Abstract
ST22-methicillin-resistant Staphylococcus aureus type IV (ST22-MRSA-IV) is endemic in Irish hospitals and is designated antibiogram-resistogram type-pulsed-field group (AR-PFG) 06-01. Isolates of this highly clonal strain exhibit limited numbers of pulsed-field gel electrophoresis (PFGE) patterns and spa types. This study investigated whether combining PFGE and spa typing with DNA sequencing of the staphylococcal cassette chromosome mec element (SCCmec)-associated direct repeat unit (dru typing) would improve isolate discrimination. A total of 173 MRSA isolates recovered in one Irish hospital during periods in 2007 and 2008 were investigated using antibiogram-resistogram (AR), PFGE, spa, dru, and SCCmec typing. Isolates representative of each of the 17 pulsed-field group 01 (PFG-01) spa types identified underwent multilocus sequence typing, and all isolates were ST22. Ninety-seven percent of isolates (168 of 173) exhibited AR-PFG 06-01 or closely related AR patterns, and 163 of these isolates harbored SCCmec type IVh. The combination of PFGE, spa, and dru typing methods significantly improved discrimination of the 168 PFG-01 isolates, yielding 65 type combinations with a Simpson's index of diversity (SID) of 96.53, compared to (i) pairwise combinations of spa and dru typing, spa and PFGE typing, and dru and PFGE typing, which yielded 37, 44, and 43 type combinations with SIDs of 90.84, 91.00, and 93.57, respectively, or (ii) individual spa, dru, and PFGE typing methods, which yielded 17, 17, and 21 types with SIDs of 66.9, 77.83, and 81.34, respectively. Analysis of epidemiological information for a subset of PFG-01 isolates validated the relationships inferred using combined PFGE, spa, and dru typing data. This approach significantly enhances discrimination of ST22-MRSA-IV isolates and could be applied to epidemiological investigations of other highly clonal MRSA strains.
Staphylococcus aureus is an important human pathogen, due largely to its ability to express a wide variety of virulence factors and antimicrobial resistance determinants which are often encoded by mobile genetic elements (7, 10, 21, 24, 25, 39, 56). Methicillin-resistant S. aureus (MRSA) infections are a major public health problem worldwide, both in hospitals and in the community, although the incidence varies. Ireland has one of the highest prevalence rates of nosocomial MRSA infection in Europe and also has an emerging problem with community-acquired MRSA (CA-MRSA) infections (http://www.rivm.nl/earss/result/Monitoring_reports/Annual_reports.jsp) (49).
MRSA first emerged in Irish hospitals in 1971 (22) and, following a major increase in prevalence in the late 1970s, 1980s, and 1990s, has now been endemic in Ireland for three decades (8, 9, 45-48). Molecular typing showed that each decade since the 1970s has been associated with a major shift in the predominant MRSA clonal type in Irish hospitals (51). The clone that predominated in the 1970s and early 1980s, ST250-MRSA-I (or staphylococcal cassette chromosome mec element I [SCCmec I variant]), was replaced by the ST239-MRSA-III (or SCCmec III variant) clone in the mid-1980s, and this clone was in turn displaced by the ST8-MRSA-II clone (harboring SCCmec IIA to IIE) in the 1990s (51). Since the late 1990s, a strain designated locally as antibiogram-resistogram type-pulsed field group (AR-PFG) 06-01, belonging to the international MRSA clone classification ST22-MRSA-IV, which is similar to the United Kingdom epidemic strain EMRSA-15, has predominated in Irish hospitals, and its incidence increased from 22% in 1999 to 80% in 2003 (46, 47).
EMRSA-15 (ST22-MRSA-IV) was first reported in England in 1991 (44) and has since been described as a pandemic MRSA strain due to the predominance of ST22-MRSA-IV among nosocomial MRSA strains in many countries (1, 17, 23, 26, 33, 35, 47, 50, 53). ST22-MRSA-IV has also been identified among patients with hospital-acquired (HA) MRSA infections (11, 42, 43, 60, 63) and CA-MRSA infections (5, 15, 32, 49) in several countries, among health care workers (2, 55), and among companion animals (3, 18, 36, 41).
Informative molecular typing is essential for investigating MRSA strains and populations in individual institutions, countries, and wider geographic areas. This approach permits the genetic relatedness of isolates to be determined, which in turn allows the spread of different MRSA strains to be monitored both locally and globally. However, differentiating among isolates of some MRSA strains is very difficult, particularly in a setting where a single strain is endemic, due to the limited genetic diversity exhibited by MRSA strains such as ST22-MRSA-IV (17, 58). ST22-MRSA-IV isolates yield indistinguishable or closely related pulsed-field gel electrophoresis (PFGE) patterns (17). In Ireland, ST22-MRSA-IV isolates belong to a PFGE group described as pulsed-field group 01 (PFG-01) and exhibit the non-multiantibiotic-resistant antibiogram-resistogram (AR) type AR06 or closely related AR patterns (47). ST22-MRSA-IV isolates also yield a limited number of spa types following DNA sequencing of the protein A (spa) gene (27, 55).
Sequencing of the SCCmec-associated direct repeat unit (dru) of MRSA isolates has shown potential for differentiating MRSA isolates exhibiting limited diversity in PFGE analyses, including EMRSA-15 isolates from Scotland (17, 62). The dru region is a noncoding DNA segment consisting of imperfect 40-bp variable-number tandem repeats (VNTRs) located in the hypervariable region between mecA and IS431mec of SCCmec (17, 38). The majority of MRSA isolates investigated harbor the dru region, which ranges in size from 1 to 15 repeat units (17, 38, 58, 59, 62) (http://www.dru-typing.org). The dru region has been shown to be stable over time by dru typing of individual MRSA isolates following repeated subculture (59), and there is now an internationally agreed-upon dru typing nomenclature and a Web-based dru database (17) (http://www.dru-typing.org).
Currently, there is no effective method for subtyping of ST22-MRSA-IV isolates. The objective of the present study was to investigate the efficacy of dru typing in combination with PFGE and spa typing to discriminate among the highly clonal ST22-MRSA-IV (PFG-01) isolates in an Irish hospital where ST22-MRSA-IV is endemic and to investigate the potential of the combined integrated typing approach to facilitate epidemiological tracking of this MRSA strain.
MATERIALS AND METHODS
Isolates and experimental design.
MRSA isolates (n = 173) from 90 patients and 83 environmental sites in four wards in a 700-bed acute care hospital in Dublin, Ireland, were investigated. The isolates were recovered over two 6-week study periods in each of the four wards between May 2007 and September 2008. Isolates recovered from individual patients and their immediate ward environments during the same 6-week study period are referred to as pairs or triplets of isolates. In the majority of cases, one isolate per patient or patient-associated environmental site was investigated.
The validity of inferences drawn from the typing data was confirmed with epidemiological evidence during a pilot study in one ward. Epidemiological data collected included the numbers for the bed and bed bay corresponding to the patient or the environmental site from whom/which the sample was taken and the sample date and source (i.e., a patient or an environmental site). For patient isolates only, the probable source of the patient's MRSA (whether it was HA or whether the patient was MRSA positive on admission or had a previously known MRSA-positive status) was also recorded. An isolate was deemed to be HA if the patient was negative for MRSA upon admission screening but upon subsequent screening was found to be positive for MRSA.
Isolates were identified as S. aureus and stored in bacterial preserver vials at −70°C and methicillin resistance was confirmed, all as described previously (49). All isolates were typed by AR typing against a panel of 23 antimicrobial agents as described previously (47, 49).
Molecular typing.
All isolates were typed by DNA macrorestriction digestion analysis using SmaI and PFGE, spa, dru, and SCCmec typing. One representative isolate of each spa type identified among the 173 MRSA isolates investigated was typed by multilocus sequence typing (MLST). PFGE was performed as described previously (47). Each PFGE pattern was assigned a 5-digit pulsed-field type (PFT) to allow for future variation in PFGE patterns, and related 5-digit PFTs that differed by ≤6 bands were abbreviated to 2-digit PFGs (47). PFGs were combined with AR typing results to give AR-PFGs (47).
Genomic DNA for use in spa, dru, and SCCmec typing and MLST was extracted using a DNeasy kit according to the instructions of the manufacturer (Qiagen, Crawley, United Kingdom). spa typing was performed using the primers and thermal cycling conditions described by the European Network of Laboratories for Sequence Based Typing of Microbial Pathogens (SeqNet [http://www.seqnet.org]). Analysis of spa sequences and assignment of spa types were performed using the Spa typing plug-in tool of the BioNumerics software package (version 5.1; Applied Maths, Ghent, Belgium). For dru typing, the dru region was amplified and sequenced as described previously (17). The BioNumerics tandem-repeat sequence typing (TRST) plug-in tool was used for dru sequence analysis and assignment of dru types. dru types were assigned using an alphanumeric nomenclature (17). SCCmec typing was performed using four multiplex PCR assays to identify (i) the mec complex type (class A, B, or C) (28), (ii) the ccr complex type (ccrAB1, ccrAB2, ccrAB3, ccrAB4, or ccrC) (28), (iii) the various J regions and mecI (40), and (iv) the SCCmec IV subtype (34). Previously described MRSA control strains were used as positive controls for multiplex PCR assays i to iii (52). The following S. aureus reference strains and clinical isolates were used as positive controls for SCCmec IV subtyping: CA05 (SCCmec IV.1/IVa) (31), 8/63P (SCCmec IV.2/IVb) (31), JCSC4788 (SCCmec IV.3/IVc) (30), JCSC4469 (SCCmec IV.4/IVd) (30), M04/0177 (SCCmec IV.5/IVg) (52), and E1749 (SCCmec IV.6/IVh) (52). MLST was performed and sequences were analyzed as described previously (13, 52).
Investigating the stability of dru types.
