. [DOI] [PubMed] [Google Scholar]
- 23.Lilley, C. E., R. A. Schwartz, and M. D. Weitzman. 2007. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol. 15:119-126. [DOI] [PubMed] [Google Scholar]
- 24.Lindner, S. E., and B. Sugden. 2007. The plasmid replicon of Epstein-Barr virus: mechanistic insights into efficient, licensed, extrachromosomal replication in human cells. Plasmid 58:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lonskaya, I., V. N. Potaman, L. S. Shlyakhtenko, E. A. Oussatcheva, Y. L. Lyubchenko, and V. A. Soldatenkov. 2005. Regulation of poly(ADP-ribose) polymerase-1 by DNA structure-specific binding. J. Biol. Chem. 280:17076-17083. [DOI] [PubMed] [Google Scholar]
- 26.Lu, C. C., C. W. Wu, S. C. Chang, T. Y. Chen, C. R. Hu, M. Y. Yeh, J. Y. Chen, and M. R. Chen. 2004. Epstein-Barr virus nuclear antigen 1 is a DNA-binding protein with strong RNA-binding activity. J. Gen. Virol. 85:2755-2765. [DOI] [PubMed] [Google Scholar]
- 27.Mackey, D., and B. Sugden. 1999. The linking regions of EBNA1 are essential for its support of replication and transcription. Mol. Cell. Biol. 19:3349-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marechal, V., A. Dehee, R. Chikhi-Brachet, T. Piolot, M. Coppey-Moisan, and J. C. Nicolas. 1999. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J. Virol. 73:4385-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Norseen, J., F. B. Johnson, and P. M. Lieberman. 2009. Role for G-quadruplex RNA binding by Epstein-Barr virus nuclear antigen 1 in DNA replication and metaphase chromosome attachment. J. Virol. 83:10336-10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Norseen, J., A. Thomae, V. Sridharan, A. Aiyar, A. Schepers, and P. M. Lieberman. 2008. RNA-dependent recruitment of the origin recognition complex. EMBO J. 27:3024-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohsaki, E., K. Ueda, S. Sakakibara, E. Do, K. Yada, and K. Yamanishi. 2004. Poly(ADP-ribose) polymerase 1 binds to Kaposi's sarcoma-associated herpesvirus (KSHV) terminal repeat sequence and modulates KSHV replication in latency. J. Virol. 78:9936-9946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paddison, P. J., A. A. Caudy, E. Bernstein, G. J. Hannon, and D. S. Conklin. 2002. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 16:948-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palm, W., and T. de Lange. 2008. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42:301-334. [DOI] [PubMed] [Google Scholar]
- 33a.Rickinson, A. B., and E. Kieff. 2007. Epstein-Barr virus, p. 2655-2700. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 34.Ritzi, M., K. Tillack, J. Gerhardt, E. Ott, S. Humme, E. Kremmer, W. Hammerschmidt, and A. Schepers. 2003. Complex protein-DNA dynamics at the latent origin of DNA replication of Epstein-Barr virus. J. Cell Sci. 116:3971-3984. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Saridakis, V., Y. Sheng, F. Sarkari, M. N. Holowaty, K. Shire, T. Nguyen, R. G. Zhang, J. Liao, W. Lee, A. M. Edwards, C. H. Arrowsmith, and L. Frappier. 2005. Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization. Mol. Cell 18:25-36. [DOI] [PubMed] [Google Scholar]
- 37.Schepers, A., M. Ritzi, K. Bousset, E. Kremmer, J. L. Yates, J. Harwood, J. F. Diffley, and W. Hammerschmidt. 2001. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 20:4588-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sears, J., M. Ujihara, S. Wong, C. Ott, J. Middeldorp, and A. Aiyar. 2004. The amino terminus of Epstein-Barr Virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J. Virol. 78:11487-11505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, S. 2001. The world according to PARP. Trends Biochem. Sci. 26:174-179. [DOI] [PubMed] [Google Scholar]
- 40.Snudden, D. K., J. Hearing, P. R. Smith, F. A. Grasser, and B. E. Griffin. 1994. EBNA-1, the major nuclear antigen of Epstein-Barr virus, resembles “RGG” RNA binding proteins. EMBO J. 13:4840-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wacker, D. A., K. M. Frizzell, T. Zhang, and W. L. Kraus. 2007. Regulation of chromatin structure and chromatin-dependent transcription by poly(ADP-ribose) polymerase-1: possible targets for drug-based therapies. Subcell. Biochem. 41:45-69. [DOI] [PubMed] [Google Scholar]
- 42.Wang, Y., H. Li, Q. Tang, G. G. Maul, and Y. Yuan. 2008. Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: involvement of host cellular factors. J. Virol. 82:2867-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams, D. L., and D. Kowalski. 1993. Easily unwound DNA sequences and hairpin structures in the Epstein-Barr virus origin of plasmid replication. J. Virol. 67:2707-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woodhouse, B. C., and G. L. Dianov. 2008. Poly ADP-ribose polymerase-1: an international molecule of mystery. DNA Repair (Amst.) 7:1077-1086. [DOI] [PubMed] [Google Scholar]
- 45.Yates, J. L., S. M. Camiolo, and J. M. Bashaw. 2000. The minimal replicator of Epstein-Barr virus oriP. J. Virol. 74:4512-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young, L. S., and A. B. Rickinson. 2004. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4:757-768. [DOI] [PubMed] [Google Scholar]