The stability of the dru region was investigated using three MRSA isolates that had previously been subjected to dru typing. These comprised two Irish ST8-MRSA-IV isolates, M05/0028 (49) and M06/0376, both of which exhibited dru type dt9g, and one EMRSA-15 isolate from the Harmony collection with dru type dt10h (17, 37). Each isolate was cultured on brain heart infusion (BHI) agar (Becton Dickinson and Company, Sparks, MD) and incubated at 37°C for 24 to 48 h. Several colonies from each isolate were subsequently subcultured on fresh BHI agar plates and incubated at 37°C for 24 to 48 h. This procedure was repeated for a minimum of 10 subcultures over a 14-day period. For each isolate, several colonies from the original and final subculture plates were analyzed by dru typing as described above.
Cluster analyses of spa and dru types.
The BioNumerics Spa typing and TRST plug-in tools were used for cluster analyses of spa and dru types, respectively. With both of these plug-ins, sequences are compared and aligned using an algorithm based on the DSI (duplication, substitution, and indels) model for pairwise alignment of repeats, which considers that modification of sequences can occur through duplication of tandem repeats, substitutions, insertions, and deletions (the latter two events are collectively termed indels) (4). A similarity matrix is generated based on the DSI model and used to construct a minimum spanning tree (MST); the type with the greatest number of related types is assigned as the root node, and the other types derive from this node. In the present study, the default parameters were used for alignment of sequences. The software creates groups of certain distance intervals or similarity values (which BioNumerics terms bins) and converts the data into distance units. Because of the highly clonal nature of the MRSA isolates investigated in the present study, the bin distance was set to 0.5%, i.e., the distance between two entries with >99.5% similarity was 0 (a distance interval of 99.5 to 100% similarity equals a distance of 0) on the MST, and the distance between two entries with 99 to 99.5% similarity was 1 (a distance interval of 99 to 99.5% similarity equals a distance of 1). Using the MSTs, the following criteria were established for clustering of dru types and for clustering of spa types: spa types and dru types were deemed to belong to different clusters if they were separated by an MST distance of >2 (i.e., if they showed <98.5% similarity). Therefore, if two spa types or two dru types were at an MST distance of ≤2, they were considered to be closely related (i.e., they formed a subgroup).
Clustering of isolates.
Each isolate was assigned a 3-digit cluster code with the first number representing the spa type, the second representing the dru type, and the third representing the PFT (Table 1). For example, spa type t032, dru type dt10a, and PFT 01018 were assigned the codes 01, 01 and 1, respectively, and isolates with this spa, dru, and PFGE type combination were assigned the 3-digit cluster code 01.01.1. Subtypes recognized by each typing method were designated by alphabetic suffixes after the relevant numerical element of the cluster code (Table 1). To investigate the overall relatedness of isolates, a composite dendrogram for all PFG-01 (ST22-MRSA-IV) isolates identified during the present study was constructed in BioNumerics by using the averages of the similarity matrices from the individual experiments (PFGE, spa, and dru typing) and clustering by the unweighted-pair group method using average linkages (UPGMA).
TABLE 1.
Cluster code nomenclature used to describe clusters identified by spa, dru, and PFGE typinga
| Method | Type | Repeat succession | Type cluster code | Type subcluster code |
|---|---|---|---|---|
| spa typing | t032 | 26-23-23-13-23-31-29-17-31-29-17-25-17-25-16-28 | 01 | Founder |
| t022 | 26-23-13-23-31-29-17-31-29-17-25-17-25-16-28 | 01 | 01a | |
| t557b | 26-23-23-13-23-31-31-29-17-31-29-17-25-17-25-16-28 | 01 | 01bc | |
| t628 | 26-23-23-13-23-31-29-17-31-29-17-31-29-17-25-17-25-16-28 | 01 | 01c | |
| t1214 | 26-23-23-13-23-31-29-17-31-29-17-25-16-28 | 01 | 01d | |
| t515 | 26-23-23-13-23-31-29-17-31-29-17-25-16-16-28 | 01 | 01da | |
| t4622 | 26-23-23-13-23-31-31-29-17-31-29-17-25-16-16-28 | 01 | 01daa | |
| t018 | 15-12-16-02-16-02-25-17-24-24-24 | 02 | NSC | |
| t1802 | 26-16-16-28 | 03 | NSC | |
| t025 | 26-23-23-13-23-29-17-31-29-17-25-17-25-16-28 | 04 | NSC | |
| t578 | 26-23-23-13-23-31-29-17-31-29-17-25-17-25-28 | 05 | NSC | |
| t4623 | 26-23-13-23-31-29-132-17-31-29-17-25-17-25-16-28 | 06 | NSC | |
| t1865 | 26-23-23-17-31-29-17-25-17-25-16-28 | 07 | NSC | |
| t2951 | 26-16-31-29-17-25-17-25-16-28 | 08 | NSC | |
| t2978 | 04-20-17-20-17-31-16-16-34 | 09 | NSC | |
| t3185 | 26-23-23-20-31-29-17-31-29-17-25-17-25-16-28 | 10 | NSC | |
| t3213 | 26-23-23-13-23-31-29-17-31 | 11 | NSC | |
| t4122 | 26-23-23-13-23-31-29-23-31-29-17-25-17-16-28 | 12 | NSC | |
| t4267 | 26-23-13-23-31-36-25-28 | 13 | NSC | |
| t4765 | 26-23-23-13-23-31-29-17-31-17-25-16-28 | 14 | NSC | |
| t190 | 11-17-34-24-34-22-25 | 15 | NSC | |
| dru typing | dt10a | 5a-2d-4a-0-2d-5b-3a-2g-3b-4e | 01 | NSC |
| dt10j | 5a-2d-4a-0-2d-7a-3a-2g-3b-4e | 01 | 01a | |
| dt10af | 5a-2d-4a-0-2d-2c-3a-2g-3b-4e | 01 | 01b | |
| dt10n | 5a-2d-4a-0-2d-3b-3a-2g-3b-4e | 01 | 01c | |
| dt10i | 5a-2d-4a-0-2d-4f-3a-2g-3b-4e | 01 | 01d | |
| dt10o | 5a-2d-4a-0-2d-4f-3a-2g-2c-4e | 01 | 01da | |
| dt10p | 5a-2d-4a-1b-2d-7a-3a-2g-3b-4e | 01 | 01aa | |
| dt11a | 5a-2d-4a-0-2d-5b-3a-2g-3b-4e-3e | 02 | NSC | |
| dt11o | 5a-2d-4a-0-2d-5b-3a-2g-3b-4e-4e | 02 | 02a | |
| dt11j | 5a-2d-2d-4a-0-2d-5b-3a-2g-3b-4e | 02 | 02aa | |
| dt5b | 5a-2d-4a-5b-3a | 03 | NSC | |
| dt6e | 5a-7a-3a-2g-3b-4e | 04 | NSC | |
| dt7c | 5a-2d-2d-4a-0-3e-3e | 05 | NSC | |
| dt7g | 5a-2d-7a-3a-2g-3b-4e | 06 | NSC | |
| dt7i | 5a-2d-4a-0-2d-3b-4e | 07 | NSC | |
| dt8a | 5a-2d-4a-0-2d-2g-3b-4e | 08 | NSC | |
| dt8p | 6d-0-2d-7a-3a-2g-3b-4e | 09 | NSC | |
| dt9j | 5a-2d-4a-0-2d-5b-3a-2g-3b | 10 | NSC | |
| dt9p | 5a-2d-4a-0-7a-3a-2g-3b-4e | 11 | NSC | |
| PFGE | 01018 | 1 | Founder | |
| 01002 | 1 | 1a | ||
| 01006 | 1 | 1b | ||
| 01022 | 1 | 1c | ||
| 01024 | 1 | 1d | ||
| 01030 | 1 | 1e | ||
| 01032 | 1 | 1f | ||
| 01039 | 1 | 1g | ||
| 01042 | 1 | 1h | ||
| 01047 | 1 | 1i | ||
| 01049 | 1 | 1j | ||
| 01063 | 1 | 1k | ||
| 01075 | 1 | 1l | ||
| 01077 | 1 | 1m | ||
| 01088 | 1 | 1n | ||
| 01114 | 1 | 1o | ||
| 01126 | 1 | 1p | ||
| 01146 | 1 | 1q | ||
| 01151 | 1 | 1r | ||
| 01154 | 1 | 1s | ||
| 01156 | 1 | 1t | ||
| 00041 | 2 | Founder | ||
| 00080 | 2 | 2a | ||
| 00216 | 2 | 2b | ||
| 02017 | 3 | NSC | ||
| 99083 | 4 | NSC |
spa and dru types at a distance of >2 on the MSTs (i.e., types that showed <98.5% similarity) were assigned distinct cluster codes (Fig. 1). spa and dru types that showed >98.5% similarity on the MSTs (i.e., types at an MST distance of ≤2) were assigned spa subcluster codes. Subclusters were assigned alphabetic suffixes following the relevant numerical element of the cluster code. spa types and dru types that were identified as subgroups of spa/dru types that were already assigned to spa/dru subclusters were assigned additional alphabetic suffixes, e.g., spa type t515 was assigned the spa cluster code 01da, as it is a subgroup of t1214 (spa cluster code 01d), which is a subgroup of spa type t032 (spa cluster code 01). For each cluster that consisted of more than one spa or dru type, the type that was assigned as the founder by using the MSTs was not assigned a subcluster code but retained the original cluster code designation. PFTs were designated with distinct cluster codes if they differed by >6 bands. PFTs that differed by ≤6 bands were assigned subcluster codes. For PFT clusters that were represented by more than one PFT, the most frequently occurring PFT was assigned the numerical value for that cluster code and all other PFTs were assigned alphabetic suffixes. NSC, no subcluster code.
Isolates exhibiting spa type t557 were recovered from staff members only during a wider investigation and were assigned spa cluster code 01b but were not included in the present study.
Discriminatory powers of and concordance of data from spa, dru, and PFGE typing methods.
The abilities of PFGE, spa, and dru typing methods alone and in every combination to discriminate among the PFG-01 (ST22-MRSA-IV) isolates investigated were assessed quantitatively by calculating Simpson's indices of diversity (SIDs) with 95% confidence intervals (CI) using an online tool developed by Faria et al. (14) (available at http://www.comparingpartitions.info). SID provides an objective assessment of the discriminatory power of a typing method (14, 20).
The concordance among the data from the typing methods was determined by calculating the adjusted Rand index (ARI) using the online tool mentioned above (14). The ARI indicates the overall concordance between data from two typing methods and includes a correction factor to take into account the possibility that concordance may have arisen by chance. The online tool was also used to calculate the Wallace (W) coefficient (14), which indicates the probability that two isolates classified as the same type by one method will also be classified as the same type by another method. Hence, the W coefficient gives a quantitative estimate of the value of including additional typing methods. A high W coefficient suggests that including a particular additional method does not yield further information. The W coefficient also provides directional information about the concordance of data from typing methods in that it quantifies the probability that isolates clustered by one typing method (e.g., PFGE) will be assigned to the same cluster by a second typing method (e.g., spa typing) and vice versa (14). Where the value of the W coefficient is low when comparing one method to another and results are similar in both directions (e.g., spa to dru versus dru to spa), the inference is that isolates clustered by one typing method may be subdivided by the other typing method (6).
RESULTS
MRSA isolates (n = 173) were recovered from one Dublin hospital during two 6-week study periods in four different wards. The PFTs and AR-PFGs as well as the spa, dru, and SCCmec typing and MLST results for the 173 MRSA isolates are shown in Table 2.
TABLE 2.
DGs, cluster codes, data from spa, dru, PFGE, SCCmec typing, MLST, and AR-PFGs for 173 MRSA isolates
| DG | Cluster codea | spa type | dru type | PFT | AR-PFGb (no. of isolates) | SCCmec type | STc |
|---|---|---|---|---|---|---|---|
| 1a | 01.01.1 | t032 | dt10a | 01018 | 06-01 (8) | IVh | ST22 |
| 1a | 01.01c.1 | t032 | dt10n | 01018 | 06-01 (14) | IVh | ND |
| 1a | 01.01a.1 | t032 | dt10j | 01018 | 06-01 (10) | IVh | ND |
| 1a | 01.01a.1 | t032 | dt10j | 01018 | Unf-01 (5) | IVh | ND |
| 1a | 01da.01a.1 | t515 | dt10j | 01018 | 06-01 (1) | IVh | ND |
| 1a | 04.01a.1 | t025 | dt10j | 01018 | 06-01 (1) | IVh | ST22 |
| 1a | 01.10.1 | t032 | dt9j | 01018 | 06-01 (1) | IVh | ND |
| 1b | 01a.01a.1 | t022 | dt10j | 01018 | 06-01 (1) | IVh | ND |
| 2 | 01c.01.1 | t628 | dt10a | 01018 | 06-01 (9) | IVh | ST22 |
| 3 | 01.06.1 | t032 | dt7g | 01018 | 06-01 (2) | IVh | ND |
| 4a | 01d.01.1g | t1214 | dt10a | 01039 | 06-01 (6) | IVh | ST22 |
| 4a | 01daa.01.1g | t4622 | dt10a | 01039 | 06-01 (1) | IVh | ST22 |
| 4a | 01da.01.1g | t515 | dt10a | 01039 | 06-01 (4) | IVh | ST22 |
| 4a | 01da.01.1g | t515 | dt10a | 01039 | NT-01 (1) | IVh | ND |
| 4a | 01d.02a.1g | t1214 | dt11o | 01039 | 06-01 (1) | IVh | ND |
| 4a | 01da.02a.1g | t515 | dt11o | 01039 | 06-01 (1) | IVh | ND |
| 4b | 01.01a.1g | t032 | dt10j | 01039 | 06-01 (1) | IVh | ND |
| 4b | 01.01a.1g | t032 | dt10j | 01039 | NT-01 (1) | IVh | ND |
| 4b | 01.01.1g | t032 | dt10a | 01039 | 06-01 (3) | IVh | ND |
| 4b | 01.01.1g | t032 | dt10a | 01039 | NT-01 (1) | IVh | ND |
| 4b | 01.01b.1g | t032 | dt10af | 01039 | 06-01 (6) | IVh | ND |
| 4b | 01.01da.1g | t032 | dt10o | 01039 | 06-01 (1) | IVh without dcs | ND |
| 4b | 12.01a.1g | t4122 | dt10j | 01039 | 06-01 (1) | IVh | ST22 |
| 5 | 01da.01.1k | t515 | dt10a | 01063 | 06-01 (1) | IVh | ND |
| 6 | 01a.01.1g | t022 | dt10a | 01039 | 06-01 (4) | IVh | ST22 |
| 6 | 01a.01aa.1g | t022 | dt10p | 01039 | 06-01 (2) | IVh | ND |
| 7 | 01da.07.1g | t515 | dt7i | 01039 | 06-01 (1) | IVh | ND |
| 8 | 01da.08.1g | t515 | dt8a | 01039 | 06-01 (1) | IVh | ND |
| 9 | 07.01a.1 | t1865 | dt10j | 01018 | 06-01 (3) | IVh | ST22 |
| 10 | 07.01a.1a | t1865 | dt10j | 01002 | 06-01 (1) | IVh | ND |
| 11 | 01.01c.1i | t032 | dt10n | 01047 | 06-01 (2) | IVh | ND |
| 12 | 06.01.1 m | t4623 | dt10a | 01077 | 06-01 (1) | IVh | ST22 |
| 13 | 01.01.1b | t032 | dt10a | 01006 | Unf-01 (1) | IVh | ND |
| 13 | 01.01c.1b | t032 | dt10n | 01006 | 06-01 (1) | IVh | ND |
| 13 | 01.01a.1b | t032 | dt10j | 01006 | Unf-01 (1) | IVh | ND |
| 14 | 01d.01.1b | t1214 | dt10a | 01006 | 06-01 (1) | IVh | ND |
| 15 | 01.01a.1e | t032 | dt10j | 01030 | 06-01 (1) | IVh | ND |
| 16 | 01.01a.1h | t032 | dt10j | 01042 | NT-01 (1) | IVh | ND |
| 16 | 01da.01.1h | t515 | dt10a | 01042 | 06-01 (3) | IVh | ND |
| 16 | 01.01.1h | t032 | dt10a | 01042 | 06-01 (1) | IVh | ND |
| 16 | 01.01c.1h | t032 | dt10n | 01042 | 06-01 (1) | IVh | ND |
| 16 | 01d.01.1h | t1214 | dt10a | 01042 | 06-01 (1) | IVh | ND |
| 17 | 01.01a.1d | t032 | dt10j | 01024 | 06-01 (2) | IVh | ND |
| 17 | 01.01a.1d | t032 | dt10j | 01024 | NT-01 (1) | IVh | ND |
| 17 | 01.01a.1d | t032 | dt10j | 01024 | Unf-01 (7) | IVh | ND |
| 17 | 01.01c.1d | t032 | dt10n | 01024 | 06-01 (4) | IVh | ND |
| 18 | 01.01a.1q | t032 | dt10j | 01146 | Unf-01 (1) | IVh | ND |
| 19 | 01da.01.1c | t515 | dt10a | 01022 | 06-01 (2) | IVh (n = 1) | ND |
| IV, nonsubtypeable (n = 1) | ND | ||||||
| 20 | 01.01.1l | t032 | dt10a | 01075 | 06-01 (1) | IVh | ND |
| 20 | 01.02aa.1l | t032 | dt11j | 01075 | 06-01 (1) | IVh | ND |
| 21 | 01.03.1 | t032 | dt5b | 01018 | 06-01 (2) | IVh | ND |
| 22 | 10.01da.1s | t3185 | dt10o | 01154 | 06-01 (2) | IVh | ND |
| 22 | 10.01da.1s | t3185 | dt10o | 01154 | Unf-01 (13) | IVh | ST22 |
| 23 | 01.01.1j | t032 | dt10a | 01049 | 06-01 (2) | IVh | ND |
| 23 | 05.01.1j | t578 | dt10a | 01049 | 06-01 (1) | IVh | ST22 |
| 23 | 01da.02aa.1j | t515 | dt11j | 01049 | 06-01 (1) | IVh | ND |
| 24 | 01.01a.1a | t032 | dt10j | 01002 | 06-01 (3) | IVh | ND |
| 24 | 01da.01.1a | t515 | dt10a | 01002 | 06-01 (1) | IVh | ND |
| 25 | 01.11.1j | t032 | dt9p | 01049 | 06-01 (1) | IVh | ND |
| 26a | 01a.09.1j | t022 | dt8p | 01049 | 06-01 (3) | IVh | ND |
| 26b | 01.09.1j | t032 | dt8p | 01049 | 06-01 (1) | IVh | ND |
| 27 | 11.01.1g | t3213 | dt10a | 01039 | 06-01 (1) | IVh | ST22 |
| 28 | 11.01.1h | t3213 | dt10a | 01042 | 06-01 (1) | IVh | ND |
| 29 | 13.01.1g | t4267 | dt10a | 01039 | 06-01 (1) | IVa | ST22 |
| 30 | 14.01.1k | t4765 | dt10a | 01063 | 06-01 (1) | IVh | ST22 |
| 31 | 01.01.1f | t032 | dt10a | 01032 | 06-01 (1) | IVh | ND |
| 32 | 01.01.1r | t032 | dt10a | 01151 | 06-01 (1) | IVh | ND |
| 33 | 01.01a.1t | t032 | dt10j | 01156 | 06-01 (1) | IVh with ccrAB4 | ND |
| 34 | 01.02.1p | t032 | dt11a | 01126 | 06-01 (1) | IVh with ccrC and Tn554 to orfX | ND |
| 35 | 03.01.1g | t1802 | dt10a | 01039 | 06-01 (1) | IVh | ST22 |
| 36 | 01.01.1o | t032 | dt10a | 01114 | 06-01 (1) | IVh | ND |
| 37 | 08.04.1n | t2951 | dt6e | 01088 | 06-01 (1) | IVh | ST22 |
| 38 | 02.05.3 | t018 | dt7c | 02017 | NT-02 (1) | II without pUB110 | ST36 |
| 39 | 15.01a.2 | t190 | dt10j | 00041 | New03-00 (1) | Characterized by ccrAB4, class A mec, mecI, and dcs | ST8 |
| 40 | 15.01.02a | t190 | dt10a | 00080 | 14-00 (1) | Characterized by ccrAB2, ccrAB4, J1 type IVb, dcs, and novel mec complex | ND |
| 41 | 15.01.2b | t190 | dt10j | 00216 | 13-00 (1) | IIE | ND |
| 42 | 09.01d.4 | t2978 | dt10i | 99083 | Unf-99 (1) | IVb | ST87 |
Based on the nomenclature presented in Table 1, each isolate was assigned a 3-digit cluster code in which the first number represents the spa type, the second represents the dru type, and the third represents the PFT.
Unf, unfamiliar (these isolates exhibited a hitherto unfamiliar AR pattern); NT, nontypeable (these isolates exhibited AR patterns that differed from the AR06 group of patterns only with regard to resistance to lincomycin [see Table S1 in the supplemental material for further details]).
One isolate representative of each spa type identified in the present study underwent MLST. ND, not determined.
PFGE and AR typing.
Twenty-six PFTs representing four PFGs were identified among the 173 isolates (Table 2). PFG-01 predominated, accounting for 97% of isolates (168 of 173). The 168 PFG-01 isolates exhibited 21 highly similar PFTs, with the two most predominant patterns (PFT 01018 [n = 57] and PFT 01039 [n = 39]), which accounted for 57.1% of all PFG-01 isolates (96 of 168), differing by only a single band.
The majority of PFG-01 isolates (135 of 168 [80.4%]) exhibited AR type AR06 and were assigned to AR-PFG 06-01 (Table 2). The AR types and subtypes and the antimicrobial resistance patterns for all isolates investigated are shown in Table S1 in the supplemental material.
spa typing.
Seventeen spa types were identified among the 168 isolates classified into PFG-01, but 55.4% of these isolates (93 of 168) belonged to spa type t032. The proportions of isolates of other spa types among the PFG-01 isolates were as follows: t515, 16 of 168 (9.5%); t3185, 15 of 168 (8.9%); t1214, 10 of 168 (6%); t022, 10 of 168 (6%); t628, 9 of 168 (5.4%); t1865, 4 of 168 (20.4%); and t3213, 2 of 168 (1.2%). spa types t025, t578, t1802, t2951, t4122, t4267, t4622, t4623, and t4765 were exhibited by single isolates only (Table 2).
dru typing.
The stability of the dru region in three MRSA isolates was confirmed by the finding that several colonies from original cultures of each isolate on BHI agar plates and from growth following a minimum of 10 sequential subcultures exhibited the same dru types originally assigned in earlier studies (i.e., dt9g for M06/0376 and M05/0028 and dt10h for the Harmony EMRSA-15 isolate).
Seventeen dru types were identified among the 168 PFG-01 isolates, with dt10a isolates (61 of 168 [36.3%]) predominating. Proportions of isolates of other types were as follows: dt10j, 43 of 168 (25.6%); dt10n, 22 of 168 (13%); dt10o, 16 of 168 (9.5%); dt10af, 6 of 168 (3.6%); dt8p, 4 of 168 (2.4%); dt5b, 2 of 168 (1.2%); dt7g, 2 of 168 (1.2%); dt10p, 2 of 168 (1.2%); dt11j, 2 of 168 (1.2%); and dt11o, 2 of 168 (1.2%). The dru types dt6e, dt7i, dt8a, dt9j, dt9p, and dt11a were exhibited by single isolates among the remaining PFG-01 isolates (Table 2).
MLST and SCCmec typing.
By MLST, four distinct sequence types (STs) were detected among 20 isolates representative of each of the 20 spa types identified (Table 2). Isolates exhibiting the 17 spa types found among the 168 PFG-01 isolates were all identified as ST22 (i.e., MLST allelic profile 7-6-1-5-8-8-6) and belonged to clonal complex 22 (i.e., CC22).
SCCmec typing revealed that the majority of PFG-01 isolates (163 of 168 [97%]) harbored SCCmec IVh (with ccrAB2, class B mec, dcs, and J1 region type IVh). Of the remaining five PFG-01 isolates, one harbored SCCmec IVa (with ccrAB2, class B mec, dcs, and J1 region type IVa) and four harbored novel SCCmec IV variants (Table 2).
Clustering of isolates.
The 173 MRSA isolates were divided into clusters based on spa, dru, and PFGE typing data. Each isolate was assigned a 3-digit cluster code with the first number representing the spa type, the second representing the dru type, and the third representing the PFT (Tables 1 and 2). Subgrouping among spa and dru types was investigated by constructing MSTs (Fig. 1), and subgrouping among PFGE types was based on the numbers of band differences, as described below.
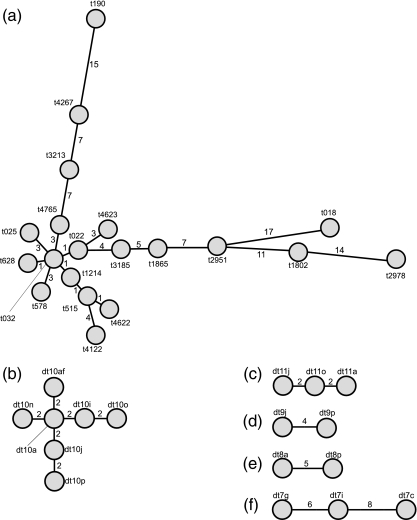
FIG. 1.
MSTs generated using the BioNumerics software program representing the 20 spa types (a) and the dru types with 10 (b), 11 (c), 9 (d), 8 (e), and 7 (f) repeat units identified among the 173 MRSA isolates investigated. Each individual circle represents a different spa or dru type, and the numerical values on the branches represent the similarity (expressed as the MST distance) between two spa or two dru types. The BioNumerics software creates groups of certain distance intervals or similarity values (termed bins) and converts these data into distance units. The bin distance was set to 0.5% (i.e., two entries at a distance of 1 on the MST have between 99 and 99.5% similarity, and two entries at a distance of 2 have between 98.5 and 99% similarity, etc.). spa types and dru types were assigned the same cluster code if they were separated by an MST distance of <2 (i.e., if they showed >98.5% similarity) (Table 1).
Cluster analysis of spa types.
An MST constructed from all spa types identified is shown in Fig. 1a. spa types were deemed to be distinct if they differed from all others identified by an MST distance of >2 (corresponding to <98.5% similarity) and were assigned different spa cluster codes with numerical values ranging from 01 to 15 (Fig. 1a and Table 1). spa types that showed >98.5% similarity on the MST (i.e., those at an MST distance of ≤2) were assigned spa subcluster codes (Table 1). Each spa type within a subcluster, apart from the founder of a subgroup, was assigned an additional alphabetic suffix (Table 1). The MST showed that some spa type subgroups contained further subgroups, so additional alphabetic suffixes were added to the alphanumeric spa subcluster codes (Table 1).
Cluster analysis of dru types.
For dru typing, individual MSTs were generated for all groups of distinct dru types harboring the same number of dru repeat units. MSTs for dru types with 11 (dt11a, dt11j, and dt11o), 10 (dt10a, dt10i, dt10j, dt10n, dt10o, dt10p, and dt10af), 9 (dt9j and dt9p), 8 (dt8a and dt8p), and 7 (dt7c, dt7g, and dt7i) repeat units are shown in Fig. 1c, b, d, e, and f, respectively.
dru type cluster codes were based on the number of dru repeat units present (e.g., dru types with 10 repeats were assigned dru cluster code 01) (Table 1). Thereafter, subgroups of closely related dru types were identified using the same criteria used for subgrouping of spa types (Table 1).
Cluster analysis of PFTs.
For cluster analysis, each of the four PFGs was assigned a PFGE cluster code consisting of a number ranging from 1 to 4. Where PFGs were represented by more than one PFT, the most frequently occurring PFT was assigned the numerical value for that PFG and all other PFTs were assigned additional alphabetic suffixes (Table 1).
Discriminatory powers of and concordance of data from spa, dru, and PFGE typing methods.
The abilities of PFGE, spa, and dru typing methods to discriminate among the 168 PFG-01 (ST22-MRSA-IV) isolates were determined quantitatively using SID for each individual typing method and for all combinations of the three methods (Table 3). The combination of spa, dru, and PFGE typing yielded the largest number of type combinations (65 types) and the greatest discriminatory power (SID, 96.53) with the narrowest 95% CI (Table 3). Of the three individual methods, PFGE was the most discriminatory (SID, 81.34) (Table 3).
TABLE 3.
Discriminatory powers of spa, dru, and PFGE typing methodsa
| Typing method(s) | No. of types | SID | 95% CI |
|---|---|---|---|
| spa typing | 17 | 66.90 | 59.36-74.44 |
| dru typing | 17 | 77.83 | 73.86-81.80 |
| PFGE | 21 | 81.34 | 77.38-85.31 |
| spa and dru typing | 37 | 90.84 | 88.66-93.02 |
| spa typing and PFGE | 44 | 91.00 | 88.12-93.89 |
| dru typing and PFGE | 43 | 93.57 | 92.08-95.66 |
| spa, dru, and PFGE typing | 65 | 96.53 | 95.53-97.52 |
Discriminatory powers of the methods used individually and in combination for the 168 PFG-01 (ST22-MRSA-IV) isolates investigated were measured by using SID (with 95% CIs).
The enhanced discrimination obtained by combining all three typing methods was confirmed by the ARI and W coefficient values (Table 4). Based on the ARI, the probability that the isolate clustering patterns obtained using the combination of spa, dru, and PFGE typing methods would be similar to those obtained using any one of the typing methods individually or pairwise combinations of the methods was <69% (range, ca. 13 to 69%) (Table 4). In addition, the low W coefficients obtained for the comparison of individual methods suggest that no method is redundant and that each method contributes additional information. The highest value for the comparison between a pair of individual methods was obtained for PFGE and spa typing (W coefficient, 0.482), but the value for the comparison between spa typing and PFGE was much lower (W coefficient, 0.272) (Table 4). Hence, the PFGE type could predict the spa type with 48% probability whereas the probability with which the spa type predicted the PFGE type was only 27%. High W coefficients were obtained for comparisons between a combination of two or three methods and one of the methods individually (e.g., the combination of spa typing and PFGE and spa typing alone) (Table 4).
TABLE 4.
Concordance of data from spa, dru, and PFGE typing methods used individually and in combination for the 168 PFG-01 (ST22 MRSA-IV) isolates
| Typing method(s) | ARI for comparison with: |
W coefficient for comparison with: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| spa typing | dru typing | PFGE | spa and dru typing | spa typing and PFGE | dru typing and PFGE | spa typing | dru typing | PFGE | spa and dru typing | spa typing and PFGE | dru typing and PFGE | spa, dru, and PFGE typing | |
| spa typing | 0.277 | 0.272 | 0.277 | 0.272 | 0.105 | 0.105 | |||||||
| dru typing | 0.090 | 0.413 | 0.290 | 0.413 | 0.157 | 0.290 | 0.157 | ||||||
| PFGE | 0.143 | 0.141 | 0.482 | 0.345 | 0.186 | 0.482 | 0.345 | 0.186 | |||||
| spa and dru typing | 0.339 | 0.523 | 0.144 | 1.000 | 1.000 | 0.379 | 0.379 | 0.379 | 0.379 | ||||
| spa typing and PFGE | 0.333 | 0.109 | 0.602 | 0.321 | 1.000 | 0.386 | 1.000 | 0.386 | 0.386 | 0.386 | |||
| dru typing and PFGE | 0.076 | 0.389 | 0.461 | 0.400 | 0.406 | 0.540 | 1.000 | 1.000 | 0.540 | 0.540 | 0.540 | ||
| spa, dru, and PFGE typing | 0.136 | 0.224 | 0.271 | 0.526 | 0.534 | 0.687 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
Cluster analysis of isolates based on the combination of spa, dru, and PFGE typing results.
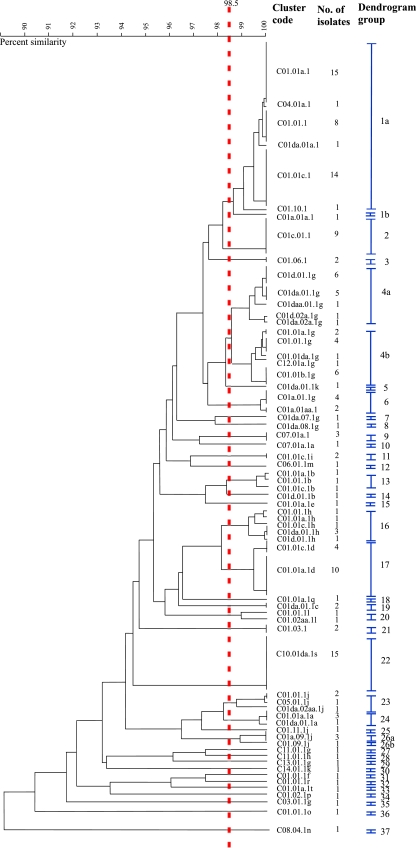
Seventy cluster codes representative of each different combination of spa, dru, and PFGE types were identified for the 173 isolates investigated (Table 2). For the 168 PFG-01 isolates, 65 cluster codes were identified (Table 2). Many of the isolates with different cluster codes exhibited only minor differences by combinations of spa, dru, and/or PFGE typing methods. To further investigate the relationship among PFG-01 isolates, a dendrogram was generated from the averages of the similarity matrices for spa, dru, and PFGE typing data for all PFG-01 isolates (Fig. 2). Isolates that showed ≥98.5% similarity on the dendrogram were deemed to be very closely related and were assigned to the same dendrogram group (DG), while those with <98.5% similarity were deemed to be distinguishable and were assigned to different DGs (Fig. 2). By using these criteria, a total of 37 DGs were identified among the 168 PFG-01 isolates (Table 2 and Fig. 2). Three of these DGs (DG-1, DG-4, and DG-26) were divided into subgroups because they included isolates that showed between 98.5 and 99% similarity (Fig. 2). DG-1 and DG-4 were the largest groups identified and consisted of 41 of 168 isolates (24.4%) and 28 of 168 isolates (16.7%), respectively (Fig. 2).
FIG. 2.
Composite dendrogram generated using UPGMA clustering and the averages of the similarity matrices from spa, dru, and PFGE typing data for the 168 PFG-01 MRSA (ST22 MRSA-IV) isolates investigated during the present study. Isolates were assigned 3-digit cluster codes (C) with the first number representing the spa type, the second representing the dru type, and the third representing the PFT. Isolate cluster codes were then assigned to DGs as follows: isolates with cluster codes that showed ≥98.5% similarity on the dendrogram were deemed to be very closely related and were assigned to the same DG. Those isolates with cluster codes showing <98.5% similarity were deemed to be distinguishable and were assigned to different DGs. The dendrogram demonstrates that the PFG-01 isolates were assigned to 65 cluster codes that were divided into 37 DGs. The red, dashed vertical line marks 98.5% similarity.
Five distinct cluster codes were identified among the five non-PFG-01 isolates; these isolates showed <90% similarity to one another and to all other isolates investigated according to a dendrogram generated from the averages of the similarity matrices for spa, dru, and PFGE typing data for all isolates investigated in the present study (data not shown). Therefore, these five isolates were deemed to be distinguishable and were assigned to distinct DGs (DG-38 to DG-42) (Table 2).
Combining the dendrogram groupings with epidemiological evidence.
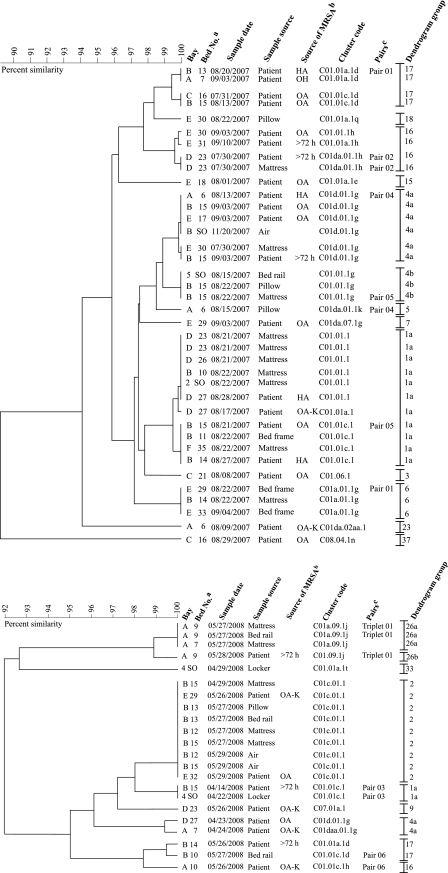
The dendrogram groupings for one hospital ward during two 6-week study periods were analyzed in the context of available epidemiological data. Dendrograms were generated from the averages of the similarity matrices for spa, dru, and PFGE typing data for all PFG-01 isolates recovered from patients and environmental sites in one ward (ward 1) from (i) July to September 2007, with one additional isolate recovered in November 2007 (study period I; n = 38) (Fig. 3, top), and (ii) April to May 2008 (study period II; n = 22) (Fig. 3, bottom).
FIG. 3.
Composite dendrogram generated using UPGMA clustering and the averages of the similarity matrices from spa, dru, and PFGE typing data for 38 PFG-01 MRSA isolates investigated during study period I (top) and 22 PFG-01 MRSA isolates investigated during study period II (bottom). Available epidemiological information for each isolate, as well as the cluster code and DG, is indicated. DGs were assigned to the different cluster codes determined from the dendrogram depicting all PFG-01 isolates identified in the present study (Fig. 2). The dendrogram shows that the PFG-01 isolates from study period I (n = 38) and study period II (n = 22) were differentiated into 12 and 8 DGs, respectively.a, the abbreviation SO in place of a bed number indicates a single-occupancy room.b, OA, the patient was MRSA positive on admission; OA-K, the patient's MRSA-positive status was known at the time of admission to the ward; >72 h, the patient's MRSA status was determined 72 h after admission to the ward.c, the pairs or triplets of isolates recovered during study periods I and II are indicated and include isolates recovered from patients and from their immediate environments during the same study period. Each pair or triplet consists of one isolate from a patient and at least one environmental isolate.
Twelve DGs were identified among the 38 PFG-01 isolates recovered during study period I (Fig. 3, top). The largest DG recognized was DG-1a, consisting of 11 isolates. The earliest DG-1a isolate was recovered from a patient who was MRSA positive upon admission to the ward (Fig. 3, top). Over the next 10 days, DG-1a isolates were recovered from three patients and seven environmental sites; MRSA isolates from two of these patients were deemed to have been HA (Fig. 3, top).
The second largest DG (DG-4) consisted of nine isolates belonging to DG-4a (n = 6) or DG-4b (n = 3) (Fig. 3, top). The earliest DG-4 isolate was recovered from a bed mattress and belonged to DG-4a (Fig. 3, top). Two weeks later, a second DG-4a isolate was recovered from a patient whose MRSA infection was considered to have been HA. Subsequently, DG-4a and DG-4b isolates were recovered from additional patients and environmental sites, but none of the MRSA isolates from the patients were deemed to have been HA (Fig. 3, top). Four pairs of isolates were recovered during study period I, but isolates from one pair only (pair 02) were assigned to the same DG (DG-16) (Fig. 3, top).
During study period II, eight DGs were identified among the 22 PFG-01 isolates recovered, with DG-2 isolates (9 of 22 [40.9%]) predominating (Fig. 3, bottom). The earliest DG-2 isolate came from a bed mattress. Subsequently, DG-2 isolates were recovered from six environmental sites and from two patients, both of whom were MRSA positive upon admission (Fig. 3, bottom). Four DG-26 isolates, including a triplet of isolates (triplet 01) recovered from a patient and the patient's mattress and bed rail, were identified during study period II. Two DG-1a, two DG-4a, and two DG-17 isolates were also identified during study period II (Fig. 3, bottom). The two DG-1a isolates (pair 03) were recovered from a patient and, 8 days later, from that patient's locker, while the DG-4a isolates were recovered from two patients within 24 h of each other. The first DG-17 isolate was from a patient, and 1 day later, the second isolate was recovered from a bed rail of a different bed (bed 10) in the same bed bay occupied by the patient (Fig. 3, bottom). A DG-16 isolate had been recovered the previous day from the patient in bed 10. This DG-16 isolate and the DG-17 isolate recovered from the rail of bed 10 (pair 06) differ by one PFGE band only, and while they showed 98.1% similarity on the dendrogram generated for all PFG-01 isolates (Fig. 2), they showed ca. 98.4% similarity on the dendrogram for PFG-01 isolates recovered in ward 1 during study period II (Fig. 3, bottom).
DISCUSSION
Epidemiological tracking of ST22-MRSA-IV isolates is a major challenge, as they exhibit limited diversity by PFGE and spa typing, the most frequently used epidemiological typing methods available for MRSA. The present study investigated whether integration of PFGE, spa, and dru typing data would provide improved discrimination among ST22-MRSA-IV isolates recovered in a large tertiary-referral hospital in Ireland.
A total of 168 ST22-MRSA-IV isolates were investigated using the three typing methods. The combined use of the PFGE, spa, and dru typing data yielded the highest number of type combinations (65 types) and the greatest discriminatory power (SID, 96.53) with the narrowest 95% CI. Faria et al. (14) compared the abilities of several typing methods, including PFGE and spa typing, to discriminate among a diverse collection of MRSA and methicillin-susceptible S. aureus isolates. They reported that spa typing and PFGE differentiated their MRSA isolates with SIDs of 95.85 and 94.27, respectively. In contrast, the SIDs for spa and PFGE typing of the ST22-MRSA-IV isolates obtained in the present study were 66.9 and 81.34, respectively. Faria et al. (14) also found that the combination of PFGE and spa typing had a discriminatory power yielding a SID of 98.32, whereas the SID for this combination of typing methods in the present study was 91.00. These findings indicate that while the combination of PFGE and spa typing is highly discriminatory for collections of diverse MRSA isolates, it is inadequate in local epidemiological studies where strain diversity is limited, as with ST22-MRSA-IV. While the 168 ST22-MRSA-IV isolates investigated here were differentiated into 65 type combinations by integrating spa, dru, and PFGE typing data, some of the types identified using each individual method exhibited only minor differences and were assigned to subgroups. All 21 PFTs identified among the 168 ST22-MRSA-IV isolates were assigned to a single group (PFG-01) and were deemed to be possibly related according to the criteria of Tenover et al. (57), as they all differed by ≤6 bands. These criteria were originally devised for a range of bacterial species, including S. aureus, but can present problems with clonal populations of MRSA exhibiting limited genetic diversity (16). A cutoff of 80% similarity for grouping clusters of MRSA isolates, with a cutoff of 95% similarity for recognition of subtypes, has been proposed previously (14, 37). In the present study, dendrogram clustering of PFGE data showed >80% similarity among all 168 ST22-MRSA-IV isolates. The most frequently occurring PFTs (PFT 01018 and PFT 01039) differed from each other by a single band and showed 98% similarity to each other (data not shown). With PFGE, undue weight cannot be placed on a single band difference, so for the majority of the isolates in the study population, PFGE alone could not provide reliable differentiation. Unlike PFGE, for which there are agreed-upon interpretive criteria (57), spa and dru typing currently have no criteria defined by international consensus for interpreting the significance of differences in results. Consequently, isolates with different spa types or dru types are deemed to be distinct even though they may be closely related if the types differ by changes consistent with a single genetic event, such as duplication of a tandem repeat, a point mutation, or a base insertion or deletion. To investigate the significance of such differences objectively, spa and dru types were assigned as subgroups by using MSTs if they showed ≥98.5% similarity. This cutoff value grouped 7 of the 17 spa types identified among the 168 ST22-MRSA-IV isolates investigated into six spa subgroups (Table 1). Each spa type within each subgroup differed from the other members of the subgroup by the presence or absence of one to three tandem repeats. With dru typing, 10 of the 17 dru types identified were grouped into eight subgroups (Table 1). The dru types within each subgroup differed by nucleotide changes in one or two repeat units only.
While spa typing and PFGE are well established methods for typing of MRSA isolates, few studies have investigated the usefulness of dru typing for MRSA. Smyth et al. (54) identified 42 dru types among 111 isolates of the pandemic nosocomial strain ST239-MRSA-III. Goering et al. (17) identified 13 and 12 dru types among 47 EMRSA-15 (ST22-MRSA-IV) and 57 EMRSA-16 (ST36-MRSA-II) isolates, respectively. In contrast, the majority of CA-MRSA USA300 (ST8-MRSA-IV) and CC80 isolates from patients from various geographical locations exhibited dt9g and dt10a, respectively (58, 29). The results of these studies indicate that CA-MRSA strains exhibit less genetic diversity within the dru region than nosocomial MRSA strains. This difference may reflect the fact that the nosocomial strains have been extant longer than CA-MRSA strains (12, 44, 54, 61). In the present study, 17 spa types were identified among 168 ST22-MRSA-IV isolates, of which 4, t032, t515, t022, and t1214, were further differentiated into 12, 6, 4, and 2 dru types, respectively. Nevertheless, dru typing cannot be used as a stand-alone method for typing MRSA isolates, as the two predominant types recognized among the ST22-MRSA-IV isolates (dt10a and dt10j) were also identified among the three ST8-MRSA-II variant isolates in the present study and among CC80-MRSA-IV isolates described in a previous study (29). Three dru types identified among ST22-MRSA-IV isolates in the present study (dt10a, dt11a, and dt8a) were also identified previously among ST239-MRSA-III isolates (54), while dt10i has been identified in unrelated MRSA lineages, including EMRSA-15 (17) and the ST87-MRSA-IV isolate in the present study. These data indicate that an isolate's dru type is not lineage or SCCmec type specific. However, unrelated MRSA lineages sharing indistinguishable dru types may reflect the presence of related SCCmec elements in diverse genetic backgrounds.
The composite dendrogram generated from the combined spa, dru, and PFGE typing data for all 168 ST22-MRSA-IV isolates provides a visual representation of the overall relatedness of isolates. Using a cutoff of 98.5% similarity, isolates were differentiated into 37 DGs, 17 (46%) of which contained more than one isolate and 10 of which contained isolates representing more than one cluster code, further indicating the close relatedness of the isolates. Of the 10 DGs consisting of isolates belonging to different cluster codes, 4 contained isolates with spa and/or dru types that were not assigned as subgroups (Fig. 2 and Table 2), including DG-1a (t025 and dt9j), DG-4 (for DG-4b, t4122, and for DG-4a, dt11o), DG-23 (t578 and dt11j), and DG-20 (dt11j). In each case, the PFGE patterns were indistinguishable from those of other isolates within that DG. In addition, where the spa type was distinct, the dru type was indistinguishable from or closely related to that of other isolates within that DG; where the dru type was distinct, the spa type was indistinguishable from or closely related to that of other isolates within that DG (Table 2). These findings highlight the need for caution when interpreting data from individual typing methods and show how combining data from the three typing methods permits a more informative evaluation of the relationship among isolates.
In the present study, analysis of available epidemiological information for a selected subset of ST22-MRSA-IV isolates was used to confirm the validity of the relationships inferred from the combined PFGE, spa, and dru typing data. Six pairs and one triplet of isolates were recovered from individual patients and their immediate ward environments during the same time periods, and by using the combination of all three typing methods, isolates in four of these pairs (pairs 01, 04, 05, and 06) were differentiated into distinct DGs. Isolates in pair 01 differed by all three typing methods and exhibited <96% similarity on the composite dendrogram (Fig. 3, top). Interestingly, the environmental isolate in pair 01 was recovered from a bed bay 2 days after the patient isolate was obtained while the patient was in a different bed bay. Pair 04 isolates belonged to DG-4a and DG-5 and differed in their spa type and PFT, and the composite dendrogram showed that these isolates had <98.5% similarity (Fig. 3, top). Pair 05 isolates belonged to DG-1a and DG-4b, and although they shared the same spa type and belonged to dru types and PFTs that were assigned as subtypes, the composite dendrogram showed that they had <98% similarity (Fig. 3, top), suggesting that they are distinguishable. However, pair 06 isolates belonged to DG-16 and DG-17, differed only with regard to the PFGE patterns (exhibiting a one-band difference), and showed ca. 98.4% similarity (Fig. 3, bottom), suggesting that these isolates should be considered to be very closely related. Isolates in each of the two remaining pairs (pair 02, consisting of DG-16 isolates, and pair 03, consisting of DG-1a isolates) and the triplet (triplet 01, consisting of DG-26 isolates) were indistinguishable from each other (Fig. 3). This analysis revealed that certain patient and environmental ST22-MRSA-IV isolates could be differentiated while others remained indistinguishable and showed that the combination of typing methods used in the present study significantly improves isolate discrimination and therefore can be used for epidemiological tracking of isolates of this highly clonal strain.
Different DGs predominated among isolates from the two study periods (DG-1a in study period I and DG-2 in study period II). Particular isolates within DG-1a and DG-2 differed in the spa type only, with DG-1a isolates exhibiting spa type t032 while DG-2 isolates belonged to spa type t628. While these spa types were assigned to the same subgroup, only nine isolates exhibiting spa type t628 were recovered during the study and all nine were recovered from ward 1 during study period II, suggesting that the difference between t032 and t628 is significant (Fig. 3).
Based on the results of this study, it is recommended that in performing epidemiological investigations of a highly clonal MRSA strain, such as ST22-MRSA-IV, in a hospital setting where the strain is endemic, optimal tracking can be achieved by combining spa and PFGE typing data with dru typing data. This approach has revealed a previously unrecognized level of diversity among ST22-MRSA-IV isolates that can be used to provide data fundamental to epidemiological tracking of isolates of this pandemic MRSA strain. PFGE and spa typing are routinely used for typing of MRSA isolates (19, 37), and while dru typing may not be as well established or widely used for typing of MRSA isolates, it involves the same techniques and principles as spa typing (DNA sequencing of a VNTR unit). Therefore, use of dru typing by a laboratory that currently uses DNA-based sequencing methods for routine epidemiological typing of MRSA isolates should not require additional expertise or result in a major increase in costs. In addition, all spa, dru, and PFGE data analyses can be performed with a commonly used software package (i.e., BioNumerics), and if required, statistical analysis of results can be readily and easily attained using a previously published free online tool (http://www.comparingpartitions.info). However, the ability of dru typing in combination with PFGE and spa typing to discriminate among isolates of highly clonal strains of MRSA other than ST22-MRSA-IV remains to be determined.
Acknowledgments
This work was supported by Irish Health Research Board grant TRA/2006/4 and by the Microbiology Research Unit, Dublin Dental School and Hospital.
We thank the staff of the National MRSA Reference Laboratory, including Gráinne Brennan, Emma Gibbons, Paul Grier, and Pamela Morgan for AR typing and PFGE. MRSA control isolates were kindly provided by Teuryo Ito, Juntendo University, Japan; Herminia de Lencastre, Rockfeller University, New York; and Robert Daum, University of Chicago.
Footnotes
Published ahead of print on 24 March 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Aires-de-Sousa, M., B. Correia, and H. de Lencastre. 2008. Changing patterns in frequency of recovery of five methicillin-resistant Staphylococcus aureus clones in Portuguese hospitals: surveillance over a 16-year period. J. Clin. Microbiol. 46:2912-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amorim, M. L., C. Vasconcelos, D. C. Oliveira, A. Azevedo, E. Calado, N. A. Faria, M. Pereira, A. P. Castro, A. Moreira, E. Aires, J. M. Cabeda, M. H. Ramos, J. M. Amorim, and H. de Lencastre. 2009. Epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) nasal colonization among patients and healthcare workers in a Portuguese hospital: a pre-intervention study toward the control of MRSA. Microb. Drug Resist. 15:19-26. [DOI] [PubMed] [Google Scholar]
- 3.Baptiste, K. E., K. Williams, N. J. Willams, A. Wattret, P. D. Clegg, S. Dawson, J. E. Corkill, T. O'Neill, and C. A. Hart. 2005. Methicillin-resistant staphylococci in companion animals. Emerg. Infect. Dis. 11:1942-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson, G. 1997. Sequence alignment with tandem duplication. J. Comput. Biol. 4:351-367. [DOI] [PubMed] [Google Scholar]
- 5.Boakes, E., A. M. Kearns, M. Ganner, C. Perry, M. Warner, R. L. Hill, and M. J. Ellington. 17 February 2010. Molecular diversity within CC22 meticillin-resistant Staphylococcus aureus encoding Panton-Valentine Leukocidin in England and Wales. Clin. Microbiol. Infect. [Epub ahead of print.] doi: 10.1111/j.1469-0691.2010.03199.x. [DOI] [PubMed]
- 6.Carrico, J. A., C. Silva-Costa, J. Melo-Cristino, F. R. Pinto, H. de Lencastre, J. S. Almeida, and M. Ramirez. 2006. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J. Clin. Microbiol. 44:2524-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carroll, D., M. A. Kehoe, D. Cavanagh, and D. C. Coleman. 1995. Novel organization of the site-specific integration and excision recombination functions of the Staphylococcus aureus serotype F virulence-converting phages phi 13 and phi 42. Mol. Microbiol. 16:877-893. [DOI] [PubMed] [Google Scholar]
- 8.Carroll, J. D., H. M. Pomeroy, R. J. Russell, J. P. Arbuthnott, C. T. Keane, O. M. McCormick, and D. C. Coleman. 1989. A new methicillin- and gentamicin-resistant Staphylococcus aureus in Dublin: molecular genetic analysis. J. Med. Microbiol. 28:15-23. [DOI] [PubMed] [Google Scholar]
- 9.Coleman, D. C., H. Pomeroy, J. K. Estridge, C. T. Keane, M. T. Cafferkey, R. Hone, and T. J. Foster. 1985. Susceptibility to antimicrobial agents and analysis of plasmids in gentamicin- and methicillin-resistant Staphylococcus aureus from Dublin hospitals. J. Med. Microbiol. 20:157-167. [DOI] [PubMed] [Google Scholar]
- 10.Collery, M. M., D. S. Smyth, J. M. Twohig, A. C. Shore, D. C. Coleman, and C. J. Smyth. 2008. Molecular typing of nasal carriage isolates of Staphylococcus aureus from an Irish university student population based on toxin gene PCR, agr locus types and multiple locus, variable number tandem repeat analysis. J. Med. Microbiol. 57:348-358. [DOI] [PubMed] [Google Scholar]
- 11.Coombs, G. W., G. R. Nimmo, J. C. Pearson, K. J. Christiansen, J. M. Bell, P. J. Collignon, and M. L. McLaws. 2009. Prevalence of MRSA strains among Staphylococcus aureus isolated from outpatients, 2006. Commun. Dis. Intell. 33:10-20. [PubMed] [Google Scholar]
- 12.Cox, R. A., C. Conquest, C. Mallaghan, and R. R. Marples. 1995. A major outbreak of methicillin-resistant Staphylococcus aureus caused by a new phage-type (EMRSA-16). J. Hosp. Infect. 29:87-106. [DOI] [PubMed] [Google Scholar]
- 13.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faria, N. A., J. A. Carrico, D. C. Oliveira, M. Ramirez, and H. de Lencastre. 2008. Analysis of typing methods for epidemiological surveillance of both methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J. Clin. Microbiol. 46:136-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faria, N. A., D. C. Oliveira, H. Westh, D. L. Monnet, A. R. Larsen, R. Skov, and H. de Lencastre. 2005. Epidemiology of emerging methicillin-resistant Staphylococcus aureus (MRSA) in Denmark: a nationwide study in a country with low prevalence of MRSA infection. J. Clin. Microbiol. 43:1836-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goering, R. V. 2004. Molecular microbiology: diagnostic principles and practice. ASM Press, Washington, DC.
- 17.Goering, R. V., D. Morrison, Z. Al-Doori, G. F. Edwards, and C. G. Gemmell. 2008. Usefulness of mec-associated direct repeat unit (dru) typing in the epidemiological analysis of highly clonal methicillin-resistant Staphylococcus aureus in Scotland. Clin. Microbiol. Infect. 14:964-969. [DOI] [PubMed] [Google Scholar]
- 18.Grinberg, A., D. D. Kingsbury, I. R. Gibson, B. M. Kirby, H. J. Mack, and D. Morrison. 2008. Clinically overt infections with methicillin-resistant Staphylococcus aureus in animals in New Zealand: a pilot study. N. Z. Vet. J. 56:237-242. [DOI] [PubMed] [Google Scholar]
- 19.Grundmann, H., D. M. Aanensen, C. C. van den Wijngaard, B. G. Spratt, D. Harmsen, and A. W. Friedrich. 2010. Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: a molecular-epidemiological analysis. PLoS Med. 7:e1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grundmann, H., S. Hori, and G. Tanner. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanssen, A. M., and J. U. Ericson Sollid. 2006. SCCmec in staphylococci: genes on the move. FEMS Immunol. Med. Microbiol. 46:8-20. [DOI] [PubMed] [Google Scholar]
- 22.Hone, R., and C. T. Keane. 1974. Characteristics of methicillin resistant Staphylococcus aureus. Ir. J. Med. Sci. 143:145-154. [DOI] [PubMed] [Google Scholar]
- 23.Hsu, L. Y., T. H. Koh, K. Singh, M. L. Kang, A. Kurup, and B. H. Tan. 2005. Dissemination of multisusceptible methicillin-resistant Staphylococcus aureus in Singapore. J. Clin. Microbiol. 43:2923-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Humphreys, H., C. T. Keane, R. Hone, H. Pomeroy, R. J. Russell, J. P. Arbuthnott, and D. C. Coleman. 1989. Enterotoxin production by Staphylococcus aureus isolates from cases of septicaemia and from healthy carriers. J. Med. Microbiol. 28:163-172. [DOI] [PubMed] [Google Scholar]
- 25.Ito, T., K. Okuma, X. X. Ma, H. Yuzawa, and K. Hiramatsu. 2003. Insights on antibiotic resistance of Staphylococcus aureus from its whole genome: genomic island SCC. Drug Resist. Updat. 6:41-52. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, A. P., H. M. Aucken, S. Cavendish, M. Ganner, M. C. Wale, M. Warner, D. M. Livermore, and B. D. Cookson. 2001. Dominance of EMRSA-15 and -16 among MRSA causing nosocomial bacteraemia in the UK: analysis of isolates from the European Antimicrobial Resistance Surveillance System (EARSS). J. Antimicrob. Chemother. 48:143-144. [DOI] [PubMed] [Google Scholar]
- 27.Khandavilli, S., P. Wilson, B. Cookson, J. Cepeda, G. Bellingan, and J. Brown. 2009. Utility of spa typing for investigating the local epidemiology of MRSA on a UK intensive care ward. J. Hosp. Infect. 71:29-35. [DOI] [PubMed] [Google Scholar]
- 28.Kondo, Y., T. Ito, X. X. Ma, S. Watanabe, B. N. Kreiswirth, J. Etienne, and K. Hiramatsu. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51:264-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larsen, A. R., M. Stegger, S. Bocher, M. Sorum, D. L. Monnet, and R. L. Skov. 2009. Emergence and characterization of community-associated methicillin-resistant Staphylococcus aureus infections in Denmark, 1999 to 2006. J. Clin. Microbiol. 47:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma, X. X., T. Ito, P. Chongtrakool, and K. Hiramatsu. 2006. Predominance of clones carrying Panton-Valentine leukocidin genes among methicillin-resistant Staphylococcus aureus strains isolated in Japanese hospitals from 1979 to 1985. J. Clin. Microbiol. 44:4515-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chongtrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marchese, A., L. Gualco, E. Maioli, and E. Debbia. 2009. Molecular analysis and susceptibility patterns of meticillin-resistant Staphylococcus aureus (MRSA) strains circulating in the community in the Ligurian area, a northern region of Italy: emergence of USA300 and EMRSA-15 clones. Int. J. Antimicrob. Agents. 34:424-428. [DOI] [PubMed] [Google Scholar]
- 33.Melter, O., P. Urbaskova, V. Jakubu, B. Mackova, and H. Zemlickova. 2006. Emergence of EMRSA-15 clone in hospitals throughout the Czech Republic. Euro Surveill. 11:E060803.6. [DOI] [PubMed] [Google Scholar]
- 34.Milheirico, C., D. C. Oliveira, and H. de Lencastre. 2007. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: ‘SCCmec IV multiplex’. J. Antimicrob. Chemother. 60:42-48. [DOI] [PubMed] [Google Scholar]
- 35.Monecke, S., L. Jatzwauk, S. Weber, P. Slickers, and R. Ehricht. 2008. DNA microarray-based genotyping of methicillin-resistant Staphylococcus aureus strains from Eastern Saxony. Clin. Microbiol. Infect. 14:534-545. [DOI] [PubMed] [Google Scholar]
- 36.Moodley, A., M. Stegger, A. F. Bagcigil, K. E. Baptiste, A. Loeffler, D. H. Lloyd, N. J. Williams, N. Leonard, Y. Abbott, R. Skov, and L. Guardabassi. 2006. spa typing of methicillin-resistant Staphylococcus aureus isolated from domestic animals and veterinary staff in the UK and Ireland. J. Antimicrob. Chemother. 58:1118-1123. [DOI] [PubMed] [Google Scholar]
- 37.Murchan, S., M. E. Kaufmann, A. Deplano, R. de Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nahvi, M. D., J. E. Fitzgibbon, J. F. John, and D. T. Dubin. 2001. Sequence analysis of dru regions from methicillin-resistant Staphylococcus aureus and coagulase-negative staphylococcal isolates. Microb. Drug Resist. 7:1-12. [DOI] [PubMed] [Google Scholar]
- 39.Noto, M. J., B. N. Kreiswirth, A. B. Monk, and G. L. Archer. 2008. Gene acquisition at the insertion site for SCCmec, the genomic island conferring methicillin resistance in Staphylococcus aureus. J. Bacteriol. 190:1276-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Mahony, R., Y. Abbott, F. C. Leonard, B. K. Markey, P. J. Quinn, P. J. Pollock, S. Fanning, and A. S. Rossney. 2005. Methicillin-resistant Staphylococcus aureus (MRSA) isolated from animals and veterinary personnel in Ireland. Vet. Microbiol. 109:285-296. [DOI] [PubMed] [Google Scholar]
- 42.Pearman, J. W., G. W. Coombs, W. B. Grubb, and F. O'Brien. 2001. A British epidemic strain of methicillin-resistant Staphylococcus aureus (UK EMRSA-15) in Western Australia. Med. J. Aust. 174:662. [DOI] [PubMed] [Google Scholar]
- 43.Perez-Roth, E., F. Lorenzo-Diaz, N. Batista, A. Moreno, and S. Mendez-Alvarez. 2004. Tracking methicillin-resistant Staphylococcus aureus clones during a 5-year period (1998 to 2002) in a Spanish hospital. J. Clin. Microbiol. 42:4649-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richardson, J. F., and S. Reith. 1993. Characterization of a strain of methicillin-resistant Staphylococcus aureus (EMRSA-15) by conventional and molecular methods. J. Hosp. Infect. 25:45-52. [DOI] [PubMed] [Google Scholar]
- 45.Rossney, A., and S. O'Connell. 2008. Emerging high-level mupirocin resistance among MRSA isolates in Ireland. Euro Surveill. 13:pii=8084. [PubMed] [Google Scholar]
- 46.Rossney, A. S., and C. T. Keane. 2002. Strain variation in the MRSA population over a 10-year period in one Dublin hospital. Eur. J. Clin. Microbiol. Infect. Dis. 21:123-126. [DOI] [PubMed] [Google Scholar]
- 47.Rossney, A. S., M. J. Lawrence, P. M. Morgan, M. M. Fitzgibbon, A. Shore, D. C. Coleman, C. T. Keane, and B. O'Connell. 2006. Epidemiological typing of MRSA isolates from blood cultures taken in Irish hospitals participating in the European Antimicrobial Resistance Surveillance System (1999-2003). Eur. J. Clin. Microbiol. Infect. Dis. 25:79-89. [DOI] [PubMed] [Google Scholar]
- 48.Rossney, A. S., P. McDonald, H. Humphreys, G. M. Glynn, and C. T. Keane. 2003. Antimicrobial resistance and epidemiological typing of methicillin-resistant Staphylococcus aureus in Ireland (North and South), 1999. Eur. J. Clin. Microbiol. Infect. Dis. 22:379-381. [DOI] [PubMed] [Google Scholar]
- 49.Rossney, A. S., A. C. Shore, P. M. Morgan, M. M. Fitzgibbon, B. O'Connell, and D. C. Coleman. 2007. The emergence and importation of diverse genotypes of methicillin-resistant Staphylococcus aureus (MRSA) harboring the Panton-Valentine leukocidin gene (pvl) reveal that pvl is a poor marker for community-acquired MRSA strains in Ireland. J. Clin. Microbiol. 45:2554-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scicluna, E. A., A. C. Shore, A. Thumer, R. Ehricht, P. Slickers, M. A. Borg, D. C. Coleman, and S. Monecke. 2010. Characterisation of MRSA from Malta and the description of a Maltese epidemic MRSA strain. Eur. J. Clin. Microbiol. Infect. Dis. 29:163-170. [DOI] [PubMed] [Google Scholar]
- 51.Shore, A., A. S. Rossney, C. T. Keane, M. C. Enright, and D. C. Coleman. 2005. Seven novel variants of the staphylococcal chromosomal cassette mec in methicillin-resistant Staphylococcus aureus isolates from Ireland. Antimicrob. Agents Chemother. 49:2070-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shore, A. C., A. S. Rossney, B. O'Connell, C. M. Herra, D. J. Sullivan, H. Humphreys, and D. C. Coleman. 2008. Detection of staphylococcal cassette chromosome mec-associated DNA segments in multiresistant methicillin-susceptible Staphylococcus aureus (MSSA) and identification of Staphylococcus epidermidis ccrAB4 in both methicillin-resistant S. aureus and MSSA. Antimicrob. Agents Chemother. 52:4407-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith, J. M., and G. M. Cook. 2005. A decade of community MRSA in New Zealand. Epidemiol. Infect. 133:899-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smyth, D. S., L. K. McDougal, F. W. Gran, A. Manoharan, M. C. Enright, J. H. Song, H. de Lencastre, and D. A. Robinson. 2010. Population structure of a hybrid clonal group of methicillin-resistant Staphylococcus aureus, ST239-MRSA-III. PLoS One 5:e8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soliman, R. S., G. Phillips, P. Whitty, and D. H. Edwards. 2009. Distribution of meticillin-resistant Staphylococcus aureus spa types isolated from health-care workers and patients in a Scottish university teaching hospital. J. Med. Microbiol. 58:1190-1195. [DOI] [PubMed] [Google Scholar]
- 56.Tallent, S. M., T. B. Langston, R. G. Moran, and G. E. Christie. 2007. Transducing particles of Staphylococcus aureus pathogenicity island SaPI1 are comprised of helper phage-encoded proteins. J. Bacteriol. 189:7520-7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tenover, F. C., L. K. McDougal, R. V. Goering, G. Killgore, S. J. Projan, J. B. Patel, and P. M. Dunman. 2006. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J. Clin. Microbiol. 44:108-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tohda, S., M. Maruyama, and N. Nara. 1997. Molecular typing of methicillin-resistant Staphylococcus aureus by polymerase chain reaction: distribution of the typed strains in hospitals. Intern. Med. 36:694-699. [DOI] [PubMed] [Google Scholar]
- 60.Udo, E. E., F. G. O'Brien, N. Al-Sweih, B. Noronha, B. Matthew, and W. B. Grubb. 2008. Genetic lineages of community-associated methicillin-resistant Staphylococcus aureus in Kuwait hospitals. J. Clin. Microbiol. 46:3514-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. E. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Witte, W., G. Werner, and C. Cuny. 2001. Subtyping of MRSA isolates belonging to a widely disseminated clonal group by polymorphism of the dru sequences in mec-associated DNA. Int. J. Med. Microbiol. 291:57-62. [DOI] [PubMed] [Google Scholar]
- 63.Wolter, D. J., A. Chatterjee, M. Varman, and R. V. Goering. 2008. Isolation and characterization of an epidemic methicillin-resistant Staphylococcus aureus 15 variant in the central United States. J. Clin. Microbiol. 46:3548-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]